Significance
RNA polymerase (RNAP) carries out transcription from DNA to RNA. The double-stranded helical nature of DNA necessitates RNAP rotation of DNA during active elongation, leading to DNA supercoiling and the accumulation of torsional stress, which may ultimately stall transcription. As a torsional motor, RNAP generates and works against torsion, but it remains unclear whether and how RNAP’s torque generation capacity may be regulated. In this work, real-time single-molecule transcriptional assays revealed that, in response to increased torsion, RNAP extensively backtracks along DNA and ultimately becomes stalled. A transcription factor, GreB, can effectively increase RNAP’s stall torque by limiting RNAP backtracking. This work provides the first illustration of how a transcription factor can regulate the capacity of transcriptional torque generation.
Keywords: RNA polymerase, DNA supercoiling, transcription factors, optical trapping, modeling
Abstract
During transcription, RNA polymerase (RNAP) supercoils DNA as it translocates. The resulting torsional stress in DNA can accumulate and, in the absence of regulatory mechanisms, becomes a barrier to RNAP elongation, causing RNAP stalling, backtracking, and transcriptional arrest. Here we investigate whether and how a transcription factor may regulate both torque-induced Escherichia coli RNAP stalling and the torque generation capacity of RNAP. Using a unique real-time angular optical trapping assay, we found that RNAP working against a resisting torque was highly prone to extensive backtracking. We then investigated transcription in the presence of GreB, a transcription factor known to rescue RNAP from the backtracked state. We found that GreB greatly suppressed RNAP backtracking and remarkably increased the torque that RNAP was able to generate by 65%, from 11.2 pN⋅nm to 18.5 pN·nm. Variance analysis of the real-time positional trajectories of RNAP after a stall revealed the kinetic parameters of backtracking and GreB rescue. These results demonstrate that backtracking is the primary mechanism by which torsional stress limits transcription and that the transcription factor GreB effectively enhances the torsional capacity of RNAP. These findings suggest a broader role for transcription factors in regulating RNAP functionality and elongation.
RNA polymerase (RNAP) is a powerful biological motor, capable of generating both forces and torques. Although the force generation capacity has been well characterized for both Escherichia coli RNAP (1) and Pol II (2), much less is known about RNAP’s capacity to generate torque (3). During transcription, RNAP overtwists the downstream DNA and undertwists the upstream DNA, as described by the twin-supercoiled domain model (4). Increasing evidence is emerging that transcription induces genome-wide supercoiling that propagates thousands of base pairs from transcription start sites (5–7), and the accumulation of torsional stress may hinder further translocation of RNAP if not relaxed in a timely fashion via DNA rotation or through the action of topoisomerases. Such torsion may also up- or down-regulate transcription by creating highly supercoiled DNA (8–10), encouraging the formation of non-B DNA (11, 12), recruiting regulatory proteins (9, 10, 13), and/or removing roadblock proteins (14).
Although RNAP-generated torsion is widely recognized as an essential component of fundamental cellular processes and has been shown to have significant impact on gene expression (9, 10), the torsional properties of RNAP have yet to be fully understood. As a torsional motor, RNAP may be characterized by its torque generation capacity, and this valuable parameter was long sought after (4, 9, 10, 15, 16). Historically, the most prevalent approach has been biochemical studies using 2D gels which have provided a wealth of information on the degree of DNA supercoiling generated by transcription. Recent torque measurements during real-time transcription have allowed a more complete understanding of RNAP’s torsional mechanics. In particular, studies using the angular optical trap (AOT) showed that excessive torque accumulation during transcription stalls an elongating E. coli RNAP, with a measured RNAP stall torque of ∼11 pN·nm (17). This value represents the intrinsic capacity of RNAP for torque generation and establishes a baseline for a physiologically relevant torque scale.
Intriguingly, under (−) DNA supercoiling behind a transcribing RNAP, the mean stall torque value, 11 pN·nm, coincides with the torque required for DNA melting, where DNA undergoes a phase transition from B-DNA to melted, denatured DNA (12, 18), and this transition plays critical roles in gene regulation, such as facilitating transcription initiation (19–21) and binding of transcription factors (9, 10). Conversely, (+) DNA supercoiling ahead of the RNAP may dissociate protein roadblocks (14) and thus facilitate the passage of an elongating RNAP. Thus, perturbations to the torque generation capacity should lead to sensitive tuning of the level of gene expression. Although, under (−) supercoiling, the torque RNAP can generate is limited to ∼11 pN·nm due to the DNA melting transition, under (+) supercoiling, DNA is able to sustain a much greater torque without undergoing any structural denaturation (22–25). However, it is unclear whether and how RNAP may further increase its torque generation capacity under (+) supercoiling.
In vivo, RNAP functions closely with many transcription factors which regulate RNAP activities, and therefore it is possible that these transcription factors might also regulate the torsional properties of RNAP. Our previous studies suggested that torque-induced stalling might be due to backtracking (17), during which RNAP reverse translocates along DNA with the catalytic site disengaged from the 3′ end of the RNA, inactivating transcription (26, 27). A universal class of transcription elongation factors, including TFIIS in eukaryotic cells (28, 29) and GreB in prokaryotic cells (30–32), rescues backtracked complexes and promotes transcription through obstructive regions of DNA such as regulatory transcription pause sites. These proteins act either as auxiliary factors or, in the case of RNA Polymerase I and RNA Polymerase III, as parts of the core RNAP (33, 34). They stimulate the intrinsic cleavage activities of RNAP, leading to the removal of the 3′ end of the RNA. The newly generated RNA 3′ end then becomes aligned with the catalytic site, reactivating transcription. Thus, here we have systematically investigated whether and how the interaction of E. coli GreB with RNAP allows RNAP to more efficiently transcribe through regions of DNA under torsional stress.
Results
Single-Molecule Assay for Transcription Stalling Under Torque.
To investigate RNAP’s stall torque regulation, we must first fully understand the nature of the torque-induced stalling. We thus employed a single-molecule AOT assay (17, 24, 35) to closely examine transcription stalling in real time under (+) DNA supercoiling in the presence or absence of the transcription factor GreB.
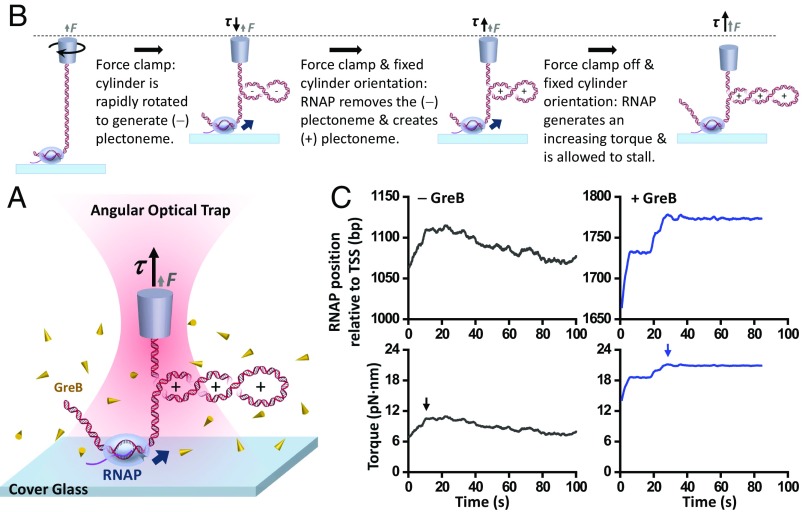
As shown in Fig. 1A, an RNAP was torsionally anchored to the surface of a microscope coverslip, with the downstream end of its transcribing DNA torsionally anchored to the bottom of a nanofabricated quartz cylinder held in an AOT. Subsequently, RNAP elongation was monitored as torque accumulated on downstream DNA (Fig. 1 B and C). Under this configuration, RNAP forward translocation introduced (+) supercoiling into the DNA, buckling the DNA to form a plectoneme and inducing a rapid shortening of DNA extension. As torque increased, RNAP was ultimately stalled, which we define here as <1 bp/s forward translocation for ∼60 s. This velocity threshold is significantly below the pause-free elongation velocity under zero torque (∼20 bp/s) (1, 36) while well above the drift velocity of the instrument (∼0.06 bp/s) (SI Appendix, Fig. S1). In addition, the force applied here was typically <2 pN, much smaller than the force required to stall the RNAP (about 25 pN) (1, 37), and thus stalling was primarily a result of the resisting torque.
Fig. 1.
Measuring RNAP stall torque in the presence of GreB. (A) Experimental configuration of transcription stalling under torsion using an AOT. RNAP was torsionally anchored to the surface of a microscope coverslip. A quartz cylinder was attached to the downstream end of the DNA and was held in the AOT. RNAP transcription induced (+) supercoiling in the downstream DNA. (B) Experimental steps of the stall torque measurements. (C) Representative traces of RNAP stall torque measurements in the absence or presence of GreB during the last step of stalling experiments. RNAP template position was defined as the RNAP’s physical position relative to the transcription start site (TSS). Arrows indicate the beginning of stalling.
When RNAP worked on its own and became stalled, it moved bidirectionally but with an overall motion in the reverse direction, reminiscent of a biased random walk. There are some finer features of motion during stalling, such as apparent transient pauses, which may be a result of the stochastic nature of translocation (38). It is also possible that RNA secondary structures restrict further backtracking (39, 40).
In the presence of 1 µM of GreB, upon stalling, RNAP showed minimal motion around the stalling position. Under this concentration of GreB, the probability of RNAP having a bound GreB should be ∼91% with a binding rate of ∼10/s, estimated using binding parameters from Tetone et al. (41), who also observed a short dwell time of GreB in RNAP (0.3 s to 0.5 s). Therefore, during the course of our experiment, RNAP should experience rapid GreB binding and unbinding, but remain in a GreB-bound state for the vast majority of time.
GreB Limits Backtracking After a Torque-Induced Stall.
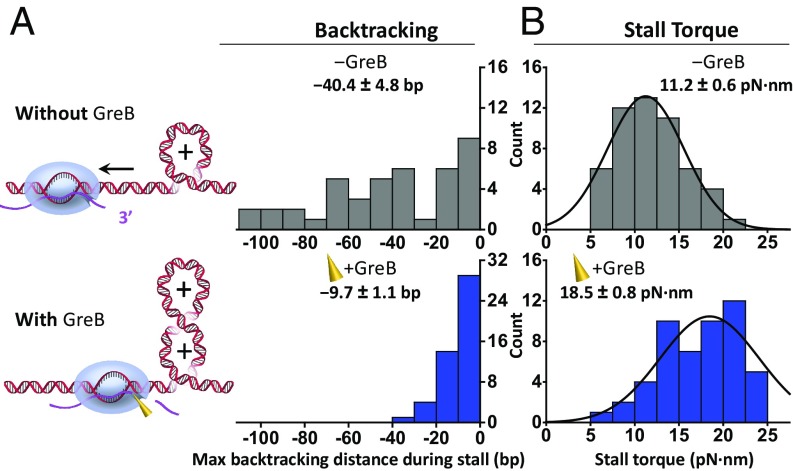
To characterize RNAP behavior at a stall, we examined RNAP movement during stalling by characterizing its maximum backtracking distance, which is defined as the maximum reverse distance during the 60 s of stalling. In the absence of GreB (Fig. 2A, Top), RNAP became significantly backtracked upon stalling. The mean maximum backtracking distance was −40.4 ± 4.8 bp (mean ± SEM), with 70% backtracked over 20 bp and some even exceeding 100 bp. The extent of backtracking is significantly greater than force-induced backtracking reported earlier for E. coli RNAP (1, 42–44). We speculate that this may be due to the fact that, during force-induced stalling experiments, upon backtracking, force decreases rapidly, thus reducing the bias toward backtracking. In contrast, under our torque-induced stalling experiments, upon backtracking, torque decreases gradually, thus retaining the bias toward backtracking. This difference is a direct result of the differential response of DNA’s linear and torsional properties to RNAP motion. These results demonstrate that a resisting torsional stress can be highly effective in inducing RNAP backtracking.
Fig. 2.
RNAP backtracking distance and stall torque in the absence (Top) and presence (Bottom) of GreB. (A) The distributions of the maximum backtracking distance during stalling in the absence and presence of GreB, with the means and SEMs indicated. (B) The distributions of the stall torque in the absence and presence of GreB. Each distribution was fit with a Gaussian function with its mean and SEM indicated.
In contrast, the presence of GreB greatly reduced the backtracking distance. Approximately 94% of traces showed a maximum backtracking distance of −9.7 ± 1.1 bp (mean ± SEM) (Fig. 2A, Bottom), which is a fourfold reduction from that seen with RNAP alone. The remaining 6% of traces showed a much greater backtracking distance of around 40 bp, comparable to those of RNAP alone. We interpret these traces as corresponding to RNAP molecules that were unable to interact with GreB. Indeed, a recent single-molecule study of GreB by fluorescence visualization also found that a minority fraction of RNAP molecules was incapable of binding to GreB (41). We thus excluded this fraction from further analysis.
These results demonstrate that, even when RNAP is under substantial torsion, GreB can effectively limit backtracking, likely by stimulating the cleavage activity of RNAP. Subsequent resumption of transcription provides a renewed opportunity for RNAP to work against a greater torque.
GreB Greatly Enhances Stall Torque Capacity.
To examine the effect of GreB on the torque generation capacity of RNAP, we measured the stall torque of RNAP for each trace and examined the resulting stall torque histogram. In the absence of GreB, the stall torque distribution was fit by a single Gaussian function, yielding a mean torque of 11.2 ± 0.6 pN·nm (mean ± SEM) (Fig. 2B, Top), consistent with our previous measurements (17). In the presence of GreB, the stall torque distribution shifts to greater values with a mean torque of 18.5 ± 0.8 pN·nm, 65% greater than the stall torque generated by RNAP alone (Fig. 2B, Bottom).
These data demonstrate that GreB can play an important role in up-regulating RNAP’s torsional capacity during transcription. Although the torque enhancement is attributable to the presence of GreB, torque generation capacity of RNAP stems from the RNAP itself, while GreB serves to catalyze the conversion of a backtracked complex to an active elongation complex.
Variance Analysis Theory.
To better understand the stalling behavior in the presence of GreB, we must determine the kinetic rates of backtracking and GreB rescue during a stall. Since RNAP showed little movement during stalling in the presence of GreB (e.g., Fig. 1C), this suggests a short backtracking distance before rescue that is difficult to experimentally discern. Fortuitously, if backtracking and rescue events are stochastic in nature, the position variance of RNAP should increase with time, and thus contains valuable information on the kinetics of backtracking and GreB rescue. In contrast, the variance of the Brownian noise remains the same over a long time scale and contributes minimally to the overall variance (SI Appendix).
To extract these kinetic parameters via a variance analysis, we present unique analytical solutions to a general two-state transition problem, derived from rigorous statistical mechanics and Eigenvector analysis (SI Appendix). Here the process of torque-induced stalling in the presence of GreB is modeled as reversible first-order transitions between two states: backtracking and forward elongation. We show how the mean RNAP position and the variance of the RNAP position evolve with time : (SI Appendix, Eq. S17) and (SI Appendix, Eq. S19), where is the backtracking velocity, is the active elongation velocity, is the GreB rescue rate, and is the rate to enter backtracking.
Kinetics of RNAP Backtracking and RNA Cleavage.
To determine the GreB rescue rate and the rate to enter backtracking using the variance analysis, we must first measure the backtracking velocity and active elongation velocity .
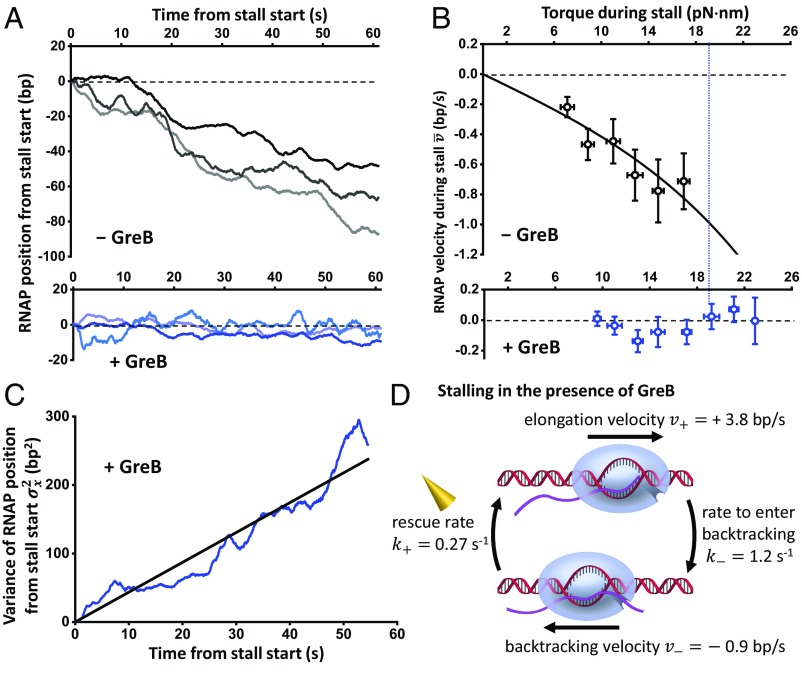
We determined the backtracking velocity from the real-time motion of RNAP during a stall in the absence of GreB. These data also provide a unique window of opportunity to dissect the backtracked states. Fig. 3A, Top shows representative traces with a torque of ∼18 pN·nm upon stalling, close to the mean stall torque with GreB. RNAP’s reverse motion due to backtracking was readily detectable, and the backtracking velocity increased with torque (Fig. 3B, Top). We model the backtracking process as a 1D Brownian diffusion process (36, 38, 45), with an effective hopping rate between backtracking states in the absence of torque and the net motion biased by torque (SI Appendix). Analysis of the RNAP velocity dependence on torque yielded 0.26 s−1. Thus, we were able to make a rather direct measurement of the backtracking hopping rate. To our knowledge, this rate has not been determined for E. coli RNAP. Our value is comparable to those estimated for Pol II using pause duration analysis (45). Our analysis of E. coli RNAP yielded a backtracking velocity of 0.9 bp/s at 18.5 pN·nm (the measured mean stall torque with GreB).
Fig. 3.
Kinetics of backtracking and GreB rescue. (A) Representative traces showing the RNAP position versus time after stalling in the absence (Top) and presence (Bottom) of GreB. For clarity, all traces are aligned with respect to the start of stall in both time and position. These traces have a torque ∼18 pN·nm upon stalling, close to the mean stall torque with GreB. (B) RNAP velocity versus torque during stall in the absence (Top) and presence (Bottom) of GreB. The error bars of torque are SD, and the error bars for RNAP velocity are SEMs. The black solid line is a fit to SI Appendix, Eq. S1, yielding a backtracking hopping rate in the absence of torque 0.26 s−1. The blue vertical dashed line indicates the mean stall torque in the presence of GreB (18.5 pN·nm). (C) Measured variance of RNAP position from the stall start versus time in the presence of GreB (blue). The black solid line is a linear fit passing through origin, yielding a slope of 4.4 bp2/s. (D) A two-state model of the RNAP during a stall in the presence of GreB (SI Appendix). We model the transitions between backtracking and elongation states as reversible first-order reactions.
We also determined the active elongation velocity as a function of torque (SI Appendix, Fig. S2) by measuring the velocity between pauses before stalling. The active elongation velocity decreased with an increase in resisting torque, and, previously, a similar decrease in velocity was observed in response to an increase in resisting force (36). We interpret this decrease as a result of a resisting torque biasing the pretranslocation state over the posttranslocation state. In the absence of torque, RNAP translocated at around 20 bp/s (17, 36), whereas, under a resisting torque of 18.5 pN·nm, RNAP forward translocation slowed down to 3.8 bp/s (SI Appendix, Fig. S2).
In the presence of GreB and during stalling, RNAP’s motion fluctuated slightly around the initial stalling position (Fig. 3A, Bottom), with the RNAP velocity having minimal dependence on torque (Fig. 3B, Bottom). The mean velocity was 0.05 bp/s (SI Appendix, Fig. S3), whose magnitude is much slower than that of backtracking or active elongation and is comparable to the instrument’s drift velocity (SI Appendix, Fig. S1C). However, the variance of the RNAP position grew with time, with a slope to a linear fit being 4.4 bp2/s (Fig. 3C). Therefore, we were able to obtain the GreB rescue rate 0.27 s−1 and the rate to enter backtracking 1.2 s−1.
Fig. 3D summarizes translocation velocities and transition rates. It shows that, in the presence of GreB and upon stalling, RNAP backtracks by ∼3 bp over ∼4 s before being rescued by GreB. RNAP then actively forward elongates for ∼3 bp over ∼1 s before backtracking again. In addition, the 4-s time to rescue suggests that a single rescue event requires multiple rounds (∼10) of GreB binding and unbinding events, in agreement with observations from a recent study of GreB performed under zero torsion and tension (41). This agreement suggests that GreB’s ability to rescue a backtracked RNAP may be insensitive to the torsion that RNAP experiences.
Although GreB is most well-known for its role in stimulating RNA cleavage, GreB may also limit the mobility of RNAP’s trigger loop and thereby inhibit transcription elongation (46). We have thus examined the pause-free, active elongation rate (SI Appendix, Fig. S2), and our data do not show any slowdown of the rate by the presence of GreB. Gre factors and TFIIS have also been suggested to have antiarrest activity that is independent of RNA cleavage (47, 48). The presence of GreB in the secondary channel of RNAP may hinder RNA extrusion through the channel and thus slow the backtracking rate (49). We therefore have ascertained how the value of the GreB rescue rate obtained via the variance analysis may be altered if the rate of active elongation and the rate of backtracking are different from what we have used for analysis. We found that value is rather insensitive to the elongation rate but more strongly depends on the backtracking rate, decreasing with a decrease in the backtracking rate (SI Appendix, Fig. S4).
Taken together, our theoretical model and the use of variance analysis provide a framework to obtain kinetic parameters from complex translocation trajectories. This framework is general and may be applied to many other two-state transition problems.
GreB Facilitates Transcription Resumption After a Stall.
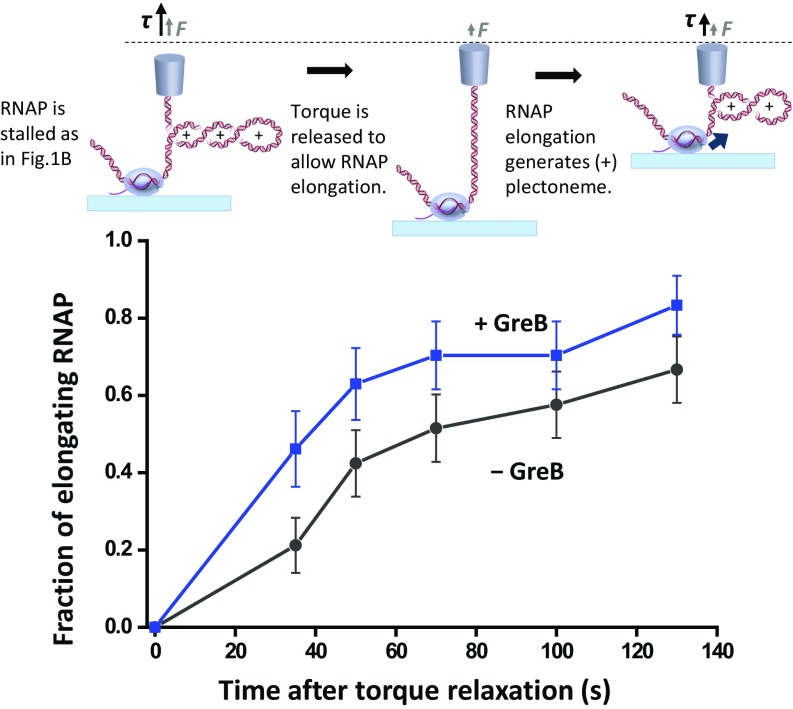
Upon torque relaxation, a stalled RNAP may exit the backtracked state to resume transcription, but how does the presence of GreB alter the rate of transcription resumption? To investigate, we first stalled the RNAP under torque, and then relaxed the torque to examine how fast transcription resumed, both in the absence and presence of GreB. As shown in Fig. 4, in the absence of GreB, 21% of stalled RNAPs resumed transcription within 35 s, consistent with our previous findings (17). In the presence of GreB, 46% resumed within the same time period. GreB also increased the fraction of RNAPs that resumed transcription at longer time scales. Therefore, upon torque relaxation, GreB expedites transcription resumption. This is likely due to a combined effect of GreB shortening the backtracking distance during the initial stall and GreB rescuing a backtracked complex upon torque relaxation.
Fig. 4.
Transcription resumption. After RNAP was stalled, torque on the RNAP was relaxed to allow transcription resumption. Subsequent transcription was detected by repeating the RNAP stall torque measurement as shown in Fig. 1B. Error bars indicate SEMs.
However, the extent that GreB facilitates the resumption is less than anticipated given GreB’s rescue rate determined in Fig. 3. We have thus examined whether this might have been a result of photodamage to RNAP by the trapping laser. We found that the transcription resumption time course was not significantly altered when the experiments were carried out in the presence of protocatechuic acid/protocatechuate-3,4-dioxygenase, a commonly employed oxygen scavenger system (SI Appendix, Fig. S5), suggesting that photodamage might not have been the primary cause. It is possible that the high torque afforded by the presence of GreB might induce some conformational changes in the RNAP, putting RNAP into an off-pathway (50, 51) that may be slow to transition back to the main pathway. These off-pathways may even require concurrent presence of high torque and GreB.
Torque Enhancement Model.
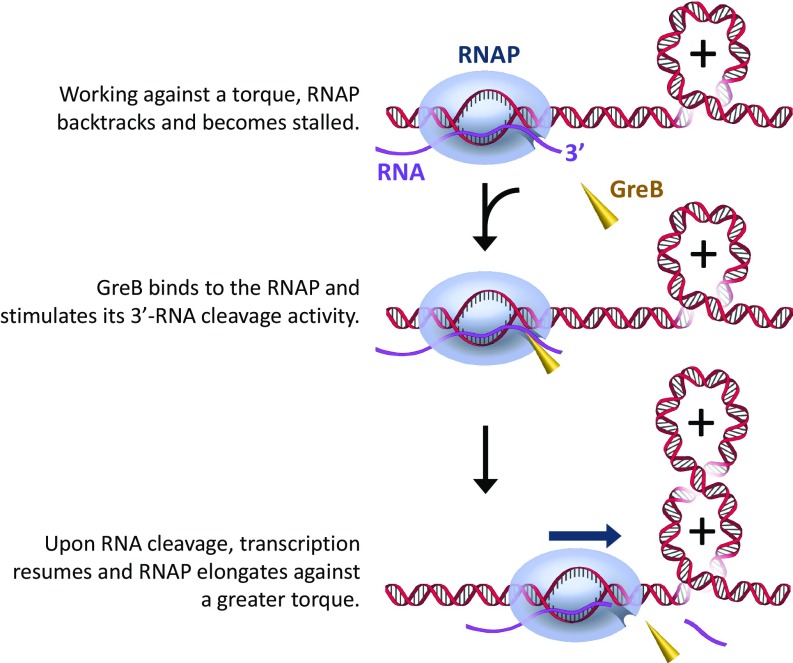
Our experimental results support the following torque enhancement model for transcription under torsion in the presence of GreB (Fig. 5). Transcription-generated torsional stress hinders RNAP forward translocation and is highly effective in inducing RNAP backtracking, which may lead to transcription arrest. GreB binding to RNAP stimulates 3′ RNA cleavage and thus prevents RNAP from extensive backtracking. Consequently, the stalled RNAP can be rescued, and transcription can be resumed. As this cycle repeats, RNAP works against an increasingly greater torque which further hinders forward transcription elongation. RNAP ultimately stalls when forward elongation is exactly reverted by backtracking. Upon stalling, RNAP repeats the cycle of forward elongation and backtracking, with no or minimal net motion.
Fig. 5.
A proposed model for RNAP’s enhanced torsional capacity by GreB. During transcription, torsional stress accumulates with transcription, and RNAP backtracks and is ultimately stalled. GreB can bind to the backtracked RNAP, stimulate its intrinsic RNA cleavage activity, and bring the RNAP back to elongation. Once transcription is resumed, the RNAP will be able to work against a stronger torsional barrier.
Discussion
Comparison of Force and Torque During Transcription.
As a DNA-based motor, RNAP generates not only force but also torque during transcription. Previous studies found that E. coli RNAP can generate ∼25 pN of force (1) and ∼11 pN·nm of torque (17). Assuming the same thermodynamic efficiency for conversion of chemical energy to carry out translocation and rotation, a stall force of 25 pN will convert to a stall torque of 14 pN·nm, greater than the measured torque. The difference may be due to multiple causes that are not mutually exclusive. It is possible that RNAP does not generate force and torque with the same thermodynamic efficiency. It is also possible that torque and force induce different conformational changes in RNAP that differentially impact RNAP’s functions.
Differential Consequences of Force and Torque.
The difference in RNAP’s response to mechanical force as opposed to torsional stress also suggests that force and torque may differentially regulate transcription. Consider the displacement of a bound protein that is a roadblock to an advancing RNAP. In order for RNAP to use force to displace the protein, RNAP may need to come into direct contact with the protein, making force a local/short distance regulator of transcription. In contrast, torsional stress can rapidly reach even a distant region of DNA via “action at a distance” such that RNAP can remotely perturb and displace without the need for direct contact with the protein, making torque a global/long distance regulator.
In addition, since torque alters DNA topology, torque can create DNA structures that are difficult to attain using force. For example, the force required to melt DNA or form DNA bubbles was found to be larger than 45 pN (52, 53), beyond what RNAP can generate. The melting torque of DNA, however, is ∼11 pN·nm, which is well attainable by RNAP. Torque, not force, has also been found to encourage formation of other nucleic acid structures, such as Z-DNA and R loops (54, 55).
Therefore, torque has diverse roles in how it impacts transcription and is potentially more important than force in the regulation of transcription. It is thus essential to understand what limits and regulates torque generation capacity.
Torque Generation Regulation.
The work presented here demonstrates that backtracking is the primary mechanism limiting RNAP’s torsional capacity. This limitation can be mitigated by the transcription factor GreB, which can decrease backtracking and enhance the torsional capacity of RNAP by 65%. In vivo, many other transcription factors also interact with E. coli RNAP and could potentially regulate RNAP’s torsional capacity as well. For example, NusA increases RNAP pausing (56) and thus may potentially decrease RNAP’s stall torque, while factors like GreA (30), DksA (57), NusG (58), and Mfd (59) prevent backtracking and may increase RNAP’s stall torque. These factors may work in coordination to down- or up-regulate RNAP’s torsional capacity to achieve a broad range of regulation. In this way, backtracking could serve as a general mediator for transcription factors to regulate RNAP torsional properties.
In the cell, the enhanced torsional capacity of RNAP may aid in overcoming obstacles, such as removing roadblock proteins or transcribing a long gene under high torsional stress while ensuring continuous elongation. Indeed, although cells are still viable under normal growth conditions when antibacktracking factors such as GreB and GreA are absent (60), these factors are necessary for efficient RNAP transcription through template-encoded arresting sites. For example, E. coli RNAP must transcribe through various bound proteins such as HU and H-NS, which create and maintain specific DNA conformations, such as DNA bridges or loops. Recently it has been shown that bridged, but not linear, H-NS filaments slow down transcription and strongly increase pausing by E. coli RNA polymerase at backtracking-prone pausing sites (61). These results favor a model in which the bridged H-NS filaments create topologically fixed domains that prevent dissipation of transcription-generated DNA supercoiling. Interestingly, the presence of antibacktracking factors, such as GreA and GreB, greatly facilitates transcription in these regions and reduces pausing. These observations are entirely consonant with our findings that GreB can promote transcription through torsional stress.
Although this work focuses on a prokaryotic RNAP, the findings here have implications for its eukaryotic counterparts. Eukaryotic Pol II must transcribe through nucleosomes to access genetic information, and nucleosomes form formidable barriers to transcription. It has recently been shown that transcription-generated (+) torsional stress increases nucleosome turnover within genes (7), and, indeed, single-molecule studies found that a (+) torque of ∼19 pN·nm or larger can disrupt nucleosome structures, leading to significant loss of H2A-H2B dimers (14). In addition, TFIIS, a functional analog of GreB in eukaryotic cells, has been shown to enhance the stall force of Pol II (2) and thus may also increase the torsional capacity of Pol II and facilitate its passage through the nucleosome barrier.
This work has demonstrated that RNAP’s torsional capacity can be regulated by transcription factors. The interaction kinetics of GreB and RNAP is on the scale of base pairs and seconds. This type of transient interaction between proteins and factors must occur broadly in the cell but is difficult to capture, due to their stochastic nature. The theoretical framework and experimental approach established here can potentially be applied to elucidate many other kinetic and mechanical processes that are important for cellular functions.
Materials and Methods
Protocols for E. coli RNAP purification (59) and GreB purification (32), as well as transcription and single-molecule assays (17), were performed as previously described. The detailed description of the methods and data analysis is provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank members of the M.D.W. laboratory for critical discussion and comments, Drs. James T. Inman and Jessica L. Killian for technical assistance, Dr. Eric Strobel for sharing the GreB plasmid and purification protocol, and Drs. James P. Sethna and Anatoly Kolomeisky for helpful discussions on biased random walks. This work was supported by HHMI (M.D.W.), National Science Foundation Grants MCB-0820293 and MCB-1517764 (to M.D.W.), National Natural Science Foundation of China Grant NSFC-11674403 (to J.M.), and the Fundamental Research Funds for the Central Universities Grant 18lgzd16 (to J.M.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. R.L. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1807031116/-/DCSupplemental.
References
- 1.Wang MD, et al. Force and velocity measured for single molecules of RNA polymerase. Science. 1998;282:902–907. doi: 10.1126/science.282.5390.902. [DOI] [PubMed] [Google Scholar]
- 2.Galburt EA, et al. Backtracking determines the force sensitivity of RNAP II in a factor-dependent manner. Nature. 2007;446:820–823. doi: 10.1038/nature05701. [DOI] [PubMed] [Google Scholar]
- 3.Ma J, Wang MD. RNA polymerase is a powerful torsional motor. Cell Cycle. 2014;13:337–338. doi: 10.4161/cc.27508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu LF, Wang JC. Supercoiling of the DNA template during transcription. Proc Natl Acad Sci USA. 1987;84:7024–7027. doi: 10.1073/pnas.84.20.7024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kouzine F, et al. Transcription-dependent dynamic supercoiling is a short-range genomic force. Nat Struct Mol Biol. 2013;20:396–403. doi: 10.1038/nsmb.2517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Naughton C, et al. Transcription forms and remodels supercoiling domains unfolding large-scale chromatin structures. Nat Struct Mol Biol. 2013;20:387–395. doi: 10.1038/nsmb.2509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Teves SS, Henikoff S. Transcription-generated torsional stress destabilizes nucleosomes. Nat Struct Mol Biol. 2014;21:88–94. doi: 10.1038/nsmb.2723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chong S, Chen C, Ge H, Xie XS. Mechanism of transcriptional bursting in bacteria. Cell. 2014;158:314–326. doi: 10.1016/j.cell.2014.05.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kouzine F, Liu J, Sanford S, Chung HJ, Levens D. The dynamic response of upstream DNA to transcription-generated torsional stress. Nat Struct Mol Biol. 2004;11:1092–1100. doi: 10.1038/nsmb848. [DOI] [PubMed] [Google Scholar]
- 10.Kouzine F, Sanford S, Elisha-Feil Z, Levens D. The functional response of upstream DNA to dynamic supercoiling in vivo. Nat Struct Mol Biol. 2008;15:146–154. doi: 10.1038/nsmb.1372. [DOI] [PubMed] [Google Scholar]
- 11.Leng F, Amado L, McMacken R. Coupling DNA supercoiling to transcription in defined protein systems. J Biol Chem. 2004;279:47564–47571. doi: 10.1074/jbc.M403798200. [DOI] [PubMed] [Google Scholar]
- 12.Oberstrass FC, Fernandes LE, Bryant Z. Torque measurements reveal sequence-specific cooperative transitions in supercoiled DNA. Proc Natl Acad Sci USA. 2012;109:6106–6111. doi: 10.1073/pnas.1113532109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ding Y, et al. DNA supercoiling: A regulatory signal for the λ repressor. Proc Natl Acad Sci USA. 2014;111:15402–15407. doi: 10.1073/pnas.1320644111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sheinin MY, Li M, Soltani M, Luger K, Wang MD. Torque modulates nucleosome stability and facilitates H2A/H2B dimer loss. Nat Commun. 2013;4:2579. doi: 10.1038/ncomms3579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Harada Y, et al. Direct observation of DNA rotation during transcription by Escherichia coli RNA polymerase. Nature. 2001;409:113–115. doi: 10.1038/35051126. [DOI] [PubMed] [Google Scholar]
- 16.Wu HY, Shyy SH, Wang JC, Liu LF. Transcription generates positively and negatively supercoiled domains in the template. Cell. 1988;53:433–440. doi: 10.1016/0092-8674(88)90163-8. [DOI] [PubMed] [Google Scholar]
- 17.Ma J, Bai L, Wang MD. Transcription under torsion. Science. 2013;340:1580–1583. doi: 10.1126/science.1235441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sheinin MY, Forth S, Marko JF, Wang MD. Underwound DNA under tension: Structure, elasticity, and sequence-dependent behaviors. Phys Rev Lett. 2011;107:108102. doi: 10.1103/PhysRevLett.107.108102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lilley DMJ, Higgins CF. Local DNA topology and gene expression: The case of the leu-500 promoter. Mol Microbiol. 1991;5:779–783. doi: 10.1111/j.1365-2958.1991.tb00749.x. [DOI] [PubMed] [Google Scholar]
- 20.Revyakin A, Ebright RH, Strick TR. Promoter unwinding and promoter clearance by RNA polymerase: Detection by single-molecule DNA nanomanipulation. Proc Natl Acad Sci USA. 2004;101:4776–4780. doi: 10.1073/pnas.0307241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Revyakin A, Liu C, Ebright RH, Strick TR. Abortive initiation and productive initiation by RNA polymerase involve DNA scrunching. Science. 2006;314:1139–1143. doi: 10.1126/science.1131398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bryant Z, et al. Structural transitions and elasticity from torque measurements on DNA. Nature. 2003;424:338–341. doi: 10.1038/nature01810. [DOI] [PubMed] [Google Scholar]
- 23.Deufel C, Forth S, Simmons CR, Dejgosha S, Wang MD. Nanofabricated quartz cylinders for angular trapping: DNA supercoiling torque detection. Nat Methods. 2007;4:223–225. doi: 10.1038/nmeth1013. [DOI] [PubMed] [Google Scholar]
- 24.Forth S, et al. Abrupt buckling transition observed during the plectoneme formation of individual DNA molecules. Phys Rev Lett. 2008;100:148301. doi: 10.1103/PhysRevLett.100.148301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Forth S, Sheinin MY, Inman J, Wang MD. Torque measurement at the single-molecule level. Annu Rev Biophys. 2013;42:583–604. doi: 10.1146/annurev-biophys-083012-130412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Komissarova N, Kashlev M. Transcriptional arrest: Escherichia coli RNA polymerase translocates backward, leaving the 3′ end of the RNA intact and extruded. Proc Natl Acad Sci USA. 1997;94:1755–1760. doi: 10.1073/pnas.94.5.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nudler E, Mustaev A, Lukhtanov E, Goldfarb A. The RNA-DNA hybrid maintains the register of transcription by preventing backtracking of RNA polymerase. Cell. 1997;89:33–41. doi: 10.1016/s0092-8674(00)80180-4. [DOI] [PubMed] [Google Scholar]
- 28.Izban MG, Luse DS. SII-facilitated transcript cleavage in RNA polymerase II complexes stalled early after initiation occurs in primarily dinucleotide increments. J Biol Chem. 1993;268:12864–12873. [PubMed] [Google Scholar]
- 29.Reinberg D, Roeder RG. Factors involved in specific transcription by mammalian RNA polymerase II. Purification and functional analysis of initiation factors IIB and IIE. J Biol Chem. 1987;262:3310–3321. [PubMed] [Google Scholar]
- 30.Marr MT, Roberts JW. Function of transcription cleavage factors GreA and GreB at a regulatory pause site. Mol Cell. 2000;6:1275–1285. doi: 10.1016/s1097-2765(00)00126-x. [DOI] [PubMed] [Google Scholar]
- 31.Stepanova E, Wang M, Severinov K, Borukhov S. Early transcriptional arrest at Escherichia coli rplN and ompX promoters. J Biol Chem. 2009;284:35702–35713. doi: 10.1074/jbc.M109.053983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Strobel EJ, Roberts JW. Regulation of promoter-proximal transcription elongation: Enhanced DNA scrunching drives λQ antiterminator-dependent escape from a σ70-dependent pause. Nucleic Acids Res. 2014;42:5097–5108. doi: 10.1093/nar/gku147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lisica A, et al. Mechanisms of backtrack recovery by RNA polymerases I and II. Proc Natl Acad Sci USA. 2016;113:2946–2951. doi: 10.1073/pnas.1517011113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chédin S, Riva M, Schultz P, Sentenac A, Carles C. The RNA cleavage activity of RNA polymerase III is mediated by an essential TFIIS-like subunit and is important for transcription termination. Genes Dev. 1998;12:3857–3871. doi: 10.1101/gad.12.24.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.La Porta A, Wang MD. Optical torque wrench: Angular trapping, rotation, and torque detection of quartz microparticles. Phys Rev Lett. 2004;92:190801. doi: 10.1103/PhysRevLett.92.190801. [DOI] [PubMed] [Google Scholar]
- 36.Bai L, Fulbright RM, Wang MD. Mechanochemical kinetics of transcription elongation. Phys Rev Lett. 2007;98:068103. doi: 10.1103/PhysRevLett.98.068103. [DOI] [PubMed] [Google Scholar]
- 37.Yin H, et al. Transcription against an applied force. Science. 1995;270:1653–1657. doi: 10.1126/science.270.5242.1653. [DOI] [PubMed] [Google Scholar]
- 38.Bai L, Shundrovsky A, Wang MD. Sequence-dependent kinetic model for transcription elongation by RNA polymerase. J Mol Biol. 2004;344:335–349. doi: 10.1016/j.jmb.2004.08.107. [DOI] [PubMed] [Google Scholar]
- 39.Klopper AV, Bois JS, Grill SW. Influence of secondary structure on recovery from pauses during early stages of RNA transcription. Phys Rev E Stat Nonlin Soft Matter Phys. 2010;81:030904. doi: 10.1103/PhysRevE.81.030904. [DOI] [PubMed] [Google Scholar]
- 40.Zamft B, Bintu L, Ishibashi T, Bustamante C. Nascent RNA structure modulates the transcriptional dynamics of RNA polymerases. Proc Natl Acad Sci USA. 2012;109:8948–8953. doi: 10.1073/pnas.1205063109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tetone LE, et al. Dynamics of GreB-RNA polymerase interaction allow a proofreading accessory protein to patrol for transcription complexes needing rescue. Proc Natl Acad Sci USA. 2017;114:E1081–E1090. doi: 10.1073/pnas.1616525114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shaevitz JW, Abbondanzieri EA, Landick R, Block SM. Backtracking by single RNA polymerase molecules observed at near-base-pair resolution. Nature. 2003;426:684–687. doi: 10.1038/nature02191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Herbert KM, et al. Sequence-resolved detection of pausing by single RNA polymerase molecules. Cell. 2006;125:1083–1094. doi: 10.1016/j.cell.2006.04.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Shundrovsky A, Santangelo TJ, Roberts JW, Wang MD. A single-molecule technique to study sequence-dependent transcription pausing. Biophys J. 2004;87:3945–3953. doi: 10.1529/biophysj.104.044081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Depken M, Galburt EA, Grill SW. The origin of short transcriptional pauses. Biophys J. 2009;96:2189–2193. doi: 10.1016/j.bpj.2008.12.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mishanina TV, Palo MZ, Nayak D, Mooney RA, Landick R. Trigger loop of RNA polymerase is a positional, not acid-base, catalyst for both transcription and proofreading. Proc Natl Acad Sci USA. 2017;114:E5103–E5112. doi: 10.1073/pnas.1702383114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borukhov S, Sagitov V, Goldfarb A. Transcript cleavage factors from E. coli. Cell. 1993;72:459–466. doi: 10.1016/0092-8674(93)90121-6. [DOI] [PubMed] [Google Scholar]
- 48.Cipres-Palacin G, Kane CM. Cleavage of the nascent transcript induced by TFIIS is insufficient to promote read-through of intrinsic blocks to elongation by RNA polymerase II. Proc Natl Acad Sci USA. 1994;91:8087–8091. doi: 10.1073/pnas.91.17.8087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Vassylyeva MN, et al. The carboxy-terminal coiled-coil of the RNA polymerase β’-subunit is the main binding site for Gre factors. EMBO Rep. 2007;8:1038–1043. doi: 10.1038/sj.embor.7401079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Kireeva ML, Kashlev M. Mechanism of sequence-specific pausing of bacterial RNA polymerase. Proc Natl Acad Sci USA. 2009;106:8900–8905. doi: 10.1073/pnas.0900407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Landick R. Transcriptional pausing without backtracking. Proc Natl Acad Sci USA. 2009;106:8797–8798. doi: 10.1073/pnas.0904373106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Smith SB, Cui Y, Bustamante C. Overstretching B-DNA: The elastic response of individual double-stranded and single-stranded DNA molecules. Science. 1996;271:795–799. doi: 10.1126/science.271.5250.795. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X, et al. Revealing the competition between peeled ssDNA, melting bubbles, and S-DNA during DNA overstretching by single-molecule calorimetry. Proc Natl Acad Sci USA. 2013;110:3865–3870. doi: 10.1073/pnas.1213740110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kouzine F, et al. Permanganate/S1 nuclease footprinting reveals non-B DNA structures with regulatory potential across a mammalian genome. Cell Syst. 2017;4:344–356. doi: 10.1016/j.cels.2017.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Scott S, et al. Visualizing structure-mediated interactions in supercoiled DNA molecules. Nucleic Acids Res. 2018;46:4622–4631. doi: 10.1093/nar/gky266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhou J, Ha KS, La Porta A, Landick R, Block SM. Applied force provides insight into transcriptional pausing and its modulation by transcription factor NusA. Mol Cell. 2011;44:635–646. doi: 10.1016/j.molcel.2011.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zhang Y, et al. DksA guards elongating RNA polymerase against ribosome-stalling-induced arrest. Mol Cell. 2014;53:766–778. doi: 10.1016/j.molcel.2014.02.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Herbert KM, et al. E. coli NusG inhibits backtracking and accelerates pause-free transcription by promoting forward translocation of RNA polymerase. J Mol Biol. 2010;399:17–30. doi: 10.1016/j.jmb.2010.03.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Le TT, et al. Mfd dynamically regulates transcription via a release and catch-up mechanism. Cell. 2018;172:344–357. doi: 10.1016/j.cell.2017.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Orlova M, Newlands J, Das A, Goldfarb A, Borukhov S. Intrinsic transcript cleavage activity of RNA polymerase. Proc Natl Acad Sci USA. 1995;92:4596–4600. doi: 10.1073/pnas.92.10.4596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kotlajich MV, et al. Bridged filaments of histone-like nucleoid structuring protein pause RNA polymerase and aid termination in bacteria. eLife. 2015;4:e04970. doi: 10.7554/eLife.04970. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.