Significance
Mutants of RAS are major oncogenes and occur in many human cancers, but efforts to develop drugs that directly inhibit the corresponding constitutively active RAS proteins have failed so far. We therefore focused on SOS1, the guanine nucleotide exchange factor (GEF) and activator of RAS. A combination of high-throughput and fragment screening resulted in the identification of nanomolar SOS1 inhibitors, which effectively down-regulate active RAS in tumor cells. In cells with wild-type KRAS, we observed complete inhibition of the RAS-RAF-MEK-ERK pathway. In a mutant KRAS cell line, SOS1 inhibition resulted in a reduction of phospho-ERK activity by 50%. Together, the data indicate that inhibition of GEFs may represent a viable approach for targeting RAS-driven tumors.
Keywords: RAS, SOS, fragment screen, crystal structure, small-molecule inhibitor
Abstract
Since the late 1980s, mutations in the RAS genes have been recognized as major oncogenes with a high occurrence rate in human cancers. Such mutations reduce the ability of the small GTPase RAS to hydrolyze GTP, keeping this molecular switch in a constitutively active GTP-bound form that drives, unchecked, oncogenic downstream signaling. One strategy to reduce the levels of active RAS is to target guanine nucleotide exchange factors, which allow RAS to cycle from the inactive GDP-bound state to the active GTP-bound form. Here, we describe the identification of potent and cell-active small-molecule inhibitors which efficiently disrupt the interaction between KRAS and its exchange factor SOS1, a mode of action confirmed by a series of biophysical techniques. The binding sites, mode of action, and selectivity were elucidated using crystal structures of KRASG12C–SOS1, SOS1, and SOS2. By preventing formation of the KRAS–SOS1 complex, these inhibitors block reloading of KRAS with GTP, leading to antiproliferative activity. The final compound 23 (BAY-293) selectively inhibits the KRAS–SOS1 interaction with an IC50 of 21 nM and is a valuable chemical probe for future investigations.
First linked to human cancer in 1982 (1–3), members of the RAS family of GTPases (which comprises KRAS, NRAS, and HRAS) have since been recognized as major oncogenes, occurring in up to 20 to 30% of human cancers (4–6). RAS proteins act as molecular switches that cycle between an active, GTP-bound state and an inactive, GDP-bound state. Activated by guanine nucleotide exchange factors (GEFs), RAS in its GTP-bound state interacts with a number of effectors. Return to the inactive state is driven by GTPase-activating proteins (GAPs), which down-regulate active RAS by accelerating the weak intrinsic GTPase activity by up to 5 orders of magnitude. For oncogenic RAS mutants, however, the GAP activity is impaired or greatly reduced, resulting in permanent activation, which is the basis of oncogenic RAS signaling (7); for example, through the RAS-RAF-MEK-ERK and RAS-PI3K-PDK1-AKT pathways, both essential to cell survival and proliferation (8). Direct inhibition of RAS has proved extremely challenging due to the picomolar affinity of GTP for its binding site, the lack of other well-defined pockets, and the interaction of RAS with GEFs, GAPs, and effectors via extended and flat protein–protein interaction surfaces that are difficult to drug by small molecules. Additionally, attempts to inhibit RAS indirectly by targeting farnesyl transferases have not yet yielded approved drugs (9). Based on the failure of all direct and indirect approaches so far, RAS has been generally considered undruggable. Recent strategies to directly inhibit RAS have focused on (i) targeting Cys12 of the oncogenic mutant KRASG12C with covalent inhibitors, (ii) RAS–effector interactions to disrupt downstream signaling, or (iii) inhibiting the RAS–GEF interactions to prevent reloading with GTP (10). While the first two strategies have seen recent encouraging successes (11–14), targeting the RAS–GEF interactions has not yet generated potent inhibitors. Furthermore, whether mutant RAS proteins require GEF activity for full activation remains to be fully explored and may differ depending on the specific mutation (15). The most-studied GEF for RAS is the protein Son of Sevenless (SOS) for which two human isoforms, SOS1 and SOS2, are known (16). Attempts to inhibit the RAS–SOS interaction via peptides mimicking an orthosteric SOS helix identified hydrocarbon-stapled peptides with nanomolar affinity, but only low cellular activity (17, 18). Fragment-based screening, rational design, and high-throughput screening approaches led to identification of small molecules addressing the KRAS–SOS1 interaction, resulting in compounds with moderate micromolar affinity (19–22). Surprisingly, rather than inhibition, some of these binders activated the SOS1-mediated nucleotide exchange, resulting in biphasic modulation of RAS signaling through negative feedback on SOS1 (23).
Here, we report the identification of small molecules that efficiently inhibit the activation of KRAS by SOS1. We focused on the oncogenic G12C mutant of KRAS because of its clinical importance in lung cancer (24). Taking a dual approach supported by structure-guided design, we combined results from fragment-based and high-throughput screening. This included elucidation of crystal structures of the KRASG12C–SOS1 complex, of SOS1 in complex with inhibitors, and of apo SOS2. We present selective and potent compounds with double-digit nanomolar affinity to SOS1, submicromolar antiproliferative activity in tumor cell lines, and synergistic combination potential with the covalent KRASG12C inhibitor ARS-853 (12, 13).
Results
In our efforts to identify inhibitors of mutant RAS for cancer treatment, we initiated two parallel approaches: (i) a fragment screen was performed to identify inhibitors via KRAS–SOS1 complex stabilization, in analogy to the inhibition of the small GTPase ARF by brefeldin A (25); and (ii) a high-throughput screen (HTS) was designed to search for inhibitors of the enzymatic SOS1 nucleotide exchange activity, via binding either to KRAS or to SOS1.
Fragment Screen for Stabilizers of the KRASG12C–SOS1 Interaction.
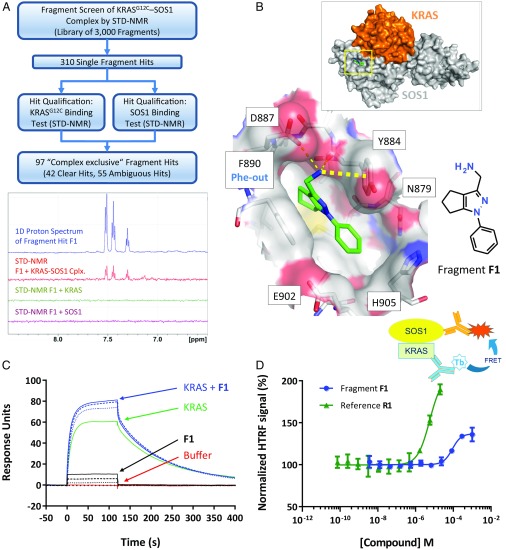
To identify dead-end stabilizers of the KRASG12C–SOS1 interaction, a ligand-observed NMR fragment screen for binders of the complex of KRASG12C and the catalytic domain of wild-type SOS1 (SOS1cat) was performed (Fig. 1A). A library of 3,000 fragments was screened by saturation transfer difference (STD)-NMR in pools of eight, resulting in 310 single hits that were then counterscreened with KRASG12C and SOS1cat alone. Of 97 fragments binding exclusively to the KRASG12C–SOS1cat complex, 42 were selected for crystallization based on their STD-NMR signals. Signals in the STD spectra indicated binding of fragment hit F1 exclusively to the preformed KRASG12C–SOS1cat complex, and not to SOS1cat or KRASG12C alone (Fig. 1A).
Fig. 1.
KRASG12C–SOS1cat NMR fragment screen. (A) Screening cascade (Top) and example spectra for F1 (Bottom): 1D proton spectrum (blue) and STD-NMR spectrum with KRASG12C–SOS1 complex (Cplx.; red), KRASG12C_C118S (green), and SOS1cat (purple). (B) Cocrystal structure of F1 bound to KRASG12C–SOS1cat. (Top) Overall complex with location of the fragment binding site (yellow box); Inset is the area in the yellow box enlarged, showing hydrogen bonds as thin dashed lines and cation–π interaction as a thick dashed line. (C) SPR assay with immobilized SOS1cat. Green line: KRASG12C_C118S; dotted, dashed, and solid blue lines: KRASG12C_C118S in the presence of 100, 250, and 500 µM F1, respectively; dotted, dashed, and solid black lines: respective addition of 100, 250, and 500 µM F1 alone, showing unspecific binding of F1 to SOS1cat; and red line: buffer. (D) F1 and the SOS-activator R1 increase the interaction between KRASG12C_C118S and SOS1cat. (Top) Assay scheme showing RAS, SOS1, and detection antibodies with fluorescent labels (Tb, terbium). (Bottom) Data points represent mean ± SD (n = 4). Normalization: 100% HTRF signal, DMSO control; 0% HTRF signal, without SOS1cat.
Crystals of the KRASG12C–SOS1cat complex were obtained using KRASG12C_SB, a KRASG12C construct containing the mutation C118S to increase stability (26), as well as a triple mutation (D126E-T127S-K128R) identified in a surface mutation screen (SI Appendix, Supplementary Materials and Methods). These mutations enabled KRASG12C_SB–SOS1cat to crystallize in the same crystal form as reported for HRAS–SOS1 (27). Soaking of the 42 fragments into KRASG12C_SB–SOS1cat crystals resulted in 13 cocrystal structures, of which four (fragments F1 to F4) are presented (Fig. 1B and SI Appendix, Fig. S1 and Table S1). Surprisingly, all 13 fragment hits did not bind within the KRAS–SOS1 interface but into a mainly hydrophobic pocket on SOS1 located immediately adjacent to KRAS (Fig. 1B). The same pocket was recently reported for fragment hits targeting HRAS–SOS1 (19) and for SOS1 activators (21, 22). Remarkably, fragments F1, F3, and F4 induced a conformational shift in the binding pocket by triggering a side-chain rotation of Phe890, thereby opening a new back pocket (SI Appendix, Fig. S1D). This Phe-out conformation was also observed by Winter et al. (19) for some fragment hits and by Burns et al. (21) for HTS-derived activators of the KRAS–SOS interaction. The other 10 fragment hits, represented by F2 (SI Appendix, Fig. S1C), left Phe890 in its Phe-in conformation.
SOS1 features two distinct RAS binding sites: a catalytic site and an additional allosteric RAS binding site (28). Superimposition of the cdc25 domains of the fragment-bound KRAS–SOS1 crystal structures reported here with the HRAS–SOS1 complex that has an additional HRAS molecule bound to the allosteric site (PDB ID code 1NVU) revealed that RAS engagement at the allosteric site does not affect the fragment binding site.
All fragment hits were characterized for their stabilizing or disrupting effect on the KRASG12C–SOS1cat complex using a 2D protein-observed NMR assay (29), a surface plasmon resonance (SPR) assay, and a biochemical assay that quantifies the equilibrium binding interaction of KRASG12C and SOS1cat (see detailed assay descriptions in SI Appendix, Supplementary Materials and Methods). Fragment F1 stabilized the KRASG12C–SOS1cat complex in all three assays: In the 2D NMR assay, this was indicated by the reduction of signals for 15N-labeled KRASG12C upon addition of F1 (SI Appendix, Fig. S2A). In the SPR assay, addition of F1 increased the amount of KRASG12C binding to immobilized SOS1cat (Fig. 1C). In the interaction assay, F1 resulted in an increased homogeneous time-resolved fluorescence (HTRF) signal (Fig. 1D) similar to reference R1 (SI Appendix, Table S2), a compound previously shown to bind and activate SOS1 (22). Only fragment F3 behaved similarly to F1, whereas F2 and all other fragments showed no effect in the KRASG12C–SOS1cat interaction assay, no stabilizing effect in the SPR assay, and no (or only weak) disruption effects in the NMR assay. Fragment F1 was therefore chosen as the starting point for optimization (see also SI Appendix, Supplementary Results for further details on the fragment hit prioritization and fragment binding modes).
F1 interacts with SOS1 via a π–π interaction with Phe890 in its new Phe-out position and forms two hydrogen bonds to Tyr884 and Asp887 (Fig. 1B). The aminomethyl moiety additionally forms a cation–π interaction with the side chain of Tyr884. In an attempt to optimize F1, synthesis of a broad set of variants was undertaken; however, none of the variants yielded any significant improvement in potency.
HTS and Initial Optimization.
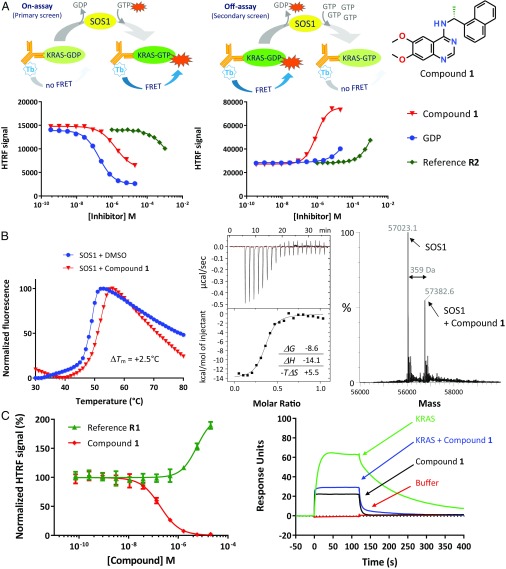
To screen the Bayer library, consisting of over 3 million compounds, we developed a miniaturized enzymatic assay quantifying the SOS1-mediated loading of a fluorescently labeled GTP analog onto KRASG12C, which results in an increased HTRF signal (“On-assay” in Fig. 2A). Hits were retested using a secondary assay monitoring the SOS1-catalyzed HTRF signal decrease by the deloading of a fluorescently tagged GDP analog preloaded onto KRASG12C (“Off-assay” in Fig. 2A). This secondary assay efficiently removed not only artificial hits that inhibit the primary assay by quenching but also GTP-competitive hits that are inactive in the Off-assay due to the requirement of excess GTP for nucleotide exchange. All hits were characterized for their selectivity for mutant KRASG12C and against wild-type KRAS (KRASWT). The SOS1-dependence of inhibition was tested using an assay measuring intrinsic nucleotide exchange of KRASG12C in the absence of SOS1. We further checked whether hits impact the interaction between KRASWT–GTP and the RAS binding domain (RBD) of its downstream effector CRAF kinase (CRAFRBD). Finally, we used thermal shift assays (TSAs) to analyze the interaction of the small molecules with either KRASWT, KRASG12C, or SOS1cat as indicated by a shift of the protein melting point to higher temperature compared with the protein alone.
Fig. 2.
Discovery of quinazolines as direct SOS1 inhibitors that disrupt the KRAS–SOS1 complex. (A, Top) Assay schemes (showing KRAS, fluorescently labeled GDP and GTP nucleotides, SOS1, and detection antibodies with fluorescent terbium label), and (A, Bottom) dose–response curves for GDP, compound 1, and KRAS reference compound R2 (SI Appendix, Table S2), shown for the On-assay (Left) and the secondary Off-assay (Middle). Data points represent mean ± SD (n = 4). (B, Left) TSA. Compound 1 stabilizes SOS1cat with a ΔTm of 2.5 °C. (B, Middle, Top) ITC of the interaction of 1 with SOS1cat. (B, Middle, Bottom) Heat curve of titration of SOS1cat into a solution of 1 and integrated enthalpies plotted against the protein-to-compound molar ratio. Inset shows thermodynamic values obtained from fitting a Wiseman isotherm to the measured calorimetric data. (B, Right) Native MS analysis confirmed binding of 1 to SOS1cat with a 1:1 stoichiometry. (C, Left) HTRF-based KRASG12C–SOS1cat interaction assay showing disruption of the KRASG12C–SOS1cat complex by 1 (red curve); the SOS activator R1 (green curve, SI Appendix, Table S2) leads to a stabilization of the KRASG12C–SOS1cat complex. Data points represent mean ± SD (n = 4). Normalization as in Fig. 1D. (C, Right) SPR assay with immobilized SOS1cat. Green: 250 nM KRASG12C_C118S; blue: 250 nM KRASG12C_C118S in the presence of 10 µM compound 1; black: 10 µM 1 alone, showing unspecific binding of 1 to SOS1cat; and red: buffer. Compound 1 resulted in reduced binding of KRASG12C to immobilized SOS1cat.
We focused on a quinazoline series, represented by initial-hit compound 1 (Fig. 2A). Biochemical characterization revealed that 1 inhibited SOS1-mediated loading of KRASG12C with GTP much more efficiently than the direct KRAS inhibitor, reference R2 (29) (Fig. 2A and SI Appendix, Table S2). In contrast to GDP, compound 1 was equipotent in the On-assay and Off-assay. It did not affect intrinsic KRASG12C nucleotide exchange or the nucleotide exchange of another small GTPase, CDC42, by its GEF DBS (SI Appendix, Table S3). Compound 1 inhibited KRASWT and KRASG12C activation with promising submicromolar potency and did not affect the KRAS interaction with CRAFRBD (SI Appendix, Table S3). Together, these initial biochemical data suggested that compound 1 could be a non–GDP-competitive inhibitor of KRAS or a SOS1 inhibitor.
To elucidate the mechanism of action, we performed a set of biophysical assays. TSA, isothermal titration calorimetry (ITC), and native mass spectrometry (native MS) showed binding to SOS1cat (Fig. 2B) rather than a direct interaction with KRAS (SI Appendix, Fig. S2). Compound 1 stabilized SOS1cat, but not KRASG12C or KRASWT, in the TSA. ITC confirmed a strong enthalpy-driven binding of compound 1 to SOS1cat, with a binding enthalpy, ΔH, of −14.1 kcal/mol, suggesting a favorable hydrogen bond network. The entropic penalty upon binding, −TΔS, contributes +5.5 kcal/mol, resulting in a KD of 450 nM. Native MS confirmed binding of 1 to SOS1cat, with a 1:1 stoichiometry, but not to KRASWT. Additional experiments revealed the mode of action of this series as disruption of the KRASG12C–SOS1cat interaction (Fig. 2C): Compound 1 addition led to a reduced FRET signal in the KRASG12C–SOS1cat interaction assay in contrast to the SOS1 activator R1, which increased the FRET signal. Furthermore, addition of compound 1 resulted in a decreased amount of KRASG12C binding to immobilized SOS1cat, as measured by SPR. Also, addition of compound 14 (a close derivative of 1, see Fig. 3D) led to increased NMR signals for 15N-labeled KRASG12C_C118S, indicative of disruption of the KRASG12C–SOS1cat complex (SI Appendix, Fig. S2B).
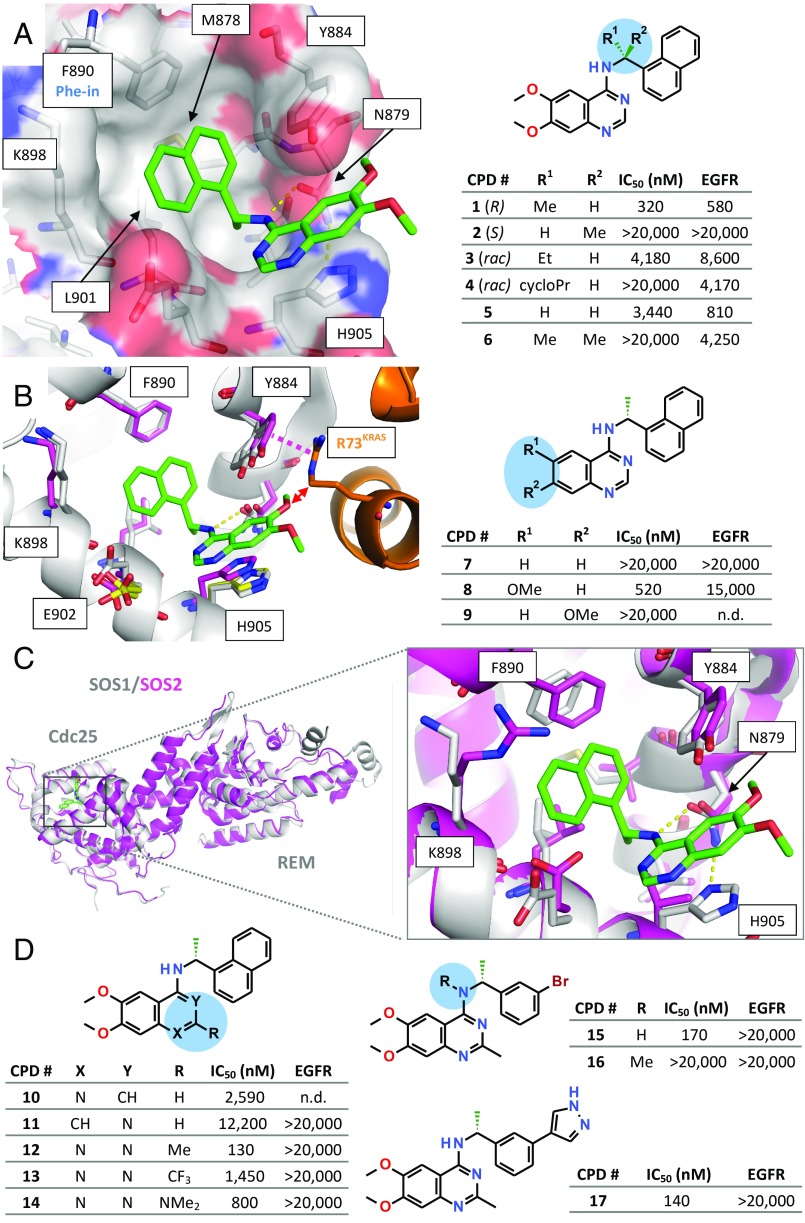
Fig. 3.
SOS1–compound 1 cocrystal structure, SAR, and crystal structure of SOS2. (A, Left) Cocrystal structure of SOS1SB (carbon atoms in gray) in complex with 1 (stick model, carbon atoms in green). (B, Left) Crystal structure of SOS1SB in complex with 1 (protein carbon atoms in gray, inhibitor carbon atoms in green), superimposed with the crystal structures of apo SOS1SB (selected binding site residues shown, carbon atoms in yellow) and KRASG12C_SB–SOS1cat (KRAS in orange, SOS1 carbon residues in magenta). Magenta dashed line indicates a stacking interaction between the side chain of Tyr884 and KRAS residue Arg73. Red arrow highlights a predicted clash between one of the two methoxy groups of the inhibitor with Arg73KRAS. (C) Superimposition of the crystal structures of SOS1SB (gray ribbon) in complex with 1 and apo SOS2SB (magenta). Overall view (Left) and Inset view (Right) into the inhibitor binding site. (A, Right and B, Right and D) Initial SAR data for the SOS1 inhibitor series. IC50 values measured with the KRASG12C–SOS1cat interaction assay and EGFR kinase inhibition assay (mean values; see SI Appendix, Table S8 for SD and biological replicates).
To understand the molecular basis for the interaction with SOS1, we determined the cocrystal structure of SOS1 with compound 1 (Fig. 3A and SI Appendix, Table S4). Crystals were obtained with a variant of SOS1cat with four additional N-terminal residues (termed SOS1SB), first described by Freedman et al. (30). The inhibitor binds into a surface pocket on SOS1, which is located immediately adjacent to the KRAS binding site. The quinazoline scaffold is sandwiched between His905 and Tyr884 (π–π stacking). The naphthyl moiety occupies a hydrophobic pocket (formed by Leu901 and Phe890) and is in T-stacking contact with Tyr884. The central aniline NH function forms a hydrogen bond to the side chain of Asn879, an interaction shown to be essential, with complete loss of activity upon methylation of the free NH (see compound 16, Fig. 3D). The key roles of Asn879, His905, and Leu901 were confirmed in a study in which mutations of these residues reduced the inhibitory effect of compound 1 (SI Appendix, Fig. S3). The methyl substituent at the stereocenter optimally occupies a small subpocket, which explains the observed eudysmic ratio, with the (R)-enantiomer 1 being active and the (S)-enantiomer 2 being inactive (Fig. 3A). The hydrophobic subpocket addressed by the naphthyl moiety is identical to the binding site reported above for the Phe-in binding fragment hits (SI Appendix, Fig. S1C). Comparison of the SOS1SB–1 complex with apo SOS1SB and with KRASG12C_SB–SOS1cat indicated that the binding site for compound 1 is mostly already preformed in the absence of the ligand (Fig. 3B). The structures reveal how compound 1 weakens the KRAS–SOS1 interaction: Compared with the native KRASG12C–SOS1cat complex, compound 1 triggers a movement of the side chain of Tyr884 away from KRAS. This weakens the stacking interaction between this side chain and Arg73KRAS. Remarkably, the two stabilizing fragments, F1 and F3, also interact with the side chain of Tyr884 but, in doing so, stabilize this side chain in the conformation required for the interaction with Arg73KRAS (SI Appendix, Fig. S4). Also, the methoxy group at position 6 of compound 1 would clash with the side chain of Arg73KRAS. This prediction from the structural data of the importance of the methoxy substitution at position 6 of the quinazoline core was experimentally confirmed by the synthesis and testing of compounds 7 to 9 (Fig. 3B). Thus, 7 and 9, lacking substitution at position 6, are inactive, whereas 8, with a single 6-methoxy group is as active as the initial hit 1. The molecular basis for the observed disrupting mode of action of this inhibitor series therefore appears to be a combination of steric hindrance by the methoxy group and an indirect effect via the side chain of Tyr884.
SOS1, however, is only one of several exchange factors known to target the RAS family (16, 31). Its closest relative is SOS2 (80% identity in the catalytic domain), whereas the other known GEFs (e.g., RASGRF1/2 and RASGRP1 to RASGRP4) are less than 30% identical to SOS1. Despite the high sequence identity, selectivity assays with the quinazoline series revealed a strong selectivity against SOS2 (SI Appendix, Table S3). We were able to solve the crystal structure of the catalytic domain of SOS2 using a surface mutation approach (construct SOS2SB). The overall fold is conserved between SOS1 and SOS2 (Fig. 3C). Comparison of the inhibitor-bound SOS1SB and apo SOS2SB crystal structures indicated that the observed selectivity can most likely be attributed to the exchange of His905SOS1 to a valine residue in SOS2, which prevents the essential stacking interaction with the quinazoline core of compound 1 (Fig. 3C). Considering the much larger sequence differences between SOS1 and the other known RASGEFs, the quinazoline series of inhibitors is most likely also selective against the other exchange factors of RAS.
After identification of SOS1 as the molecular target and elucidation of the binding mode of the selected hit series, the structure–activity relationship (SAR) was explored. Consistent with the cocrystal structure of SOS1 with compound 1 (Fig. 3A), a methyl substituent at the benzylic position seems to be ideal. Thus, a sharp drop in potency was observed when methyl was replaced by a larger residue (compounds 3 and 4) or, indeed, by hydrogen (compound 5). Disubstitution at the benzylic position was not tolerated at all (compound 6). The quinazoline motif proved to be essential for activity (Fig. 3D, 10 and 11). Compound 1 shows structural similarity with known kinase inhibitors, such as the epidermal growth factor receptor (EGFR) kinase inhibitor erlotinib (32), and indeed inhibits EGFR kinase with an IC50 of 580 nM (SI Appendix, Table S3). To prevent interaction with the hinge region of kinases, an additional substituent was introduced at position 2, leading to the identification of 2-methyl-substituted quinazolines as compounds devoid of kinase inhibitory activity. Substituents in position 2 result in a steric clash with the hinge region of protein kinases and thereby abrogate the crucial interaction of the aminoquinazoline core with the hinge region, as illustrated for the case of the EGFR kinase inhibitor erlotinib (33) (SI Appendix, Fig. S5). The “dehinged” compound 12 indeed still strongly inhibited SOS1cat but also exhibited good selectivity against EGFR kinase (Fig. 3D), and the related compound 17 was inactive against a large panel of other kinases (SI Appendix, Table S5), including all kinases of the RAS-RAF-MEK-ERK pathway.
Guided by the cocrystal structure of SOS1 with compound 1, we further optimized the quinazoline inhibitor series, culminating in compound 17. Replacement of the naphthyl moiety by a pyrazolylphenyl group resulted in good potency and improved aqueous solubility, and the cocrystal structure (SI Appendix, Fig. S6A) revealed an additional water-bridged hydrogen bond to Glu902. However, the IC50 values of this series could not be optimized to better than 130 nM.
Linking of the HTS Hit Series with the Fragment Hit.
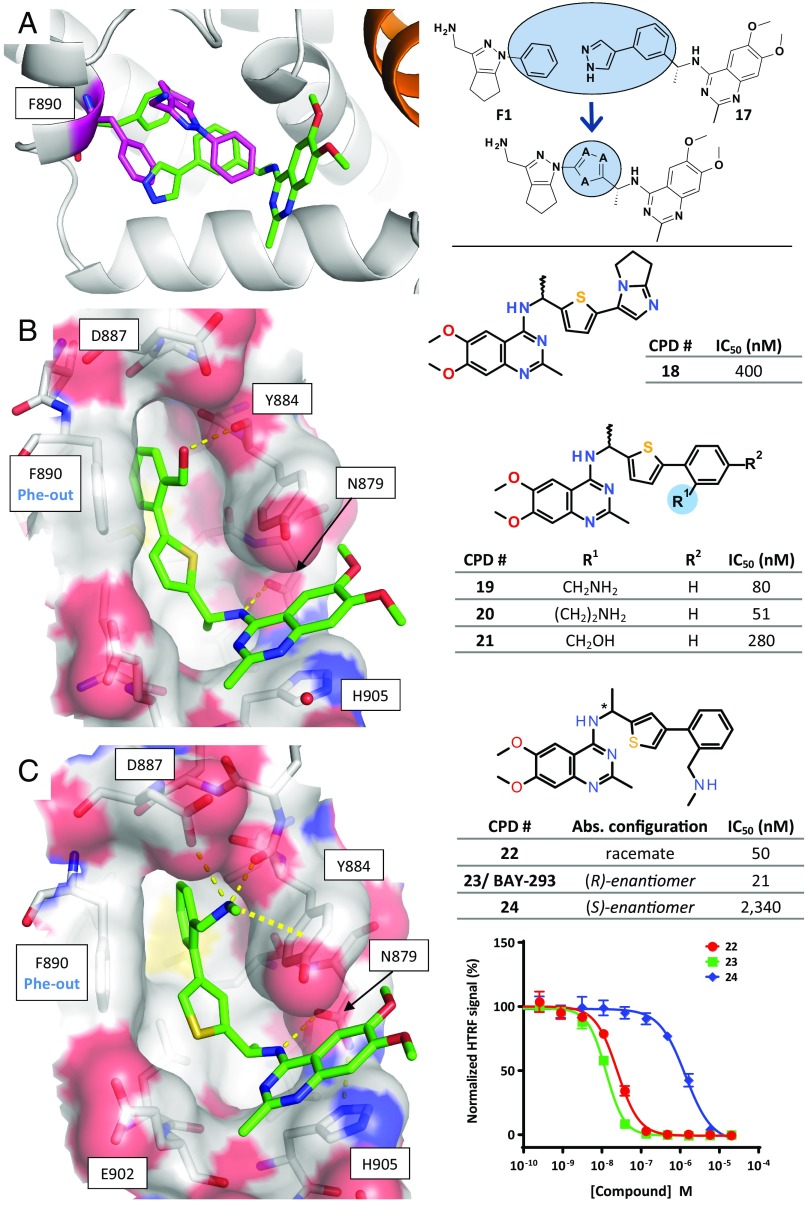
As the fragment screen had identified a new subpocket that was not yet addressed by the HTS series, we aimed at further improving the potency by combining both ligand series. A superimposition of the cocrystal structures of compound 17 and fragment F1 (Fig. 4A) suggested that hybrid compounds generated by linking the quinazoline inhibitor series to F1 may show increased potency. Appropriate linkers that could orientate both the tetrahydrocyclopenta[c]pyrazole core of fragment F1 and the aminoquinazoline core of compound 17 in their respective binding sites were designed by a computational approach using the software Spark (34). The overlapping aromatic groups were cut out, and appropriate replacements were identified in 3D databases of common building blocks and scored with respect to steric, geometric, and electrostatic properties (Fig. 4A). Most of the top-scoring linker candidates contained a five-membered aromatic heterocycle. For synthetic reasons, thiophene was selected as linker to investigate this hybrid approach.
Fig. 4.
Structure-based linking of the fragment and HTS hits. (A, Left) Superimposition of the crystal structures of F1 bound to KRASG12C_SB–SOS1cat (KRAS in orange, SOS1 in gray, F1 and Phe890 with carbon atoms in magenta) and 17 bound to SOS1SB (only inhibitor and Phe890 shown; carbon atoms in green). (Right) Schematic depiction of the merging approach, with 18 as an initial example of the resulting hybrid compounds. (B, Left and C, Left) Crystal structures of SOS1SB in complex with 21 (B) and 23 (C); hydrogen bonds shown as thin dashed lines, cation–π interaction as a thick dashed line. (B, Right and C, Right) IC50 values measured with the KRASG12C–SOS1cat interaction assay (mean values; see SI Appendix, Table S8 for SD and biological replicates). (C, Bottom Right) Dose–response curves for compounds 22 to 24.
Optimization of the hybrid series followed a two-pronged approach: initially, variants of the original fragment core of F1 were fused to the thiophene linker; however, these hybrids failed to trigger the Phe-out conformation of Phe890 (see compound 18, SI Appendix, Fig. S6B). In contrast, addition of moieties that mimic the hydrogen bonds of the amino side chain of F1 resulted in improved IC50 values (Fig. 4B), and subsequent cocrystal structures revealed that both amino and hydroxyl groups trigger the Phe-out conformation: In the cocrystal structure with compound 21 (Fig. 4B), the hydroxyl group forms a new hydrogen bond to the backbone carbonyl of Tyr884. Cocrystallization with amino-containing racemate 22 revealed, unambiguously, that only the (R)-enantiomer 23 had bound in the crystal (Fig. 4C and SI Appendix, Table S4). The side-chain amino group of 23 forms two new hydrogen bonds, to Asp887 and Tyr884, and is in a favorable position for a cation–π interaction with the side chain of Tyr884 (see SI Appendix, Supplementary Results for a detailed analysis of the observed SAR of this hybrid series). Compound 23 was initially tested as a racemate (compound 22), and later separated into the active (R)-enantiomer (23) and the weakly active (S)-enantiomer (24, eudysmic ratio ∼111; Fig. 4C). Biophysical characterization of the active and less active enantiomers was performed. TSA confirmed binding of the racemate 22 and the active enantiomer 23 to SOS1cat, while the less active variant 24 showed no stabilization of SOS1cat. None of the three compounds stabilized KRASWT or KRASG12C (SI Appendix, Fig. S2E). The interaction of SOS1 with compounds 22, 23, and 24 was characterized by ITC. Binding was observed for racemate 22 and the active enantiomer 23, but not for the less active enantiomer 24 (SI Appendix, Fig. S2F). The KD values of 18 nM for 22 and 36 nM for 23 derived from the ITC binding curves were in line with the IC50 data obtained by the KRAS–SOS1 interaction assay (50 nM and 21 nM for 22 and 23, respectively; Fig. 4C). Native MS analysis with SOS1cat showed a mass shift of 449 units with compound 23, but not with compound 24 (SI Appendix, Fig. S2G). All optimized compounds of the HTS series showed a disrupting effect on the KRAS–SOS1 interaction, as shown for 22 and 23 in the interaction assay (Fig. 4C, Right). Compounds 22 and 23 were chosen as the best representatives of this inhibitor series before and after fusion with the fragment-derived moiety, respectively.
Cellular Characterization.
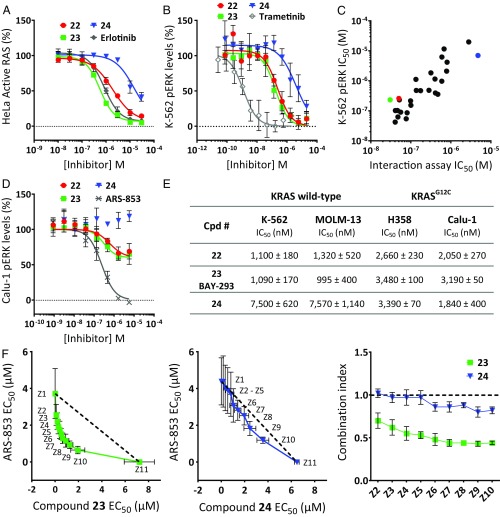
The cellular activity of the quinazoline series was assessed by incubating HeLa cells with the SOS1 inhibitors, followed by quantification of the amount of activated, GTP-loaded total RAS from cellular lysates. Compounds 22 and 23 inhibited the activation of RAS in HeLa cells, with IC50 values in the submicromolar range, whereas the (S)-enantiomer 24 showed significantly lower activity (Fig. 5A and SI Appendix, Table S6).
Fig. 5.
Cellular characterization of compounds 22 to 24. (A) Inhibition of active RAS levels in HeLa cells. (B) pERK levels in K-562 cells. (C) Correlation of IC50 data for pERK inhibition in K-562 cells with biochemical KRAS–SOS1 interaction. (D) pERK levels in Calu-1 cells. The colored dots represent compounds 22 (red), 23 (green), and 24 (blue). Reference compounds (SI Appendix, Table S2) in A, B, and D are indicated in gray. Data points in A, B, and D represent mean ± SD (n = 4). The IC50 values of 22 to 24 for these assays are summarized in SI Appendix, Table S6. (E) Antiproliferative activity against wild-type KRAS cell lines (K-562, MOLM-13) and cell lines with KRASG12C mutation (NCI-H358, Calu-1). Mean IC50 values ± SD, n = 4. (F) Antiproliferative activity of 23 (Left) and 24 (Middle) was assessed in combination with the covalent KRASG12C inhibitor ARS-853 in NCI-H358 cells. IC50 isobologram plots of SOS1-inhibitors (Z1) and ARS-853 (Z11) alone and of fixed combinations (Z2 to Z10) of both compounds are shown. The dotted line represents the line of additivity. Data points represent mean ± SD of biological independent experiments (n = 3). (Right) The combination index was calculated according to the median-effect model of Chou–Talalay (43); a value below 0.8 indicates a more-than-additive (i.e., a synergistic) interaction.
Next, the downstream effects of the SOS1 inhibitors were analyzed by assays quantifying phospho-ERK (pERK) levels in K-562 cells, a tumor cell line for which sensitivity to SOS1 inhibition by CRISPR knockout has been reported (35). The racemate 22 and the (R)-enantiomer 23 efficiently inhibited pERK levels in K-562 cells after incubation for 60 min without affecting total protein levels of ERK, whereas the (S)-enantiomer 24 again showed significantly lower activity (Fig. 5B and SI Appendix, Fig. S8 for total ERK levels). Data from an extended set of compounds of our quinazoline inhibitor series revealed a significant correlation of the IC50 values measured by cellular pERK inhibition with biochemical inhibition of the KRAS–SOS1 interaction, indicating that the observed cellular effects of this inhibitor series are mediated by intracellular target engagement of SOS1 (Fig. 5C).
There is a common understanding that cells carrying mutant KRAS alleles are less dependent on their exchange factors than wild-type cells. To directly test this not-yet-fully explored hypothesis with our SOS1 inhibitors, we chose Calu-1 cells, which carry two KRASG12C alleles. Analysis of total and pERK levels in these cells revealed that compounds 22 and 23 are able to inhibit pERK levels in a concentration-dependent manner. In contrast to the covalent KRASG12C inhibitor ARS-853 (12, 13), the SOS1 inhibitors are not able to fully suppress downstream signaling, with ∼50% of the pERK levels remaining after treatment for 24 h (Fig. 5D and SI Appendix, Fig. S8 for total ERK levels).
Compound 22 was profiled for antiproliferative activity in 60 cell lines derived from lung, liver, and hematopoietic tissue (SI Appendix, Fig. S9). Compound 22 displayed a relatively broad inhibition spectrum, with hematopoietic cells (K-562, KG-1, MOLM-13, and THP-1) being most vulnerable to SOS1 inhibition. This is consistent with the previously identified SOS dependency of K-562 cells after SOS knockout by CRISPR (35). The antiproliferative activity of compounds 22 and 23 versus the less active enantiomer 24 was further studied in cells with wild-type KRAS (K-562, MOLM-13) and in cells with a KRASG12C mutation (NCI-H358, Calu-1). In cells with wild-type KRAS, we found seven- to eightfold reduced activity of the less active enantiomer 24 compared with the racemate 22 and the pure active enantiomer 23 (Fig. 5E). In contrast, there was no significant difference between the active and less active compounds in cells carrying a KRASG12C mutation.
Recent data suggest that SOS1 inhibition may have synergistic antiproliferative potential when combined with direct covalent KRASG12C inhibitors. This is based on findings that (i) these covalent inhibitors selectively bind to the GDP-bound, but not to the GTP-bound, KRASG12C protein and (ii) compared with other mutant KRAS proteins, KRASG12C undergoes nucleotide cycling within cells and, therefore, requires reactivation by exchange factors (11, 13). With our potent SOS1 inhibitors, we were now able to test this hypothesis. We selected NCI-H358 cells, which are heterozygous for KRASG12C, and treated them with a combination of the covalent KRASG12C inhibitor ARS-853 and either compound 23 or 24. In contrast to the less active enantiomer 24, synergy between 23 and ARS-853 was indeed observed (Fig. 5F, Left), with a combination index significantly below 0.8 over a wide range of combinations. These data suggest that parallel inhibition of SOS1 and KRASG12C leads to synergistic antiproliferative activity and may therefore offer a viable option for the treatment of KRASG12C-mutant cancers in patients.
Discussion
This work describes a successful approach to identify nanomolar, selective, and cell-active inhibitors of SOS1, the exchange factor of RAS. The inhibitor design was enabled by a dual screening approach and structure-guided design. An HTS identified a core scaffold for which the potency was optimized to an IC50 of 130 nM (Figs. 2 and 3). A fragment screen identified an induced fit that opened a subpocket directly adjacent to the binding site of the HTS series (Fig. 1), also reported recently by Winter et al. (19) in a similar fragment-screening approach. Combination of both approaches was essential and led to the design of compounds addressing this subpocket, with an improved IC50 of 21 nM.
Remarkably, this binding site on the surface of SOS1, targeted by both the HTS hit series and the fragment hits, was reported initially as the binding site for activators of the SOS1-catalyzed nucleotide exchange of RAS (22). Consistent with this observation, two of the fragment hits acted as stabilizers and not disruptors of the KRASG12C–SOS1 interaction (Fig. 1). Fusing the binding functionality of one of these stabilizing fragments to the HTS-derived inhibitor series resulted in improved disruptors, not activators (Fig. 4). We suggest that the molecular basis for this functional flip is the stabilization of the side chain of Tyr884 in either a conformation optimal for π–π stacking with the KRAS residue Arg73, generating stabilizers (SI Appendix, Fig. S4), or in a conformation that is no longer able to engage in this stacking with Arg73KRAS, generating disruptors (i.e., inhibitors, Fig. 3B).
Our nanomolar SOS1 inhibitors have allowed investigations of the effect of chemical SOS1 inhibition in cells. We could demonstrate that selective inhibition of SOS1 effectively down-regulates the levels of active RAS in tumor cells. In cells with wild-type KRAS, we observed complete inhibition of the RAS-RAF-MEK-ERK pathway (Fig. 5B). In a tumor cell line with mutated KRAS alleles, chemical SOS1 inhibition resulted in a reduction of pERK activity by ∼50% (Fig. 5D). We investigated whether this still-limited downstream effect could be further improved by co-inhibition of additional targets. Indeed, covalent KRASG12C inhibitors are known to require GDP-bound inactive KRASG12C for binding, and potential combination therapies by upstream inhibition of RAS activation (e.g., by inhibition of receptor tyrosine kinase or RASGEF activity) have been discussed (11–13). We have shown that the combination of our SOS1 inhibitor with ARS-853, a covalent inhibitor of KRASG12C, results in synergistic antiproliferative activity in a KRASG12C-mutated cell line (Fig. 5F).
We therefore present compound 23 (BAY-293) as a tool for the further investigation of RAS–SOS1 biology in vitro. Improvements in the bioavailability of the inhibitor series will be required for in vivo experiments. Together, the data presented here indicate that inhibition of GEFs may represent a viable approach for targeting RAS-driven tumors. Of particular note is the synergistic effect between our inhibitors and ARS-853 observed in a KRASG12C-mutated cancer cell line, which highlights the potential for combination therapy between a direct KRAS and a SOS1 inhibitor.
Materials and Methods
DNA sequences for the recombinant proteins used in this study were optimized for expression in Escherichia coli, synthesized by GeneArt technology at Life Technologies, expressed in E. coli, and purified via affinity chromatography and size exclusion chromatography. All details of the cloning, expression, and purification steps are described in SI Appendix, Supplementary Materials and Methods. All expression constructs are listed in SI Appendix, Table S7. Quantification of SOS1cat-mediated loading of human KRASG12C–GDP with a fluorescent GTP analog was carried out by measuring energy transfer from anti-GST-terbium (FRET donor) bound to GST-KRASG12C after binding of a fluorescent GTP analog (FRET acceptor). Details of this assay and all secondary biochemical assays for SOS1cat, SOS2cat, KRASWT, CRAFRBD, CDC42, and EGFR kinase are described in SI Appendix, Supplementary Materials and Methods.
Biophysical methods (TSAs, ITC, native MS, SPR complex assay, NMR methods) and crystallization methods are described in SI Appendix, Supplementary Materials and Methods. SOS1 inhibitors were cocrystallized with SOS1SB. Fragments were soaked into pregrown crystals of KRASG12C_SB–SOS1cat. Datasets were collected at the Helmholtz-Zentrum Berlin in Germany, at the European Synchrotron Radiation Facility in Grenoble, France, or at the PETRA III synchrotron in Hamburg, Germany; processed using XDS (36) and XDSAPP (37); and solved using Molecular Replacement with Phaser (38) from the CCP4 suite (39). Models were rebuilt using Coot (40) and refined using REFMAC (41). Ligand models were generated using BIOVIA Discovery Studio (Dassault Systèmes) and parameter files calculated with PRODRG (42). Figures were generated using PyMOL (Schrödinger, LLC).
The cell lines NCI-H358 and K-562 were obtained from the American Type Culture Collection. Calu-1 cells were obtained from CLS Cell Lines Service. MOLM-13 and HeLa cells were obtained from the German Collection of Microorganisms and Cell Cultures. Detailed information on the cellular assays is provided in SI Appendix, Supplementary Materials and Methods.
Fragments are commercially available from Enamine (F1, F2, and F3) and Asinex (F4). Detailed synthetic routes, procedures, and characterizations (compounds 1 to 45, fragment F1) are available in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dagmar Zeggert-Springer, Carmen Kropp, Marion Slezewski, Anne Sparmann, Jutta Hoffmann, Simone Thalheim, Linda Caparusagi, Katja Kauffeldt, Daniel Seifert, Patrick Zielinski, and Julian Plaga for technical support; the staffs at beamline BL14.1 at Helmholtz-Zentrum Berlin, at beamline P11 at PETRA III at the Deutsches Elektronen-Synchrotron (a member of the Helmholtz Association), and at beamline ID29 at the European Synchrotron Radiation Facility in Grenoble, for access to synchrotron radiation and support during data collection; the staffs at moloX GmbH for data collection services and at Crown Bioscience for performing the viability cell panel; and Hans Schick (Angewandte Synthesechemie Adlershof GmbH) and Qiuwen Wang (Pharmaron Beijing Co. Ltd.) and their teams for synthesis support.
Footnotes
Conflict of interest statement: R.C.H. B.S., J.S., D.M., A.H., C.M.S., N.D.W., H.B., U.B., J.W., V.B., J.M., K.P., G.S., N.B., K.E., K.G., L.W., F.v.N., and B.B. are or have been employees and stockholders of Bayer AG. J.D.K. and D.W. are employees of Evotec AG. R.C.H., B.S., J.S., D.M., H.B., K.P., N.B., K.E., L.W., F.v.N., and B.B. are coauthors of a patent application.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.wwpdb.org [PDB ID codes 5OVD, 5OVE, 5OVF, 5OVG, 5OVH, and 5OVI (SOS1 complexes); 6EIE (SOS2); and 6EPL, 6EPM, 6EPN, 6EPO, and 6EPP (KRAS–SOS1 complexes)].
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812963116/-/DCSupplemental.
References
- 1.Der CJ, Krontiris TG, Cooper GM. Transforming genes of human bladder and lung carcinoma cell lines are homologous to the ras genes of Harvey and Kirsten sarcoma viruses. Proc Natl Acad Sci USA. 1982;79:3637–3640. doi: 10.1073/pnas.79.11.3637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Parada LF, Tabin CJ, Shih C, Weinberg RA. Human EJ bladder carcinoma oncogene is homologue of Harvey sarcoma virus ras gene. Nature. 1982;297:474–478. doi: 10.1038/297474a0. [DOI] [PubMed] [Google Scholar]
- 3.Santos E, Tronick SR, Aaronson SA, Pulciani S, Barbacid M. T24 human bladder carcinoma oncogene is an activated form of the normal human homologue of BALB- and Harvey-MSV transforming genes. Nature. 1982;298:343–347. doi: 10.1038/298343a0. [DOI] [PubMed] [Google Scholar]
- 4.Cox AD, Fesik SW, Kimmelman AC, Luo J, Der CJ. Drugging the undruggable RAS: Mission possible? Nat Rev Drug Discov. 2014;13:828–851. doi: 10.1038/nrd4389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hobbs GA, Der CJ, Rossman KL. RAS isoforms and mutations in cancer at a glance. J Cell Sci. 2016;129:1287–1292. doi: 10.1242/jcs.182873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Simanshu DK, Nissley DV, McCormick F. RAS proteins and their regulators in human disease. Cell. 2017;170:17–33. doi: 10.1016/j.cell.2017.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Haigis KM. KRAS alleles: The devil is in the detail. Trends Cancer. 2017;3:686–697. doi: 10.1016/j.trecan.2017.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Downward J. Targeting RAS signalling pathways in cancer therapy. Nat Rev Cancer. 2003;3:11–22. doi: 10.1038/nrc969. [DOI] [PubMed] [Google Scholar]
- 9.Berndt N, Hamilton AD, Sebti SM. Targeting protein prenylation for cancer therapy. Nat Rev Cancer. 2011;11:775–791. doi: 10.1038/nrc3151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ostrem JM, Shokat KM. Direct small-molecule inhibitors of KRAS: From structural insights to mechanism-based design. Nat Rev Drug Discov. 2016;15:771–785. doi: 10.1038/nrd.2016.139. [DOI] [PubMed] [Google Scholar]
- 11.Janes MR, et al. Targeting KRAS mutant cancers with a covalent G12C-specific inhibitor. Cell. 2018;172:578–589.e17. doi: 10.1016/j.cell.2018.01.006. [DOI] [PubMed] [Google Scholar]
- 12.Lito P, Solomon M, Li LS, Hansen R, Rosen N. Allele-specific inhibitors inactivate mutant KRAS G12C by a trapping mechanism. Science. 2016;351:604–608. doi: 10.1126/science.aad6204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Patricelli MP, et al. Selective inhibition of oncogenic KRAS output with small molecules targeting the inactive state. Cancer Discov. 2016;6:316–329. doi: 10.1158/2159-8290.CD-15-1105. [DOI] [PubMed] [Google Scholar]
- 14.Sautier B, Nising CF, Wortmann L. Latest advances towards ras inhibition: A medicinal chemistry perspective. Angew Chem Int Ed Engl. 2016;55:15982–15988. doi: 10.1002/anie.201608270. [DOI] [PubMed] [Google Scholar]
- 15.Hunter JC, et al. Biochemical and structural analysis of common cancer-associated KRAS mutations. Mol Cancer Res. 2015;13:1325–1335. doi: 10.1158/1541-7786.MCR-15-0203. [DOI] [PubMed] [Google Scholar]
- 16.Vigil D, Cherfils J, Rossman KL, Der CJ. Ras superfamily GEFs and GAPs: Validated and tractable targets for cancer therapy? Nat Rev Cancer. 2010;10:842–857. doi: 10.1038/nrc2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patgiri A, Yadav KK, Arora PS, Bar-Sagi D. An orthosteric inhibitor of the Ras-Sos interaction. Nat Chem Biol. 2011;7:585–587. doi: 10.1038/nchembio.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Leshchiner ES, et al. Direct inhibition of oncogenic KRAS by hydrocarbon-stapled SOS1 helices. Proc Natl Acad Sci USA. 2015;112:1761–1766. doi: 10.1073/pnas.1413185112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Winter JJ, et al. Small molecule binding sites on the Ras:SOS complex can be exploited for inhibition of Ras activation. J Med Chem. 2015;58:2265–2274. doi: 10.1021/jm501660t. [DOI] [PubMed] [Google Scholar]
- 20.Evelyn CR, et al. Rational design of small molecule inhibitors targeting the Ras GEF, SOS1. Chem Biol. 2014;21:1618–1628. doi: 10.1016/j.chembiol.2014.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Burns MC, et al. High-throughput screening identifies small molecules that bind to the RAS:SOS:RAS complex and perturb RAS signaling. Anal Biochem. 2018;548:44–52. doi: 10.1016/j.ab.2018.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Burns MC, et al. Approach for targeting Ras with small molecules that activate SOS-mediated nucleotide exchange. Proc Natl Acad Sci USA. 2014;111:3401–3406. doi: 10.1073/pnas.1315798111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Howes JE, et al. Small molecule-mediated activation of RAS elicits biphasic modulation of phospho-ERK levels that are regulated through negative feedback on SOS1. Mol Cancer Ther. 2018;17:1051–1060. doi: 10.1158/1535-7163.MCT-17-0666. [DOI] [PubMed] [Google Scholar]
- 24.Stephen AG, Esposito D, Bagni RK, McCormick F. Dragging ras back in the ring. Cancer Cell. 2014;25:272–281. doi: 10.1016/j.ccr.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 25.Renault L, Guibert B, Cherfils J. Structural snapshots of the mechanism and inhibition of a guanine nucleotide exchange factor. Nature. 2003;426:525–530. doi: 10.1038/nature02197. [DOI] [PubMed] [Google Scholar]
- 26.Mott HR, Carpenter JW, Campbell SL. Structural and functional analysis of a mutant Ras protein that is insensitive to nitric oxide activation. Biochemistry. 1997;36:3640–3644. doi: 10.1021/bi962790o. [DOI] [PubMed] [Google Scholar]
- 27.Boriack-Sjodin PA, Margarit SM, Bar-Sagi D, Kuriyan J. The structural basis of the activation of Ras by Sos. Nature. 1998;394:337–343. doi: 10.1038/28548. [DOI] [PubMed] [Google Scholar]
- 28.Margarit SM, et al. Structural evidence for feedback activation by Ras.GTP of the Ras-specific nucleotide exchange factor SOS. Cell. 2003;112:685–695. doi: 10.1016/s0092-8674(03)00149-1. [DOI] [PubMed] [Google Scholar]
- 29.Sun Q, et al. Discovery of small molecules that bind to K-Ras and inhibit Sos-mediated activation. Angew Chem Int Ed Engl. 2012;51:6140–6143. doi: 10.1002/anie.201201358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Freedman TS, et al. A Ras-induced conformational switch in the Ras activator Son of sevenless. Proc Natl Acad Sci USA. 2006;103:16692–16697. doi: 10.1073/pnas.0608127103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rojas JM, Oliva JL, Santos E. Mammalian son of sevenless guanine nucleotide exchange factors: Old concepts and new perspectives. Genes Cancer. 2011;2:298–305. doi: 10.1177/1947601911408078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cruz-López O, et al. Novel substituted quinazolines for potent EGFR tyrosine kinase inhibitors. Curr Med Chem. 2011;18:943–963. doi: 10.2174/092986711794940824. [DOI] [PubMed] [Google Scholar]
- 33.Stamos J, Sliwkowski MX, Eigenbrot C. Structure of the epidermal growth factor receptor kinase domain alone and in complex with a 4-anilinoquinazoline inhibitor. J Biol Chem. 2002;277:46265–46272. doi: 10.1074/jbc.M207135200. [DOI] [PubMed] [Google Scholar]
- 34.Slater M, Vinter A. XED force field and spark. In: Brown N, editor. Scaffold Hopping in Medicinal Chemistry. Vol 58. Wiley-VCH; Weinheim: 2013. pp. 195–213. [Google Scholar]
- 35.Wang T, et al. Identification and characterization of essential genes in the human genome. Science. 2015;350:1096–1101. doi: 10.1126/science.aac7041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kabsch W. XDS. Acta Crystallogr D Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sparta KM, Krug M, Heinemann U, Mueller U, Weiss MS. XDSAPP2.0. J Appl Crystallogr. 2016;49:1085–1092. [Google Scholar]
- 38.McCoy AJ, et al. Phaser crystallographic software. J Appl Cryst. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Winn MD, et al. Overview of the CCP4 suite and current developments. Acta Crystallogr D Biol Crystallogr. 2011;67:235–242. doi: 10.1107/S0907444910045749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schüttelkopf AW, van Aalten DM. PRODRG: A tool for high-throughput crystallography of protein-ligand complexes. Acta Crystallogr D Biol Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- 43.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–446. doi: 10.1158/0008-5472.CAN-09-1947. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.