One hundred years ago, the devastating 1918–1919 Spanish influenza pandemic took the lives of 50 to 100 million people, or 3 to 5% of the world population (1). Influenza A virus (IAV) pandemics occur when an animal IAV crosses the species barrier, usually by acquiring a new genetic trait by reassortment (2). According to the 2017 National Risk Register of Civil Emergencies of the United Kingdom, the predicted impact severity of a full-blown influenza pandemic is at the highest level; greater than that of coastal flooding (tsunami), major industrial accidents, and attacks on crowded places or transport. Has the advent of vaccines and antivirals truly increased our preparedness for the next influenza outbreak, as we struggle to predict which seasonal strains will circulate next winter? Needless to say, much is unknown about the cell biology of influenza infection and how the virus interacts with the multitude of host cellular processes that enable infection. Virus entry mechanics can be explored as a target for antiinfluenza therapy. To complement virus–host interaction studies using cell biology, biochemistry, and structural biology, robust live-imaging strategies that offer high temporal and spatial resolution are essential. However, the influenza RNA genome is intolerant to large genetic insertions, and efforts to rescue viruses using GFP fused to viral core proteins have had limited success (3). This has delayed progress in the field of influenza-entry live-imaging studies. In PNAS, Qin et al. (4) develop a nanotechnology that labels IAV viral ribonucleoprotein complexes (vRNPs) with quantum dots (QDs)—an approach that will advance the mechanistic understanding of influenza virus entry using live fluorescence microscopy.
Influenza virus is a member of the Orthomyxoviridae family. It is an enveloped, single-stranded negative-strand RNA virus with eight pinlike genomic segments called vRNPs protected by a single M1 matrix shell. Each vRNP consists of oligomeric nucleoprotein (NP), viral RNA, and viral polymerase made of PA, PB1, and PB2 subunits (5). A single NP molecule binds 24 to 27 nucleotides in vivo (6, 7) and has two sets of nuclear localization signals (NLSs), ensuring robust nuclear import of uncoated vRNPs during entry (8, 9). vRNPs are 10 nm in width and 30 to 110 nm in length (10) and package themselves into a revolving cylinderlike orientation, adopting a typical 7+1 arrangement with seven vRNPs surrounding a central one (11, 12). All eight viral RNA segments contain unique packaging sequences residing mainly in the 3′ and 5′ termini of each viral RNA (13), ensuring the packaging of eight distinct vRNPs into budding particles. It is the swapping of these vRNP segments that contributes to genetic reassortment between strains that coinfect the same cell (2).
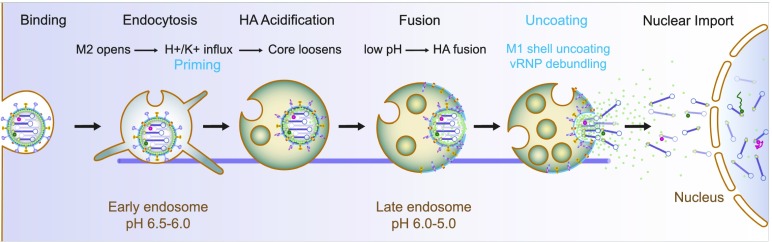
Influenza virus uncoating can be broken down into three steps: priming, M1 shell uncoating, and vRNP debundling (Fig. 1, highlighted in cyan). In the priming step, the decreasing pH during endocytic trafficking renders the viral core “uncoating competent” (14, 15). Opening of the M2 ion channels in the viral envelope allows influx of protons and potassium ions into the viral core, altering the core so that M1 and the vRNPs can dissociate once the core is delivered to the cytosol by fusion (14, 16–19). Priming is critical for infection, because if the M2 channel is blocked by amantadine, viral fusion still happens, but the viral particle remains stuck in late endosomes (15). After HA activation and fusion, M1 shell disassembly and vRNP debundling take place on the cytosolic surface of late endosomes (20, 21). Following fusion, unanchored ubiquitin chains are exposed from the viral core and activate histone deacetylase 6 (HDAC6) and the aggresome processing machinery (20). HDAC6 links the M1 shell to actomyosin, dynein, and the microtubule network, and shearing forces from molecular motors break apart the shell. After M1 shell uncoating, transportin 1 (TNPO1), a member of the importin-β family proteins (22, 23), removes residual M1 from vRNPs by binding a PY-NLS signal at the M1 N terminus (21). This debundles vRNPs and facilitates their nuclear import by importin-α/β via the classical NLS pathway (8).
Fig. 1.
Stepwise entry and uncoating of IAV. Influenza virus is an enveloped single-stranded negative strand RNA virus. It contains within its M1 matrix shell eight genomic segments called vRNPs that are bundled together. When infecting a new cell, the IAV virion binds to sialic acids on cell surface receptors and triggers its own endocytosis into early endosomes via clathrin-mediated or macropinocytic uptake. Endosome maturation is key to successful virus entry and uncoating. The decreasing pH in endosomes opens the M2 ion channels on the viral membrane, allowing influx of protons and potassium ions into the viral core, altering the core so that M1 and the vRNPs can dissociate once the core is delivered to the cytosol by HA-activated fusion. After fusion, the cytosolic uncoating factors HDAC6 and TNPO1 promote further disassembly of the M1 shell and vRNPs, respectively. The real-time entry and uncoating of vRNPs can be tracked and dissected in detail, by imaging two individual vRNPs labeled with a QD (shown inside the particle as dark green and pink dots). Adapted with permission from ref. 28.
Previously, microinjection of dye-labeled vRNPs into the cytoplasm and fluorescence in situ hybridization (FISH) have been used to study the behavior of incoming vRNPs (24, 25). FISH studies of incoming vRNPs suggested that the separate viral RNAs travel together until they reach the nucleus (25). Superresolution imaging of incoming particles showed that vRNPs debundle before reaching the nucleus (21). When purified influenza vRNPs free of M1 were microinjected into cells, their nuclear accumulation was observed after 1 h of injection (26). Cores that consisted of vRNPs and M1 could not be imported into the nucleus. Thus, the removal of M1 causes the dissociation of vRNPs from each other, ensuring their passage through the nuclear pore (14, 26). Single-particle tracking of microinjected, fluorescently labeled vRNPs showed that vRNPs undergo multiple rounds of binding and release before finally being translocated through the nuclear pore (24).
In PNAS, Qin et al. (4) tackle the vRNP debundling puzzle by generating infectious influenza virions that package vRNPs noncovalently labeled with a QD via the PA subunit of the polymerase. Their approach enables long-term imaging of individual vRNP segments in living cells from viral attachment to nuclear import. Live imaging of single incoming vRNPs over an extended period of time requires dye brightness, photostability, and a high signal-to-noise ratio, and QDs are superior compared with GFP in all of these requirements. How are QDs incorporated into infectious influenza virions? Qin et al. genetically manipulated the IAV PR8 strain to express a biotin acceptor peptide (AP) fused to the polymerase PA subunit. The rescued rPR8-PA-AP virus was genetically stable over multiple passages, and was thus used to replicate in BirA-expressing Madin-Darby Canine Kidney cells in the presence of biotin. Shortly after, streptavidin-coated QDs (SA-QDs) were introduced into these cells by lipofection to allow noncovalent binding of the QDs to biotinylated PA.Influenza virions generated using this method packaged up to five QDs in a single particle. Many (60%) virus particles, however, contained only one QD. Those that contained two QDs of different color were used to live image vRNP debundling.
In PNAS, Qin et al. develop a nanotechnology that labels IAV viral ribonucleoprotein complexes (vRNPs) with quantum dots (QDs)—an approach that will advance the mechanistic understanding of influenza virus entry using live fluorescence microscopy.
Codetection with Rab7-enhanced cyan fluorescent protein revealed that the two different segments that colocalized in endosomes separated into single entities as they left the Rab7 compartment. If combined with viral fusion detection, the dual QD virions will prove even more informative. It is likely that soon after fusion, M1 shell breakage and vRNP debundling take place efficiently on the cytosolic surface of late endosomes by host factors such as HDAC6 (20) and TNPO1 (21). After debundling, the vRNPs undergo a three-stage movement as they import into the nucleus (4).
In summary, Qin et al.’s (4) study is important because it provides continuous documentation of the vRNP’s journey from late endosomal escape to nuclear entry. The approach, however, has limitations. The separated QD signals during uncoating can indicate two subbundles (e.g., in a 7+1, 6+2, 5+3, or 4+4 arrangement) or several subbundles (e.g., a 2+3+3 arrangement), and not necessarily eight single segments. Further, QDs are 2 to 10 nm in diameter and their virion incorporation decreases viral replication significantly. The physical presence of QDs will influence the properties of vRNP–vRNP interactions that regulate debundling mechanics. Thus, a multifaceted approach using alternative labeling techniques such as FlAsH (27), wild-type viruses, and cellular loss-of-function experiments using siRNA or CRISPR/Cas is warranted for deeper investigation before we conclude the precise mechanism of influenza vRNP debundling. However, this study expands the toolbox that allows researchers to visualize virus–host interactions during virus entry at single-molecule resolution in combination with host factors of mechanistic interest.
Footnotes
The author declares no conflict of interest.
See companion article on page 2577.
References
- 1.Medina RA. 1918 influenza virus: 100 years on, are we prepared against the next influenza pandemic? Nat Rev Microbiol. 2018;16:61–62. doi: 10.1038/nrmicro.2017.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lowen AC. It’s in the mix: Reassortment of segmented viral genomes. PLoS Pathog. 2018;14:e1007200. doi: 10.1371/journal.ppat.1007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lakdawala SS, et al. Influenza a virus assembly intermediates fuse in the cytoplasm. PLoS Pathog. 2014;10:e1003971. doi: 10.1371/journal.ppat.1003971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Qin C, et al. Real-time dissection of dynamic uncoating of individual influenza viruses. Proc Natl Acad Sci USA. 2019;116:2577–2582. doi: 10.1073/pnas.1812632116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Moeller A, Kirchdoerfer RN, Potter CS, Carragher B, Wilson IA. Organization of the influenza virus replication machinery. Science. 2012;338:1631–1634. doi: 10.1126/science.1227270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Area E, et al. 3D structure of the influenza virus polymerase complex: Localization of subunit domains. Proc Natl Acad Sci USA. 2004;101:308–313. doi: 10.1073/pnas.0307127101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Martín-Benito J, et al. Three-dimensional reconstruction of a recombinant influenza virus ribonucleoprotein particle. EMBO Rep. 2001;2:313–317. doi: 10.1093/embo-reports/kve063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Eisfeld AJ, Neumann G, Kawaoka Y. At the centre: Influenza A virus ribonucleoproteins. Nat Rev Microbiol. 2015;13:28–41. doi: 10.1038/nrmicro3367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu WW, Sun YH, Panté N. Nuclear import of influenza A viral ribonucleoprotein complexes is mediated by two nuclear localization sequences on viral nucleoprotein. Virol J. 2007;4:49. doi: 10.1186/1743-422X-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zheng W, Tao YJ. Structure and assembly of the influenza A virus ribonucleoprotein complex. FEBS Lett. 2013;587:1206–1214. doi: 10.1016/j.febslet.2013.02.048. [DOI] [PubMed] [Google Scholar]
- 11.Noda T, et al. Three-dimensional analysis of ribonucleoprotein complexes in influenza A virus. Nat Commun. 2012;3:639. doi: 10.1038/ncomms1647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fournier E, et al. Interaction network linking the human H3N2 influenza A virus genomic RNA segments. Vaccine. 2012;30:7359–7367. doi: 10.1016/j.vaccine.2012.09.079. [DOI] [PubMed] [Google Scholar]
- 13.Goto H, Muramoto Y, Noda T, Kawaoka Y. The genome-packaging signal of the influenza A virus genome comprises a genome incorporation signal and a genome-bundling signal. J Virol. 2013;87:11316–11322. doi: 10.1128/JVI.01301-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bui M, Whittaker G, Helenius A. Effect of M1 protein and low pH on nuclear transport of influenza virus ribonucleoproteins. J Virol. 1996;70:8391–8401. doi: 10.1128/jvi.70.12.8391-8401.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Martin K, Helenius A. Transport of incoming influenza virus nucleocapsids into the nucleus. J Virol. 1991;65:232–244. doi: 10.1128/jvi.65.1.232-244.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stauffer S, et al. Stepwise priming by acidic pH and a high K+ concentration is required for efficient uncoating of influenza A virus cores after penetration. J Virol. 2014;88:13029–13046. doi: 10.1128/JVI.01430-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pinto LH, Holsinger LJ, Lamb RA. Influenza virus M2 protein has ion channel activity. Cell. 1992;69:517–528. doi: 10.1016/0092-8674(92)90452-i. [DOI] [PubMed] [Google Scholar]
- 18.Zhirnov OP. Solubilization of matrix protein M1/M from virions occurs at different pH for orthomyxo- and paramyxoviruses. Virology. 1990;176:274–279. doi: 10.1016/0042-6822(90)90253-n. [DOI] [PubMed] [Google Scholar]
- 19.Martin K, Helenius A. Nuclear transport of influenza virus ribonucleoproteins: The viral matrix protein (M1) promotes export and inhibits import. Cell. 1991;67:117–130. doi: 10.1016/0092-8674(91)90576-k. [DOI] [PubMed] [Google Scholar]
- 20.Banerjee I, et al. Influenza A virus uses the aggresome processing machinery for host cell entry. Science. 2014;346:473–477. doi: 10.1126/science.1257037. [DOI] [PubMed] [Google Scholar]
- 21.Miyake Y, et al. Influenza A virus uses transportin 1 for vRNP debundling during cell entry. Nat Microbiol. 2019 doi: 10.1038/s41564-018-0332-2. [DOI] [PubMed] [Google Scholar]
- 22.Pollard VW, et al. A novel receptor-mediated nuclear protein import pathway. Cell. 1996;86:985–994. doi: 10.1016/s0092-8674(00)80173-7. [DOI] [PubMed] [Google Scholar]
- 23.Fridell RA, Truant R, Thorne L, Benson RE, Cullen BR. Nuclear import of hnRNP A1 is mediated by a novel cellular cofactor related to karyopherin-beta. J Cell Sci. 1997;110:1325–1331. doi: 10.1242/jcs.110.11.1325. [DOI] [PubMed] [Google Scholar]
- 24.Babcock HP, Chen C, Zhuang X. Using single-particle tracking to study nuclear trafficking of viral genes. Biophys J. 2004;87:2749–2758. doi: 10.1529/biophysj.104.042234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chou YY, et al. Colocalization of different influenza viral RNA segments in the cytoplasm before viral budding as shown by single-molecule sensitivity FISH analysis. PLoS Pathog. 2013;9:e1003358, and erratum (2013). doi: 10.1371/annotation/8f53e7f2-2348-436f-b37e-a883a01e9bbd. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemler I, Whittaker G, Helenius A. Nuclear import of microinjected influenza virus ribonucleoproteins. Virology. 1994;202:1028–1033. doi: 10.1006/viro.1994.1432. [DOI] [PubMed] [Google Scholar]
- 27.Vahey MD, Fletcher DA. Low-fidelity assembly of influenza A virus promotes escape from host cells. Cell. 2019;176:281–294.e19. doi: 10.1016/j.cell.2018.10.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Banerjee I, Yamauchi Y, Helenius A, Horvath P. High-content analysis of sequential events during the early phase of influenza A virus infection. PLoS One. 2013;8:e68450. doi: 10.1371/journal.pone.0068450. [DOI] [PMC free article] [PubMed] [Google Scholar]