Abstract
Introduction
Schizophrenia and bipolar disorder account for a large proportion of the global burden of disease. Despite their enormous impact, little is known about their pathophysiology. Given the high heritability of schizophrenia and bipolar disorder, unbiased genetic studies offer the opportunity to gain insight into their neurobiology. However, advances in understanding the genetic architecture of schizophrenia and bipolar disorder have been based almost exclusively on subjects of Northern European ancestry. The Neuropsychiatric Genetics of African Populations-Psychosis (NeuroGAP-Psychosis) project aims to expand our understanding of the causes of schizophrenia and bipolar disorder through large-scale sample collection and analyses in understudied African populations.
Methods and analysis
NeuroGAP-Psychosis is a case-control study of 34 000 participants recruited across multiple sites within Ethiopia, Kenya, South Africa and Uganda. Participants will include individuals who are at least 18 years old with a clinical diagnosis of schizophrenia or bipolar disorder (‘psychosis’) or those with no history of psychosis. Research assistants will collect phenotype data and saliva for DNA extraction. Data on mental disorders, history of physical health problems, substance use and history of past traumatic events will be collected from all participants.
DNA extraction will take place in-country, with genotyping performed at the Broad Institute. The primary analyses will include identifying major groups of participants with similar ancestry using the computation-efficient programme single nucleotide polymorphisms (SNP) weights. This will be followed by a GWAS within and across ancestry groups.
Ethics and dissemination
All participants will be assessed for capacity to consent using the University of California, San Diego Brief Assessment of Capacity to Consent. Those demonstrating capacity to consent will be required to provide informed consent. Ethical clearances to conduct this study have been obtained from all participating sites. Findings from this study will be disseminated in publications and shared with controlled access public databases, such as the database of Genotypes and Phenotypes, dbGaP.
Keywords: genetics, mental Health, Africa
Strengths and limitations of this study.
This will be the first psychiatric genetics study at this scale in Africa and across different African populations.
Significant phenotypic data will be collected with the goal of a looking at a transdiagnostic category, without necessarily being limited to Diagnostic and Statistical Manual of Mental Disorders/International Statistical Classification of Diseases and Related Health Problems diagnoses.
We will extract DNA from saliva samples which are logistically easier to manage than blood. However, there is less DNA in saliva than in blood, and there is a higher risk of contamination.
As we have chosen not to exclude based on race, some of the participants are likely to be of European descent.
Although we obtain permission to follow-up enrolled participants, this is a case-control study, and thus will only have phenotypic data at one point in time.
Introduction
Neuropsychiatric disorders are the leading cause of years lived with disability in the world.1 Within mental, neurological and substance use disorders, schizophrenia and bipolar disorder account for more than 14% of years of life lost to premature mortality and years lived with disability.2 In the past several years, there have been great strides in our understanding of the genetic architecture of schizophrenia and bipolar disorder. A landmark paper in Nature from 2014 discovered 108 genome-wide significant loci for schizophrenia in ~37 000 cases and ~113 000 controls.3 In the intervening years, the number of genome-wide significant hits for schizophrenia has grown to 145.4 Successes in schizophrenia research have demonstrated that extremely large scale meta-analyses are necessary to identify genetic variants associated with neuropsychiatric disorders.5 The hope is that these breakthroughs in neuropsychiatric genetics will lead to new pharmacological targets, and ultimately treatments to reduce the global burden of psychiatric disorders.
However, for historical, cultural, financial and practical reasons, these genetic findings are based predominantly on subjects of Northern European ancestry, with a growing but still small proportion on populations of East Asian ancestry.6 7 Currently, there are major limitations in our knowledge of the genetic and environmental risk architecture of psychiatric disorders in persons of African descent.8 As a result, we are limited in our ability to understand biological mechanisms, predict genetic risk9 and produce optimal therapy for African populations. Moreover, African genomes are characterised by shorter haplotype blocks and contain almost a million more variants per individual than populations outside Africa.10 Further, genetic studies of under-represented populations afford the opportunity to discover novel loci that are invariant in European populations.11 Thus, including data from African populations in genetic studies of neuropsychiatric disorders may accelerate genetic discovery and could be useful for fine mapping of disease causing alleles.12 Studies of psychiatric genetics are in their infancy in Africa and are not yet at a scale necessary for variant discovery.
Aim and objective
The Neuropsychiatric Genetics of African Populations-Psychosis (NeuroGAP-Psychosis) project aims to expand knowledge of the genetic and environmental risk factors for neuropsychiatric disorders in Africa through large-scale sample collection and analysis, so that future advances in science and therapeutics can account for and be applicable to African populations.
Methods and analysis
Study design
The design will be a case-control study. This project is structured around two diagnostic categories: schizophrenia and bipolar I disorder (grouped under the heading ‘psychotic disorders’). The rationale for grouping schizophrenia and bipolar disorder stems from literature showing a high level of genetic correlation between schizophrenia and bipolar disorder, which may indicate that some of the same genetic variants confer risk for both phenotypes.13
Cases will be individuals with a diagnosis of schizophrenia or bipolar disorder, referred to subsequently as psychosis. Controls will be individuals from the same geographical location, without psychosis, who will be matched to cases for age, sex and ancestry.
Study sites
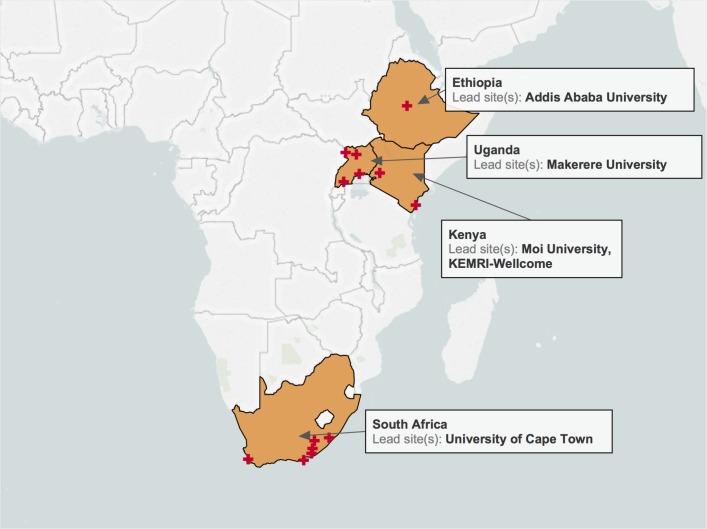
The study will be conducted over 4 years, starting in September 2018 and ending in 2022, at several sites in Ethiopia, Kenya, South Africa and Uganda (figure 1).14 DNA extraction will be performed on site, and genotyping will performed at the Broad Institute in the USA using the Illumina Global Screening Array. Sites were selected on the basis of the following criteria: (1) proven track record of psychiatric research; (2) availability of research personnel and the necessary research infrastructure to be able to recruit thousands of participants and (3) existing trusted relationships from prior collaborations. Each of the countries where participants will be recruited from has enormous genetic diversity within and between them, which is likely to improve the ability of this research to answer the study objectives. Pilot studies are currently underway in all four countries.
Figure 1.
Proposed collection sites for Neuropsychiatric Genetics of African Populations-Psychosis.
Ethiopia
Participants will be recruited from the Amanuel Mental Specialized Hospital (cases) and Black Lion Hospital also known as the Tikur Anbessa Hospital (controls).
Kenya
In Kenya, participants will be recruited from the Moi Teaching and Referral Hospital and affiliated sites in Webuye, Kapenguria, Kitale, Kapsabet, Iten and Kakamega and the KEMRI-Wellcome Trust Research Programme with recruiting sites in Kilifi County, Malindi sub-County, Port Reitz and Coast General Provincial Hospitals.
South Africa
Participants will be recruited from the Western Cape and Eastern Cape provinces. In the Western Cape, participants will be recruited from the Valkenberg, Lentegeur, Khayelitsha District and Groote Schuur Hospitals as well as a number of community clinics. In the Eastern Cape, participants (cases) will be recruited from the Fort England Psychiatric, Elizabeth Donkin, Tower Psychiatric and Komani Hospitals while controls will be recruited from the Nelson Mandela Academic and Dora Nginza Hospitals as well as five affiliated health clinics.
Uganda
Study participants in Uganda will be recruited from the Butabika National Mental Health Referral Hospital (cases and controls), Naguru (controls only), Arua (cases and controls), Mbarara (cases and controls) and Gulu Regional Referral Hospitals (cases and controls).
Inclusion criteria
Individuals with a clinical diagnosis of psychosis (cases) as confirmed by clinician referral and/or medical record review, and those without a clinical diagnosis of psychosis (controls) will be eligible to participate. All participants (cases and controls) will be required to provide written informed consent or a fingerprint in case of illiteracy and must be at least 18 years old. To ensure that participants (cases and controls) have sufficient capacity and autonomy to consent to the study, we will use the University of California, San Diego Brief Assessment of Capacity to Consent (UBACC),15 which has been used as an iterative learning tool in similar populations in South Africa.16
Exclusion criteria for cases
Individuals (cases) will be excluded if the following are present:
Absence of a diagnosis of a psychotic disorder.
Severe, intrusive levels of psychiatric symptoms at the time of consent.
Intoxication or withdrawal from alcohol or substance abuse at the time of consent.
A current psychiatric hospitalisation (inpatients).
Involuntary detention at the time of consent.
Lack of fluency in one of the languages the consent form has been translated into.
Lack of capacity to consent to the study as determined by the UBACC.
Potential cases may be approached at a later date once their symptoms are controlled if they meet other inclusion criteria. There will be no exclusions for cases or controls based on sex, ancestry/ethnicity, religious affiliations or sexual orientation.
Exclusion criteria for controls
Potential controls will be excluded if they:
Have current psychotic symptoms or a past diagnosis of a psychotic disorder.
Are currently taking medication for psychosis.
Have acute levels of alcohol or substance misuse as demonstrated by being a current inpatient or under acute medical care for substance misuse.
Lack of fluency in one of the languages the consent form has been translated into.
Lack of capacity to consent to the study as determined by the UBACC.
Participant recruitment
Eligible participants (cases) will be identified by clinical staff through review of their medical records. A research assistant (RA), who is a nurse, clinical officer, clinical assistant or bachelor’s level accredited RA and who has received study-specific training and human subjects training, will approach the prospective participant, carefully explaining to them in the local language the purpose and procedures of the study and emphasising that participation is entirely voluntary and will not impact any medical care they receive. Languages include: Acholi, Afrikaans, Amharic, English, Kigiryama, Kiswahili, Luganda, Lugbara, Runyankole and isiXhosa.
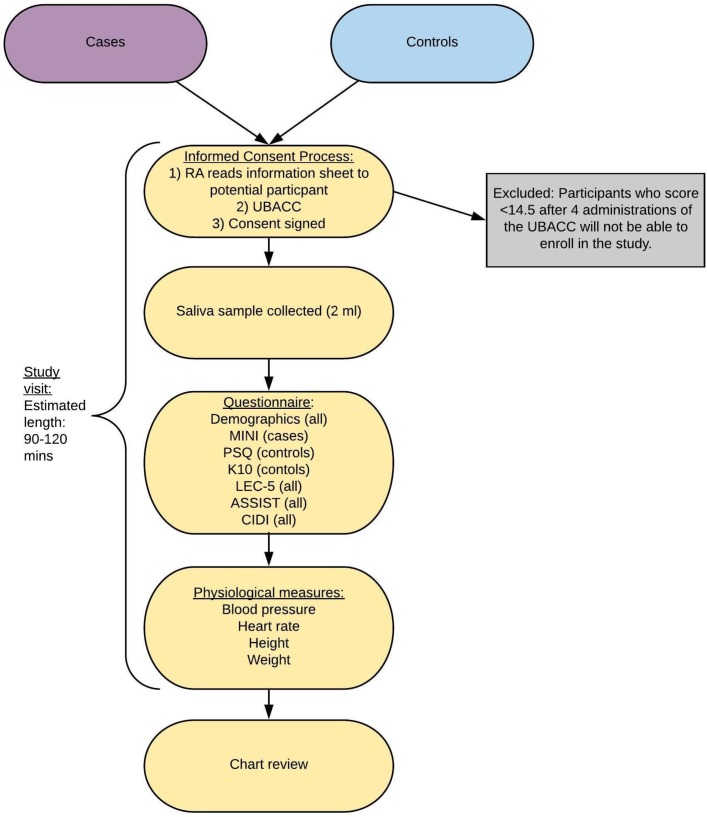
After reading the information sheet out loud to the subject, the RA will administer the UBACC, a 10-item questionnaire that evaluates the potential participant’s understanding of different components of the study. Each response will be scored on a range of 0–2, with 0 representing no understanding and 2 representing a clear understanding. In the NeuroGAP-Psychosis study, the RA will administer the UBACC over a maximum of four trials. The RA will re-explain and re-administer any items the subject answered incorrectly. The process will end if the full score of 20 is obtained. After the fourth trial, participants who are unable to achieve a score of 14.5, the cut-off originally developed for screening decisional capacity using the UBACC,15 will be excluded.
The RA will proceed to obtain informed consent from those who express interest in participation, and ask participants to provide saliva into an Oragene tube, from which their DNA will be extracted. To maximise the quality of the collected DNA, participants will be asked not to eat, drink, smoke or chew gum for 30 min prior to providing the sample during the consenting process.
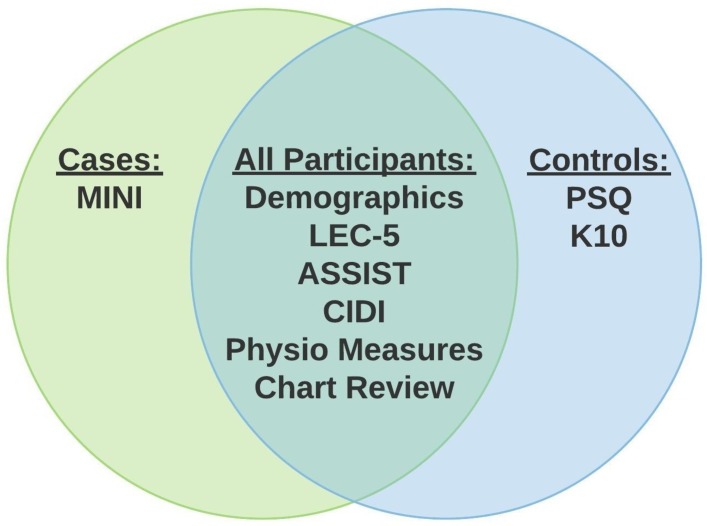
Phenotypic assessments were selected on the basis of the following criteria: (1) the particular domain assessed; (2) cross-cultural validity; (3) non-proprietary measures, when possible; (4) investigators’ prior experience using the tool and (5) the time length of administering the tool. The study will collect the following battery of instruments (see figure 2).
Figure 2.
Phenotyping tools for study participants. ASSIST, Alcohol, Smoking and Substance Involvement Screening Test, V.3.0; CIDI, Composite International Diagnostic Interview screener; K10, Kessler Psychological Distress Scale; LEC-5, Life Events Checklist for Diagnostic and Statistical Manual of Mental Disorders-5; MINI, Mini International Neuropsychiatric Interview, Standard 7.0.2 for Diagnostic and Statistical Manual of Mental Disorders-5. PSQ, Psychosis Screening Questionnaire.
Mini International Neuropsychiatric Interview, Standard 7.0.2 for Diagnostic and Statistical Manual of Mental Disorders-5 (MINI): modules A, C, K and O on major depressive episode, manic and hypomanic episodes and psychotic disorders and mood disorder with psychotic features, respectively.17
Life Events Checklist for Diagnostic and Statistical Manual of Mental Disorders-5: a 17-item scale covering exposures to potentially traumatic events.18
Alcohol, Smoking and Substance Involvement Screening Test, V.3.0 (ASSIST): a subset of the ASSIST on substance type and use over a participant’s lifetime and over the past 3 months.19
Composite International Diagnostic Interview screener (CIDI): a checklist within the CIDI on chronic physical conditions20 including diabetes, HIV/AIDS, epilepsy/seizures and tuberculosis.
Participant phenotypic data will be collected using encrypted tablets and uploaded to a secure cloud-based server. Other sources of data will include chart reviews to ascertain clinical diagnoses and medication use (both psychotropic and non-psychotropic) by the participants; as well as measurements of blood pressure, heart rate, height and weight.
Recruitment of controls will take place from general medical facilities (some of these facilities are located within the premises where cases will be identified). Controls will be approached in the same manner as cases and will likely consist of people who are seeking clinical care for themselves, accompanying a friend or family member to a clinic visit, or picking up a medication refill. In one site, at the KEMRI-Wellcome Trust Research Programme in Kenya, controls may also be recruited from homes through the Kilifi Health and Demographic Surveillance System (KHDSS).21 [In KHDSS, census data is recorded every 4 months through household surveys in the catchment area and linked with healthcare data from Kilifi District Hospital.]
The study visit for controls will match the process for cases except for two phenotypic batteries, the Kessler Psychological Distress Scale (K10) and the Psychosis Screening Questionnaire (PSQ) (see figure 3); instead of using the MINI for controls, the RA will administer:
Figure 3.
The study process for cases and controls. ASSIST, Alcohol, Smoking and Substance Involvement Screening Test, V.3.0; CIDI, Composite International Diagnostic Interview screener; K10, Kessler Psychological Distress Scale; LEC-5, Life Events Checklist for Diagnostic and Statistical Manual of Mental Disorders-5; MINI, Mini International Neuropsychiatric Interview, Standard 7.0.2 for Diagnostic and Statistical Manual of Mental Disorders-5; PSQ, Psychosis Screening Questionnaire; RA, research assistant; UBACC, University of California, San Diego Brief Assessment of Capacity to Consent.
K10: a 10-item questionnaire to measure anxiety and depression.22
PSQ: a brief screening tool for psychotic conditions.23
All participants will receive compensation to offset the time and inconvenience of participating in the study based on recommendations from the local ethics committees and principal investigators. The estimated length of a study visit is 90 to 120 min.
Patient and public involvement
Patients’ priorities and experiences were taken into account in the design of the study. Prior research and clinical work with people with neuropsychiatric disorders at each of the sites informed the research question and study design, including limiting the length of the study battery, minimising intrusive sample collection (saliva instead of blood) and incorporating a tool as part of the consent process to assess understanding of the research and study visit. In addition, most of the sites have a precedent of genetics research and we engaged with teams from those other genetic studies to incorporate patient concerns emerging from genetic research. Study findings will be shared with local community advisory boards.
Sample size and power calculation
Over a period of 4 years, the study team expects to recruit a total of 17 000 schizophrenia and bipolar disorder cases and 17 000 controls across four countries: Ethiopia, Kenya, South Africa and Uganda. The geographic and demographic sampling distribution of the controls will follow that of the cases, and cases and controls will be matched for ethnic ancestry in all analyses. These numbers, described in more detail below, will give adequate statistical power to independently examine the genetic architecture of schizophrenia and bipolar disorder in African populations, the distribution and influence of rare copy number variation on neuropsychiatric disease risk in African populations and determine the consistency of genetic influences on neuropsychiatric disease risk between populations of European and African descent. Ultimately, these data will be analysed in combination with other studies of psychiatric genetics, most notably from the Psychiatric Genomics Consortium, which will further increase our power to discover new and validate current variants with small effect sizes.
With 17 000 cases and controls, using a threshold of 2.5×10-8 for genome-wide significance, we have >90% power to find common variants (Minor Allele Frequency (MAF) >5%) with an OR of 1.15 or greater.24 For more common variants with MAF=25%, we have 89% power to a see significant OR as low as 1.06. These effect sizes are within the range of what was found in a recent Genome-wide Association Study (GWAS) of schizophrenia in a European population.3
DNA extraction and processing
All study participants will provide a 2.0 mL sample of saliva by spitting into a funnel in an Oragene OR-500 kit. DNA extraction will take place in-country, with one aliquot of the extract sent to the Broad Institute in Cambridge, MA, USA for genotyping. The remaining DNA will be frozen and stored in-country within designated biobanks and will be governed by the rules of its institution. In Ethiopia, DNA will be stored at the Black Lion Hospital. In Kenya, DNA will be stored at Moi University and KEMRI-Wellcome Trust Research Programme Biorepository Laboratories. In South Africa, DNA will be stored in a biobank at the Division of Human Genetics at the University of Cape Town, while in Uganda, DNA will be stored at the Integrated Biorepository Lab of H3Africa in Uganda (IBR-H3A-U).
All samples will be assayed on the same genotyping array at the same facility. To maximise information and allow for comparisons across studies genotyped on different platforms, we will impute SNP genotypes not present on specific arrays. Genotype imputation of untyped markers will be performed using default parameters in IMPUTE2 using appropriate reference panels.
Training
To ensure research staff understand the study and their responsibilities, teams will undergo multiple trainings. Study staff who have contact with research participants will complete the online Human Subject Training offered by the US National Institutes of Health. Site leadership and Harvard staff will provide in-person training to the teams by providing an overview of the project, leading discussions, providing role playing on ethics and confidentiality, on recruiting and consenting participants, on reportable events, and on storing forms and data properly. Additionally, there will be hands-on training using the electronic tablets, collecting saliva using Oragene kits, administering the phenotypic batteries, and taking physiological measures, such as height and weight. A checklist will be created of key skills staff need before they can begin interacting with participants one-on-one (figure 4). This form will need to be signed and dated by the staff member and the staff member’s supervisor before he or she can begin working with participants; it will then be uploaded to the regulatory binder. Harvard teams will reinforce these skills with multiple site visits per year and weekly video calls.
Figure 4.
NeuroGAP-Psychosis training checklist. ASSIST, Alcohol, Smoking and Substance Involvement Screening Test, V.3.0; CIDI, Composite International Diagnostic Interview screener; K10, Kessler Psychological Distress Scale; LEC-5, Life Events Checklist for Diagnostic and Statistical Manual of Mental Disorders-5; MINI, Mini International Neuropsychiatric Interview, Standard 7.0.2 for Diagnostic and Statistical Manual of Mental Disorders-5; NeuroGAP-Psychosis, Neuropsychiatric Genetics of African Populations-Psychosis; PI, Principal Investigator; PSQ, Psychosis Screening Questionnaire; UBACC, University of California, San Diego Brief Assessment of Capacity to Consent.
In addition, Broad Institute wet lab staff will work in partnership with the lab teams in Africa to standardise the DNA extraction process across the five sites and to increase the quality of the extractions through visits, video calls and email. Where needed, the Broad staff will train lab members on DNA extraction from saliva and establish a sample chain of custody process in order to decrease the likelihood of sample swaps.
Statistical analysis plans
(a) Ancestry analyses
In contrast to most samples previously analysed by the psychiatric genetics community—which were predominately of European ancestry25—the NeuroGAP-Psychosis samples will include individuals of predominantly African ancestry and will be drawn from multiple populations across East and South Africa where genetic variation will be more structured and demographically complex.26 27 Therefore, before proceeding with any phenotype analyses we will first analyse genetic ancestry in our cohorts to identify large, homogeneous groups as described below to conserve optimal power in the GWAS and ultimately control for additional population stratification within these groups to avoid spurious results.
We will take advantage of the large amount of genotypic information available and control for potential population stratification in a standard and validated two-step process. First, we will compute principal components analysis (PCA) to adjust for population substructure. We will also include principal components (PCs) when computing relatedness using PC-AiR.28 We will define clusters of ancestries using ADMIXTURE29 and choose the optimal number of clusters with fivefold cross-validation to identify major groups of participants with similar ancestry. The inclusion of extensive population reference samples compiled by our group and available from other studies, such as the 1000 Genomes Project30 and the African Genome Variation Project,27 will increase the power of this approach and lead to stable individual ancestry calling across studies. Second, if significant differences are found between populations in our cohorts, as expected, we will define more homogeneous populations within several standard deviations of these reference data, selecting each population by constructing multidimensional ellipses across the PCs. Our initial analyses will then be conducted separately on more homogeneous ancestry groups, and later meta-analysed. Within each group, we will conduct PCA again using PC-AiR, and the PCs that explain the most variation within the cohort will be included as covariates in the regression analyses. To adjust for cryptic relatedness that considers ancestry, we will compute a genetic relatedness matrix using PC-Relate,31 which can be used as a random effect in a linear or logistic mixed model. In addition, as new methods are developed by our group and others to account for population structure, we will modify our analyses when appropriate.
(b) Genome-wide Association Study
The primary GWAS discovery analysis will be conducted for the ‘psychotic disorders’ diagnosis, as well as through subsets stratified by primary disorder (schizophrenia, bipolar disorder) using logistic regression. All analyses will be performed using PLINK32 and R.33 To correct for multiple comparisons and the large number of variants in the African genome, we will apply a more conservative significance threshold of p<2.5×10-8.34 Using these subsets, we will assess the genetic correlation and diagnostic stratification among phenotypes.
(c) Heritability estimates
Estimation of heritability, the fraction of disease risk attributable to inherited genetic differences, is an important initial analytic goal. Heritability estimates speak to the anticipated architecture and power of a GWAS, a primary endpoint of the NeuroGAP-Psychosis project. We will use genome-wide complex trait analysis to estimate SNP-based heritability within Africa.25 35 We will assess genetic correlation with existing GWAS in other globally diverse populations using POPCORN.36
Ethics and dissemination
Ethics
Ethical and safety considerations will be taken across multiple levels. Since the subjects the study aims to recruit are deemed vulnerable populations, additional measures will be taken to protect them. Potential participants will be excluded if they are presenting with severe, intrusive levels of psychiatric symptoms at the time of consent. In addition, as described previously, the RAs will use the UBACC15 16 during the consent process to make sure participants understand the study, what is required of them, and that they can withdraw at any point. Participants who pass the UBACC and who want to continue will be required to provide written informed consent or a fingerprint in lieu of a signature.
All steps will be taken to mitigate the potential loss of confidentiality for participants. A two-part ID system will be instituted in order to limit the possibility of connecting the participant with their data. There will be one ID for the phenotypic information and one for genetic information. A single encrypted database linking the phenotypic ID and genetic ID will be stored in-country. The participant’s name and contact information will be on the consent form, which will be stored in a locked cabinet. This information will be put in a separate secure database and will never leave the country of origin. No data that is considered identifiable, by Health Insurance Portability and Accountability Act standards, will ever be collected on the electronic tablets.
Dissemination
We aim to share results through a number of mechanisms in addition to conferences and peer-reviewed publications. In order to maximise scientific utility of the samples/data and to minimise data waste, both genomic and phenotypic data from NeuroGAP-Psychosis will be deposited in controlled access public databases, such as the database of Genotypes and Phenotypes (dbGaP), the Psychiatric Genomics Consortium and/or the European Genome-Phenome Archive. By pooling NeuroGAP-Psychosis’ data with that from other studies, it will be possible to maximise statistical power. Sharing the data will also contribute to the ‘genomic revolution’ currently underway in Africa due to the pioneering work of the African Genome Variation Project, MalariaGEN, African Society of Human Genetics and Human Hereditary and Health in Africa (H3Africa).27 37–40
To help ensure all collaborators have the capacity to work with all the data that will be created, a unique training programme has been created, the Global Initiative for Neuropsychiatric Genetics Education in Research (GINGER),41 that will run in parallel to the NeuroGAP-Psychosis study. Over a 2-year period, 17 early-career researchers from Ethiopia, Kenya, South Africa and Uganda will take part in online and in-person trainings on topics including biostatistics, genetic analysis and epidemiology. Ultimately, the goal of GINGER is to build the next generation of neuropsychiatric geneticists who will be able to work on the NeuroGAP-Psychosis data and continue its legacy in the future.
Supplementary Material
Footnotes
Patient consent for publication: Not required.
AS and DA contributed equally.
Contributors: KCK, DS: conceptualised and designed the study. AS, DA, RES, LA, EK, SMK, VdM, CRJCN, DS, ST, ZZ, KCK: wrote the protocol, selected the phenotype questionnaires and added site-specific information. LC, ARM: wrote the sample size, power calculations and statistical analysis plans. MMC, GS: contributed to the UBACC process. AS, DA: wrote the draft of the manuscript and incorporated the revisions by the co-authors; they contributed equally to this paper. All authors reviewed the manuscript for intellectual content, contributed to revisions and approved the final version for publication.
Funding: This study is funded by the Stanley Center for Psychiatric Research at Broad Institute. The sponsor was involved in the study design, in the writing of the report and in the decision to submit the paper for publication. The sponsor will be involved in the analysis and interpretation of the data. ARM was supported by funding from the National Institutes of Health (K99MH117229).
Competing interests: DS has received research grants and/or consultancy honoraria from Biocodex, Lundbeck, Servier and Sun.
Ethics approval: Ethical clearances to conduct this study have been obtained from all participating sites, including: Ethiopia: Addis Ababa University College of Health Sciences (#014/17/Psy) and the Ministry of Science and Technology National Research Ethics Review Committee (#3.10/14/2018). Kenya: Moi University School of Medicine Institutional Research and Ethics Committee (#IREC/2016/145, approval number: IREC 1727), Kenya National Council of Science and Technology (#NACOSTI/P/17/56302/19576) KEMRI Centre Scientific Committee (CSC# KEMRI/CGMRC/CSC/070/2016), KEMRI Scientific and Ethics Review Unit (SERU#KEMRI/SERU/CGMR-C/070/3575). South Africa: The University of Cape Town Human Research Ethics Committee (#466/2016) and Walter Sisulu University Research and Ethics Committee (# 051/2016). Uganda: The Makerere University School of Medicine Research and Ethics Committee (SOMREC #REC REF 2016-057) and the Uganda National Council for Science and Technology (UNCST #HS14ES). USA: The Harvard T.H. Chan School of Public Health (#IRB17-0822).
Provenance and peer review: Not commissioned; externally peer reviewed.
References
- 1. Whiteford HA, Ferrari AJ, Degenhardt L, et al. . Global Burden of Mental, Neurological, and Substance Use Disorders: An Analysis from the Global Burden of Disease Study 2010 : Patel V, Chisholm D, Dua T, Mental, neurological, and substance use disorders: disease control priorities. 3rd edn Washington (DC): The International Bank for Reconstruction and Development / The World Bank (c) 2016 International Bank for Reconstruction and Development / The World Bank, 2016. [PubMed] [Google Scholar]
- 2. Whiteford HA, Degenhardt L, Rehm J, et al. . Global burden of disease attributable to mental and substance use disorders: findings from the Global Burden of Disease Study 2010. Lancet 2013;382:1575–86. 10.1016/S0140-6736(13)61611-6 [DOI] [PubMed] [Google Scholar]
- 3.Schizophrenia Working Group of the Psychiatric Genomics Consortium. Biological insights from 108 schizophrenia-associated genetic loci. Nature 2014;511:421–7. 10.1038/nature13595 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pardiñas AF, Holmans P, Pocklington AJ, et al. . Common schizophrenia alleles are enriched in mutation-intolerant genes and in regions under strong background selection. Nat Genet 2018;50:381–9. 10.1038/s41588-018-0059-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Zeggini E, Ioannidis JP. Meta-analysis in genome-wide association studies. Pharmacogenomics 2009;10:191–201. 10.2217/14622416.10.2.191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Popejoy AB, Fullerton SM. Genomics is failing on diversity. Nature 2016;538:161–4. 10.1038/538161a [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Li Z, Chen J, Yu H, et al. . Genome-wide association analysis identifies 30 new susceptibility loci for schizophrenia. Nat Genet 2017;49:1576–83. 10.1038/ng.3973 [DOI] [PubMed] [Google Scholar]
- 8. Dalvie S, Koen N, Duncan L, et al. . Large scale genetic research on neuropsychiatric disorders in African populations is needed. EBioMedicine 2015;2:1259–61. 10.1016/j.ebiom.2015.10.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Martin AR, Gignoux CR, Walters RK, et al. . Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet 2017;100:635–49. 10.1016/j.ajhg.2017.03.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Auton A, Brooks LD, Durbin RM, et al. . A global reference for human genetic variation. Nature 2015;526:68–74. 10.1038/nature15393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Wojcik G, Graff M, Nishimura KK, et al. . Genetic diversity turns a new page in our understanding of complex traits. bioRxiv 2017. [Google Scholar]
- 12. Campbell MC, Tishkoff SA. African genetic diversity: implications for human demographic history, modern human origins, and complex disease mapping. Annu Rev Genomics Hum Genet 2008;9:403–33. 10.1146/annurev.genom.9.081307.164258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cardno AG, Owen MJ. Genetic relationships between schizophrenia, bipolar disorder, and schizoaffective disorder. Schizophr Bull 2014;40:504–15. 10.1093/schbul/sbu016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ulrich T. Nations Represented in NeuroGAP-Psychosis: broad communications. 2018.
- 15. Jeste DV, Palmer BW, Appelbaum PS, et al. . A new brief instrument for assessing decisional capacity for clinical research. Arch Gen Psychiatry 2007;64:966–74. 10.1001/archpsyc.64.8.966 [DOI] [PubMed] [Google Scholar]
- 16. Campbell MM, Susser E, Mall S, et al. . Using iterative learning to improve understanding during the informed consent process in a South African psychiatric genomics study. PLoS One 2017;12:e0188466 10.1371/journal.pone.0188466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Sheehan DV, Lecrubier Y, Sheehan KH, et al. . The Mini-International Neuropsychiatric Interview (M.I.N.I.): the development and validation of a structured diagnostic psychiatric interview for DSM-IV and ICD-10. J Clin Psychiatry 1998;59:22–33. [PubMed] [Google Scholar]
- 18. Weathers FW, Blake DD, Schnurr PP, et al. . The Life Events Checklist for DSM-5 (LEC-5). Instrument available from the National Center for PTSD. 2013. wwwptsdvagov
- 19. Humeniuk R, Henry-Edwards S, Ali R, et al. . The Alcohol, Smoking and Substance Involvement Screening Test (ASSIST): Amanual for use in primary care, 2010. [Google Scholar]
- 20. Kessler RC, Ustün TB. The World Mental Health (WMH) Survey Initiative Version of the World Health Organization (WHO) Composite International Diagnostic Interview (CIDI). Int J Methods Psychiatr Res 2004;13:93–121. 10.1002/mpr.168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Scott JA, Bauni E, Moisi JC, et al. . Profile: The Kilifi Health and Demographic Surveillance System (KHDSS). Int J Epidemiol 2012;41:650–7. 10.1093/ije/dys062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kessler RC, Barker PR, Colpe LJ, et al. . Screening for serious mental illness in the general population. Arch Gen Psychiatry 2003;60:184–9. 10.1001/archpsyc.60.2.184 [DOI] [PubMed] [Google Scholar]
- 23. Bebbington P, Nayani T. Psychosis screening questionnaire. Int J of methods in psychiatric research 1995;5:11–19. [Google Scholar]
- 24. Skol AD, Scott LJ, Abecasis GR, et al. . Joint analysis is more efficient than replication-based analysis for two-stage genome-wide association studies. Nat Genet 2006;38:209–13. 10.1038/ng1706 [DOI] [PubMed] [Google Scholar]
- 25. de Candia TR, Lee SH, Yang J, et al. . Additive genetic variation in schizophrenia risk is shared by populations of African and European descent. Am J Hum Genet 2013;93:463–70. 10.1016/j.ajhg.2013.07.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Busby GB, Band G, Si Le Q, et al. . Admixture into and within sub-Saharan Africa. Elife 2016;5:e15266 10.7554/eLife.15266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Gurdasani D, Carstensen T, Tekola-Ayele F, et al. . The African Genome Variation Project shapes medical genetics in Africa. Nature 2015;517:327–32. 10.1038/nature13997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Conomos MP, Miller MB, Thornton TA. Robust inference of population structure for ancestry prediction and correction of stratification in the presence of relatedness. Genet Epidemiol 2015;39:276–93. 10.1002/gepi.21896 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Alexander DH, Novembre J, Lange K. Fast model-based estimation of ancestry in unrelated individuals. Genome Res 2009;19:1655–64. 10.1101/gr.094052.109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Abecasis GR, Auton A, Brooks LD, et al. . An integrated map of genetic variation from 1,092 human genomes. Nature 2012;491:56–65. 10.1038/nature11632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Conomos MP, Reiner AP, Weir BS, et al. . Model-free estimation of recent genetic relatedness. Am J Hum Genet 2016;98:127–48. 10.1016/j.ajhg.2015.11.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Purcell S, Neale B, Todd-Brown K, et al. . PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet 2007;81:559–75. 10.1086/519795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Statistical Computing. R: A language and environment for statistical computing [program. Vienna, Austria: Statistical Computing, 2018. [Google Scholar]
- 34. Kanai M, Tanaka T, Okada Y. Empirical estimation of genome-wide significance thresholds based on the 1000 Genomes Project data set. J Hum Genet 2016;61:861–6. 10.1038/jhg.2016.72 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Yang J, Lee SH, Goddard ME, et al. . GCTA: a tool for genome-wide complex trait analysis. Am J Hum Genet 2011;88:76–82. 10.1016/j.ajhg.2010.11.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Brown BC, Ye CJ, Price AL, et al. . Transethnic genetic-correlation estimates from summary statistics. Am J Hum Genet 2016;99:76–88. 10.1016/j.ajhg.2016.05.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Beiswanger CM, Abimiku Alash’le, Carstens N, et al. . Accessing Biospecimens from the H3Africa Consortium. Biopreserv Biobank 2017;15:95–8. 10.1089/bio.2017.0008 [DOI] [Google Scholar]
- 38.Consortium THA. Enabling the genomic revolution in Africa: H3Africa is developing capacity for health-related genomics research in Africa. Science 2014;344:1346–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Mulder N, Abimiku A, Adebamowo SN, et al. . H3Africa: current perspectives. Pharmgenomics Pers Med 2018;11:59–66. 10.2147/PGPM.S141546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Band G, Rockett KA, Spencer CC, et al. . A novel locus of resistance to severe malaria in a region of ancient balancing selection. Nature 2015;526:253–7. 10.1038/nature15390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. van der Merwe C, Mwesiga EK, McGregor NW, et al. . Advancing neuropsychiatric genetics training and collaboration in Africa. Lancet Glob Health 2018;6:e246–7. 10.1016/S2214-109X(18)30042-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.