SUMMARY
Histidine kinases (HKs) funnel diverse environmental stimuli into a single autophosphorylation event at a conserved histidine residue. The HK EnvZ is a global sensor of osmolality and cellular acid pH. In previous studies, we discovered that osmosensing in EnvZ was mediated through osmolyte-induced stabilization of the partially disordered helical backbone spanning the conserved histidine autophosphorylation site (His243). Here we describe how backbone stabilization leads to changes in microenvironment of His243 resulting in enhanced autophosphorylation through relief of inhibition and repositioning of critical side chains and imidazole rotamerization. The conserved His-Asp/Glu dyad within the partially structured helix is equally geared to respond to acid pH, an alternative environmental stimulus in bacteria. This high-resolution ‘double clamp’ switch model proposes that a His-Asp/Glu dyad functions as an integrative node for regulating autophosphorylation in HKs. Because the His-Asp/Glu dyad is highly conserved in HKs, this study provides a universal model for describing HK function.
eTOC Blurb:
A His-Asp/Glu dyad forms the core of EnvZ histidine-kinase osmosensing wherein osmolytes mediate changes in backbone and sidechain microenvironment leading to enhanced histidine autophosphorylation. This His-Asp/Glu dyad is highly conserved across diverse HKs, suggesting a unified model for integrating diverse environmental stimuli to a singular histidine phosphorylation response.
INTRODUCTION
Bacterial adaptation to diverse extracellular stimuli is mediated through a network of well-conserved stimulus-response coupled mechanisms known as two-component signaling systems (TCS), consisting of a histidine kinase (HK) and a response regulator (RR) (see (Goulian, 2010) for a review). Input stimuli alter the rate of autophosphorylation at a conserved histidine on a membrane-anchored HK, and the phosphoryl group is subsequently transferred to its cognate RR on a conserved aspartic acid. These histidyl-aspartyl phosphorelays are responsible for eliciting specific responses to a wide range of environmental stimuli, including nutrient fluxes and osmolality, and are critical for regulating processes essential for bacterial survival and virulence (Beier and Gross, 2006; Grebe and Stock, 1999; Hoch, 2000; Krell et al., 2010).
A particularly well-characterized TCS is the EnvZ/OmpR system that is responsible for sensing changes in intracellular osmotic and acid environments, resulting in changes in the transcription profile (Chakraborty et al., 2015; Chakraborty et al., 2017; Wang et al., 2012). EnvZ is autophosphorylated (Aiba et al., 1989; Igo and Silhavy, 1988) on a conserved histidine residue, His243 (Forst et al., 1989) and osmolality enhances EnvZ autophosphorylation (Wang et al., 2012). Transfer of the phosphoryl group to the cognate RR OmpR at a conserved Asp, Asp55 (Delgado et al., 1993) modulates expression of outer membrane porin genes ompF and ompC (Kenney et al., 1995). This strategy is one of several mechanisms employed by bacteria for cellular osmoregulation (Csonka and Hanson, 1991; Sleator and Hill, 2002).
Each monomer of the homodimeric EnvZ consists of two transmembrane segments (residues 16–35 and 159–179), a periplasmic domain (residues 36–158), an intracellular HAMP domain (present in Histidine kinase, Adenylate cyclase, Methyl-accepting proteins and Phosphatases; 180–232) and a large C-terminal cytoplasmic domain (residues 233–450) (Forst et al., 1987). The cytoplasmic domain is further divided into two subdomains: a dimerization subdomain forms a four-helix bundle containing the autophosphorylation site His243, which is connected through a flexible linker to the C-terminal ATP-binding subdomain. A soluble cytoplasmic construct of EnvZ (EnvZc; residues 180–450) lacking the transmembrane and periplasmic domains was wholly capable of osmosensing and autophosphorylation in vitro and in vivo (Wang et al., 2012). This observation underscored the modularity of HKs and the under-recognized two-way allostery whereby stimulus sensing is achieved by generating cytoplasmic signals from extracellular stimuli (Figure 1A). Membrane-anchored HKs are also regulated by integral and peripheral lipid interactions (Ghosh et al., 2017). These studies challenged the common notion of a unidirectional relay that coupled sensing by extra-cytoplasmic domains of a receptor to the cytoplasmic functional site through large-scale conformational changes in the transmembrane segments (Chervitz and Falke, 1996; Gushchin et al., 2017; Hall et al., 2011; Hulko et al., 2006; Kitanovic et al., 2011; Ottemann et al., 1999). Indeed, transmembrane signaling is also regulated via phospholipid interactions with transmembrane and cytoplasmic domains (daCosta et al., 2009; Dowhan, 1997; Ghosh et al., 2017; Lee, 2003, 2004; Leonard and Hurley, 2011; Phillips et al., 2009; Tsaloglou et al., 2011).
Figure 1. (A) Mechanism of osmosensing by EnvZ.
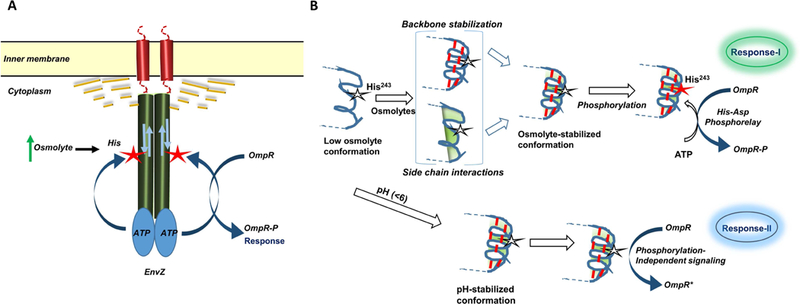
EnvZ is an inner membrane anchored dimeric receptor that responds to changes in osmolyte concentrations by altering the conformation of a cytoplasmic helical subdomain. These conformational changes lead to enhanced autophosphorylation at a conserved His residue (red asterix) that are transferred to OmpR. This signaling relay can also be modulated through alternate allosteric mechanisms (bidirectional arrow). The phospholipid membrane in particular, is a critical regulator of EnvZ receptor function effected through changes in mechanical and physicochemical forces such as membrane curvature, fluidity, lateral pressure and lipid composition. Alternately, allosteric regulation of the cytoplasmic kinase domain can also be achieved through peripheral interactions between the phospholipid membrane and the non-embedded soluble cytoplasmic kinase domain (yellow dashed lines). Mutations in the cytoplasmic domain can in turn modulate the interactions between the phospholipid bilayer, the transmembrane segments and the cytoplasmic domain thereby altering EnvZ response to stimuli.
(B) Deconstructing the osmosensing switch in EnvZ
Cartoon representation of the His243 locus shows local disorder in helical structure under low osmolality conditions. High osmolality promotes stabilization through backbone H-bonds (red dashed lines) measured by HDXMS (Wang et al. (2012)) and side chain microenvironment (shaded green) leading to enhanced autophosphorylation at His243 denoted by star, phosphorylation in red. In brackets are the varying and integrative effects that osmolytes might induce on the His243 locus. Direct osmolyte-mediated changes in His side chain microenvironment (shaded green), backbone H-bonds (red) and how stabilization of backbone might indirectly the His243 rotamer and its microenvironment. Dual pathways for osmolyte and low pH signaling by EnvZ:OmpR. Osmolyte-stabilized conformation of the cytoplasmic subdomain enhances His243 autophosphorylation, which is transferred to OmpR to trigger changes in porin gene expression (Response-I). The same locus also operated to detect lowering of pH through non-canonical activation of OmpR when acid pH does not favor phosphorylation (Response-II).
Although the His-Asp phosphorelay has been well characterized across TCS proteins, how changes in a physical environmental cue such as osmolality are translated into a chemical phosphorylation output signal remained a mystery until recently. Application of amide hydrogen-deuterium exchange mass spectrometry (HDXMS) led to a breakthrough in understanding the mechanism of osmosensing and signaling in EnvZ (Wang, et al., 2012). HDXMS determined that His243 was in a partially unfolded region of the four-helix bundle. High concentrations of osmolytes decreased deuterium exchange at the His243 locus. Osmolyte-induced H-bonding stabilized the four helix bundle, enhancing autophosphorylation at His243 (Wang et al., 2012) (Figure 1B). In this study, we describe the next step in signal transduction, how a chemical signal in the form of enhanced His243 autophosphorylation is driven by osmolyte-mediated helical backbone stabilization and concomitant side chain effects.
HDXMS monitors exchange of hydrogens solely at the backbone amide position, offering a highly sensitive readout of H-bonding propensities and/or solvent accessibility. Although it is highly sensitive to side chain interactions, contacts and microenvironments, HDXMS does not offer a direct readout of side chain interactions. We previously demonstrated that osmolytes stabilize helical backbone H-bonds (Wang et al., 2012), but osmolytes might also impact side chain interactions around His243 (Figure 1B). Our observation that osmolyte-mediated stabilization of backbone interactions was abolished in the EnvZc H243A mutant suggested that both main chain and side chain interactions work in tandem to transduce osmolality signals into a phosphorylation response (Wang et al., 2012). We applied a combination of phosphorylation assays of osmosensing mutants, HDXMS-mediated backbone dynamics measurements and in silico models in order to deconstruct the function of the phosphorylation switch, which constitutes the mechanistic core of HK signaling.
The core of the osmosensing apparatus involves a network of coupled backbone and side chain interactions. The heart of the switch consists of a conserved His-Asp/Glu dyad motif within a region of local disorder. A His-Asp dyad is found in HKs EnvZ, KdpD and CheA (Figure 2). Interestingly, KdpD regulates potassium homeostasis in Escherichia coli and most other bacteria by altering expression of the kdpFABC operon encoding potassium ion pumps (Freeman et al., 2013). KdpD activity is stimulated by changes in potassium ion concentrations, osmolarity, and ATP concentrations (Heermann and Jung, 2010). These signals are similar to the signals sensed by EnvZ (salt/sugar concentrations, osmolarity, pH, and temperature). The other HK with a His-Asp dyad is CheA, which is involved in chemotaxis and responds to numerous osmolytes, including amino acids and sugars. Most other HKs have a conservative His-Glu dyad (Figure 2).
Figure 2: Alanine and threonine flanking the invariant histidine are evolutionarily conserved.
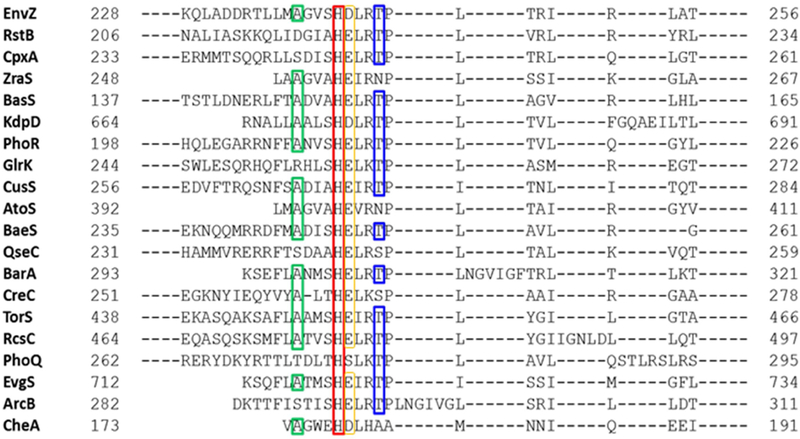
Sequence alignment of various bacterial histidine kinases using Basic Local Alignment Search Tool (BLAST) (54) showed that Ala (green box) and Thr residues (blue box) at flanking positions one helical turn away from the conserved His (red box) are common in most histidine kinases. An Asp/Glu is conserved in all but one HK (yellow box). This highlights an evolutionary conservation in histidine kinases that may have an important role for enzymatic functions. Sensor histidine kinases shown in the search with known functions are: ArcB, anaerobic respiration control; AtoS, conversion of short-chain fatty acids to acetoacetate; BasS, iron (Fe2+) sensor; CheA, chemotaxis; CpxA, envelope stress sensor; CreC, catabolite regulation; CusS, cuprite (Cu2+) and silver (Ag2+) sensor; EnvZ, osmosensor; EvgS, homolog of virulence gene; KdpD, potassium (K+) sensor; PhoQ, sensor for magnesium (Mg2+) and acid resistance genes; PhoR, phosphate regulon gene expression; QseC, quorum sensor and flagellum regulation; RcsC, biofilm production; RstB, sensor for RstA; TorS, sensor for trimethylamine oxide (TMAO); and ZraS, zinc (Zn2+) and lead (Pb2+) sensor. BarA, BaeS, and GlrK do not have a known function assigned currently.
The histidine sidechain that is autophosphorylated has unique features essential to the signaling process. Two imidazole nitrogen atoms mediate dual inhibitory interactions: a poorly defined H-bond mediated by a carbonyl moiety of the helical backbone at the n-4 position and the side-chain carboxyl of an adjacent conserved Asp residue provide a ‘double negative’ clamp. These two interactions chemically dampen autophosphorylation of His243 at low osmolality. High osmolality disrupts these inhibitory contacts, enhancing autophosphorylation at His243. EnvZ also responds to changes in pH (Chakraborty et al., 2015; Chakraborty et al., 2017), as acidic pH in the 4.5–6 range promotes helicity (Scholtz and Baldwin, 1992), and favors the activated state. These results emphasize the versatility of HK cytoplasmic domains as integrators of diverse stimuli across the HK superfamily.
RESULTS
Backbone stabilization by osmolytes resolves a helical kink at the His243 locus
We were interested in probing how helical backbone stabilization altered the His243 sidechain interactions. Backbone stabilization would directly impact side chain interactions (Figure 1B). To model these changes in side chain interactions in the immediate neighborhood of the His243 in the presence of high osmolality, we generated in silico models of the EnvZc four helix bundle. The aim was to capture the large-scale conformational changes within the osmosensing core of EnvZ in response to osmolytes (Figure 3). The low osmolality model was based on the solution NMR structure of the homodimeric domain of EnvZ as a template (residues 223–289; PDB ID: 1JOY), which was resolved at low osmolality. The high osmolality form was modeled on the X-ray crystal structure of a chimera of E. coli EnvZ histidine kinase catalytic domain fused to the Archaeoglobus fulgidus Af1503 HAMP domain (PDB ID: 4CTI), solved at high osmolality (see STAR Methods for details).
Figure 3: Molecular dynamics simulations demonstrate osmolyte-induced secondary structural stabilization of EnvZc.
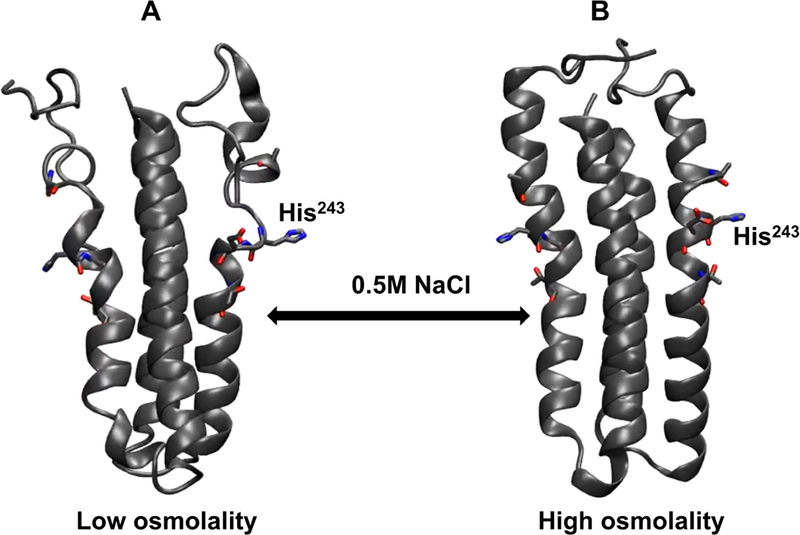
In silico models of EnvZ four helix bundle subdomain (residues 223–289) were generated under conditions mimicking low and high osmolality. It is evident from these structures that under low osmolality (left), the four-helix bundle subdomain is highly flexible particularly within the His243-containing region. In presence of high osmolality (right), there is reduced flexibility of the peptide backbone with more defined secondary structures being observed across the subdomain. These models further reinstate the model of EnvZ osmosensing proceeding through osmolyte-mediated peptide backbone stabilization of the four helix bundle.
At low osmolality, the four helix bundle was conformationally dynamic (Figure 3A and Movie S1), as we reported previously (Wang et al., 2012). At high osmolality, an enhanced overall stabilization of the peptide backbone was observed (Figure 3B and Movie S2). In particular, the kink observed in the α-helix at the 239–240 position resolved into a continuous α-helix, consistent with the HDXMS-derived helix stabilization model (Wang et al., 2012). These computational simulations emphasize that osmolytes stabilize backbone H-bonding, leading to increased ordering of the four-helix subdomain in EnvZ. These events occur on a nanosecond timescale.
His243 autophosphorylation is sensitive to both helical backbone stabilization and its microenvironment
We were interested in determining how secondary structure stabilization of the four-helix bundle subdomain was coupled to autophosphorylation of His243. We thus monitored the dynamics of exchange and the functional consequences of two EnvZ mutants, A239T and T247R. The T247R mutant was an activating mutant that exhibited high phosphorylation and signaling even at low osmolality, while phosphorylation and signaling by A239T was absent at all osmolalities. These substitutions are spatially symmetric, located one helical turn N- and C-terminal to His243 (Figure S1), respectively, and are highly conserved across the HK superfamily (Figure 2). EnvZ-dependent transcriptional activation was monitored using an ompC-lacZ fusion. We measured β-galactosidase activity as a function of osmolality in the wild-type and various EnvZ mutants (Figure 4). In the absence of envZ, β-galactosidase activity was essentially background, and activity was complemented with a plasmid expressing the cytoplasmic domain of envZc (EnvZc). The EnvZc T247R mutant exhibited much greater osmolality-dependent ompC-lacZ activity compared to the wildtype, while EnvZc A239T activity was at background levels. The effects of the above substitutions on EnvZc autophosphorylation were consistent with their β-galactosidase activity (Figure 4A). Since substitution of threonine for alanine (A239T) does not alter the backbone structure, backbone stabilization by osmolytes would remain intact. But because autophosphorylation was no longer stimulated by osmolytes, this result indicates that sidechain effects contribute significantly to signaling (i.e. autophosphorylation).
Figure 4: EnvZc autophosphorylation is highly sensitive to mutations in the His243 microenvironment.
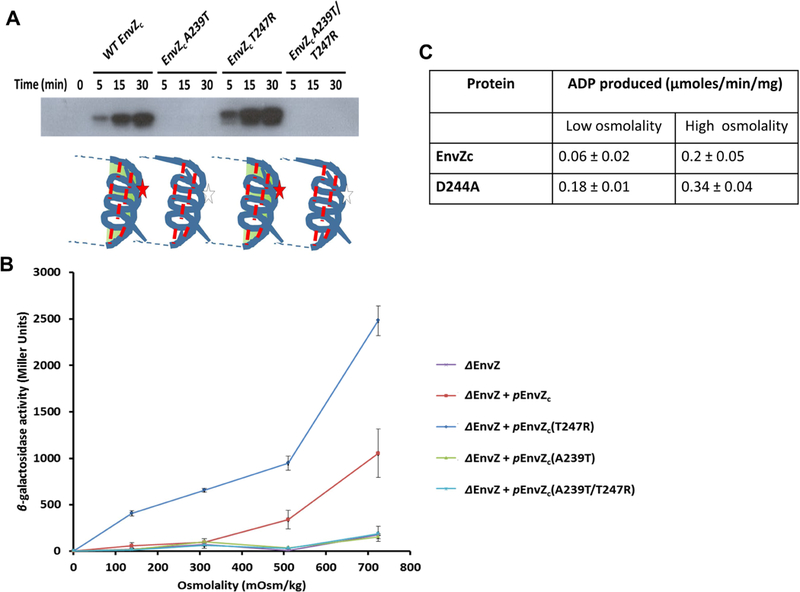
(A) Wild-type (WT), A239T, T247R and A239T/T247R EnvZc were tested for their ability to undergo autophosphorylation using a [γ−32P]-ATP kinase assay for kinase reaction times 0, 5, 15 and 30 min. WT EnvZc showed increased levels of autophosphorylation over time. The point mutants A239T and T247R revealed contrasting phenotypes with the A239T mutant showing negligible autophosphorylation while the T247R mutant showing constitutively higher levels of autophosphorylation in comparison to WT EnvZc. These results were consistent with the phenotypes observed for the same mutants in intact EnvZ (Matsuyama et al., 1986; Russo and Silhavy, 1991). The A239T/T247R EnvZc double mutant was surprisingly unable to autophosphorylate, indicating that the Ala239 residue has a significant role in modulating autophosphorylation in EnvZc (see Discussion). (B) WT EnvZc and the mutants were also examined for their ability to phosphotransfer to OmpR. Phosphorylated OmpR (OmpR~P) enhances transcription of the reporter gene fusion ompC-lacZ. In the envZ deletion (ΔenvZ) strain of E. coli PG189, a low level of ompC-lacZ expression (purple curve) is observed for cells grown in minimal A medium across a range of sucrose concentrations (0, 5, 10, and 15% (w/v), indicated as 139, 311, 510, and 724 mOsm/kg on the x-axis). When a plasmid carrying envZc (penvZc) was transformed into the ΔenvZ E. coli strain, expression of ompC-lacZ was rescued (red curve). EnvZc A239T (orange curve) and the EnvZc A239T/T247R double mutant (DM) (green curve) failed to rescue ompC-lacZ expression while EnvZc T247R (blue curve) showed higher OmpC-LacZ expression than WT EnvZc at all osmolyte concentrations. The results were reported as mean ± SEM from at least three independent measurements. (C) Autophosphorylation by EnvZ was measured using the in vitro ADP-Glo™ Kinase assay (Promega, Madison, WI), which quantifies the amount of ADP formed as a product in a kinase reaction using a luminescence-based detection method in low and high osmolality conditions as described in STAR methods.
To determine whether the activating and inhibitory effects of T247R and A239T were compensatory, we measured osmolality-dependent autophosphorylation of the EnvZc double mutant A239T/T247R. Interestingly, in the double mutant, autophosphorylation was undetectable and it was unable to induce ompC expression when it was expressed in a ΔenvZ strain (Figures 4A and 4B). Thus, the behavior of A239T prevailed in the double A239T/T247R mutant.
Helical backbone stabilization precedes autophosphorylation at His243.
We next measured HDXMS of EnvZc A239T at low and high osmolality. This mutant showed decreased deuterium exchange at high osmolality, with a profile identical to wild-type EnvZc. However, it was not phosphorylated and β-galactosidase activity was at background levels (Figures 4A and 4B). This result highlighted the importance of side chain interactions in addition to backbone stabilization. HDXMS of the double mutant EnvZc A239T/T247R presented an interesting result. Its HDXMS profile was similar to T247R, where osmolyte-dependent helical backbone stabilization was apparent (Figure 5), but it was not phosphorylated (Figure 4A). Impairment of side chain interactions of the A239T mutant negatively interfered with phosphorylation (Figure 4A), despite retaining the main chain backbone H-bond elements of the EnvZ osmosensing switch. Thus, helical backbone stabilization by osmolytes is a necessary first step in signaling, but it is not sufficient for signaling.
Figure 5: Mutation-induced altered peptide backbone dynamics can be correlated to His243 autophosphorylation.
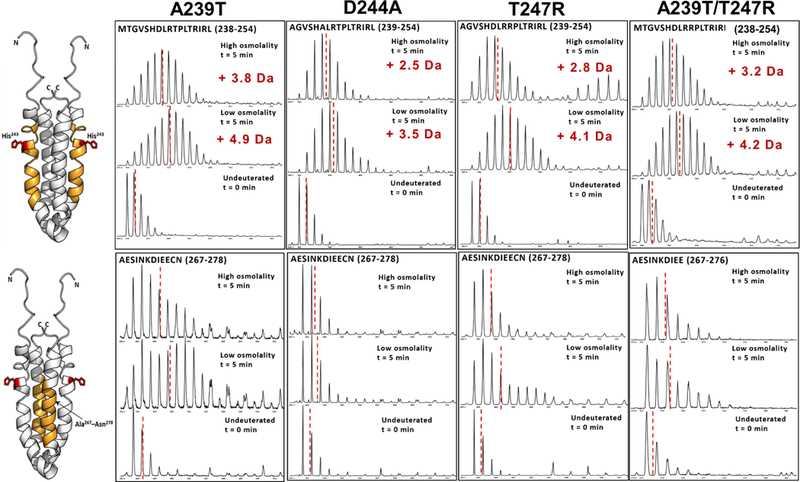
ESI-Q-TOF mass spectra shown here compare backbone dynamics of A239T, D244A, T247R and A239T/T247R EnvZc, for two critical regions within the four helix bundle. (A) In the His243-spanning locus (highlighted in orange in the NMR structure of the four helix bundle; His243 shown as sticks; PDB ID: 1JOY) A239T mutant undergoes deuterium exchange similar to WT under low osmolality, while a significantly reduced deuterium exchange is observed for the other mutants. However, osmolyte-induced reduction in deuterium exchange is observed for all mutants. (B) In the putative OmpR-binding region (highlighted in yellow), the A239T mutant populates a conformational ensemble, evident from the characteristic bimodal feauture of the mass spectral envelope, in the low osmolality condition, with an osmolality-induced predominance of the lower exchanging conformer. The lower exchanging conformer dominates in the other mutants with a minor population of the higher exchanging conformer observed in T247R and A239T/T247R EnvZc. Reduced backbone dynamics in the four-helix bundle of mutants D244A and T247R correlates with their constitutive kinase activity.
Helical backbone stabilization enhances autophosphorylation at His243
To further test the link between helical backbone stabilization and enhanced His243 autophosphorylation, we carried out HDXMS of an activating mutant, EnvZc T247R. In our previous study, we identified two regions within the four-helix bundle subdomain that were osmosensitive (Wang et al., 2012). One flanked the phosphorylatable histidine (residues 238–254) and the other was the OmpR binding site (residues 267–278) (Foo et al., 2015). Deuterium exchange in these two peptides served as reporters for osmolyte-dependent changes in H-bonding (Figure S3A, B). We measured HDXMS of EnvZ mutants to directly monitor how backbone stabilization (stimulated by osmolytes) enhanced chemical autophosphorylation. Substitution of the invariant His243 to Ala (H243A) eliminated autophosphorylation, and osmolyte-dependent backbone stabilization was no longer observed (Figure S3 C, D). Figures S3 A, B, C, D are published with permission (Wang et al., 2012) as described in supplemental information. Overall, all mutants showed osmolality-specific changes in the kinase subdomain (residues 320–450) that were not observed in WT EnvZc (Wang et al., 2012).
It was therefore of interest to examine the HDXMS profile of T247R. T247R was fully capable of backbone stabilization in response to osmolytes (Figure 5; Table 1). Although T247R occupied the active state at low osmolality (as evident by its reduced exchange rate), it was still osmosensitive (Figure 5). Changes were also observed in the ATP-containing phosphotransferase subdomain of all of the EnvZc mutants (Figures S4–7), although such changes were not observed in wildtype EnvZc (Wang et al., 2012) (see Discussion).
Table 1: The numbers of deuterons exchanged after 5 min deuteration in the His243-containing peptide under low and high osmolality conditions.
Mutation site(s) are underlined. Values are reported as mean ± standard deviation from at least two independent experiments.
| EnvZc construct | Reporter peptide | Deuterons exchanged in the His243-containing peptide (5 min deuteration) | |
|---|---|---|---|
| Low osmolality | High osmolality | ||
| Wild type* | MAGVSHDLRTPLTRIRL | 5.4 ± 0.1 | 4.3 ± 0.1 |
| H243A* | ADLRTPLTRIRL | 5.3 ± 0.1 | 5.2 ± 0.1 |
| A239T | MTGVSHDLRTPLTRIRL | 4.9 ± 0.0 | 3.9 ± 0.1 |
| T247R | MAGVSHDLRRPLTRIRL | 3.2 ± 0.2 | 2.7 ± 0.0 |
| A239T/T247R | TGVSHDLRRPLTRIRL | 4.3 ± 0.2 | 3.3 ± 0.1 |
| D244A | AGVSHALRTPLTRIRL | 3.5 ± 0.2 | 2.5 ± 0.2 |
Data were reproduced from Wang et al. (2012) with permission.
For EnvZc H243A mutant, two peptic peptides were identified (238–245 and 243–254). The deuterium uptake value reported here is for the peptide spanning residues 243–254.
Backbone stabilization is coupled to OmpR binding
Peptides within the OmpR binding site of EnvZc (residues 267–278) exhibited a bimodal spectrum, representative of an ensemble of conformations. In wildtype EnvZc, two populations of conformations were observed with distinctly different deuterium exchange rates (Wang et al., 2012). At low osmolality, the conformation with faster deuterium exchange dominated, with a minor population of the lower exchanging conformer observed towards the left of the spectral profile. An increase in osmolality shifted the population to the lower exchanging, more active conformer. These two conformational profiles were also observed in the EnvZc mutants (Figure 5).
The bimodal spectrum was observed with A239T EnvZc at low osmolality, consistent with wildtype EnvZc (Wang et al., 2012). The higher-exchanging spectral envelope diminished in the presence of osmolytes (Figure 5). Thus, in the A239T mutant, both reporter regions exhibited osmolyte-dependent stabilization, but the inability to undergo the appropriate sidechain interactions prevented autophosphorylation (Figure 4).
Analysis of the same region in the mutant T247R showed a distinctly different deuterium exchange profile (Figure 5). The bimodal profile of deuterium exchange of the OmpR-binding site peptide showed a predominance of the lower exchanging population, even at low osmolality (Figure 5). A further increase in intensity of the lower exchanging conformer was evident at high osmolality, indicating the mutant was still capable of osmosensing, but a higher percentage of the mutant populated the active conformation at low osmolality. This result was consistent with a higher level of autophosphorylation and ompC-lacZ activity (Figures 4A, B).
Modulation of His243 autophosphorylation coordinates Ala239 backbone and Asp244 sidechain interactions
It was evident from the autophosphorylation assays and HDXMS experiments that osmosensing and kinase activity in EnvZc was modulated by a coordinated mechanism involving both the peptide backbone and the side chains in the neighborhood of His243. We hypothesized that this mechanism was primarily mediated through H-bonding and electrostatic contacts in the His243 microenvironment and therefore set out to quantify these interactions in silico in WT and EnvZc mutants. The propensity of bond formation was quantified by molecular dynamics simulations to identify their contribution to osmosensing and autophosphorylation.
Bonding propensities of two interactions were of particular interest (Figure 6). The indispensability of the Ala239 residue for kinase activity has been unambiguously demonstrated by our in vitro and in vivo kinase assays (Figure 4). The solution NMR structure of the EnvZ four helix bundle (PDB ID: 1JOY; Figure 7C) solved at low osmolality shows the backbone carbonyl oxygen of Ala239 positioned in close proximity to the hydrogen atom attached to the Nδ of the His243 imidazole, potentially forming an H-bond. In the high osmolality X-ray crystallographic structure (PDB ID: 4KP4; Figure 7D), the H-bond between Ala239 carbonyl oxygen and the backbone amide hydrogen of His243 appears more favorable, based on distance and geometrical alignment. This would result in the formation of a stronger peptide backbone H-bond. Therefore, the propensity for backbone H-bond formation between Ala239 and His243 were computed for WT and EnvZc mutants A239T, T247R and A239T/T247R. Strengthening of the H-bond between the backbone Ala239 carbonyl oxygen and His243 amide hydrogen by osmolytes was inferred from bond propensities calculated from molecular dynamics simulations in WT EnvZc. This interaction (Figure 7) enhances phosphorylation at high osmolality. In the constitutively active mutant T247R, this H-bond is as probable at low osmolality as it is in WT EnvZc at high osmolality. Thus, T247R increases the proportion of protein in the activated conformation at low osmolality (Figure 5). An increase in the probability of Ala239-His243 H-bond formation by osmolytes was observed in WT EnvZ and all of the mutants. This increase was irrespective of their basal kinase activity.
Figure 6: H-bonding propensities show that local secondary structure is highly sensitive to mutations in the His243 locus.
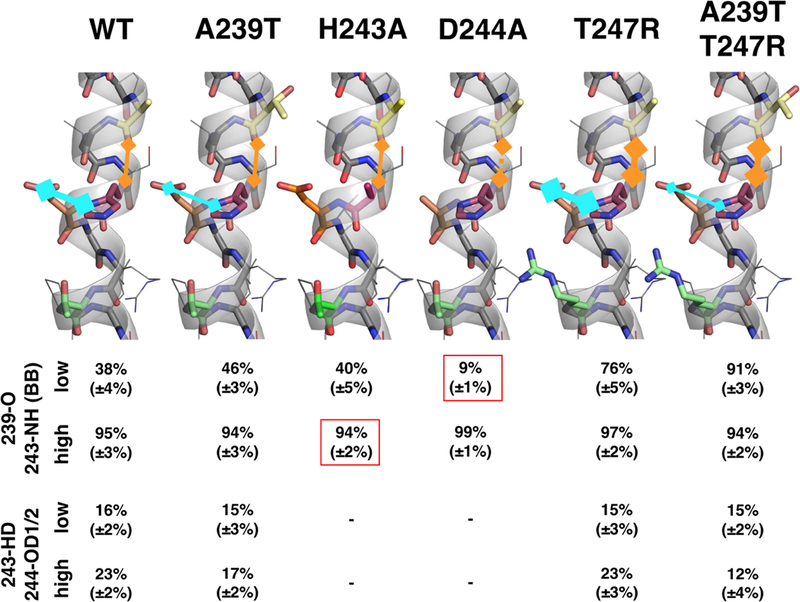
This figure compares bonding propensities of two critical bonds in the His243 locus in WT EnvZc and EnvZc mutants A239T, D244A, T247R and A239T/T247R, as measured by molecular dynamics simulations. In the wild-type, hydrogen bonding propensitiesincrease with increased osmolality . In all mutants, the sensitivity of this bonding behavior is abolished. A239T and the double mutant stabilize bonding propensity at a constant low level, whereas the T247R mutant locks the bonding propensity in a high state. All values are given as mean of both simulations with errors stated in brackets. The backbone H-bond propensity between Ala239 and His243 is indicated by 239-O/243-NH (carbonyl oxygen of Ala239/amide backbone of His243) (orange) while side chain interaction between His243 and Asp244 are indicated by 243-HD/244-OD1/2 (H on Nδ of His243/O on Asp244) (blue). The boxed values indicate the H-bonding propensities not matching experimental HDXMS measurements.
Figure 7: His243 autophosphorylation and osmosensing is controlled by His243 rotamerization, coordinated side-chain and backbone interactions in the His243 microenvironment.
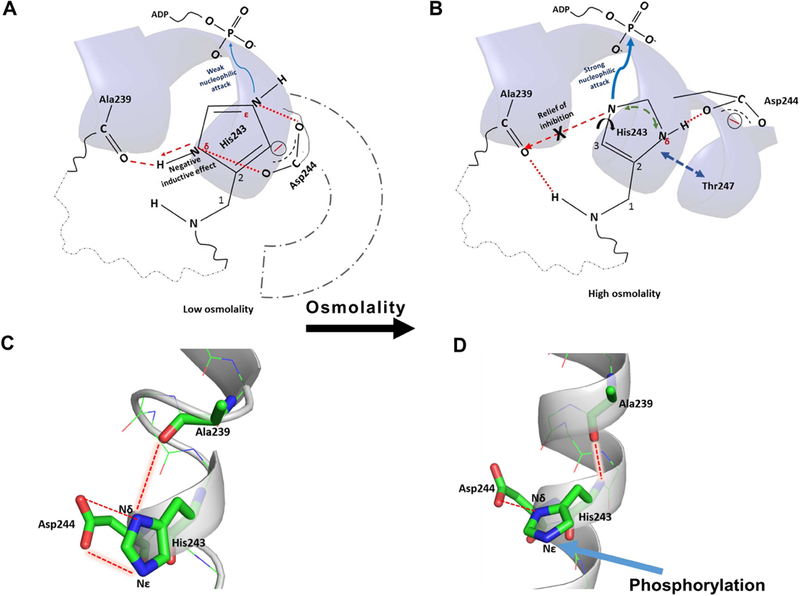
(A) Under low osmolality, there is local disorder in the His243 helix and two cooperative interactions mediated by Asp244 side chain with the His243 imidazole nitrogen atoms that suppress the nucleophilicity of Nε, by promoting delocalization of the imidazole protons between Nε and Nδ (red dashed lines). Presence of the Asp244 side chain also impedes free rotamerization of the imidazole side chain, essential for effective His243 phosphorylation. The Ala239 backbone carbonyl group forms mutually exclusive and competing H-bonds; one with the H on Nδ of His243 and the backbone amide proton of His243, further drawing the electron density away from the imidazole ring (red dashed arrow). Substitution of Asp244 by alanine leads to enhanced autophosphorylation in D244A EnvZc, in low osmolality, through relief of inhibition. (B) Osmolality removes the impeding effect of the Asp244 carboxylate group and osmolyte-induced backbone stabilization strengthens the backbone H-bond between Ala239 carbonyl and amide H of His243, weakening the H-bond between Ala239 carbonyl and Nδ of His243 (marked ‘X’). Relief of Asp244 inhibitory interactions frees His243 rotamerization. Relief from these two inhibitory interactions enhances the nucleophilicity of the Nε promoting enhanced phosphorylation. High resolution structures of the His243 neighborhood under conditions of low ((C); PDB ID: 1JOY) and high osmolality ((D); PDB ID: 4KP4) show that the residues His243, Ala239 and Asp244 are aligned in a particular geometry to favor the interactions described above.
Asp244 negatively regulates His243 autophosphorylation at low osmolality, but enhances autophosphorylation at high osmolality
Histidine-aspartic/glutamic acid dyads are critical components of the active sites in the esterase superfamily. The aspartate/glutamate side chain coordinates a proton relay among the imidazole nitrogens, which is critical for catalysis (Birktoft and Banaszak, 1983). To identify whether there was a role for Asp244 in the EnvZ reaction cycle, we generated an EnvZc D244A mutant. Interestingly, this mutant exhibited high turnover in both low and high osmolality (Figure 4C). The deuterium exchange profile of D244A at low osmolality was similar to the T247R mutant, suggesting that Asp244 functions as a brake to reduce the kinase activity at low osmolality and D244A shifts the ensemble to favor a constitutively active conformation. Further, in the D244A mutant, the bimodal spectral profile for the OmpR-binding site peptide was barely detectable, indicating the near absence of the higher exchanging conformation in solution (Figure 5). This suggests that the side chain of Asp244 functions to increase local unfolding of both the His243 containing helix as well as the two flanking helices (OmpR binding sites) in the dimer. Correspondingly, an acidic cluster of residues (Asp273, Glu275 and Glu 276) in the OmpR binding helix modulates the stability of the His243 locus through Asp244. The D244A mutant abolishes this allosteric relay between the two sites. Substitution of the adjacent Asp244 residue with Ala dramatically reduced the Ala239-His243 backbone H-bond propensity at low osmolality (Figure 6). The H-bond propensity of the His243 helix occurs on a faster (ns) time scale, whereas the bimodal deuterium exchange kinetics for the flanking OmpR binding site helix) indicate a slower (msec) time scale for conformational changes in this helix (Wang et al., 2012). Changes at this helix are allosterically coupled through a negative induction between the acidic cluster of residues (Asp273, Glu275 and Glu 276) and the side chains of the His243-Asp244 dyad. Allosteric coupling through the side chains of the His-Asp dyad are not captured by our MD simulations. Therefore, for the two mutants, D244A and H243A, the H-bond propensities predicted do not correlate with our HDXMS results due to the differing time scales (see below).
Peptide backbone stabilization, as modelled by MD simulations of the H-bond propensities, occurs on a nanosecond time scale. HDXMS monitors dynamics across nanosecond and slower time scales. However, conformational relays from the OmpR binding site (as measured by HDXMS) occur on a slower (milliseconds) time scale. Asp244 functions as an integrative node for mediating a two-way cross-talk between the OmpR binding helix and the His243-containing helix. High osmolality disrupts the destabilizing effects of Asp244 as reflected in the high values for Ala239-His243 backbone H-bond propensity (Figure 6). Removing the negative influence of Asp244 shifted the equilibrium to a nearly complete population in the active state, which would drive OmpR activation. Interestingly, H243A exhibited Ala239-His243 backbone H-bond propensities that were consistent with the HDXMS results at low osmolality (Wang et al. 2012), but showed higher values at high osmolality, which were inconsistent with the HDXMS measurements (Figure 6). The inconsistency highlights the additional importance of side chain interactions.
DISCUSSION
The first step in HK activation involves helical backbone stabilization
Ensemble behavior is a hallmark of intrinsically disordered proteins (IDPs) (Dunker et al., 2002; Dyson and Wright, 2001). Modulation of a conformational ensemble in response to perturbants via stabilization of secondary structural elements forms the basis of stimulus-response coupling in signaling proteins (Baron and McCammon, 2013; Boehr et al., 2009). This mechanism is especially relevant to EnvZ, a sensor HK that senses changes in osmolality and acid pH (Chakraborty et al., 2015; Chakraborty et al., 2017) and communicates the impact of diverse convergent perturbations as a phosphorylation output signal. This is achieved through step-wise osmolyte-mediated stabilization of an intrinsically disordered osmosensing locus (Figure 3A, Figure 7A and Movie S1)(Wang et al., 2012). The helical backbone is stabilized, and autophosphorylation at His243 is enhanced (Figure 3B, Figure 7B and Movie S2) (Wang et al., 2012). This change is then propagated as a downstream signal through phosphotransfer to the EnvZ cognate RR OmpR, to mediate a cellular response (Response-I, Figure 1B).
Backbone stabilization is concomitant with changes in the RR OmpR binding site
The step-wise activation of an HK through an initial helical backbone stabilization step makes it a highly versatile switch that funnels the effects of diverse stimuli-changes in osmolyte concentration and pH-to divergent signaling outputs, through phosphorylation-dependent or independent modes. In the phosphorylation-dependent mode, osmolyte-stabilization of the His243 locus enhances autophosphorylation, and increases OmpR phosphorylation. Higher levels of OmpR~P result in differential regulation of porin gene expression (Response-I, Figure 1B).
The osmosensing locus with a central, critical His is additionally engineered to respond to acidic stimuli. A decrease in pH also mediates backbone stabilization (Scholtz and Baldwin, 1992) and concomitant changes in the OmpR binding site. This fits with previous experimental observations that OmpR binds to EnvZ with higher affinity compared to OmpR~P (Mattison et al., 2002). The EnvZ-OmpR interaction drives OmpR dimerization, mimicking the activated state (Desai and Kenney, 2017) and inducing a phosphorylation-independent alternative response (Response-II, Figure 1B). This explains how the EnvZ-OmpR TCS system has evolved to additionally enable non-canonical phosphorylation-independent activation of OmpR, even when acid pH does not favor phosphorylation (Chakraborty et al., 2017).
Osmolyte-dependent rotamerization of His243 relieves backbone carbonyl inhibition by Ala239 and promotes autophosphorylation at Nε
Based on our results, we propose a model for His243 autophosphorylation in EnvZ (Figure 7) showing a coordinated network of residues with His243 operating as the integrative node for backbone and side chain interactions to coalesce, coordinating osmosensing and autophosphorylation. A strategic positioning of the phosphorylatable histidine in a locally unfolded locus within the phosphotransfer domain thus defines the basis for the osmosensing switch. Substituting His243 in the EnvZc mutant H243A abolished the focal point of this network and prevented autophosphorylation and osmolyte-mediated stabilization (Wang et al., 2012). The unique role of histidine residues at functional sites of enzymes is attributed to the pKa of ionization of the imidazole side chain, which is close to physiological pH (Li and Hong, 2011; Puttick et al., 2008). At neutral pH, either of the imidazole nitrogen atoms are protonated, allowing the side chain to assume alternate Nδ- or Nε-protonated tautomeric forms (Figure S2) (Hass et al., 2008; Huang et al.,1984; Puttick et al., 2008). The nitrogen atom that is not covalently bonded to hydrogen in either tautomeric form can subject high-energy phosphates such as ATP to nucleophilic attack, resulting in histidine phosphorylation. The predominant tautomeric form of the histidine is highly sensitive to its electrostatic microenvironment and even minimal modifications to the His243 neighborhood have dramatic consequences for its function (Klumpp and Krieglstein, 2002). Although either Nδ or Nε atoms on the His imidazole ring can be phosphorylated, Nε phosphorylation is more common for bacterial HK proteins (Shi et al., 2011; Stock et al., 2000; Zhou and Dahlquist, 1997).
Lower levels of EnvZ autophosphorylation are attributed to the inherent flexibility of the peptide backbone at low osmolality, which fails to position the Nε atom for efficient nucleophilic attack on the γ-phosphate of ATP. However, apart from unfavorable geometry, interactions of the imidazole ring with residues in its immediate neighborhood also contribute to diminished nucleophilic efficiency of Nε at low osmolality (Figure 7A). The Ala239 carbonyl can potentially toggle between mediating H-bonds to either His243 Nδ-H of the imidazole ring or the backbone amide hydrogen. We postulate that the H-bond between Ala239 carbonyl oxygen and Nδ-H imposes a negative inductive effect on the imidazole ring, withdrawing electron density delocalized between the Nδ and Nε atoms, leading to weak nucleophilicity at Nε (Figure 7A). High osmolality switches the Ala239-NδH H-bond to favor the Ala239-His243 H-bond, stabilizing the backbone. A concomitant removal of the negative inductive effect allows the electron density on the imidazole ring and Nε to be readily available for a stronger nucleophilic attack on ATP. This backbone stabilization can also be promoted through substitution with helix-promoting residues (Pace and Scholtz, 1998) such as Thr247 distal to the His243 (Figure S1).
In addition to regulating nucleophilic efficiency at the Nε atom, Ala239 is also an essential determinant of EnvZ autophosphorylation. (Figure 2). Substitution of the highly conserved Ala239 to Thr in the A239T mutant (Figure 2) greatly diminished kinase activity (Figure 4A), but did not affect osmolyte-mediated backbone stabilization. Interestingly, mutations showed osmolality-dependent changes in the kinase subdomain not observed in WT EnvZc suggesting that changes in the microenvironment of His243 in the mutants induced allosteric changes in the kinase subdomain.
The His-Asp/Glu dyad functions as a brake at low osmolality
Substitution of Asp244 in EnvZc leads to high ATP turnover, even at low osmolality (Figure 4C). This result implies that Asp244 has an impeding effect on the nucleophilicity of Nε in WT EnvZc. Thus, the defining motif of HKs is not only a phosphorylatable His residue within an intrinsically disordered locus, but also a His-Asp/Glu motif. This motif is also present in the active sites of hydrolases, including nucleases and phosphodiesterases (Wang et al., 2007). The acidic residue following the phosphorylatable histidine is highly conserved in HKs and was proposed to perform a crucial role in regulating kinase activity (Bhate et al., 2015; Casino et al., 2014), although it was never tested experimentally. Closer examination of the structures of the four-helix bundle subdomain at low (PDB ID: 1JOY) and high osmolality (PDB ID: 4KP4) identifies the backbone carbonyl of Ala239 as within H-bonding distance to Nδ of His243. The two oxygens of the Asp244 carboxyl group are equidistant from the two imidazole nitrogens of His243. The imidazole nitrogen atom that is not H-bonded undergoes autophosphorylation (Klumpp and Krieglstein, 2002). We postulate that autophosphorylation requires free rotamer movement, and Asp244 impedes the free rotamer flipping of His243. Together with the negative inductive effect of Ala239 on His243, these interactions form a ‘double negative clamp’ that weakens the propensity of Nε to facilitate nucleophilic attack of the γ-phosphate of ATP (Figure 7A). High osmolality relieves inhibition of Asp244 and promotes main chain H-bonding between the carbonyl of Ala239 and the backbone amide of His243. The more stable helical conformation reorients the carboxyl side chain of Asp244 to anchor the proximal H-bond on Nδ. This allows a stronger nucleophilic attack on the γ-phosphate of ATP, enhancing autophosphorylation. Evidence for a negative regulatory role for Asp244 comes from the observation that aspartates destabilize helices containing histidine residues (Huyghues-Despointes and Baldwin, 1997). Histidines mediate stable H-bonds/electrostatic interactions with aspartic acids, but preferentially when they are 3–4 residues apart (Huyghues-Despointes and Baldwin, 1997). Asp244 likely stretches the helical backbone to favor its i,i+3 favored geometry of Histidine-Aspartate pairing. High osmolality relieves the inhibitory constraints of Asp244 to promote backbone stabilization.
Coordinated dynamics at the OmpR binding site with the His243 helix
Our HDXMS analysis showed bimodal kinetics of deuterium exchange only at the OmpR binding site, but not at the His243 locus (Wang et al., 2012). The appearance of bimodal kinetics (EX1) indicated a slower timescale of the conformational ensemble at the OmpR binding site (~ milliseconds) because the rates of interconversion of conformations was equivalent to the intrinsic rates of exchange in milliseconds (reviewed in (Hoofnagle et al., 2003) ). MD simulations (100 ns timescale) determined the H-bonding propensity for the A239 carbonyl to the amino group of H243 that closely matched the HDXMS results for WT, A239T, T247R and the A239T/T247R double mutant at low and high osmolality (Figure 6). However, for H243A and D244A mutants the H-bond propensity did not correlate with the HDXMS. The side chain interactions mediated by the His243-Asp244 dyad function to integrate osmolyte-mediated helical stabilization with the OmpR binding site. Since the OmpR binding site dynamics are at slower (msec) timescales, our H-bond propensities did not correlate with HDXMS for these two mutants (Figure 6). This discrepancy provides insight regarding the time scale of the dynamics of the osmosensing locus and the OmpR binding helix, and indicates a central linchpin role of Asp244 in integrating these two events. Asp244 relays changes due to His243 phosphorylation and osmolyte effects on backbone H-bonding to the OmpR binding site. Examination of the structure reveals that the side chain Glu275 from the trans monomer peptide is in close proximity to the critical Asp244 leading to interhelical electrostatic repulsion (Kohn et al., 1995). We predict that Glu275 functions as a critical relay point to communicate changes in the His243-containing helix to the OmpR-binding site. This explains the absence of bimodal spectra in the OmpR-binding site of the D244A mutant (Figure 5) and its constitutive activity, even in the absence of osmolytes. In the bacterial chemoreceptor CheA HK, activity is regulated by reversible methylation, which similarly serves to modulate glutamate electrostatic repulsion (Antommattei and Weis, 2006).
Conclusion
Based on our results, we describe the regulation of His243 autophosphorylation in EnvZ by the following series of events: stimuli (osmolytes or acid pH) stabilize the helical backbone around His243 and promote OmpR binding via changes in the acidic cluster of the OmpR binding locus (SINKDIEE). This step is followed by His243 sidechain interactions that promote relief of Asp244 inhibition and drive EnvZ autophosphorylation. The altered conformation of His243 and the relief of inhibitory contacts with Asp244 promote phosphorylation of His243 and phosphotransfer to OmpR to mediate the effector response. Thus, Asp55, the phosphorylated aspartic acid of OmpR, serves to displace aspartic acid Asp244 in EnvZ to promote phosphoryl transfer. This ‘double-negative clamp’ model also explains how EnvZ can also function as a low pH sensor (Chakraborty et al., 2015; Chakraborty et al., 2017). In this scenario, low pH promotes helical stabilization, enhances OmpR binding and relieves inhibitory contacts with Asp244, without phosphorylation. Low pH also favors protonation of Glu275 and Asp244 to alleviate electrostatic repulsion in the OmpR binding site, promoting OmpR binding without phosphotransfer. Thus, low pH sensory stimuli could be detected by the same His243 locus and relayed without enhanced autophosphorylation (Chakraborty et al., 2017). Based on our previous results (Mattison et al., 2002) and those reported herein, we suggest that EnvZ is already bound to OmpR prior to EnvZ phosphorylation and is thus poised to accept the phosphate from His243 on activated EnvZ~P to the acceptor Asp55 on OmpR. This work provides a mechanistic explanation for how EnvZ sensing of diverse sensory stimuli such as osmolality and acid pH is mediated by the same locus, but employs phosphorylation-dependent and phosphorylation-independent signaling pathways. Such roles for TCS in phosphorylation-independent pathways have only recently been described (Chakraborty et al., 2017; Desai and Kenney, 2017; Desai et al., 2016) and are of enormous significance in understanding the multiplicity of signaling outcomes mediated by TCS. The universality of such phosphorylation-independent conformational switches are akin to pseudokinases or dead kinases in eukaryotic systems (Taylor et al., 2013). Understanding how the output functions are altered as a result of phosphorylation-dependent or independent outputs is the next challenge in understanding two-component signal transduction.
STAR Methods
Contact for Reagent and Resource Sharing
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact: Ganesh S. Anand (dbsgsa@nus.edu.sg)
Experimental Model and Subject Details
The experimental model is the cytoplasmic fragment of EnvZ, EnvZ(Δ1–179) or EnvZc a known histidine kinase from E. coli that is part of the EnvZ-OmpR Two component system responsible for osmosensing in E. coli and other bacterial species. Recombinant E. coli with plasmid pET11a-envZc expressing EnvZc with an N-terminal hexahistidine tag was a kind gift from Dr. Masayori Inouye from the University of Medicine and Dentistry of New Jersey for overexpressing EnvZc and were grown in LB medium (10 g tryptone, 5 g yeast extract and 10 g NaCl per liter) containing a final concentration of 100 μg/ml ampicillin for 1.5–2 h with vigorous aeration at 37 °C. At OD600 ~0.5, 1 mM IPTG was added (final concentration) and the culture was incubated for a further 3 h.
Method Details
• EnvZc mutagenesis
The three EnvZc mutants were generated using inverse PCR method. pET11a-envZc expressing EnvZc with an N-terminal hexahistidine tag was a kind gift from Dr. Masayori Inouye from the University of Medicine and Dentistry of New Jersey and used as the template. Double mutation (A239T/T247R) was achieved by using the EnvZc A239T mutant plasmid (pET11a-envZc (A239T)) generated in this study as the template. Amplification for all three mutants was achieved using Phusion® Hot Start II DNA polymerase (Thermo Fisher Scientific, Lafayette, CO). Mutagenesis was confirmed by DNA sequencing.
• Expression and purification of EnvZc mutants
Expression and purification of the EnvZc mutants follow the same protocol as described previously with some modifications (Skarphol et al., 1997). Briefly, the plasmids containing the mutant genes were used to transform E. coli BL21 (DE3) competent cells for protein overexpression. The cells were grown in LB medium (10 g tryptone, 5 g yeast extract and 10 g NaCl per liter) containing a final concentration of 100 μg/ml ampicillin for 1.5–2 h with vigorous aeration at 37 °C. At OD600 ~0.5, 1 mM IPTG was added (final concentration) and the culture was incubated for a further 3 h. Induced cells were harvested at 7,500 × g for 15 min and stored at −20 °C until purification. The frozen pellet was resuspended in ice-cold lysis buffer (20 mM Tris-HCl, pH 7.6, 10 μM EDTA, 5% (v/v) glycerol) complemented with protease inhibitor cocktail and lysed by sonication (1 s pulse every 2 s for 5 min). The lysate was centrifuged at 17,600 × g for 30 min at 4 °C to remove non-soluble debris. The supernatant was then incubated with TALON® cobalt affinity resin for at least 30 min at 4 °C before being placed into an empty chromatography column. The resin was washed extensively with two bed volumes of lysis buffer followed by lysis buffer containing 5 mM and 10 mM imidazole (twice per buffer) to remove non-specific proteins. EnvZc was finally eluted with lysis buffer containing 250 mM imidazole. The eluate was further purified using HiLoad 16/60 Superdex™ 75 gel filtration column with imidazole-free 20 mM Tris-HCl, pH 7.6 on an AKTA™ FPLC system (GE Healthcare, Chicago, IL). Protein purity was determined from the fractions with the highest concentration using 15% denaturing SDS-PAGE.
• EnvZc [γ−32P]-phosphorylation assay
The phosphorylation assay was carried out as described (Wang et al., 2012). Briefly, each sample (WT and the mutants) was phosphorylated in a buffer containing 50 mM KCl and 50 mM MgCl2 with EnvZc at a final concentration of 4 μM. The reactions were initiated by the addition of 2 μCi of [γ−32P]-ATP followed by incubation at various times at room temperature. Reactions were terminated by addition of 3 μl of denaturing solution (124 mM Tris-HCl, pH 6.8, 20% (v/v) glycerol, 4% (w/v) SDS, 8% (v/v) β-mercaptoethanol, and 0.025% (w/v) bromophenol blue) and 10 μl of the 20 μl reaction was separated by 15% SDS-PAGE. The gel was dried for 1 h and exposed to a phosphoimager screen and visualized on a Molecular Dynamics Storm 860 imager (GMI, Inc., Ramsey, MN). The osmolality of the reaction medium was altered by adjusting the salt concentration and measured using a Wescor Vapor Pressure Osmometer. The autophosphorylation assay was only carried out under low osmolality condition to measure basal level activity. The low osmolality buffer contained mM Tris-HCl, 12.5 mM KCl, 17 mM NaCl, and 10 mM MgCl2.
• β-galactosidase assay
Using the protocol as described previously (Mattison et al., 2002), plasmid carrying the mutant genes was transformed into E. coli MH225 and PG189 strains. E. coli MH225 contains a chromosomal ompC-lacZ fusion while PG189 is the isogenic envZ-null (ΔenvZ) strain. β-galactosidase assays were performed in triplicate with at least three independent cultures as described previously to monitor the transcription of ompC-lacZ (Mattison et al., 2002). Briefly, an overnight culture of bacteria was diluted 1:100 and cells were grown to mid-exponential phase (OD600 ~0.4) in minimal A medium at varying osmolality. Measurement of OmpC-LacZ levels was performed by adding one volume of cells (0.1 ml) to nine volumes of the reaction buffer (0.9 ml) (60 mM Na2HPO4, 40 mM NaH2PO4, pH 7.0, 10 mM KCl, 1 mM MgSO4, 2.7 μl/ml β-mercaptoethanol) before being disrupted by addition of 0.1% (w/v) SDS and chloroform and incubated with 0.2 ml of 4 mg/ml ortho-nitrophenyl-β-galactoside (ONPG). β-galactosidase activity was expressed in Miller units and calculated using the formula below:
• Kinase autophosphorylation assay using ADP-Glo® assay
Autophosphorylation by EnvZ was measured using the in vitro ADP-Glo™ Kinase assay (Promega, Madison, WI), which quantifies the amount of ADP formed as a product in a kinase reaction using a luminescence-based detection method (Zegzouti et al., 2009). All reactions were carried out following manufacturer’s recommendations and were performed at least in duplicates. Kinase reactions were performed for the following EnvZc samples: (1) Wild-type (WT) (2) D244A (3) A239T and (4) T247R under both low and high osmolality conditions. The protein samples were diluted to a final concentration of 5 μM EnvZ in the low osmolality buffer containing 20 mM Tris-HCl, 2 mM MgCl2, 100 μM ATP, pH 7.6 to a final volume of 10 μL. The high osmolality buffer additionally contained a final concentration of 20% sucrose. Luminescence recordings were taken in a Glomax® Discover Multimode Detection System (Promega, Madison, WI). Amount of ADP produced was calculated based on standard curves constructed according to the manufacturer’s recommendations. Enzyme activity is represented as micromoles of ADP produced per milligrams of enzyme per minute (Figure 4C).
• Amide HDXMS
Amide HDXMS on EnvZc mutants follows the same protocol as described previously (Wang et al., 2012). To summarize, EnvZc mutants were concentrated using a Vivaspin ultrafiltration centrifugal device (Sartorius Stedim Biotech GmbH, Göttingen, Germany) to at least 50 μM, as measured by Bradford assay (Bradford, 1976). Filter-sterilized Tris buffer (20 mM Tris-HCl, pH 7.6) was the low osmolality buffer. High osmolality solution contained 20% (w/v) sucrose. Water was removed from all three solutions using a centrifugal vacuum concentrator and replaced with 99.9% D2O prior to the experiment. Amide HDX was carried out by incubating 2 μl of protein with 18 μl of perdeuterated buffer at 20 °C for various times (1, 2, 5, 10 and 30 min), yielding a final concentration of 90% D2O. The undeuterated sample was included as a negative control. The reaction mixture was quenched by the addition of 30 μl of pre-chilled 0.1% (v/v) TFA solution that lowered the pH of the solution to 2.5. This serves to minimize exchange-in and back-exchange of the amides with the solvent (Englander and Poulsen, 1969). The quenched sample (50 μl) was injected into a chilled nanoACQUITY UltraPerformance LC® (UPLC) system (Waters Corporation, Milford, MA) as previously described (Wales et al., 2008). In the UPLC system, the sample was trapped and digested with a 2.1 × 30 mm2 Poroszyme® immobilized pepsin column (Applied Biosystems, Foster City, CA) supplied with 0.1% (v/v) formic acid in water at a flow rate of 100 μl/min. Peptides were eluted using an organic solvent gradient of 8–40% (v/v) acetonitrile in 0.1% (v/v) formic acid at 40 μl/min and resolved with a 1.0 × 100 mm2 ACQUITY UPLC BEH C18 reversed-phase column (Waters Corporation, Milford, MA). Both the immobilized pepsin cartridge and C18 reversed-phase column were housed in a refrigerated HDX manager module maintained at 0 °C to minimize deuterium back-exchange during analysis. Peptide signals were detected and their masses were measured using a SYNAPT® High Definition Mass Spectrometer™ (HDMS™) G2-Si (Waters Corporation, Milford, MA). The mass spectrometer was continuously calibrated with 200 fmol/μl of [Glu1]-fibrinogen peptide B ([Glu1]-Fib) as a standard at a flow rate of 5 μl/min. All data were collected using MSE data acquisition mode.
Peptides were first identified from MSE data of undeuterated samples using ProteinLynx Global Server software (PLGS v3) (Waters Corporation, Milford, MA) (Geromanos et al., 2009; Li et al., 2009) by searching the peptides against a database containing the EnvZc primary sequence (with specific residues altered to corresponding mutation) cleaved by a non-specific protease. HDXMS analysis was carried out using DynamX (v3.0) software (Waters Corporation, Milford, MA) with cutoffs of minimum intensity of 1000, a tolerance of 10 ppm (parts per million) from the theoretical mass-to-charge (m/z) and a maximum peptide length of 25 residues to generate DynamX files for each mutant. Peptides with non-overlapping and favorable signal-to-noise ratio spectra were identified on the software by visual inspection and subjected to quantitative analysis. In a second step, a list of 49 peptides generated by pepsin cleavage of EnvZc A239T outside the His243 and mutation sites – Ala239, Asp244 and Thr247 locus, was used as a master list to visually scan the DynamX output files of T247R and A239T/T247R to identify additional peptides with optimal signal to noise for analysis. That generated a total of 53 pepsin fragment peptides representing 84.0 % primary sequence coverage of EnvZc A239T (Figure S4); 44 peptides representing 86.3% coverage of EnvZc T247R (Figure S5); 50 peptides representing 82.0% coverage in EnvZc A239T/T247R (Figure S6). A greater number of peptides with optimal signal intensities (83 peptides) were obtained for EnvZc D244A representing 93% coverage (Figure S7). 31 peptides analyzed were common across all EnvZc mutants examined in this study (Table 1). The software generated a centroid value for the isotopic envelope of each peptide, which reflected the average mass of the peptide. The difference in the average masses of the undeuterated and experimental peptide represented the average number of deuterons exchanged. A difference of half a deuteron (0.5 D) exchanged represents the significance threshold in HDXMS experiments (Houde et al., 2011). Although the N-terminus primary amine for each peptide could also undergo HDX, the reaction was too rapid to measure and was thus excluded from the calculation (Zhang and Smith, 1993). As regions sensitive to osmotic perturbations in EnvZc have been identified in the previous study (Wang et al., 2012), the analyses were focused on these regions directly and the results are summarized in Figure 5 and Table 1.
• Molecular dynamics simulations
The osmolality-sensing domain of EnvZ, a homodimeric bundle consisting of residues 223–289 was extracted from high- and low-osmolality structures of EnvZ in the PDB. From our previous study, titration of NaCl or sucrose elicited a differential response at ~500–600 mOsm/kg (Wang et al., 2012). Thus, the NMR condition is considered to be of low osmolality because the solution contained low amounts of excipients(Tomomori et al., 1999), giving an estimated osmolality of <250 mOsm/kg. A crystallographic structure of EnvZc (PDB ID: 4KP4) was solved after crystallization through vapor diffusion (10 mg/ml EnvZ, 4 mM AMPPNP and 4 mM MgCl2 in a reservoir solution (2%PEG1000, 2–6% PEG4000, 1–1.7 M ammonium sulfate, 0.1 M Hepes pH 7.5 and 2—40 mM sodium acetate). Crystals were further cryoprotected in mother liquor with increased concentrations of polyethylene glycol (4%, PEG1000 and 12%, PEG4000), 22% sucrose and decreased ammonium sulfate (0.85M) concentration (Casino et al. 2014). This structure thus represents a snapshot of the high osmolyte-stabilized conformation. A second crystal structure of a chimera of E. coli EnvZ fused to the HAMP domain of A. fulgidus Af1503 receptor (PDB ID: 4CTI) was solved in 0.1 MMT buffer supplemented with 25% (w/v) polyethylene glycol (PEG)-1500 (Ferris et al., 2014). 25% (w/v) PEG-1500 (~0.17 M) alone corresponds to ~1.5 MPa in osmotic pressure based on measurements by Money (1989) for different PEG molecules, which translates to ~600 mOsm/kg, a high osmolality condition. Low-osmolality models were constructed using 1JOY (Tomomori et al., 1999) as a template, while high-osmolality structures were modeled on 4CTI (Ferris et al., 2014). Besides the wild-type EnvZ structure, the mutants A239T, T247R, D244A and the double mutant A239T/T247R were constructed using the mutagenesis feature in PyMol. Subsequently, molecular interactions were modeled using the CHARMM36 force field (Huang and MacKerell, 2013). All systems were solvated in TIP3P water to a final cubic box size of approximately 9 nm edge length (Jorgensen et al., 1983). Low osmolality systems were neutralized by adding sodium ions. High osmolality systems were prepared with sodium and chloride ions to a concentration of 0.5 M. All systems were minimized using GROMACS 5.1.46 followed by 1 ns of NpT equilibration at 1.0 bar and 300 K while keeping the protein part restrained by a harmonic potential of 1000 kJ nm−2 (Berendsen et al., 1995). Temperature was held constant using a velocity rescale algorithm while pressure was maintained by a Parrinello-Rahman barostat (Bussi et al., 2007) (Parrinello and Rahman, 1980). Two replicate simulations of each system were performed to a total length of 100 ns each. All systems were analyzed using the ‘gmx hbond’ utility included in GROMACS. Additionally, we conducted two simulations where we used the high-osmolality conformation in a low-osmolality solvent and the low-osmolality starting state in a high-osmolality solvent in order to stimulate the conformational transition between the two states.
Quantification and Statistical Analysis
HDXMS analysis was carried out using DynamX (v3.0) software (Waters Corporation, Milford, MA) with cutoffs of minimum intensity of 1000, a tolerance of 10 ppm (parts per million) from the theoretical mass-to-charge (m/z) and a maximum peptide length of 25 residues to generate DynamX files for each mutant. Peptides with non-overlapping and favorable signal-to-noise ratio spectra were identified on the software by visual inspection and subjected to quantitative analysis. Values are reported as mean ± standard deviation from at least two (n ≥ 2) independent experiments.
β-galactosidase assays were performed in triplicate with at least three independent cultures (n ≥ 3) as described previously to monitor the transcription of ompC-lacZ.
For the MD simulations, two replicate (n = 2) simulations of each system were performed to a total length of 100 ns each.
Data and Software Availability
Pepsin fragment peptides used in the analysis were first identified from MSE data of undeuterated samples using ProteinLynx Global Server software (PLGS v3) (Waters Corporation, Milford, MA) (Geromanos et al., 2009; Li et al., 2009) by searching the peptides against a database containing the EnvZc primary sequence (with specific residues altered to corresponding mutation) cleaved by a non-specific protease.
HDXMS analysis was carried out using DynamX (v3.0) software (Waters Corporation, Milford, MA).
Structures were examined by Pymol software (Schrodinger, USA). Molecular interactions were modeled using CHARMM36 force field and minimized using GROMACS 5.1.46.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| Escherichia coli BL21 (DE3) Competent cells | Novagen/Merck KGaA | Cat# 69449–3CN |
| Escherichia coli MH225 strain | Mattison et. al. (2002) | N/A |
| Escherichia coli PG189 strain | Mattison et. al. (2002) | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| TALON® metal affinity resin | ClonTech/Takara Bio | Cat# 635504 |
| Isopropyl β-d-1-thiogalactopyranoside (IPTG) | Bio Basic Inc. | Cat# IB0168 |
| cOmplete™, EDTA-free Protease Inhibitor Cocktail | Roche/Merck KGaA | Cat# 11873580001 |
| Trifluoroacetic Acid for HPLC | Sigma/Merck KGaA | Cat#302031 |
| Water, HPLC grade | Sigma/Merck KGaA | Cat#270733 |
| Acetonitrile, HPLC grade | Sigma/Merck KGaA | CAS# 75–05-8 |
| Sucrose, UltraPure Grade | 1st Base | Cat# BIO-1090–500g |
| Critical Commercial Assays | ||
| ADP-Glo™ Kinase assay | Promega Corporation | Cat# V6930 |
| Recombinant DNA | ||
| pET11a plasmid | Novagen/Merck | Cat# 69436 |
| Software and Algorithms | ||
| ProteinLynxGlobalServer (PLGS 3.0.1) | Waters Corporation | http://www.waters.com/waters/en_SG/ProteinLynx-Global-SERVER-%28PLGS%29/nav.htm?cid=513821&locale=en_SG |
| DynamX v3.0 | Waters Corporation | N/A |
| CHARMM36 | Huang and MacKerell, 2013 | N/A |
| GROMACS | Berendsen et al., 1995 | N/A |
Highlights:
Stabilization of disordered backbone helix promotes histidine autophosphorylation
His-Asp dyad acts as integrative node for backbone and side chain interactions
This ‘double-clamp’ switch allows dual cellular pH and osmolality sensing
ACKNOWLEDGEMENTS
This work was supported by grants from the Singapore Ministry of Education Academic Research Fund Tier 3 to G.S.A. (MOE2012-T3–1-008), the Research Centre of Excellence in Mechanobiology at the National University of Singapore, funded by the Ministry of Education), Singapore, and, Veterans Affairs Grant IOBX-000372 and NIH AI-123640 to LJK. We additionally wish to thank both reviewers of this manuscript for their insightful feedback.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
DECLARATION OF INTEREST
The authors report no conflict of interest with the findings reported in this manuscript.
REFERENCES
- Antommattei FM, and Weis RM (2006). 12 Reversible methylation of glutamate residues in the receptor proteins of bacterial sensory systems. Enzymes 24, 325–382. [DOI] [PubMed] [Google Scholar]
- Baron R, and McCammon JA (2013). Molecular recognition and ligand association. Annual Review of Physical Chemistry 64, 151–175. [DOI] [PubMed] [Google Scholar]
- Beier D, and Gross R (2006). Regulation of bacterial virulence by two-component systems. Curr Opin Microbiol 9, 143–152. [DOI] [PubMed] [Google Scholar]
- Berendsen HJC, van der Spoel D, and van Drunen R (1995). GROMACS: A message-passing parallel molecular dynamics implementation. Computer Physics Communications 91, 43–56. [Google Scholar]
- Bhate MP, Molnar KS, Goulian M, and DeGrado WF (2015). Signal transduction in histidine kinases: insights from new structures. Structure 23, 981–994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Birktoft JJ, and Banaszak LJ (1983). The presence of a histidine-aspartic acid pair in the active site of 2-hydroxyacid dehydrogenases. X-ray refinement of cytoplasmic malate dehydrogenase. J Biol Chem 258, 472–482. [DOI] [PubMed] [Google Scholar]
- Boehr DD, Nussinov R, and Wright PE (2009). The role of dynamic conformational ensembles in biomolecular recognition. Nat Chem Biol 5, 789–796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradford MM (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Analytical Biochemistry 72, 248–254. [DOI] [PubMed] [Google Scholar]
- Bussi G, Donadio D, and Parrinello M (2007). Canonical sampling through velocity rescaling. The Journal of Chemical Physics 126, 014101. [DOI] [PubMed] [Google Scholar]
- Casino P, Miguel-Romero L, and Marina A (2014). Visualizing autophosphorylation in histidine kinases. Nat Commun 5, 3258. [DOI] [PubMed] [Google Scholar]
- Chakraborty S, Mizusaki H, and Kenney LJ (2015). A FRET-based DNA biosensor tracks OmpR-dependent acidification of Salmonella during macrophage infection. PLoS Biol 13, e1002116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakraborty S, Winardhi RS, Morgan LK, Yan J, and Kenney LJ (2017). Non-canonical activation of OmpR drives acid and osmotic stress responses in single bacterial cells. Nat Commun 8, 1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chervitz SA, and Falke JJ (1996). Molecular mechanism of transmembrane signaling by the aspartate receptor: a model. Proc Natl Acad Sci U S A 93, 2545–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Csonka LN, and Hanson AD (1991). Prokaryotic osmoregulation: genetics and physiology. Annual Review of Microbiology 45, 569–606. [DOI] [PubMed] [Google Scholar]
- daCosta CJ, Medaglia SA, Lavigne N, Wang S, Carswell CL, and Baenziger JE (2009). Anionic lipids allosterically modulate multiple nicotinic acetylcholine receptor conformational equilibria. J Biol Chem 284, 33841–33849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado J, Forst S, Harlocker S, and Inouye M (1993). Identification of a phosphorylation site and functional analysis of conserved aspartic acid residues of OmpR, a transcriptional activator for ompF and ompC in Escherichia coli. Mol Microbiol 10, 1037–1047. [DOI] [PubMed] [Google Scholar]
- Desai SK, and Kenney LJ (2017). To approximately P or Not to approximately P? Non-canonical activation by two-component response regulators. Mol Microbiol 103, 203–213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desai SK, Winardhi RS, Periasamy S, Dykas MM, Jie Y, and Kenney LJ (2016). The horizontally-acquired response regulator SsrB drives a Salmonella lifestyle switch by relieving biofilm silencing. Elife 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dowhan W (1997). Molecular basis for membrane phospholipid diversity: why are there so many lipids? Annu Rev Biochem 66, 199–232. [DOI] [PubMed] [Google Scholar]
- Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, and Obradovic Z (2002). Intrinsic disorder and protein function. Biochemistry 41, 6573–6582. [DOI] [PubMed] [Google Scholar]
- Dyson HJ, and Wright PE (2001). Nuclear magnetic resonance methods for elucidation of structure and dynamics in disordered states. Methods Enzymol 339, 258–270. [DOI] [PubMed] [Google Scholar]
- Englander SW, and Poulsen A (1969). Hydrogen-tritium exchange of the random polypeptide chain. Biopolymers 7, 379–393. [Google Scholar]
- Ferris HU, Coles M, Lupas AN, and Hartmann MD (2014). Crystallographic snapshot of the Escherichia coli EnvZ histidine kinase in an active conformation. Journal of Structural Biology 186, 376–379. [DOI] [PubMed] [Google Scholar]
- Forst S, Comeau D, Norioka S, and Inouye M (1987). Localization and membrane topology of EnvZ, a protein involved in osmoregulation of OmpF and OmpC in Escherichia coli. J Biol Chem 262, 16433–16438. [PubMed] [Google Scholar]
- Forst SA, and Roberts DL (1994). Signal transduction by the EnvZ-OmpR phosphotransfer system in bacteria. Res Microbiol 145, 363–373. [DOI] [PubMed] [Google Scholar]
- Freeman ZN, Dorus S, and Waterfield NR (2013). The KdpD/KdpE two-component system: integrating K(+) homeostasis and virulence. PLoS Pathog 9, e1003201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geromanos SJ, Vissers JP, Silva JC, Dorschel CA, Li GZ, Gorenstein MV, Bateman RH, and Langridge JI (2009). The detection, correlation, and comparison of peptide precursor and product ions from data independent LC-MS with data dependant LC-MS/MS. Proteomics 9, 1683–1695. [DOI] [PubMed] [Google Scholar]
- Ghosh M, Wang LC, Ramesh R, Morgan LK, Kenney LJ, and Anand GS (2017). Lipid-Mediated Regulation of Embedded Receptor Kinases via Parallel Allosteric Relays. Biophys J 112, 643–654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goulian M (2010). Two-component signaling circuit structure and properties. Current Opinion in Microbiology 13, 184–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grebe TW, and Stock JB (1999). The histidine protein kinase superfamily. Adv Microb Physiol 41, 139–227. [DOI] [PubMed] [Google Scholar]
- Gushchin I, Melnikov I, Polovinkin V, Ishchenko A, Yuzhakova A, Buslaev P, Bourenkov G, Grudinin S, Round E, Balandin T, et al. (2017). Mechanism of transmembrane signaling by sensor histidine kinases. Science 356. [DOI] [PubMed] [Google Scholar]
- Hall BA, Armitage JP, and Sansom MS (2011). Transmembrane helix dynamics of bacterial chemoreceptors supports a piston model of signalling. PLoS Comput Biol 7, e1002204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hass MA, Hansen DF, Christensen HE, Led JJ, and Kay LE (2008). Characterization of conformational exchange of a histidine side chain: protonation, rotamerization, and tautomerization of His61 in plastocyanin from Anabaena variabilis. J Am Chem Soc 130, 8460–8470. [DOI] [PubMed] [Google Scholar]
- Heermann R, and Jung K (2010). The complexity of the ‘simple’ two-component system KdpD/KdpE in Escherichia coli. FEMS Microbiol Lett 304, 97–106. [DOI] [PubMed] [Google Scholar]
- Hoch JA (2000). Two-component and phosphorelay signal transduction. Curr Opin Microbiol 3, 165–170. [DOI] [PubMed] [Google Scholar]
- Hoofnagle AN, Resing KA, and Ahn NG (2003). Protein analysis by hydrogen exchange mass spectrometry. Annual Review of Biophysics and Biomolecular Structure 32, 1–25. [DOI] [PubMed] [Google Scholar]
- Houde D, Berkowitz SA, and Engen JR (2011). The utility of hydrogen/deuterium exchange mass spectrometry in biopharmaceutical comparability studies. Journal of Pharmaceutical Sciences 100, 2071–2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J, and MacKerell AD Jr. (2013). CHARMM36 all-atom additive protein force field: validation based on comparison to NMR data. J Comput Chem 34, 2135–2145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang TH, Bachovchin WW, Griffin RG, and Dobson CM (1984). High-resolution nitrogen-15 nuclear magnetic resonance studies of alpha-lytic protease in solid state. Direct comparison of enzyme structure in solution and in the solid state. Biochemistry 23, 5933–5937. [DOI] [PubMed] [Google Scholar]
- Hulko M, Berndt F, Gruber M, Linder JU, Truffault V, Schultz A, Martin J, Schultz JE, Lupas AN, and Coles M (2006). The HAMP domain structure implies helix rotation in transmembrane signaling. Cell 126, 929–940. [DOI] [PubMed] [Google Scholar]
- Huyghues-Despointes BM, and Baldwin RL (1997). Ion-pair and charged hydrogen-bond interactions between histidine and aspartate in a peptide helix. Biochemistry 36, 1965–1970. [DOI] [PubMed] [Google Scholar]
- Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, and Klein ML (1983). Comparison of simple potential functions for simulating liquid water. The Journal of Chemical Physics 79, 926–935. [Google Scholar]
- Kenney LJ, Bauer MD, and Silhavy TJ (1995). Phosphorylation-dependent conformational changes in OmpR, an osmoregulatory DNA-binding protein of Escherichia coli. Proc Natl Acad Sci U S A 92, 8866–8870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitanovic S, Ames P, and Parkinson JS (2011). Mutational analysis of the control cable that mediates transmembrane signaling in the Escherichia coli serine chemoreceptor. J Bacteriol 193, 5062–5072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klumpp S, and Krieglstein J (2002). Phosphorylation and dephosphorylation of histidine residues in proteins. European Journal of Biochemistry 269, 1067–1071. [DOI] [PubMed] [Google Scholar]
- Kohn WD, Monera OD, Kay CM, and Hodges RS (1995). The effects of interhelical electrostatic repulsions between glutamic acid residues in controlling the dimerization and stability of two-stranded alpha-helical coiled-coils. J Biol Chem 270, 25495–25506. [DOI] [PubMed] [Google Scholar]
- Krell T, Lacal J, Busch A, Silva-Jimenez H, Guazzaroni ME, and Ramos JL (2010). Bacterial sensor kinases: diversity in the recognition of environmental signals. Annu Rev Microbiol 64, 539–559. [DOI] [PubMed] [Google Scholar]
- Lee AG (2003). Lipid-protein interactions in biological membranes: a structural perspective. Biochim Biophys Acta 1612, 1–40. [DOI] [PubMed] [Google Scholar]
- Lee AG (2004). How lipids affect the activities of integral membrane proteins. Biochim Biophys Acta 1666, 62–87. [DOI] [PubMed] [Google Scholar]
- Leonard TA, and Hurley JH (2011). Regulation of protein kinases by lipids. Curr Opin Struct Biol 21, 785–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li GZ, Vissers JP, Silva JC, Golick D, Gorenstein MV, and Geromanos SJ (2009). Database searching and accounting of multiplexed precursor and product ion spectra from the data independent analysis of simple and complex peptide mixtures. Proteomics 9, 1696–1719. [DOI] [PubMed] [Google Scholar]
- Li S, and Hong M (2011). Protonation, tautomerization, and rotameric structure of histidine: a comprehensive study by magic-angle-spinning solid-state NMR. J Am Chem Soc 133, 1534–1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuyama S, Mizuno T, and Mizushima S (1986). Interaction between two regulatory proteins in osmoregulatory expression of ompF and ompC genes in Escherichia coli: a novel ompR mutation suppresses pleiotropic defects caused by an envZ mutation. Journal of Bacteriology 168, 1309–1314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mattison K, Oropeza R, Byers N, and Kenney LJ (2002). A phosphorylation site mutant of OmpR reveals different binding conformations at ompF and ompC. J Mol Biol 315, 497–511. [DOI] [PubMed] [Google Scholar]
- Money NP (1989). Osmotic Pressure of Aqueous Polyethylene Glycols : Relationship between Molecular Weight and Vapor Pressure Deficit. Plant Physiol 91, 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ottemann KM, Xiao W, Shin YK, and Koshland DE Jr. (1999). A piston model for transmembrane signaling of the aspartate receptor. Science 285, 1751–1754. [DOI] [PubMed] [Google Scholar]
- Pace CN, and Scholtz JM (1998). A helix propensity scale based on experimental studies of peptides and proteins. Biophys J 75, 422–427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrinello M, and Rahman A (1980). Crystal Structure and Pair Potentials: A Molecular-Dynamics Study. Phys Rev Lett 45, 1196–1199. [Google Scholar]
- Phillips R, Ursell T, Wiggins P, and Sens P (2009). Emerging roles for lipids in shaping membrane-protein function. Nature 459, 379–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Puttick J, Baker EN, and Delbaere LT (2008). Histidine phosphorylation in biological systems. Biochim Biophys Acta 1784, 100–105. [DOI] [PubMed] [Google Scholar]
- Russo FD, and Silhavy TJ (1991). EnvZ controls the concentration of phosphorylated OmpR to mediate osmoregulation of the porin genes. J Mol Biol 222, 567–580. [DOI] [PubMed] [Google Scholar]
- Scholtz JM, and Baldwin RL (1992). The mechanism of alpha-helix formation by peptides. Annu Rev Biophys Biomol Struct 21, 95–118. [DOI] [PubMed] [Google Scholar]
- Shi T, Lu Y, Liu X, Chen Y, Jiang H, and Zhang J (2011). Mechanism for the autophosphorylation of CheA histidine kinase: QM/MM calculations. J Phys Chem B 115, 11895–11901. [DOI] [PubMed] [Google Scholar]
- Skarphol K, Waukau J, and Forst SA (1997). Role of His243 in the phosphatase activity of EnvZ in Escherichia coli. Journal of Bacteriology 179, 1413–1416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleator RD, and Hill C (2002). Bacterial osmoadaptation: the role of osmolytes in bacterial stress and virulence. FEMS Microbiol Rev 26, 49–71. [DOI] [PubMed] [Google Scholar]
- Stock AM, Robinson VL, and Goudreau PN (2000). Two-component signal transduction. Annu Rev Biochem 69, 183–215. [DOI] [PubMed] [Google Scholar]
- Taylor SS, Shaw A, Hu J, Meharena HS, and Kornev A (2013). Pseudokinases from a structural perspective. Biochem Soc Trans 41, 981–986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomomori C, Tanaka T, Dutta R, Park H, Saha SK, Zhu Y, Ishima R, Liu D, Tong KI, Kurokawa H, et al. (1999). Solution structure of the homodimeric core domain of Escherichia coli histidine kinase EnvZ. Nat Struct Biol 6, 729–734. [DOI] [PubMed] [Google Scholar]
- Tsaloglou MN, Attard GS, and Dymond MK (2011). The effect of lipids on the enzymatic activity of 6-phosphofructo-1-kinase from B. stearothermophilus. Chem Phys Lipids 164, 713–721. [DOI] [PubMed] [Google Scholar]
- Wales TE, Fadgen KE, Gerhardt GC, and Engen JR (2008). High-speed and high-resolution UPLC separation at zero degrees Celsius. Analytical Chemistry 80, 6815–6820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Robinson H, and Ke H (2007). The molecular basis for different recognition of substrates by phosphodiesterase families 4 and 10. J Mol Biol 371, 302–307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang LC, Morgan LK, Godakumbura P, Kenney LJ, and Anand GS (2012). The inner membrane histidine kinase EnvZ senses osmolality via helix-coil transitions in the cytoplasm. EMBO Journal 31, 2648–2659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zegzouti H, Zdanovskaia M, Hsiao K, and Goueli SA (2009). ADP-Glo: A Bioluminescent and homogeneous ADP monitoring assay for kinases. Assay Drug Dev Technol 7, 560–572. [DOI] [PubMed] [Google Scholar]
- Zhang Z, and Smith DL (1993). Determination of amide hydrogen exchange by mass spectrometry: a new tool for protein structure elucidation. Protein Science 2, 522–531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, and Dahlquist FW (1997). Phosphotransfer site of the chemotaxis-specific protein kinase CheA as revealed by NMR. Biochemistry 36, 699–710. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Pepsin fragment peptides used in the analysis were first identified from MSE data of undeuterated samples using ProteinLynx Global Server software (PLGS v3) (Waters Corporation, Milford, MA) (Geromanos et al., 2009; Li et al., 2009) by searching the peptides against a database containing the EnvZc primary sequence (with specific residues altered to corresponding mutation) cleaved by a non-specific protease.
HDXMS analysis was carried out using DynamX (v3.0) software (Waters Corporation, Milford, MA).
Structures were examined by Pymol software (Schrodinger, USA). Molecular interactions were modeled using CHARMM36 force field and minimized using GROMACS 5.1.46.


