SUMMARY
Accumulation of visceral adiposity is directly linked to the morbidity of obesity, while subcutaneous body fat is considered more benign. We have identified an unexpected role for B cell lymphoma 6 (BCL6), a critical regulator of immunity, in the developmental expansion of subcutaneous adipose tissue. In adipocyte-specific knockout mice (Bcl6AKO), we found that Bcl6 deletion results in strikingly increased inguinal, but not perigonadal, adipocyte size and tissue mass in addition to marked insulin sensitivity. Genome-wide RNA expression and DNA binding analyses revealed that BCL6 controls gene networks involved in cell growth and fatty acid biosynthesis. Using deuterium label incorporation and comprehensive adipokine and lipid profiling, we discovered that ablation of adipocyte Bcl6 enhances subcutaneous adipocyte lipogenesis, increases levels of adiponectin and fatty acid esters of hydroxy fatty acids (FAHFAs), and prevents steatosis. Thus, our studies identify BCL6 as a negative regulator of subcutaneous adipose tissue expansion and metabolic health.
Graphical Abstract

In Brief
Senagolage et al. identify BCL6 as a key regulator of body fat distribution. BCL6 directly represses fatty acid biosynthetic and growth genes in adipocytes. Mice constitutively lacking adipocyte Bcl6 exhibit expansion of their subcutaneous adipose tissue, enhanced insulin sensitivity, and protection from hepatic steatosis.
INTRODUCTION
Although obesity is linked to metabolic dysregulation, the distribution of body fat strongly influences its risk to health (Lee et al., 2013; Rosen and Spiegelman, 2014; Tchkonia et al., 2013). Visceral adiposity in humans is associated with insulin resistance, dyslipidemia, nonalcoholic fatty liver disease (NAFLD), and atherosclerosis, while subcutaneous fat distributions may be benign or even protective (McLaughlin et al., 2011; Neeland et al., 2018; Ross et al., 2002). Up to 50% of obese adults are considered ‘‘metabolically healthy’’ based on varying criteria, and these individuals are typified by expanded subcutaneous, but reduced visceral and intrahepatic, fat distributions (Fabbrini et al., 2015). Although women generally have higher percentages of total body fat than men, pre-menopausal women exhibit predominantly peripheral fat distributions, improved insulin sensitivity, and reduced susceptibility to type 2 diabetes mellitus (T2DM) (Palmer and Clegg, 2015). Overall, these observations suggest a healthful, sexually dimorphic role for subcutaneous adiposity to buffer overnutrition, which can be associated with reduced visceral or ectopic lipid accumulation and metabolic disease (Fabbrini et al., 2015; Tran et al., 2008).
Adipose tissue expands through adipocyte hypertrophy and hyperplasia in a depot-specific manner to preserve whole-body metabolic homeostasis in the setting of nutrient excess (Rutkowski et al., 2015). In mice, recent cell tracking studies have revealed that high-fat feeding in males expands the subcutaneous adipose tissue (ScAT) exclusively via adipocyte hypertrophy, whereas perigonadal fat may also expand via adipogenesis but only after several weeks of dietary challenge (Jeffery et al., 2015; Wang et al., 2013b). By contrast, female mice exhibit both adipocyte hypertrophy and adipogenesis in subcutaneous and perigonadal fat pads in response to high-fat feeding (Jeffery et al., 2016). The extent to which these different modes of expansion contribute to metabolically ‘‘healthy’’ or ‘‘unhealthy’’ obesity is incompletely defined.
Major advances in our understanding of adiposity and insulin sensitivity have emerged from studies of transcription factors (Rosen et al., 2009; Siersbæk et al., 2012). PPARg and C/EBPs function as the central, pioneering factors to establish the early chromatin state necessary for white adipocyte differentiation, and many others play important collaborative roles, including SREBPs, STAT5, GR, and EBF1, by modulating their activity. Beyond adipogenesis, dynamic transcriptional programming in differentiated adipocytes by PPARg, SREBP, ChREBP, IRFs, LXRs, and other transcriptional activators impacts lipid storage and response to insulin as well as whole-body physiology. Thus, further work to decipher transcriptional regulators of the adipocyte may provide new insights into obesity and targets for metabolic therapy.
Although much is known about transcriptional activators, the role for repressors in metabolic tissue programming is less understood. Among these, our attention turned to the transcriptional repressor B cell lymphoma 6 (BCL6) as a potential regulator of adiposity. BCL6 is a member of the ZBTB family of C2H2-type zinc-finger transcription factors. Although first discovered for its roles in B and T cell development, immunity, and cancer (Basso and Dalla-Favera, 2012), BCL6 is broadly expressed in metabolic organs. In liver and adipocytes, BCL6 is not only a gene target of growth hormone signaling, but BCL6 protein can physically compete with the nuclear effector of growth hormone, STAT5, at a subset of genomic binding sites (Chen et al., 2009; Lin et al., 2014; Meyer et al., 2009; Zhang et al., 2012). Although cell-type-specific functions for BCL6 outside of the immune system remain undefined, analysis of whole-body knockout mice indicated a potential role for BCL6 in lipid regulation but was confounded by co-occurring defects in immunity and severe inflammatory pathology (LaPensee et al., 2014). Finally, a recent report using short hairpin RNA (shRNA) knockdown in C3H10T1/2 cells indicated a possible role for BCL6 in promoting adipogenesis (Hu et al., 2016). These observations together raised the question of the in vivo role for BCL6 in adipose tissue.
Here, we use adipocyte-specific Bcl6 knockout (Bcl6AKO) mice to discover a role for Bcl6 ablation to increase ScAT mass during development and to enhance whole-body insulin sensitivity. Using a combination of transcriptomics, chromatin immunoprecipitation sequencing (ChIP-seq) in primary adipocytes, stable isotopic labeling in vivo, and lipidomics, we reveal that loss of BCL6 directly upregulates genes controlling growth pathways as well as fatty acid saturation, elongation, and thioesterase enzymes to enhance the production of fatty acids, phospholipids, glycerolipids, sphingolipids, and fatty acid esters of hydroxy fatty acids (FAHFAs) in ScAT. Despite their enhanced adiposity, Bcl6AKO animals exhibit reduced ectopic fat deposition and enhanced hepatic insulin signaling. These results identify BCL6 as a developmental determinant between subcutaneous and visceral adipose tissue expansion and regulator of metabolic health.
RESULTS
BCL6 Is Highly Induced, but Not Required, during Preadipocyte Differentiation
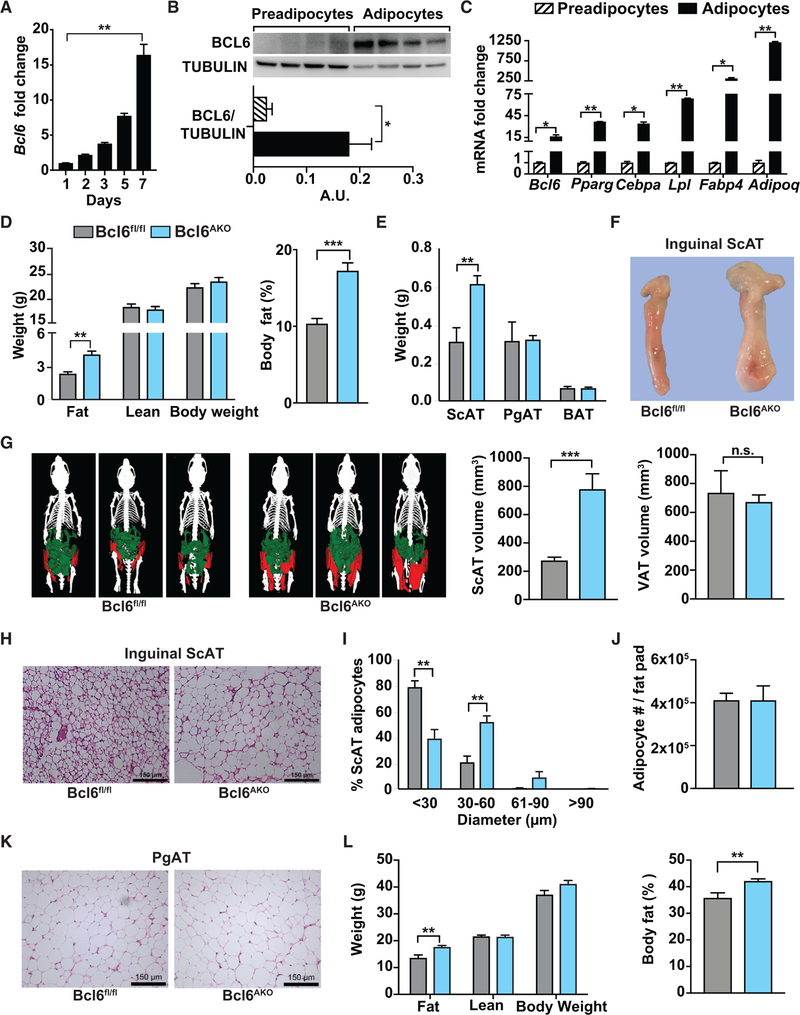
To begin to understand the role for BCL6 in adipocytes, we interrogated BCL6 expression over the course of preadipocyte differentiation. Bcl6 RNA was upregulated more than 15-fold during a 1 week in vitro transition of 3T3L1 preadipocytes to mature adipocytes, with peak Bcl6 RNA levels corresponding to 7-fold increased BCL6 protein levels and the expression of mature adipocyte markers, including Pparg, Cebpa, Lpl, Fabp4, and Adipoq (Figures1A–1C). We observed similar profiles in stromal vascular fraction (SVF)-derived primary preadipocytes from subcutaneous and perigonadal fat pads, as Bcl6 expression was low in progenitors but increased by 10-fold in fully differentiated primary adipocytes (Figure S1A).
Figure 1. BCL6 Is Expressed in Mature Adipocytes and Limits Subcutaneous Adiposity.
(A) qRT-PCR expression of Bcl6 over a 7-day time course of 3T3L1 pre-adipocyte differentiation.
(B) BCL6 and tubulin protein in 3T3L1 pre-adipocytes and differentiated (day 7) adipocytes. Western blot (top) and protein densitometry (below) are shown.
(C) qRT-PCR expression analysis of Bcl6 and mature adipocyte markers in 3T3L1 pre-adipocytes and differentiated (day 7) adipocytes.
(D) Fat, lean, and total body mass in chow-fed control and Bcl6AKO male mice at 8 weeks of age (left) and percent body fat (right). n = 10–11 per group.
(E) Inguinal subcutaneous, perigonadal, and interscapular brown adipose tissue masses in chow-fed male Bcl6fl/fl and Bcl6AKO mice at 9 weeks of age. n = 8–9 per group.
(F) Representative inguinal subcutaneous fat pads from chow-fed Bcl6fl/fl and Bcl6AKO male mice.
(G) Representative three-dimensional CT scan reconstructions comparing inguinal subcutaneous (ScAT, pseudo-colored red) and visceral (VAT, pseudo-colored green) adipose tissues in chow-fed Bcl6fl/fl and Bcl6AKO male mice. Volumes for each are shown (right).
(H) H&E staining of ScAT from chow-fed control and Bcl6AKO male mice. Scale bar, 150 µM.
(I) Adipocyte diameters determined from H&E stained sections of ScAT from chow-fed Bcl6fl/fl and Bcl6AKO male mice. n = 4 per group.
(J) Adipocyte numbers per inguinal ScAT fat pad. n = 5 per group.
(K) H&E stained sections of representative PgAT from chow-fed Bcl6fl/fl and Bcl6AKO male mice. Scale bar, 150 µM.
(L) Fat, lean, and total body mass in control and Bcl6AKO male mice fed a high-fat diet for 12 weeks (left) and percent body fat (right) in chow-fed Bcl6fl/fl and Bcl6AKO male mice. n = 9–12 per group.
Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant. See also Figures S1 and S2.
Next, to test the impact of BCL6 on adipogenesis, we isolated preadipocytes from SVFs of mice harboring floxed alleles of Bcl6 (Bcl6fl/fl) and a tamoxifen-inducible Cre recombinase (CAG-Cre-ER™) (Hayashi and McMahon, 2002). We exposed Bcl6fl/fl and Bcl6fl/fl;CAG-CreER™ SVF-derived preadipocytes to tamoxifen or ethanol control solvent prior to initiating differentiation, which resulted in over 80% deletion of Bcl6 in drug-treated cells from Bcl6fl/fl;CAG-Cre mice (Figure S1B). Genetic knockout of Bcl6 had a neutral or positive effect on the subsequent expression of mature adipocyte markers, including Pparg, Cebpa, Lpl, Fabp4, and Adipoq (Figure S1B), and no impact on in vitro lipid accrual (Figure S1C) compared to tamoxifen-exposed preadipocytes from Bcl6fl/fl mice. Together, these findings led us to conclude that Bcl6 deletion had no apparent functional impact on adipogenesis.
Ablation of Adipocyte Bcl6 during Fetal Adipose Development in Mice Expands ScAT
To explore the impact of BCL6 in adipocytes, in which BCL6 levels are high, we next crossed Bcl6 floxed mice (Bcl6fl/fl) to mice expressing Cre recombinase from the Adipoq promoter (Eguchi et al., 2011) and generated animals constitutively deleted of Bcl6 in adipocytes (Bcl6AKO mice) (Figure S1D). Remarkably, we found that regular chow-fed adult male and female Bcl6AKO mice exhibited increased mass and percentage of fat despite having body weights comparable to Bcl6fl/fl or Adipoq-Cre+ control animals (Figures 1D, S1E, and S1F). Using necropsy analysis and computed tomography (CT) scanning, we found that this expanded fat mass in Bcl6AKO mice was due to a 2-fold increase in subcutaneous, but not perigonadal or brown, adipose tissue (Figures 1E–1G and S1G).
To determine whether the expansion was due to adipocyte hypertrophy, hyperplasia, or both, we quantified adipocyte sizes and numbers. Notably, subcutaneous adipocyte diameters in H&E stained tissue sections were dramatically larger in male Bcl6AKO animals (Figure 1H). Over 60% of subcutaneous adipocytes in Bcl6AKO mice were 30 to 60 microns or larger in diameter, while more than 75% of adipocytes in control animals were under 30 microns in diameter (Figure 1I). Similar results were observed in female Bcl6AKO mice (Figures S1H and S1I). Additionally, this size difference was limited to ScAT and not seen in perigonadal adipose tissue (PgAT) (Figures 1K and S1J). Consistent with our finding that BCL6-deficient preadipocytes undergo normal in vitro differentiation (Figures S1B and S1C), we found no evidence for hypercellularity in the adipose tissues of Bcl6AKO mice (Figure 1J).
High-Fat Diet Provokes Adipose Tissue Inflammation in Bcl6AKO Mice
To assess the impact of BCL6 on fat accrual in response to overnutrition, we provided Bcl6AKO mice with a high-fat diet (HFD) containing 45% of kcal from fat. Total fat mass more than quadrupled in all mice but remained significantly higher in Bcl6AKO mice (average 17.6 g body fat) than Bcl6fl/fl animals (average of 13.6 g body fat) after 12 weeks of diet (Figure 1L). Upon necropsy, we found that progressive adiposity in Bcl6AKO mice was predominantly due to increased ScAT, whereas PgAT was reduced by 30% compared to controls (Figure S1K). Although HFD caused adipocyte hypertrophy in all fat depots, subcutaneous adipocytes of Bcl6AKO mice achieved significantly larger sizes, with nearly 50% of cells exhibiting cell diameters of over 60 microns in Bcl6AKO mice compared to 25% of adipocytes in controls (Figures S2A and S2B). Interestingly, although the size distribution of perigonadal adipocytes was comparable between genotypes, the PgAT of Bcl6AKO mice had increased interstitial cell infiltration and crown-like structures (Figures S2C and S2D). We examined inflammatory gene expression in high-fat-fed Bcl6AKO and control mice, which demonstrated increased levels of the macrophage markers F4/80 and Cd11b, elevated levels of M1 macrophage activation genes Ccl2, IL-6, and Tnfa, and reduced expression of the M2 markers Retnla and Mgl2 in PgAT of Bcl6AKO animals (Figure S2E). A similar but less robust expression pattern was observed in ScAT of Bcl6AKO mice (Figure S2F). Additionally, we assessed vascularization markers since angiogenesis is critical for adipose tissue expansion and insufficiency leads to adipocyte hypoxia, death, and inflammation (Sun et al., 2011). Under chow-fed conditions, there was no qualitative difference in PECAM staining or quantitative difference in the expression of Cd31, Cdh5, Tek, and Vegfa (Figures S2G and S2H), but in HFD-fed Bcl6AKO mice these transcripts were significantly reduced in both ScAT and PgAT compared to HFD-fed controls (Figure S2I). Thus, lack of BCL6 promoted the expansion of subcutaneous adipocytes and overall adiposity, while it simultaneously increased adipose tissue inflammation, attenuated angiogenic markers, and limited visceral fat expansion in the setting of overnutrition.
Loss of Adipocyte Bcl6 Results in Whole-Body Insulin Sensitivity
Subcutaneous body fat distribution is linked to improved glucose metabolism, yet hypertrophic adipocytes and adipose tissue inflammation have been correlated with insulin resistance (Glass and Olefsky, 2012). Given these opposing associations, we sought to determine whether Bcl6 deletion in adipocytes would influence insulin sensitivity. In either regular chow-fed or HFD-fed male and female Bcl6AKO mice, we observed significantly reduced fasting insulin and glucose levels compared to controls (Figures 2A and 2B; data not shown). Moreover, calculated insulin resistance based on the Homeostasis Model Assessment was significantly reduced in Bcl6AKO mice, suggesting improved glucose metabolism (Figure 2C) (Berglund et al., 2008). Next, we performed hyperinsulinemic-euglycemic clamps in HFD-fed Bcl6AKO and Bcl6fl/fl control mice. Remarkably, Bcl6AKO mice required glucose infusion rates nearly double those of Bcl6fl/fl controls to maintain steady-state glucose levels, indicating that Bcl6 ablation in adipocytes dramatically enhanced overall insulin sensitivity (Figure 2D). The rate of glucose disappearance (Rd) trended higher in Bcl6AKO mice but did not reach statistical significance (Figure 2E), while tissue-specific glucose uptake of 14C-deoxyglucose tracer revealed increased accumulation in gastrocnemius and vastus lateralis muscles as well as PgAT but decreased uptake into brown adipose tissue (BAT) (Figure 2F). Although fasting free fatty acid levels were comparable between Bcl6fl/fl and Bcl6AKO mice, clamped levels were significantly lower in Bcl6AKO animals (Figure 2G), reflecting enhancement in their insulin-mediated suppression of lipolysis. More remarkably, however, the calculated rate of endogenous glucose production in Bcl6AKO mice was reduced at all examined time points (Figure 2H, left panel). When considered in the context of plasma insulin concentrations before and during the clamp (Figures 2H, right panel), these results indicated that gluconeogenesis is markedly more sensitive to insulin-mediated suppression in Bcl6AKO animals compared to controls. It is also notable that despite receiving equal rates of exogenous insulin infusion during the procedure, both pre-clamp and clamped plasma insulin levels were significantly lower in Bcl6AKO mice (Figure 2I). Since insulin signaling is coupled to hepatic insulin clearance (Duckworth et al., 1998), this result suggested dramatically enhanced insulin sensitivity in the livers of Bcl6AKO mice. Thus, deletion of Bcl6 in adipocytes, despite rendering mice more obese, improves insulin sensitivity predominantly through enhanced suppression of gluconeogenesis.
Figure 2. Ablation of BCL6 in Adipocytes Promotes Insulin Sensitivity.
(A and B) Fasting serum insulin (A) and blood glucose levels (B) in chow-fed or high-fat diet (HFD)-fed Bcl6fl/fl and Bcl6AKO mice. n = 9–11 per group.
(C) Calculated insulin resistance based on Homeostasis Model Assessment-2 for chow-fed and HFD-fed Bcl6fl/fl and Bcl6AKO mice. n = 9–11 per group.
(D) Glucose infusion rates during hyperinsulinemic-euglycemic clamps in Bcl6fl/fl and Bcl6AKO mice performed after 8 weeks of HFD. n = 9 per group.
(E) Total glucose flux (Rd) before and during insulin clamps. n = 9 per group.
(F) Tissue-specific glucose uptake under clamp conditions. n = 9 per group.
(G) Plasma free fatty acids under basal and insulin clamped conditions. n = 9 per group.
(H) Endogenous glucose production plotted before and during clamping (left) or in relation to plasma insulin levels (right) in Bcl6fl/fl and Bcl6AKO mice. n = 9 per group.
(I) Plasma insulin concentrations under basal and insulin clamped conditions. n = 9 per group.
Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, n.s., not significant. See also Figure S3.
Expansion of Subcutaneous Fat Is Independent of Brown Fat Activity
Bcl6AKO animals exhibited decreased insulin-stimulated glucose uptake into BAT (Figure 2F) and a slight tendency for increased body weight (Figure 1D), raising the possibility of diminished brown fat activity and positive energy balance. To further examine this, we performed PET/CT scanning with [18F]fluoro-deoxyglucose (FDG) and identified a 25% reduction in basal BAT uptake in Bcl6AKO mice (Figure S3A). Consistent with reduced BAT activity, RNA levels of some thermogenic markers, including Ucp1, Prdm16, and Cidea, were significantly reduced in Bcl6AKO animals (Figure S3B). To determine changes in energy balance, we next compared Bcl6AKO and Bcl6fl/fl control mice using metabolic chambers to quantify VO2 and VCO2 by indirect calorimetry, calculate energy expenditure (EE) and respiratory exchange ratio (RER), and determine food intake and activity. Virtually no significant differences were revealed in any of these parameters (Figures S3C–S3H), although RER was lower in Bcl6AKO animals at some time points (Figure S3F). Finally, to address whether decreased thermogenesis could influence the adiposity of Bcl6AKO mice, we bred and continuously housed Bcl6AKO and Bcl6fl/fl control mice at 30°C. Even under thermo-neutral conditions, Bcl6AKO mice exhibited increased total body fat as well as subcutaneous adipocyte cell size and tissue mass (Figures S3I–S3L). Overall, these findings indicated that Bcl6 deletion in adipocytes may diminish BAT activity and energy expenditure, but short-term indirect calorimetry may lack the sensitivity to detect small differences at the whole-body level. However, reduced thermogenesis is unlikely to account for the white adipose tissue expansion of Bcl6AKO mice, since thermoneutral conditions failed to minimize their increased body fat and its marked subcutaneous distribution compared to controls.
Constitutive Bcl6 Ablation Increases ScAT De Novo Lipogenesis
We next investigated the functional impact of Bcl6 ablation on adipocyte lipid metabolism to better understand why Bcl6AKO mice develop subcutaneous adipocyte hypertrophy. To analyze the impact of BCL6 on lipogenesis, we followed the incorporation of 2H from a bolus of deuterated water into C-H bonds of newly formed palmitate and glycerol backbones of adipose tissue triglycerides in Bcl6AKO and Bcl6fl/fl mice. We identified 2-fold increased rates of de novo synthesis of triglyceride-bound palmitate (Figure 3A) and triglyceride-bound glycerol (Figure 3B) in the ScAT of Bcl6AKO mice compared to Bcl6fl/fl controls. Although trending higher, there was no significant difference in lipogenesis in the PgAT between genotypes (Figures 3A and 3B).
Figure 3. BCL6 Governs Healthful Adipose Tissue Gene Expression.
(A and B) Rates of palmitate synthesis (A) and triglycerogenesis (B) in Bcl6fl/fl and Bcl6AKO mice in ScAT, and PgAT determined by 2H incorporation and normalized to DNA. n = 7 per group.
(C) mRNA sequencing data from inguinal subcutaneous adipose tissue and perigonadal adipose tissue plotted as log2 fold change in expression in male Bcl6AKO mice over WT (wild-type) controls (Bcl6fl/fl). Correlation coefficient (r) for all significantly regulated genes in either fat depot is listed. n = 4 per group.
(D and E) Metascape gene list ontology analysis of genes upregulated (D) and downregulated (E) in ScAT and PgAT of Bcl6AKO mice compared to controls.
(F) qRT-PCR quantification of growth and lipid-regulatory genes in ScAT of Bcl6AKO and control mice.
(G) Metascape gene list ontology analysis comparing differentially upregulated genes in ScAT of Bcl6AKO mice to gene expression changes in the subcutaneous adipose tissues of metabolically abnormal obese (MAO) and metabolically normal obese (MNO) human subjects following ~6% weight gain (p < 0.1).
Data are represented as mean ± SEM. n = 4 per group. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figure S4.
To determine the impact of adipocyte BCL6 on lipolysis, we treated chow-fed Bcl6AKO and Bcl6fl/fl mice with the beta-3 adrenergic receptor agonist CL316,243 and sampled circulating free fatty acids (non-esterified fatty acids [NEFAs]) and glycerol over time (Figure S4A). We observed no difference in the concentrations of serum NEFAs or glycerol in Bcl6AKO mice compared to controls. Additionally, we tested isoproterenol-stimulated glycerol and free fatty acid release from isolated ScAT depots ex vivo, which were also not significantly different between genotypes (Figure S4B). Finally, we exposed ScAT lysates to 14C-labeled palmitate and performed beta oxidation assays to determine changes in fatty acid oxidation. We detected comparable total rates of fatty acid oxidation in tissues from Bcl6AKO and Bcl6fl/fl mice based on combined quantities of completely oxidized 14C-labeled CO2 and incompletely oxidized acid soluble metabolites (Figure S4C). Thus, loss of BCL6 in adipocytes increased cell-intrinsic lipogenesis but did not functionally impact fatty acid oxidation or acute lipolytic responses to beta adrenergic agonists.
BCL6 Represses Growth and Lipid Biosynthetic Genes in Adipocytes
To determine the transcriptional basis for subcutaneous adipocyte expansion following Bcl6 ablation, we next determined the BCL6-regulated gene expression networks in inguinal ScAT and PgAT (Figure 3C). Using mRNA sequencing to compare Bcl6AKO and Bcl6fl/fl mice, we identified 1,074 differentially regulated ScAT transcripts (adjusted p value < 0.05) in males consisting of 570 upregulated and 504 downregulated genes. In PgAT, we identified 1,488 differentially regulated transcripts comprised of 759 upregulated and 729 downregulated genes. Although embryonic Bcl6 ablation selectively expands subcutaneous adipocytes, we found that expression changes within the PgAT and ScAT of Bcl6AKO mice were strongly correlated both in males (r = 0.78) and females (r = 0.72) (Figures 3C and S4D). ScAT and PgAT expression changes due to Bcl6 ablation were also highly correlated across sexes (ScAT, r = 0.80 and PgAT, r = 0.76) (Figure S4E; data not shown). Furthermore, integrative analysis of upregulated and downregulated transcripts in ScAT and PgAT revealed similar gene ontologies in both depots (Figures 3D and 3E). We identified tyrosine kinase and developmental growth pathways among the top-scoring terms (Figures 3D), which contained insulin or IGF signaling components implicated in growth, including Igf2 and Akt1, and glycolytic regulators that drive glycerogenesis and NADPH production for lipid synthesis such as Pdk3. Carbohydrate metabolic process and biosynthesis of unsaturated fatty acids were other high-scoring ontologies for upregulated genes in the ScAT and PgAT of Bcl6AKO mice, which consisted of Fads1, Fads2, Elovl6, Hacd1, Asah1, Acot3, Acot4, Lpgat1, Cds2, and other genes linked to lipid synthesis and adipose tissue expansion. Although ScAT and PgAT were categorically similar in many respects, ontologies related to immunity were selectively enriched among genes upregulated in PgAT (Figure 3D), consistent with the proclivity for Bcl6AKO mice to develop immune cell infiltration following a HFD (Figures S2C and S2E). Top-ranking ontologies for genes downregulated in ScAT and PgAT of Bcl6AKO mice were related to fatty acid catabolism (Figure 3E), which included genes involved in beta oxidation (Acox1, Acsl5, Cpt2, and Ehhadh) and lipolysis (Adrb3, Lipe, and Pnpla2). PTEN regulation was also significantly overrepresented as a term but limited to male ScAT, featuring Pten and genes encoding components of the proteasome.
Using qPCR, we confirmed enhanced transcription of unsaturated fatty acid biosynthesis and growth signaling genes, as well as reduced expression of lipolysis and fatty acid oxidation genes in ScAT, PgAT, and BAT of Bcl6AKO mice (Figures 3F, S4H, and S4I). We also interrogated established regulators of de novo saturated fatty acid synthesis (Glut4, Scd1, Fasn, and Dgat) and fatty acid uptake (Cd36, Lpl, and Fatp1/4), which were unchanged in both ScAT and PgAT (Figures S4F–S4H; data not shown) but significantly reduced in BAT of Bcl6AKO animals (Figure S4I; data not shown). Thus, our data overall suggested that ablation of adipocyte Bcl6 enhanced expression of anabolic mediators of growth and lipid synthesis while suppressing mRNAs for catabolic regulators of lipid oxidation and hydrolysis in ScAT, PgAT, and BAT.
Gene Signatures in BCL6-Deficient ScAT Overlap with Healthful Human Patterns
Since Bcl6AKO mice exhibit improved metabolism despite increased adiposity, we compared their gene networks to those in humans with metabolically healthy obesity (Fabbrini et al., 2015). Using expression data from metabolically abnormal or metabolically normal obese (MNO) subjects who underwent subcutaneous fat biopsy before and after moderate weight gain, we found that gene pathways associated with metabolically healthy obese subjects clustered more closely with orthologous gene ontologies enriched in the subcutaneous fat of Bcl6AKO mice, including categories of lipid and monocarboxylic acid metabolism, biosynthesis of unsaturated fatty acids, metabolism of carbohydrates, extracellular matrix organization, and vascular development (Figure 3G). In particular, upregulation of genes involved in fatty acid biosynthesis, including FADS1, FADS2, ELOV6, and HACD1, was common to both MNO humans and Bcl6AKO mice. Overall, these expression data linked Bcl6 ablation in ScAT to lipid synthesis and tissue remodeling genes reminiscent of expression changes in humans with metabolically healthy obesity.
BCL6 Binds to DNA near Lipid Biosynthetic and Growth Regulatory Genes
BCL6 has been characterized as a transcriptional repressor in B cell and other immune lineages, but its direct regulatory impact on gene expression in metabolic tissues is unknown (Basso and Dalla-Favera, 2012). We sought to examine the genomic context through which BCL6 governs adipocyte expansion by generating an adipocyte-specific form of the mammalian INTACT (isolation of nuclei tagged in specific cell types) system (Adipo-Sun1-sfGFP-myc mice) for in vivo ChIP-seq (Figure 4A) (Mo et al., 2015). In primary inguinal subcutaneous adipocytes, we identified 1,784 BCL6 binding sites using a false discovery rate (FDR) cut-off of 0.1%. Ontology analysis for genes near BCL6 binding sites identified mouse phenotype terms that included decreased susceptibility to hepatic steatosis, increased insulin sensitivity, increased adiponectin level, decreased gonadal fat pad weight, abnormal white fat cell size, and others consistent with the ScAT phenotype of Bcl6AKO animals (Figure 4B).
Figure 4. BCL6 Directly Represses Lipid Biosynthetic and Growth Genes.
(A) Scheme for adipocyte-specific ChIP-seq in vivo using Adipo-Sun1-sfGFP-myc mice. Fat pads were harvested, fixed, and homogenized. Adipocyte nuclei were precipitated using myc antibody-coated beads, lysed, and sonicated. Sheared chromatin was immunoprecipitated using BCL6-specific antibody, and the isolated DNA was sequenced.
(B) Mouse phenotype gene ontologies for BCL6 binding sites in subcutaneous adipocytes.
(C) Motif analysis in BCL6 ChIP-seq identified peaks from subcutaneous and perigonadal adipocytes.
(D) Bar graph showing number of targets transcriptionally altered (up or down) and directly bound by BCL6 in ScAT and PgAT.
(E) Representative genome browser tracks along the Fads1 (top) and Igf2 (bottom) genes for BCL6 ChIP-seq in subcutaneous adipocytes and RNA-seq in ScAT of Bcl6fl/fl and Bcl6AKO mice.
(F and G) Directly upregulated (F) and downregulated (G) target gene ontology analysis of ChIP-seq binding sites associated with differentially expressed genes in Bcl6AKO ScAT and PgAT.
See also Figure S4.
In perigonadal adipocytes, we identified 3,956 BCL6 binding sites using the same FDR cut-off, with 510 binding sites in common with ScAT. Overall, BCL6 binding peaks were moderately correlated across depots (rs = 0.23), and peaks common to both ScAT and PgAT exhibited the highest and most correlated signal strengths (rs = 0.69) (Figure S5A). Ontologies and associated mouse phenotypes for genes in vicinity of these common binding sites included regulation of glucose and carbohydrate metabolism, generation of metabolites and energy, response to insulin stimulus, decreased susceptibility to hepatic steatosis, and decreased triglyceride level (Figure S5B). Both in ScAT and PgAT, BCL6 genome-wide DNA binding sites were strongly enriched for consensus BCL6 motifs and to a lesser extent recognition sequences for C/EBP, EBF, NF1, TEAD, STAT, and others (Figure 4C).
Next, we integrated our RNA sequencing (RNA-seq) and ChIP-seq datasets using the Binding and Expression Target Analysis (BETA) pipeline to determine putative direct target genes for BCL6 and define its regulatory potential (Wang et al., 2013c). We found that among genes that were transcriptionally altered by adipocyte BCL6 ablation and located within 100 kb of a BCL6 binding site, 134 genes (out of a total of 177) and 195 (out of 336) were upregulated by loss of BCL6 in ScAT and PgAT, respectively (Figure 4D; Table S1), corresponding to a repressive regulatory potential (p value = 1.56e—7 [ScAT]; p value = 1.44e—6 [PgAT]) for BCL6 in adipocytes (Figure S5C). Gene ontologies for these directly bound, upregulated targets in Bcl6AKO mice, including the Fads1 and Igf2 genes (Figures 4E and S5D), were dominated by unsaturated fatty acid biosynthesis, kinase signaling, and cell growth pathways (Figure 4F). In contrast, genes downregulated in Bcl6AKO mice were not enriched in BCL6 binding sites compared to statically expressed genes (p value = 0.913 [ScAT], p value = 0.804 [PgAT]) (Figure S5C). Ontologies for directly bound downregulated genes included cholesterol metabolism, peroxisome organization, and water-soluble vitamins and cofactor metabolism that were common in both depots (Figure 5G). Using ChIP-qPCR, we validated several binding peaks along directly regulated genes in ScAT and PgAT (Igf2, Fads1, Acot1, Elovl5, Ccnd2/3) and confirmed the absence of BCL6 at genomic regions lacking BCL6 ChIP-seq peaks (Adipoq, Scd1, Fasn) (Figure S5D). Over-all, our genomic studies indicated a direct role for BCL6 in the repression of anabolic signaling and lipid biosynthetic genes in ScAT and PgAT.
Figure 5. Adipocyte Bcl6 Ablation Produces an Insulin-Sensitizing Milieu of Lipokines and Adipokines.
(A) Fatty acid esters of hydroxy fatty acid (FAHFA) in ScAT (left, n = 3 per group) and serum (right, n = 6 per group) of Bcl6fl/fl and Bcl6AKO mice.
(B) ScAT (left, n = 3 per group) and serum (right, n = 6 per group) FAHFAs in Bcl6AKO and Bcl6fl/fl mice.
(C) Serum ceramide levels of chow-fed Bcl6AKO and Bcl6fl/fl mice. n = 6 per group.
(D) Serum adiponectin in chow-fed (left) and HFD-fed (right) mice. n = 9–10 per group.
Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001. See also Figures S5 and S6.
Ablation of Adipocyte BCL6 in Adult Mice Expands All White Adipose Tissue Depots
Transcriptomic and genome-wide binding analyses revealed similar BCL6-regulated networks in visceral and ScAT, yet increased fat mass and adipocyte size was only observed in the ScAT of Bcl6AKO mice (Figures 1E and 1I). To test whether developmental timing of Bcl6 deletion influenced adipose tissue expansion, we developed a doxycycline (Dox)-inducible model to delete Bcl6 in adiponectin-expressing adipocytes, herin denoted as i-Bcl6AKO mice (Wang et al., 2013b). At 8 weeks of age, i-Bcl6AKO and control mice were treated with Dox for 7 days. Although baseline (pre-Dox) body compositions were comparable, fat mass became significantly higher in i-Bcl6AKO mice within 4 weeks after treatment (Figure S5E). In contrast to Bcl6AKO mice, Dox-treated i-Bcl6AKO mice developed increased white adipose tissue in all depots, including the inguinal, inter-scapular, perigonadal, perinephric, and interscapular adipose tissues but not BAT (Figure S5F). Further, adipocyte size was increased in all white adipose tissue (WAT) depots, including ScAT and PgAT (Figures S5G and S5H). Together, these data indicated that de-repression of the BCL6 regulatory network has the potential to expand all white adipocytes but is compensated for during development within visceral depots.
Loss of Adipocyte BCL6 Increases Adipose Tissue and Circulating Lipokines and Adiponectin
To analyze the impact of BCL6 ablation in adipocytes on cellular and circulating lipid composition, we applied shotgun lipidomics to quantify ~ 250 individual species from the ScAT and serum of Bcl6AKO and Bcl6fl/fl animals. In ScAT of Bcl6AKO mice, quantities of the reported insulin-sensitizing lipokines known as FAHFAs and C16:1n7-palmitoleate were more than twice as high compared to controls (Figures 5A, 5B, and S6A) (Cao et al., 2008; Yore et al., 2014). Additionally, we observed increased levels of other triglyceride-associated fatty acyl chains and total triglycerides (Figures S6A and S6B), various membrane-associated and signaling glycerophospholipids, and sphingomyelin (Figures S6C and S6D). In contrast, we found no difference in the level of acylcarnitines in the adipose tissue of Bcl6AKO mice (Figure S6E, left), in line with their unaltered fatty acid oxidation rate. In serum, we similarly detected 2.5-fold elevated levels of FAHFAs (Figure 5A), including the 16:0H18:0 form (PAHSA), which has been directly correlated with human insulin sensitivity (Yore et al., 2014) (Figure 5B, right). In contrast, circulating free palmitoleate levels were unchanged in Bcl6AKO mice (Figure S6A, center), and levels of many other lipids were significantly reduced, including triglyceride-associated fatty acyl chains (Figure S6A, right), total triglycerides (Figure S6B, right), ceramides (Figure 5C), and several glycerophospholipids (Figure S6D, right). As in adipose tissue, levels of acylcarnitines in the serum were unchanged in Bcl6AKO animals (Figure S6E, right).
Additionally, we quantified circulating adipokines and inflammatory mediators. Consistent with the observed 50% reduction in ceramides, we detected ∼30% increased serum levels of adiponectin in regular chow-fed or HFD-fed Bcl6AKO mice (Figure 5D), which is known to induce ceramide degradation(Holland et al., 2011). Levels of leptin were also significantly higher in chow-fed, but not HFD-fed, Bcl6AKO animals (Figure S6F). Despite heightened inflammation in the adipose tissues of Bcl6AKO mice (Figures S2C, S2E, and S2F), circulating adipo-kines associated with insulin resistance, including PAI-1, resistin, interleukin-6 (IL-6), TNFa, and CCL2, were unchanged in chow-fed or HFD-fed mice (Figures S6G–S6K). Thus, Bcl6 ablation broadly altered adipocyte lipid content and conferred an insulin-sensitive pattern of circulating FAHFAs, ceramides, and adiponectin.
Adipocyte BCL6 Deficiency Potentiates Insulin Responses while Reducing Lipid Signaling, Steatosis, and Inflammation in the Liver
Based on our insulin clamp findings and the sensitizing profile of lipokines and adipokines in Bcl6AKO mice, we next examined insulin signaling and gene regulation in liver to further define how loss of Bcl6 conferred insulin sensitivity. Following insulin injection into chow-fed Bcl6AKO and Bcl6fl/fl animals, we observed enhanced liver phosphorylation of the insulin receptor (p-IR) and serine-473 of AKT (p-AKT), with 1.8-and 2.8-fold increased ratios of p-IR to total insulin receptor and p-AKT to total AKT, respectively, in Bcl6AKO mice (Figure 6A). Insulin-induced AKT signaling is known to direct de novo lipogenesis in the liver, which is exacerbated by insulin resistance (Titchenell et al., 2016). Thus, we used in vivo deuterium incorporation to measure de novo hepatic lipogenesis in chow-fed animals, finding rates reduced by 30% in Bcl6AKO mice (Figure 6B). We also revealed a signature of reduced lipid-directed transcription in their livers. RNA-seq of chow-fed mice identified 37 differentially regulated genes, of which 28 were downregulated in livers of Bcl6AKO mice, including Plin2 and Pparg, which have been previously implicated in steatosis (Figure 6C) (Gavrilova et al., 2003; Najt et al., 2016). These altered transcripts were predominantly related to PPAR signaling and associated lipid and carbohydrate metabolism (Figure 6D), and their gene promoters were enriched in PPAR/RXR DNA binding motifs (Figure 6E), consistent with a recent report that SREBP-directed hepatic de novo lipogenesis is a source for PPARa endogenous ligands (Guan et al., 2018). Thus, the liver transcriptional signatures of chow-fed Bcl6AKO mice reflected a reduction in lipogenesis-driven, PPAR-activated gene expression.
Figure 6. Adipocyte Bcl6 Deletion Protects against Hepatic Insulin Resistance and Steatosis.
(A) Insulin-stimulated insulin receptor (IR) and AKT phosphorylation in livers of Bcl6fl/fl and Bcl6AKO mice on chow diet (left). Densitometry analysis (right). n = 4–5 per group.
(B) Rates of total liver palmitate synthesis in chow-fed Bcl6fl/fl and Bcl6AKO mice. n = 7 per group.
(C) Heatmap of differentially regulated genes in livers of ad lib chow-fed Bcl6fl/fl and Bcl6AKO mice. n = 4 per group.
(D) Gene ontology analysis of differentially expressed genes in livers of chow-fed Bcl6fl/fl and Bcl6AKO mice.
(E) Top-scoring motifs in the promoters of differentially regulated genes in livers of chow-fed Bcl6fl/fl and Bcl6AKO mice.
(F) Liver weights in chow-fed and HFD-fed Bcl6fl/f and Bcl6AKO mice. n = 7–9 per group.
(G) Liver triglyceride content in chow-fed and HFD-fed Bcl6fl/fl and Bcl6AKO mice. n = 7–9 per group.
(H) H&E staining of representative livers from Bcl6fl/fl and Bcl6AKO mice after 12 weeks of HFD. Scale bar, 150 mM.
(I) qRT-PCR expression analysis of inflammatory genes in livers of Bcl6fl/fl and Bcl6AKO mice after 12 weeks of HFD. n = 6–7 per group.
Data are represented as mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001.
At the level of tissue composition, we found that liver weights and triglyceride contents were significantly reduced by 13% and 48% in chow-fed or 20% and 44% in high-fat-fed Bcl6AKO animals, respectively (Figures 6F and 6G). Consistent with these quantitative assessments, histopathological examination of liver sections revealed that HFD-induced steatosis was strongly mitigated in Bcl6AKO mice (Figure 6H). These results indicate that loss of BCL6 in adipocytes exerts a protective effect on hepatic lipid accumulation.
Finally, since steatosis is often accompanied by hepatic inflammation, we used qPCR to interrogate the expression of macrophage markers and pro-inflammatory mediators in livers from high-fat-fed Bcl6AKO and control mice. In contrast to the pro-inflammatory changes observed in adipose tissue (Figures S2E and S2F), hepatic expression of F4/80, Ccl2, and Tnfa were each significantly reduced and Cd11b and Il1b trended lower in Bcl6AKO mice (Figure 6I). In sum, these findings revealed that loss of BCL6 in adipocytes enhances liver insulin sensitivity, which is accompanied by diminished hepatic lipid accumulation and lipid-driven PPAR and pro-inflammatory transcription.
DISCUSSION
We have described a mouse model of healthful obesity in Bcl6AKO mice characterized by expanded ScAT and marked insulin sensitivity. Adipocyte Bcl6 deletion both enhances glucose disposal and sharply reduces gluconeogenesis, indicating a potent role for this repressor pathway in whole-body metabolism. At the molecular level, genomic analysis in adipocytes revealed that BCL6 directly downregulates kinase signaling and fatty acid biosynthetic genes, similar to its actions as a transcriptional repressor in immune cells. Moreover, derepression of BCL6 in adipocytes mimics the adipose tissue transcriptomic profile of humans with ‘‘metabolically normal obesity.’’ The impact of adipocyte BCL6 ablation shown here is markedly distinguished from prior characterization of whole-body Bcl6 knockout mice, which demonstrate variable degrees of growth retardation and ill health beginning at a few days to 3 weeks of life, with half dying before 5 weeks of age (Dent et al., 1997). Over 80% of Bcl6−/− mice exhibit myocarditis and over 70% have pulmonary vasculitis with elevated levels of IL-4, −5, and −13, cytokines known to directly impact metabolism (Ricardo-Gonzalez et al., 2010; Stanya et al., 2013). Accordingly, prior metabolic analysis in total knockouts concluding that loss of BCL6 causes lipoatrophy was uninterpretable (LaPensee et al., 2014), and our work unambiguously identifies BCL6 as a direct regulator of adipocyte metabolism and insulin sensitivity.
Fat expansion occurs selectively in the ScAT of Bcl6AKO mice, yet the transcriptomic changes incurred with Bcl6 ablation and BCL6 genome-wide DNA binding are similar in the inguinal and perigonadal fat pads. Remarkably, we find that inducible deletion of adipocyte Bcl6 in adult mice causes adipose tissue hypertrophy in all white adipose tissue depots, more consistent with our genomic data. Hence, the selective subcutaneous adipocyte hypertrophy in Bcl6AKO mice reflects a distinction in depot development. The means by which in utero deletion of Bcl6 becomes ‘‘compensated’’ in visceral depots to prevent adipocyte hypertrophy is unknown, but other studies have documented cellular differences between depots that may influence developmental phenotypes in adipose tissue and favor subcutaneous over visceral growth (Baglioni et al., 2012; Macotela et al., 2012). Adi-poq-Cre targets a developmental population of proliferative and highly adipogenic progenitors present in inguinal subcutaneous, but not perigonadal, adipose tissue (Hong et al., 2015), which has been linked to depot-specific adiposity in knockouts of Zfp423 (Shao et al., 2016; Tao et al., 2017). Accordingly, the timing of Adipoq-Cre expression in utero could play a role in the depot-specific phenotype of Bcl6AKO mice. Finally, distinct transcriptional requirements have been documented in adipogenic development during embryogenesis versus adulthood even within the same depot (Wang et al., 2015), which could also contribute to the depot-selective versus global expansion of adipose tissue in Bcl6AKO and i-Bcl6AKO mice, respectively. Studies to further define the cell and molecular requirements for Bcl6 ablation to expand adipocytes may provide insight into the differences between these models and the developmental basis for body fat distribution.
Our identification of BCL6 as a repressor of adipose tissue expansion shares features with other mouse models of healthful obesity, including transgenic adipocyte overexpression of Glut4 (Shepherd et al., 1993; Yore et al., 2014), mitoNEET (Kusminski et al., 2012), adiponectin (Kim et al., 2007), and Cidea (Abreu-Vieira et al., 2015), or genetic deletion models targeting collagen VI (Khan et al., 2009) or Pten (Morley et al., 2015). Although these molecules are quite varied in their cellular roles, they commonly impact growth pathway signaling, de novo adipocyte lipogenesis, and insulin-sensitizing adipokines or lipokines. Of note, however, in most of these other models, healthy adipose tissue expansion is only observed in states of overnutrition. In contrast, Bcl6AKO mice exhibit spontaneously increased adipose tissue mass and enhanced insulin sensitivity even in the absence of a HFD.
The basis by which Bcl6 deletion in adipocytes confers insulin sensitivity to the liver and other tissues merits further inquiry, but changes in circulating adipokine and lipokines may play important roles. Indeed, increased circulating adiponectin has powerful stimulatory effects to activate ceramidase and mitigate lipotoxic mediators (Holland et al., 2011; Xia et al., 2015), consistent with the reduced levels of serum ceramides found in Bcl6AKO mice. Moreover, we observed elevated levels of FAHFAs, including PAHSA, which has been inversely associated with human insulin resistance and may contribute to the improved metabolism via activation of GPR120 and GPR40 (Syed et al., 2018; Yore et al., 2014). However, the beneficial effects of exogenous FAHFAs have recently been challenged (Pflimlin et al., 2018), raising the possibility that elevated FAHFAs in Bcl6AKO mice could instead be a marker rather than a mediator of systemic insulin sensitivity in Bcl6AKO animals.
Despite their metabolic health, Bcl6AKO mice exhibit exacerbated adipose tissue inflammation and impaired brown fat activity, features that distinguish it from many other models with heightened insulin sensitivity. Although immune cell activation has generally been linked to insulin resistance (Glass and Olefsky, 2012), other studies conclude that inflammation is required for proper adipose tissue remodeling and insulin sensitivity (Tao et al., 2017; Wernstedt Asterholm et al., 2014). Notably, circulating levels of pro-inflammatory cytokines and chemokines were unchanged in Bcl6AKO mice, indicating that inflammation within the adipose tissue did not ramify to other tissues in this model. Our findings suggest that either the benefits of Bcl6 ablation, including reduced hepatic lipid accumulation and inflammation, outweigh the negative consequences of local adipose tissue inflammation or pro-inflammatory effects in adipose tissue could even contribute to the improved metabolic state of Bcl6AKO animals. Although impaired brown fat function has generally been linked to adverse metabolic health (Bartelt and Heeren, 2014), Bcl6AKO mice manifest reduced BAT glucose uptake. These findings suggest that BAT activity and whole-body insulin sensitivity can be dissociated.
Deletion of Bcl6 in adipocytes increases de novo lipogenesis, yet BCL6 does not repress the expression of ‘‘classical’’ lipo-genic enzymes and transporters such as Scd1, Fasn, Dgat1, and Glut4. Thus, our findings indicate that the lipogenic impact of BCL6 ablation is likely to derive from post-transcriptional changes in enzyme activities or pro-anabolic changes in substrates and cofactors. Notably, previous work has demonstrated that lipogenic gene expression and lipogenesis can be uncoupled. For example, in ob/ob mice, the expression of lipogenic genes is reduced, yet de novo lipogenesis is sharply increased in their adipose tissues (Nadler et al., 2000; Soukas et al., 2000; Turner et al., 2007). BCL6-repressed growth factors and signaling components, including IGF, AKT, and PTEN, are known to enhance the activities of anabolic enzymes and the synthesis of metabolites, which can be limiting in lipogenic reactions (Solinas et al., 2015; Ward and Thompson, 2012). Future studies are warranted to determine the impact of adipocyte BCL6 ablation on intermediary metabolism and flux, which may provide further insight into its anabolic impact on adipocytes and whole-body insulin sensitivity.
Aside from PPARg (Soccio et al., 2014), BCL6 represents one of the only examples of a transcriptional regulator of healthful adiposity. BCL6 has been previously proposed to antagonize STAT5, yet growth hormone-STAT5 signaling has lipolytic effects to potently suppress adipose tissue mass (Kaltenecker et al., 2017). The unexpected lipogenic expansion of adipose tissue in Bcl6AKO mice suggests a more complex, non-binary interrelationship between these transcription factors. Given that pharmacologic inhibitors of BCL6 have been developed to target selective interactions between BCL6 and its co-repressors (Cardenas et al., 2016), these findings raise the possibility that BCL6 de-repression could represent an insulin-sensitizing strategy to be tested in future work.
STAR☆METHODS
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| BCL6 | Santa Cruz Biotechnology | Cat# sc-7388; RRID: AB_2063455 |
| BCL6 | Custom polyclonal antibody | N/A |
| Guinea pig IgG | Rockland antibodies & assays | Cat# 006–0102; RRID: AB_840780 |
| Phospho-Akt (Ser473) | Cell Signaling Technology | Cat# 5012; RRID: AB_2224726 |
| Akt (Pan) | Cell Signaling Technology | Cat# 4691; RRID: AB_915783 |
| Phospho-IR | Cell Signaling Technology | Cat# 2969; RRID: AB_11178660 |
| IR | Cell Signaling Technology | Cat# 3020; RRID: AB_2249166 |
| Beta actin | Sigma-Aldrich | Cat# A1978; RRID: AB_476692 |
| Beta tubulin | Sigma-Aldrich | Cat# T4026; RRID: AB_477577 |
| Horseradish peroxidase mouse secondary | Jackson ImmunoResearch Labs | Cat# 115–035-174; RRID: AB_2338512 |
| Horseradish peroxidase rabbit secondary | Jackson ImmunoResearch Labs | Cat# 211–032-171; RRID: AB_2339149 |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Collagenase D | Roche diagnostics | CAS#11088882001 |
| Dispase II | Sigma-Aldrich | D4693 |
| IBMX | Sigma-Aldrich | I5879 |
| Dexamethasone | Sigma-Aldrich | D4902 |
| Rosiglitazone | Sigma-Aldrich | R2408 |
| Tamoxifen | Sigma-Aldrich | T5648 |
| Oil red O | Sigma-Aldrich | O0625 |
| Humulin-100 | Eli Lilly | N/A |
| CL-316,246 | Cayman Chemicals | 17499 |
| Isoproterenol | Sigma-Aldrich | I6379 |
| Disuccinimidyl glutarate | Proteochem | C1104 |
| Formaldehyde | Polysciences, Inc. | Cat#18814 |
| Doxycycline hyclate | Sigma-Aldrich | D9891 |
| Deuterium oxide | Sigma-Aldrich | 151882 |
| Fmoc chloride | Sigma-Aldrich | 23186 |
| N- [4-(Amino methyl)phenyl] pyridinium | Wang et al., 2013a | N/A |
| 14-Fludeoxyglucose | Sofie Biosciences | N/A |
| 14C-Palmitic acid | MP Biomedicals | 011219583 |
| Critical Commercial Assays | ||
| NEFA assay kit | Wako diagnostics | 99934691 |
| Glycerol assay kit | Sigma-Aldrich | MAK117 |
| Triglyceride assay kit | Thermo Fisher Scientific | TR22421 |
| Mouse insulin ELISA kit | Crystal Chem | Cat#90080 |
| Mouse Adiponectin ELISA kit | EMD Millipore | EZMADP-60K |
| Mouse adipokine MILLIPLEX-TNFa, IL-6, MCP-1, Leptin, Resistin, and PAI-1 | EMD Millipore | MADKMAG-71K |
| KAPA hyper prep kit | KAPA biosystems | KK8504 |
| TruSeq Stranded mRNA kit | Illumina | 20020595 |
| Deposited Data | ||
| Raw and analyzed RNA-seq data | This paper | GEO: GSE113408 |
| Raw and analyzed ChIP-seq data | This paper | GEO: GSE113408 |
| Experimental Models: Cell Lines | ||
| 3T3L-1 | ATCC | CL-173 |
| Experimental Models: Organisms/Strains | ||
| B6.FVB-Tg(Adipoq-cre)1Evdr/J | The Jackson Laboratory | 028020 |
| Bcl6fl/fl | This paper | N/A |
| AdipoChaser (adnP-rtTA,TRE-cre, Rosa26-loxP- stop-loxP-lacZ) | Wang et al., 2013b | N/A |
| B6.Cg-Tg(CAG-cre/Esr1*)5Amc/J | The Jackson Laboratory | 004682 |
| B6;129-Gt(ROSA)26Sortm5(CAG-Sun1/sfGFP) Nat/J |
The Jackson Laboratory | 021039 |
| C57BL/6J mouse | The Jackson Laboratory | N/A |
| Oligonucleotides | ||
| Primers for qRT-PCR | Table S2 | N/A |
| Primers for ChIP-PCR | Table S3 | N/A |
| Software and Algorithms | ||
| Graph Pad Prism 7.0 | GraphPad Software | |
| Image Pro | Media Cybernetics | N/A |
| RNA Express | https://support.illumina.com/sequencing/sequencing_software/basespace.html | N/A |
| HOMER | http://homer.ucsd.edu/homer/ | N/A |
| Morpheus | https://software.broadinstitute.org/morpheus/ | N/A |
| Metascape | http://metascape.org/gp/index.html#/main/step1 | N/A |
| BETA | http://cistrome.org/BETA/ | N/A |
| ImageJ | https://imagej.nih.gov/ij/ | N/A |
| Cellometer K2 | Nexcelom Bioscience | N/A |
| Biorad CFX manager | Biorad | #184500 |
| Other | ||
| High fat diet | Research Diets | D12451 |
| Tissue Homogenizer | MoBio PowerLyzer 24 Homogenizer | N/A |
| Mouse MRI machine | EchoMRI | N/A |
| qPCR machine | Biorad CFX384 Touch | N/A |
| NextSeq 500 Sequencer | Illumina | N/A |
| Metabolic Chambers | TSE systems | N/A |
| Nanoscan | Mediso medical imaging systems | N/A |
| Altis triple-quadrupole and Q-Exactive mass spectrometer | Thermo Fisher Scientific | N/A |
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Grant Barish (grant.barish@northwestern.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Animals
Bcl6fl/fl mice were generated through the UC Davis Mouse Biology Program by engineering loxP sites between exons 5 and 6 of the mouse Bcl6 locus. Cre-mediated deletion creates a frameshift mutation, resulting in a protein of 138 amino acids (compared to 708 amino acids in wild-type Bcl6) lacking exons 5 – 10 and the zinc finger DNA binding domain. Bcl6fl/fl mice were crossed with Adipoq-Cre mice (Jackson Laboratories, Stock #028020) to generate Bcl6fl/fl; Adipoq-Cre mice (herein referred to as Bcl6AKO mice). Bcl6fl/fl mice were also crossed with transgenic mice encoding tetracycline transactivator (rtTA) under the control of adiponectin promoter and cre recombinase expressed from a promoter containing Tet response element (TRE-cre) (Wang et al., 2013b), which were kindly provided by Dr. Philipp Scherer (UTSW), to generate animals for doxycycline-inducible Bcl6 deletion in adipocytes (i-Bcl6AKO mice). During Dox treatment, 9-week old male mice were limited to drinking water containing 2 mg/ml doxycycline for 7 days to induce Bcl6 deletion. Additionally, Bcl6fl/fl mice were crossed to animals expressing tamoxifen-inducible Cre from the chicken beta actin promoter, CAG-Cre-ER™ mice (Jackson Laboratories, Stock #004682) for primary preadipocyte differentiation studies. Sun1-sfGFP-myc (Jackson Laboratories, Stock #021039) were further backcrossed to C57BL/6J mice for seven generations and crossed with Adipoq-Cre mice to generate Adipoq-Cre; Sun1-sfGFP-myc mice (Adipo-Sun1-sfGFP-myc mice) for ChIP-seq. Unless otherwise stated, mice were maintained on a 14:10 light: dark (LD) cycle with free access to standard chow and water. We provided either a standard chow or high fat diet (HFD; 45% of kcal from fat, 35% of kcal from carbohydrate and 20% of kcal from protein, Research Diets) ad libitum. 9–12 week old male and female littermates were used for studies conducted on standard chow, or HFD was given to mice beginning at these ages and continued for 8 – 12 weeks as indicated. All animal care and use procedures were conducted in accordance with regulations of the Institutional Animal Care and Use Committee at Northwestern University.
In vitro models
Primary adipocytes: Fat depots were dissected from mice (6–8 weeks of age), washed in PBS, minced and incubated in digestion buffer (PBS, 1.5U/ml Collagenase D, 2.4U/ml Dispase II, 10mM CaCl2) for 30–45 minutes at 37°C with shaking to isolate SVF fractions. Digested tissues were centrifuged to pellet the stromal vascular cells, which were re-suspended in growth media (DMEM/F12 containing 10% fetal bovine serum (FBS), antibiotic and antimycotic (anti-anti, ThermoFisher), added to collagen coated plates, and incubated at 5% CO2. Medium was refreshed to remove floating cells 2 hours after plating.
METHOD DETAILS
In vitro differentiation of preadipocytes into adipocytes
Once stromal vascular cells or 3T3L-1 cells reached confluency, they were differentiated by changing the medium to DMEM containing 10% fetal bovine serum 1% anti-anti, 0.5 mM IBMX, 0.25 mM dexamethasone, 2 mM rosiglitazone and 1g/ml insulin. After 2 days, the medium was changed to DMEM containing 10% fetal bovine serum 1% anti-anti 1 g/ml insulin for 48 hours. On day 5, the medium was changed to basal medium (DMEM containing 10% fetal bovine serum 1% anti-anti). Cells were harvested for assays at specified time points. For in vitro deletion studies in Bcl6fl/fl;CAG-Cre-ER™ SVF-derived preadipocytes, confluent cells were treated with 1mM tamoxifen or ethanol for 24 hours prior to induction of differentiation.
Oil Red O staining
Differentiated adipocytes were fixed in 10% formalin for 10 minutes at room temperature. Following fixation, the cells were washed twice with deionized water and incubated in 60% isopropanol for 10 minutes. Next, isopropanol was removed, Oil Red O solution (2g Oil Red O in 1L of 60% isopropanol) was added, and cells were incubated for 10 minutes. After incubation, the Oil Red O solution was removed, and cells were washed four times with deionized water and imaged.
Adipocyte size and number determination
Adipocyte size was calculated using microscopic images (Zeiss Axio) of hematoxylin and eosin (H&E) stained adipocytes in both chow-fed and HFD-fed mice using Image Pro software (Media Cybernetics). Diameters were determined for at least 300 adipo-cytes/mouse using the count/size function. Adipocyte numbers were determined using a Cellometer K2 (Nexcelom Biosciences). After whole fat pads from one side of the body were collagenase digested as described above, the top adipocyte-containing layer was re-suspended in PBS with 2% FBS cells and labeled with dual fluorescence of Acridine Orange (AO) and Propidium iodide (PI) prior to determining adipocyte number/fat pad.
Histological analysis
Tissues were dissected and fixed in 10% formaldehyde overnight and transferred to 70% ethanol. Paraffin processing, embedding, sectioning and hematoxylin and eosin (H&E) staining were performed at Northwestern University’s Mouse Histology & Phenotyping Laboratory (MHPL).
Immunoblotting
Cells or tissues were homogenized in RIPA buffer (1 mM EDTA, 0.5 mM EGTA, 1% Triton X-100, 0.1% sodium deoxycholate, 0.1% SDS, 140 mM NaCl) containing protease and phosphatase inhibitors (Roche). Total lysates were subjected to 10% SDS-PAGE and transferred to PVDF membranes. Membranes were probed with antibodies to BCL6 (Santa Cruz, sc-7388), p-AKT (Cell Signaling, D9E), total AKT (Cell Signaling, C67E7), p-IR (Cell Signaling, 19H7), b-IR (Cell Signaling, L55B10), b-ACTIN (Sigma, A1978), b-TUBULIN (Sigma, T4026). Primary antibody incubations were followed by incubations with horseradish peroxidase conjugated secondary antibodies (Jackson ImmunoResearch). Blots were visualized using enhanced chemiluminescent (ECL) horseradish peroxidase (HRP) substrate (Thermo Scientific SuperSignal West Pico or West Femto).
Blood glucose measurements
After a 5 hour fast, blood glucose levels were measured using an AlphaTRAK® Glucose Meter (Abbott Animal Health) from tail vein bleeds.
Insulin signaling assays
Insulin signaling tests were performed following a 5 hour fast. Tissues were quickly obtained and flash frozen 15 minutes after intraperitoneal injection of insulin (0.5U/kg lean mass; Humulin R U-100). Lysates were prepared for immunoblotting as described above.
Hyperinsulinemic–euglycemic clamps
Hyperinsulinemic–euglycemic clamps were performed on 8 week HFD-fed conscious, unrestrained male mice at the Mouse Metabolic Phenotyping Center (MMPC) of Vanderbilt University, https://vmmpc.org/methods-standards/.
In vivo lipolysis assay
Following a 5 hour fast, mice were given an IP injection of CL 316, 246 (1mg/kg, Cayman Chemical) and blood was collected for serum NEFA and glycerol analysis at 0, 15 and 60 min after injection.
Ex vivo lipolysis assay
Assays of inguinal subcutaneous fat explants were performed using 20 mg tissue pieces incubated in 200ml DMEM containing 2% FA-free BSA, with or without 1mM isoproterenol in 96 well plates at 37°C 5% CO2, and 95% humidified incubator for one hour. Medium was taken out after one hour and glycerol and NEFA release was quantified. DNA was extracted from the tissues as described above for data normalization.
Fatty acid oxidation
Fatty acid (FA) oxidation assay was performed as described elsewhere (Huynh et al., 2014). Briefly, tissue was dounce homogenized in sucrose/Tris/EDTA buffer. FA oxidation in isolated adipose tissue mitochondria was determined by measuring 14CO2 production and incompletely oxidized 14C-labeled fatty acids derived from14C-palmitic acid (0.4 mCi/ml) after 1 hour incubation at 37°C. The buffer was acidified with 1M perchloric acid and maintained sealed for an additional 1 hour at room temperature. Acid soluble and filter-trapped radioactivity was quantified by scintillation counting. 14CO2 and acid production rates were normalized to mg protein content.
Serum measurements
Blood samples were collected after a 5 hour fast and serum was collected using clotting activator tubes (Sarstedt). Serum insulin (Crystal Chem) and adiponectin (EMD Millipore) were measured by ELISA according to manufacturer’s instructions. TNFa, IL-6, MCP-1, Leptin, Resistin, and PAI-1 serum levels were measured using a Luminex multiplex kit (EMD Millipore) according to manufacturer’s instructions. Serum NEFA (Wako diagnostics) and glycerol (Sigma) levels were measured using colorimetric assays.
Triglyceride quantification in liver
Lipid was extracted using the Folch method. Briefly, liver tissue samples were homogenized in chloroform: methanol (2:1 by volume). After an overnight extraction, 0.7% sodium chloride was added. The aqueous layer was aspirated and triplicate aliquots of the chloroform/lipid layer were dried under nitrogen gas. The lipid was reconstituted in isopropyl alcohol and assayed for triglyceride using colorimetric assays (Thermo Fisher).
Indirect calorimetry
Mice were individually housed in metabolic chambers (PhenoMaster, TSE Systems) to monitor food intake, locomotor activity and gas exchange. Chambers were maintained with 12-hr light/dark cycles. Following 48 hours of acclimatization, O2 consumption (VO2) and CO2 production (VCO2) were measured every 30 minutes for a total of 72 hours and used to determine the respiratory exchange ratio (RER = VCO2/VO2) and energy expenditure (EE = 3.185+1.232 x RER) x VO2). VO2, VCO2, and energy expenditure values were normalized to total body weight. Total locomotor activity was simultaneously measured on the X and Y axes using an infrared photocell beam interruption technique.
DNA isolation and quantification
Tissues were flash frozen upon necropsy and stored at —80°C. DNA from frozen adipose tissues was isolated per manufacturer’s instructions using DNeasy Blood and Tissue Kits (QIAGEN). Concentrations were analyzed using a NanoDrop spectrophotometer.
RNA isolation and qPCR mRNA quantification
Tissues were flash frozen upon necropsy and stored at —80°C. RNA was isolated from 100mg of tissue using TRIzol reagent (Thermo Fisher) according to the manufacturer’s protocol and purified using RNeasy columns (QIAGEN). cDNAs were synthesized using iScript cDNA synthesis kits (Bio-Rad). Quantitative real-time PCR analysis was performed with SYBR Green Master Mix (Bio-Rad) and analyzed using a Bio-Rad CFX384 Touch Real-Time PCR System. Expression levels were determined using the relative standard curve method and normalized to the housekeeping gene 36b4. Primer sequences for qRT-PCR are listed in Table S2.
RNA-sequencing and analysis
Following RNA isolation, RNA quality was confirmed using a Bioanalyzer (Agilent), and sequencing libraries were constructed using Illumina TruSeq Stranded mRNA sample prep kits according to the manufacturer’s instructions. Libraries were quantified using Bio-analyzer (Agilent) and qPCR-based quantification (Kapa Biosystems) and sequenced on an Illumina NextSeq 500 instrument using 75 bp paired-end reads (adipose tissue samples) or 75 bp single-end reads (liver samples). Sequenced reads were aligned to the mm10 reference mouse genome using STAR and differentially expressed RNAs were identified using DESeq2 (FDR-adjusted p value < 0.05). STAR alignment and DESeq2 analysis were executed through the RNA Express BaseSpace application (Illumina). Ontology analysis was done using Metascape (http://metascape.org/), performed in March 2018. Heatmaps were generated using rlog stabilized counts. Promoter Motif analysis was done using HOMER. For Human microarray data and RNA-seq data comparison: Metascape multiple gene list analysis was performed using combined gene lists of Bcl6AKO males and females (AKO/WT) converted to human gene identifiers, MNO post/pre and MAO post/pre microarray gene lists (Fabbrini et al., 2015). The raw and processed data are deposited in the GEO database (accession number GEO: GSE113408).
Chromatin immunoprecipitation
Adipo-Sun1-sfGFP-myc mice were used for the specific isolation of adipocyte chromatin. Finely minced adipose tissue was fixed for 30 minutes in 2mM disuccinimidyl glutarate and then for 10 minutes in 1% formaldehyde. Fixed tissue was then dounced in homogenization buffer (5mM NaCl, 0.05mM EDTA pH 7.5, 5mM, 0.005% NP40, 0.01% Triton X-100) and filtered through a 30-micron mesh. Filtrates were centrifuged to pellet nuclei, which were thoroughly resuspended in homogenization buffer and affinity purified using anti-c-myc magnetic beads (Pierce). Isolated adipocyte nuclei were sonicated using a Diagenode Bioruptor. Sheared chromatin was incubated with antibodies against BCL6 (affinity-purified guinea pig IgG) and precipitated with M280 paramagnetic beads (ThermoFisher) coated with anti-guinea pig IgG. DNA was isolated using MinElute PCR purification columns (QIAGEN). ChIP qPCR assays were performed using duplicates. Primer sequences for ChIP-qPCR are listed in Table S3.
ChIP-sequencing and analysis
ChIP DNA libraries were prepared using KAPA Hyper Prep kits (Kapa Biosystems) per manufacturer’s instructions and size selected using a PippenHT (Sage Science). Libraries were assessed with a Bioanalyzer (Agilent) and qPCR-based quantification (Kapa Bio-systems), and sequenced using 75bp single-end reads on an Illumina Next-Seq 500. Raw sequence reads were aligned to the mm10 reference genome using bowtie 1.1.1 with parameters ‘‘–best’’ and ‘‘–m’’ to ensure selection of uniquely mapped reads. Aligned reads were visualized using UCSC genome browser. Analysis was performed using HOMER. Tag directories were generated using ‘‘makeTagDirectory’’ using the -tbp 1 option. ChIP-Seq peaks were designated as regions with 4-fold enrichment over both the input sample and the local background and were normalized to 10 million reads using default parameters (FDR = 0.001) for the ‘‘findPeaks’’ command and specifying ‘‘-style factor.’’ Peaks were annotated to nearest genes using ‘‘annotatePeaks.pl.’’ To identify motifs, we scanned 200 bp windows surrounding peaks using ‘‘findMotifsGenome.pl’’ with standard background. Binding site gene ontology analysis was performed using GREAT (http://great.stanford.edu/public/html/) and default parameters, and sites were integrated with RNA-seq data using BETA (Wang et al., 2013c) and default parameters.
De novo lipogenesis
The rates of tissue lipogenesis were determined following the incorporation of 2H from 2H2O into newly made TG-bound fatty acids and glycerol. Briefly, mice were given 0.7 mL intraperitoneal injections of 2H-labeled saline (9g of NaCl in 1,000 mL of 99.9% 2H2O) at 3 pm, returned to their cages and maintained on 6% 2H-labeled drinking water. After 24 hours, mice were euthanized and serum, liver, and adipose tissues were collected, weighed, and snap frozen. Sample processing and GC/MS analysis was performed as described previously (Bederman et al., 2012, Bederman et al., 2006). The rate of triglyceride formation or triglycerogenesis was based on 2H label incorporation into C-H bonds of TG-bound glycerol. Data was normalized to tissue mass or DNA content, as indicated.
Lipid extraction and lipid derivatization for lipidomic analysis
Mice were euthanized and adipose tissues were harvested, weighed, snap frozen in liquid N2, and stored at —80°C. Tissues were homogenized in 0.5 mL of ice-cold diluted PBS (0.1x) with a Potter-Elvehjem tissue grinder, or serum samples were collected as described previously. A pre-mixture of internal standards for quantification of all reported lipid classes was added based on the tissue DNA content and the volume of serum samples, respectively. Lipid extraction was performed by a modified Bligh and Dyer method as previously described (Wang and Han, 2014, Han, 2016). All the lipid extracts were flushed with N2, capped, and stored at —20°C. A portion of individual lipid extract was derivatized with fluorenylmethoxylcarbonyl (Fmoc) chloride for analysis of PE (Han et al., 2005), and another part was reacted with N-[4-(Aminomethyl)phenyl]pyridinium in order to measure the levels of FAHFAs (Wang et al., 2013a).
Mass spectrometric analysis of lipids
Mass spectrometric analysis of lipids was performed with an Altis triple-quadrupole mass spectrometer (Thermo Fisher Scientific, San Jose, CA) or a Q-Exactive mass spectrometer (Thermo Fisher Scientific, San Jose, CA) equipped with an automated nanospray device (Triversa Nanomate, Advion Biosciences, Ithaca, NY) and operated with Xcalibur software as previously described (Han et al., 2008). Identification and quantification of all reported lipid molecular species were performed using an in-house automated software program (Wang et al., 2017). All adipose tissue or serum lipid levels were normalized to sample DNA content or serum volume, respectively.
PET and CT image acquisition and analysis
PET and CT imaging studies were performed with nanoScan8 (Mediso, Budapest, Hungary). The mice were positioned head-first prone on a plastic bed, with a nose cone providing 1.5% isoflurane throughout the scan. Body-temperature was monitored and maintained stable. A tail vein catheter was placed in each mouse while anesthetized to deliver 18F labeled Fluorodeoxyglucose (18-FDG) within the scanner. Radioactive tracers were obtained through commercial sources (Sofie, Chicago) and each dose was calibrated using a Capintec dose calibrator. A whole-body CT sequence was initially used with the following parameters (energy = 35 kVp, exposure time = 300 msec, exposure = 240 µAs, slice thickness = 0.137 mm, voxel size = 0.137 mm3). The 3D whole body CT images were reconstructed from the data and a material map was also generated to be used for attenuation correction during PET reconstruction. For all subjects, a PET scanning sequence was initiated 5–10 minutes after bolus injection of an FDG dose of ~1MBq/gram (of body-weight). A total acquisition time of 75 minutes from moment of injection was used to acquire the data. A list-mode acquisition modality was used to enable dynamic reconstruction of the acquired data with different temporal resolution throughout the whole scan. PET images were generated using an OSEM reconstruction approach and corrected for attenuation using the CT material maps. The final images yielded whole body FDG uptake-maps in 3D at a resolution of 0.7 3 0.7 3 0.7 mm3
All image analysis (for both PET and CT) was performed using commercial software VivoQuant (inviCRO, Boston, MA, USA). The analysis of the images to generate quantitative results for whole-body fat-content characterization was conducted the following way: 1) abdominal subcutaneous tissue regions were detected and manually delineated with CT volumes windowed between —1000 to 1000 [HU] for optimal visual contrast. 2) visceral tissue regions were semi-automatically delineated by manually selecting the peritoneum, and automatically thresholding all voxels between —500 and —50 [HU] as visceral adipose tissue.
Selection of these ranges was based on calibration data that we obtained from fresh ex-vivo samples of different types of fat harvested from several mice (data not shown). 3D-renderings were generated from the corresponding 3D CT images using the segmentation outlines obtained with these threshold settings for each mouse. The rendered surfaces corresponding to the fat-tissue types were superimposed on corresponding skeletal maximum intensity projection images (MPI). PET was standardized across subjects to quantify glucose uptake and metabolism using standardized uptake value (SUV) maps generated from raw data using the conventional formula. To obtain an accurate measure of the regional uptake values for BAT, we used a hybrid approach for segmentation which relied on both PET and CT data. In the interscapular region, all voxels with CT values between —300 and 150 [HU] and PET-SUV values greater than 2 were classified as BAT. The resulting volume and activity was chosen to be the corresponding BAT value. This enabled us to detect BAT regions of higher activity by controlling for BAT volume.
QUANTIFICATION AND STATISTICAL ANALYSIS
All data were expressed as the mean ± SEM. We used Microsoft Excel and GraphPad Prism 7.0 (GraphPad Software, Inc.) to perform the statistical analyses. For comparisons between two independent groups, a Student’s t test was used and p < 0.05 was considered statistically significant. For comparisons between three or more groups, two-way ANOVA with Bonferroni multiple comparisons testing was performed. Correlation coefficients were calculated using Pearson correlation or nonparametric Spearman correlation. All sample sizes, statistical test methods, and p values are listed in the figure legends.
Supplementary Material
Highlights.
Bcl6 deletion in adipocytes leads to subcutaneous adipose tissue expansion
Loss of Bcl6 in adipocytes improves insulin sensitivity and protects from steatosis
BCL6 directly represses genes controlling fatty acid biosynthesis and cell growth
Bcl6 ablation in adipocytes increases adiponectin and FAHFAs
ACKNOWLEDGMENTS
We thank J. Bass, C. Peek, N. Chandel, and D. Chakravarti for helpful discussion, the UC Davis Mouse Biology Program for generating Bcl6 floxed mice, and the Vanderbilt MMPC for performing clamps. We are also grateful to Northwestern University’s Center for Translational Imaging, Comprehensive Metabolic Core, and Mouse Histology and Phenotyping Laboratory supported by NCI P30-CA060553; the Mass Spectrometry Core Facility at the UT Health Science Center at San Antonio and the Case Western Reserve University Research Institute for Children’s Health Analytical Core for services; and Illumina for support with sequencing. This work was supported by HHMI and NIH grants DK057978 and HL105278 (R.M.E.), as well as R01DK108987, P30DK020595, and K08HL092298 (G.D.B.).
Footnotes
DECLARATION OF INTERESTS
The authors declare no competing interests.
SUPPLEMENTAL INFORMATION
Supplemental Information includes six figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.11.074.
DATA AND SOFTWARE AVAILABILITY
The accession number for RNA-seq and ChIP-seq data reported in this paper is GEO: GSE113408.
REFERENCES
- Abreu-Vieira G, Fischer AW, Mattsson C, de Jong JM, Shabalina IG, Ryde´ n M, Laurencikiene J, Arner P, Cannon B, Nedergaard J, and Pet-rovic N (2015). Cidea improves the metabolic profile through expansion of adipose tissue. Nat. Commun 6, 7433. [DOI] [PubMed] [Google Scholar]
- Baglioni S, Cantini G, Poli G, Francalanci M, Squecco R, Di Franco A, Borgogni E, Frontera S, Nesi G, Liotta F, et al. (2012). Functional differences in visceral and subcutaneous fat pads originate from differences in the adipose stem cell. PLoS ONE 7, e36569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartelt A, and Heeren J (2014). Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol 10, 24–36. [DOI] [PubMed] [Google Scholar]
- Basso K, and Dalla-Favera R (2012). Roles of BCL6 in normal and transformed germinal center B cells. Immunol. Rev 247, 172–183. [DOI] [PubMed] [Google Scholar]
- Bederman IR, Dufner DA, Alexander JC, and Previs SF (2006). Novel application of the ‘‘doubly labeled’’ water method: measuring CO2 production and the tissue-specific dynamics of lipid and protein in vivo. Am. J. Physiol. En-docrinol. Metab 290, E1048–E1056. [DOI] [PubMed] [Google Scholar]
- Bederman I, Perez A, Henderson L, Freedman JA, Poleman J, Guentert D, Ruhrkraut N, and Drumm ML (2012). Altered de novo lipogenesis contributes to low adipose stores in cystic fibrosis mice. Am. J. Physiol. Gastrointest. Liver Physiol 303, G507–G518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berglund ED, Li CY, Poffenberger G, Ayala JE, Fueger PT, Willis SE, Jewell MM, Powers AC, and Wasserman DH (2008). Glucose metabolism in vivo in four commonly used inbred mouse strains. Diabetes 57, 1790–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao H, Gerhold K, Mayers JR, Wiest MM, Watkins SM, and Hotamisli-gil GS (2008). Identification of a lipokine, a lipid hormone linking adipose tissue to systemic metabolism. Cell 134, 933–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cardenas MG, Yu W, Beguelin W, Teater MR, Geng H, Goldstein RL, Oswald E, Hatzi K, Yang SN, Cohen J, et al. (2016). Rationally designed BCL6 inhibitors target activated B cell diffuse large B cell lymphoma. J. Clin. Invest 126, 3351–3362. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y, Lin G, Huo JS, Barney D, Wang Z, Livshiz T, States DJ, Qin ZS, and Schwartz J (2009). Computational and functional analysis of growth hormone (GH)-regulated genes identifies the transcriptional repressor B-cell lymphoma 6 (Bc16) as a participant in GH-regulated transcription. Endocrinology 150, 3645–3654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, and Staudt LM (1997). Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science 276, 589–592. [DOI] [PubMed] [Google Scholar]
- Duckworth WC, Bennett RG, and Hamel FG (1998). Insulin degradation: progress and potential. Endocr. Rev 19, 608–624. [DOI] [PubMed] [Google Scholar]
- Eguchi J, Wang X, Yu S, Kershaw EE, Chiu PC, Dushay J, Estall JL, Klein U, Maratos-Flier E, and Rosen ED (2011). Transcriptional control of adipose lipid handling by IRF4. Cell Metab 13, 249–259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fabbrini E, Yoshino J, Yoshino M, Magkos F, Tiemann Luecking C, Samovski D, Fraterrigo G, Okunade AL, Patterson BW, and Klein S (2015). Metabolically normal obese people are protected from adverse effects following weight gain. J. Clin. Invest 125, 787–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gavrilova O, Haluzik M, Matsusue K, Cutson JJ, Johnson L, Dietz KR, Nicol CJ, Vinson C, Gonzalez FJ, and Reitman ML (2003). Liver peroxisome proliferator-activated receptor gamma contributes to hepatic steatosis, triglyceride clearance, and regulation of body fat mass. J. Biol. Chem 278, 34268–34276. [DOI] [PubMed] [Google Scholar]
- Glass CK, and Olefsky JM (2012). Inflammation and lipid signaling in the etiology of insulin resistance. Cell Metab 15, 635–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan D, Xiong Y, Borck PC, Jang C, Doulias PT, Papazyan R, Fang B, Jiang C, Zhang Y, Briggs ER, et al. (2018). Diet-Induced Circadian Enhancer Remodeling Synchronizes Opposing Hepatic Lipid Metabolic Processes. Cell 174, 831–842.e12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X (2016). Lipidomics: comprehensive mass spectrometry of lipids (Hoboken, New Jersey: Wiley; ). [Google Scholar]
- Han X, Yang K, Cheng H, Fikes KN, and Gross RW (2005). Shotgun lipidomics of phosphoethanolamine-containing lipids in biological samples after one-step in situ derivatization. J. Lipid Res 46, 1548–1560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han X, Yang K, and Gross RW (2008). Microfluidics-based electrospray ionization enhances the intrasource separation of lipid classes and extends identification of individual molecular species through multi-dimensional mass spectrometry: development of an automated high-throughput platform for shotgun lipidomics. Rapid Commun. Mass Spectrom 22, 2115–2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, and McMahon AP (2002). Efficient recombination in diverse tissues by a tamoxifen-inducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev. Biol 244, 305–318. [DOI] [PubMed] [Google Scholar]
- Holland WL, Miller RA, Wang ZV, Sun K, Barth BM, Bui HH, Davis KE, Bikman BT, Halberg N, Rutkowski JM, et al. (2011). Receptor-mediated activation of ceramidase activity initiates the pleiotropic actions of adiponectin. Nat. Med 17, 55–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong KY, Bae H, Park I, Park DY, Kim KH, Kubota Y, Cho ES, Kim H, Adams RH, Yoo OJ, and Koh GY (2015). Perilipin+ embryonic preadipocytes actively proliferate along growing vasculatures for adipose expansion. Development 142, 2623–2632. [DOI] [PubMed] [Google Scholar]
- Hu X, Zhou Y, Yang Y, Peng J, Song T, Xu T, Wei H, Jiang S, and Peng J (2016). Identification of zinc finger protein Bcl6 as a novel regulator of early adipose commitment. Open Biol 6, 160065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huynh FK, Green MF, Koves TR, and Hirschey MD (2014). Measurement of fatty acid oxidation rates in animal tissues and cell lines. Methods En-zymol 542, 391–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Church CD, Holtrup B, Colman L, and Rodeheffer MS (2015). Rapid depot-specific activation of adipocyte precursor cells at the onset of obesity. Nat. Cell Biol 17, 376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeffery E, Wing A, Holtrup B, Sebo Z, Kaplan JL, Saavedra-Pen˜ a R, Church CD, Colman L, Berry R, and Rodeheffer MS (2016). The Adipose Tissue Microenvironment Regulates Depot-Specific Adipogenesis in Obesity. Cell Metab 24, 142–150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaltenecker D, Mueller KM, Benedikt P, Feiler U, Themanns M, Schlederer M, Kenner L, Schweiger M, Haemmerle G, and Moriggl R (2017). Adipocyte STAT5 deficiency promotes adiposity and impairs lipid mobilisation in mice. Diabetologia 60, 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan T, Muise ES, Iyengar P, Wang ZV, Chandalia M, Abate N, Zhang BB, Bonaldo P, Chua S, and Scherer PE (2009). Metabolic dysregulation and adipose tissue fibrosis: role of collagen VI. Mol. Cell. Biol 29, 1575–1591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim JY, van de Wall E, Laplante M, Azzara A, Trujillo ME, Hofmann SM, Schraw T, Durand JL, Li H, Li G, et al. (2007). Obesity-associated improvements in metabolic profile through expansion of adipose tissue. J. Clin. Invest 117, 2621–2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusminski CM, Holland WL, Sun K, Park J, Spurgin SB, Lin Y, Askew GR, Simcox JA, McClain DA, Li C, and Scherer PE (2012). MitoNEET-driven alterations in adipocyte mitochondrial activity reveal a crucial adaptive process that preserves insulin sensitivity in obesity. Nat. Med 18, 1539–1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- LaPensee CR, Lin G, Dent AL, and Schwartz J (2014). Deficiency of the transcriptional repressor B cell lymphoma 6 (Bcl6) is accompanied by dysregulated lipid metabolism. PLoS ONE 9, e97090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MJ, Wu Y, and Fried SK (2013). Adipose tissue heterogeneity: implication of depot differences in adipose tissue for obesity complications. Mol. Aspects Med 34, 1–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin G, LaPensee CR, Qin ZS, and Schwartz J (2014). Reciprocal occupancy of BCL6 and STAT5 on Growth Hormone target genes: contrasting transcriptional outcomes and promoter-specific roles of p300 and HDAC3. Mol. Cell. Endocrinol 395, 19–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macotela Y, Emanuelli B, Mori MA, Gesta S, Schulz TJ, Tseng YH, and Kahn CR (2012). Intrinsic differences in adipocyte precursor cells from different white fat depots. Diabetes 61, 1691–1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaughlin T, Lamendola C, Liu A, and Abbasi F (2011). Preferential fat deposition in subcutaneous versus visceral depots is associated with insulin sensitivity. J. Clin. Endocrinol. Metab 96, E1756–E1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RD, Laz EV, Su T, and Waxman DJ (2009). Male-specific hepatic Bcl6: growth hormone-induced block of transcription elongation in females and binding to target genes inversely coordinated with STAT5. Mol. Endocrinol 23, 1914–1926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mo A, Mukamel EA, Davis FP, Luo C, Henry GL, Picard S, Urich MA, Nery JR, Sejnowski TJ, Lister R, et al. (2015). Epigenomic Signatures of Neuronal Diversity in the Mammalian Brain. Neuron 86, 1369–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morley TS, Xia JY, and Scherer PE (2015). Selective enhancement of insulin sensitivity in the mature adipocyte is sufficient for systemic metabolic improvements. Nat. Commun 6, 7906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nadler ST, Stoehr JP, Schueler KL, Tanimoto G, Yandell BS, and At-tie AD (2000). The expression of adipogenic genes is decreased in obesity and diabetes mellitus. Proc. Natl. Acad. Sci. USA 97, 11371–11376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Najt CP, Senthivinayagam S, Aljazi MB, Fader KA, Olenic SD, Brock JR, Lydic TA, Jones AD, and Atshaves BP (2016). Liver-specific loss of Perilipin 2 alleviates diet-induced hepatic steatosis, inflammation, and fibrosis. Am. J. Physiol. Gastrointest. Liver Physiol 310, G726–G738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neeland IJ, Poirier P, and Despre´ s JP (2018). Cardiovascular and Metabolic Heterogeneity of Obesity: Clinical Challenges and Implications for Management. Circulation 137, 1391–1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer BF, and Clegg DJ (2015). The sexual dimorphism of obesity. Mol. Cell. Endocrinol 402, 113–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pflimlin E, Bielohuby M, Korn M, Breitschopf K, Lohn M, Wohlfart P, Konkar A, Podeschwa M, Barenz F, Pfenninger A, et al. (2018). Acute and Repeated Treatment with 5-PAHSA or 9-PAHSA Isomers Does Not Improve Glucose Control in Mice. Cell Metab 28, 217–227.e13. [DOI] [PubMed] [Google Scholar]
- Ricardo-Gonzalez RR, Red Eagle A, Odegaard JI, Jouihan H, Morel CR, Heredia JE, Mukundan L, Wu D, Locksley RM, and Chawla A (2010). IL-4/STAT6 immune axis regulates peripheral nutrient metabolism and insulin sensitivity. Proc. Natl. Acad. Sci. USA 107, 22617–22622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen ED, and Spiegelman BM (2014). What we talk about when we talk about fat. Cell 156, 20–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosen E, Eguchi J, and Xu Z (2009). Transcriptional targets in adipocyte biology. Expert Opin. Ther. Targets 13, 975–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross R, Aru J, Freeman J, Hudson R, and Janssen I (2002). Abdominal adiposity and insulin resistance in obese men. Am. J. Physiol. Endocrinol. Metab 282, E657–E663. [DOI] [PubMed] [Google Scholar]
- Rutkowski JM, Stern JH, and Scherer PE (2015). The cell biology of fat expansion. J. Cell Biol 208, 501–512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao M, Ishibashi J, Kusminski CM, Wang QA, Hepler C, Vishvanath L, MacPherson KA, Spurgin SB, Sun K, Holland WL, et al. (2016). Zfp423 Maintains White Adipocyte Identity through Suppression of the Beige Cell Thermogenic Gene Program. Cell Metab 23, 1167–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shepherd PR, Gnudi L, Tozzo E, Yang H, Leach F, and Kahn BB (1993). Adipose cell hyperplasia and enhanced glucose disposal in transgenic mice overexpressing GLUT4 selectively in adipose tissue. J. Biol. Chem 268, 22243–22246. [PubMed] [Google Scholar]
- Siersbæk R, Nielsen R, and Mandrup S (2012). Transcriptional networks and chromatin remodeling controlling adipogenesis. Trends Endocrinol. Metab 23, 56–64. [DOI] [PubMed] [Google Scholar]
- Soccio RE, Chen ER, and Lazar MA (2014). Thiazolidinediones and the promise of insulin sensitization in type 2 diabetes. Cell Metab 20, 573–591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solinas G, Bore´ n J, and Dulloo AG (2015). De novo lipogenesis in metabolic homeostasis: More friend than foe? Mol. Metab 4, 367–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukas A, Cohen P, Socci ND, and Friedman JM (2000). Leptin-specific patterns of gene expression in white adipose tissue. Genes Dev 14, 963–980. [PMC free article] [PubMed] [Google Scholar]
- Stanya KJ, Jacobi D, Liu S, Bhargava P, Dai L, Gangl MR, Inouye K, Barlow JL, Ji Y, Mizgerd JP, et al. (2013). Direct control of hepatic glucose production by interleukin-13 in mice. J. Clin. Invest 123, 261–271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun K, Kusminski CM, and Scherer PE (2011). Adipose tissue remodeling and obesity. J. Clin. Invest 121, 2094–2101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Syed I, Lee J, Moraes-Vieira PM, Donaldson CJ, Sontheimer A, Aryal P, Wellenstein K, Kolar MJ, Nelson AT, Siegel D, et al. (2018). Palmitic Acid Hydroxystearic Acids Activate GPR40, Which Is Involved in Their Beneficial Effects on Glucose Homeostasis. Cell Metab 27, 419–427.e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao C, Holland WL, Wang QA, Shao M, Jia L, Sun K, Lin X, Kuo YC, Johnson JA, Gordillo R, et al. (2017). Short-Term Versus Long-Term Effects of Adipocyte Toll-Like Receptor 4 Activation on Insulin Resistance in Male Mice. Endocrinology 158, 1260–1270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tchkonia T, Thomou T, Zhu Y, Karagiannides I, Pothoulakis C, Jensen MD, and Kirkland JL (2013). Mechanisms and metabolic implications of regional differences among fat depots. Cell Metab 17, 644–656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Titchenell PM, Quinn WJ, Lu M, Chu Q, Lu W, Li C, Chen H, Monks BR, Chen J, Rabinowitz JD, and Birnbaum MJ (2016). Direct Hepatocyte Insulin Signaling Is Required for Lipogenesis but Is Dispensable for the Suppression of Glucose Production. Cell Metab 23, 1154–1166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran TT, Yamamoto Y, Gesta S, and Kahn CR (2008). Beneficial effects of subcutaneous fat transplantation on metabolism. Cell Metab 7, 410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner SM, Roy S, Sul HS, Neese RA, Murphy EJ, Samandi W, Roohk DJ, and Hellerstein MK (2007). Dissociation between adipose tissue fluxes and lipogenic gene expression in ob/ob mice. Am. J. Physiol. Endocri-nol. Metab 292, E1101–E1109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, and Han X (2014). Multidimensional mass spectrometry-based shotgun lipidomics. Methods Mol. Biol 1198, 203–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Han RH, and Han X (2013a). Fatty acidomics: global analysis of lipid species containing a carboxyl group with a charge-remote fragmentation-assisted approach. Anal. Chem 85, 9312–9320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Tao C, Gupta RK, and Scherer PE (2013b). Tracking adipogenesis during white adipose tissue development, expansion and regeneration. Nat. Med 19, 1338–1344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S, Sun H, Ma J, Zang C, Wang C, Wang J, Tang Q, Meyer CA, Zhang Y, and Liu XS (2013c). Target analysis by integration of transcriptome and ChIP-seq data with BETA. Nat. Protoc 8, 2502–2515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang QA, Tao C, Jiang L, Shao M, Ye R, Zhu Y, Gordillo R, Ali A, Lian Y, Holland WL, et al. (2015). Distinct regulatory mechanisms governing embryonic versus adult adipocyte maturation. Nat. Cell Biol 17, 1099–1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M, Wang C, and Han X (2017). Selection of internal standards for accurate quantification of complex lipid species in biological extracts by electrospray ionization mass spectrometry-What, how and why? Mass Spectrom. Rev 36, 693–714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ward PS, and Thompson CB (2012). Signaling in control of cell growth and metabolism. Cold Spring Harb. Perspect. Biol 4, a006783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernstedt Asterholm I, Tao C, Morley TS, Wang QA, Delgado-Lopez F, Wang ZV, and Scherer PE (2014). Adipocyte inflammation is essential for healthy adipose tissue expansion and remodeling. Cell Metab 20, 103–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia JY, Holland WL, Kusminski CM, Sun K, Sharma AX, Pearson MJ, Sifuentes AJ, McDonald JG, Gordillo R, and Scherer PE (2015). Targeted Induction of Ceramide Degradation Leads to Improved Systemic Metabolism and Reduced Hepatic Steatosis. Cell Metab 22, 266–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yore MM, Syed I, Moraes-Vieira PM, Zhang T, Herman MA, Homan EA, Patel RT, Lee J, Chen S, Peroni OD, et al. (2014). Discovery of a class of endogenous mammalian lipids with anti-diabetic and anti-inflammatory effects. Cell 159, 318–332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Laz EV, and Waxman DJ (2012). Dynamic, sex-differential STAT5 and BCL6 binding to sex-biased, growth hormone-regulated genes in adult mouse liver. Mol. Cell. Biol 32, 880–896. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.