Abstract
Aberrant activation of Akt disturbs proliferation, survival and metabolic homeostasis of various human cancers. Thus, it is critical to understand upstream signaling pathways governing Akt activation. Here, we report that Akt undergoes SETDB1-mediated lysine-methylation to promote its activation, which is antagonized by the Jumonji-family demethylase, KDM4B. Notably, compared with wild-type mice, mice harboring non-methylated mutant Akt1 not only exhibited reduced body size, but also were less prone to carcinogen-induced skin tumors in part due to reduced Akt activation. Mechanistically, Phosphatidylinositol (3,4,5)-trisphosphate (PIP3) interaction with Akt facilitates its interaction with SETDB1 for subsequent Akt methylation, which in turn sustains Akt phosphorylation. Pathologically, genetic alterations including SETDB1 amplification aberrantly promote Akt methylation to facilitate its activation and oncogenic functions. Thus, Akt methylation is an important step synergizing with PI3K signaling to control Akt activation, suggesting that targeting the SETDB1 signaling could be a potential therapeutic strategy for combatting hyperactive Akt-driven cancers.
INTRODUCTION
Epigenetic regulation, such as DNA methylation and histone modifications, plays important roles in governing gene expression patterns during human development and disease progression 1, 2. Inhibitors targeting epigenetic factors have been explored for cancer therapies and have undergone clinical trials, including DNA methyltransferase 1 (DNMT1) inhibitors, histone deacetylases (HDAC) inhibitors, and histone methyltransferase inhibitors 3–5. Among these epigenetic inhibitors, histone methyltransferase (such as EZH2 and DOT1L) inhibitors display impressive efficacy in cancer patients 6, 7. This robust efficacy may also be attributed to regulating methylation of non-histone proteins such as Rb and p53, in addition to regulating histone methylation 8–11. However, it remains largely unknown whether predominant oncogenic signaling pathways that are frequently activated in human cancers, such as PI3K/Akt signaling pathway, are subjected to methylation-dependent regulation. Thus the identification of the major oncogenic proteins governed by methylation is critical to identify new therapeutic targets.
Hyperactivation of PI3K/Akt signaling is a central module of cell proliferation, survival and metabolic homeostasis in human cancers 12, 13. Physiologically, stimulations derived from various types of growth factors tend to activate Akt, which in turn phosphorylates distinct substrates to perform different biological processes 13, 14. Recently, emerging evidence has demonstrated that distinct signals govern Akt kinase activity e.g. TRAF6/Skp2-mediated positive regulation of Akt in an ubiquitination-dependent manner 15, CDK2/Cyclin A-mediated positive regulation of Akt in a tail phosphorylation-dependent manner 16, and pVHL-mediated negative regulation of Akt in a hydroxylation-dependent manner 17. However, the regulation of Akt as a non-histone substrate by histone methyltransferases is not well defined.
Here, we described that Akt1 methylation in its Linker-region is mediated by the histone methyltransferase SETDB1, which is antagonized by the demethylase KDM4B. Biologically, absence of Akt1 methylation attenuates its kinase activity, represses cell growth, glucose uptake, and tumorigenesis. As a result, deficiency in Akt methylation can physiologically decrease mouse body size and can protect mice from developing carcinogen-induced skin tumors. Thus, our data unravel a profound role for the SETDB1/KDM4B axis in manipulating Akt activity and highlight histone methyltransferase SETDB1 as a potential target for combating hyperactive Akt-driven tumors.
RESULTS
Methylation of Akt enhances its kinase activity
To identify important non-histone proteins regulated in a methylation-dependent manner, we used a specific pan-lysine tri-methylation (K-me3) antibody and performed a mass spectrometry (MS) based screening on cell lysates derived from ovarian cancer cells (Fig. 1a and Supplementary Fig. 1a) 10. Notably, an Akt1-derived peptide was identified, containing methylated modifications at two nearby evolutionarily conserved lysine residues (K140 and K142) in the Akt1 Linker region (Table S1, Fig. 1b). Furthermore, Akt1 methylation was validated using the K-me3 antibody in cells treated with a global histone methylation inhibitor, 3-Deazaneplanocin A (DZneP) (Supplementary Fig. 1b).
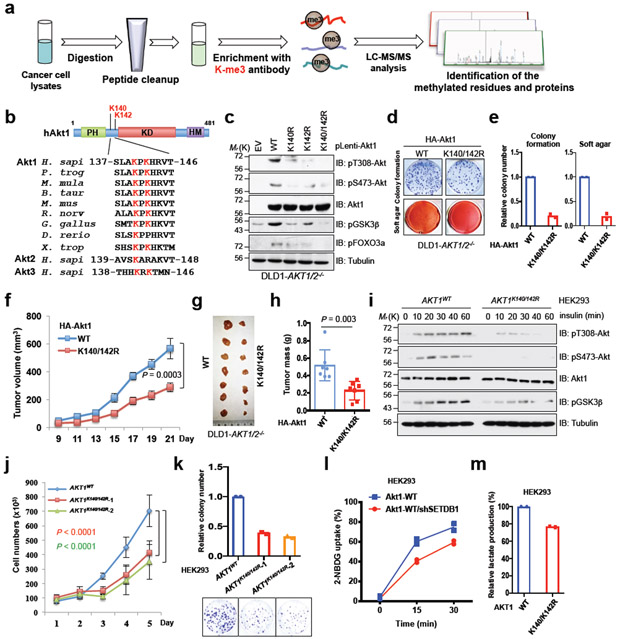
Fig. 1. Akt methylation promotes its activity and oncogenic functions.
a, A schematic workflow of IAP-LC-mass spectrometry (MS)/MS experiments. OVCAR5 cell lysates were proteolytically digested to perform IAP-LC-MS/MS assays. b, Alignment of MS-characterized Akt1 putative methylation residues among different species, Akt2 and Akt3. c, Immunoblot (IB) analysis of whole cell lysates (WCL) derived from DLD1-AKT1/2−/− cells infected with indicated Akt1 encoding virus and selected with hygromycin (200 μg/ml) for 72 hrs before harvesting. Data shown represent two independent experiments. d-e, cells generated in c were subjected to colony formation and soft agar assays. The experiment was performed twice independently with three repeats, and exhibited similar results (d). Representative images were shown in d and relative colony numbers derived from two independent experiments were plotted in e. f-h, Cells generated in (c) were subjected to mouse xenograft assays. Tumor sizes were monitored (f), and dissected tumors were weighed (g, h). Error bars in f and h are mean ± s.e.m, n = 7 mice. i-k, K140R and K142R mutations of Akt1 were genetically engineered in HEK293 cells by the CRISPR/CAS9-based technique. Resulting cells were serum-starved for 12 hrs, and harvested for IB analysis at different time points after stimulation with insulin (100 nM) (i). The experiment was performed twice independently with similar results (i). Cells generated in (i) were assessed for proliferation (j) assays. The experiment in j was performed three times independently and exhibited similar results. Error bars in j are mean ± s.e.m, n = 3 independent experiments. k,l,m, Cells generated in i were subjected to colony formation (k), glucose uptake (l) and lactate production (m) assays. The experiment was performed twice independently with three repeats, and exhibited similar results (k,l,m). Relative colony numbers, glucose and lactate levels derived from two independent experiments were plotted in (k,l,m). Two-way ANOVA analysis was performed in (f,j) to calculate the P value. Detailed statistical tests are described in the Methods. Source data for e, f, h and j-m are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
Additional MS analyses on ectopically expressed Akt1 further confirmed the tri-methylation of K140 and K142, where mono- and di-methylation events were also identified (Supplementary Fig. 1c). Moreover, K64, another lysine residue in the PH domain of Akt1, was also identified to be mono-methylated in our system. Interestingly, mutation of both K140 and K142 residues led to a dramatic reduction of Akt1 K-me3 signal in cells (Supplementary Fig. 1d), indicating that K140 and K142 might be the major Akt1 K-me3 residues. In addition, consistent with a previous report 18, deletion of Smyd3, a methyltransferase targeting MAP3K2, did not disturb Akt kinase activity in cells, although the interaction of SMYD3 with Akt1 has been previously reported (Supplementary Fig. 1e-g) 19. Given that both K140 and K142 sites were identified at endogenous levels through our large-scale non-biased MS approach, we chose to mainly focus on understanding the contribution of K140 and K142 tri-methylation to Akt activity and oncogenic functions in the remainder of this study.
Absence of Akt methylation represses its oncogenic functions in cells and increases resistance to carcinogen-induced skin tumors in vivo
In order to reveal the potential biological functions of tri-methylation within its Linker region of Akt1, a methylation-deficient variant of Akt1 (K140R and/or K142R) was ectopically expressed in AKT1 and AKT2 double knockout DLD1 cells (DLD1-AKT1/2−/−). Compared to Akt1-WT, the Akt1-K140/142R mutant, and to a lesser extent, Akt1-K140R or -K142R, exhibited a significantly reduced activation, as shown by a marked reduction in Akt-pT308 and Akt-pS473, as well as its downstream targets, pGSK3β and pFOXO3a (Fig. 1c and Supplementary Fig. 1h-j). This observation is coupled with a reduction of Akt1 kinase activity observed in vitro (Supplementary Fig. 1k). As a consequence, absence of methylation on K140 and K142 led to reduced cell colony formation, anchorage-independent growth, glucose uptake, and lactate production in vitro (Fig. 1d, e and Supplementary Fig. 1l, m) and tumor growth in vivo (Fig. 1f-h and Supplementary Fig. 1n, o).
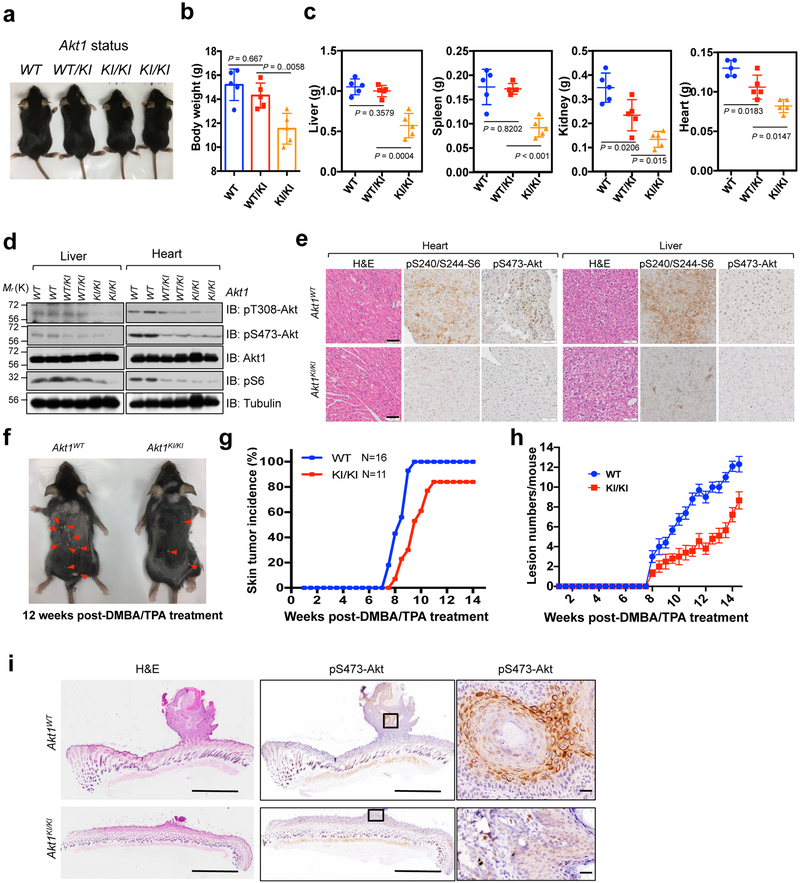
To explore the role of Akt methylation under physiological conditions, we generated Akt1-K140/142R double mutations in HEK293 cells (termed AKT1K140/142R) using CRISPR/Cas9 (Supplementary Fig. 1p-r) 20. Notably, K140/142R mutation of endogenous Akt1 decreased Akt phosphorylation in response to various stimuli (Fig. 1i and Supplementary Fig. 1s-u), and also compromised cell growth, colony formation, glucose uptake, and lactate production (Fig. 1j-m). Next, we generated Akt1-K140/142R double mutation knock-in mice (termed Akt1KI/KI) (Supplementary Fig. 2a-c). Compared with the WT mice, Akt1KI/KI mice exhibited a decrease in body size/weight (Fig. 2a, b and Supplementary Fig. 2d-f), organ size/weight (Fig. 2c and Supplementary Fig. 2g) and Akt kinase activity (Fig. 2d, e and Supplementary Fig. 2h), phenocopying Akt1−/− mice 21. To pinpoint whether methylation deficient mutants of Akt1 abrogate tumorigenesis in vivo, we utilized a two-step chemical carcinogen (DMBA followed by TPA)-induced skin tumor model 22. In this model, we observed that Akt1KI/KI mice displayed a lower incidence of skin tumors and reduced papilloma burden compared with WT mice (Fig. 2f-h), which correlated with decreased Akt signaling (Fig. 2i). These results together support the notion that methylation of Akt in its Linker region likely promotes Akt kinase activity and oncogenic functions both in vitro and in vivo.
Fig. 2. Methylation deficient Akt1 knock-in mice display reduced body size/weight and resist to chemical carcinogen-induced skin tumorigenesis in vivo.
a, Mice derived from the same litter were imaged at age of 4 weeks old. b, The mice derived from the age of 4 weeks were weighed (including 15 male mice with 5 WT, 5 heterogeneous, and 5 homogeneous-Akt1K140/142R genetic background status). Error bars are mean ± s.e.m, n = 5 mice. P values were calculated using two-tailed unpaired Student’s t test. c, The mice in (b) were euthanized and their organs were dissected and weighed. Error bars are mean ± s.e.m, n = 5 mice. P values were calculated using two-tailed unpaired Student’s t test. d, IB analysis of WCL derived from livers or hearts of WT (+/+), Akt1K140/142R-knock-in heterogeneous (+/KI) or homogeneous (KI/KI) mice from the same litter at age of 4 weeks. The experiment was performed twice, independently, with similar results. e, Graphic representation of H&E and IHC staining of heart and liver tissues derived from WT or Akt1K140/142R-KI mice. Scale bar, 50 μm. This experiment was performed twice, independently, with similar results. f, The side view of 12-weeks old mice derived from WT or Akt1K140/142R knock-in mice were treated with chemical carcinogen (DMBA following with TPA) (n = 16 for WT mice; n = 11 for Akt1-K140/142-KI mice). The neoplasm lesions were arrowed. The tumor incidence (g) and lesion numbers (h) of the mice described in (f) were calculated and plotted. Error bars are mean ± s.e.m (for WT n = 16 mice; for Akt1-K140/142-KI n=11 mice). After treatment 12 weeks with DMBA/TPA, the mice were euthanized and the H&E and IHC staining were performed (i). Scale bar, 1 mm. The experiment in (i) was performed twice, independently, with similar results. Statistical source data for b,c,g and h are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
SETDB1 methylates and activates Akt
Next, we set out to identify the physiological upstream methyltransferase(s) for Akt. Consistent with previous reports 23–25, we found that there is a relatively strong physical interaction between Akt1 and two tri-methyltransferases, SETDB1 and EZH2 (Fig. 3a and Supplementary Fig. 2i). Interestingly, genetic alterations of both SETDB1 and EZH2 tended to mutually exclusive of alterations in PI3K/Akt pathway-related genes including PTEN deficiencies, EGFR, PIK3CA or AKT1 amplification/mutations in human melanoma and breast cancer (Supplementary Fig. 2j) 26. Furthermore, SETDB1, but not other methyltransferases including EZH2, could enhance Akt tri-methylation in cells (Supplementary Fig. 2i). This result indicates that unlike EZH2 as a downstream substrate of Akt 25, SETDB1 may be an upstream regulator of Akt methylation (Supplementary Fig. 2k).
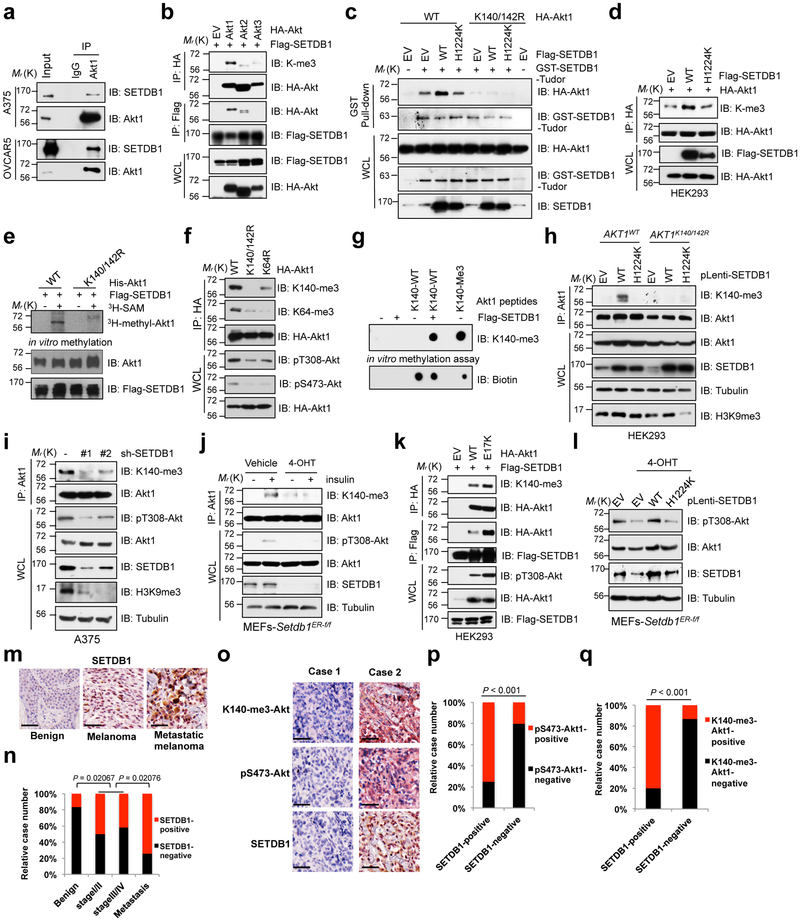
Fig. 3. SETDB1 methylates Akt on K140 and K142 to promote its kinase activity.
a-d,f,h, IB analysis of immunoprecipitates (IP), GST pulldown products and WCL derived from A375 and OVCAR5 (a) or HEK293 cells transfected with indicated constructs (b-d, f) or AKT1K140/142R-KI cells stably expressed WT or catalytically inactive SETDB1-H1224K (d). IgG was used as a negative control. e, In vitro methylation assays were performed with recombinant His-Akt1 proteins purified from insect cells as substrates and purified Flag-SETDB1 from HEK293T cells as the source of methyltransferase in the presence/absence of 3H-SAM. g, In vitro methylation assays were performed with IP Flag-SETDB1 derived from HEK293T cells as the source of methyltransferase, and the synthetic Akt1 peptides containing K140 and K142 as the substrate. Akt1-K140-me3 peptides were used as a positive control. h, IB analysis of IP products and WCL derived from A375 cells lentivirally transfected with shRNAs against SETDB1. Resulting cells were selected with puromycin for 72 hrs before harvesting. j, Setdb1 conditional knockout MEFs were treated with or without 4-OHT (500 nM) for 48 hrs to deplete endogenous Setdb1, then resulting cells were serum-starved for another 20 hrs and stimulated with insulin (100 nM) for 15 min before being harvested and subjected to IP and IB analysis. k, IB analysis of IP products and WCL derived from HEK293 cells transfected with indicated constructs. l, Setdb1 conditional knockout MEFs were infected with WT or H1224K-SETDB1 encoding virus and selected with puromycin for 72 hrs, then treated with or without 4-OHT (500 nM) for another 48 hrs before harvesting for IB analysis. m-q, Image illustration of the immunohistochemistry (IHC) staining for SETDB1, pS473-Akt1 and K140-me3-Akt1 in melanoma TMA (m,o). Scale bar, 50 μm. The distribution of SETDB1 staining was plotted in (n) (n = 97 tissue specimens). The correlation of pS473-Akt or K140-me3-Akt with SETDB1 were plotted in (p,q) (n = 95 tissue specimens). Western-blots in (a-l) were performed twice, independently, with similar results. P values were calculated using Chi-Square test in (n-q). Statistical source data for n-q are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
Notably, SETDB1 specifically interacted with the PH domain of Akt1, and to a lesser extent, Akt2 and Akt3 (Fig. 3b, Supplementary Fig. 2l, m). Interestingly, mutation of Akt1 methylated residues (K140/142R) could markedly decrease the interaction of Akt1 with SETDB1 (Supplementary Fig. 3a). Importantly, the Tudor domain of SETDB1 was identified to bind Akt1, which was enhanced by enforcing expression of WT, but not the SETDB1-H1224K catalytic inactive mutant (Fig. 3c, Supplementary Fig. 3b). These results indicate that SETDB1 could be a potential reader of the Akt1-K140/142 methylation events through its Tudor domain, and by enhancing its interaction with Akt1, SETDB1 could promote Akt1 methylation on additional residues (such as K64). In addition, SETDB1 promoted Akt1 tri-methylation in an enzymatic activity-dependent manner in cells (Fig. 3d), and mutation of either K140 or/and K142 residue could decrease SETDB1-mediated Akt tri-methylation in cells (Supplementary Fig. 3c). Moreover, 3H-SAM-mediated in vitro methylation assays also indicate SETDB1 could directly methylate Akt1 at K140 and K142 residues (Fig. 3e). Hence, these results coherently support the notion that SETDB1 serves as a putative Akt1 methyltransferase.
SETDB1 promotes cell growth and glycolysis through methylation of Akt
As histone H3 methyltransferase capable of catalyzing tri-methylation on the lysine 9 of histone H3 (H3K9me3) 27, SETDB1 is amplified in various cancers 28–31. Notably, we identified that, in addition of being in the nucleus and modifying histone marks, SETDB1 was also observed in the cytoplasm of cells (Supplementary Fig. 3d). To further elucidate the physiological functions attributed to methylated-Akt1, we generated specific antibodies against tri-methylation of Akt1 at K140 (termed K140-me3), which was validated by dot blot and immunoprecipitation assays (Supplementary Fig. 3e, f), and recognized SETDB1-mediated tri-methylation of Akt1 at K140 in cells (Fig. 3f, Supplementary Fig. 3g) and in vitro (Fig. 3g, Supplementary Fig. 3h). Additionally, genetic depletion of SETDB1 markedly reduced Akt1-K140-me3 in cells (Fig. 3h-j and Supplementary Fig. 3i). Compared with Akt1-WT, a cancer patient-derived Akt1-E17K mutant 32 displayed an enhanced interaction with SETDB1, coupled with an increase of its K140 methylation level (Fig. 3k and Supplementary Fig. 3j, k). These findings suggest that Akt1 is a bona fide methylation substrate of SETDB1.
Importantly, genetic deletion of Setdb1 in cells diminished Akt activation (Fig. 3i, j, l and Supplementary Fig. 3l-o). Furthermore, depletion of endogenous SETDB1 by shRNA also reduced Akt-pT308 in different cancer cell lines (Supplementary Fig. 4a, b), leading to suppression of colony formation, anchorage-independent growth, and glucose uptake in cells (Supplementary Fig. 4c-f). Conversely, ectopic expression of SETDB1-WT, but not SETDB1-H1224K 27, induced Akt-pT308 in Setdb1-deficient MEFs (Fig. 3l). Collectively, these results indicate that the activation of Akt by SETDB1 appears to be methylation-dependent. Of note, consistent with a previous report 28, IHC staining revealed that compared to nevus tissues, SETDB1 is highly expressed in melanoma, especially in metastatic malignancies (Fig. 3m, n), and positively correlates with Akt1 K140-me3 and Akt-pS473 IHC signals (Fig. 3o-q).
SETDB1 is suggested to accelerate melanoma onset in cooperation with BRAFV600E 28. Given that coordinated actions of BRAFV600E with activated Akt are required to promote melanomagenesis 33, 34, we employed a well-established human immortalized melanocyte (HIM) model 35 to examine whether SETDB1 modulates the activation of Akt. To this end, we found that concomitant activation of Akt and ERK signaling by SETDB1 and BRAFV600E led to a marked enhancement of anchorage-independent cell growth, coupled with an increased Akt phosphorylation (Fig. 4a-c). Furthermore, deletion of endogenous AKT1 in HEK293 cells compromised the oncogenic capability of SETDB1 to promote cell colony formation compared with parental cells (Fig. 4d-f).
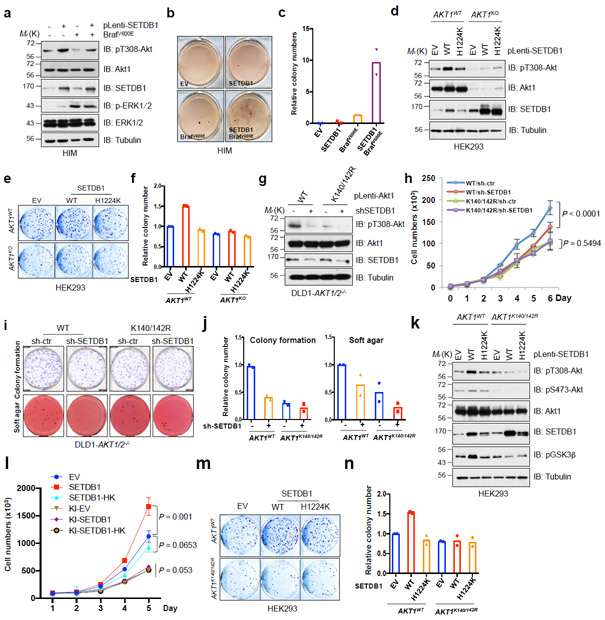
Fig. 4. Oncogenic function of SETDB1 depends on the activation of Akt.
a-c, Human immortalized melanocytes (HIM) were lentivirally infected with indicated constructs, and selected with puromycin and hygromycin for 72 hrs before harvesting for IB analysis (a). Resulting cells were subjected to soft agar assays (b). d, IB analysis of CRISPR/CAS9-mediated AKT1 knockout and parental HEK293 cells which were lentivirally infected with the constructs encoding SETDB1. e,f, Cells described in d were subjected to colony formation assays (e). g-j, DLD1-AKT1/2−/− cells were infected with virus encoding WT or mutated Akt1, and selected with hygromycin for 72 hrs. Resulting cells were lentivirally infected with shRNA against SETDB1 (with shCtr as a negative control) and selected with puromycin for 72 hrs, and were harvested for IB analysis (g), cell proliferation (h), colony formation and soft agar (i) assays. k-n, AKT1 K140/142R-edited and parental HEK293 cells were lentivirally infected with SETDB1-WT or SETDB1-H1224K encoding constructs and selected with puromycin for 72 hrs before harvesting for IB analysis (k). Resulting cells were subjected to proliferation (l) and colony formation (m) assays. The experiments in (b,e,i,m) were performed twice independently with three repeats, and exhibited similar results. Relative colony numbers derived from two independent experiments were plotted in (c,f,j,n). The cell proliferation assays in (h,l) were performed three times independently, and cell numbers were quantified in (h,l). Error bars are mean ± s.e.m, n = 3 independent experiments. Two-way ANOVA analysis was performed in (h,l) to calculate the P value. Detailed statistical tests are described in the Methods. Source data for c,f,h,j,l and n are shown in Supplementary Table 2. Western-blots in (a,g,k) were performed twice, independently, with similar results. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
More importantly, depletion of SETDB1 resulted in decreased Akt phosphorylation and cellular malignant phenotypes in Akt1-WT expressing DLD1-AKT1/2−/− cells, but did not in Akt1-K140/142R expressing cells that already displayed relatively lower oncogenic capacity than WT cells (Fig. 4g-j). These results indicate that, Akt is likely one of the major downstream pathways through which SETDB1 exerts its oncogenic role. Consistently, re-introduction of constitutively active Akt1 (myr-Akt1) partially restored the colony formation capability of SETDB1-depleted A375 cells (Supplementary Fig. 4g, h). Furthermore, SETDB1 was capable of accelerating cell growth and colony formation in AKT1WT, but not in the methylation-deficient AKT1K140/142R expressing HEK293 cells (Fig. 4k-n). Hence, our results suggest that SETDB1 may exert its oncogenic roles largely through Akt activation in a methylation-dependent manner.
SETDB1-mediated Akt methylation crosstalks with PI3K-mediated Akt phosphorylation
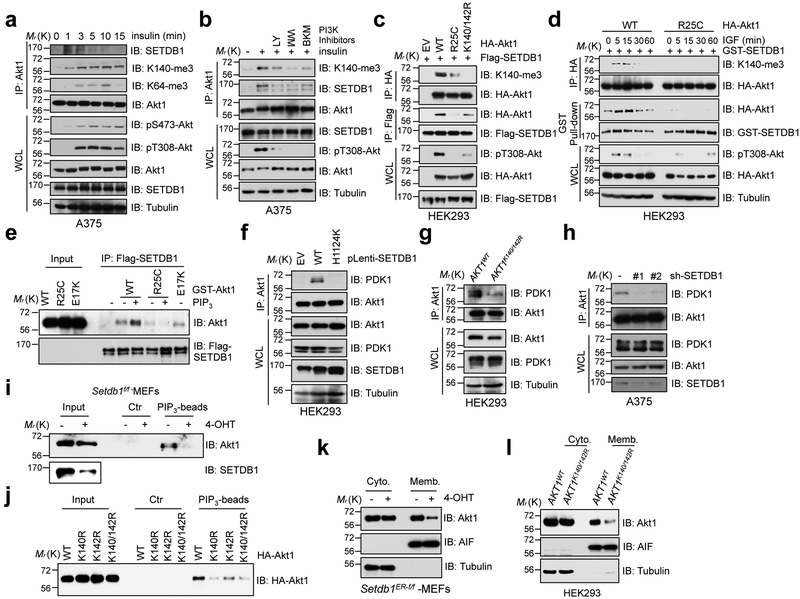
It is well established that Akt activation requires PI3K-dependent generation of PIP3, which binds with the PH-motif of Akt at the plasma membrane 36–38. We explored the temporal relationship between methylation-dependent and PI3K-dependent activation of Akt in cells. We showed that the interaction between Akt1 and SETDB1, and subsequent methylation of Akt, temporally coincided with Akt-pT308 following insulin or IGF stimulation (Fig. 5a and Supplementary Fig. 4i-k). Interestingly, PI3K inhibitors, but not Akt1 mutated variants (T308A and/or S473A), diminished the interaction of Akt1 with SETDB1, and decreased Akt1-K140 methylation in cells (Fig. 5b and Supplementary Fig. 4l, m), indicating that the PI3K pathway may function upstream of SETDB1-mediated Akt methylation to govern Akt activation. In further support of this notion, the non-PIP3 binding mutation of Akt1-R25C 39, largely impaired the interaction between Akt and SETDB1, leading to a marked decrease in Akt1-K140 methylation (Fig. 5c, d).
Fig. 5. SETDB1-medidated methylation of Akt synergizes with PI3K to activate Akt.
a,b, A375 cells were serum-starved for 20 hrs, then stimulated with insulin (50 nM) at different time points (a) or post-treatment of various PI3K inhibitors (b) before harvesting for IP and IB analysis. c,d, IB analysis of IP products and WCL derived from HEK293 cells transfected with indicated constructs stimulated without (c) or with IGF (100 ng/ml) (d) before harvesting. e, In vitro binding assays were performed with recombinant GST-Akt1 protein purified from mammalian cells, and flag beads bound SETDB1. The binding was performed in 4°C for 4 hrs incubated with or without PIP3 (20 μM) and subjected to IB analysis. f-h, IB analysis of Akt1-IP and WCL derived from HEK293 cells infected with indicated SETDB1 encoding constructs (f), AKT1K140/142R and its parental HEK293 cells (g) and SETDB1 depleted A375 cells (h). i,j, IB analysis of PIP3 pull-down products and WCL derived from Setdb1 conditional knockout MEFs treated with or without 4-OHT (500 nM) for 48 hrs (i) or from HEK293 cells transfected with indicated constructs (j). Where indicated, empty beads (Ctr) serve as a negative control. k,l, IB analysis of cell fractionations separated from Setdb1 conditional knockout MEFs treated with or without 4-OHT (500 nM) for 48 hrs (k) or from AKT1K140/142R-edited and parental HEK293 cells (l). All Western-blots above were performed twice, independently, with similar results. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
To further pinpoint the potential role of PIP3 in mediating the interaction between SETDB1 with Akt1-PH motif, we performed in vitro binding assays. Results suggest that the interaction of SETDB1 with Akt1-WT, but not Akt1-R25C, could be enhanced by PIP3 in vitro (Fig. 5e and Supplementary Fig. 5a-e), indicating that PIP3 binding could induce a conformation change of the PH motif, which facilitates Akt1 binding to SETDB1. Importantly, PTEN deficiency dramatically increased Akt1-K140 methylation coupled with increased Akt-pT308 (Supplementary Fig. 5f). Together, these data suggest that SETDB1 mediates Akt methylation likely in a PI3K catalytic activity-dependent manner.
Consistent with the finding that methylation of Akt1 could promote Akt-pT308, we observed that WT, but not the catalytically inactive SETDB1-H1224K, could induce the interaction of Akt1 with PDK1 in cells (Fig. 5f and Supplementary Fig. 5g). Moreover, the methylation-deficient Akt1 mutant (K140/142R) or depletion of SETDB1 displayed an attenuated interaction with PDK1 compared with Akt1-WT (Fig. 5g, h and Supplementary Fig. 5h). In further support of the role for Akt methylation in controlling its activation, reducing Akt methylation by deleting Setdb1 or introducing the Akt1-K140/142R mutation diminished the interaction of Akt1 with PIP3 (Fig. 5i, j), and subsequently led to a marked decrease in association of Akt1 with the cell membrane (Fig. 5k, l and Supplementary Fig. 5i-m). These results indicate that methylated Akt1 may have a greater propensity to bind PIP3 and translocate to the cellular membrane to achieve full activation. Thus, there is likely an intrinsic interplay between PI3K-mediated and SETDB1-mediated pathways to tightly control the spatial and temporal activation of Akt (Supplementary Fig. 5n).
Given that TRAF6-mediated Akt ubiquitination has a critical role in regulating Akt membrane translocation 15, 40, 41, we also found that WT, but not SETDB1-H1224K mutant, increased the interaction between Akt1 and TRAF6 (Supplementary Fig. 5o), resulting in an increase of Akt ubiquitination (Supplementary Fig. 5p). Furthermore, in comparison with Akt1-WT, Akt1-K140/142R displayed a reduced interaction with TRAF6 (Supplementary Fig. 5q), resulting in decreased Akt ubiquitination (Supplementary Fig. 5r). These findings suggest that SETDB1-induced methylation likely mediates the interaction of Akt1 with TRAF6, and subsequent Akt ubiquitination may promote membrane translocation of methylated Akt (Supplementary Fig. 5n).
KDM4B demethylates and attenuates Akt kinase activity
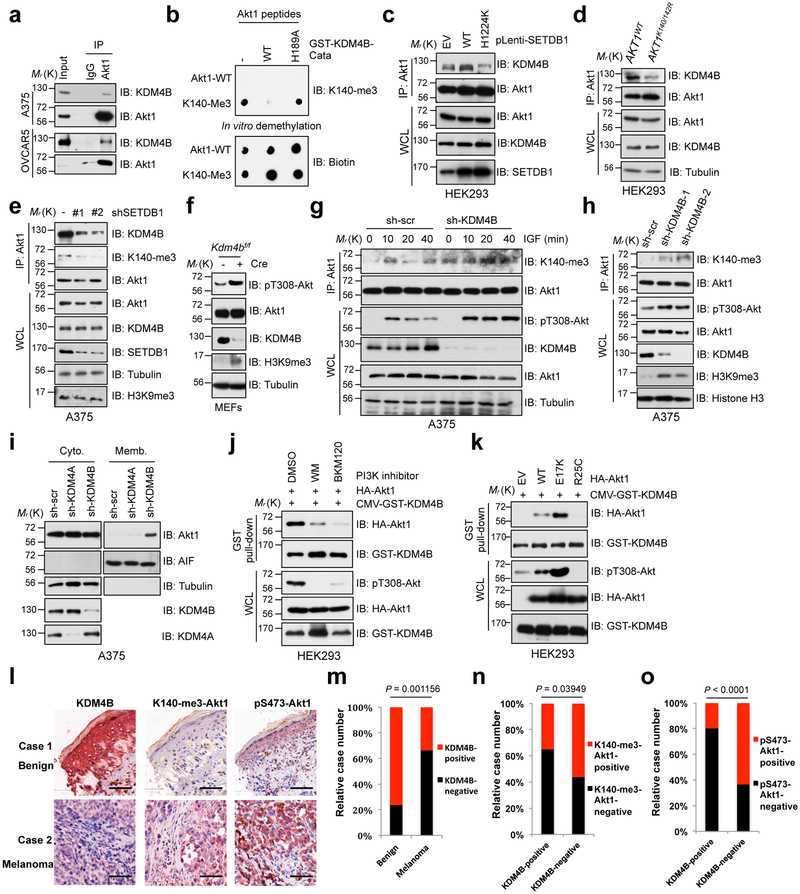
Importantly, the Jumonji 2 (JMJD2, also termed KDM4) family proteins have been shown to function as specific erasers of SETDB1-mediated H3K9me3 42, therefore, we assessed their ability to demethylate Akt1-K140me3. Notably, among all members of KDM4 family, KDM4A and KDM4B, and to a lesser extent KDM4C, were observed to interact with Akt1 (Fig. 6a and Supplementary Fig. 6a). Furthermore, in vitro demethylation assays 42 demonstrated that KDM4B-WT, but not the catalytically inactive KDM4B-H189A or other KDM4 members could efficiently erase the tri-methylation of Akt1 at K140 in vitro (Fig. 6b and Supplementary Fig. 6b-e) or in cells (Supplementary Fig. 6f). Moreover, the interaction between KDM4B and Akt1 was readily induced by enforcing expression of SETDB1-WT, but not its catalytic mutant (Fig. 6c). We further showed that Akt interacts with KDM4B through its Tudor domain (Supplementary Fig. 6g-j) in a methylation-dependent manner (Fig. 6d, e and Supplementary Fig. 6k, l). Hence, these data indicate that KDM4B might be a bona fide demethylase of Akt1.
Fig. 6. KMD4B demethylates Akt to inhibit Akt kinase activity.
a,c-e, IB analysis of Akt1-IP products and WCL derived from A375, OVACR5 cells (a), SETDB1 expressing HEK293 cells (c), AKT1K140/142R-edited and parental HEK293 cells (d) and SETDB1-depleted A375 cells (e). IgG was used as a negative control. b, IB analysis of in vitro de-methylation assays performed with synthetic Akt1-K140-me3 peptides as substrate, and bacterially purified catalytic domain of KDM4B as the source of demethylase. f, IB analysis of WCL derived from primary Kdm4b conditional knockout MEFs infected with or without phage-Cre for 48 hrs before harvesting. g, A375 cells were lentivirally infected with shRNA against KDM4B. Resulting cells were serum starved for 20 hrs, then stimulated with IGF (100 ng/ml) before harvesting for IP and IB analysis. h, IB analysis of IP products and WCL derived from A375 cells infected with lentivirus against KDM4B. i, IB analysis of cell fractionations separated from A375 cells lentivirally infected with shRNA against KDM4B or KDM4A. j,k, HEK293 cells were transfected with indicated constructs and treated with different PI3K inhibitors for 1 hr (j) before subjected to GST pull-down assay and IB analysis. l-o, IHC staining of KDM4B, pS473-Akt1 and K140-me3-Akt1 in melanoma TMA (l). Scale bar, 50 μm. The distribution of KDM4B staining was plotted in (m, n = 97 tissue specimens). The correlations of pS473-Akt or K140-me3-Akt1 with KDM4B were plotted in (n, n = 96 tissue specimens; o, n = 95 tissue specimens). All P values were calculated using Chi-Square test. Detailed statistical tests were described in Methods. Western-blots were performed twice, independently, with similar results. Statistical source data for m-n are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
KDM4B-depletion concomitantly increased Akt-pT308 and K140-me3 in multiple cell lines with Akt1-WT, but not those with AKT1K140/142R cells (Fig. 6f-h and Supplementary Fig. 6m-p). Strikingly, depletion of KDM4A displayed a reduction in Akt phosphorylation level (Supplementary Fig. 6q, r). In addition, depletion of KDM4B, but not KDM4A, largely enhanced Akt1 membrane translocation (Fig. 6i and Supplementary Fig. 6s). Furthermore, the interaction between Akt1 and KDM4B primarily occurred in the later phase of insulin stimulation (Supplementary Fig. 6t). This interaction was abolished by PI3K inhibitors (Fig. 6j). Consistent with the finding that KDM4B interacts with methylated-Akt, the patient-derived mutation (Akt1-E17K) increased the interaction between Akt1 and KDM4B, while the non-PIP3 binding mutation (Akt1-R25C) had the opposite effect (Fig. 6k). Clinically, KDM4B was expressed at relatively high levels in benign nevus compared with malignant melanoma (Fig. 6l, m). This observation correlated with decreased Akt-pS473 and K140-me3 IHC signals (Fig. 6l, n, o). Taken together, these data suggest that KDM4B might function as a negative regulator, antagonizing SETDB1-mediated methylation and activation of Akt.
SETDB1 is a potential therapeutic target of Akt-driven cancers
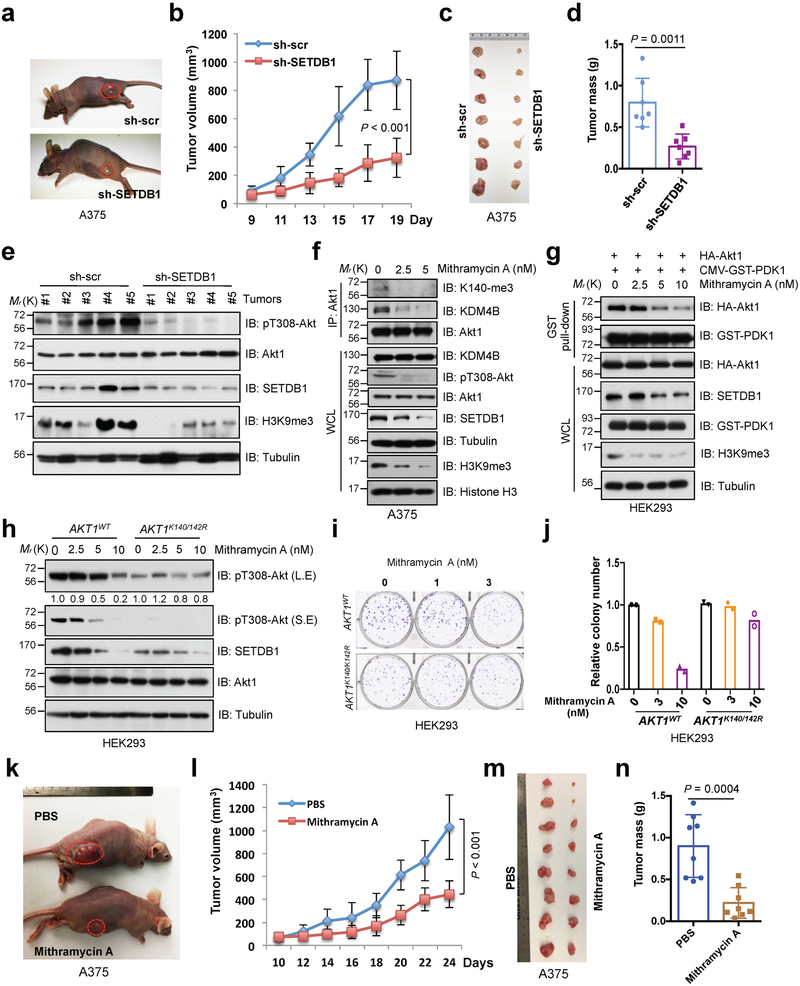
Given the critical role for the PI3K/Akt pathway in facilitating tumorigenesis, targeting hyperactive PI3K/Akt signaling by PI3K and/or Akt inhibitors have been pursued as promising anti-cancer therapies. However, relatively high-dose of these inhibitors induced cellular toxicity restricting their potential use as a treatment option in cancer patients 43, 44. Thus, our finding that SETDB1 synergizes with PI3K to activate Akt kinase (Supplementary Fig. 7a) indicates that SETDB1 specific inhibitors could benefit cancer patients by targeting both H3K9-mediated epigenetic and PI3K/Akt oncogenic pathways. In support of this hypothesis, we showed that depletion of SETDB1 significantly decrease A375 tumor growth in vivo (Fig. 7a-d), coupled with a reduction in H3K9me3 and Akt phosphorylation (Fig. 7e).
Fig. 7. Deficiency of SETDB1 inhibits Akt kinase activity and oncogenic function.
a-d, SETDB1-depleted A375 and control cells were subjected to mouse xenograft assays. Tumor sizes were monitored (a,b). Tumors were dissected (c) and tumor mass were weighed (d). Error bars are mean ± s.e.m, n = 7 mice. P values were calculated by using two-way ANOVA analysis (b) and two-tailed unpaired Student’s t test (d). e, The phosphorylation status of Akt1 (pT308-Akt) and methylation of H3K9 (H3K9me3) were detected by IB analysis with WCL derived from recovered xenografted tumors. f, IB analysis of IP products and WCL derived from A375 cells treated with different doses of Mithramycin A for 72 hrs before harvesting. g, HEK293 cells were transfected with indicated constructs and treated with different doses of Mithramycin A for 72 hrs before harvesting for GST pull-down assays and IB analysis. h-j, AKT1K140/142R-edited and parental HEK293 cells were treated with different doses of Mithramycin A for 72 hrs and subjected to IB analysis (h). Meantime, resulting cells were subjected to colony formation (i) assay. The experiment was performed twice independently with three repeats, and exhibited similar results (i). Representative images were shown in (i) and relative colony numbers derived from two independent experiments were plotted in (j). k-n, Mithramycin A treatment reduced in vivo tumorigenesis of xenografted A375 cells. When the tumors of xenografted A375 cells reached 100 mm3, the mice were treated with Mithramycin A (0.2 mg/kg) or PBS (as a negative control). Tumor sizes were monitored in (k,l) and tumor mass were weighed and presented in (m,n). Error bars are mean ± s.e.m, n = 8 mice. P values were calculated by using two-way ANOVA analysis (l) and two-tailed unpaired Student’s t test (n). Detailed statistical tests are described in the Methods. Source data for b, d, j, l and n are shown in Supplementary Table 2. Scanned images of unprocessed blots are shown in Supplementary Fig. 8.
Consistent with a previous report 45, we found that Mithramycin A, an anti-neoplastic antibiotic, could markedly decrease SETDB1 expression and H3K9me3 in different cells. Mithramycin A subsequently led to reduced Akt methylation and phosphorylation (Fig. 7f and Supplementary Fig. 7b) and decreased the interaction of Akt1 and PDK1 (Fig. 7g). Biologically, Mithramycin A attenuated the phosphorylation and colony formation of cells expressing Akt1-WT, but not in AKT1/2-depleted DLD1 cells or AKT1K140/142R-edited HEK293 cells (Fig. 7h-j and Supplementary Fig. 7c-e). To explore the potential anti-tumor roles of Mithramycin A in vivo, we treated nude mice bearing A375 or DLD1 xenografted tumors, and showed that Mithramycin A could significantly decrease tumor growth compared with the vehicle treatment (Fig. 7k-n, and Supplementary Fig. 7f-i). Conceivably, the reduction of SETDB1 expression mediated by Mithramycin A decreased H3K9me3 and Akt phosphorylation (Supplementary Fig. 7j). Taken together, these results suggest that SETDB1 inhibition could possibly benefit cancer patients by repressing Akt oncogenic signaling in addition to its well characterized role in reprogramming the epigenome (Supplementary Fig. 7k).
DISCUSSION
In order to identify novel non-histone methylated proteins involved in oncogenic signaling pathways, we performed a mass spectrometry based high-throughput screen and detected numerous proteins modified by tri-methylation. We found that Akt1 could be methylated in its Linker region, which was enhanced during physiological conditions such as growth factor (Insulin or IGF) stimuli or pathological conditions such as Akt1-E17K mutation or PTEN deficiency. Absence of Akt methylation represses its kinase activity, and markedly decreases cell growth, glucose uptake, and tumorigenesis. More strikingly, methylation-deficient Akt1 knock-in mice exhibit decreased body weight/size, which phenocopies Akt1 knockout mice 46. Importantly, we also observed that methylation-deficient Akt1 knock-in could attenuate tumorigenesis in a carcinogen-induced skin tumor mouse model. Further investigations will be needed to determine whether knock-in methylation-deficient Akt1 in genetic alteration models, such as KRasG12D mutation and p53 depletion-mediated lung cancer models (KP mice) 47, could efficiently compromise the lung tumorigenesis through the decrease of Akt activity.
Among the known upstream signals, Akt1-T308 phosphorylation mediated by PDK1 is pivotal for Akt activation in a PIP3 dependent fashion 48. Here, we observed that SETDB1-mediated Akt methylation crosstalks with PI3K-mediated Akt phosphorylation. We suspect that PIP3 binding to the PH domain of Akt could “unlock” the Akt auto-inhibition confirmation. The conformation change could be necessary for SETDB1 to interact with the Akt PH domain, and to methylate Akt in its exposed Linker region, in turn enhancing the interaction of Akt with its upstream kinase PDK1 (Supplementary Fig. 5n). We also observed that SETDB1 could enhance the interaction of Akt with TRAF6 to facilitate Akt ubiquitination, subsequent PIP3 binding and membrane translocation (Supplementary Fig. 5n). Our model suggests a loop of Akt activation triggered by PIP3 accumulation or Akt1-E17K mutation, which could be antagonized by the demethylase KDM4B (Supplementary Fig. 5n). Additionally, the SETDB1-Tudor domain interacts with Akt1 mainly in the context of Akt1 Linker region being methylated. Thus, we speculate that methylation of the Linker region could be the primary modification that enhances SETDB1 interaction with Akt1, and sequentially promotes methylation of Akt on other lysine residues (such as K64 in Akt1 PH domain). Consistent with our findings, another group also revealed that SETDB1-mediated Akt1 tri-methylation at K64 plays important role for Akt interacting with and being ubiquitinated by TRAF6 to facilitate Akt membrane translocation and kinase activation 49. Therefore, the complicated connection and crosstalk of SETDB1-mediate different lysine methylation on Akt including K140/K142 in linker domain and K64 in PH domain warrants further investigation.
Finally, our study reveals that SETDB1 accelerates tumorigenesis largely through Akt activation in a methylation-dependent manner. As such, the function of SETDB1 is not restricted to the nucleus to repress gene transcription, but SETDB1 also methylates non-histone proteins in the cytoplasm such as Akt to activate its oncogenic functions.
METHODS
Cell culture, transfection and cell fractionations.
Cell line sources are detailed in the Reporting Summary. All cell lines were free of mycoplasma contamination (tested by the vendor). HEK293, HEK293T, DLD1, OVCAR5 and A375 cells were cultured in DMEM medium supplemented with 10% FBS, 100 units of penicillin and 100 μg/ml streptomycin. HCT116 PTEN+/+ and PTEN−/− cells (as gifts from Dr. Todd Waldman in School of Medicine, Georgetown University). Setdb1f/f-ER-Cre mouse embryonic fibroblasts (MEFs) were obtained as gifts from Drs. Yoich Shinakai and Matthew C. Lorincz 50. DLD1-AKT1−/−AKT2−/− (termed AKT1/2−/−) and counterpart cells were kindly provided by Dr. Bert Vogelstein (Johns Hopkins University School of Medicine), and these cells were also maintained in DMEM medium supplemented with 10% FBS. The human immortalized melanocyte HIM was cultured in keratinocyte serum-free medium (K-SFM, Gibco Life Technologies), supplemented with epidermal growth factor (EGF; 5 ng/ml) and bovine pituitary extract (40 pg/ml). Cell transfection was performed using Lipofectamine and Plus reagents, as described previously 17. Packaging of lentiviral shRNA or cDNA expressing viruses and retroviral cDNA expressing viruses, as well as subsequent infection of various cell lines were performed according to the protocols described previously 17. Following viral infection, cells were maintained in the presence of hygromycin (200 μg/ml) or puromycin (1 μg/ml), depending on the viral vectors used to infect cells.
To prepare primary MEFs, Jmjd2bflox/flox mice were crossed and fibroblasts were established from E13.5 embryos according to standard procedures. In brief, the embryos were dissociated and then trypsinized to produce single-cell suspensions. The cells were maintained in DMEM supplemented with 10% fetal calf serum (FCS), L-glutamine, and antibiotics. The cells were infected with phage-Cre lentiviruses for 2 days to deletion Jmjd2b. Kras;p53 and Kras;p53;sMYD3−/− cells derived from mouse lung and pancreas were obtained from Pawel K. Mazur at Stanford University School of Medicine 18.
Cell fractionations were performed with Cell Fractionation Kit (CST9038). Kinase inhibitors Wortmannin (Selleck S2758), BKM120 (Selleck S2247) and LY294002 (Selleck S1105) were used at the indicated doses. Mithramycin A (Sigma M6891), growth factors including EGF (Sigma E9644), insulin (Invitrogen 41400–045) and IGF (Sigma SRP3069), were used at the indicated doses. PIP3 beads (P-B00Ss) and free PIP3 (P-3908) were purchased from Echelon Biosciences.
Plasmid construction.
Constructs of pcDNA3-HA-Akt1, pcDNA3-HA-Akt2, pcDNA3-HA-Akt3 and pcDNA3-HA-myr-Akt1, pcDNA3-HA-Akt1-E17K, pET-GSK3β, pCMV-Flag-PDK1 were previously described 17. pCMV-flag-EZH2, pCMV-flag-SET7, pCMV-flag-SET8 and pCMV-flag-MMSET were bought from Addgene. pCDNA-flag-SETDB1 and pCDNA-flag-SETDB1-H1224K were as gift from Dr. Gerd P. Pfeifer 51. pCMV-Flag-KDM4A, 4B, 4C and 4D were obtained from Dr. Yang Shi (Harvard Medical School). pCMV-GST-PDK1, pCMV-GST-SETDB1, pCMV-GST-Akt1, pCMV-GST-Akt1-PH (aa1–108), pCMV-GST-Akt1-Linker (aa109–150), pCMV-GST-Akt1-KD (aa151–408), pCMV-GST-Akt1-HM (aa409–481), pCMV-GST-SETDB1-Tudor (aa257–318), pCMV-GST-KDM4A, pCMV-GST-KDM4C, pCMV-GST-KDM4D, pCMV-GST-KDM4B, pCMV-GST-KDM4B-jmj (aa1–180), pCMV-GST-KDM4B-Linker (aa181–425) and pCMV-GST-KDM4B-PH-Tudor (aa181–425) were cloned into mammalian expression GST-fusion vectors. Details of plasmid constructions are available upon request.
Various Akt1 and KDM4B mutants were generated using the QuikChange XL Site-Directed Mutagenesis Kit (Stratagene) according to the manufacturer’s instruction. All mutants were generated using mutagenesis PCR and the sequences were verified by DNA sequencing. sh-Akt1-resistant mutants were generated with the specific primers as previously described 16.
Antibodies.
All antibodies were used at a 1:1000 dilution in TBST buffer with 5% non-fat milk for western blot. Anti-phospho-Ser473-Akt antibody (4060), anti-phospho-Thr308-Akt antibody (2965), anti-Akt1 antibody (2938), anti-Akt total antibody (4691), anti-PDK1 antibody (13037), anti-SMYD3 antibody (12859), anti-H3K9me3 antibody (13969), anti-Pan-Kme3 antibody (14680), anti-AIF antibody (5318), anti-Histone H3 antibody (4499), anti-H3K4me2 antibody (9725), anti-phospho-Ser9-GSK3β antibody (5558), anti-GSK3β antibody (12456), anti-phospho-FOXO1 (Thr24)/FOXO3A (Thr32) antibody (9464), anti-FOXO3A antibody (2497), anti-GST antibody (2625), anti-pS6K1 (Thr389) antibody (9205), anti-S6K1 antibody (2708) and anti-pS240/244-S6 antibody (5364) were obtained from Cell Signaling Technology. Anti-SETDB1 antibody (11231) was obtained from Proteintech. Anti-KDM4A (A300–860A) and anti-KDM4B (A301–478A) antibodies were bought from Bethyl. Anti-Akt1 agarose beads (sc-5298) and polyclonal anti-HA antibody (sc-805) were obtained from Santa Cruz. Polyclonal anti-Flag antibody (F-2425), monoclonal anti-Flag antibody (F-3165, clone M2), anti-Tubulin antibody (T-5168), anti-Flag agarose beads (A-2220), anti-HA agarose beads (A-2095), peroxidase-conjugated anti-mouse secondary antibody (A-4416) and peroxidase-conjugated anti-rabbit secondary antibody (A-4914) were obtained from Sigma. Monoclonal anti-HA antibody (MMS-101P) was obtained from Covance.
The polyclonal Akt1-K140-me3 antibodies generated by Cell Signaling Technology (CTS) were derived from rabbit, with each hydroxylation residue produced four clones. The antigen sequence used for immunization was Akt1 aa130–152 (GAEEMEVSLAKPKHRVTMNEFEY). K stands for tri-methylation residue in this synthetic peptide. The antibodies were affinity purified using the antigen peptide column, but they were not counter selected on unmodified antigen.
shRNAs, sgRNAs and CRISPR/CAS9-mediated knock-in and knockout assay.
shRNAs against SETDB1 as gift obtained from Dr. H. Phillip Koeffler 52, and shRNAs against KDM4A as gift obtained from Dr. Cun-yu Wang 53. shRNAs against KDM4B were reported previously 54.
The lentiviruses for CRISPR/Cas9-based editing of AKT1 were generated by cloning the annealed short guide RNAs (sgRNAs) into BsmBI-digested lentiCRISPR V2 vector, which encodes both Cas9 and an sgRNA of interest, as previously described 20. The sgRNAs were designed by CRISPR Design tool (crispr.mit.edu) as listed below:
sghAKT1#1 Forward: 5’-CACCGTGAAGAGATGGAGGTGTCCC-3’;
Reverse: 5’-AAACGGGACACCTCCATCTCTTCAG-3’;
sghAKT1#2 Forward: 5’-CACCGTGGGGACAGGCCTCACCACG-3’;
Reverse: 5’-AAACCGTGGTGAGGCCTGTCCCCAC-3’;
For knock-in experiments, the lentiCRISPR V2 constructs containing sgRNA (as mentioned above) were co-transfected with template DNA into HEK293 cells. After selected with puromycin for two days, resulting cells were seeded into 96 well plates with 1 cell/well. After culture around 2 weeks, the single clone was expanded to 12 well-plate, at the same time, the genomic DNA of each clone was harvested by using DNA Extract Solution (Epicentre), and PCR was performed with the primers covered the guild RNA sequence (around 600bp). The PCR products were digested with SacII, which can digest the knock-in (KI) genome but not the wild-type genome AKT1. Following that, the genomic DNAs of digestible clone were subjected to sequence to confirm the knock-in sequence. The examined clones were expanded and prepared for further studies.
AKT-K140/142R guild DNA sequence:
GGAGATGGACTTCCGGTCGGGCTCACCCAGTGACAACTCAGGGGCTGAAGAGATGGAGGTGTCCCTCGCCAGGCCGCGGCATCGCGTGGTGAGGCCTGTCCCCACTTCTGCCTGTGCCTGGGGCTGCCTTGGACTGTGGAGGGCTGGGTG (the bold indicated mutant nucleotides to restitute K140/142 to R140/142, and at the same time to generate a new Sac II digest site. The shadow nucleotides indicated the destruction of sgRNA-recognized site.).
Generation and validation of Akt1K140/142R knock-in mice.
sgRNAs were synthesized by using in vitro transcription (IVT) method (PNA Bio Company) with the sequence: TGAAGAGATGGAGGTGTCCC and TGTGGTGAGTCTGGGCTCTG. Cas9 enzyme protein was obtained from PNA Bio. Mutated mouse genomic single strand template DNA (ssODN) was synthesized from IDT with the sequence: GGACTCAAGAGGCAGGAAGAAGAGACGATGGACTTCCGATCAGGCTCACCCAGTGACAACTCAGGGGCTGAAGAGATGGAGGTGTCCCTCGCCAGGCCGCGGCATCGTGTGGTGAGTCTGGGCTCTGCTTCTGCTGGGGCTGCCAGGGGTTGTGGAGGGACCTGGTAGGTCCTGGTATTCCTGCCTGGCTTCCTGTATGG (the bold indicated mutant nucleotides to restitute K140/142 to R140/142, and at the same time to generate a new Sac II digest site. The shadow nucleotides indicated the destruction of sgRNA-recognized site.). The mixture of Cas9 protein (200 ng/μl), sgRNA(s) (100 ng/μl) and ssODN DNA (10–70 ng/μl) in 50 μl was utilized for generating knock-in mice by BIDMC transgenic facility.
The genomic DNAs derived from the tail of edited mice were extracted with QuickExtract DNA extraction solution (Epicentre, QE09050), and the PCR was performed with primers: mAkt1-geno-5’ 5-AGGCCCAGGATCTGAGTGG and mAkt1-geno-3’ 5-TCAGCGGGCATCTTCATATTAC. The PCR products were digested with Sac II for 1 hr or subjected for Sanger sequencing with the primer mAkt1-seq-3’ 5-GACAAGCACTCTGCCAACTG. The validated heterogeneous knock-in mice (termed founder mice, P1) were selected and crossed to each other, and got the WT, heterogeneous and homogenous Akt1K140/142R knock-in mice (termed P2). Furthermore, we crossed homogenous knock-in mice with wild-type C57BL6 mice (Jackson Lab), and got the heterogeneous Akt1K140/142R knock-in mice (termed P3). Then we crossed these heterogeneous knock-in mice to each other and got the WT, heterogeneous and homogenous knock-in mice (termed P4). In this time point, we monitored the mice body size, organ size as well as Akt signaling among WT, heterogeneous and homogeneous Akt1K140/142R knock-in mice. To further diminish the nonspecific effect of genomic editing, we would cross back the homogenous knock-in mice (P4) with wild-type C57BL6 mice again to generate heterogeneous knock-in mice (P5) for further crossing and phenotype monitoring.
All experimental procedures were approved by the Institutional Animal Care & Use Committee (IACUC, RN150D) at Beth Israel Deaconess Medical Center with protocol #043–2015. The research projects that are approved by the IACUC are operated according the applicable Institutional regulations. The Institute is committed to the highest ethical standards of care for animals used for the purpose of continued progress in the field of human cancer research.
IAP-LC-MS/MS screening.
The experiments were performed as previously reported 10. Briefly, OVCAR5 cell lysates were proteolytically digested, then purified over a Sep-Pak C18 cartridge. After dissolving in IAP buffer, the recovered peptides were purified with tri-methylation-specific antibodies. Finally, the enriched peptides were subjected to LC-MS/MS to identify the tri-methylation sites in corresponding proteins.
Two-step chemical carcinogen-induced skin tumors.
The method of two-step chemical carcinogen inducing skin tumors were described previously 22,56. In brief, the shaved naked back of 4–6 week WT or Akt1K140/142R knock-in mice were painted with 150 μg DMBA (Sigma D3254) resolved in 200 μl acetone. After recovered two weeks, the mice were treated with 20 μg TPA (LC laboratories, P1680) in 200 μl acetone twice/week for 15 weeks. The neoplasms were monitored and then dissected after mice euthanized, and subjected to HE and IHC staining.
Immunoblot (IB) and immunoprecipitation (IP) analyses.
Cells were lysed in EBC buffer (50 mM Tris pH 7.5, 120 mM NaCl, 0.5% NP-40) supplemented with protease inhibitors (Complete Mini, Roche) and phosphatase inhibitors (phosphatase inhibitor cocktail set I and II, Calbiochem). The protein concentrations of whole cell lysates were measured by the Beckman Coulter DU-800 spectrophotometer using the Bio-Rad protein assay reagent. Equal amounts of whole cell lysates were resolved by SDS-PAGE and immunoblotted with indicated antibodies. For immunoprecipitation analysis, 1000 μg lysates were incubated with the indicated antibody (1–2 μg) for 3–4 hrs at 4°C followed by 1 hr incubation with Protein A/G sepharose beads (GE Healthcare). The recovered immuno-complexes were washed five times with NETN buffer (20 mM Tris, pH 8.0, 150 mM NaCl, 1 mM EDTA and 0.5% NP-40) before being resolved by SDS-PAGE and immunoblotted with indicated antibodies. Quantification of the immunoblot band intensity was performed with Image J software.
Melanoma and Nevus TMA and IHC staining.
Tissue microarray (ME1004f) containing 18 nevi, 33 metastatic melanomas and 49 malignant melanomas were obtained from Biomax.
Immunohistochemistry was performed on four micron-thick, FFPE sections. FFPE sections were deparaffinized using xylene and rehydrated in graded ethanol. Sections were heated with a pressure cooker to 125°C for 30 seconds and 90°C for 10 seconds in citrate buffer (pH 6.0) for antigen retrieval. All sections were incubated with peroxidase (Dako #S2003) and protein blocking reagents (Dako #X0909) for 5 minutes each. Sections were then incubated with anti-pS473-Akt (CST #4060, 1:50), anti-K140-me3-Akt (1:200), anti-pS420/424-S6 (CST #5364, 1:1500) and anti-SETDB1 (CST #93212, 1:100) antibody diluted in Dako diluent with background reducing components (Dako #S3022) for 1 hr at room temperature. Following primary antibody incubation, sections were incubated with monoclonal mouse anti-rabbit immunoglobulins (Dako #M0737) for 30 minutes at room temperature. Afterwards, sections were incubated with Envision+ System-HRP Labeled Polymer Anti-Rabbit (Dako #K4003) for 30 minutes. All sections were developed using the DAB chromogen kit (Dako #K3468) and lightly counterstained with hematoxylin. The score of the IHC signals was judged by two independent pathologists blindly.
Purification of GST- and His-tagged proteins from bacteria.
Recombinant GST-conjugated KDM4B catalytic domain and His-conjugated GSK3β were generated by transforming the BL21 (DE3) E. coli strain with pGEX-KDM4B-Cata and pET-28a-GSK3β, respectively. Starter cultures (5 ml) grown overnight at 37°C were inoculated (1%) into larger volumes (500 ml). Cultures were grown at 37°C until an O.D. of 0.8, following which protein expression was induced for 12–16 hrs using 0.1 mM IPTG at 16°C with vigorous shaking. Recombinant proteins were purified from harvested pellets. Pellets were re-suspended in 5 ml EBC buffer and sonicated (5 cycles of 10 sec each at 50% output). Insoluble proteins and cell debris were discarded following centrifugation in a table-top centrifuge (13000 rpm/4°C/15 min). Each 1 ml supernatant was incubated with 50 µl of 50% Glutathione-sepharose slurry (Pierce) or Ni-beads (Qiagen) for 3 hrs at 4°C. The Glutathione beads were washed 3 times with PBS buffer (1 ml per wash) and stored at 4°C in PBS buffer containing 10% glycerol or eluted by elution buffer. The Ni-beads were washed 3 times with 50 mM Tris-Cl (pH 8.0) containing 20 mM Imidazole, and eluted by Tris-buffers containing 100 mM Imidazole. Recovery and yield of the desired proteins (or complexes) was confirmed by analyzing 10 µl of beads by Coomassie blue staining, and quantified against BSA standards.
Purification of His-Akt1 protein from insect S9 cells.
Akt1 cDNA was cloned into an insect cell expression vector, pFastBac-HT A (Invitrogen), in frame with a Histidine (His) tag at the N-terminus. Constructs were transformed into DH10Bac bacteria (Invitrogen) to generate bacmid DNAs. Akt baculoviruses were produced in Sf9 insect cells according to the manufacturer’s specification. His-tagged proteins were purified on HisPur Cobalt resin (Thermo Scientific) and eluted by Imidazole (Sigma). In brief, Sf9 insect cells expressing His-tagged Akt proteins were collected and washed once with ice-cold Phosphate Buffered Saline (PBS). Cell pellet was then re-suspended for 30 minutes at 4°C in lysis buffer containing 10 mM HEPES pH 7.6, 3 mM MgCl2, 300 mM KCl, 5% glycerol, 0.5% NP-40, 10 mM Imidazole, 1 mM Na2VO4, 20 mM NaF, 1 mM sodium pyrophosphate, 25 mM β-glycerophosphate and complete EDTA-free protease inhibitors (Roche). The cell lysate was cleared by centrifugation for 20 min at 21,000× g and 4 °C. While centrifuging the supernatant a second time in the same condition, the HisPur Cobalt resin (Thermo Scientific) was equilibrated in the lysis buffer. The cell lysate was incubated with the resin for at least one hour and afterward extensively washed with the lysis buffer. His-tagged Akt proteins were then eluted from the resin in several fractions using a buffer containing 10 mM HEPES (pH 7.6), 3 mM MgCl2, 300 mM KCl, 10% glycerol and 250 mM imidazole.
Peptide synthesis and dot immunoblot assays.
N-terminal biotinylated peptides used for dot blot assays were synthesized at Tufts Medical School in 0.1mmol scale. The sequences were listed below:
Akt1-K140-WT (aa131–151): AEEMEVSLAKPKHRVTMNEFE;
Akt1-K140-me3: AEEMEVSLA*KPKHRVTMNEFE;
Akt1-K140/142-me3: AEEMEVSLA*KP*KHRVTMNEFE (*Denoted tri-methylation).
Peptides were diluted into 1 μM for further biochemical assays. For dot blot assays, peptides were spotted onto nitrocellulose membrane allowing solution to penetrate (usually 3–4 mm diameter) by applying it slowly as a volume of 1 μl. The membrane was dried, and blocked in non-specific sites by soaking in TBST buffer with 5% non-fat milk for immunoblot analysis as described previously 17.
Akt in vitro kinase assays.
Akt in vitro kinase assays were adapted from a protocol described previously 16. Briefly, 1 μg of the bacterially purified His-GSK3β fusion proteins were incubated with immunoprecipitated Akt1 from HEK293 cells transfected with various mutant Akt1 in the presence of 100 ng [γS]ATP (abcam 138911) in the kinase reaction buffer (50 mM Tris pH 7.5, 1 μM MnCl2, 2 mM DTT) for 30 minutes at 30°C. The reaction was subsequently stopped by adding in 0.1 mM EDTA, and further react for another 1 hour by adding in the pNitrobenzyl mesylate (abam 138910) to alkylate the thiophosphorylation site on the substrates. The reaction was stopped by adding 3 x SDS loading buffer and resolved by SDS-PAGE. Phosphorylation of His-GSK3β was detected by specific antibody to recognize thiophosphate ester as previously reported 57.
In vitro methylation and demethylation assays.
The in vitro methylation assays were performed as described 58. In briefly, Flag-tagged SETDB1 were expressed and purified from HEK293T cells serving as the source of methyltransferase. The Akt1 proteins including GST-tagged Akt1 expressed and purified from HEK293T cells, recombinant his-Akt1 obtained from Thermofisher Scientific, or the Akt1 peptides synthesis from Tufts were used as substrates. The reactions were performed in the condition of 50 mM Tris-HCl (pH 8.5), 5 mM MgCl2, 4 mM DTT and 10 μM S-adenosyl-methionine (SAM) at 37°C for 6 hrs.
Demethylation assays were performed as reported previously 59. In briefly, the bacterially purified KDM4B catalytic domain (1–400aa) were used as the source of demethylase, and the synthetic Akt1-K140-me3 peptides were used as substrates. The reactions were performed in the condition of 50 mM Tris-HCl (pH 8.2), 2% glycerol, 0.1% MgCl2, 0.1% KCl and 0.1 mM PMSF at 37°C for 12 hrs.
In vivo ubiquitination assays.
Akt ubiquitination assays were performed as reported previously 41. Briefly, His-ubiquitin along with different variants of Akt1 were transfected into cells, and resulting cells were lysed in buffer A (6 M guanidine-HCl, 0.1 M Na2HPO4/NaH2PO4, and 10 mM imidazole [pH 8.0]) and subjected to sonication. After high-speed centrifuged, the supernatants were incubated with nickel-beads (Ni-NTA) (QIAGEN) for 3 hrs at room temperature. The products were washed twice with buffer A, twice with buffer A/TI (1 volume buffer A and 3 volumes buffer TI), and one time with buffer TI (25 mM Tris-HCl and 20 mM imidazole [pH 6. 8]). The pull-down proteins were resolved in 8% SDS-PAGE for immunoblot analysis.
Mass spectrometry analyses.
For mass spectrometry analysis, anti-HA immunoprecipitations (IP) were performed with the whole cell lysates derived from three 10 cm dishes of HEK293 cells co-transfected with Flag-SETDB1 and HA-Akt1. The IP proteins were resolved by SDS-PAGE, and identified by Coomassie staining. The band containing Akt1 was reduced with 10 mM DTT for 30 minutes, alkylated with 55 mM iodoacetamide for 45 minutes, and in-gel-digested with trypsin enzymes. The resulting peptides were extracted from the gel and analyzed by microcapillary reversed-phase (C18) liquid chromatography-tandem mass spectrometry (LC-MS/MS), using a high resolution QExactive HF Orbitrap (Thermo Fisher Scientific) in positive ion DDA mode (Top 8) via higher energy collisional dissociation (HCD) coupled to a Proxeon EASY-nLc II nano-HPLC 60. MS/MS data were searched against the Uniprot Human protein database (version 20151209 containing 21,024 entries) using Mascot 2.5.1 (Matrix Science) and data analysis was performed using the Scaffold 4.4.8 software (Proteome Software). Peptides and modified peptides were accepted if they passed a 1% FDR threshold.
Colony formation assays.
Cells were seeded into 6-well plates (300 or 600 cells/well) and left for 8–12 days until formation of visible colonies. Colonies were washed with PBS and fixed with 10% acetic acid/10% methanol for 20 minutes, then stained with 0.4% crystal violet in 20% ethanol for 20 minutes. After staining, the plates were washed and air-dried, and colony numbers were counted.
Soft agar assays.
The anchorage-independent cell growth assays were performed as described previously 16. Briefly, the assays were preformed using 6-well plates where the solid medium consists of two layers. The bottom layer contains 0.8% noble agar and the top layer contains 0.4% agar suspended with 1 × 104 or 3 ×104 cells. 500 μl complete DMEM medium was added every 7 days to keep the top layer moisture and 4 weeks later the cells were stained with iodonitrotetrazolium chloride (1 mg/ml) (sigma I10406) for colony visualization and counting.
Glucose uptake and lactate production assays.
For glucose uptake assays, cells were cultured in 60 cm plates, and were starved for 24 hrs with serum-free and glucose-free DMEM, then subjected to 20 μM 2-NBDG (Sigma 72987) containing glucose-free DMEM for different time points. Last, the cellular glucose uptake was quantified by FACS analysis using BD FACSDiva 6.1.3 software.
For lactate production assays, cells were planted into 60 cm plates, and were cultured for 24 hrs, then the cultured medium was collected, and lactate concentration was measured with lactate test strips and Accutrend Lactate analyzer (Accutrend Lactate, Nova Biomedical). At the same time, the viable cell numbers were calculated under the microscope. Finally, the relative lactate production was calculated with the formula (relative lactate production = lactate concentration/cell numbers) and normalized with the ratio of control cells.
Mouse xenograft assays.
Mouse xenograft assays were performed as described previously 4. Briefly, 2×106 DLD1-AKT1/2−/− cells stably expressing WT or K140/142R mutant form of Akt1, or 1×106 A375 cells knocked down SETDB1 were injected into the flank of 8 female nude mice (NCRNU-M-M from Taconic, 4–5 weeks of age). Tumor size was measured every three days with a caliper, and the tumor volume was determined with the formula: LxW2×0.5, where L is the longest diameter and W is the shortest diameter. In the drug treatment group, when the tumor reached 100 mm3, the mice were i.p. injected with Mithramycin A (0.2 mg/kg) or PBS (as a negative control) in the other day. After around 18 days, mice were sacrificed and xenografted solid tumors were dissected, then tumor weights were measured and recorded post-necropsy.
Statistics and reproducibility
Statistical analysis (two-tailed Student’s t test) was performed using Prism 8 (GraphPad Software) for Figs. 1h; 2b, 2c and 7n and Supplementary Fig. 7i in this manuscript. To calculate the P value between groups in Figs. 1f, 1j; 2h, 2l; 7b, 7l and Supplementary Figs. 7g, two-way analysis of variance (ANOVA) analysis was performed with Prism 8. Chi-square test was also performed to analyze the P value for Figs. 3j, 3l, 3m and Figs. 6m, 6n, 6o by using Prism 8. All the P values calculated above are indicated in the corresponding figures, and derived from three independent experiments with 3 technical replicates for each experiment. MS screens were performed twice successfully. All the western blots were performed twice independently with similar results. In vitro methylation assays were performed once in Fig. 3e, and performed twice in Fig. 3g and supplementary Fig. 3h independently with similar results. In vitro demethylation , ubiquitination, soft agar, colony formation, glucose uptake and lactate production assays were performed twice independently with similar results. To knock out or knock down genes of interest, at least two independent sgRNAs or shRNAs were employed to generate cell lines, and similar results were found in all cell lines.
Reporting Summary
Further information on experimental design is available in the Nature Research Reporting Summary linked to this article.
Data availability
The mass spectrometry-based screening data generated in this study have been deposited in ProteomeXchange under the accession code PXD011657. The SETDB1, EZH2, PTEN, EGFR, PIK3CA and AKT1 genetic alterations in TCGA datasets was integrated from cBioPortal database (www.cbioportal.org), with the query of gene “ESET”, “EZH2”, “PTEN”, “EGFR”, “PIK3CA” and “AKT1” for both mutation and copy number alterations (CNA) in different cancer types, such as BRCA and Melanoma. Following are the information of each cancer studies. One dataset for BRCA: TCGA, provisional. One dataset for Melanoma: TCGA, provisional.
Source data for Fig. 1e-f, 1h, 1j-m; 2b-c, 2g-h; 3n, 3p-q; 4c, 4f, 4h, 4j, 4l, 4n; 6m-o; 7b, 7d, 7j, 7l, 7n and Supplementary Fig. 1l-m; 2e-f; 3o; 4c-f, 4h; 7e, 7g, 7i have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.
Supplementary Material
ACKNOWLEDGMENTS
We thank B. North, J. Zhang, F. Dang and other Wei lab members for critical reading of the manuscript, as well as members of Dr. Pandofi and Toker laboratory for helpful discussions. We thank Dr. Hitoshi Okada (Kindai University of Medicine) for the generation of Kdm4bf/f MEFs. W.G. is supported by K99CA207867 from National Cancer Institute. W.W. is an LLS research scholar. This work was supported in part by the NIH grants (WW. and A.T., CA177910). The MS work was partially supported by NIH grants P01CA120964 (J.A.) and P30CA006516 (J.A.)
Footnotes
FINANCIAL AND NON-FINANCIAL COMPETING INTERESTS
We have no financial and non-financial competing interests.
REFERENCES
- 1.Jones PA, Issa JP & Baylin S Targeting the cancer epigenome for therapy. Nat Rev Genet 17, 630–641 (2016). [DOI] [PubMed] [Google Scholar]
- 2.You JS & Jones PA Cancer genetics and epigenetics: two sides of the same coin? Cancer Cell 22, 9–20 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Glaser KB HDAC inhibitors: clinical update and mechanism-based potential. Biochem Pharmacol 74, 659–671 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Fahy J, Jeltsch A & Arimondo PB DNA methyltransferase inhibitors in cancer: a chemical and therapeutic patent overview and selected clinical studies. Expert Opin Ther Pat 22, 1427–1442 (2012). [DOI] [PubMed] [Google Scholar]
- 5.Delmore JE et al. BET bromodomain inhibition as a therapeutic strategy to target c-Myc. Cell 146, 904–917 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kelly TK, De Carvalho DD & Jones PA Epigenetic modifications as therapeutic targets. Nat Biotechnol 28, 1069–1078 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Daigle SR et al. Potent inhibition of DOT1L as treatment of MLL-fusion leukemia. Blood 122, 1017–1025 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Biggar KK & Li SS Non-histone protein methylation as a regulator of cellular signalling and function. Nat Rev Mol Cell Biol 16, 5–17 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Chuikov S et al. Regulation of p53 activity through lysine methylation. Nature 432, 353–360 (2004). [DOI] [PubMed] [Google Scholar]
- 10.Guo A et al. Immunoaffinity enrichment and mass spectrometry analysis of protein methylation. Mol Cell Proteomics 13, 372–387 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saddic LA et al. Methylation of the retinoblastoma tumor suppressor by SMYD2. J Biol Chem 285, 37733–37740 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vanhaesebroeck B & Alessi DR The PI3K-PDK1 connection: more than just a road to PKB. Biochem J 346 Pt 3, 561–576 (2000). [PMC free article] [PubMed] [Google Scholar]
- 13.Manning BD & Toker A AKT/PKB Signaling: Navigating the Network. Cell 169, 381–405 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ozes ON et al. NF-kappaB activation by tumour necrosis factor requires the Akt serine-threonine kinase. Nature 401, 82–85 (1999). [DOI] [PubMed] [Google Scholar]
- 15.Yang WL et al. The E3 ligase TRAF6 regulates Akt ubiquitination and activation. Science 325, 1134–1138 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu P et al. Cell-cycle-regulated activation of Akt kinase by phosphorylation at its carboxyl terminus. Nature 508, 541–545 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo J et al. pVHL suppresses kinase activity of Akt in a proline-hydroxylation-dependent manner. Science 353, 929–932 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazur PK et al. SMYD3 links lysine methylation of MAP3K2 to Ras-driven cancer. Nature 510, 283–287 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoshioka Y et al. SMYD3-mediated lysine methylation in the PH domain is critical for activation of AKT1. Oncotarget 7, 75023–75037 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cho H, Thorvaldsen JL, Chu Q, Feng F & Birnbaum MJ Akt1/PKBalpha is required for normal growth but dispensable for maintenance of glucose homeostasis in mice. J Biol Chem 276, 38349–38352 (2001). [DOI] [PubMed] [Google Scholar]
- 22.Nasti TH et al. A murine model for the development of melanocytic nevi and their progression to melanoma. Mol Carcinog 55, 646–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gao H et al. Akt/PKB interacts with the histone H3 methyltransferase SETDB1 and coordinates to silence gene expression. Mol Cell Biochem 305, 35–44 (2007). [DOI] [PubMed] [Google Scholar]
- 24.Liu T et al. Histone methyltransferase SETDB1 maintains survival of mouse spermatogonial stem/progenitor cells via PTEN/AKT/FOXO1 pathway. Biochim Biophys Acta 1860, 1094–1102 (2017). [DOI] [PubMed] [Google Scholar]
- 25.Cha TL et al. Akt-mediated phosphorylation of EZH2 suppresses methylation of lysine 27 in histone H3. Science 310, 306–310 (2005). [DOI] [PubMed] [Google Scholar]
- 26.Cerami E et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2, 401–404 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schultz DC, Ayyanathan K, Negorev D, Maul GG & Rauscher FJ 3rd SETDB1: a novel KAP-1-associated histone H3, lysine 9-specific methyltransferase that contributes to HP1-mediated silencing of euchromatic genes by KRAB zinc-finger proteins. Genes Dev 16, 919–932 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ceol CJ et al. The histone methyltransferase SETDB1 is recurrently amplified in melanoma and accelerates its onset. Nature 471, 513–517 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Macgregor S et al. Genome-wide association study identifies a new melanoma susceptibility locus at 1q21.3. Nat Genet 43, 1114–1118 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fei Q et al. Histone methyltransferase SETDB1 regulates liver cancer cell growth through methylation of p53. Nat Commun 6, 8651 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wong CM et al. Up-regulation of histone methyltransferase SETDB1 by multiple mechanisms in hepatocellular carcinoma promotes cancer metastasis. Hepatology 63, 474–487 (2016). [DOI] [PubMed] [Google Scholar]
- 32.Carpten JD et al. A transforming mutation in the pleckstrin homology domain of AKT1 in cancer. Nature 448, 439–444 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Cao J et al. MC1R is a potent regulator of PTEN after UV exposure in melanocytes. Mol Cell 51, 409–422 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dankort D et al. Braf(V600E) cooperates with Pten loss to induce metastatic melanoma. Nat Genet 41, 544–552 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Garraway LA et al. Integrative genomic analyses identify MITF as a lineage survival oncogene amplified in malignant melanoma. Nature 436, 117–122 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Cantley LC The phosphoinositide 3-kinase pathway. Science 296, 1655–1657 (2002). [DOI] [PubMed] [Google Scholar]
- 37.Alessi DR et al. Characterization of a 3-phosphoinositide-dependent protein kinase which phosphorylates and activates protein kinase Balpha. Curr Biol 7, 261–269 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Stephens L et al. Protein kinase B kinases that mediate phosphatidylinositol 3,4,5-trisphosphate-dependent activation of protein kinase B. Science 279, 710–714 (1998). [DOI] [PubMed] [Google Scholar]
- 39.Franke TF, Kaplan DR, Cantley LC & Toker A Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate. Science 275, 665–668 (1997). [DOI] [PubMed] [Google Scholar]
- 40.Chan CH et al. The Skp2-SCF E3 ligase regulates Akt ubiquitination, glycolysis, herceptin sensitivity, and tumorigenesis. Cell 149, 1098–1111 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cederquist CT et al. Systemic insulin sensitivity is regulated by GPS2 inhibition of AKT ubiquitination and activation in adipose tissue. Mol Metab 6, 125–137 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Whetstine JR et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell 125, 467–481 (2006). [DOI] [PubMed] [Google Scholar]
- 43.Luo J, Manning BD & Cantley LC Targeting the PI3K-Akt pathway in human cancer: rationale and promise. Cancer Cell 4, 257–262 (2003). [DOI] [PubMed] [Google Scholar]
- 44.Hennessy BT, Smith DL, Ram PT, Lu Y & Mills GB Exploiting the PI3K/AKT pathway for cancer drug discovery. Nat Rev Drug Discov 4, 988–1004 (2005). [DOI] [PubMed] [Google Scholar]
- 45.Ryu H et al. ESET/SETDB1 gene expression and histone H3 (K9) trimethylation in Huntington’s disease. Proc Natl Acad Sci U S A 103, 19176–19181 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chen WS et al. Growth retardation and increased apoptosis in mice with homozygous disruption of the Akt1 gene. Genes Dev 15, 2203–2208 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.DuPage M, Dooley AL & Jacks T Conditional mouse lung cancer models using adenoviral or lentiviral delivery of Cre recombinase. Nat Protoc 4, 1064–1072 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Downward J Mechanisms and consequences of activation of protein kinase B/Akt. Curr Opin Cell Biol 10, 262–267 (1998). [DOI] [PubMed] [Google Scholar]
- 49.Wang GH et al. SETDB1-mediated methylation of Akt promotes its K63-linked ubiquitination and activation leading to tumorigenesis. Nat Cell Biol 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Liu S et al. Setdb1 is required for germline development and silencing of H3K9me3-marked endogenous retroviruses in primordial germ cells. Genes & development 28, 2041–2055 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Li H et al. The histone methyltransferase SETDB1 and the DNA methyltransferase DNMT3A interact directly and localize to promoters silenced in cancer cells. J Biol Chem 281, 19489–19500 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Sun QY et al. SETDB1 accelerates tumourigenesis by regulating the WNT signalling pathway. J Pathol 235, 559–570 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Ding X et al. Epigenetic activation of AP1 promotes squamous cell carcinoma metastasis. Sci Signal 6, ra28 21–13, S20–15 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kawazu M et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One 6, e17830 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cong L et al. Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Nasti TH et al. A murine model for the development of melanocytic nevi and their progression to melanoma. Mol Carcinog 55, 646–658 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Allen JJ et al. A semisynthetic epitope for kinase substrates. Nat Methods 4, 511–516 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Shi X et al. Modulation of p53 function by SET8-mediated methylation at lysine 382. Mol Cell 27, 636–646 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Huang J et al. p53 is regulated by the lysine demethylase LSD1. Nature 449, 105–108 (2007). [DOI] [PubMed] [Google Scholar]
- 60.Breitkopf SB, Yuan M, Helenius KP, Lyssiotis CA & Asara JM Triomics Analysis of Imatinib-Treated Myeloma Cells Connects Kinase Inhibition to RNA Processing and Decreased Lipid Biosynthesis. Anal Chem 87, 10995–11006 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The mass spectrometry-based screening data generated in this study have been deposited in ProteomeXchange under the accession code PXD011657. The SETDB1, EZH2, PTEN, EGFR, PIK3CA and AKT1 genetic alterations in TCGA datasets was integrated from cBioPortal database (www.cbioportal.org), with the query of gene “ESET”, “EZH2”, “PTEN”, “EGFR”, “PIK3CA” and “AKT1” for both mutation and copy number alterations (CNA) in different cancer types, such as BRCA and Melanoma. Following are the information of each cancer studies. One dataset for BRCA: TCGA, provisional. One dataset for Melanoma: TCGA, provisional.
Source data for Fig. 1e-f, 1h, 1j-m; 2b-c, 2g-h; 3n, 3p-q; 4c, 4f, 4h, 4j, 4l, 4n; 6m-o; 7b, 7d, 7j, 7l, 7n and Supplementary Fig. 1l-m; 2e-f; 3o; 4c-f, 4h; 7e, 7g, 7i have been provided as Supplementary Table 2. All other data supporting the findings of this study are available from the corresponding author upon reasonable request.