Significance
Most plants resist most plant pathogens. Barley resists wheat-infecting powdery mildew races (and vice versa), and both barley and wheat resist potato late blight. Such “nonhost” resistance could result because the pathogen fails to suppress defense or triggers innate immunity due to failure to evade detection. Albugo candida causes white rust on most Brassicaceae, and we investigated Arabidopsis NHR to Brassica-infecting races. Transgressive segregation for resistance in Arabidopsis recombinant inbred lines revealed genes encoding nucleotide-binding, leucine-rich repeat (NLR) immune receptors. Some of these NLR-encoding genes confer resistance to white rust in Brassica sp. This genetic method thus provides a route to reveal resistance genes for crops, widening the pool from which such genes might be obtained.
Keywords: Arabidopsis thaliana, oomycete, Albugo candida, nonhost resistance, Brassicaceae
Abstract
Arabidopsis thaliana accessions are universally resistant at the adult leaf stage to white rust (Albugo candida) races that infect the crop species Brassica juncea and Brassica oleracea. We used transgressive segregation in recombinant inbred lines to test if this apparent species-wide (nonhost) resistance in A. thaliana is due to natural pyramiding of multiple Resistance (R) genes. We screened 593 inbred lines from an Arabidopsis multiparent advanced generation intercross (MAGIC) mapping population, derived from 19 resistant parental accessions, and identified two transgressive segregants that are susceptible to the pathogen. These were crossed to each MAGIC parent, and analysis of resulting F2 progeny followed by positional cloning showed that resistance to an isolate of A. candida race 2 (Ac2V) can be explained in each accession by at least one of four genes encoding nucleotide-binding, leucine-rich repeat (NLR) immune receptors. An additional gene was identified that confers resistance to an isolate of A. candida race 9 (AcBoT) that infects B. oleracea. Thus, effector-triggered immunity conferred by distinct NLR-encoding genes in multiple A. thaliana accessions provides species-wide resistance to these crop pathogens.
Plants and animals are colonized by diverse pathogens and parasites, and their mechanisms of immunity are of broad significance. Plants have two layers of cell-autonomous innate immunity (1–3). Pathogen molecules such as flagellin and chitin are perceived by cell surface pattern recognition receptors (PRRs). Activation of PRRs results in pattern-triggered immunity (PTI) that restricts microbial growth (4, 5). Most plant pathogens translocate pathogenicity proteins, called effectors, into host cells; many of these suppress PTI, facilitating colonization (6–8). Genetic variation for disease resistance within a plant species is often explained by allelic variation in Resistance (R) genes that encode nucleotide-binding, leucine-rich repeat (NLR) immune receptors. Effector recognition leads to effector-triggered immunity (ETI) (1). Many NLRs carry either Toll/Interleukin-1 receptor/Resistance (TIR-NLRs) or coiled-coil (CC) domains at their N-termini (CC-NLRs) (9–11) and can activate ETI either by directly detecting an effector (12–19) or indirectly through “guarding” host proteins that are modified by effectors (20–22). Unlike CC-NLRs, the function of TIR-NLR proteins requires EDS1 (ENHANCED DISEASE SUSCEPTIBILITY 1), which encodes a lipase-like protein, and forms functional heterodimers in Arabidopsis with the related proteins PAD4 (PHYTOALEXIN-DEFICIENT 4) or SAG101 (SENESCENCE-ASSOCIATED GENE 101) (23–25).
Plants are challenged by many potential pathogens but most plants are resistant to most pathogens, and disease is rare. Resistance of a particular plant species against all isolates of a pathogen that can infect other plant species is known as nonhost resistance (NHR) (26). The molecular mechanisms underlying NHR are poorly understood; if all accessions of a species are resistant, genetic analysis of NHR is difficult (27, 28). Conceivably, NHR or species-level resistance could involve PTI (if effectors cannot suppress PTI), ETI (if effectors do not evade detection), and/or other mechanisms (28, 29). Fundamental insights into this question are of broad interest. NHR genes that confer complete immunity in a nonhost might confer resistance in susceptible crops and elevate resistance to important crop diseases.
To investigate NHR, we studied Albugo candida, an obligate biotrophic oomycete plant pathogen that causes white blister rust disease in Brassicaceae. In contrast to A. candida, Albugo laibachii has specialized to cause white rust only on Arabidopsis (30). The asexual life cycle of A. candida starts with the release of biflagellate motile zoospores from sporangia. Zoospores target host stomata where they encyst and germinate into a germ tube followed by colonization of mesophyll cells by branched hyphae, which also give rise to a specialized feeding structure called an haustorium. Infection culminates in formation of zoosporangia-bearing white pustules that rupture the epidermis; these constitute the visible symptoms of the disease (31). A. candida forms many physiological races, each of which specialize on different host species (32–36). Some races of A. candida such as Race 2 cause severe annual losses of oilseed mustard (Brassica juncea) in India, Canada, and Australia. Albugo spp. infection induces a strongly immuno-compromised state in host plants, which can enable avirulent races to colonize and reproduce in the same tissue (37). Sex between different cocolonizing races in the same host could be an important source of new recombinant races (32). Comparative genomics has revealed extensive genetic exchange between races of A. candida (34), and this genetic exchange could result in races with novel repertoires of effector alleles that, in turn, might enable colonization of new hosts. Therefore, understanding the underlying mechanism of NHR in different Brassica species could inform breeding for resistance to A. candida.
Here, we investigate adult plant resistance to A. candida Race 2 (Ac2V) in diverse Arabidopsis thaliana accessions. While all Arabidopsis accessions are resistant to Ac2V, some A. candida strains can grow on Arabidopsis, but although this pathosystem does not involve NHR to the whole A. candida species complex, it is nonetheless instructive. We hypothesized that resistance in A. thaliana to Ac2V is due to multiple R genes, but the R gene repertoire in different Arabidopsis accessions might be distinct, creating the potential for transgressive segregation for susceptibility in recombinant inbreds or other segregating progeny from interaccession crosses. We screened a population of “MAGIC” inbred lines (38). These lines result from intercrosses of 19 parents, followed by random intercrossing, and then selfing. These lines have been extensively genotyped (39). We inoculated 593 lines and identified two transgressive segregant inbreds (MAGIC.329 and MAGIC.23) that are susceptible in true leaves to Ac2V. However, none of the MAGIC lines tested, nor the 19 parental accessions, are fully susceptible to Race 9 (AcBoT) collected from Brassica oleracea.
We defined three loci that contribute resistance to Ac2V, including a known locus, White Rust Resistance 4 (WRR4) on chromosome 1 (40). WRR4 carries two paralogs, WRR4A and WRR4B, that can each confer resistance. We also defined WRR8 and WRR9. To investigate AcBoT resistance in Arabidopsis, we intercrossed MAGIC.329 with MAGIC.23. Screening of selfed progeny from this cross revealed fully susceptible plants at a frequency suggesting that resistance in the two parents is conferred by distinct genes. Using RenSeq (Resistance gene Enrichment Sequencing) (41), we identified WRR12 (previously reported as SOC3) as a gene on chromosome 1 that confers AcBoT resistance (42). These data provide insights into the genetic basis of resistance that restricts pathogen host range and open up a greater subset of the gene pool of crop relatives as a source of genes for crop protection.
Results
Identification of Ac2V-Susceptible MAGIC Lines.
All of 107 previously tested wild-type Arabidopsis accessions are resistant to B. juncea-infecting A. candida race Ac2V, but a Ws-2-eds1 mutant is susceptible (34). To test if resistance in different Arabidopsis accessions is due to distinct resistance gene loci, we evaluated MAGIC lines derived from 19 different Arabidopsis accessions (38). We tested Ac2V resistance in 593 MAGIC lines at adult leaf stage with four replicates and identified 10 MAGIC lines that showed either a chlorotic phenotype or different levels of susceptibility. Eight of these 10 lines showed strong chlorotic as well as necrotic patches on infected leaves, although two of these eight lines (MAGIC.453 and MAGIC.485) supported occasional pustule formation (Fig. 1). We regularly observed pustules on the two most susceptible MAGIC lines (MAGIC.23 and MAGIC.329) with Ac2V (Fig. 1). After inoculation with Ac2V, pustules appear 7–10 d after infection (dpi) with MAGIC.329 but later (12–14 dpi) with MAGIC.23 (Fig. 1). However, MAGIC.23 and MAGIC.329 are not as susceptible as Ws-2-eds1 or Col-eds1-2 plants.
Fig. 1.
Identification of transgressive segregant MAGIC lines showing different susceptibility to B. juncea-infecting A. candida race Ac2V. Different levels of susceptibility to Ac2V are observed in an eds1-2 mutant and in four of 593 MAGIC recombinant inbred lines. Adaxial (Left) and abaxial (Right) sides of the leaves are presented. Examples of pustules (arrows) and necrotic patches (arrowheads) are indicated. Susceptibility was scored in 4-wk-old plants at 14 dpi. (Scale bars: 3 mm.)
Genetic Segregation of Resistance and Susceptibility Phenotypes in F2 Progeny Derived from Crosses Between MAGIC Parents and Susceptible MAGIC.329 Line.
Identification of susceptible lines enables genetic analysis of resistance in Arabidopsis against Ac2V. We crossed MAGIC.329 with each of the 19 MAGIC parents and selfed F1 plants to obtain F2 populations. We also analyzed Ws-2 (also known as Ws, Ws-1, Ws-3, and Ws-4, but different from accession Ws-0 that is one of the MAGIC parents) (43) because of its adult plant resistance but seedling susceptibility to Ac2V. All F1 progeny were resistant. F2 populations were inoculated with Ac2V, and resistance or susceptibility was scored at 14 dpi. We classified F2 progeny into three phenotypes: resistant (Green Resistant, GR), partially resistant with chlorosis or necrosis but no pustules (Necrotic-Chlorotic Resistant, NCR), and susceptible, with pustules (Susceptible, S) (Table 1). Segregation ratios ranged from 13R:3S to 255R:1S, suggesting that different Arabidopsis accessions carry two to four unlinked WRR genes against Ac2V. All tested F2 plants from the MAGIC.329 × Wu-0 cross were resistant, suggesting >4 resistance loci.
Table 1.
Genetic segregation of resistance and susceptibility phenotypes in F2 populations between MAGIC.329 and MAGIC parents as well as Ws-2
| Interaction | ||||||
| F2 population | R, GR | R, CNR | S | Expected ratio, (R:S) | No. of loci | P |
| MAGIC.329 x Bur-0 | 135 | 24 | 1 | 63:1 | 3 | 0.34 |
| MAGIC.329 x Can-0 | 155 | 41 | 4 | 63:1 | 3 | 0.61 |
| MAGIC.329 x Col-0 | 147 | 10 | 30 | 13:3 | 2* | 0.34 |
| MAGIC.329 x Ct-1 | 140 | 18 | 4 | 63:1 | 3 | 0.35 |
| MAGIC.329 x Edi-0 | 500 | 16 | 2 | 255:1 | 4 | 0.98 |
| MAGIC.329 x Hi-0 | 151 | 32 | 23 | 15:1 | 2† | 0.0036 |
| MAGIC.329 x Kn-0 | 76 | 79 | 10 | 15:1 | 2 | 0.92 |
| MAGIC.329 x Ler-0 | 228 | 11 | 16 | 15:1 | 2 | 0.20 |
| MAGIC.329 x Mt-0 | 154 | 10 | 3 | 63:1 | 3 | 0.81 |
| MAGIC.329 x No-0 | 53 | 60 | 1 | 63:1 | 3 | 0.55 |
| MAGIC.329 x Oy-0 | 206 | 27 | 11 | 15:1 | 2 | 0.26 |
| MAGIC.329 x Po-0 | 74 | 26 | 4 | 15:1 | 2 | 0.31 |
| MAGIC.329 x Rsch-4 | 165 | 25 | 32 | 13:3 | 2* | 0.1 |
| MAGIC.329 x Sf-2 | 134 | 115 | 16 | 15:1 | 2 | 0.07 |
| MAGIC.329 x Tsu-0 | 223 | 23 | 21 | 15:1 | 2 | 0.27 |
| MAGIC.329 x Wil-2 | 205 | 69 | 5 | 63:1 | 3 | 0.75 |
| MAGIC.329 x Ws-0 | 126 | 32 | 11 | 15:1 | 2 | 0.89 |
| MAGIC.329 x Ws-2 | 170 | 58 | 46 | 13:3 | 2* | 0.40 |
| MAGIC.329 x Wu-0 | 200 | 0 | 0 | NT | NT | NT |
| MAGIC.329 x Zu-0 | 110 | 9 | 2 | 63:1 | 3 | 0.93 |
GR, green resistant; NCR, necrotic-chlorotic resistant; NT, not tested; P, probability value following χ2 test; R, resistant; S, susceptible.
One dominant and one recessive gene.
Two linked genes.
Most MAGIC Parents Carry Resistance That Maps to the WRR4 Locus.
The Arabidopsis WRR4Col-0 gene (At1g56510) confers resistance against multiple races of A. candida in Arabidopsis and in B. juncea (33, 40). WRR4 encodes a TIR-NLR protein. A. candida infects by entry of a germ tube into stomata and production of a primary vesicle under an epidermal cell. WRR4 arrests the development of the pathogen in this epidermal cell, which undergoes a hypersensitive response (HR) (40). As these HR symptoms are not visible macroscopically, we classify this phenotype as GR. We scored susceptible F2 individuals using markers at the WRR4 locus (Dataset S1) and observed cosegregation between Ac2V resistance and WRR4 for all of the Arabidopsis accessions tested except Sf-2 and Wil-2 (SI Appendix, Table S1). Cosegregation of Ws-2 resistance with the WRR4 locus was unexpected, as the WRR4 gene is absent in Ws-2 (SI Appendix, Fig. S1 and Dataset S2), suggesting that at least one more gene at the WRR4 locus could confer Ac2V resistance.
Ac2V Resistance in Ws-2 Is Conferred by the WRR4A Paralog WRR4B.
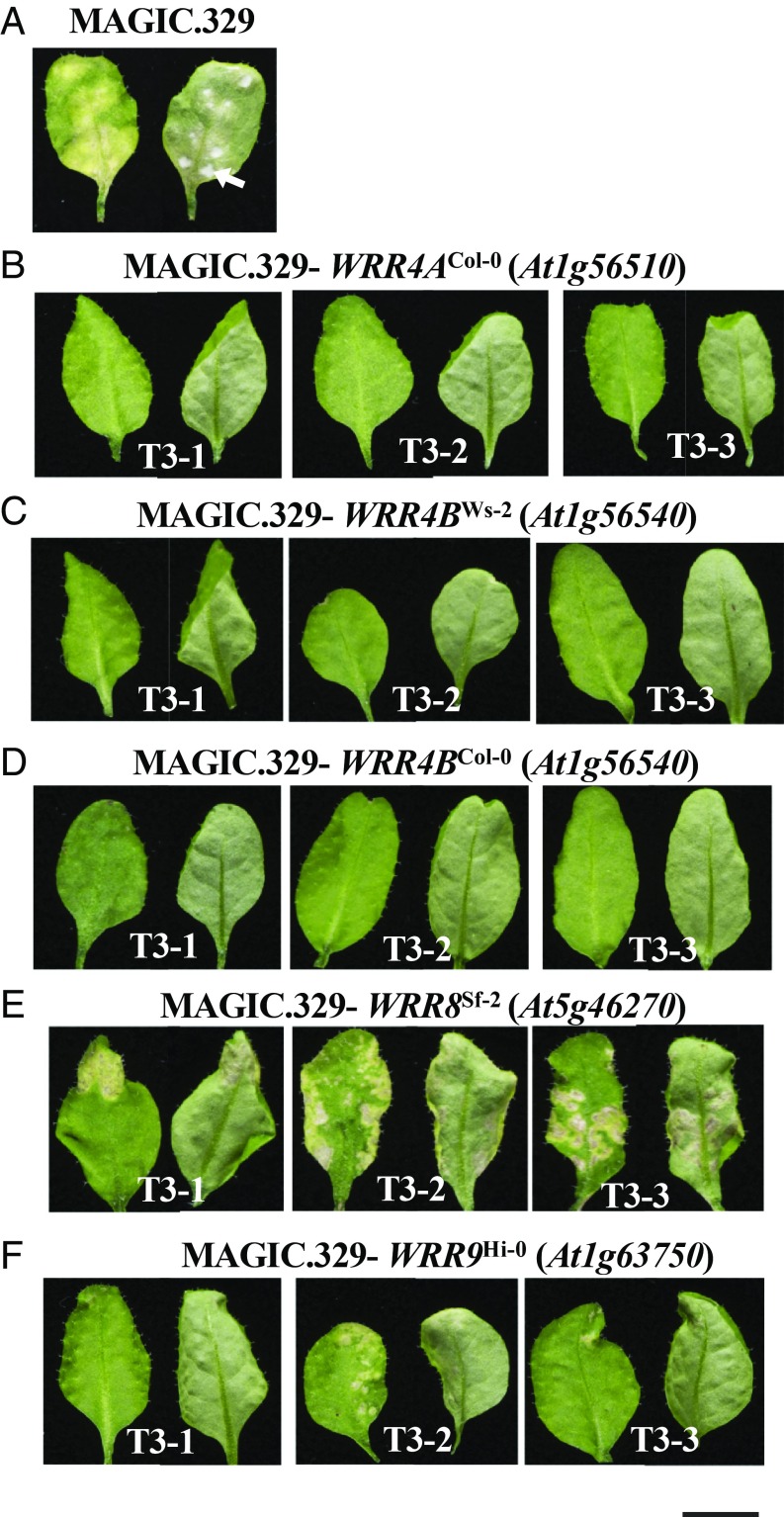
Previously, cotyledons of Ws-2 seedlings were found to be susceptible to Ac2V and Ac7V (a Brassica rapa-infecting race) but not to AcBoT (a B. oleracea-infecting race) (40). However, Ws-2 leaves are fully resistant (GR) to Ac2V and Ac7V. An F2 population derived from MAGIC.329 × Ws-2 segregated as 13 GR/NCR: 3 S (P = 0.40), suggesting one dominant and one recessive or haplo-insufficient Ac2V resistance gene in Ws-2. All Ac2V-susceptible individuals from the MAGIC.329 × Ws-2 F2 lacked the Ws-2 alleles of the markers at the WRR4 locus. By screening susceptible F2 individuals with additional molecular markers (Dataset S1), we found no other loci linked to Ac2V resistance. To improve definition of the resistance locus, we identified 672 Ac2V-susceptible F2 plants. We found two recombinants with the molecular marker corresponding to At1g56040 and only one recombinant with the marker corresponding to At1g57670. These markers delineated the locus to ∼397 kb (SI Appendix, Fig. S2A). WRR4 maps to this interval in Col-0 but is deleted in Ws-2 (SI Appendix, Fig. S1). We therefore cloned two other WRR4 paralogs At1g56520 and At1g56540 from Ws-2 and transformed them into MAGIC.329. For each construct, we tested Ac2V resistance in 48 independent T1 plants and in homozygous T3 lines. All plants transformed with At1g56520Ws-2 were susceptible to Ac2V (SI Appendix, Fig. S3A), but plants with At1g56540Ws-2 were all resistant (GR) (Fig. 2C). We named this gene WRR4B. We also cloned the Col-0 allele of WRR4B, transformed it into MAGIC.329, and found it also confers resistance to Ac2V (Fig. 2D). This suggests that in addition to the broad-spectrum A. candida resistance gene WRR4Col-0 (hereafter WRR4ACol-0), the WRR4B allele of Col-0 functions against Ac2V.
Fig. 2.
Distinct WRR genes confer resistance to Ac2V in the susceptible MAGIC.329 line. (A) Nontransformed MAGIC.329 line. (B–F) Independent homozygous T3 MAGIC.329 lines transformed with the genomic clones of WRR4ACol-0 (At1g56510) (B), WRR4BWs-2 (At1g56540) (C), WRR4BCol-0 (At1g56540) (D), WRR8Sf-2 (At5g46270) (E), and WRR9Hi-0 (At1g63750) (F). Interaction phenotypes were assayed at 12 dpi. Examples of pustules (arrows) are indicated. (Scale bar: 5 mm.)
Ac2V Resistance in Sf-2 Is Conferred by a Resistance Gene, WRR8.
Analysis of MAGIC line DNA sequences indicates that the MAGIC.329 WRR4 haplotype derives from Sf-2 (39). As MAGIC.329 is susceptible to Ac2V, this suggests that Sf-2 lacks functional WRR4A and WRR4B alleles. Screening of susceptible MAGIC.329 × Sf-2 F2 progeny confirmed that resistance is unlinked to WRR4. We genotyped susceptible F2 individuals derived from a MAGIC.329 × Sf-2 cross. A single locus was revealed on chromosome 5 between molecular markers derived from At5g45400 and At5g47130 (SI Appendix, Fig. S2B). Fine mapping using 576 additional susceptible F2 individuals revealed an interval between markers derived from At5g46250 (one recombinant) and At5g46310 (four recombinants) that carries two TIR-NLR–encoding genes At5g46260 and At5g46270 in Col-0. We cloned both genes from Arabidopsis accession Sf-2, transformed them into MAGIC.329, inoculated T1 plants with Ac2V, and found that transgenic plants carrying At5g46260Sf-2 were all susceptible (48 of 48), but most plants carrying At5g46270Sf-2 showed chlorotic resistance (40 of 48) to Ac2V (Fig. 2E and SI Appendix, Fig. S3B). At5g46270 thus corresponds to WRR8 in Sf-2.
Cloning of WRR9 from Arabidopsis Accession Hi-0.
The WRR4 locus in the Arabidopsis accession Hi-0 is linked to Ac2V resistance. Using 352 susceptible F2 individuals derived from a MAGIC.329 × Hi-0 cross, we found an additional resistance locus (WRR9) on chromosome 1, distinct from WRR4. WRR9 lies between At1g57670 (one recombinant in 352 plants) and At1g63820 (one recombinant in 352 plants) (SI Appendix, Fig. S2C). We thus defined three TIR-NLR WRR9 candidate genes At1g63730, At1g63740, and At1g63750. We cloned all three genes from Hi-0, transformed into MAGIC.329, and tested T1 plants with Ac2V. All of the plants transformed with At1g63730Hi-0 and At1g63740Hi-0 were susceptible, but 43 of 48 transgenic T1 plants with At1g63750Hi-0 were resistant to Ac2V (Fig. 2F). We infer WRR9 corresponds to At1g63750.
WRR4B but Not WRR8 and WRR9 Confer Resistance to Ac2V in B. juncea.
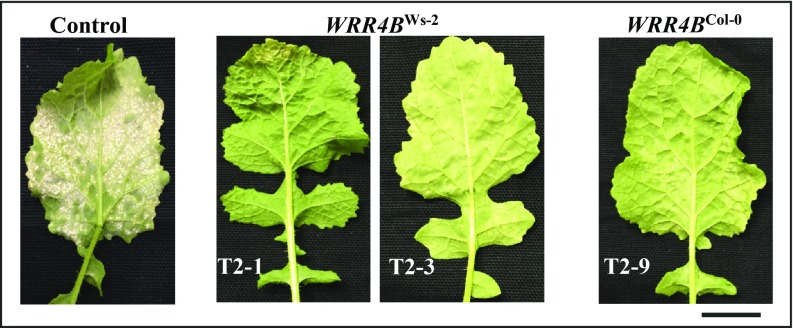
WRR4ACol-0 confers resistance to two different races of A. candida in B. juncea and Brassica napus (33). We transformed WRR4B, WRR8, and WRR9 into B. juncea, obtained two independent transgenic B. juncea plants with WRR4BWs-2 but only one transgenic plant with the WRR4BCol-0, and tested T2 plants derived from these lines. WRR4B transgenic B. juncea lines showed green to chlorotic resistance to Ac2V (Fig. 3), resembling the Arabidopsis phenotype (Fig. 2 C and D). We obtained two and four independent transgenic B. juncea plants with WRR8Sf-2 and WRR9Hi-0, respectively. Following inoculation with Ac2V, the T2 plants obtained from these independent transgenic lines were all fully susceptible to the pathogen (SI Appendix, Fig. S4), although reverse transcription–PCR (RT-PCR) revealed that WRR8Sf-2 and WRR9Hi-0 were expressed in these lines (SI Appendix, Fig. S5).
Fig. 3.
Arabidopsis WRR genes provide resistance to A. candida race Ac2V in B. juncea. Col-0 and Ws-2 alleles of WRR4B provide resistance to Ac2V in transgenic B. juncea. Nontransgenic control plants and independent T2 plants transformed with the indicated WRR genes were inoculated with Ac2V. The pictures were taken at 15 dpi. (Scale bar: 10 mm.)
Transgressive Segregation for AcBoT Susceptibility in a MAGIC.329 × MAGIC.23 F2 Reveals WRR12, an Additional TIR-NLR for AcBoT Resistance.
MAGIC.329 and MAGIC.23 are resistant or partially resistant, respectively, to B. oleracea-infecting A. candida race AcBoT. To identify potential transgressive segregants susceptible to AcBoT, we crossed MAGIC.329 × MAGIC.23 and obtained F2 progeny. Inoculation of this F2 with AcBoT revealed fully susceptible individuals. The F2 population segregated as 15 GR or NCR: 1 S (200GR+34CR:19S) (P = 0.41), suggesting a single dominant WRR gene is present in each parent. To test if AcBoT-susceptible F2 lines are also susceptible to other Brassica-infecting A. candida races, we obtained F4 plants derived from independent susceptible F2 lines. We named these plants as “Double MAGIC” (DM) lines. We found that DM lines are also fully susceptible to A. candida races Ac2V and Ac7V (SI Appendix, Fig. S6).
To identify the underlying genes conferring resistance to AcBoT in MAGIC.329 and MAGIC.23, we collected ∼200 fully susceptible F2 individuals following AcBoT inoculation. To accelerate the cloning, we conducted RenSeq (41) on DNA of the resistant parents MAGIC.329 and MAGIC.23 as well as bulked susceptible DNA (BS) obtained from the fully susceptible F2 individuals. MiSeq reads obtained from the parents and from BS were used to identify polymorphisms and linkage by mapping the reads to the Col-0 reference genome. This revealed a single locus where the resistance gene from MAGIC.329 is located (SI Appendix, Table S2). We named this gene WRR12 and found that, in MAGIC.329, this genomic region was introgressed from Ler-0, whereas the nonfunctional allele in MAGIC.23 was introgressed from Wu-0. We found no additional locus linked to the resistance in MAGIC.23, suggesting that its partial resistance could be multigenic. Three genes within the WRR12 locus cosegregate with resistance (SI Appendix, Table S2): the TIR-NLR gene At1g17600 and TIR-NB–only genes At1g17610 and At1g17615 (SI Appendix, Fig. S7). At1g17600 was previously designated SUSA1 or SOC3 and implicated in cold-induced activation of defense by an allele of At1g17610 (CHS1) (42, 44, 45). Recently, SOC3/CHS1 was proposed to “guard” an immune-regulating E3 ligase SAUL1 (46). TN2 (At1g17615) was reported to be required for the enhanced disease resistance phenotype in exo70B1 mutant Arabidopsis plants (47).
The At1g17600 allele from Wu-0 in MAGIC.23 (and only this allele; Dataset S2) carries a ∼4-kb transposon insertion (SI Appendix, Fig. S7 and Dataset S2), suggesting that it is nonfunctional, and that the Ler-0 allele in MAGIC.329 is a strong candidate for WRR12-mediated resistance. We cloned At1g17600 from MAGIC.329 and transformed into line DM10, one of the DM lines. Independent T1 transgenic plants were screened with A. candida race AcBoT. All 24 T1 transgenic DM10 plants were resistant to AcBoT. This suggests that At1g17600Ler-0 corresponds to WRR12 (Fig. 4). We also transformed WRR4BCol-0, WRR8Sf-2, and WRR8Hi-0 into DM10 to determine if these genes confer resistance to AcBoT in Arabidopsis. We found all WRR4BCol-0 transgenic T1 plants (eight of eight) were resistant to AcBoT, while seven of eight WRR8Sf-2 transgenic plants showed resistance to the pathogen. In contrast, WRR9Hi-0 transgenic DM10 lines (nine of nine) were fully susceptible to AcBoT (Fig. 4).
Fig. 4.
WRR12Ler-0, WRR4BCol-0, WRR8Sf-2, but not WRR9Hi-0 confer resistance to B. oleracea-infecting A. candida race AcBoT in Arabidopsis. MAGIC.329 and MAGIC.23 are resistant or partially resistant, respectively, to AcBoT. DM10 lines were transformed with WRR12Ler-0 (At1g17600), WRR4BCol-0, WRR8Sf-2, and WRR9Hi-0 and interaction phenotypes were assayed in independent T1 plants at 20 dpi. (Scale bar: 10 mm.)
In addition, we transformed B. oleracea DH1012 with WRR4ACol-0, WRR4BCol-0, and WRR4BWs-2, as well as At1g56520Col-0 as a negative control and inoculated independent T1 transgenic B. oleracea lines with AcBoT. T1 transgenic plants with WRR4ACol-0 (15 of 16), WRR4BWs-2 (13 of 19), and WRR4BCol-0 (two of two) were fully resistant to AcBoT, whereas transgenic plants with At1g56520Col-0 (four of four) were fully susceptible (SI Appendix, Fig. S8).
WRR Gene Haplotypes in MAGIC Parents.
To determine the distribution and sequence variation of WRR4A, WRR4B, WRR8, WRR9, and WRR12 genes, the MAGIC parents as well as Ws-2 were sequenced using SMRT RenSeq (48). The sequences of the WRR alleles from each accession were identified by blastn (49) against the SMRT RenSeq assemblies. Blastn hits showing less than 95% identity were not considered to be alleles of the WRR genes. We used the Augustus gene prediction server (50) to obtain predicted protein sequences of the WRR alleles. We identified WRR4A alleles in all MAGIC parent accessions except Ws-2, Edi-0, and No-0 (Dataset S2). We also identified WRR4B alleles in all Arabidopsis accessions except Tsu-0 in the RenSeq assemblies. Both Sf-2 and Wil-2 (source of the WRR4 haplotypes in MAGIC.329 and MAGIC.23, respectively) lack functional WRR4A and WRR4B genes. We identified RenSeq assemblies for WRR4A and WRR4B in both Arabidopsis accessions, and the lack of functional WRR4A and WRR4B in Sf-2 and Wil-2 is not due to deletion (Dataset S2). Although the Sf-2 WRR4A region was not clearly resolved in the de novo assembly, by aligning the RenSeq reads to the Col-0 genome, we confirmed a single base deletion, also observed in the Arabidopsis 1001 genomes browser, at nucleotide position 177 that results in an early stop codon, explaining why the Sf-2 WRR4A allele is nonfunctional.
Blastn analysis revealed that all of the Arabidopsis accessions contain WRR8 and WRR9 alleles except for WRR9 in Ler-0. However, as for the WRR4A and WRR4B alleles, some of the assemblies did not cover full-length WRR8 and WRR9. This is most likely due to partial SMRT RenSeq assemblies or incomplete capture.
We also identified WRR12 alleles in MAGIC parents and Ws-2. All lines carried an apparently functional allele, except for Wu-0.
Discussion
NHR in one plant species can be defined as complete resistance to pathogens that infect another species (26). Multiple mechanisms, such as preformed antimicrobial metabolites, and induced defenses such as PTI and ETI, could contribute to NHR (51, 52). A better understanding of the mechanisms of NHR could reveal additional genes that confer resistance in crops to plant pathogens.
We investigated NHR in Arabidopsis against Brassica-infecting A. candida races. All Arabidopsis accessions tested are resistant to B. juncea-infecting race Ac2V, B. rapa-infecting race Ac7V, and B. oleracea-infecting race AcBoT (ref. 34, this study). However, we found that both Col-0-eds1-2 (53) and Ws-2-eds1 (34) are susceptible to all three A. candida races, suggesting that NHR to these races might involve TIR-NLR genes (23). We further hypothesized that resistance in different Arabidopsis accessions could be mediated by distinct resistance genes. Therefore, we screened MAGIC lines derived from 19 different Arabidopsis parents (38) and identified transgressive segregant lines that are susceptible to Ac2V. These susceptible plants enabled us to perform genetic analysis to identify resistance genes in multiple Arabidopsis accessions.
We defined three WRR (WRR4BCol-0, WRR8Sf-2, and WRR9Hi-0) genes against Ac2V, and a gene, WRR12 (SOC3), conferring NHR to AcBoT, in addition to the previously identified broad spectrum resistance gene WRR4ACol-0. Other investigations have revealed additional WRR genes, but we focus in this paper on resistances at the WRR4, 8, 9, and 12 loci. A point mutation in At1g17610, the neighboring gene of WRR12 encoding a TIR-NB protein, results in chilling sensitive 1 (CHS1), with an autoactive defense phenotype (44). This phenotype could be suppressed by mutations in WRR12, which was therefore named suppressor of chilling sensitive 1–3 (SOC3). SOC3 and CHS1 can associate physically (42).
A phylogenetic analysis using an alignment of the NB-ARC region of TNLs in Arabidopsis accession Col-0 reveals that WRR4, WRR4B, and WRR9 are monophyletic, suggesting they shared a more recent common ancestor than with WRR8 (SI Appendix, Fig. S9). This analysis also reveals that WRR12 and CHS1 are located in neighboring expanded clades, many members of which are part of divergently transcribed pairs in the Col-0 genome (SI Appendix, Fig. S9). This suggests that multiple duplications of an ancestral WRR12/CHS1 pair occurred, similar to the expansion that occurred of RPS4/RRS1-like pairs (refs. 54 and 55 and SI Appendix, Fig. S9).
Neither WRR8 nor WRR9 confer resistance to Ac2V in B. juncea, although these genes confer resistance in Arabidopsis. WRR8 also confers resistance to AcBoT in Arabidopsis. This could be due to the fact that WRR8- and WRR9-mediated resistance involves a guardee or decoy that is present in Arabidopsis but absent or divergent in Brassica sp. Indeed, recent publications show that WRR12/SOC3 and CHS1 form a gene pair and that WRR12/SOC3, together with CHS1, monitors the homeostasis of E3 ligase SAUL1, a potential guardee that we hypothesize might be targeted by A. candida effector(s) (42, 46).
F2 individuals from crosses between MAGIC.329 and Col-0, Rsch-4, or Ws-2 segregated at a ratio of 13:3, suggesting one dominant and one recessive or haplo-insufficient gene. Identification of a second resistance locus in these F2s will require genotyping fully resistant individuals that lack resistant WRR4 haplotypes. Crosses between MAGIC.329 and Oy-0 or Sf-2 show a 15:1 segregation in the F2, suggesting two independent dominant resistance loci, but genotyping susceptible plants revealed only one locus. How many more WRR genes might there be in Arabidopsis? All F2 individuals resulting from selfing the F1 between MAGIC.329 × Wu-0 are resistant, suggesting that Wu-0 likely contains >4 resistance loci, so additional loci for resistance to Ac2V and AcBoT likely remain to be discovered.
Our data suggest that Arabidopsis NHR against Brassica-infecting A. candida races is primarily mediated via ETI, consistent with the expectation that ETI is more likely to contribute to NHR if there is a close evolutionary relationship between the host and nonhost plant species (29). ETI may contribute to NHR in other plant pathosystems. For example, various RxLR effectors from Phytophthora infestans trigger a HR in the nonhost pepper (56). NHR to P. capsici in various Nicotiana species likely involves PcAvr3a1 effector recognition (57). Pseudomonas syringae AvrRps4 homologs (HopK1DC3000 and AvrRps4Pph1448A) trigger HR in lettuce, and this HR phenotype cosegregates with a NLR locus RGC4 (58). NHR against wheat stripe rust (Puccinia striiformis f. sp. tritici) in barley or in Brachypodium distachyon was mapped to Rps6 or Yrr2 loci, respectively. Both intervals were shown to contain NLR genes, suggesting that NLRs may contribute to NHR against wheat stripe rust in barley and B. distachyon (59–61). Furthermore, nonhost resistance to Lolium and Avena isolates of Pyricularia oryzae in wheat was shown to be mediated by two resistance genes, Rwt3 and Rwt4, and the emergence of wheat blast was attributed to a host jump as a result of widespread growth of rwt3 wheat (62).
NLR-encoding resistance genes recognize pathogen effectors. When A. candida races of Ac2V and Ac7V were intercrossed, and F2 individuals obtained and inoculated on B. rapa (host for Ac7V but nonhost for Ac2V), a segregation ratio of three avirulent to one virulent was obtained. This supports the hypothesis that resistance to Ac2V in B. rapa involves resistance gene-dependent recognition of an Ac2V effector allele that is absent from or different in Ac7V (63).
Specific races of A. candida, usually considered a generalist pathogen, colonize a particular host species (34). Why might resistance genes in nonhost plants recognize effectors from nonadapted pathogens? Conceivably, host and nonhost plants share a common ancestor that was a host for the pathogen (56). Our data suggest that host/race specificity of A. candida is determined by the NLR repertoire of the host plant and the recognized effectors of the pathogen race, rather than host compatibility factors. Therefore, some of the NLRs recognizing specific races or multiple races are maintained in different Brassicaceae species. This, in turn, provides an excellent resource to identify WRR genes for different Brassica species. In summary, by using transgressive segregation to reveal susceptible lines, we were able to reveal genes that underpin resistance in Arabidopsis to Brassica-infecting A. candida races and show that some of these genes might be useful for elevating crop disease resistance. This strategy could also be applied to identify useful new resistance genes in other crop relatives that show NHR to crop-adapted pathogen races.
Materials and Methods
All Arabidopsis accessions used in this study were obtained from the Nottingham Arabidopsis Stock Centre. Col-0-eds1-2 and Ws-2-eds1 were described in refs. 32 and 48. MAGIC lines were described in ref. 38. Arabidopsis seeds were sown on Scotts Levington F2 compost (Scotts) and vernalized for 1 wk at 5–6 °C. Seedlings were subsequently grown in a controlled environment room (CER) with a 10-h day and a 14-h night photoperiod and at a constant temperature of 22 °C for 2 wk and then pricked-out into “Arabidopsis mix” [Scotts Levington F2 compost-grit (6:1, vol/vol), 0.03% (m/v) Intercept insecticide] and returned to the CER. B. juncea seeds were sown on Scotts Levington F2 compost. Seedlings were subsequently grown in a controlled environment room (CER) with a 10-h day and a 14-h night photoperiod and at a constant temperature of 22 °C for 1 wk and then pricked-out into Arabidopsis mix and returned to the CER. Detailed information is provided in SI Appendix, Supplementary Materials and Methods.
Supplementary Material
Acknowledgments
We thank Matthew Smoker and Jodie Taylor for help with Arabidopsis and Brassica transformation and the members of the Gordon and Betty Moore Foundation (GBMF)-funded Ren-seq Team—Felix Bemm, Anna-Lena Van de Weyer, Freddy Monteiro, Jeff Dangl, and Detlef Weigel—for their help in generating the RenSeq NLR database. F.B., W.A., A.R.-S., and D.C.P. were supported by European Research Council Advanced Investigator Grant 233376 (ALBUGON) (to J.D.G.J.); V.C. was supported by the Biotechnology and Biological Sciences Research Council (BBSRC) Grant BB/L011646/1; O.J.F. was supported by BBSRC Grant BB/M003809/1; A.R. was supported by European Molecular Biology Organization Long-Term Fellowship ALTF-842-2015. The authors also were supported by GBMF Grant GBMF4725.
Footnotes
Conflict of interest statement: J.D.G.J., O.J.F., and G.V.d.A. with 31 others are coauthors on a 2014 review article. J.D.G.J. and R.P. are coauthors on a 2014 research article (PubMed identifier: 25211078) but did not collaborate directly for this article.
Data deposition: The sequences reported in this paper have been deposited in the GenBank database: WRR4BWs-2 (accession no. MK034466), WRR4BCol-0 (MK034465), WRR8Sf-2 (MK034463), WRR9Hi-0 (MK034464), and WRR12Ler-0 (MK034462). Illumina reads for REN-Seq data produced for this study have been deposited in the European Nucleotide Archive (ENA) under accession no. PRJEB26457. SMRT RenSeq sequence reads for Arabidopsis accession Can-0 for this study have been deposited in the ENA under accession no. PRJEB26457.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1812911116/-/DCSupplemental.
References
- 1.Jones JD, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Dangl JL, Jones JD. Plant pathogens and integrated defence responses to infection. Nature. 2001;411:826–833. doi: 10.1038/35081161. [DOI] [PubMed] [Google Scholar]
- 3.Dodds PN, Rathjen JP. Plant immunity: Towards an integrated view of plant-pathogen interactions. Nat Rev Genet. 2010;11:539–548. doi: 10.1038/nrg2812. [DOI] [PubMed] [Google Scholar]
- 4.Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cell. 2014;54:263–272. doi: 10.1016/j.molcel.2014.03.028. [DOI] [PubMed] [Google Scholar]
- 5.Boutrot F, Zipfel C. Function, discovery, and exploitation of plant pattern recognition receptors for broad-spectrum disease resistance. Annu Rev Phytopathol. 2017;55:257–286. doi: 10.1146/annurev-phyto-080614-120106. [DOI] [PubMed] [Google Scholar]
- 6.Rovenich H, Boshoven JC, Thomma BP. Filamentous pathogen effector functions: Of pathogens, hosts and microbiomes. Curr Opin Plant Biol. 2014;20:96–103. doi: 10.1016/j.pbi.2014.05.001. [DOI] [PubMed] [Google Scholar]
- 7.Dangl JL, Horvath DM, Staskawicz BJ. Pivoting the plant immune system from dissection to deployment. Science. 2013;341:746–751. doi: 10.1126/science.1236011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Deslandes L, Rivas S. Catch me if you can: Bacterial effectors and plant targets. Trends Plant Sci. 2012;17:644–655. doi: 10.1016/j.tplants.2012.06.011. [DOI] [PubMed] [Google Scholar]
- 9.Maekawa T, Kufer TA, Schulze-Lefert P. NLR functions in plant and animal immune systems: So far and yet so close. Nat Immunol. 2011;12:817–826. doi: 10.1038/ni.2083. [DOI] [PubMed] [Google Scholar]
- 10.Cui H, Tsuda K, Parker JE. Effector-triggered immunity: From pathogen perception to robust defense. Annu Rev Plant Biol. 2015;66:487–511. doi: 10.1146/annurev-arplant-050213-040012. [DOI] [PubMed] [Google Scholar]
- 11.Jacob F, Vernaldi S, Maekawa T. Evolution and conservation of plant NLR functions. Front Immunol. 2013;4:297. doi: 10.3389/fimmu.2013.00297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Césari S, et al. The NB-LRR proteins RGA4 and RGA5 interact functionally and physically to confer disease resistance. EMBO J. 2014;33:1941–1959. doi: 10.15252/embj.201487923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Le Roux C, et al. A receptor pair with an integrated decoy converts pathogen disabling of transcription factors to immunity. Cell. 2015;161:1074–1088. doi: 10.1016/j.cell.2015.04.025. [DOI] [PubMed] [Google Scholar]
- 14.Sarris PF, et al. A plant immune receptor detects pathogen effectors that target WRKY transcription factors. Cell. 2015;161:1089–1100. doi: 10.1016/j.cell.2015.04.024. [DOI] [PubMed] [Google Scholar]
- 15.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ueda H, Yamaguchi Y, Sano H. Direct interaction between the tobacco mosaic virus helicase domain and the ATP-bound resistance protein, N factor during the hypersensitive response in tobacco plants. Plant Mol Biol. 2006;61:31–45. doi: 10.1007/s11103-005-5817-8. [DOI] [PubMed] [Google Scholar]
- 17.Dodds PN, et al. Direct protein interaction underlies gene-for-gene specificity and coevolution of the flax resistance genes and flax rust avirulence genes. Proc Natl Acad Sci USA. 2006;103:8888–8893. doi: 10.1073/pnas.0602577103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Krasileva KV, Dahlbeck D, Staskawicz BJ. Activation of an Arabidopsis resistance protein is specified by the in planta association of its leucine-rich repeat domain with the cognate oomycete effector. Plant Cell. 2010;22:2444–2458. doi: 10.1105/tpc.110.075358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Catanzariti AM, et al. The AvrM effector from flax rust has a structured C-terminal domain and interacts directly with the M resistance protein. Mol Plant Microbe Interact. 2010;23:49–57. doi: 10.1094/MPMI-23-1-0049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 21.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 22.Wang G, et al. The decoy substrate of a pathogen effector and a pseudokinase specify pathogen-induced modified-self recognition and immunity in plants. Cell Host Microbe. 2015;18:285–295. doi: 10.1016/j.chom.2015.08.004. [DOI] [PubMed] [Google Scholar]
- 23.Wiermer M, Feys BJ, Parker JE. Plant immunity: The EDS1 regulatory node. Curr Opin Plant Biol. 2005;8:383–389. doi: 10.1016/j.pbi.2005.05.010. [DOI] [PubMed] [Google Scholar]
- 24.Feys BJ, et al. Arabidopsis SENESCENCE-ASSOCIATED GENE101 stabilizes and signals within an ENHANCED DISEASE SUSCEPTIBILITY1 complex in plant innate immunity. Plant Cell. 2005;17:2601–2613. doi: 10.1105/tpc.105.033910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rietz S, et al. Different roles of enhanced disease susceptibility1 (EDS1) bound to and dissociated from phytoalexin Deficient4 (PAD4) in Arabidopsis immunity. New Phytol. 2011;191:107–119. doi: 10.1111/j.1469-8137.2011.03675.x. [DOI] [PubMed] [Google Scholar]
- 26.Heath MC. Nonhost resistance and nonspecific plant defenses. Curr Opin Plant Biol. 2000;3:315–319. doi: 10.1016/s1369-5266(00)00087-x. [DOI] [PubMed] [Google Scholar]
- 27.Mysore KS, Ryu CM. Nonhost resistance: How much do we know? Trends Plant Sci. 2004;9:97–104. doi: 10.1016/j.tplants.2003.12.005. [DOI] [PubMed] [Google Scholar]
- 28.Niks RE, Marcel TC. Nonhost and basal resistance: How to explain specificity? New Phytol. 2009;182:817–828. doi: 10.1111/j.1469-8137.2009.02849.x. [DOI] [PubMed] [Google Scholar]
- 29.Schulze-Lefert P, Panstruga R. A molecular evolutionary concept connecting nonhost resistance, pathogen host range, and pathogen speciation. Trends Plant Sci. 2011;16:117–125. doi: 10.1016/j.tplants.2011.01.001. [DOI] [PubMed] [Google Scholar]
- 30.Kemen E, et al. Gene gain and loss during evolution of obligate parasitism in the white rust pathogen of Arabidopsis thaliana. PLoS Biol. 2011;9:e1001094. doi: 10.1371/journal.pbio.1001094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Holub EB, et al. Phenotypic and genotypic variation in the interaction between Arabidopsis thaliana and Albugo candida. Mol Plant Microbe Interact. 1995;8:916–928. doi: 10.1094/mpmi-8-0916. [DOI] [PubMed] [Google Scholar]
- 32.Saharan GSVP, Meena PD, Kumar A. White Rust of Crucifers: Biology, Ecology and Management. Springer; New Delhi: 2014. [Google Scholar]
- 33.Borhan MH, et al. WRR4, a broad-spectrum TIR-NB-LRR gene from Arabidopsis thaliana that confers white rust resistance in transgenic oilseed Brassica crops. Mol Plant Pathol. 2010;11:283–291. doi: 10.1111/j.1364-3703.2009.00599.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McMullan M, et al. Evidence for suppression of immunity as a driver for genomic introgressions and host range expansion in races of Albugo candida, a generalist parasite. eLife. 2015;4:e04550. doi: 10.7554/eLife.04550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ploch S, et al. Evolution of diversity in Albugo is driven by high host specificity and multiple speciation events on closely related Brassicaceae. Mol Phylogenet Evol. 2010;57:812–820. doi: 10.1016/j.ympev.2010.06.026. [DOI] [PubMed] [Google Scholar]
- 36.Jouet A, et al. Albugo candida race diversity, ploidy and host-associated microbes revealed using DNA sequence capture on diseased plants in the field. New Phytol. October 5, 2018 doi: 10.1111/nph.15417. [DOI] [PubMed] [Google Scholar]
- 37.Cooper AJ, et al. Basic compatibility of Albugo candida in Arabidopsis thaliana and Brassica juncea causes broad-spectrum suppression of innate immunity. Mol Plant Microbe Interact. 2008;21:745–756. doi: 10.1094/MPMI-21-6-0745. [DOI] [PubMed] [Google Scholar]
- 38.Kover PX, et al. A multiparent advanced generation inter-cross to fine-map quantitative traits in Arabidopsis thaliana. PLoS Genet. 2009;5:e1000551. doi: 10.1371/journal.pgen.1000551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gan X, et al. Multiple reference genomes and transcriptomes for Arabidopsis thaliana. Nature. 2011;477:419–423. doi: 10.1038/nature10414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Borhan MH, et al. WRR4 encodes a TIR-NB-LRR protein that confers broad-spectrum white rust resistance in Arabidopsis thaliana to four physiological races of Albugo candida. Mol Plant Microbe Interact. 2008;21:757–768. doi: 10.1094/MPMI-21-6-0757. [DOI] [PubMed] [Google Scholar]
- 41.Jupe F, et al. Resistance gene enrichment sequencing (RenSeq) enables reannotation of the NB-LRR gene family from sequenced plant genomes and rapid mapping of resistance loci in segregating populations. Plant J. 2013;76:530–544. doi: 10.1111/tpj.12307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, et al. Temperature-dependent autoimmunity mediated by chs1 requires its neighboring TNL gene SOC3. New Phytol. 2017;213:1330–1345. doi: 10.1111/nph.14216. [DOI] [PubMed] [Google Scholar]
- 43.Anastasio AE, et al. Source verification of mis-identified Arabidopsis thaliana accessions. Plant J. 2011;67:554–566. doi: 10.1111/j.1365-313X.2011.04606.x. [DOI] [PubMed] [Google Scholar]
- 44.Wang Y, Zhang Y, Wang Z, Zhang X, Yang S. A missense mutation in CHS1, a TIR-NB protein, induces chilling sensitivity in Arabidopsis. Plant J. 2013;75:553–565. doi: 10.1111/tpj.12232. [DOI] [PubMed] [Google Scholar]
- 45.Zbierzak AM, et al. A TIR-NBS protein encoded by Arabidopsis Chilling Sensitive 1 (CHS1) limits chloroplast damage and cell death at low temperature. Plant J. 2013;75:539–552. doi: 10.1111/tpj.12219. [DOI] [PubMed] [Google Scholar]
- 46.Tong M, et al. E3 ligase SAUL1 serves as a positive regulator of PAMP-triggered immunity and its homeostasis is monitored by immune receptor SOC3. New Phytol. 2017;215:1516–1532. doi: 10.1111/nph.14678. [DOI] [PubMed] [Google Scholar]
- 47.Zhao T, et al. A truncated NLR protein, TIR-NBS2, is required for activated defense responses in the exo70B1 mutant. PLoS Genet. 2015;11:e1004945. doi: 10.1371/journal.pgen.1004945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Witek K, et al. Accelerated cloning of a potato late blight-resistance gene using RenSeq and SMRT sequencing. Nat Biotechnol. 2016;34:656–660. doi: 10.1038/nbt.3540. [DOI] [PubMed] [Google Scholar]
- 49.Camacho C, et al. BLAST+: Architecture and applications. BMC Bioinformatics. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hoff KJ, Stanke M. WebAUGUSTUS–A web service for training AUGUSTUS and predicting genes in eukaryotes. Nucleic Acids Res. 2013;41:W123–W128. doi: 10.1093/nar/gkt418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Lee HA, et al. Current understandings on plant nonhost resistance. Mol Plant Microbe Interact. 2017;30:5–15. doi: 10.1094/MPMI-10-16-0213-CR. [DOI] [PubMed] [Google Scholar]
- 52.Fan J, Doerner P. Genetic and molecular basis of nonhost disease resistance: Complex, yes; silver bullet, no. Curr Opin Plant Biol. 2012;15:400–406. doi: 10.1016/j.pbi.2012.03.001. [DOI] [PubMed] [Google Scholar]
- 53.Bartsch M, et al. Salicylic acid-independent ENHANCED DISEASE SUSCEPTIBILITY1 signaling in Arabidopsis immunity and cell death is regulated by the monooxygenase FMO1 and the Nudix hydrolase NUDT7. Plant Cell. 2006;18:1038–1051. doi: 10.1105/tpc.105.039982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Narusaka M, Kubo Y, Shiraishi T, Iwabuchi M, Narusaka Y. A dual resistance gene system prevents infection by three distinct pathogens. Plant Signal Behav. 2009;4:954–955. doi: 10.4161/psb.4.10.9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Di Donato A, Andolfo G, Ferrarini A, Delledonne M, Ercolano MR. Investigation of orthologous pathogen recognition gene-rich regions in solanaceous species. Genome. 2017;60:850–859. doi: 10.1139/gen-2016-0217. [DOI] [PubMed] [Google Scholar]
- 56.Lee HA, et al. Multiple recognition of RXLR effectors is associated with nonhost resistance of pepper against Phytophthora infestans. New Phytol. 2014;203:926–938. doi: 10.1111/nph.12861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vega-Arreguín JC, Jalloh A, Bos JI, Moffett P. Recognition of an Avr3a homologue plays a major role in mediating nonhost resistance to Phytophthora capsici in Nicotiana species. Mol Plant Microbe Interact. 2014;27:770–780. doi: 10.1094/MPMI-01-14-0014-R. [DOI] [PubMed] [Google Scholar]
- 58.Wroblewski T, et al. Comparative large-scale analysis of interactions between several crop species and the effector repertoires from multiple pathovars of Pseudomonas and Ralstonia. Plant Physiol. 2009;150:1733–1749. doi: 10.1104/pp.109.140251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dawson AM, et al. Isolation and fine mapping of Rps6: An intermediate host resistance gene in barley to wheat stripe rust. Theor Appl Genet. 2016;129:831–843. doi: 10.1007/s00122-015-2659-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Li K, et al. Fine mapping of barley locus Rps6 conferring resistance to wheat stripe rust. Theor Appl Genet. 2016;129:845–859. doi: 10.1007/s00122-015-2663-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Gilbert B, et al. Components of Brachypodium distachyon resistance to nonadapted wheat stripe rust pathogens are simply inherited. PLoS Genet. 2018;14:e1007636. doi: 10.1371/journal.pgen.1007636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Inoue Y, et al. Evolution of the wheat blast fungus through functional losses in a host specificity determinant. Science. 2017;357:80–83. doi: 10.1126/science.aam9654. [DOI] [PubMed] [Google Scholar]
- 63.Adhikari TB, Liu JQ, Mathur S, Wu CX, Rimmer SR. Genetic and molecular analyses in crosses of race 2 and race 7 of Albugo candida. Phytopathology. 2003;93:959–965. doi: 10.1094/PHYTO.2003.93.8.959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.