Significance
Influenza A virus (IAV) is one of the most important human pathogens, and it is crucial to understand its life cycle to develop antiviral strategies. However, IAV uncoating, an essential step in viral infection, has remained incomprehensible. Here, via the construction of infectious IAV virions encapsulating quantum dots, we tracked the uncoating and viral ribonucleoprotein complex (vRNP) dynamics of single IAV virions. Our results reveal that after viral fusion and uncoating, IAV vRNP segments are released separately into the cytosol, and individual vRNPs undergo a three-stage active nuclear import process and display two diffusion patterns within the nucleus. These findings reveal uncoating and vRNP trafficking mechanisms which may assist in developing new strategies to block IAV infection.
Keywords: influenza virus, single-particle tracking, quantum dots, uncoating, vRNP dynamics
Abstract
Uncoating is an obligatory step in the virus life cycle that serves as an antiviral target. Unfortunately, it is challenging to study viral uncoating due to methodology limitations for detecting this transient and dynamic event. The uncoating of influenza A virus (IAV), which contains an unusual genome of eight segmented RNAs, is particularly poorly understood. Here, by encapsulating quantum dot (QD)-conjugated viral ribonucleoprotein complexes (vRNPs) within infectious IAV virions and applying single-particle imaging, we tracked the uncoating process of individual IAV virions. Approximately 30% of IAV particles were found to undergo uncoating through fusion with late endosomes in the “around-nucleus” region at 30 to 90 minutes postinfection. Inhibition of viral M2 proton channels and cellular endosome acidification prevented IAV uncoating. IAV vRNPs are released separately into the cytosol after virus uncoating. Then, individual vRNPs undergo a three-stage movement to the cell nucleus and display two diffusion patterns when inside the nucleus. These findings reveal IAV uncoating and vRNP trafficking mechanisms, filling a critical gap in knowledge about influenza viral infection.
During virus infection, uncoating is an obligatory step to release the viral genome into the host cell, and this step is an attractive antiviral target. To date, however, virus uncoating has been a poorly understood process due to limitations in applicable methodology for detecting this transient and dynamic event. Here, we visualized the uncoating of individual influenza A viruses (IAVs) by using quantum dot (QD)-based single-particle tracking.
IAV is one of the most important human viral pathogens; it causes severe morbidity and mortality and poses a serious financial burden (1). The IAV genome consists of eight negative-sense, single-stranded RNAs encapsulated by nucleoprotein (NP) and associated with the polymerase complex comprising PB1, PB2, and PA subunits to form viral ribonucleoprotein complexes (vRNPs) (2). Viral endocytosis entry, intracellular transport, and virus–endosome fusion have been widely studied by various methods, including single-particle tracking (3, 4). However, after endocytosis, the virus uncoating process remains elusive. Although some factors, such as cellular aggresome processing machinery and the ubiquitin ligase Itch, have been found to be involved in uncoating (5, 6), the dynamic uncoating process has not been directly observed in real time. Some critical aspects of the uncoating process, such as its timing, location, and dynamics, are unknown. It is also unknown whether different vRNP segments are released separately or as a package from a single virion.
IAV RNA transcription and replication occur in the nucleus (2). After viral uncoating, vRNPs must be transported into the nucleus. Some cellular factors, such as importin-α and importin-β1, likely participate in the nuclear import of vRNPs (2, 7). Microinjection of dye-labeled vRNPs into cells and fluorescence in situ hybridization (FISH) analysis have been attempted to study the intracellular behavior of vRNPs (8, 9). However, because it has been difficult to record viral uncoating in real time, the dynamics of vRNPs released from virions remain to be elucidated.
Single-particle tracking of virions with QD-labeled genetic material provides an opportunity for tracking viral uncoating in real time. Semiconductor QDs have unique optical properties such as remarkable brightness and superior photostability and are well-suited to single-particle tracking of viral infection (10–12). Here, by assembling IAV particles with QD-labeled vRNPs (QD-vRNPs) and applying single-particle tracking, we studied IAV uncoating and vRNP dynamics in live infected cells in real time. QDs were site-specifically conjugated to the vRNP and encapsulated in IAV particles during virus assembly. We also constructed dual-color IAV particles by combining internal QD encapsulation with QD surface decoration or by incorporating differently colored QD-vRNP segments into single virions. By infecting cells with these fluorescent IAV particles, virus entry and uncoating and vRNP dynamic behaviors were able to be observed and clarified at the single-particle level in real time. Our findings fill a critical gap in knowledge about the early events of influenza viral infection.
Results
Construction and Characterization of IAV Virions Encapsulating QD-vRNPs.
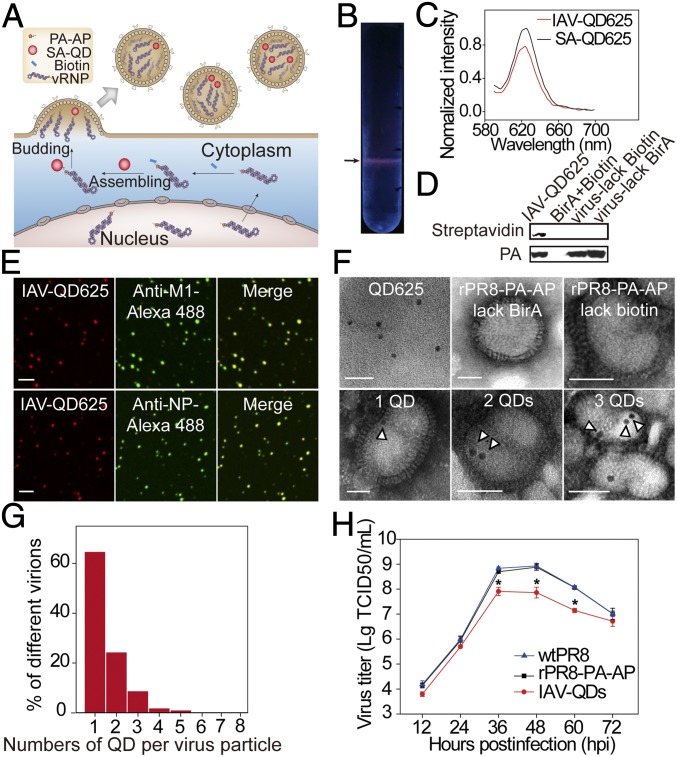
Our strategy to construct IAV particles encapsulating QD-vRNPs is shown in Fig. 1A. A biotin acceptor peptide (AP) tag was fused to the PA protein that is associated with vRNP segments. The PA–AP chimera protein can be specifically biotinylated. A streptavidin-coated QD (SA-QD) was then conjugated to PA. Finally, the QD-vRNPs were encapsulated into progeny virions during virus assembly.
Fig. 1.
Construction and characterization of IAV-QDs. (A) Schematic of construction of IAV-QDs. (B) A representative SDGC tube. Arrow indicates IAV-QD625 band. (C) Fluorescence spectra of IAV-QDs. (D) Western blot analysis of IAV-QD625 using HRP-SA and anti-PA antibody. (E) Immunofluorescence assay of IAV-QD625 using anti-M1 or anti-NP antibody on coverslips. (Scale bar: 2 µm.) (F) TEM images. (Scale bar: 50 nm.) The hollow arrowheads indicate QD625. (G) The percentage of QD numbers per virus particle. (H) Multiple-cycle growth curves of WT PR8 (wtPR8), rPR8-PA–AP, and IAV-QD625 on MDCK cells; n = 3 (*P < 0.05). TCID50, 50% tissue culture infectious dose.
To construct a QD-containing IAV (IAV-QD), we first generated the recombinant IAV rPR8-PA–AP in which the PA protein is fused with an AP tag (SI Appendix, Fig. S1A). The recombinant virus rPR8-PA–AP was rescued by cotransfection with the other seven wild-type (WT) PR8 plasmids into 293T cells. rPR8-PA–AP virus was then isolated by plaque purification (SI Appendix, Fig. S1B) and detected by RT-PCR (SI Appendix, Fig. S1C). These purified clonal stocks were amplified in chicken embryos for further use. The recombinant rPR8-PA–AP virus has an infectivity similar to that of WT PR8 virus (SI Appendix, Fig. S1D) and exhibited spheroidal morphology and looked the same as the WT virus (SI Appendix, Fig. S1E). The recombinant PA–AP segment could be readily detected from passages 1 to 6 and no mutations were present (SI Appendix, Fig. S1F). These data indicate that we successfully constructed a recombinant virus possessing an AP tag fused to the C terminus of PA and retaining WT-like infectivity.
The vRNPs were then labeled by QDs during viral assembly in rPR8-PA–AP virus-infected cells. Specifically, pcDNA3.1(+)-BirA was transfected into Madin-Darby Canine Kidney (MDCK) cells, and 6 h later, cells were infected with rPR8-PA–AP viruses and biotin was added. After a 2-h incubation, SA-QDs were transfected into the MDCK cells by Lipofectamine. The suspension containing IAV particles encapsulating QDs (IAV-QDs) was collected and purified by sucrose density gradient ultracentrifugation (SDGC). As shown in Fig. 1B, a band with a fluorescence signal could be detected in the SDGC tube. The fluorescence spectrum of the IAV-QDs was similar to that of SA-QDs (Fig. 1C). Western blot results verified that the PA protein was biotinylated in IAV-QDs (Fig. 1D). Purified IAV-QDs were overlaid onto coverslips and immunostained with specific antibodies against IAV M1 protein and NP. The majority of QDs (∼80.1%) were colocalized with M1 or NP (Fig. 1E). When there was no biotin addition or BirA coexpression during the assembly procedure, as controls, no QD fluorescence was observed in the resulting IAV particles. These results demonstrate that QDs were successfully incorporated into IAV virions.
IAV-QDs were then characterized by transmission electron microscopy (TEM). In our constructed IAV-QDs, dark electron-dense QDs were clearly detected inside IAV virions. No such dots were observed in WT PR8 virions, rPR8-PA–AP virions, or virions assembled without BirA coexpression or biotin (Fig. 1F). During our TEM imaging of IAV-QDs, 66.3% of the 821 analyzed virions carried one QD, 23.6% carried two QDs, and 10.1% carried three to five QDs (Fig. 1G). These viruses showed typical IAV morphology, indicating that QD-vRNPs did not interfere with viral maturation morphogenesis. TEM imaging confirmed that, as designed, the QDs were specifically encapsulated into IAV particles by labeling vRNPs.
We evaluated the infectivity of IAV-QDs by comparing the growth curves of IAV-QDs, rPR8-PA–AP, and WT PR8 in MDCK cells from 50% tissue culture infectious dose assays. The growth rate of IAV-QDs was only slightly affected compared with those of rPR8-PA–AP and WT PR8 (Fig. 1H). The IAV-QDs also had similar entry kinetics to WT PR8 virus (SI Appendix, Fig. S2).
Tracking Infection of Individual Virions Using IAV-QDs.
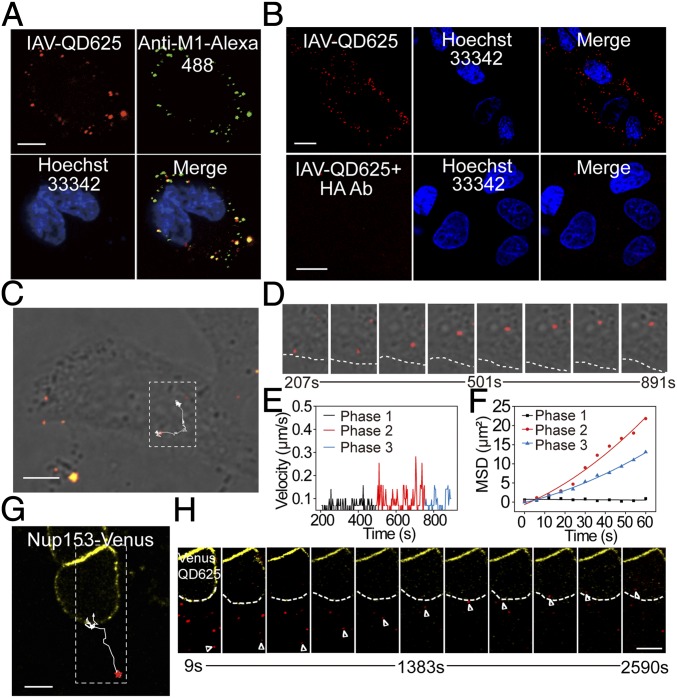
IAV-QDs were added to host cells for tracking the virus infection processes. When IAV-QDs were incubated with MDCK cells at 4 °C, they attached on the cell membrane (Fig. 2A). When the cells were immunostained with specific antibodies against the IAV M1 protein, most of the QD signals colocalized with the anti-M1 antibody signals. This further verified that the QD signals represent virus particles.
Fig. 2.
Real-time tracking analysis of IAV-QD infection. (A) Immunofluorescence assay of IAV-QD625 attachment on MDCK cells. (B) Images of MDCK cells infected with IAV-QD625 or IAV-QD625 that were neutralized by anti-HA antibody. (C) Image showing the trajectory of an IAV-QD625 virion in cytoplasm. (D) Sequential images of the amplified rectangular region in C. The cellular boundary is highlighted by a dashed line. (E) Analysis of mean velocities of IAV-QD625 in C. (F) MSD plots of IAV-QD625 in C. (G) Image showing the trajectory of an individual IAV-QD625 virion from the cytoplasm to the nucleus. The nuclear membrane is highlighted by Nup153-Venus. (H) Snapshots of real-time tracking of IAV-QD625 in G. Images were taken at 60× magnification objective lens under a confocal microscope. (Scale bar: A–C and G, 10 µm; H, 10 µm.)
When IAV-QDs were incubated with MDCK cells at 37 °C, they entered the cells, and QD signals were observed in the cytoplasm (Fig. 2B). We also performed a viral neutralization test to confirm that the cytoplasmic QD signals were indeed due to virus infection. The resulting QD signals indicated that when neutralized by anti-HA antibody, the IAV-QD particles could barely attach to and enter the cells (Fig. 2B). These results indicate that QD internalization was directed by IAV-QD infection.
We next tracked the infection process of individual IAV-QDs under an UltraView Vox spinning disk confocal laser scanning system. We recorded the virus dynamic behaviors at 3-s intervals for 20 min postinfection. Single IAV-QD particles could be tracked in real time. The dynamic intracellular behavior of a representative virus particle is shown in Fig. 2C and Movie S1. We recorded 630 individual trajectories, from which we calculated velocities and mean square displacements (MSDs) (Fig. 2 D–F). These IAV-QDs exhibited a typical three-phase transport process before fusion, as reported previously (3). Briefly, IAV-QDs first move slowly in the cell periphery (phase 1) and then transport rapidly toward the nucleus (phase 2), followed by slow and intermittent movement in the perinuclear region (phase 3).
In addition to their use in tracking the infection process before viral fusion, IAV-QDs could also be tracked for over 1 hour postinfection (hpi) to monitor the whole early infection process from viral entry to vRNP trafficking in the nucleus. As shown in Fig. 2 G and H and Movie S2, a virion encapsulating QD625-labeled vRNPs (IAV-QD625) in a cell with a Nup153-Venus–labeled nuclear membrane was tracked for a long time. The processes of cytoplasmic transport, nuclear entry, and intranuclear trafficking for this virus particle were captured in real time. Thus, our constructed IAV-QDs are a powerful tool for studying viral infection.
Visualizing the Uncoating of Individual IAV-QD Virions by Decorating the Surface with Differently Colored QDs.
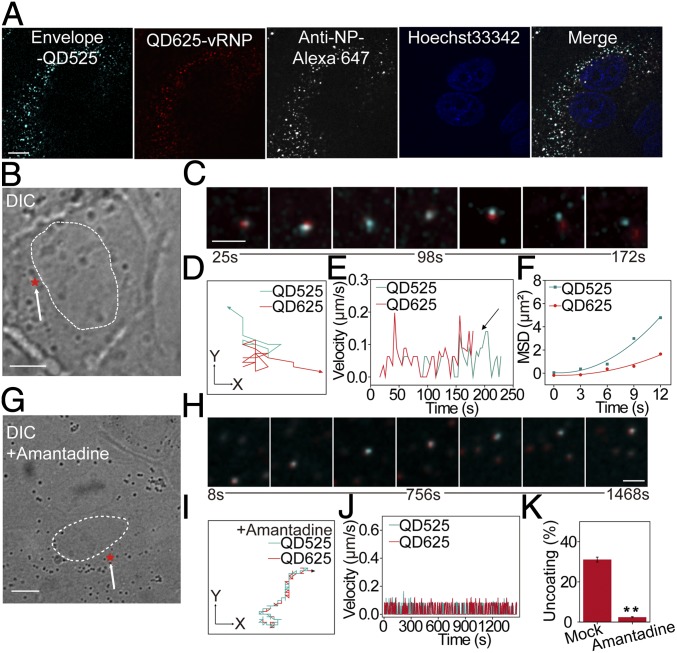
We next studied the virus uncoating process by combining internal encapsulation and surface decoration using differently colored QDs. IAV-QD625 virions were used for this dual-color labeling. The surface envelope was labeled with QD525s as reported previously (10). Colocalization of the dual-color QD signals with anti-NP antibody signals verified the QD525 surface decoration of IAV-QD625 (Fig. 3A).
Fig. 3.
Dual-color visualization of IAV uncoating in live cells. (A) MDCK cells with dual-color IAVs (encapsulating QD625 and decorated with QD525 on surface) and immunostained with anti-NP antibody. (Scale bar: 5 µm.) (B) Differential interference contrast (DIC) image of an infected cell. The nucleus boundary is highlighted by a dashed line, and the arrow marks the uncoating site (red star). (Scale bar: 10 µm.) (C) Snapshots of the separation of QD625-vRNP (red) and QD525-viral envelope (cyan). (Scale bar: 2 µm.) (D) Trajectories of the two QD fluorescent dots in C. (E) Analysis of the mean velocities of these two fluorescent dots. The arrow marks the significant difference in the velocities during separation. (F) MSD plots of these two fluorescent dots. (G) DIC image of an infected cell treated with amantadine. The nucleus boundary is highlighted by a dashed line, and the arrow marks the initiation site (red star). (Scale bar: 10 µm.) (H) Snapshots of the virus shown in G. (Scale bar: 2 µm.) Trajectories (I) and the mean velocities (J) of the colocalized fluorescent dots from H. (K) Analysis of the effect of amantadine treatment on IAV uncoating. A total of 1,055 trajectories in drug-treated cells and 1,160 trajectories in untreated cells were analyzed. **P < 0.01.
We tracked the uncoating of individual virions in living cells from 20 min postinfection. The particles with fluorescent signals from both QD525 and QD625 were counted as dual-color IAV virions and tracked as merged gray fluorescent dots. During cytoplasmic transportation of a dual-color virion, the QD625 dot (red) separated from the QD525 dot (cyan) in the perinuclear region, indicating that the vRNPs were released from the viral envelope (Fig. 3 B and C and Movie S3). The dynamic trajectories, velocities, and MSDs of the two QD signals are shown in Fig. 3 D–F. These real-time tracking results describe the dynamic uncoating process by which vRNPs egress from the viral envelope and are released into the cytosol. For this experiment, 1,160 individual trajectories of dual-color QD-labeled IAV virions in cells were tracked and analyzed. Approximately 31.1% of these IAV particles underwent uncoating at 30 to 90 min postinfection.
Virus uncoating was also observed in amantadine-treated cells (13). The inhibition of amantadine on viral replication was first tested under our experimental conditions (SI Appendix, Fig. S3). In MDCK cells treated with amantadine, the QD625 dots barely separated from the QD525 dots of dual-color virions (Fig. 3 G–J and Movie S4), suggesting that the uncoating process was inhibited. Our statistical analysis of 1,055 trajectories shows that the efficiency of the uncoating process decreased sharply (2.4%) after cell treatment with amantadine (Fig. 3K).
Visualization of QD-vRNP Release from Late Endosomes.
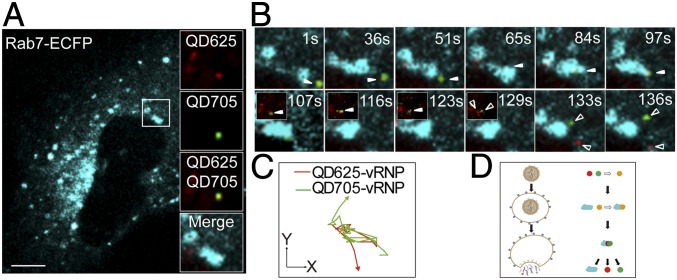
Because IAV uses the endocytic entry pathway and endosomal fusion for uncoating, we also tracked the vRNP release from late endosomes by using IAV-QDs and MDCK cells transiently expressing Rab7-enhanced cyan fluorescent protein (Rab7-ECFP) to mark late endosomes. Fig. 4 A and C and Movie S5 show a typical sequential dynamic interaction process between an IAV-QD virion and late endosome. The sequential images show that the QD625 signal (red) initially overlapped with the signal from the late endosome marker Rab7 (cyan), after which they were transported together from the cytoplasm to the perinuclear region. During the cytoplasmic transportation, endosomal fusion events between vRNP-positive and -negative endosomes happened several times. Finally, the QD625-vRNP was released from the Rab7-marked late endosome (Rab7-endosome) in the perinuclear region. This single-particle tracking clearly uncovered a sequential event in which endosomes containing QD-vRNPs are transported and fused with other endosomes before finally releasing the QD-vRNPs into the cytoplasm. The vRNP release from a late endosome mainly occurs in the perinuclear region at 30 to 90 min postinfection.
Fig. 4.
Real-time imaging of vRNP release from a Rab7-positive endosome. (A) IAV-QD625 (red) colocalization with a Rab7-ECFP–positive endosome (cyan) was tracked. (Scale bar: 5 µm.) (B) Sequential snapshots are shown for the separation of the QD signal and Rab7 signal in the rectangular region of A. Filled arrowheads indicate the colocalization of QD and Rab7 signals, arrows indicate virus-negative endosomes, and hollow arrowheads indicate the released vRNP. (C) Trajectories of the two fluorescent dots from B. (D) Fluorescence image of an infected cell treated with NH4Cl. (Scale bar: 5 µm.) (E) Sequential images of the rectangular regions of D. (F) Trajectories of the QD625 and ECFP fluorescent signals in NH4Cl-treated cells. Images were taken at 60× magnification objective lens under a confocal microscope.
We also tracked the QD-vRNP release in cells treated with ammonium chloride (NH4Cl), which blocks endosome acidification and maturation. In NH4Cl-treated MDCK cells, IAV-QD virions rarely colocalized with Rab7-endosomes: 912 virions were randomly selected for statistical analyses, and only 2.6% of IAV-QD virions were colocalized with Rab7-endosomes in NH4Cl-treated cells. Single-particle tracking showed that IAV-QD625s did not enter into Rab7-endosomes under these conditions (Fig. 4 D–F and Movie S6). Coexpression of Rab5 and Rab7 showed that IAV-QDs were trapped in Rab5-endosomes when cells were treated with NH4Cl (SI Appendix, Fig. S4 C and D and Movie S7). Coimaging of dual-color (QD625-envelope and QD705-vRNP) IAV virions and Rab7-positive endosomes further showed that dual-color virions fused and penetrated from these endosomes (SI Appendix, Fig. S5 A and B and Movie S8). This process was also inhibited by NH4Cl (SI Appendix, Fig. S5 C and D and Movie S9).
Visualization of IAV vRNP Separation into Individual Parts by Using IAV Encapsulating Dual-Color QD-vRNP Segments.
To investigate whether vRNP segments are released separately or as a package during uncoating, IAV virions encapsulating dual-color QD-vRNP segments were constructed (Fig. 5A). Because each vRNP segment is ligated with only one QD, we were able to incorporate differently colored QD-vRNP segments into single virions. Here, SA-QD625 and SA-QD705 were incorporated into IAV particles by our labeling procedure. A proportion of IAV particles encapsulating both QD625 and QD705, named IAV-QD(625+705), were acquired (26.2%), although there were also IAV particles encapsulating only QD625 or QD705. When characterized by negative staining in TEM, individual QD625s are round and QD705s appear triangular. Accordingly, inside virions, QD625 looks dark and round, whereas QD705 looks triangular (Fig. 5B). Colocalization of QD625 and QD705 with NP was observed for dual-color QD-labeled IAV particles by immunofluorescence staining (Fig. 5C). In our study, the stability of IAV-QDs was also tested through in vitro disassembly experiments. Virion acidification and potassium treatment assay (14) showed that IAV-QD(625+705) had disassembly properties similar to WT virus (SI Appendix, Fig. S6), indicating that QD incorporation did not affect virus stability.
Fig. 5.
Characterization of IAV-QD(625+705) and visualization of IAV vRNP separation into individual parts at the AN region. (A) Schematic representation of IAV-QD(625+705). (B) TEM images. (Scale bar: 50 nm.) The white arrowheads indicate QD625, and the hollow arrowheads indicate QD705. (C) Immunofluorescence assay of IAV-QD(625+705) on coverslips. (Scale bar: 2 µm.) (D) Image of an infected cell. The nucleus boundary, labeled by Nup153-Venus, of a host cell is highlighted by a dashed line. The arrow marks the separation site. (Scale bar: 10 µm.) (E) Colocalization of QD625 and QD705. (F) Trajectories of QD625 and QD705. (G) Snapshot of the real-time separation of the QD625-vRNP and QD705-vRNP from D. (Scale bar: 1 µm.) (H) Differential interference contrast (DIC) image of an infected cell. The nucleus boundary is highlighted, and the arrow marks the uncoating site (red star). (Scale bar: 10 µm.) (I) Snapshots of the separation of QD625-vRNP (red), QD705-vRNP (green), and QD525-viral envelope (cyan). After the shift of cyan from green and red, the green and red were separated from each other. (Scale bar: 2 µm.) (Insets) Unmerged panels for QD625-vRNP and QD705-vRNP. Arrowheads indicate the separation of three QD signals. (J) Diagram of the two regions of cytoplasm. (Scale bar: 10 µm.) (K) Analysis of IAV vRNP separation in the AN region with and without drug treatment. A total of 1,017 trajectories in drug-treated cells and 1,130 trajectories in untreated cells were analyzed. **P < 0.01. p.i., postinfection. Images were taken at 60× magnification objective lens under a confocal microscope.
These virus particles were added to MDCK cells for tracking of uncoating. The particles with fluorescent signals from both QD625 and QD705 were considered as virions encapsulating dual-color QD-vRNP segments (Fig. 5 D–G). The uncoating process of individual IAV-QD(625+705) virions was tracked in real time. Fig. 5G and Movie S10 show a typical dynamic uncoating process for a dual-color IAV virion. During imaging, QD625-vRNP (red) and QD705-vRNP (green) were separated from each other. Fig. 5F shows the trajectories of QD625 and QD705 for a viral particle during this process. When IAV-QD(625+705) virions were further decorated with QD525 on the viral envelope, triple-color virion imaging was carried out. As shown in Fig. 5 H and I and Movie S11, the triple-color virion was tracked in the cytoplasm, and QD625-vRNP and QD705-vRNP were separated from each other and were released from the QD525 envelope. We observed separation events for 28.5% of the IAV-QD(625+705) virions in our experiments (total of 1,130 trajectories). When host cells were treated with amantadine to inhibit viral uncoating, the separation ratio reduced significantly (5.1% from 1,017 trajectories) (Fig. 5K). These single-particle tracking results indicate that the QD625-vRNP segment and the QD705-vRNP segment were released separately during uncoating.
To determine where the vRNPs separate into individual components, we labeled the nuclear pore complex (NPC) using Nup153-Venus to mark the nuclear membrane. We divided the cytoplasm into two parts: the “around-nucleus” (AN) region within 1 µm of the nucleus, and the “other cellular” (OC) region, which consisted of the areas from the cell membrane to the AN region (Fig. 5J). Through tracking the merged fluorescent dots in live cells, we found that the separation of QD625 and QD705 always occurred in the AN region. These separations took place mostly at 30 to 90 min postinfection, which is consistent with the timing of the vRNP release from the late endosome.
vRNP Segments Are Released Separately from Late Endosomes.
We further tracked the release process of dual-color QD-vRNP segments from the late endosome. IAV-QD(625+705) virions were used to infect MDCK cells with Rab7-ECFP–marked late endosomes. The merged QD625 and QD705 fluorescent dots initially colocalized with the Rab7 fluorescent signals, where they stayed and moved together for a while, and then finally moved away from the Rab7 signals (Fig. 6A and Movie S12). At the same time, the merged fluorescent dot separated into two fluorescent dots, which moved in different directions for the next transportation stage (Fig. 6B). These results further demonstrate that the vRNPs from a single virion separate into individual units soon after fusion and uncoating at the late endosome. About 24.3% of separation events were observed in 1,240 trajectories. In NH4Cl-treated MDCK cells, IAV-QD(625+705) virions could barely enter the late endosomes. Only 2.4% of IAV-QD(625+705) virions (1,190 trajectories) colocalized with Rab7-endosomes at 30 to 90 min postinfection, and no separation events were observed.
Fig. 6.
Visualization of IAV vRNP segments released separately from late endosomes. (A) Representative image of an IAV-QD(625+705) particle localized with a Rab7-ECFP–marked endosome. (Scale bar: 10 µm.) (Right) Magnified views of the colocalized fluorescent signals.(B) Sequential snapshots of the colocalization of an IAV-QD(625+705) particle (yellow) with an Rab7-ECFP–marked endosome (cyan) and the subsequent egress and separation of QD625-vRNP (red) and QD705-vRNP (green) from the endosome (cyan) shown in the rectangular region of A. (Insets) Unmerged panels for IAV-QDs. It was likely that the green/red spots were retained on the surface of Rab7-endosomes before eventually separating into two individual units. Hollow arrowheads indicate QD signals, and filled arrowheads indicate colocalized QD signals. (C) Trajectories of the two fluorescent dots from B. (D) Schematic presentation of the release of individual vRNPs from Rab7-positive endosomes into the cytosol. Images were taken at 60× magnification objective lens under a confocal microscope.
Visualization of Nuclear Import and Intranuclear Dynamics of QD-vRNPs.
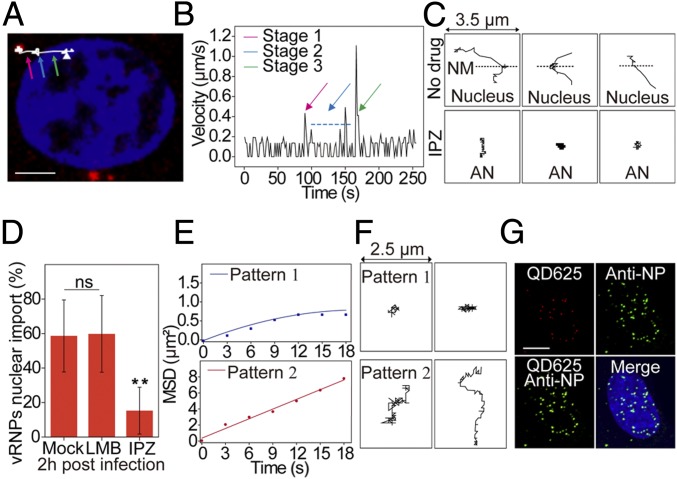
After uncoating and late endosome escape, QD-vRNPs were delivered into the AN region and transported into the cell nucleus. The vRNP dynamics of this process were tracked in real time. Fig. 7A and Movie S13 show a typical trajectory of QD625-vRNP movement from the AN region to the nucleus. The time trajectories of the virus velocities are shown in Fig. 7B. These results illustrate a three-stage transport pattern: stage 1, QD-vRNPs move with a mean velocity of 0.34 ± 0.07 µm/s from the AN region to the nucleus boundary; stage 2, they associate with and cross the nuclear membrane with a vibratory movement, taking 33 to 89 s; and stage 3, they undergo rapid and unidirectional movement in the nucleus for several seconds (SI Appendix, Fig. S7D) with an instantaneous velocity of 0.2 to 1.1 µm/s (127 measurements) to arrive at their nuclear loci. A portion of QD-vRNPs (41.4%) did not complete nuclear entry (Fig. 7C and SI Appendix, Fig. S7 A and B): some of them (23.5%) moved only in the cytoplasmic AN region without progressing to stages 2 and 3, and the others (17.9%) anchored on the nuclear membrane but failed to enter into the nucleus (SI Appendix, Fig. S7C). When the cells were treated with importazole, which blocks the nuclear import pathway, the majority of QD-vRNPs were limited to the cytoplasm, and the nuclear import efficiency was significantly reduced (Fig. 7 C and D).
Fig. 7.
Characterization of vRNP nuclear import and intranuclear movement. (A) Trajectory of a representative QD625-vRNP transport from the AN region to the nucleus. The colored arrows indicate stage 1 (pink), stage 2 (blue), and stage 3 (green). (Scale bar: 5 µm.) (B) Analysis of the mean velocities of QD625-labeled vRNPs shown in A. (C) Trajectories of the QD signals in untreated and importazole (IPZ)-treated cells. NM, nuclear membrane. (D) Statistical analysis of IAV vRNP nuclear import with drug treatment. ns, no significant difference; n = 3; **P < 0.01. (E) Analysis of the MSD plots of two vRNP diffusive patterns in the nucleus. (F) Trajectories of two vRNP diffusive patterns. (G) Immunofluorescence assay of QD625-vRNPs in the nucleus at 2 hpi. (Scale bar: 5 µm.)
We next tracked the diffusion of QD-vRNPs within the nucleus after their three-stage active transport there. Analysis revealed two different patterns of QD-vRNP diffusion in the nucleus (Fig. 7 E and F). Pattern 1 (41.7%) had movement typical of corralled diffusion, suggesting that the vRNPs were constrained in the distance they could reach, with a mean velocity of 0.050 ± 0.006 µm/s (diffusion coefficient: 0.011 ± 0.007 µm2/s) (15). Pattern 2 is a simple diffusive movement pattern with a mean velocity of 0.091 ± 0.028 µm/s (diffusion coefficient: 0.071 ± 0.029 µm2/s). We also verified the QD-vRNP colocalization in the nucleus at 2 hpi by immunofluorescence with anti-NP antibody (Fig. 7G). Our data also show that QD-vRNPs distribute randomly in the nucleus with no obvious specific regional preference.
Discussion
Historically, tracking virus uncoating during entry has been a challenge. IAV uncoating is particularly mysterious because this virus contains eight negative-sense RNA segments. Here, we developed a method to label single vRNP segments with QDs and to image IAV uncoating via single-particle tracking. We used SA-QDs to label biotinylated PA, which is bound on the terminal of vRNPs. Using this strategy, we successfully constructed IAV particles containing QD-vRNPs. This strategy offers advantages such as precise site-specific labeling and being easy to control. Furthermore, compared with other infectious reporter IAVs harboring GFP or luciferase, QD incorporation showed less impact on viral infectivity (16, 17). As there is only one copy of PA on each vRNP, each vRNP segment inside the progeny virion is labeled with only one QD. Thus, when two differently colored QDs were used during assembly, dual-color QD-vRNP–labeled IAV particles were produced. Although it is still not feasible to label all eight vRNP segments with QDs, possibly due to virion capacity and labeling efficiency, our QD–PA conjugation provides a good strategy to site-specifically label vRNP segments in infectious IAVs with QDs. In addition, the strategy of QD internal encapsulation can be combined with surface decoration to label virus particles with differently colored QDs. Due to the superior optical properties of QDs, single virions and single vRNPs were able to be tracked over extended periods of time.
Using single-virus tracking, the release of single QD-vRNPs from the IAV viral envelope during uncoating was observed in real time. Endosome recruiting and vRNP-QD release from late endosomes into the cytoplasm were also tracked during virus transport and uncoating. Approximately 30% of IAV particles undergo uncoating through fusion with late endosomes in the AN region at 30 to 90 min postinfection. Treatment with NH4Cl or amantadine decreased the uncoating efficiency, further demonstrating that viral M2 channels and cellular endosome acidification are critical for IAV uncoating.
Interestingly, when tracking IAV particles encapsulating dual-color QD-vRNPs, we directly observed that the vRNPs from a virion separate into individual units after fusion and uncoating at the late endosome. The real-time imaging results clearly show that different vRNP segments are released separately during uncoating and that vRNPs enter the nucleus separately rather than as a package. A previous FISH analysis study proposed that viral RNAs travel together until they reach the nucleus (9). Here, our direct-tracking method provides evidence supporting that IAV virions release segment vRNPs separately. We did not observe vRNPs as a bundle being released from virions and transported to the nucleus. We speculate that if vRNPs could be transported as bundles, this might be a rare event. The diameter of the NPC is ∼10 nm and can increase to 30 nm to let large complexes through (7). Based on vRNP dimensions (single rod: width, 10 to 15 nm; length, 30 to 120 nm), the passage of vRNPs through the NPC as individual rod-shaped segments rather than as a group may be more likely.
A three-stage active nuclear import process was revealed for individual vRNPs after uncoating, and the cytoplasmic–nuclear transportation of vRNPs released from infected virions was observed in real time. Nuclear import of vRNPs is critical for IAV infection because viral RNA replication takes place in the nuclei. This three-stage active transport process may correspond to the importin-α/importin-β1–mediated pathway (18, 19). After nuclear entry, vRNPs displayed two distinct diffusion patterns. The two patterns may be due to two types of microenvironments in the nucleus with distinct viscosities (20, 21). Whether these two diffusion pattern types are related to different functions suitable for further replication and transcription processes remains to be determined.
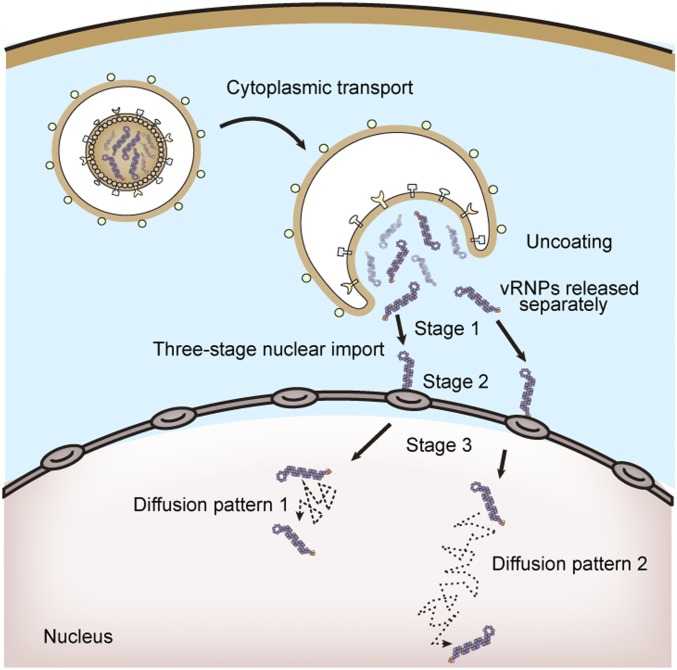
In conclusion, based on the construction of QD-containing infectious IAVs and single-particle tracking, we were able to track the uncoating and vRNP dynamics of individual influenza virions. Our results reveal important uncoating mechanisms (Fig. 8); specifically, (i) IAVs release vRNP segments separately from late endosomes into the AN region, and (ii) after uncoating, individual vRNPs undergo a three-stage active nuclear import process and have two distinct diffusion patterns within the nucleus. These findings facilitate a better understanding of the early stages of the IAV life cycle and may assist in developing new strategies to block IAV infection.
Fig. 8.
Model for IAV uncoating and vRNP dynamics. An IAV virion enters the host cell via endocytosis. It then releases vRNP segments separately from late endosomes through fusion and uncoating regulated by host factors in the AN region at 30 to 90 min postinfection. Individual vRNPs finally undergo a three-stage active transport process to arrive at the cell nucleus and display two diffusion patterns within the nucleus.
Materials and Methods
A detailed description of materials and methods is provided in SI Appendix, Materials and Methods.
Supplementary Material
Acknowledgments
This work was supported by the Strategic Priority Research Program of the Chinese Academy of Sciences (Grant XDB29050201), the National Key Research and Development Program of China (Grant 2018ZX10301405), the National Natural Science Foundation of China (Grants 31470269, 21727816, 91743108, and 31470837), and the Youth Innovation Promotion Association of the Chinese Academy of Sciences.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
See Commentary on page 2404.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812632116/-/DCSupplemental.
References
- 1.Medina RA, García-Sastre A. Influenza A viruses: New research developments. Nat Rev Microbiol. 2011;9:590–603. doi: 10.1038/nrmicro2613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Eisfeld AJ, Neumann G, Kawaoka Y. At the centre: Influenza A virus ribonucleoproteins. Nat Rev Microbiol. 2015;13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakadamyali M, Rust MJ, Babcock HP, Zhuang X. Visualizing infection of individual influenza viruses. Proc Natl Acad Sci USA. 2003;100:9280–9285. doi: 10.1073/pnas.0832269100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rust MJ, Lakadamyali M, Zhang F, Zhuang X. Assembly of endocytic machinery around individual influenza viruses during viral entry. Nat Struct Mol Biol. 2004;11:567–573. doi: 10.1038/nsmb769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Banerjee I, et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science. 2014;346:473–477. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 6.Su WC, et al. Pooled RNAi screen identifies ubiquitin ligase Itch as crucial for influenza A virus release from the endosome during virus entry. Proc Natl Acad Sci USA. 2013;110:17516–17521. doi: 10.1073/pnas.1312374110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutchinson EC, Fodor E. Nuclear import of the influenza A virus transcriptional machinery. Vaccine. 2012;30:7353–7358. doi: 10.1016/j.vaccine.2012.04.085. [DOI] [PubMed] [Google Scholar]
- 8.Babcock HP, Chen C, Zhuang X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J. 2004;87:2749–2758. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chou YY, et al. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 2013;9:e1003358, and correction (2013) 9, 10.1371/annotation/8f53e7f2-2348-436f-b37e-a883a01e9bbd. doi: 10.1371/journal.ppat.1003358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu SL, et al. Effectively and efficiently dissecting the infection of influenza virus by quantum-dot-based single-particle tracking. ACS Nano. 2012;6:141–150. doi: 10.1021/nn2031353. [DOI] [PubMed] [Google Scholar]
- 11.Joo KI, et al. Site-specific labeling of enveloped viruses with quantum dots for single virus tracking. ACS Nano. 2008;2:1553–1562. doi: 10.1021/nn8002136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li Q, et al. Single-particle tracking of human immunodeficiency virus type 1 productive entry into human primary macrophages. ACS Nano. 2017;11:3890–3903. doi: 10.1021/acsnano.7b00275. [DOI] [PubMed] [Google Scholar]
- 13.Hu J, et al. Backbone structure of the amantadine-blocked trans-membrane domain M2 proton channel from Influenza A virus. Biophys J. 2007;92:4335–4343. doi: 10.1529/biophysj.106.090183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stauffer S, et al. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J Virol. 2014;88:13029–13046. doi: 10.1128/JVI.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Shav-Tal Y, et al. Dynamics of single mRNPs in nuclei of living cells. Science. 2004;304:1797–1800. doi: 10.1126/science.1099754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Manicassamy B, et al. Analysis of in vivo dynamics of influenza virus infection in mice using a GFP reporter virus. Proc Natl Acad Sci USA. 2010;107:11531–11536. doi: 10.1073/pnas.0914994107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heaton NS, et al. In vivo bioluminescent imaging of influenza a virus infection and characterization of novel cross-protective monoclonal antibodies. J Virol. 2013;87:8272–8281. doi: 10.1128/JVI.00969-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.O’Neill RE, Jaskunas R, Blobel G, Palese P, Moroianu J. Nuclear import of influenza virus RNA can be mediated by viral nucleoprotein and transport factors required for protein import. J Biol Chem. 1995;270:22701–22704. doi: 10.1074/jbc.270.39.22701. [DOI] [PubMed] [Google Scholar]
- 19.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tseng Y, Lee JS, Kole TP, Jiang I, Wirtz D. Micro-organization and visco-elasticity of the interphase nucleus revealed by particle nanotracking. J Cell Sci. 2004;117:2159–2167. doi: 10.1242/jcs.01073. [DOI] [PubMed] [Google Scholar]
- 21.Vargas DY, Raj A, Marras SA, Kramer FR, Tyagi S. Mechanism of mRNA transport in the nucleus. Proc Natl Acad Sci USA. 2005;102:17008–17013. doi: 10.1073/pnas.0505580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.