Visualizing flu virus uncoating in infected cells
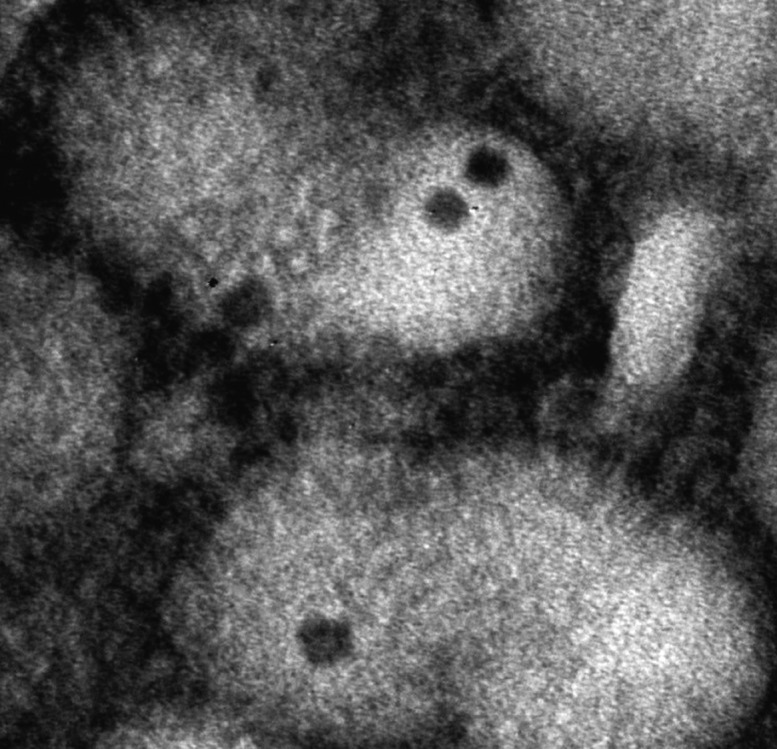
Transmission electron micrograph of IAV virions encapsulating quantum dots.
The genome of influenza A virus (IAV) comprises eight segmented RNA strands enclosed in a coat. During infection, the virus is uncoated in the cytoplasm of host cells, viral RNA is imported into the host nucleus, and viral replication ensues. Viral uncoating represents an antiviral therapeutic target, but few researchers have observed the transient and dynamic event in detail. Chong Qin et al. (pp. 2577–2582) fashioned IAV virions labeled with quantum dots and infected a canine kidney cell line with the labeled virions. Using single-particle tracking, the authors monitored viral uncoating in real time. Around 30% of IAV particles fused with late endosomes, a cellular sorting compartment, and shed their coats in the region surrounding the host nucleus within 30–90 minutes of infection. Treatment of host cells with amantadine, which blocks viral replication, or ammonium chloride, which blocks endosome maturation, sharply decreased uncoating efficiency. Viral ribonucleoprotein complexes (vRNP)—composed of proteins and viral RNA—from each virion separated into distinct units and entered the nucleus in a three-step process, indicating that vRNP segments are imported into the nucleus separately rather than as a bundle, as previously suggested. Treatment of host cells with importazole, which blocks nuclear import, greatly reduced vRNP import efficiency. According to the authors, the findings unveil a critical step in IAV infection and may aid the development of therapeutic strategies. — P.N.
Mammal foot posture and body size evolution
In mammals, foot postures reflect successful adaptations spanning a wide range of ecological niches. For example, the limb morphologies and biomechanics of whole-foot walkers, such as humans and mice, closely track underlying differences in their evolutionary trajectories, compared with toe-walkers, such as cats and dogs. Tai Kubo, Manabu Sakamoto, et al. (pp. 2618–2623) applied Bayesian statistical methods to a dataset spanning 880 extant species and reconstructed the evolutionary histories of foot postures in mammals. The authors’ analysis suggests that the ancestral mammal was likely a plantigrade animal, or flat-footed walker, whose descendants evolved to digitigrade, or tip-toed postures, and later to unguligrade, or hooved postures. Furthermore, the reconstructions revealed that transitions from one posture to another coincided with sharp increases in rates of body size evolution, suggesting links between foot posture and the emergence of large descendants. Although the current findings do not establish that foot posture drove increases in body size or vice versa, the macroevolutionary statistical approach can be applied to datasets spanning broader ranges of living and extinct terrestrial tetrapods to address this question, according to the authors. — T.J.


