Abstract
Mutation of the isocitrate-dehydrogenase (IDH) enzymes is one of the central research topics regarding gliomagenesis. Indeed, 70% of gliomas are associated with a gain-of-function IDH mutation and consequently synthesize the oncometabolite, 2-hydroxyglutarate (2-HG). This review aims to elucidate the effects of 2-HG on gliomagenesis. 2-HG promotes tumorigenesis by impacting metabolism, vascularization and altering the epigenome of glioma cells. Glioma metabolism and vascularization is altered by 2-HG’s effect on the stability of hypoxia-inducible factor (HIF) and inhibition of endostatin. However, 2-HG’s impacts on epigenetic mechanisms are more profound to gliomagenesis. Through competitive inhibition of JHDMs and TET proteins, 2-HG orchestrates histone and DNA hypermethylation, which is associated with gene silencing and dedifferentiation of cells. The hypermethylator phenotype induced by 2-HG also results in alterations of the interaction of the immune system with the tumour. Additionally, this study reviews 2-HG promotion of tumorigenesis by inhibiting repair of DNA alkylation damage through competitive inhibition of AlkB proteins.
Keywords: glioblastoma, cancer metabolism, epigenome, IDH1 mutation, 2-hydroxyglutarate
Introduction
Gliomas, tumors derived from neural stem cells or glial progenitor cells, are a heterogenous group of brain tumors [1]. They are the most common type brain tumor, representing 50% of all brain cancers [2]. Median survival time for World Health Organization (WHO) classified grade IV malignant gliomas is 15 months, even with aggressive treatment [3, 4]. Glioma research was revolutionized in 2008, when the genetic sequencing of glioblastomas revealed that the mutation of isocitrate dehydrogenase 1 and 2 (IDH1/2) is a prevalent mutation in various glioma entities [5]. The most common IDH1 mutation is R132H, while the most prevalent IDH2 mutations are: R172G, R172K, and R172M [6]. IDH1/2 catalyze the reaction isocitrate and NADP+ to α-ketoglutarate (α-KG) and NADPH+H. The gain of function IDH1/2 mutations however allow the enzymes to reduce α-KG to the oncometabolite, 2-hydroxyglutarate (2-HG) [7]. In the last years, 2-HG has become the center of research, as this oncometabolite changes the tumor metabolism and alters the glioma epigenome [7, 8]. Different glioma entities have different rates of IDH1 mutations. The most common IDH1 mutated gliomas are anaplastic oligodendrogliomas WHO grade III and anaplastic astrocytomas WHO grade III with a 81.6% and 82% IDH1 mutation rate respectively [9]. However, only 3.1% of gliomas carry IDH2 mutations [9]. Interestingly, it was also established that IDH1 and IDH2 mutations are mutually exclusive in gliomas; barely any gliomas harbor both, IDH1 and IDH2 mutations [9]. This indicates at the severity of IDH1/2 mutation, as only one mutation is enough to mediate tumorigenesis.
The relevance of IDH1/2 mutations is also demonstrated by the WHO classification system, which was updated in 2016 to accommodate the genetic makeup of the different glioma subtypes. Relevant to this review is the division between low-grade gliomas (LGG) (grade I and II) and grade III and IV gliomas, also known as high-grade gliomas [10]. Whereas grade I gliomas are benign, grade IV tumors, also known as IDH-mutant or IDH-wild-type glioblastoma, are refractory to chemotherapy and show more advanced characteristics of malignancy [11]. Grade II gliomas almost universally transform to high-grade gliomas [12]. Regarding grade II–IV gliomas, the updated WHO classification of 2016 subdivides lesions according to molecular markers, such as the presence of IDH mutations or 1p/19q deletion [10]. Of importance for this review is also the differentiation between primary and secondary glioblastomas. While primary glioblastomas are de-novo tumors, secondary glioblastomas develop from low-grade diffuse astrocytomas or anaplastic astrocytoma [13]. The distinctive genetic marker of secondary glioblastomas is the IDH1 mutation, as over 80% of secondary glioblastomas illustrate IDH1 mutations compared to only 5% of primary glioblastomas [14]. Secondary glioblastomas are also associated with a hypermethylator phenotype and exhibit a better prognosis than primary glioblastomas [14]. Due to the strong association with IDH1 mutations, this review will focus particularly on the pathogenesis of secondary glioblastomas.
2-hydroxyglutarate
IDH1/2 mutations are one of the earliest known mutations occurring during glioma formation [15]. When measuring the catalytic activity of mutant IDH1, Dang et al. [7] observed a “1,000 fold decrease in catalytic turnover” from isocitrate to α-KG. Conversely, studies suggest that cancers harboring IDH1/2 mutations produced 2-HG concentrations 10 to 100 times the levels of cancers with wild type IDH [16]. Further, while wild-type IDH1/2 produces the reducing agent NADPH+H+ during the reaction isocitrate to α-KG, mutant IDH1/2 consumes NADPH+H+ during the catalyzation of α-KG to 2-HG [7] (Fig. 1). This is significant, as NADPH+H+ is an important metabolite for macromolecule synthesis and defending cells against reactive oxygen species [17]. IDH1/2 mutations could offset the cellular redox reactions, promoting tumorigenesis [17]. 2-HG is a chiral molecule, so D- and L-enantiomer forms exist (Fig. 1). In vivo and in vitro experiments established that mutant IDH1/2 produces almost exclusively D-2-HG [16]. Therefore, D-2-HG seems to be the enantiomer most relevant to tumorigenesis. Intriguingly, while cancer cells produce almost only D-2-HG, numerous studies observed that in vitro, L-2-HG is a more potent inhibitor of various enzymes and has a greater effect on cell proliferation [18–20]. While IDH1/2 mutations have a variety of consequences on tumorigenesis, the correlation between remarkably high levels of 2-HG in gliomas led to much investigation of the specific effect of 2-HG on gliomagenesis.
Fig. 1.
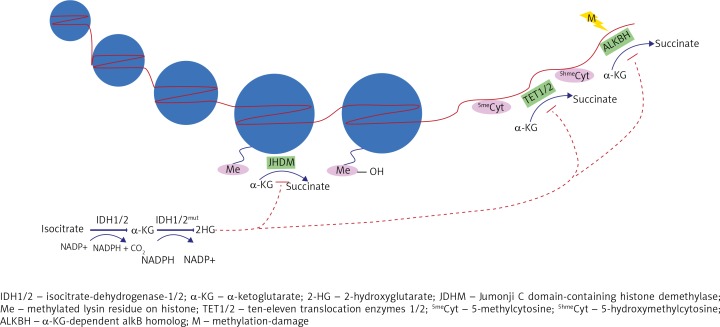
In physiological conditions, IDH1/2 metabolizes isocitrate to a-KG with the production of NADPH and CO2. Cancerous mutated IDH1/2 synthesizes 2-HG and NADP+ from a-KG. 2-HG competitively inhibits numerous a-KG-dependent-dixoxygenases. Here illustrated is 2-HG‘s competitive inhibition of JHDMs and TET1/2, which results in histone and DNA methylation, respectively. 2-HG also inhibits ALKBHs, leading to less DNA repair of methylation damage. Overall, these effects induced by 2-HG alter the epigenome of glioma cells and promote tumorigenesis Isocitrate
α-ketoglutarate dependent dioxygenases
Vital to the understanding of 2-HG’s role in tumorigenesis is the consideration of 2-HG’s similar structure compared to α-KG. The keto group of α-KG is simply exchanged for a hydroxyl group in 2-HG [21]. Due to the structural similarity of both metabolites, 2-HG acts as a competitive inhibitor to α-KG dependent dioxygenases [18]. There are over 60 known α-KG dependent dioxygenases which could potentially be inhibited by 2-HG [22]. Especially relevant to cancerogenesis are the α-KG dependent dioxygenases which catalyze hydroxylation reactions, such as DNA demethylation, histone demethylation, sensing of hypoxia and DNA repair of alkylation damage [22, 23]. For α-KG dependent enzymes, 2-HG acts as a competitive inhibitor and can thereby influence glioma tumorigenesis.
Hypoxia-inducible factors
α-KG dependent dioxygenases also play an important role during O2-sensing in cells. Hypoxia-inducible transcription factors (HIF 1–3) coordinates the body’s response to hypoxia, ranging from the formation of new blood vessels to increased synthesis of red blood cells [24]. HIF-α is degraded through post-translational hydroxylation, catalyzed by prolyl hydroxylase (also called EGLN), a α-KG dependent dioxygenase [22]. All α-KG dependent dioxygenases require oxygen as a co-substrate. In the case of HIF-α, if oxygen is present, prolyl hydroxylase can hydroxylate the prolyl residues on the HIF-α subunit. Von Hippel-Lindau protein can now bind to the hydroxylated HIF-α subunit, which leads to ubiquitination and the breakdown of HIF-α in proteasomes [22]. There is conflicting research regarding the impact of 2-HG on HIF.
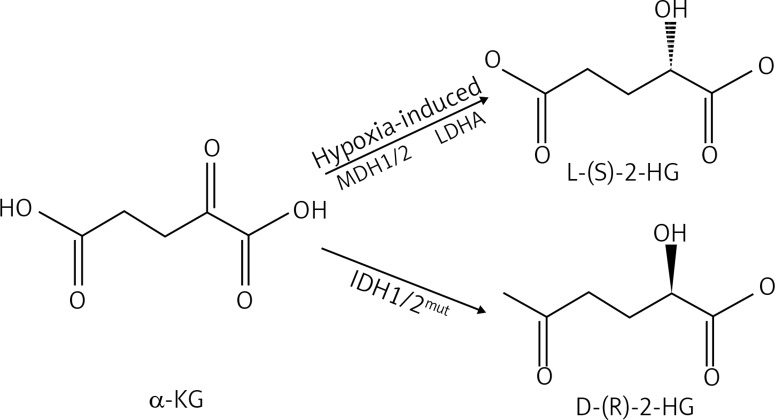
On one hand, research has shown that 2-HG inhibits prolyl hydroxylase. A study conducted by Xu et al. [18] illustrated that in vivo, HIF-α1 levels increased after the knockdown of wild type IDH, introduction of mutant IDH or addition of 2-HG. Other research groups also noticed the correlation between IDH1/2 mutations and elevated HIF-1α expression [25–27]. Interestingly, it was also observed that L-2-HG is a more potent competitive inhibitor than D-2-HG [18]. This would make sense, as it has been reported that L-2-HG is synthesized primarily by cells in hypoxic microenvironments and oxygen starved tissue has a special need for angiogenesis to combat hypoxia [28]. Under these physiological hypoxic conditions, L-2-HG is produced predominantly by lactatedehydrogenase A (LDHA) and to a lesser extent by malatedehydrogenase 1 and 2 (MDH1/2) (Fig. 2) [28]. Nonetheless, both enantiomers stabilize HIF-α through competitive inhibition of prolyl hydroxylase and thereby promote tumorigenesis. This is because the same mechanisms that allow cells to survive during hypoxia help tumor growth, as HIF regulated genes increase angiogenic factors, glycolysis, glucose transporters, invasion factors and survival factors [29].
Fig. 2.
2-hydroxyglutarate (2-HG) is a chiral molecule, which exists in a L-(S)-2-HG and D-(R)-2-HG form. In cancers, mutated IDH1/2 generates almost exclusively D-(R)-2-HG from α-ketoglutarate (α-KG). L-(S)-2-HG is synthesised during hypoxic conditions through lactatedehydrogenase A (LDHA) and malatedehydrogenase (MDH1/2)
On the other hand, research also suggests that D-2-HG promotes HIF-1α degradation by stimulating prolyl hydroxylase activity [19]. Koivunen et al. [19] obtained similar results as the papers mentioned above in regard to L-2-HG, as they observed that L-2-HG inhibited prolyl hydroxylase. Surprisingly however, this research group also describes that instead of inhibiting, D-2-HG potentiated prolyl hydroxylase catalytic activity, resulting in HIF-1α degradation. The paper reports that HIF-1α levels were reduced in human IDH-mutated proneural tumors, compared to IDH-wild type samples [19]. Other research groups also concluded that the addition of D-2-HG but not L-2-HG, reduced HIF-1α concentration in cells and that IDH mutation was not sufficient to upregulate HIF-1α concentrations in gliomas [30, 31]. Supporting, but not proving the higher prolyl 4-hydroxylase activity, IDH-mutated cancers are associated with heightened expressions of prolyl 4-hydroxylases, which tag HIF-1α for ubiquitination and subsequent proteasomal degradation [32]. The consequence of this mechanism on tumor growth was also tested, as D-2-HG stimulates activity of prolyl 4-hydroxylase and subsequent HIF-1α degradation which reduced colony formation of immortalized human astrocytes [19].
Concluding, on the one hand 2-HG was associated with HIF-1 stabilisation while other studies have also reported that D-2-HG induces HIF-1α degradation. In part, these conflicting results can be resolved by the discovery that D- and L-2-HG suppress factor-inhibiting HIF1 (FIH1) [33, 34]. If FIH1 suppresses activation of HIF-1α, then an inhibition of FIH1 through 2-HG is an alternative mechanism which can lead to HIF-1α stabilisation [33, 34]. Nevertheless, the opposing results show that the 2-HG impact on HIF-1α has to be clarified.
Endostatin
Unlike HIF, endostatin is a natural inhibitor of angiogenesis, which reduces tumor vascularization and suppresses tumor growth [35]. Upregulation of tumor vascularization is an important step during gliomagenesis, as glioblastomas are characterized by extensive angiogenesis [36]. Further, neovascularization is associated with higher malignancy grade and reduced post-operative survival in patients suffering from gliomas [36]. Similarly to HIF, endostatin synthesis is also affected by 2-HG accumulation in IDH-mutated gliomas. Endostatin is synthesized through prolyl-hydroxylation of collagen prolyl 4-hydroxylase [35]. As prolyl 4-hydroxylase is α-KG dependent, researchers postulate that 2-HG can competitively inhibit this enzyme, leading to lower concentrations of endostatin in IDH-mutated gliomas [37]. In fact, IDH-mutated gliomas were associated with reduced levels of endostatin and higher rates of blood vessel densities [37]. The effect of 2-HG on endostatin was more directly confirmed by the observation that 2-HG injection into cells decreased endostatin levels [18]. Thereby, 2-HG promotes tumorigenesis by inhibiting endostatin, which then leads to tumor vascularization and proliferation.
Epigenetics during cancerogenesis
While traditionally, 2-HG’s impacts on gliomagenesis have been associated with alterations to tumor metabolism and vascularization, more recent research suggests that glioma formation is driven to a greater extent by epigenetic alterations induced by 2-HG. In essence, epigenetics describes modifications of chromatin structure which consequently alter gene expression. The chromatin configuration influences whether DNA transcription machinery can access DNA to facilitate transcription [38]. Regarding gliomagenesis, the two most examined epigenetic mechanisms altering chromatin structure are DNA methylation and histone modification [39]. Depending on the histone methylated, chromatin structure is loosened to promote transcription, or tightened to suppress transcriptional activities. For instance, trimethylation of lysin 4 on histone 3 (H3K4me3) is associated with genes that are transcriptionally active, while trimethylation of H3K9 (H3K9me3) at gene promoters is one of the chief epigenetic silencing mechanisms in mammalian cells [39]. DNA methylation occurs on the GpG Islands of the genome; these are repetitive sequences of DNA and comprise 60% of the promoters [40]. While histone modifications can either enhance or repress transcription, DNA methylation leads to gene silencing [39]. Adding to the complexity of epigenetic mechanisms, DNA methylation can impact histone methylation and vice versa. While DNA methylation and histone methylation are often described as independent entities, they can interact with each other to modify cellular transcription status [39].
α-KG dependent histone demethylase
In 2009, Tsukada et al. [41] discovered a group of histone demethylases named Jumonji C (JmjC) domain-containing histone demethylase (JHDM), which in presence of α-KG and iron, demethylate lysin residues on histones. JHDMs are also known as Lysin (K)-specific demethylases. This group contains a variety of JHDMs, which demethylate different lysin residues. Due to 2-HG’s similar structure to α-KG, 2-HG competitively inhibits JHDMs (Fig. 2). The three most important JHDMs during gliomagenesis are JHDM1A (official symbol KDM2A), JMJD2A (KDM4A), and JMJD2C (KDM4C). Xu et al. [18] reported that elevated 2-HG levels from mutant IDH1 can inhibit multiple α-KG dependent histone demethylases in vivo and in vitro. As seen in other α-KG dependent dioxygenases, L-2-HG was found to be significantly more potent than D-2-HG [18, 42]. The effect of 2-HG on histone methylation was further demonstrated in vivo by the observation that IDH-mutated glioma samples with abnormally high 2-HG levels exhibited increased methylation marks of the following histones: H3K4, H3K9, H3K27 and H3K79 [18]. This observation was supported by other research groups as well [43]. The methylation of histones explored by these papers do not all orchestrate the same epigenetic effects; mono-methylation of H3K4, H3K27 and H3K79 are associated with gene activation, while trimethylation of H3K9, H3K27 and H3K79 suppresses transcription [44]. Current research is especially focused on the trimethylation of H3K9 (H3K9me3). While H3K9me3 has long been known as an epigenetic silencing mechanism, it also plays an important role during tumorigenesis. H3K9me3 inhibits transcription and promotes heterochromatin formation [45]. Intriguingly, H3K9me3 is associated with IDH1 mutation in different types of gliomas [43, 46, 47]. Supporting this observation, in vitro experiments on cells of the central nervous system showed that IDH-mutated astrocytes or murine neurosphere cultures exhibited enhanced H3K9me3 [43, 47]. Evidence further suggests that other mechanisms of histone methylation, other than inhibition of JHDMs, are important for gliomagenesis. The methyltransferase, Enhancer-of-zeste homolog 2 (EZH2), which catalyzes the methylation of H3K27, is overexpressed in cancers such as lung, breast, prostate, blood cancers and glioblastomas [48]. EZH2 was also found to be vital in glioblastoma cells to maintain stem-cell-like properties and therefore promote tumorigenesis [49]. This suggests that histone methylation in general is an important epigenetic mechanism orchestrating carcinogenesis, which delineates the importance of 2-HG induced histone methylation for glioma formation. The significance of histone methylation to tumorigenesis is also evident from much research showing that histone methylation inhibits cellular differentiation. For instance, a study reported that the “introduction of mutant but not wild-type IDH1 into astrocytes resulted in the upregulation of nestin (and other genes associated with stem cell identity) at the time of DNA methylation increase and the adoption of a neurosphere/stem-like phenotype” [43]. Regarding gliomas, IDH-mutated gliomas with methylated H3K9 exhibit lower levels of glial fibrillary acid protein (a protein important for central nervous system cell differentiation) and less cellular differentiation [47]. Moreover, the cancerogenic effect of histone methylation is supported by the consideration that histone methylation can impact DNA methylation [46]. For instance, Turcan et al. [43] observed that specifically H3K9me3 is associated with DNA methylation, which could in turn promote cellular proliferation.
Ten-eleven translocation enzymes
DNA methylation is an important characteristic of tumorigenesis, as cancer cells often exhibit atypical DNA methylation patterns, including global DNA hypomethylation and promoter hypermethylation [50]. Both effects are pro-tumorigenic, as hypomethylation leads to genome instability, while promoter hypermethylation, amongst other things, silences tumor suppressor genes [50]. While DNA methylation is usually catalyzed by DNA-methyl-transferases, de-methylation is initiated by the three ten-eleven translocation enzymes (TETs 1/2/3) [51]. TETs are another type of α-KG dependent dioxygenases [51]. De-methylation occurs in a two-step process. First, TET enzymes successively oxidize 5-methylcytosine to 5-hydroxymethylcytosine (5hmC), to 5-formylcytosine (5fC) and finally to 5-carboxylcytosine (5caC). The three oxidized methyl-cytosines, 5hmC, 5fC and 5caC, are collectively coined “oxi-methyl-cytosines (oxi-mCs)”. Next, the modified cytosines of 5fC and 5caC are exchanged for non-methylated cytosines through base excision, thereby restoring the un-methylated cytosine. Out of the three oxi-mCs, 5hmC is the most prevalent because only 1-10% of 5hmCs are further oxidized to 5fC and 5aC [52]. This is why 5hmC is often used as a marker to measure TET-enzyme activity. Explored more extensively in the following reviews, 5hmC is an important metabolite that contributes to cancer development [53–55].
In fact, TET2 is described as a tumor suppressor because studies suggest that many solid cancers and 15% of myeloid cancers carry TET2 mutations [56–58]. The catalytic activity of TET enzymes is dependent on α-KG, thereby researchers hypothesized that 2-HG from IDH-mutated gliomas competitively inhibits TET proteins. In a study by Xu et al. [18], D-2-HG inhibited TET1 up to 47% and TET2 up to 83%. Similar to the inhibition of prolyl hydroxylase, L-2-HG was more potent than D-2-HG. As expected from inhibition of a demethylating enzyme, high 2-HG and inhibition of TET de-methylation activity was associated with DNA hypermethylation (Fig. 2) [18]. Xu et al. [18] demonstrated that in vivo, 5-hmC was much lower in mutated IDH1 glioma samples compared to wild-type IDH1, indicating the impaired function of TET to demethylase DNA in the presence of abnormally high 2-HG levels. These findings are supported by observations that IDH-mutated acute myeloid leukemia cells showed impaired TET2 functions and hypermethylated phenotypes [21]. Not only is there a clear correlation between 2-HG dependent TET inhibition and DNA hypermethylation, but there is also evidence that 2-HG impairment of TET enzymes induces cancer formation. 2-HG induced hypermethylation silences tumor suppressors and inhibits cellular differentiation [43, 59–61]. Turcan et al. [43] observed that the introduction of mutant IDH1 into astrocytes coincided with increasing DNA methylation and the adoption of a stem-cell-like phenotype. Another study compared TET2 mutant mice to IDH-mutant mice exhibiting high levels of 2-HG. Both types, TET2 mutant mice and IDH-mutant mice, showed “stem cell expansion and impaired hematopoietic differentiation” [60]. Finally, in a very recent study, induction of TET2 expression in glioblastoma cells upregulated genes associated with differentiation of neural cells, such as the brain fatty acid-binding protein [61]. This suggests that inhibition of TET2 through 2-HG could impair nerve cell differentiation.
Immune system
The relationship between cancer and the immune system is another important factor driving tumorigenesis. Cancer cells apply various methods to circumvent host immune surveillance in order to proliferate.
Current research provides evidence of novel immune evasive mechanisms specific to IDH-mutated gliomas. Using orthotopic syngeneic low-grade glioma models, Kohanbasch et al. [62] discovered that 2-HG reduces the expression of STAT1. STAT1 is a regulator of the chemokine, CXCL10, which in turn attracts CD8+ cytotoxic T-cells. Thereby, a 2-HG induced abatement of STAT1 reduces CD8+ T-cell accumulation at the tumor site. When implanting IDH-mutated low-grade glioma cell lines in syngeneic mice, the developing tumors showed lower rates of CXCL10 and less CD8+ T-Cell infiltration than tumors from IDH-wild type cells. The effects of 2-HG were directly tested, as inhibiting 2-HG synthesis in IDH-mutated glioma cell lines resulted in an increase in CXCL10 levels and elevated expression of STAT1. Conversely, treatment of IDH-wild type glioma cells lines with 2-HG resulted in reduced STAT1 expression [62]. A supplementary review of these findings can be found in the commentary by Lucca and Hafler [63]. Kohanbasch’s et al. conclusion is supported by another study revealing a negative correlation between pathological glioma grade and STAT1 gene expression [64]. Overall, healthy brain tissue shows significantly higher average expression of STAT1 than glioma tissue [64].
However, evidence suggest that rather than stimulating immunoevasion of glioma cells, 2-HG reduces the immunosuppression usually associated with cancer and thereby impedes cancerous cell proliferation. IDH-mutated gliomas are associated with a more active immune system than non-IDH-mutated gliomas [65, 66]. Studies observed that IDH-wild type gliomas show a greater infiltration of immunosuppressive cells, such as T-regulatory cells (T-Regs) and tumor-associated macrophages (TAMs), than IDH-mutant gliomas [65–67]. T-Regs infiltrate tumor tissue and inhibit T-cell toxicity while simultaneously suppressing tumor antigen presentation by dendritic cells [68]. Moreover, while macrophages usually exhibit anti-tumorigenic functions, in response to the local tumor microenvironment, TAMs alter their function and facilitate tumor growth instead [68]. This conclusion is supported by a recent study by Rahman et al. [69], who investigated expression of immunogenic markers in IDH-mutated glioblastomas. CD163, a member of the scavenger receptor of the cysteine-rich (SRCR) superfamily, is a marker for M2 Macrophages, which are a subtype of anti-inflammatory TAMs [70]. IDH-mutated glioblastomas are associated with a significantly lower expression of CD163, which could reduce immunosuppression and suppress tumor growth [69]. For instance, CD163-positive cancer cells are potentially associated with a higher malignant potential in clear cell renal cell carcinoma [71]. In the tumor microenvironment, CD70, member of the tumor necrosis factor (TNF) family, functions as an immunosuppressive factor [72]. CD70 expression is also decreased in IDH-mutated glioblastomas, resulting in more anti-tumor immunoactivity [69]. This difference in tumor immune cell infiltration is particularly evident when comparing secondary glioblastomas to primary glioblastomas. Amankulor et al. [66] explored the reasons why secondary glioblastomas tumors exhibit less immunosuppressive tumor-associated cell infiltration than primary glioblastomas. They posit that, through mechanisms not quite clear, 2-HG increases methylation and thereby silences genes associated with the tumor-derived immune system. Consequently, IDH-mutated gliomas with high levels of 2-HG exhibit less immunosuppressive cell infiltration and a more active host immune system. As a result, the host immune system can induce an anti-tumor immune response, inhibiting tumorigenesis. Amankulor et al. postulate that the milder cancer progression of secondary glioblastoma patients is partly related to patients high 2-HG levels, which translate into a more immune-active phenotype that prohibits cancer proliferation [66]. Additionally, some studies reported that through 2-HG induced promoter methylation, IDH-mutated gliomas illustrate a lower expression of the transmembrane protein, programmed death ligand-1 (PD-L1), than IDH-wild type glioma cells [65, 67]. PD-L1, expressed on cancer cells, binds to the programmed death receptor-1 on T-cells, and consequently inhibits T-cell immunoactivity and leads to immunosuppression [73]. However, PD-L1 is only amplified in 0.3% of glioblastomas [74]. Thus, glioblastomas do not rely on PD-L1 as their main immunoevasive strategy.
AlkB homolog, α-ketoglutarate-dependent dioxygenase
Cancer cells steadily have to adapt to a changing environment. Therefore, elevated mutation rates in cancer cells increase the genetic diversity of the tumor and allow cancer cells to express a phenotype which is best suited to their environment. Thus, it can be argued that DNA mutations within existing cancerous tissue can promote tumorigenesis [75]. These DNA mutations can result from methylation of DNA bases through environmental or endogenous alkylating agents. The described microevolution of cancer is suppressed if DNA mutations, such as alkylated DNA bases, are repaired. In mammalian cells, DNA alkylation is corrected by the de-alkylating enzymes, AlkB proteins, which remove methyl groups from 1-methyl-adenine and 3-methyl-cytosine [76, 77].
AlkB proteins are α-KG and iron dependent di-oxygenases [76]. Wang et al. [77] observed that D-2-HG inhibits the repair of methyl-adenine by ALKBH2 and ALKBH3 in vitro (Fig. 2). They point out that D-2-HG is a weak inhibitor of ALKBH at physiological concentrations. However in higher concentrations as of 0.5 mM, D-2-HG results in a 50% inhibition of ALKBH2 [77]. Xu et al. [18] support this hypothesis, as they write that “the molar ratio of R-2-HG (D-2-HG) and α-KG has been calculated as an average 373-fold in glioma patients, enabling R-2-HG (D-2-HG) to exert its inhibitory effects”. From this it can be hypothesized that D-2-HG’s competitive inhibition of ALKBH elevates intra-cancerous mutation rate and promotes glioma microevolution.
Intriguingly however, the inhibition of DNA repair mechanisms through D-2-HG enhances the effectiveness of chemotherapy. Wang et al. [77] observed that IDH1/2 mutated glioma cells are more sensitive to alkylating chemotherapy and this sensitivity was abolished when D-2-HG production was disrupted. Another study explored the performance of the alkylating chemotherapy, PCV (procarbazine, lomustine and vincristine), on patients with IDH-mutated anaplastic oligodendrogliomas and IDH-wild type anaplastic oligoastrocytomas [78]. Here, patients with IDH mutations responded better to the alkylating agents than patients with wild-type IDH [78]. The study concludes that screening patients for IDH mutations is beneficial to decipher if they would benefit from neo-adjuvant PCV treatment [78]. In another paper, the overexpression of ALKBH2 in glioblastomas resulted in a resistance to the alkylating agent, temozolomide. Knockdown of ALKBH2 increased sensitivity to temozolomide [79].
Conclusions
Ultimately, the oncometabolite 2-HG promotes cellular cancerogenesis in gliomas. While tumor metabolism and angiogenesis are also affected by 2-HG, epigenetic consequences stemming from the 2-HG induced hypermethylator phenotype, especially associated with secondary glioblastomas, are chiefly responsible for tumor progression. Histone and DNA methylation promote gene silencing and lead to cellular dedifferentiation. A prominent example of the epigenetic alterations induced by 2-HG are the impacts on the immune system. However, this review has also elucidated some controversies in current research. Some argue that 2-HG does not directly promote tumorigenesis, as certain publications observed 2-HG induced destabilization of HIF and reduction of immunosuppressive cell infiltration. However, on the whole, these anti-tumorigenic effects of 2-HG are largely dispensable, because it seems they only translate into a slightly milder tumor progression of secondary glioblastoma patients; gliomagenesis occurs nonetheless. While the rough outlines of 2-HGs effects on gliomagenesis are becoming clearer, more extensive research is necessary to fully elucidate the pathogenesis of IDH-mutated gliomas. Especially the impact of epigenome alterations and the interactions between histone and DNA methylation is still vague. Understanding the pathogenesis of gliomas is imperative in order to generate effective therapies for IDH-mutated gliomas.
The authors declare no conflict of interest.
Footnotes
The authors declare no conflict of interest.
References
- 1.Sanai N, Alvarez-Buylla A, Berger MS. Neural Stem Cells and the Origin of Gliomas. N Engl J Med. 2005;353:811–822. doi: 10.1056/NEJMra043666. [DOI] [PubMed] [Google Scholar]
- 2.Hofer S, Bullinger L, Dierlamm J, et al. Gliome im Erwachsenenalter – Onkopedia. Berlin: 2018. www.onkopedia.com/de/onkopedia/guidelines/gliome-im-erwachsenenalter/@@view/html/index.html (access: 07.12.2018) [Google Scholar]
- 3.Martinez R, Rohde V, Schackert G. Different molecular patterns in glioblastoma multiforme subtypes upon recurrence. J Neurooncol. 2010;96:321–329. doi: 10.1007/s11060-009-9967-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Omuro A, DeAngelis LM. Glioblastoma and other malignant gliomas: A clinical review. JAMA. 2013;310:1842–1850. doi: 10.1001/jama.2013.280319. [DOI] [PubMed] [Google Scholar]
- 5.Parsons DW, Jones S, Zhang X, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yan H, Parsons DW, Jin G, et al. IDH1 and IDH2 Mutations in Gliomas. N Engl J Med. 2009;360:765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dang L, White DW, Gross S, et al. Cancer-associated IDH1 mutations produce 2-hydroxyglutarate. Nature. 2009;462:739–744. doi: 10.1038/nature08617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yang H, Ye D, Guan K-L, Xiong Y. IDH1 and IDH2 Mutations in Tumorigenesis: Mechanistic Insights and Clinical Perspectives. Clin Cancer Res. 2012;18:5562–5571. doi: 10.1158/1078-0432.CCR-12-1773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hartmann C, Meyer J, Balss J, et al. Type and frequency of IDH1 and IDH2 mutations are related to astrocytic and oligodendroglial differentiation and age: a study of 1,010 diffuse gliomas. Acta Neuropathol. 2009;118:469–474. doi: 10.1007/s00401-009-0561-9. [DOI] [PubMed] [Google Scholar]
- 10.Louis DN, Perry A, Reifenberger G, et al. The 2016 World Health Organization Classification of Tumors of the Central Nervous System: a summary. Acta Neuropathol. 2016;131:803–820. doi: 10.1007/s00401-016-1545-1. [DOI] [PubMed] [Google Scholar]
- 11.Gao L. Glioma an overview of current classifications characteristics molecular biology and target therapies. Front Biosci. 2015;20:4362. doi: 10.2741/4362. [DOI] [PubMed] [Google Scholar]
- 12.Claus EB, Walsh KM, Wiencke JK, et al. Survival and low-grade glioma: the emergence of genetic information. Neurosurg Focus. 2015;38:E6. doi: 10.3171/2014.10.FOCUS12367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Scherer HJ. A critical review the pathology of cerebral gliomas. J Neurol Neurosurg Psychiatry. 1940;3:147–177. doi: 10.1136/jnnp.3.2.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohgaki H, Kleihues P. The Definition of Primary and Secondary Glioblastoma. Clin Cancer Res. 2013;19:764–772. doi: 10.1158/1078-0432.CCR-12-3002. [DOI] [PubMed] [Google Scholar]
- 15.Leeper HE, Caron AA, Decker PA, et al. IDH mutation, 1p19q codeletion and ATRX loss in WHO grade II gliomas. Oncotarget. 2015;6:30295–30305. doi: 10.18632/oncotarget.4497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ward PS, Patel J, Wise DR, et al. The Common Feature of Leukemia-Associated IDH1 and IDH2 Mutations Is a Neomorphic Enzyme Activity Converting α-Ketoglutarate to 2-Hydroxyglutarate. Cancer Cell. 2010;17:225–234. doi: 10.1016/j.ccr.2010.01.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cairns RA, Mak TW. Oncogenic Isocitrate Dehydrogenase Mutations: Mechanisms, Models, and Clinical Opportunities. Cancer Discov. 2013;3:730–741. doi: 10.1158/2159-8290.CD-13-0083. [DOI] [PubMed] [Google Scholar]
- 18.Xu W, Yang H, Liu Y, et al. Oncometabolite 2-Hydroxyglutarate Is a Competitive Inhibitor of α-Ketoglutarate-Dependent Dioxygenases. Cancer Cell. 2011;19:17–30. doi: 10.1016/j.ccr.2010.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Koivunen P, Lee S, Duncan CG, et al. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature. 2012;483:484–488. doi: 10.1038/nature10898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chowdhury R, Yeoh KK, Tian Y-M, et al. The oncometabolite 2-hydroxyglutarate inhibits histone lysine demethylases. EMBO Rep. 2011;12:463–469. doi: 10.1038/embor.2011.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Figueroa ME, Abdel-Wahab O, Lu C, et al. Leukemic IDH1 and IDH2 Mutations Result in a Hypermethylation Phenotype, Disrupt TET2 Function, and Impair Hematopoietic Differentiation. Cancer Cell. 2010;18:553–567. doi: 10.1016/j.ccr.2010.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Loenarz C, Schofield CJ. Expanding chemical biology of 2-oxoglutarate oxygenases. Nat Chem Biol. 2008;4:152–156. doi: 10.1038/nchembio0308-152. [DOI] [PubMed] [Google Scholar]
- 23.Hausinger RP. Fe(II)/α-Ketoglutarate-Dependent Hydroxylases and Related Enzymes. Crit Rev Biochem Mol Biol. 2004;39:21–68. doi: 10.1080/10409230490440541. [DOI] [PubMed] [Google Scholar]
- 24.Semenza GL. Hypoxia-inducible factor 1: master regulator of O2 homeostasis. Curr Opin Genet Dev. 1998;8:588–594. doi: 10.1016/s0959-437x(98)80016-6. [DOI] [PubMed] [Google Scholar]
- 25.Zhao S, Lin Y, Xu W, et al. Glioma-derived mutations in IDH1 dominantly inhibit IDH1 catalytic activity and induce HIF-1α. Science. 2009;324:261–265. doi: 10.1126/science.1170944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Fu Y, Zheng Y, Li K, et al. Mutations in isocitrate dehydrogenase 2 accelerate glioma cell migration via matrix metalloproteinase-2 and 9. Biotechnol Lett. 2012;34:441–446. doi: 10.1007/s10529-011-0800-8. [DOI] [PubMed] [Google Scholar]
- 27.Yalaza C, Ak H, Cagli MS, et al. R132H mutation in IDH1 gene is associated with increased tumor HIF1-alpha and serum VEGF levels in primary glioblastoma multiforme. Ann Clin Lab Sci. 2017;47:362–364. [PubMed] [Google Scholar]
- 28.Intlekofer AM, Dematteo RG, Venneti S, et al. Hypoxia Induces Production of L-2-Hydroxyglutarate. Cell Metab. 2015;22:304–311. doi: 10.1016/j.cmet.2015.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Semenza GL. Targeting HIF-1 for cancer therapy. Nat Rev Cancer. 2003;3:721–732. doi: 10.1038/nrc1187. [DOI] [PubMed] [Google Scholar]
- 30.Böttcher M, Renner K, Berger R, et al. D-2-hydroxyglutarate interferes with HIF-1α stability skewing T-cell metabolism towards oxidative phosphorylation and impairing Th17 polarization. Oncoimmunology. 2018;7:e1445454. doi: 10.1080/2162402X.2018.1445454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Williams SC, Karajannis MA, Chiriboga L, et al. R132H-mutation of isocitrate dehydrogenase-1 is not sufficient for HIF-1α upregulation in adult glioma. Acta Neuropathol. 2011;121:279–281. doi: 10.1007/s00401-010-0790-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kickingereder P, Sahm F, Radbruch A, et al. IDH mutation status is associated with a distinct hypoxia/angiogenesis transcriptome signature which is non-invasively predictable with rCBV imaging in human glioma. Sci Rep. 2015;5:16238. doi: 10.1038/srep16238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lando D. FIH-1 is an asparaginyl hydroxylase enzyme that regulates the transcriptional activity of hypoxia-inducible factor. Genes Dev. 2002;16:1466–1471. doi: 10.1101/gad.991402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mahon PC, Hirota K, Semenza GL. FIH-1: a novel protein that interacts with HIF-1alpha and VHL to mediate repression of HIF-1 transcriptional activity. Genes Dev. 2001;15:2675–2686. doi: 10.1101/gad.924501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.O’Reilly MS, Boehm T, Shing Y, et al. EndostatIn: An Endogenous Inhibitor of Angiogenesis and Tumor Growth. Cell. 1997;88:277–285. doi: 10.1016/s0092-8674(00)81848-6. [DOI] [PubMed] [Google Scholar]
- 36.Fischer I, Gagner J-P, Law M, et al. Angiogenesis in Gliomas: Biology and Molecular Pathophysiology. Brain Pathol. 2006;15:297–310. doi: 10.1111/j.1750-3639.2005.tb00115.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu Y, Jiang W, Liu J, et al. IDH1 mutations inhibit multiple α-ketoglutarate-dependent dioxygenase activities in astroglioma. J Neurooncol. 2012;109:253–260. doi: 10.1007/s11060-012-0914-4. [DOI] [PubMed] [Google Scholar]
- 38.Jones PA. Overview of Cancer Epigenetics. Semin Hematol. 2005;42:S3–S8. doi: 10.1053/j.seminhematol.2005.05.001. [DOI] [PubMed] [Google Scholar]
- 39.Sharma S, Kelly TK, Jones PA. Epigenetics in cancer. Carcinogenesis. 2010;31:27–36. doi: 10.1093/carcin/bgp220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Leung FCC. An evaluation of new criteria for CpG islands in the human genome as gene markers. Bioinformatics. 2004;20:1170–1177. doi: 10.1093/bioinformatics/bth059. [DOI] [PubMed] [Google Scholar]
- 41.Tsukada Y, Fang J, Erdjument-Bromage H, et al. Histone demethylation by a family of JmjC domain-containing proteins. Nature. 2006;439:811–816. doi: 10.1038/nature04433. [DOI] [PubMed] [Google Scholar]
- 42.Füllgrabe J, Kavanagh E, Joseph B. Histone onco-modifications. Oncogene. 2011;30:3391–3403. doi: 10.1038/onc.2011.121. [DOI] [PubMed] [Google Scholar]
- 43.Turcan S, Rohle D, Goenka A, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483:479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong X, Weng Z. The correlation between histone modifications and gene expression. Epigenomics. 2013;5:113–116. doi: 10.2217/epi.13.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krishnan S, Horowitz S, Trievel RC. Structure and Function of Histone H3 Lysine 9 Methyltransferases and Demethylases. Chem BioChem. 2011;12:254–263. doi: 10.1002/cbic.201000545. [DOI] [PubMed] [Google Scholar]
- 46.Venneti S, Felicella MM, Coyne T, et al. Histone 3 Lysine 9 Trimethylation Is Differentially Associated With Isocitrate Dehydrogenase Mutations in Oligodendrogliomas and High-Grade Astrocytomas. J Neuropathol Exp Neurol. 2013;72:298–306. doi: 10.1097/NEN.0b013e3182898113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lu C, Ward PS, Kapoor GS, et al. IDH mutation impairs histone demethylation and results in a block to cell differentiation. Nature. 2012;483:474–478. doi: 10.1038/nature10860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Song Y, Wu F, Wu J. Targeting histone methylation for cancer therapy : enzymes, inhibitors, biological activity and perspectives. J Hematol Oncol. 2016;9:1–21. doi: 10.1186/s13045-016-0279-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Suva M-L, Riggi N, Janiszewska M, et al. EZH2 Is Essential for Glioblastoma Cancer Stem Cell Maintenance. Cancer Res. 2009;69:9211–9218. doi: 10.1158/0008-5472.CAN-09-1622. [DOI] [PubMed] [Google Scholar]
- 50.Robertson KD. DNA methylation and human disease. Nat Rev Genet. 2005;6:597–610. doi: 10.1038/nrg1655. [DOI] [PubMed] [Google Scholar]
- 51.Tahiliani M, Koh KP, Shen Y, et al. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science. 2009;324:930–935. doi: 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.An J, Rao A, Ko M. TET family dioxygenases and DNA demethylation in stem cells and cancers. Exp Mol Med. 2017;49:e323. doi: 10.1038/emm.2017.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ye C, Li L. 5-hydroxymethylcytosine: a new insight into epigenetics in cancer. Cancer Biol Ther. 2014;15:10–15. doi: 10.4161/cbt.27144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Pfeifer GP, Xiong W, Hahn MA, Jin S-G. The role of 5-hydroxymethylcytosine in human cancer. Cell Tissue Res. 2014;356:631–41. doi: 10.1007/s00441-014-1896-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Vasanthakumar A, Godley LA. 5-hydroxymethylcytosine in cancer: significance in diagnosis and therapy. Cancer Genet. 2015;208:167–177. doi: 10.1016/j.cancergen.2015.02.009. [DOI] [PubMed] [Google Scholar]
- 56.Delhommeau F, Dupont S, Della Valle V, et al. Mutation in TET2 in Myeloid Cancers. N Engl J Med. 2009;360:2289–2301. doi: 10.1056/NEJMoa0810069. [DOI] [PubMed] [Google Scholar]
- 57.Yang H, Liu Y, Bai F, et al. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene. 2013;32:663–669. doi: 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mercher T, Quivoron C, Couronné L, Bastard C, Vainchenker W, Bernard OA. TET2, a tumor suppressor in hematological disorders. Biochim Biophys Acta. 2012;1825:173–177. doi: 10.1016/j.bbcan.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Jones PA, Laird PW. Cancer-epigenetics comes of age. Nat Genet. 1999;21:163–167. doi: 10.1038/5947. [DOI] [PubMed] [Google Scholar]
- 60.Huang Y, Rao A. Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 2014;30:464–474. doi: 10.1016/j.tig.2014.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.García MG, Carella A, Urdinguio RG, et al. Epigenetic dysregulation of TET2 in human glioblastoma. Oncotarget. 2018;9:25922–25934. doi: 10.18632/oncotarget.25406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kohanbash G, Carrera DA, Shrivastav S, et al. Isocitrate dehydrogenase mutations suppress STAT1 and CD8+ T cell accumulation in gliomas. J Clin Invest. 2017;127:1425–1437. doi: 10.1172/JCI90644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lucca LE, Hafler DA. Resisting fatal attraction: A glioma oncometabolite prevents CD8+T cell recruitment. J Clin Invest. 2017;127:1218–1220. doi: 10.1172/JCI93565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yang W, Wang H, Ju H, Dou C. A study on the correlation between STAT-1 and mutant p53 expression in glioma. Mol Med Rep. 2018;17:7807–7812. doi: 10.3892/mmr.2018.8796. [DOI] [PubMed] [Google Scholar]
- 65.Berghoff AS, Kiesel B, Widhalm G, et al. Correlation of immune phenotype with IDH mutation in diffuse glioma. Neuro Oncol. 2017;19:1460–1468. doi: 10.1093/neuonc/nox054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Amankulor NM, Kim Y, Arora S, et al. Mutant IDH1 regulates the tumor-associated immune system in gliomas. Genes Dev. 2017;31:774–786. doi: 10.1101/gad.294991.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Mu L, Long Y, Yang C, et al. The IDH1 Mutation-Induced Oncometabolite, 2-Hydroxyglutarate, May Affect DNA Methylation and Expression of PD-L1 in Gliomas. Front Mol Neurosci. 2018;11:82. doi: 10.3389/fnmol.2018.00082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Quail DF, Joyce JA. Microenvironmental regulation of tumor progression and metastasis. Nat Med. 2013;19:1423–1437. doi: 10.1038/nm.3394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Rahman M, Kresak J, Yang C, et al. Analysis of immunobiologic markers in primary and recurrent glioblastoma. J Neurooncol. 2018;137:249–257. doi: 10.1007/s11060-017-2732-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Komohara Y, Ohnishi K, Kuratsu J, Takeya M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J Pathol. 2008;216:15–24. doi: 10.1002/path.2370. [DOI] [PubMed] [Google Scholar]
- 71.Ma C, Horlad H, Ohnishi K, et al. CD163-positive cancer cells are potentially associated with high malignant potential in clear cell renal cell carcinoma. Med Mol Morphol. 2018;51:13–20. doi: 10.1007/s00795-017-0165-8. [DOI] [PubMed] [Google Scholar]
- 72.Friese MA, Wischhusen J, Wick W, Weiler M, Eisele G, Steinle A, Weller M. RNA Interference Targeting Transforming Growth Factor-β Enhances NKG2D-Mediated Antiglioma Immune Response, Inhibits Glioma Cell Migration and Invasiveness, and Abrogates Tumorigenicity In vivo. Cancer Res. 2004;64:7596–7603. doi: 10.1158/0008-5472.CAN-04-1627. [DOI] [PubMed] [Google Scholar]
- 73.Freeman GJ, Long AJ, Iwai Y, et al. Engagement of the Pd-1 Immunoinhibitory Receptor by a Novel B7 Family Member Leads to Negative Regulation of Lymphocyte Activation. J Exp Med. 2000;192:1027–1034. doi: 10.1084/jem.192.7.1027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Goodman AM, Piccioni D, Kato S, et al. Prevalence of PDL1 Amplification and Preliminary Response to Immune Checkpoint Blockade in Solid Tumors. JAMA Oncol. 2018;4:1237–1244. doi: 10.1001/jamaoncol.2018.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gerlinger M, McGranahan N, Dewhurst SM, Burrell RA, Tomlinson I, Swanton C. Cancer: Evolution Within a Lifetime. Annu Rev Genet. 2014;48:215–236. doi: 10.1146/annurev-genet-120213-092314. [DOI] [PubMed] [Google Scholar]
- 76.Falnes PØ, Johansen RF, Seeberg E. AlkB-mediated oxidative demethylation reverses DNA damage in Escherichia coli. Nature. 2002;419:178–182. doi: 10.1038/nature01048. [DOI] [PubMed] [Google Scholar]
- 77.Wang P, Wu J, Ma S, et al. Oncometabolite D-2-Hydroxyglutarate Inhibits ALKBH DNA Repair Enzymes and Sensitizes IDH Mutant Cells to Alkylating Agents. Cell Rep. 2015;13:2353–2361. doi: 10.1016/j.celrep.2015.11.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Cairncross JG, Wang M, Jenkins RB, et al. Benefit From Procarbazine, Lomustine, and Vincristine in Oligodendroglial Tumors Is Associated With Mutation of IDH. J Clin Oncol. 2014;32:783–790. doi: 10.1200/JCO.2013.49.3726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Johannessen T-CA, Prestegarden L, Grudic A, Hegi ME, Tysnes BB, Bjerkvig R. The DNA repair protein ALKBH2 mediates temozolomide resistance in human glioblastoma cells. Neuro Oncol. 2013;15:269–278. doi: 10.1093/neuonc/nos301. [DOI] [PMC free article] [PubMed] [Google Scholar]