Significance
The accumulation of senescent cells over a lifetime causes age-related pathologies; however, the inability to reliably identify senescent cells in vivo has hindered clinical efforts to employ this knowledge as a means to ameliorate or reverse aging. Here, we describe a reporter allele, p16tdTom, enabling the in vivo identification and isolation of cells featuring high-level activation of the p16INK4a promoter. Our findings provide an insight into the functional and molecular characteristics of p16INK4a-activated cells in vitro and in vivo. We show that such cells accumulate with aging or other models of injury, and that they exhibit clinically targetable features of cellular senescence.
Keywords: senescence, cdkn2a, aging
Abstract
The activation of cellular senescence throughout the lifespan promotes tumor suppression, whereas the persistence of senescent cells contributes to aspects of aging. This theory has been limited, however, by an inability to identify and isolate individual senescent cells within an intact organism. Toward that end, we generated a murine reporter strain by “knocking-in” a fluorochrome, tandem-dimer Tomato (tdTom), into exon 1α of the p16INK4a locus. We used this allele (p16tdTom) for the enumeration, isolation, and characterization of individual p16INK4a-expressing cells (tdTom+). The half-life of the knocked-in transcript was shorter than that of the endogenous p16INK4a mRNA, and therefore reporter expression better correlated with p16INK4a promoter activation than p16INK4a transcript abundance. The frequency of tdTom+ cells increased with serial passage in cultured murine embryo fibroblasts from p16tdTom/+ mice. In adult mice, tdTom+ cells could be readily detected at low frequency in many tissues, and the frequency of these cells increased with aging. Using an in vivo model of peritoneal inflammation, we compared the phenotype of cells with or without activation of p16INK4a and found that tdTom+ macrophages exhibited some features of senescence, including reduced proliferation, senescence-associated β-galactosidase (SA-β-gal) activation, and increased mRNA expression of a subset of transcripts encoding factors involved in SA-secretory phenotype (SASP). These results indicate that cells harboring activation of the p16INK4a promoter accumulate with aging and inflammation in vivo, and display characteristics of senescence.
Cellular senescence refers to a specific form of highly durable cell cycle arrest of previously proliferation-competent cells that is resistant to mitogenic stimulation and accompanied by persistent DNA damage response. Senescence is an important tumor-suppressor mechanism, and is believed to contribute to organismal aging (1, 2). A senescence response is triggered by a variety of genotoxic stresses, including shortened telomeres, exposure to DNA damaging agents, and oncogenic insult (1, 3). While senescence is primarily characterized in replication-competent cells, recent studies have suggested that largely postmitotic cell types can also initiate a senescence program (4, 5). In addition to growth arrest, senescence is variably associated with the expression of cyclin-dependent kinase (CDK) inhibitors (especially p16INK4a), senescence-associated β-galactosidase (SA-β-gal) activity, and the elaboration of cytokines that comprise the SA-secretory phenotype (SASP) (3, 6). Given the prominence of senescence in cancer and aging, there has been great interest in the identification and characterization of senescent cells in an intact adult organism.
Although senescent cells are well-characterized in culture, identifying senescent cells in vivo has been challenging (6). The inability to reliably identify senescent cells in an intact organism has impaired the study of their precise role in tumor suppression and physiological aging. To date, activation of p16INK4a expression has proven to be one of the most useful in vivo markers of senescence. As a cell cycle regulator, p16INK4a limits G1 to S-phase progression of the cell cycle through inhibition of the CDK4 and CDK6 (CDK4/6) kinases (7). Moreover, the expression of p16INK4a is highly dynamic, being largely undetectable in healthy young tissues, but rising sharply in many tissues with aging (8, 9) or after certain sorts of tissue injury (10–12). Murine studies suggest that accumulation of p16INK4a leads to an age-related loss of replicative capacity in select tissues, thereby causing some phenotypic aspects of aging (13–16). The clearance of p16INK4a-expressing cells attenuates age-associated phenotypes and improves the healthy lifespan of progeroid and physiologically aged mice (17, 18). These murine results are underscored by a remarkable string of associations of the CDKN2a/b locus (encoding the p16INK4a, ARF, and p15INK4b transcripts) with human age-related phenotypes by genome-wide association studies (19, 20).
In prior work, activation of the p16INK4a promoter has been used to suggest senescence in vivo. Our laboratory and others have placed reporter genes [e.g., luciferase (LUC)] under the control of the p16INK4a promoter by either transgenic (10, 17, 21, 22) or knockin approaches (23). These reporter alleles have been employed to demonstrate that the p16INK4a promoter activity increases during wounding, inflammation, tumorigenesis, or aging in vivo in tissues. While valuable for studies at the tissue or organ level, these alleles have been limited in their ability to detect and isolate individual cells with strong activation of the p16INK4a promoter in vivo. To study individual p16INK4a-activated cells, we have generated a fluorescence-based reporter allele with tandem-dimer Tomato (tdTom) knocked into the endogenous p16INK4a locus. This allele enables the identification and isolation of p16INK4a-activated cells (tdTom+) at the single-cell level from cultured cells and in vivo. Using this allele, we quantified tdTom+ cells in several tissues with aging or in the setting of inflammation, and isolated these cells for characterization in terms of function and gene expression.
Results
Generation and Characterization of the p16tdTom Allele.
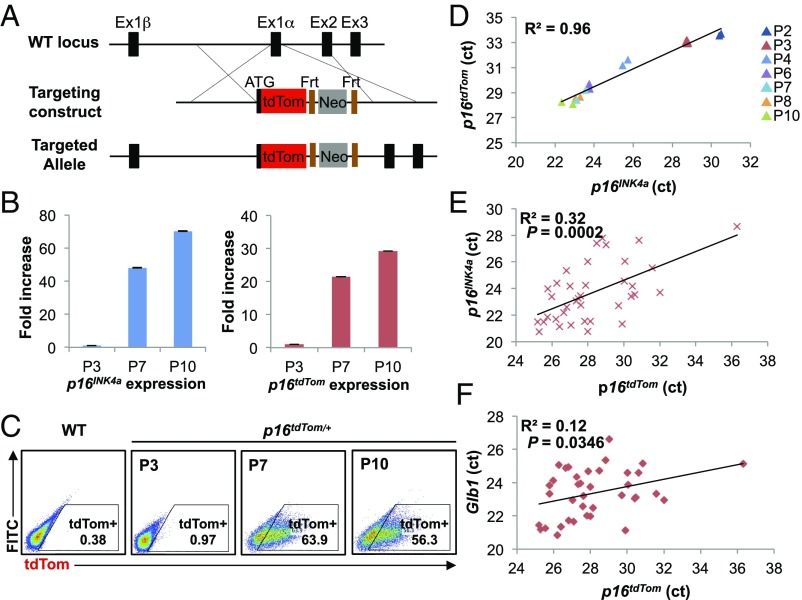
To study individual p16INK4a-expressing cells from in vivo sources, we knocked an ORF encoding a fluorescent reporter protein (tdTom) into the endogenous first exon (exon 1α) of p16INK4a through homologous recombination (Fig. 1A). The targeted allele (p16tdTom) was designed to be null for p16INK4a expression, yet with unperturbed expression of the Arf transcript, as well as retention of cis-regulatory elements around the Cdkn2a (or Ink4a/Arf) locus. A stop codon and poly-A signal were included at the end of the knocked-in tdTom ORF, and therefore the targeted mRNA would not be expected to produce a message that splices to exon 2. Importantly, a flippase recognition site (FRT)-flanked neomycin selection cassette under the regulation of a strong PGK promoter was knocked into the first intron to allow for ES cell selection (Fig. 1A). Correct homologous targeting was verified by PCR, sequencing, and Southern blot (SI Appendix, Fig. S1 A and B).
Fig. 1.
Design and validation of the p16tdTom allele. (A) Schematic of the p16tdTom knockin targeting strategy. Frt, flippase recognition site; Neo, neomycin resistance gene. (B–D) Induction of p16INK4a and tdTom expression in p16tdTom/+ MEFs over serial passage. P3, passage 3; P7, passage 7; P10, passage 10. mRNA expression of p16INK4a and tdTom by qRT-PCR. Fold-increase was calculated with respect to the mRNA levels at P3. Data shown correspond to three biological replicates. Error bars represent SEM (B). Representative flow cytometric (FACS) analysis of MEFs at indicated passage number (C). Correlation of p16INK4a and p16tdTom mRNA expression shown in the normalized threshold cycle (ct) value. Passage number is presented by different colors. Linear regression was used to calculate the coefficient of determination (R2) (D). (E and F) Linear correlations between p16INK4a, p16tdTom, and Glb1 mRNA expression on single-cell levels. Expression levels are shown in the comparative threshold cycle (ct) values. Each dot represents a single cell.
Prior efforts in our laboratory failed to produce a usable single-cell reporter allele through similar approaches. Despite correct knockin targeting, alleles featuring p16INK4a-GFP or p16INK4a-CRE recombinase (which could be used to activate Lox-STOP-Lox GFP alleles) were abandoned due to low expression of the knocked-in ORF. Therefore, to maximize reporter expression with the tdTom knockin allele, we did two additional things. First, we chose to knock in tdTom, which is six times brighter than GFP but without the predisposition to aggregation and toxicity of other red fluorescent alleles (24). Second, given prior work showing that a retained neomycin cassette can act as a local enhancer to augment reporter expression without compromising tissue reporter fidelity (25, 26), we elected to characterize the p16tdTom allele in cells with both a retained neomycin selection cassette and after flippase (FLP)-mediated excision.
We assessed the allele function in cultured murine embryonic fibroblasts (MEFs) heterozygous for the tdTom knockin (p16tdTom/+). In line with prior results (23, 27, 28), p16tdTom/+ and WT MEFs have comparable growth characteristics at early passage (SI Appendix, Fig. S1C). As expected (29, 30), serial passage induced increasing expression of endogenous p16INK4a mRNA produced from the WT locus and tdTom transcript from the p16tdTom allele (Fig. 1B). The frequency of tdTom+ cells identified by flow cytometry also increased with passage (Fig. 1C). While the frequency of tdTom+ MEFs increased with passage in cells derived from mice retaining (“Neo-in”) or without (“Neo-out”) the neomycin cassette, the frequency of tdTom+ cells in Neo-in cultures was an order-of-magnitude higher. For example, we show a comparison of MEFs at passage 7 (∼25 d in culture) where 1.9% of Neo-out cells and 19.6% of Neo-in cells were tdTom+ (SI Appendix, Fig. S2A). To determine which allele was more faithful to endogenous p16INK4a expression, we measured tdTom and p16INK4a transcript levels in heterozygous cells (Neo-in/WT or Neo-out/WT) by qRT-PCR. As was the case for tdTom protein, the tdTom mRNA increased with passage in cells derived from both Neo-in or Neo-out mice, but mRNA expression was more dynamic with passage in Neo-in cells (SI Appendix, Fig. S2B). Correspondingly, tdTom mRNA expression in Neo-in/WT cells strongly correlated with p16INK4a expression when measured in multiple independent MEF lines (n = 6) at various passages (Fig. 1D) (R2 = 0.96). In contrast, tdTom expression in Neo-out cells was more frequently below the lower limit of detection, and therefore correlated less well with endogenous p16INK4a mRNA (SI Appendix, Fig. S2C) (R2 = 0.73). Given the higher fidelity and stronger signal using the Neo-in allele, we elected to pursue all subsequent experiments using this version, and henceforth “p16tdTom” will designate the Neo-in allele. To test the functionality of this single-cell reporter, we assessed the expression of tdTom and p16INK4a on the single-cell level. By single-cell qRT-PCR, we noted a significant association between tdTom and p16INK4a expression (Fig. 1E) (R2 = 0.32; P = 0.0002). Additionally, we found tdTom levels significantly correlated with β-galactosidase (Glb1) levels as well (Fig. 1F) (R2 = 0.12; P = 0.0346).
The p16tdTom Allele Reports Promoter Activation.
Although expression of the reporter transcript strongly correlated with that of the WT allele (Fig. 1 D and E), the absolute level of tdTom was lower than that of the native p16INK4a transcript in cultured p16tdTom/+ MEFs (SI Appendix, Fig. S3A). This pattern was also noted with the previously published p16LUC allele (23). To explain this recurrent finding that expression of the endogenous allele was reproducibly 5- to 10-fold higher than the knocked-in allele in heterozygous cells, we considered the possibility that mRNA stability differs between the various knocked-in ORFs versus the endogenous p16INK4a transcript. Hara et al. (31) reported that both the p16INK4a and ARF mRNAs exhibit a remarkable stability, with half-lives that are >24 h, a finding that our group has confirmed in human and murine cells (32). Of note, >95% of linear, coding mRNAs exhibit a half-life of less than 12 h (33), indicating that this is an unusual feature of the INK4a/ARF transcripts. We used actinomycin D (ActD) treatment followed by qRT-PCR quantification to determine the mRNA half-life of the endogenous p16INK4a and Arf mRNAs, as well as the tdTom mRNA in p16tdTom/+ MEFs (SI Appendix, Fig. S3B). As expected, the p16INK4a and Arf transcripts showed little change for up to 24 h after ActD treatment, whereas the tdTom transcript exhibited a half-life of ∼12 h (SI Appendix, Fig. S3B). The decreased stability of these knocked-in transcripts occurs despite the inclusion of a 3′ poly-A signal derived from SV40 that augments transcript stability in other systems (34). These observations indicate that the lower expression of transcripts knocked into exon 1α of the Cdkn2a locus, and commensurately weak expression of reporter proteins, reflects the fact that the knocked-in transcripts do not recapitulate the extraordinary stability of the endogenous p16INK4a mRNA.
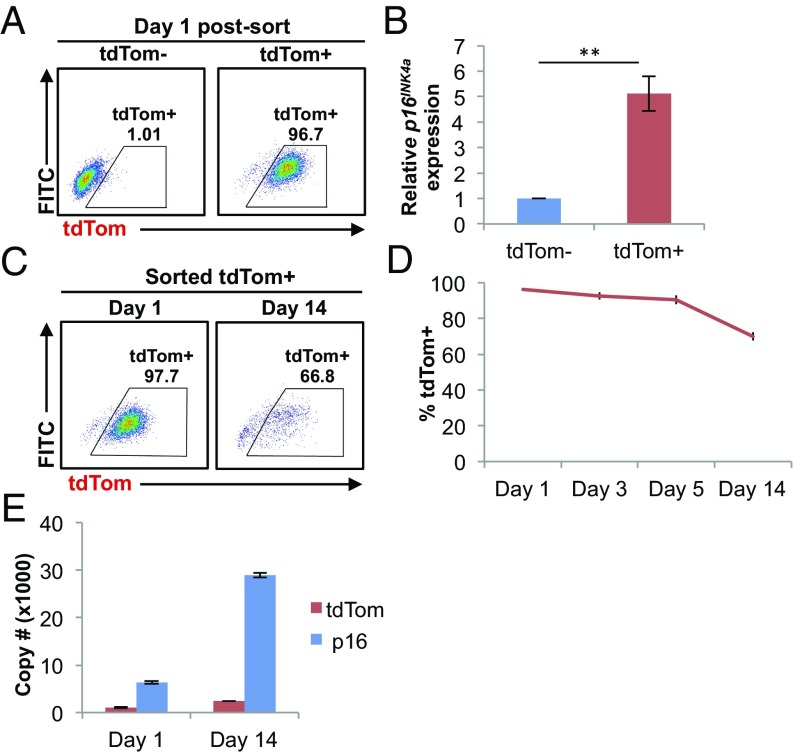
We next examined the levels of p16INK4a mRNA in p16tdTom/+ cells that were either tdTom+ or tdTom−. We sorted MEF cultures using FACS based on tdTom expression (Fig. 2A) and measured p16INK4a mRNA levels in each fraction (Fig. 2B). We noted a moderate enrichment of p16INK4a mRNA in tdTom+ MEFs (∼fivefold), but p16INK4a mRNA was readily detectable in tdTom− fractions at all passages tested. Given the observed differences in p16INK4a vs. tdTom transcript half-life, we analyzed their relative expressions in MEF cultures for up to 14 d postsorting. When analyzed in this way, we noted that expression of tdTom protein and p16INK4a mRNA began to diverge with long-term culture. At day 1 postsort, virtually all cells in the tdTom+ fraction remained tdTom+, but this fraction steadily declined with culture, such that at 14 d post-FACS, >30% of formerly tdTom+ cells had reverted to a tdTom− state (Fig. 2 C and D). We believe this largely reflects reversion of tdTom+ cells as opposed to overgrowth of the culture by tdTom− cells for two reasons. First, the proliferation rate in the tdTom+ cultures remained low for 2 wk after sorting, which would not be consistent with an expansion of proliferating tdTom− cells. Moreover, during the period of culture, tdTom+ MEFs exhibited a significant further accumulation of p16INK4a transcript (Fig. 2E), which would not be expected with expansion of tdTom− cells. Of note, levels of tdTom mRNA in cells sorted as tdTom+ on day 1 were little changed over 14 d of culture (Fig. 2E). These results suggest that while tdTom protein expression has “peaked” at the time of FACS and then decreases in some tdTom+ cells, levels of p16INK4a continues to sharply rise in the same cells for weeks after sorting. We believe these results have significant implications as to the interpretation of what the p16tdTom reporter allele actually reports. Specifically, these results suggest that tdTom positivity is a better proxy for high-level activation of the p16INK4a promoter rather than total abundance of the p16INK4a transcript. This in turn indicates that some cells with formerly strong activation of the p16INK4a promoter could appear tdTom− a few weeks later (as in Fig. 2C), despite high-level expression of the p16INK4a transcript (and presumably protein).
Fig. 2.
Promoter activity of the p16tdTom allele. (A) Representative FACS analysis of tdTom− and tdTom+ populations at day 1 postsort. (B) mRNA expression of p16INK4a by qRT-PCR. Fold-difference was calculated with respect to the mRNA levels in tdTom− MEFs (**P < 0.01). (C) Representative FACS analysis of the cultured tdTom+ MEFs at indicated time points after cell sorting. (D) Frequency of tdTom+ cells in the tdTom+ cultures at indicated time points after cell sorting. (E) Absolute copy number of p16INK4a and tdTom in cultured tdTom+ MEFs by qRT-PCR at indicated time points after cell sorting. Throughout, error bars represent SEM.
Characterization of p16INK4a Transcriptionally Active Cells in Vitro.
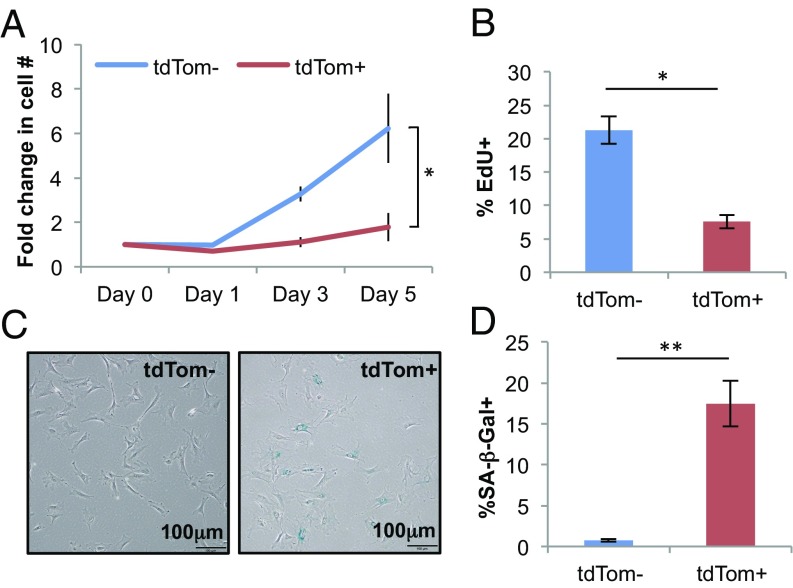
Using the p16tdTom allele, we next turned to the question of the functional properties of cultured cells featuring high-level p16INK4a promoter activation. FACS-isolated tdTom+ MEFs showed lower rates of growth (Fig. 3A) and decreased incorporation of a thymidine analog, 5-ethyl-2′-deoxyuridine (EdU) (Fig. 3B) compared with tdTom− cells, and exhibited higher activity of SA-β-gal in culture (Fig. 3 C and D). We performed an unbiased analysis of RNA expression by next-generation sequencing (RNA-seq) in tdTom+ vs. tdTom− cells with or without serum starvation (0.1% FBS for 48 h). While serum starvation induced a decline in the expression of mRNAs associated with proliferation (cell cycle, E2F) as expected, an even greater reduction in the expression of proliferative mRNAs was noted in tdTom+ compared with tdTom− cells (SI Appendix, Fig. S4A). Gene set enrichment analysis (GSEA) of tdTom+ vs. tdTom− MEFs also demonstrated enrichment of signatures associated with proliferation (e.g., cell cycle and ribosomal transcripts) in tdTom− cells consistent with their increased rates of proliferation. Additionally, GSEA demonstrated differential expression of many signatures associated with developing tissue lineages: for example, neural, cardiac, cutaneous, and hematopoietic (e.g., synaptic signaling and leukocyte development shown in SI Appendix, Fig. S4B). Given that MEF cultures are derived from disaggregated whole murine embryos, we believe this finding reflects an increased propensity to activate p16INK4a expression in certain tissue types (e.g., brain, heart, and skin), but not others (e.g., leukocytes). For the RNA-seq analyses, we developed a list of SASP transcripts compiled from several sources studying senescence in a variety of human or murine cell types (35–38) (SI Appendix, Table S1). Using this list, there was no association of SASP transcript expression with either serum starvation or tdTom expression in MEFs (SI Appendix, Fig. S4A). These findings could indicate that p16INK4a-activated, hyporeplicative MEF cultures expressing SA-β-gal are not “truly” senescent (although cells in such cultures have long been considered so in our field), or indicate that a transcriptional signature of the SASP is difficult to discern in a heterogeneous culture of mixed embryo tissues.
Fig. 3.
p16INK4a-activated cultured MEFs exhibit senescence phenotypes. (A) Growth-curve analysis of tdTom− and tdTom+ populations. Fold-increase was calculated with respect to the cell number at day 0. (B) Quantification of EdU+ cells by immunofluorescence staining. (C) Representative image of SA-β-gal staining. (D) Quantification of SA-β-gal+ cells in C. Throughout, error bars represent SEM. The statistical significance of differences was assessed using paired two-tailed Student’s t tests (*P < 0.05, **P < 0.01).
Enumeration of p16INK4a-Activated Cells in Vivo.
Although evidence suggests that the in vivo accumulation of senescent cells contributes to age-associated tissue dysfunction, the frequency of senescent cells within different aged tissues is unclear. To address this issue, we examined the percentage of tdTom+ cells from tissues harvested from young (8–12 wk) or old (100–120 wk) p16tdTom/+ mice. As p16INK4a mRNA is readily detected in murine or human peripheral blood T cells and increases with aging (9, 16), we examined the frequency of tdTom+ cells in peripheral blood CD3+ (T cells), B220+, or Mac-1+ (myeloid cells) populations every 6 mo through phlebotomy. In these compartments, there was only a minimal increase in the frequency of tdTom+ cells with aging (Fig. 4A). Of note, a subset of mice (n = 6) displayed a transient, high-level increase in the frequency of tdTom+ cells in peripheral blood at the time of routine phlebotomy (SI Appendix, Fig. S5). These transient “flares” of p16INK4a expression in peripheral blood occurred in otherwise well-appearing mice and generally resolved within 1 mo of initial observation. Up to 30–40% of mononuclear blood cells were found to be tdTom+ during these episodes, and all six observed cases showed a sharp increase within the Mac-1+ population. We have noted similar flares of luciferase activity in p16LUC/+ mice (23) and reasoned these episodes might represent a transient, subclinical inflammatory state (e.g., an occult viral infection). However, we were unable to provoke such responses by administering p16tdTom/+ mice Toll-like receptor agonists (e.g., polyinosinic:polycytidylic acid or lipopolysaccharide, LPS). These data suggest that while expression of p16INK4a mRNA is abundant in T cells from old mice, few cells in the peripheral blood exhibit high-level activation of the p16INK4a promoter even in old mice; although rare, transient promoter activation occurs in a minority of adult mice for unidentified reasons.
Fig. 4.
Age-dependent increase in the frequency of p16INK4a-activated cells in different tissues. (A–D) Representative FACS analysis of CD3+ (T cells), B220+ and Mac-1+ (myeloid cells) populations from peripheral blood (PB) (A), cartilages (B), fat progenitor cells (Sca1+CD34+) from IAT (C), and pancreatic islets (D). Tissues were harvested from young (8- to 12-wk-old) and old (100- to 120-wk-old) p16tdTom/+ mice. (E) Quantification of tdTom+ cells from the indicated tissues. Error bars represent SEM (n = 3–4 per group). The statistical significance of differences was assessed using unpaired two-tailed Student’s t tests (*P < 0.05).
Next, we examined activation of the p16INK4a promoter in nonhematopoietic tissues with aging. We focused on tissues where prior work has suggested increased p16INK4a mRNA expression with aging (4, 14, 18, 39–41). We made single-cell preparations of each tissue from young and old p16tdTom/+ mice and then employed immunophenotyping and gating schemes where appropriate to focus on specific tissue fractions of interest (e.g., CD45− cells from cartilage or pancreatic islets, and Sca1+CD34+ progenitors from adipose). We observed significant increases in the frequency of tdTom+ cells with aging in single cells derived from articular cartilage, inguinal adipose tissue (IAT), and pancreatic islets (Fig. 4 B–E). The percentage of tdTom+ cells increased ∼4- to 18-fold in these tissue compartments when comparing young and old mice, suggesting that the frequency of individual chondrocytes, white adipose progenitors, and β-cells having high-level activation of the p16INK4a promoter increases with aging.
Characterization of p16INK4a-Activated Macrophages.
While we were able to identify significant numbers of p16INK4a-expressing cells from several tissues with aging, the low frequency of tdTom+ cells (<10%) and difficulty of isolating these fractions prevented us from further functional and molecular characterization. Therefore, we turned to a recently described inflammatory model to induce high-level p16INK4a expression in activated macrophages in vivo (42). Toward that end, we implanted quiescent neonatal dermal fibroblast (NDF)-containing alginate beads into p16LUC/+ or p16tdTom/+ mice via intraperitoneal injection. Prior work has shown that these quiescent NDFs quickly acquire SA-β-gal staining and release soluble factors, including IL-6 and IL-8, in turn leading to a large influx of inflammatory cells (42). As reported, NDF beads induced a strong luminescent signal in the abdomen of p16LUC/+ mice by 3 wk postinjection (Fig. 5A). Flow cytometric analysis of cells in the peritoneal lavage of p16tdTom/+ mice 3 wk after implanting NDF beads showed a strong induction of tdTom expression in macrophage (Mac-1+F4/80+) populations (Fig. 5B), but not other lavage cell types (e.g., T cells, B220+ cells, and eosinophils) (SI Appendix, Fig. S6). To characterize peritoneal macrophages with high-level p16INK4a promoter activation, we isolated Mac-1+F4/80+ cells by FACS based on tdTom expression. Using this approach, we observed a much greater enrichment of p16INK4a mRNA expression in tdTom+ vs. tdTom− macrophages (40-fold) (Fig. 5C) compared with that seen in MEFs (fivefold) (Fig. 2B). This likely reflects much greater homogeneity among the sorted macrophages compared with mixed MEF cultures. As was the case for MEFs, tdTom+ macrophages exhibited a marked reduction in EdU incorporation (Fig. 5D) and increased SA-β-gal activity (Fig. 5 E and F). It is worth noting that SA-β-gal activity has been considered an unreliable marker of senescence in vivo, especially in this cell type (43, 44). These results show that a substantial fraction of macrophages induced in response to NDF-loaded beads exhibit features of senescence: activation of the p16INK4a promoter, reduced proliferation, and expression of SA-β-gal activity.
Fig. 5.
Reduced proliferation and high SA-β-gal activity of p16INK4a-activated peritoneal macrophages. (A) Bioluminescence imaging of p16LUC/+ mice following intraperitoneal injection with empty (control, CNTL) or quiescent human NDF-embedded alginate beads. Representative image was acquired 21 d after bead injection. (B) Representative FACS analysis of peritoneal macrophages (Mac-1+F4/80+) from p16+/+ (WT) and p16tdTom/+ mice at day 21 after NDF-bead injection. (C) mRNA expression of p16INK4a by qRT-PCR in FACS-sorted tdTom− and tdTom+ peritoneal macrophages. Fold-difference was calculated with respect to the mRNA levels in the tdTom− fraction. (D) Quantification of EdU+ cells by immunofluorescence staining. (E) Representative image of SA-β-gal staining. (F) Quantification of SA-β-gal level in E. Throughout, error bars represent SEM. The statistical significance of differences was assessed using unpaired in C and paired two-tailed Student’s t tests (*P < 0.05, **P < 0.01) in D and F.
Prior studies suggest that p16INK4a also influences cell-intrinsic properties of macrophages, such as M1/M2 polarization (45, 46). To further investigate the effect of p16INK4a on macrophage function, we examined the immunophenotype and cell-specific functions of p16INK4a-activated macrophages in more detail. We did not observe a difference in the immunophenotype of tdTom+ vs. tdTom− lavage cells with regard to macrophage polarity (e.g., CD80, CD206, and MHCII). Moreover, we did not find a modulation of p16INK4a promoter activity by M1/M2 polarizing agents including LPS and IL-4 in either tdTom+ or tdTom− macrophages (SI Appendix, Fig. S7). However, in vitro phagocytosis assays showed that tdTom+ macrophages exhibited greater phagocytic activity than tdTom− cells (Fig. 6 A and B). This demonstration of altered or even increased cellular function is reminiscent of findings in other cell types in the setting of high-level p16INK4a expression [e.g., increased insulin secretion from p16INK4a-expressing pancreatic β-cells (47) and increased cell killing in senescent T cells (46, 48)].
Fig. 6.
Gene-expression profile of p16INK4a-activated peritoneal macrophages. (A) In vitro phagocytosis of pHrodo Green Zymosan bioparticles in peritoneal macrophages. Representative FACS analysis of tdTom− and tdTom+ macrophages. (B) Quantification of phagocytosed macrophages. Error bars represent SEM. The statistical significance of differences was assessed using paired two-tailed Student’s t tests (*P < 0.05). (C and D) GSEA of tdTom− vs. tdTom+ peritoneal macrophages. Representative plots for significantly enriched gene sets at false-discovery rate < 0.01 are shown with their respective normalized enrichment score. (E) Heatmap of differentially expressed genes in tdTom− vs. tdTom+ populations. FACS sorted tdTom− samples (Left, black bar), and tdTom+ samples (Right, red bar). Representative genes in each gene set are listed to the right of the heatmap. The log2 ratio to the mean value of each gene is indicated by the color scale. GO, gene ontology; KEGG, Kyoto Encyclopedia of Genes and Genomes.
To study the underlying mechanisms and genes responsible for the response of p16INK4a-activated macrophages to NDF-beads, we performed RNA-seq of tdTom+ vs. tdTom− peritoneal macrophages. We identified 456 transcripts being up-regulated and 118 transcripts down-regulated in tdTom+ macrophages (P < 0.01). Through GSEA, we identified several gene signatures related to the cell cycle, senescence, and macrophage functions (Fig. 6 C–E). Specifically, consistent with the decreased proliferation of these cells (Fig. 5D), tdTom+ macrophages exhibited a profound decline in the expression of transcripts associated with cell cycle traversal and ribosomal proteins. Even though expression of a few “cell cycle”-classified genes was increased in tdTom+ cells, these were largely inhibitors of the cell cycle such as p16INK4a/Arf (Cdkn2a) and p15INK4b (Cdkn2b) (Fig. 6E). Macrophages with high-level activation of the p16INK4a promoter also exhibited increased expression of lysosomal mRNAs, consistent with the observed increase in β-galactosidase activity (Fig. 6 C and D). In accord with the immunophenotypic analysis, we did not observe differential expression of genes associated with M1/M2 macrophage polarization (e.g., Nos2, Arg1, and Ym1/2). On the other hand, we found increased expression of genes involved in phagocytosis in tdTom+ macrophages (Fig. 6D), consistent with the high phagocytic activity of p16INK4a-activated macrophages (Fig. 6 A and B). Additionally, we found clear up-regulation of several components and regulators of the extracellular matrix (ECM) or the “matrisome,” including collagens, matrix metalloproteinases, thrombospondins, and fibulins (Fig. 6 C and E), and these changes were highly consistent with prior studies of the ECM in senescent cells (35, 49). Finally, using the list of SASP transcripts developed for the MEF RNA-seq studies (SI Appendix, Fig. S4 and Table S1), we showed a strong enrichment by GSEA for SASP transcripts in tdTom+ cells (e.g., IL7, Mmp12, Timp2, Cxcl12/13, Hgf), while only one SASP transcript, Mif was expressed at lower levels (SI Appendix, Table S1).
Contrary to less clear results generated from heterogeneous MEFs, these studies were carried out in a well-defined in vivo hematopoietic subtype and demonstrate that p16INK4a-activated macrophages exhibit multiple features of senescent cells by gene expression analysis, including decreased replication, increased lysosomal activity, altered ECM production, and expression of many classic SASP factors.
Discussion
Here we report a p16tdTom knockin allele that allows for the purification and molecular characterization of individual cells in vivo featuring high-level activation of the p16INK4a promoter. Using this allele, we show that the p16INK4a locus is activated in cultured MEFs with serial passage, in specific tissues with aging, and in activated macrophages after inflammatory challenge. We show that these p16INK4a-activated cells exhibit features of cellular senescence including hyporeplication, increased expression of SA-related matrisomal transcripts, high activity of SA-β-gal, and expression of transcripts associated with SASP.
Prior efforts to generate a single-cell reporter of p16INK4a expression in our group have been unsuccessful due to low expression of the knocked-in reporter transcript. While other groups have reported transgenic mice featuring a fluorochrome reporter under the control of the p16INK4a promoter (10, 17), very little work on the analysis of isolated p16INK4a-expressing cells from these mice has been described, likely reflecting low level expression of the reporter in these systems as well. In the knockin reporter mice described herein, expression of p16INK4a is driven by intact cis-regulatory elements of the Cdkn2a locus, to most faithfully recapitulate the activation of p16INK4a under physiological conditions. To produce a more useful reporter strain, we used an ultrabright fluorochrome (tdTom) and also left the neomycin-selection cassette within the first intron of p16INK4a to serve as a local enhancer of expression. This latter approach has been reported by other groups to not adversely affect the fidelity of the reporter transcript (25, 26), as appears to be the case with this allele. This allele is well-suited for the detection of cells with p16INK4a activation as we detected a high frequency of tdTom+ cells in multiple settings: in vitro (up to 60% of cells in late-passaged MEFs), physiologic aging (>6% of cells in certain tissues from old mice), as well as in peritoneal macrophages from bead-injected mice (>10%). This approach allows for purification by FACS and further biological and molecular characterization.
We noted the reporter strain is a more faithful measure of p16INK4a promoter activation than transcript abundance. For example, by performing FACS to isolate tdTom+ MEFs, we could identify populations of nondividing cells with increasing p16INK4a expression and decreasing tdTom expression (Fig. 2 C–E). This observation at least in part reflects the decreased half-life of the knocked-in transcript (tdTom, ∼12 h) compared with the endogenous p16INK4a transcript (>24 h). Of note, the discovery of the extraordinary stability of the p16INK4a and Arf dates to the mid-1990s (31), but the structural basis for this finding is still unknown. While it is believed that the p16INK4a promoter is persistently active in senescent cells, the kinetics of promoter activation in individual senescent cells has not been studied. These findings suggest that activation of the p16INK4a promoter in newly senescing cells might peak at the time of cell cycle arrest, and then in some cases, decrease to a state of lower engagement. Because the tdTom allele does not label such p16INK4a-expressing cells with low promoter activity, we are confident this allele does not detect all senescent cells in vivo. We believe this observation is relevant to other systems using the p16INK4a promoter to label senescent cells in aging or stressed animals. For example, it is possible the benefits observed in aged mice relying on p16INK4a promoter activation to deplete senescent cells in vivo (17, 18) represent an underestimate of the effects of complete organismal senolysis.
The estimates of increased p16INK4a expression with aging differ using this allele as opposed to other methods. For example, whereas increased mRNA expression is readily detected in pooled murine T cells with aging, we noted little increase in tdTom+ T cells with aging. Similarly, using antibody-based approaches, others have suggested very high levels of p16INK4a-expressing pancreatic β-cells with aging (50), whereas we only observed evidence of high-level promoter activation in ∼3% of pancreatic islet cells from old mice (Fig. 4E). This could reflect technical differences (e.g., the sensitivity of qRT-PCR and immunohistochemistry compared with flow analysis), but we believe a more likely explanation relates to the function of the p16tdTom reporter allele. Specifically, we believe this discordance is further evidence that most senescent cells show high levels of p16INK4a mRNA abundance, but not necessarily high-level p16INK4a promoter activation. Presumably, these cells are able to accumulate high levels of the p16INK4a transcript despite modest promoter activity because of the marked stability of the endogenous p16INK4a transcript.
The role of p16INK4a in macrophage biology is an area of emerging interest. Sherr and coworkers (51) demonstrated that macrophages can express high levels of p16INK4a under some settings, and this induction causes a potent growth arrest in this cell type. Motivated by human genome-wide association studies linking the CDKN2a/b locus to atherosclerotic disease, we previously showed reduced INK4a/ARF expression in humans harboring risk alleles for atherosclerosis (52), a model supported by murine studies suggesting Ink4a/Arf expression in macrophages prevents atherosclerosis (53). It is unclear, however, how p16INK4a expression in macrophages would prevent atherosclerosis. It is possible p16INK4a expression limits the proliferation of macrophages or its progenitors and thereby reduces macrophage number in a nascent atherosclerotic plaque. The recent observation from Van Deursen and coworkers (54) showed that p16INK4a-expressing macrophages themselves do not protect from atherosclerosis, as the elimination of these cells actually reduced plaque size. The complex role of p16INK4a expression in macrophage function may be related to observations that p16INK4a plays a role in altering macrophage function, such as the regulation of M1/M2 polarization (45, 46).
Some debate exists as to whether macrophages expressing high levels of p16INK4a should be considered “senescent.” In fact, there are currently no universal and completely reliable markers to identify senescent cells in vivo due to the heterogeneity of senescence phenotypes in living animals (6). Gudkov and coworkers (44) have argued that SA-β-gal and p16INK4a expression in vivo are not pathognomonic for senescence [a view we share (6)] and have shown in analyses of pooled cells that p16INK4a activation can be reversible by immunomodulatory agents. In accord with that prior work (42), we noted that NDF-loaded beads potently induced an influx of macrophages featuring strong p16INK4a activation 2–3 wk after implantation. We also found these p16INK4a-activated macrophages have SA-β-gal activity and exhibit increased phagocytic ability. Using FACS-sorting to limit our analysis to p16INK4a-activated cells, we noted both functional and transcriptional features of this population that have been linked to senescence: hyporeplication, altered ECM production, and expression of proinflammatory cytokines. We did not observe a difference between cells with low and high p16INK4a promoter activity with regard to macrophage polarity, nor did we find a modulation of p16INK4a promoter activity by M1/M2 polarizing agents in these two fractions. These results suggest that previous findings on macrophage polarization may represent differences in terms of the specific population of cells studied, and differences in the reporter systems employed. While macrophages having high-level activation of the p16INK4a promoter displayed features of senescence in this study, an unambiguous claim that these cells are senescent is challenging given the physiological response of macrophages to environmental stresses. Macrophages show altered proliferation (55–57) and secretion of either pro- or antiinflammatory cytokines to promote immune response and ECM remodeling (58–60) in response to environmental cues. Regardless of the classification of these cells, the notion suggested by Gudkov and coworkers (42, 44) that such cells accumulate in response to tissue senescence and contribute to age-associated pathology remains highly intriguing and is supported by this work.
In summary, this study shows that some p16INK4a-activated cells in vivo exhibit multiple characteristics of cellular senescence, and these cells accumulate with aging in several tissues. Our findings suggest a three-component definition of senescent cells in vivo: high-level p16INK4a activation, reduced replication, and expression of senescence-associated transcripts (e.g., SASP and ECM). These data also identify an unexpected relationship between promoter activation and transcript abundance, with implications for the use of p16INK4a as an in vivo marker of senescence. Finally, this allele allows for the isolation and characterization of individual cells that are produced by the process of normal aging, feature high-level p16INK4a promoter activation, and contribute to varied, age-associated pathologies.
Methods
Animals.
Animals were housed in an Association for Assessment and Accreditation of Laboratory Animal Care-accredited facility and treated in accordance with protocols approved by the Institutional Animal Care and Use Committee for animal research at the University of North Carolina.
Generation of the p16tdTom Allele.
This allele was generated via standard homologous recombination procedures in hybrid C57BL6/129SvEv embryonic stem cells. Germline transmission was confirmed, and the allele was backcrossed seven generations to C57BL/6 before further analysis. Animals were studied with and without FlpE-mediated excision of the neomycin selection cassette, as described in Results. The primers and PCR conditions used for p16tdTom genotyping were as following: p16-tdTom 5′-AAGCCCTCGGGGAAGGACAG-3′ (0.15 mM); p16-F 5′-GGAAGGAAGGAGGGACCCACTG-3′ (0.15 mM); p16-R 5′-AGAGTTCGGGGCGTTGGGC-3′ (0.15 mM); 95 °C for 1 min, 34× (95 °C for 15 s, 65 °C for 15 s and 72 °C for 30 s), 72 °C for 10 min. The resulting PCR products are 204 (WT) and 352 (p16tdTom) base pairs long.
Isolation and Culture of MEFs.
MEFs were generated and cultured as previously described (23).
Quantitative Reverse-Transcriptase PCR.
Total RNA and cDNA were prepared from serially passaged MEFs and sorted samples using the RNeasy Mini Kit (Qiagen) and ImPro-II reverse transcriptase (Promega) according to the manufacturer’s instructions. The qRT-PCR strategy for the detection of p16INK4A was previously described (6). The primers and probe used for the detection of tdTomato were as following: tdTom-F 5′-ACCTCCCACAACGAGGACTA-3′; tdTom-R 5′-CTTGTACAGCTCGTCCATGC-3′; Probe 5′- [FAM] CCGCCACCACCTGTTCCTGT [TAM]-3′. For absolute quantification of transcript abundance, fragments of p16INK4a and tdTomato were cloned, and a standard curve was generated using serial dilution.
Single-Cell Quantitative Reverse-Transcriptase PCR.
Single tdTom+ MEFs were sorted into a 96-well plate containing cell lysis buffer. Reverse transcription and preamplification of genes of interest were performed using the Single-Cell-to-CT Kit (Invitrogen) according to the manufacturer’s guidelines, followed qRT-PCR analysis, as described above.
Tissue Dissociation and Flow Cytometry Analysis.
Blood samples were treated with ACK (ammonium-chloride-potassium) lysis buffer to remove red blood cells before the staining of select markers. Cartilage tissue was dissected from the proximal end of the femur (hip) and the end of the tibial plateau (knee), followed by predigestion with 2 mg/mL pronase (EMD Millipore) in serum-free media for 1 h at 37 °C and then digestion overnight with 0.4 mg/mL Collagenase P (Roche Diagnostics) in 10% serum media. Pancreatic islets were isolated as previously described (31) and dissociated into a single-cell suspension by 0.05% trypsin-EDTA for 3 min at 37 °C. Single cells from cartilages and islets were stained with CD45 (30-F11). Single cells derived from IAT were prepared and stained as previously described (13, 28). Cells were stained with the antibodies in HBSS with 2% FBS and analyzed in HBSS with 2% FBS and 2 mg/mL DAPI using LSRII (BD) flow cytometers. FACS data were analyzed using FlowJo software (TreeStar).
Alginate Bead Experiment.
Empty and NDF-embedded alginate beads were prepared as described previously (30). Beads were implanted into mice via intraperitoneal injection, and peritoneal lavage was collected at day 21–30 postinjection. For analysis by flow cytometry, peritoneal cells were washed with HBSS plus 2% heat-inactivated FBS and blocked with anti-CD16/CD32 (clone 93) for 10 min on ice, followed by staining with anti-F4/80 (BM8), anti–Mac-1 (M1/70), CD170 (1RNM44N), and following Biotin conjugated antibodies: anti-CD3 (145-2C11), anti-CD19 (6D5), anti-B220 (RA3-6B2), anti-Ter119 (TER-119), and anti-NK1.1 (PK136) for 30 min on ice. Cells were then washed and stained with fluorophore-conjugated streptavidin for 20 min on ice, followed by one wash, and resuspended with HBSS plus 2% heat-inactivated FBS and 2 mg/mL DAPI.
In Vivo Bioluminescent Imaging.
Isoflurane-anesthetized mice were injected intraperitoneally with d-luciferin potassium salt (15 mg/mL in PBS; PerkinElmer) and imaged using IVIS Lumina (Caliper Life Sciences). Sequential imaging was performed upon injection, 2 min in length, and 8 min in total.
In Vitro and in Vivo Cell Sorting.
Serial-passaged MEFs or bead-induced peritoneal macrophages were sorted by FACS into tdTom− and tdTom+ populations using MoFlo XDP (Beckman Coulter) or FACSAria III (Becton Dickinson). Sorted cells were used for the functional studies described below and the RNA-seq experiment.
In Vitro Cell Growth Assay.
For the in vitro cell growth assay, 5 × 104 sorted MEFs were seeded and cultured for 5 d. Total cell number was measured at days 1, 3, and 5.
In Vitro and in Vivo EdU Incorporation.
Sorted MEFs were pulsed with 5 μg/mL EdU for 2 h in a CO2 incubator at day 1 postsort, followed by fixation in 3.7% paraformaldehyde for 10 min and permeabilization in PBS plus 0.5% Triton X-100 for 15 min at room temperature. EdU staining was performed in 0.1 M Tris⋅HCl (pH 7.5), 1 mM CuSO4, 0.1 M ascorbic acid, and 1 μM AlexaFluor 555 azide (Life Technologies) for 30 min at room temperature. Stained cells were washed twice with PBS plus 0.5% Triton X-100 and then incubated with 2 mg/mL DAPI for 5 min before immunofluorescence microscopy analysis. For in vivo analyses, mice were treated with 10 mg/kg EdU via intraperitoneal injection every 12 h, five injections in total. EdU-labeled peritoneal macrophages were stained as described above.
In Vitro Phagocytosis Assay.
For in vitro phagocytosis assay, 1 × 106 peritoneal cavity cells harvested from mice were incubated with 0.2 mg/mL pHrodo Green Zymosan Bioparticles (Thermo Fisher Scientific) for 60 min at 37 °C. Treated cells were washed and stained for macrophage markers as described above.
In Vitro Treatment.
For in vitro treatment, 1–2 × 105 FACS-sorted tdTom− and tdTom+ peritoneal macrophages or 1 × 106 peritoneal cavity cells harvested from NDF bead-elicited mice were plated overnight in an uncoated 12-well plate, followed by ≤72 h treatments of LPS (Sigma) or IL-4 (Biolegend). Treated cells were analyzed by flow cytometry as described above.
SA-β-Gal Staining.
FACS-sorted samples were stained for SA-β-gal activity at day 1 postsort using Cellular Senescence Assay Kit (Millipore) according to the manufacturer’s protocol. Positive staining was quantified by ImageJ and Image Pro Premier software.
mRNA Stability Assay.
p16tdTom/+ MEFs were treated with 5 μg/mL ActD. Cells were then harvested at time 0, 2, 6, 12, and 24 h posttreatment, and RNA was purified, followed by qRT-PCR.
RNA Sequencing and Analysis.
Total RNA was isolated from sorted tdTom− and tdTom+ MEFs and peritoneal macrophages using TRIzol LS reagent (ThermoFisher) according to the manufacturer’s instructions, followed by the clean-up using NucleoSpin RNA XS (Clontech). RNA-seq libraries were constructed with the TruSeq RNA kit v2 (Illumina) and validated using the Agilent 2200 TapeStation system. The 150-cycle paired-end sequencing runs were generated with an Illumina NextSeq500 at the University of North Carolina Translational Genomics Laboratory. Purity filtered reads were aligned to the mouse reference genome (mm9) using STAR. Transcript abundance for each sample was estimated by Salmon (32). Differential expression between tdTom− and tdTom+ samples was computed using DESeq2 (33).
GSEA.
GSEA was performed as previously described (34). Enrichment of differentially expressed genes in tdTom− vs. tdTom+ cells was carried out against preranked gene lists. Default parameters were used.
Statistical Analysis.
To determine the correlation of two sample groups, linear regression was performed. Statistical comparison of two groups was performed using a two-tailed unpaired Student’s t test. For statistical comparisons of paired groups (e.g., sorted tdTom− vs. tdTom+), a two-tailed paired Student’s t test was performed. Differences were considered statistically significant at P values less than 0.05: *P < 0.05, **P < 0.01. All data presented as mean ± SEM. Sample sizes for all data are indicated in each figure legend.
Supplementary Material
Acknowledgments
The work performed relied on expertise from University of North Carolina Lineberger Comprehensive Cancer Center cores including the University of North Carolina Animal Models core, the Biomedical Research Imaging Center Small Animal Imaging Facility, and the Flow Cytometry Core. We thank Olga Chernova (Everon Biosciences) for helpful discussions. This work was funded by National Institute on Aging Grant R01-AG024379 and National Cancer Institute Grant R01 CA163896.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1818313116/-/DCSupplemental.
References
- 1.Campisi J. Aging, cellular senescence, and cancer. Annu Rev Physiol. 2013;75:685–705. doi: 10.1146/annurev-physiol-030212-183653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.He S, Sharpless NE. Senescence in health and disease. Cell. 2017;169:1000–1011. doi: 10.1016/j.cell.2017.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Campisi J, d’Adda di Fagagna F. Cellular senescence: When bad things happen to good cells. Nat Rev Mol Cell Biol. 2007;8:729–740. doi: 10.1038/nrm2233. [DOI] [PubMed] [Google Scholar]
- 4.Diekman BO, et al. Expression of p16INK4a is a biomarker of chondrocyte aging but does not cause osteoarthritis. Aging Cell. 2018;17:e12771. doi: 10.1111/acel.12771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jurk D, et al. Postmitotic neurons develop a p21-dependent senescence-like phenotype driven by a DNA damage response. Aging Cell. 2012;11:996–1004. doi: 10.1111/j.1474-9726.2012.00870.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sharpless NE, Sherr CJ. Forging a signature of in vivo senescence. Nat Rev Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- 7.Serrano M, Hannon GJ, Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- 8.Krishnamurthy J, et al. Ink4a/Arf expression is a biomarker of aging. J Clin Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Liu Y, et al. Expression of p16(INK4a) in peripheral blood T-cells is a biomarker of human aging. Aging Cell. 2009;8:439–448. doi: 10.1111/j.1474-9726.2009.00489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Demaria M, et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jun JI, Lau LF. The matricellular protein CCN1 induces fibroblast senescence and restricts fibrosis in cutaneous wound healing. Nat Cell Biol. 2010;12:676–685. doi: 10.1038/ncb2070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krizhanovsky V, et al. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Berent-Maoz B, Montecino-Rodriguez E, Signer RAJ, Dorshkind K. Fibroblast growth factor-7 partially reverses murine thymocyte progenitor aging by repression of Ink4a. Blood. 2012;119:5715–5721. doi: 10.1182/blood-2011-12-400002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen H, et al. Polycomb protein Ezh2 regulates pancreatic beta-cell Ink4a/Arf expression and regeneration in diabetes mellitus. Genes Dev. 2009;23:975–985. doi: 10.1101/gad.1742509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cosgrove BD, et al. Rejuvenation of the muscle stem cell population restores strength to injured aged muscles. Nat Med. 2014;20:255–264. doi: 10.1038/nm.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Y, et al. Expression of p16(INK4a) prevents cancer and promotes aging in lymphocytes. Blood. 2011;117:3257–3267. doi: 10.1182/blood-2010-09-304402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baker DJ, et al. Clearance of p16Ink4a-positive senescent cells delays ageing-associated disorders. Nature. 2011;479:232–236. doi: 10.1038/nature10600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Baker DJ, et al. Naturally occurring p16(Ink4a)-positive cells shorten healthy lifespan. Nature. 2016;530:184–189. doi: 10.1038/nature16932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Fortney K, et al. Genome-wide scan informed by age-related disease identifies loci for exceptional human longevity. PLoS Genet. 2015;11:e1005728. doi: 10.1371/journal.pgen.1005728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jeck WR, Siebold AP, Sharpless NE. Review: A meta-analysis of GWAS and age-associated diseases. Aging Cell. 2012;11:727–731. doi: 10.1111/j.1474-9726.2012.00871.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ohtani N, Yamakoshi K, Takahashi A, Hara E. Real-time in vivo imaging of p16gene expression: A new approach to study senescence stress signaling in living animals. Cell Div. 2010;5:1. doi: 10.1186/1747-1028-5-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamakoshi K, et al. Real-time in vivo imaging of p16Ink4a reveals cross talk with p53. J Cell Biol. 2009;186:393–407. doi: 10.1083/jcb.200904105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Burd CE, et al. Monitoring tumorigenesis and senescence in vivo with a p16(INK4a)-luciferase model. Cell. 2013;152:340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Shaner NC, et al. Improved monomeric red, orange and yellow fluorescent proteins derived from Discosoma sp. red fluorescent protein. Nat Biotechnol. 2004;22:1567–1572. doi: 10.1038/nbt1037. [DOI] [PubMed] [Google Scholar]
- 25.Scarff KL, Ung KS, Sun J, Bird PI. A retained selection cassette increases reporter gene expression without affecting tissue distribution in SPI3 knockout/GFP knock-in mice. Genesis. 2003;36:149–157. doi: 10.1002/gene.10210. [DOI] [PubMed] [Google Scholar]
- 26.Schmidt-Supprian M, Wunderlich FT, Rajewsky K. Excision of the Frt-flanked neo (R) cassette from the CD19cre knock-in transgene reduces Cre-mediated recombination. Transgenic Res. 2007;16:657–660. doi: 10.1007/s11248-007-9100-4. [DOI] [PubMed] [Google Scholar]
- 27.Sharpless NE, et al. Loss of p16Ink4a with retention of p19Arf predisposes mice to tumorigenesis. Nature. 2001;413:86–91. doi: 10.1038/35092592. [DOI] [PubMed] [Google Scholar]
- 28.Krimpenfort P, Quon KC, Mooi WJ, Loonstra A, Berns A. Loss of p16Ink4a confers susceptibility to metastatic melanoma in mice. Nature. 2001;413:83–86. doi: 10.1038/35092584. [DOI] [PubMed] [Google Scholar]
- 29.Sharpless NE, Ramsey MR, Balasubramanian P, Castrillon DH, DePinho RA. The differential impact of p16(INK4a) or p19(ARF) deficiency on cell growth and tumorigenesis. Oncogene. 2004;23:379–385. doi: 10.1038/sj.onc.1207074. [DOI] [PubMed] [Google Scholar]
- 30.Zindy F, Quelle DE, Roussel MF, Sherr CJ. Expression of the p16INK4a tumor suppressor versus other INK4 family members during mouse development and aging. Oncogene. 1997;15:203–211. doi: 10.1038/sj.onc.1201178. [DOI] [PubMed] [Google Scholar]
- 31.Hara E, et al. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol Cell Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jeck WR, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157. doi: 10.1261/rna.035667.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tani H, et al. Genome-wide determination of RNA stability reveals hundreds of short-lived noncoding transcripts in mammals. Genome Res. 2012;22:947–956. doi: 10.1101/gr.130559.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kakoki M, et al. Altering the expression in mice of genes by modifying their 3′ regions. Dev Cell. 2004;6:597–606. doi: 10.1016/s1534-5807(04)00094-2. [DOI] [PubMed] [Google Scholar]
- 35.Coppé JP, Desprez PY, Krtolica A, Campisi J. The senescence-associated secretory phenotype: The dark side of tumor suppression. Annu Rev Pathol. 2010;5:99–118. doi: 10.1146/annurev-pathol-121808-102144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Coppé JP, et al. A human-like senescence-associated secretory phenotype is conserved in mouse cells dependent on physiological oxygen. PLoS One. 2010;5:e9188. doi: 10.1371/journal.pone.0009188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Coppé JP, et al. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hernandez-Segura A, et al. Unmasking transcriptional heterogeneity in senescent cells. Curr Biol. 2017;27:2652–2660.e4. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Baker DJ, Weaver RL, van Deursen JM. p21 both attenuates and drives senescence and aging in BubR1 progeroid mice. Cell Rep. 2013;3:1164–1174. doi: 10.1016/j.celrep.2013.03.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jeon OH, et al. Local clearance of senescent cells attenuates the development of post-traumatic osteoarthritis and creates a pro-regenerative environment. Nat Med. 2017;23:775–781. doi: 10.1038/nm.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krishnamurthy J, et al. p16INK4a induces an age-dependent decline in islet regenerative potential. Nature. 2006;443:453–457. doi: 10.1038/nature05092. [DOI] [PubMed] [Google Scholar]
- 42.Hall BM, et al. Aging of mice is associated with p16(Ink4a)- and β-galactosidase-positive macrophage accumulation that can be induced in young mice by senescent cells. Aging (Albany NY) 2016;8:1294–1315. doi: 10.18632/aging.100991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Bursuker I, Goldman R. Distinct bone marrow precursors for mononuclear phagocytes expressing high and low 5′ -nucleotidase activity. J Cell Physiol. 1982;112:237–242. doi: 10.1002/jcp.1041120212. [DOI] [PubMed] [Google Scholar]
- 44.Hall BM, et al. p16(Ink4a) and senescence-associated β-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging (Albany NY) 2017;9:1867–1884. doi: 10.18632/aging.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cudejko C, et al. p16INK4a deficiency promotes IL-4-induced polarization and inhibits proinflammatory signaling in macrophages. Blood. 2011;118:2556–2566. doi: 10.1182/blood-2010-10-313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Vicente R, Mausset-Bonnefont AL, Jorgensen C, Louis-Plence P, Brondello JM. Cellular senescence impact on immune cell fate and function. Aging Cell. 2016;15:400–406. doi: 10.1111/acel.12455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Helman A, et al. p16(Ink4a)-induced senescence of pancreatic beta cells enhances insulin secretion. Nat Med. 2016;22:412–420. doi: 10.1038/nm.4054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Akbar AN, Henson SM. Are senescence and exhaustion intertwined or unrelated processes that compromise immunity? Nat Rev Immunol. 2011;11:289–295. doi: 10.1038/nri2959. [DOI] [PubMed] [Google Scholar]
- 49.Freitas-Rodríguez S, Folgueras AR, López-Otín C. The role of matrix metalloproteinases in aging: Tissue remodeling and beyond. Biochim Biophys Acta Mol Cell Res. 2017;1864:2015–2025. doi: 10.1016/j.bbamcr.2017.05.007. [DOI] [PubMed] [Google Scholar]
- 50.Tschen SI, Dhawan S, Gurlo T, Bhushan A. Age-dependent decline in beta-cell proliferation restricts the capacity of beta-cell regeneration in mice. Diabetes. 2009;58:1312–1320. doi: 10.2337/db08-1651. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Randle DH, Zindy F, Sherr CJ, Roussel MF. Differential effects of p19(Arf) and p16(Ink4a) loss on senescence of murine bone marrow-derived preB cells and macrophages. Proc Natl Acad Sci USA. 2001;98:9654–9659. doi: 10.1073/pnas.171217498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liu Y, et al. INK4/ARF transcript expression is associated with chromosome 9p21 variants linked to atherosclerosis. PLoS One. 2009;4:e5027. doi: 10.1371/journal.pone.0005027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kuo CL, et al. Cdkn2a is an atherosclerosis modifier locus that regulates monocyte/macrophage proliferation. Arterioscler Thromb Vasc Biol. 2011;31:2483–2492. doi: 10.1161/ATVBAHA.111.234492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Childs BG, et al. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354:472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xaus J, et al. Interferon gamma induces the expression of p21waf-1 and arrests macrophage cell cycle, preventing induction of apoptosis. Immunity. 1999;11:103–113. doi: 10.1016/s1074-7613(00)80085-0. [DOI] [PubMed] [Google Scholar]
- 56.Vadiveloo PK, Keramidaris E, Morrison WA, Stewart AG. Lipopolysaccharide-induced cell cycle arrest in macrophages occurs independently of nitric oxide synthase II induction. Biochim Biophys Acta. 2001;1539:140–146. doi: 10.1016/s0167-4889(01)00102-1. [DOI] [PubMed] [Google Scholar]
- 57.Jenkins SJ, et al. Local macrophage proliferation, rather than recruitment from the blood, is a signature of TH2 inflammation. Science. 2011;332:1284–1288. doi: 10.1126/science.1204351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Huang WC, Sala-Newby GB, Susana A, Johnson JL, Newby AC. Classical macrophage activation up-regulates several matrix metalloproteinases through mitogen activated protein kinases and nuclear factor-κB. PLoS One. 2012;7:e42507. doi: 10.1371/journal.pone.0042507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oishi S, et al. M2 polarization of murine peritoneal macrophages induces regulatory cytokine production and suppresses T-cell proliferation. Immunology. 2016;149:320–328. doi: 10.1111/imm.12647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Biswas SK. Metabolic reprogramming of immune cells in cancer progression. Immunity. 2015;43:435–449. doi: 10.1016/j.immuni.2015.09.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.