Abstract
Radiomics is a medical imaging analysis approach based on computer-vision. Metabolic radiomics in particular analyses the spatial distribution patterns of molecular metabolism on PET images. Measuring intratumoral heterogeneity via image is one of the main targets of radiomics research, and it aims to build a image-based model for better patient management. The workflow of radiomics using texture analysis follows these steps: 1) imaging (image acquisition and reconstruction); 2) preprocessing (segmentation & quantization); 3) quantification (texture matrix design & texture feature extraction); and 4) analysis (statistics and/or machine learning). The parameters or conditions at each of these steps are effect on the results. In statistical testing or modeling, problems such as multiple comparisons, dependence on other variables, and high dimensionality of small sample size data should be considered. Standardization of methodology and harmonization of image quality are one of the most important challenges with radiomics methodology. Even though there are current issues in radiomics methodology, it is expected that radiomics will be clinically useful in personalized medicine for oncology.
Keywords: Radiomics, Texture analysis, Intratumoral heterogeneity, FDG PET/CT, Oncology
Introduction
Tumors become heterogenous at both genetic and phenotypic levels during progression via branched evolution, which is called intratumoral heterogeneity (ITH) [1]. ITH is clinically important because it is one of the main causes of resistance to treatment due to bottle neck effect [2, 3]. Further, ITH can cause underestimation of mutational burden of tumors due to analyzing only small specimens containing limited amount of tumor cells [4]. Despite larger and multiple biopsies are needed to evaluate mutational characterization accurately, tumor sampling procedure is invasive and is limited to practice enough [5]. Therefore, the role of imaging is expected in personalized medicine, which can perform total region analysis and can be repeatedly performed by less invasive.
18F-Fluorodeoxyglucose (18F-FDG) positron emission tomography/computed tomography (PET/CT) imaging is successfully used in clinical practice, especially for oncology [6, 7]. Visual assessment of 18F-FDG PET/CT imaging by image specialists such as radiologists and nuclear medicine physicians can make diagnosis of tumor, staging, and response assessment after chemo- or radiotherapy [8]. The subjectivity of visual assessment causes large intra- and inter-observer variations of interpretation in medical imaging [9]. Quantification of imaging is increasingly highlighted to achieve objective interpretation [10]. Many studies have suggested clinical usefulness of standardized uptake value (SUV)-based PET parameters such as SUVmax, SUVpeak, and SUVavg as well as metabolic tumor volume (MTV) and total lesion glycolysis (TLG) [11–17]. However, those parameters do not reflect information of metabolic ITH which can be estimated via evaluation of spatial distribution of metabolic activity in a tumor.
One of the most highlighted methods to quantify ITH from images is texture analysis. Different mathematical methods including texture analysis has been developed for pattern recognition of images since the early 1970s [18]. Rapid development of image analysis based on high-throughput computing enables to extract many kinds of quantitative phenotype features about shape, edge, and texture from digitalized medical images including ultrasonography, CT, magnetic resonance imaging, and PET/CT [19]. This field of medical study has been termed as ‘radiomics’ which aims maximizing the information extracted from medical images and building a model using machine learning to achieve better patient management [20]. For example, radiogenomics is a subfield of radiomics, which aims to predict genomic information of tumors using macroscopic phenotypes expressed on images, and is expected to do a complementary role in personalized medicine for oncology [20]. 18F-FDG PET/CT imaging shows spatial distribution of metabolic activity in a tumor, which is thought to be related to tumor phenotype and tumor microenvironment. Recently, many studies with 18F-FDG PET/CT imaging in oncology have focused on quantification of metabolic ITH and its clinical usefulness in prediction of treatment response or prognosis as well as diagnosis.
This review aims to help researchers who are interested in starting radiomics study to carry out high quality research. Researchers should understand details of radiomics methodology, because radiomics research is more methodologically complex than conventional PET/CT imaging research. This review provides an overview of workflow in radiomics study and critical insights into the emerging methodological issues which affect results of radiomics studies.
Analysis Pipeline
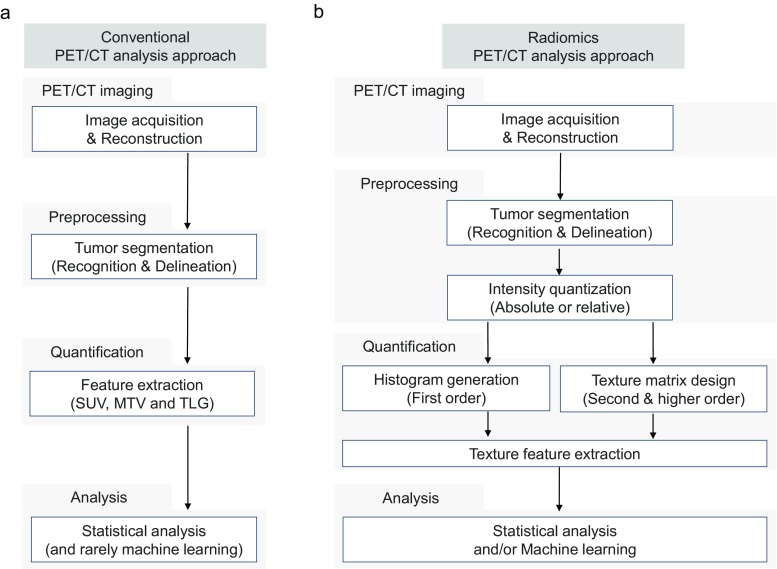
The conventional PET/CT analysis approach consists of the following: (1) PET/CT imaging: image acquisition and reconstruction; (2) preprocessing: tumor segmentation; (3) quantification: feature extraction (SUVmax, MTV, TLG, etc.); and (4) analysis: statistical analysis (Fig. 1a). The pipeline of radiomics analysis on 18F-FDG PET/CT is similar to the conventional PET/CT analysis but needs a few more steps such as intensity quantization during preprocessing and texture matrix design before feature extraction in quantification (Fig. 1b). Several user-friendly software packages provide functions for tumor segmentation and radiomics feature extraction [21–24]. Although it depends on the purpose of research, radiomics research uses machine learning in many cases while using statistical analysis. Given each step of radiomics analysis contains various options which affect results of radiomics features, it is essential to understand the method correctly and analyze them in a way that is appropriate for the purpose of the study.
Fig. 1.
The pipeline of radiomics analysis. The conventional PET/CT analysis approach is composed of imaging (acquisition and reconstruction), preprocessing (tumor segmentation), quantification (PET feature extraction: SUV, MTV and TLG) and statistical analysis (a). The pipeline of radiomics analysis is composed of imaging (acquisition and reconstruction), preprocessing (segmentation and quantization), quantification (histogram or texture matrix design and feature extraction), and analysis (statistics and/or machine learning) (b)
PET/CT Imaging
Influences of Image Acquisition
So far, most radiomics studies have been conducted in a single institution with retrospective manners. Each institution acquires PET/CT images using different machines with different imaging methods. Regardless of successive results for construction of prognosis-predicting model using texture features extracted from more than one PET/CT acquisition system [25, 26], texture features are known as being affected by settings of image acquisition and reconstruction methods. A study focusing on image acquisition setting of dual-time points, with peripheral nerve sheath tumors which had relatively low 18F-FDG uptake, showed that many texture features were changed significantly between early (101.5 ± 15.0 min) and late (251.7 ± 18.4 min) scans [27].
Several articles reported that respiratory motion affected values of the texture features significantly [28–31]. In addition, among different phase bins such as the end of inhalation, inhalation-to-exhalation, mid exhalation, end of exhalation, and exhalation-to-inhalation, the difference was low to moderate as coefficient of variation (CV) < 10% [28]. The influence of respiratory motion to the texture features of lung tumors was different according to the lesion location: e.g., minimal influence in upper lobe lesions and more significant influence in lower lobe lesions. Despite the significant difference of values in the texture features between whether respiratory gating or not, the impact on the prognostic values was not significant [30].
Influences of Image Reconstruction
The sensitivity to image reconstruction settings is feature-dependent [32–34]. Robust texture features in reconstruction settings can be considered as good parameters in radiomics analysis (Table 1) [34].Texture features were more affected by grid size than iteration number and full-width half-maximum filtration size [33]. While partial volume correction (PVC) had a smaller impact on texture features than delineation method, regional texture features were more affected by whether PVC or not than local texture features [35]. Area under curve of cumulative SUV-volume histogram (AUC-CSH) could contain more information after PVC [35, 36].
Table 1.
Robustness of texture features to reconstruction
| Classification | Texture matrix | CV ≤ 5% | 5% < CV ≤ 10% | 10% < CV ≤ 20% | CV > 20% |
|---|---|---|---|---|---|
| 1st order | Histogram | SUVavg/SULpeak/TLG/MTV/surface area/asphericity/entropy | SUVmax/SD/kurtosis | Skewness | Variance |
| 2nd order | GLCM | Entropy/homogeneity/dissimilarity | Energy/contrast/IDM/LNE | ||
| Higher order | NGTDM | Coarseness/contrast/strength | Busyness/complexity | ||
| GLRLM | SRE/LRE/RLNU/RP | HGRE/HGSRE | GLNUr/HGLRE | LGRE/LGSRE/LGLRE | |
| GLSZM | SZE/GLNUs/ZP | ZLNU/HGZE/HGSZE | LZE/LGZE/HGLZE | LGSZE/LGLZE |
Modified from Shiri (2017) [31] with formal permission
CV coefficient of variation, GLCM gray-level co-occurrence matrix, GLNUr gray-level non-uniformity for run, GLNUs gray-level non-uniformity for size, GLRLM gray-level run-length matrix, GLSZM gray-level size-zone matrix, HGLRE high gray-level long-run emphasis, HGLZE high gray-level long-zone emphasis, HGRE high gray-level run emphasis, HGSRE high gray-level short-run emphasis, HGSZE high gray-level short-zone emphasis, HGZE high gray-level zone emphasis, IDM intensity difference moment, IFH intensity frequency histogram, LGLRE low gray-level long-run emphasis, LGLZE low gray-level long-zone emphasis, LGRE low gray-level run emphasis, LGSRE low gray-level short-run emphasis, LGSZE low gray-level short-zone emphasis, LGZE low gray-level zone emphasis, LRE long run emphasis, LZE long-zone emphasis, MTV metabolic tumor volume, NGTDM neighborhood gray-tone difference matrix, RLNU run-length non-uniformity, RP run percentage, SD standard deviation, SRE short-run emphasis, SUV standardized uptake value, SZE short-zone emphasis, TLG total lesion glycolysis, ZLNU zone-length non-uniformity, ZP zone percentage
Issues: the Need for Harmonization
Harmonization is crucial for multicenter trials or obtaining compatible research results because the image reconstruction conditions of different research institutes are different. PSF reconstruction with Gaussian filtering harmonizes texture features with those obtained ordered subset expectation maximization. Since loss of information occurs in the filter process, it should be considered that the unfiltered images can be more discriminative in radiomics analysis [37]. A recent study demonstrated that using the ComBat method, initially developed for solving the problem of batch effect in genomic analysis, successfully harmonized the data of different reconstructions settings by removing the estimated center effect without loss of the pathophysiological information [38].
Tumor Segmentation
Recognition and Delineation
The process of tumor segmentation can be categorized into two sequential phases; recognition and delineation [39]. In the recognition phase, a target lesion with high uptake is identified and distinguished from other target lesion-like entities in the PET image. In the delineation phase, the target lesion is precisely separated from the background and non-significant object. The segmentation in the PET image is affected from intrinsic and extrinsic factors including spatial resolution and noise as well as shape, texture, and location of pathologies [40]. Low resolution and high smoothing make it difficult to define the precise segmentation of the tumor in PET images. Even though recent explosive use of hybrid imaging including PET/CT and PET/MR which improves tumor segmentation in the PET image mainly via improvement in the recognition phase, the challenging issues in the delineation phase are still unresolved.
Tumor Delineation Methods for PET Images
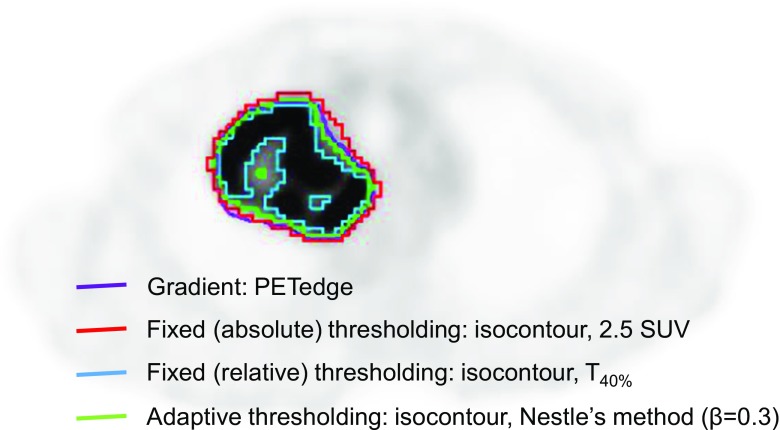
There are a wide variety of tumor delineation methods such as those which are manual, thresholding-based, stochastic and learning-based, and boundary based, which make different results [40]. So far, none have been agreed as the best tumor delineation methods for PET radiomics research. This is because, as discussed later, there is no consensus on whether to include the necrotic portion in PET radiomics studies. The manual delineation method is an intuitive and simple process, being the most common way to obtain surrogate truths if experts carry out the delineation process [41, 42]. However, it is a highly subjective and laborious method, related to high intra- and inter-operator variations as well as less reproducible products [43–47]. The thresholding methods, including fixed- and adaptive thresholding delineations, are rapid as well as simple and intuitive. A predefined cutoff value of SUV of 2.5 is one of the most commonly used fixed thresholding values for malignant tumor delineation [48–51]. Fixed relative thresholding allowing voxels with higher intensities than 40% of the SUVmax is another most commonly used fixed thresholding method in a clinical setting [48, 49, 51]. Fixed thresholding does not consider background intensities, which leads to inaccurate delineation for tumors with small volume and low metabolic activity. To correct the background-related errors in tumor delineation, it has been suggested to use various adaptive thresholding methods, which were based on phantom data. [48, 52–55]. The most commonly discussed adaptive thresholding method in the radiomics field was the Nestle’s method, determining threshold (T) by a function of tumor metabolic activity and background intensities [48, 51, 56, 57]
Regardless of simplicity and general applicability, the thresholding methods are known to be underestimating tumor volume as well as susceptible to contrast variation, noise, and heterogeneity [48, 58, 59]. Several algorithm-based delineation methods including fuzzy C-Means (FCM) [60] and fuzzy locally adaptive Bayesian (FLAB) [61] have been proposed for advanced automatic tumor delineation which showed better accuracy as well as less dependency on noise and image quality. The FLAB algorithm showed superior delineation results in small sized lesions less than 2 cm than the FCM algorithm [61]. Another advanced method is a gradient-based tumor delineation approach which showed more accurate and robust results than those of the thresholding methods [62, 63]. Meanwhile, CT-based anatomical tumor delineation showed significantly larger tumor volume than those of PET-based methods [59]. An example of different tumor delineation results is presented in Fig. 2.
Fig. 2.
Tumor segmentation using different delineation methods. Different results are presented by four different methods of tumor delineation, including gradient (PETedge, purple), fixed absolute thresholding with SUV of 2.5 (red), fixed relative thresholding with 40% of SUVmax (T40%, blue), and adaptive thresholding (the Nestle’s method, green) methods
Issues: Inclusion or Exclusion of Necrotic Portion
One specific issue regarding the tumor delineation process in radiomics research is about inclusion/exclusion of inner necrotic areas with low 18F-FDG uptake similar to background activity. For instance, contouring-based delineations such as the gradient-based methods include inner non-hypermetabolic areas [62, 64]. On the other hand, clustering-based methods exclude inner non-hypermetabolic areas [64, 65]. Thresholding-based methods exclude inner non-hypermetabolic areas while the threshold is sufficiently high. Given that more aggressive tumor makes more necrotic portions during proliferation, inclusion of necrotic portions would be helpful to assess aggressiveness of a tumor via texture analysis. On the other hand, those necrotic portions are no more metabolically active, and the approach focusing on hypermetabolic areas may be also reasonable.
Issues: Sensitive Results of Radiomics Features to Delineation Methods
Radiomics features are sensitive to different delineation methods [35]. Therefore, one tumor delineation method may not be sufficient for radiomics analysis. The recent trend of tumor segmentation in radiomics research is multiple-segmentation which uses various tumor delineation methods from manual delineation to various automatic delineations [66]. Different delineation methods can be used as a validation method as far as delineation related sensitivity issue of radiomics features [67]. To accomplish advanced level of radiomics analysis, it is recommended to applicate multiple segmentations and clarify how segmentation is performed.
Intensity Quantization
Relative and Absolute Quantization
Voxel intensity quantization, also known as SUV discretization or resampling, is the routine preprocessing step before second or higher order texture feature extraction in radiomics analysis. Quantization reduces the effect of noise in radiomics analysis, by changing the substantially continuous voxel intensity to a discontinuous value. In radiomics analysis using 18F-FDG PET/CT images, two different approaches have been suggested for SUV quantization, namely the relative quantization (RQ) known as the fixed bin number (FBN) method and the absolute quantization (AQ) also called as the fixed bin size (FBS) method.
The RQ method which resamples a voxel value, I(x), via discretization of the SUV range (SUVmax − SUVmin) inside the segmented tumor by a predetermined bin number (D; e.g., 16, 32, 64, 128, 256) to obtain a resampled voxel value (IRQ(x)) as the following formula [68]:
One of the most appropriate and widely used bin numbers for the RQ is 64, because intensity resolution would be sufficiently fine, less than 0.25 of SUV in a case with the SUV range of a tumor from 4 to 20 [68, 69]. Another study has suggested that 32 numbers of bins are appropriate because values of several texture features including entropy calculated in gray-level co-occurrence matrix (EntropyGLCM) are substantially affected by a number of bins, especially when the FBN is less than 32 [51].
Recently, the alternative quantization method, the AQ, has been suggested not to discard the absolute 18F-FDG uptake information [70, 71]. The AQ method, resampling I(x), intensities of voxel x, to IAQ(x) with a FBS (B; e.g., 0.1, 0.25, 0.5, or 1 of SUV), uses the following formula [71]:
Another option for the AQ process has been introduced using a high bound (HB; e.g., 15, 20, 25 SUV unit) with discrete values (D; e.g., 64) as [72]:
For example, when choosing a high bound of 15, 20, or 25 SUV unit with 64 discrete values with this formula, the intensity resolution is approximately 0.2, 0.3, or 0.4, respectively.
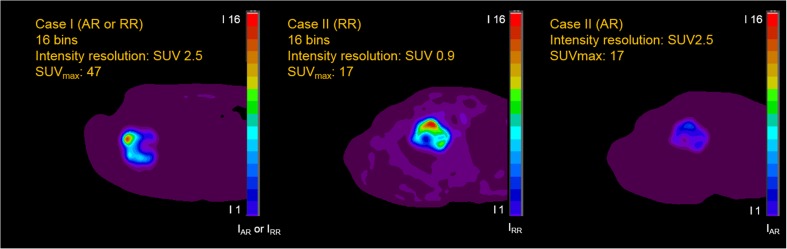
The RQ and AQ methods mostly result in discordant values of texture features to each other [71]. The RQ method treats PET images as dimensionless, which induces heterogenous intensity resolution dependent on the SUV range within tumors on different PET images. In these cases, the voxel–intensity differences within the tumor with relatively low SUVmax would be exaggerated compared to those in the tumor with higher SUVmax (Fig. 3). Therefore, values of texture features based on the RQ method may not be directly comparable with each other in different PET images. The AQ method induces identical intensity resolution among different PET images, which enables direct comparison of values of texture features in different PET images, even in a clinical response setting that the SUV range of a tumor varies during treatment [71]. While the values of texture features from the RQ method were counter-intuitive to the visual assessment of heterogeneity, those from the AQ method were more correlated in the simulated and clinical data of breast cancer patients [73]. For these reasons, the AQ method seems to be more appropriate than the RQ method for metabolic radiomics analysis using 18F-FDG PET/CT images. The AQ method also had better repeatability and lower sensitivity of texture features to the methods of delineation and reconstruction in a prospective study with double baseline 18F-FDG PET/CT scans with a 3-day interval [74].
Fig. 3.
Examples of resampled PET images using RQ and AQ methods. The 16-bin resampled voxel intensities within tumors are presented as different colors (see color bars). Even though the tumor in case II has relatively lower SUVs than the tumor in case I, the tomography of case II (RQ) is exaggerated and looks similar to that of case I (AQ or RQ). Applying the AQ method, the tumors in cases I and II show different tomographies. AQ absolute quantization, IAQ resampled intensity by the AQ method, IRQ resampled intensity by the RQ method, RQ relative quantization
Issues: Dependence on SUVmax and/or MTV
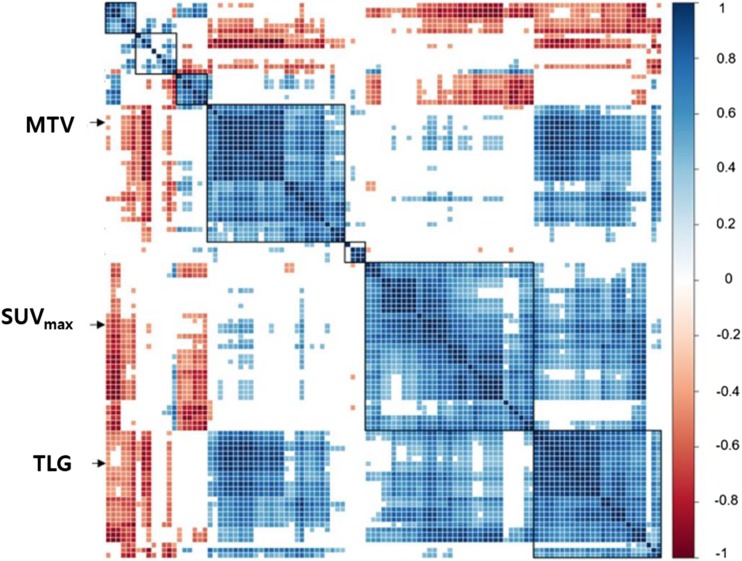
The influence of the quantization methods on values of texture features is highly related to the dependency to the conventional PET parameters, tumor SUVmax and/or MTV. Several studies commonly found a trend that most texture features based on the RQ method have the dependency of their values on MTV [64, 72, 75–77]. Because the dependence on MTV is much sensitive particularly for tumors with relatively small size, MTVs of 10 cm3, 45 cm3, or 60 cm3 have been suggested as cutoffs to find valuable complementary information to MTV in radiomics analysis using the RQ method [72, 75, 76]. Meanwhile, it has been highlighted that the dependence of AQ-based texture features on not only MTV but also SUVmax (Fig. 4) [50, 64, 72, 77]. As the higher mountain topography is more tough and complicated, the dependence on SUVmax in tumor is considered to be one of the unique characteristics of the texture features of PET images. Actually, the RQ method is regarded as a kind of intensity normalization to the maximum intensity within a tumor. Therefore, it seems artificial to normalize and eliminate the dependence on SUVmax by using the RQ method. It might be more effective to find additive meaning of texture features based on the AQ method in multivariable analysis.
Fig. 4.
Dependence on SUVmax and MTV of AQ-based texture features. The correlogram of 109 texture features shows dependence on conventional PET parameters, including SUVmax and MTV (p < 0.0005). AQ absolute quantization, MTV metabolic tumor volume, SUVmax maximum standardized uptake value, TLG total lesion glycolysis. Note: Color bar, Pearson correlation coefficient. Modified from Ha (2017) [47], which is licensed under a Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/)
Texture Matrix Design and Feature Extraction
The First Order: Histogram-Based
Texture features are categorized into first, second, and higher orders by the number of voxels involved in their texture matrix design [78]. The first-order texture features are mainly calculated from intensity frequency histogram which represents the frequency distribution of one-voxel intensity in a delineated tumor. Therefore, the first-order texture features describe global characters of a tumor on PET images. The first-order texture features include CV, skewness, kurtosis, entropyhisto, and energy as well as conventional PET parameters like the SUVmax, SUVavg, MTV, and TLG (Table 2). A commonly used heterogeneity-associated parameter, the CV which is defined as the standard deviation (SD) over SUVavg in a delineated tumor [79], is also the first-order texture features derived from intensity frequency histogram (IFH) (Fig. 5a, b). There is another option to calculate global heterogeneity using the CSH, also known as the intensity volume histogram (IVH), having a decline curve form (Fig. 5a, c) [80]. The CSH describes percent of total tumor volume above percent threshold of SUVmax, which threshold varies from 0 to 100%. Lower AUC-CSH indicates more heterogeneity in a tumor [36].
Table 2.
First-order texture features
| Feature | Definition | Description |
|---|---|---|
| SUVmax, SUVavg, MTV, TLG | – | Conventional PET parameters |
| CV | Standard deviation of SUVs/SUVavg | A global heterogeneity marker. Higher values indicate more heterogeneity |
| AUC-CSH | Percent of total tumor volume above percent threshold of SUVmax, which threshold varies from 0 to 100%. | A global heterogeneity marker. Lower values indicate more heterogeneity |
| Skewness | (with E = the total number of voxels in the VOI, = the average of gray levels in the histogram) | The asymmetry of the gray-level distribution in the intensity frequency histogram |
| Kurtosis | (with E = the total number of voxels in the VOI, = the average of gray levels in the histogram) | The sharpness of the peak of the gray-level distribution in the intensity frequency histogram |
| Entropyhisto | The randomness of the gray-level distribution in the intensity frequency histogram |
AUC-CSH, area under curve of cumulative SUV-volume histogram; CV, coefficient of variation; MTV, metabolic tumor volume; SUV, standardized uptake values; SUVavg, average of SUVs; SUVmax; maximum of SUVs; TLG, total legion glycolysis; VOI, volume of interest
Fig. 5.
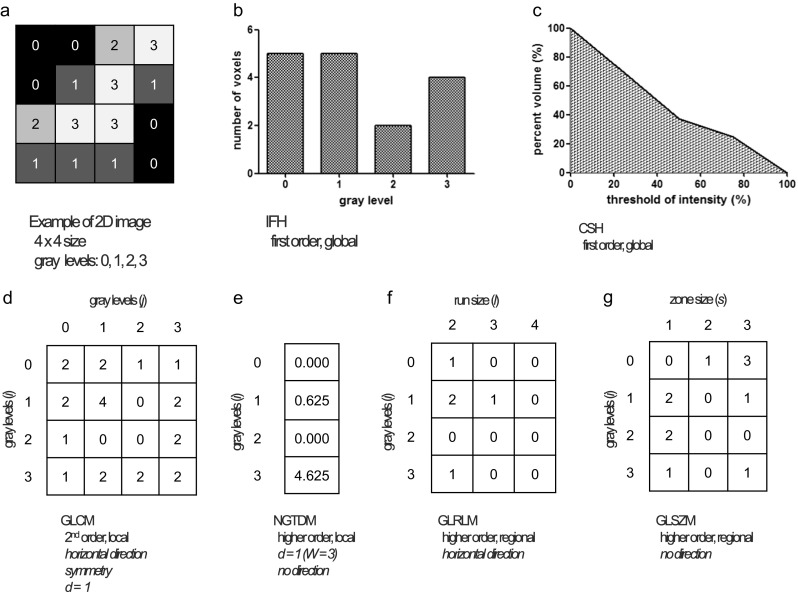
2D example of texture matrix design. Using a 2D sample matrix with four bins of pixel intensity (4 × 4 size (a)), various texture matrices including IFH (b), CSH (c), GLCM (d), NGTDM (e), GLRLM (f), and GLSZM (g) were computed. CSH cumulative SUV-volume histogram, d distance, GLCM gray-level co-occurrence matrix, GLRLM gray-level run-length matrix, GLSZM gray-level size-zone matrix, IFH intensity frequency histogram, NGTDM neighborhood gray-tone difference matrix, W window size
The Second Order: GLCM-Based
The second order texture features are calculated based on GLCMs also known as spatial gray-level dependence matrices (SGLDMs) which compute for the second order joint probabilities, namely the local association of intensities of 2 voxels in a delineated tumor [78]. The information on GLCMs is about how many pairs of voxels of resampled intensities (i, j) which satisfy a certain relationship of a defined direction and distance (Fig.5a, d) [81]. Theoretically, multiple GLCMs could be computed for a single PET case with various distances and directions. The one-voxel distance has been usually chosen to compute GLCMs in PET radiomics analysis [68, 71]. Given 26 voxels surrounding a voxel in 3D space, 13 directions are usually used to compute GLCMs in 3D space. The difference of “symmetry” and “asymmetry” methods is whether the number of paired voxels including in the reverse direction is counted or not. Two methods have been suggested for GLCM-based 3D texture feature extraction; one is using 13 GLCMs for each direction in 3D space to calculate texture features independently and then average them; the other one is using 1 GLCM in consideration of neighbor voxels for all 13 directions directly in 3D space (Fig. 6) [64, 76]. In regard to the issue of volume-dependence of the RQ-based texture features, the single GLCM method showed less volume dependence than the 13 GLCMs with averaging method [64, 76]. The normalized GLCM can be computed as following formula [23]:
Fig. 6.
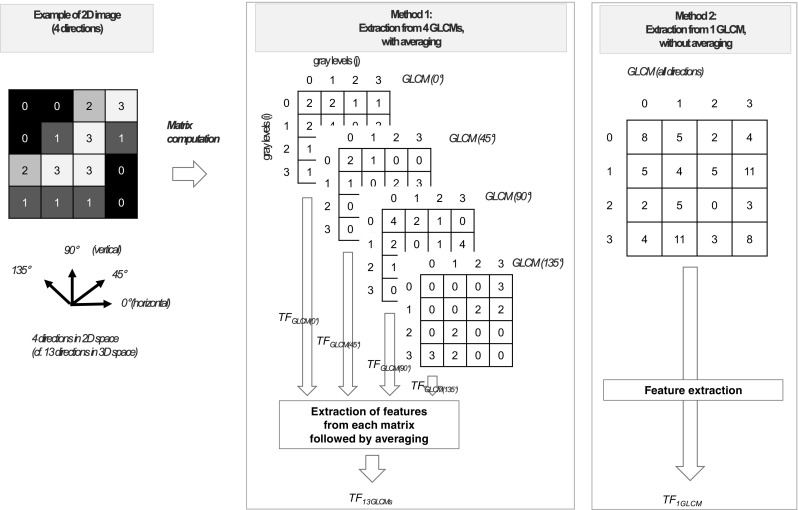
Approach to extract the multi-directional GLCM-based texture feature. Two ways extracting the GLCM-based texture feature in multi-directions are presented. The first method, with averaging, extracts a multi-directional texture feature (TF13GLCMs) by averaging every texture feature on each direction. The second method, without averaging, extracts another multi-directional texture feature (TF1GLCM) from a multi-directional GLCM. GLCM gray-level co-occurrence matrix, TF a texture feature
Table 3 summarizes GLCM-based texture features including homogeneity, energy, contrast, correlation, entropy, and dissimilarity.
Table 3.
GLCM-based features
| Name | Definition | Description |
|---|---|---|
| Normalized GLCM | Normalized |
2nd-order texture matrix |
| HomogeneityGLCM | The homogeneity of gray-level voxel pairs | |
| EnergyGLCM | The uniformity of gray-level voxel pairs | |
| ContrastGLCM | The local variations in the GLCM | |
| CorrelationGLCM | The linear dependency of gray levels in GLCM | |
| EntropyGLCM | The randomness of gray-level voxel pairs | |
| DissimilarityGLCM | The variation of gray-level voxel pairs |
GLCM gray-level co-occurrence matrix
The Higher Order: NGTDM-Based
The higher order texture features are calculated from several texture matrices computed based on interrelationships of 3 or more voxels [78]. Neighborhood gray-tone difference matrices (NGTDMs) are one of the higher order texture matrices, which compute for differences between each voxel and neighboring voxels within a certain distance (Fig. 5a, e). When a distance for NGTDMs are 1 voxel, a centered voxel and 8 surrounding voxels are constructing a 3 × 3 neighborhood group, namely 3 voxels as the window size. NGTDMs calculate sum of the absolute difference between centered voxel level and average of the neighboring voxels for g. The window size (W) of 3 voxels is mostly used [82, 83]. The NGTDM can be computed as follows [23]:
NGTDM-based texture features including coarseness, contrast, and busyness, which represent local texture within a delineated tumor, and the measures showed good correspondences with human’s visual perception [84]. Table 4 summarizes NGTDM-based features.
Table 4.
NGTDM-based features
| Name | Definition | Description |
|---|---|---|
| NGTDM | (with the average of the neighbor voxel intensities) | Higher order, local texture matrix |
| CoarsenessNGTDM | The level of spatial rate of change in intensity | |
| ContrastNGTDM | (with E = the number of voxels in VOI, G = the number of gray levels) | The intensity difference between neighboring regions of voxels |
| BusynessNGTDM | (with p(i) ≠ 0, p(j) ≠ 0) | The spatial frequency of changes in intensity |
NGTDM neighborhood gray-tone difference matrix
The Higher Order: GLRLM and GLSZM-Based
Gray-level run-length matrices (GLRLMs) are statistical higher order texture matrices and measure for runs (l) of voxels with same gray-level (i) along a given direction (Fig. 5a, f) [85]. The counting process of GLRLMs is symmetrical between a given direction and its reverse; only 13 directions in 3D space are needed to be considered, consequently [86]. Extracting GLRLM-based texture features in 3D space is similar to that of GLCM. One way to obtain a value of GLRLM-based texture feature in 3D images is that averaging the texture values separately calculated in each matrix in 13 directions [87]. The other way is that simultaneously considering 13 directions to compute 1 GLRLM [88]. Gray-level size zone matrices (GLSZMs) are alternative to GLRLMs, considering the size (s) of continued voxels with same gray level (i) rather than the run length of voxels with same gray level in a certain direction (Fig.5a, g) [89]. Therefore, the GLSZM has a variable number of columns determined by the size of the largest zone [90]. Computing GLSZMs does not need to consider directions like computing GLCMs or GLRLMs. Tables 5 and 6 summarize GLRLM and GLSZM-based features.
Table 5.
GLRLM-based features
| Name | Definition | Description |
|---|---|---|
| GLRLM | GLRLM(i, j), the number of runs with intensity i of homogenous runs of j voxels in some direction (where H corresponds to the number of homogenous runs) | Higher order, regional texture matrix |
| Short-run emphasis (SRE) | The distribution of short runs, which is larger for fine textures | |
| Long-run emphasis (LRE) | The distribution of long runs, which is larger for coarse textures | |
| Low gray-level run emphasis (LGRE) | The distribution of low gray-level runs | |
| High gray-level run emphasis (HGRE) | The distribution of high gray-level runs | |
| Short-run low gray-level emphasis (SRLGE) | The distribution of the short homogeneous runs with low gray levels | |
| Short-run high gray-level emphasis (SRHGE) | The distribution of the short homogeneous runs with high gray levels | |
| Long-run low gray-level emphasis (LRLGE) | The distribution of the long homogeneous runs with low gray levels | |
| Long-run high gray-level emphasis (LRHGE) | The distribution of The long homogeneous runs with high gray levels | |
| Gray-level non-uniformity for run (GLNUr) | The non-uniformity of the gray levels of the homogeneous runs | |
| Run length non-uniformity (RLNU) | The non-uniformity of the length of the homogeneous runs | |
| Run percentage (RP) | The homogeneity of the homogeneous runs |
GLRLM gray-level run-length matrix
Table 6.
GLSZM-based features
| Name | Definition | Description |
|---|---|---|
| GLSZM | GLSZM(i, j), the number of homogeneous zones of j voxels with the intensity i (where H corresponds to the number of homogenous zones) | Higher order, regional texture matrix |
| Short zone emphasis (SZE) | The distribution of the short homogeneous zones | |
| Long-zone emphasis (LZE) | The distribution of the long homogeneous zones | |
| Low gray-level zone emphasis (LGZE) | The distribution of the low gray-level zones | |
| High gray-level zone emphasis (HGZE) | The distribution of the high gray-level zones | |
| Short-zone low gray-level emphasis (SZLGE) | the distribution of the short homogeneous zones with low gray levels | |
| Short-zone high gray-level emphasis (SZHGE) | the distribution of the short homogeneous zones with high gray levels | |
| Long-zone low gray-level emphasis (LZLGE) | the distribution of the long homogeneous zones with low gray levels | |
| Long-zone high gray-level emphasis (LZHGE) | The distribution of the long homogeneous zones with high gray levels | |
| Gray-level non-uniformity for zone (GLNUz) | The non-uniformity of the gray levels of the homogeneous zones | |
| Zone length non-uniformity (ZLNU) | The non-uniformity of the length of the homogeneous zones | |
| Zone percentage (ZP) | The homogeneity of the homogeneous zones |
GLRLM gray-level run-length matrix
Issues: Reproducibility of Texture Features
Several studies have explored the issues of the repeatability of texture features using test–retest data sets using the same setting of image acquisition, reconstruction, segmentation, and quantization. A test–retest study, with the RQ preprocessing, proved that most GLCM texture features including entropy, homogeneity, and dissimilarity, and some GLSZM texture features including intensity variability and size zone variability had comparable reproducibility to SUVmax. However, several GLSZM-features including small area emphasis and low intensity emphasis had low reproducibility [91]. Another test–retest study, using the AQ preprocessing with 0.5 SUV intensity resolution, proved that the majority of texture features had a high reproducibility, although GLSZM texture features showed the overall lowest reproducibility compared with global and GLCM texture features [70]. A greater number of GLCM- and GLRLM texture features had better repeatability with the AQ preprocess compared with that of the RQ preprocess [74]. CV and AUC-CSH, which are ITH-related global texture features, had moderate-to-high reproducibility. Other global texture features, such as skewness and kurtosis, had various levels of reproducibility from low to high [70, 91, 92].
Analysis for Radiomics Data
Statistics
The traditional approach to the clinical image research is driven by medical statistics. This approach is still valuable while focusing on the clinical usefulness of ITH-related parameters from images. For this purpose, several ITH-related parameters carefully selected by researchers would be tested to identify certain clinical importance such as the role of prognostic markers for drug resistance [93, 94]. Multiple comparison problems should be considered when using multiple parameters such as radiomics features. To correct the multiple comparison problems, P values in statistics should be adjusted by post-hoc analysis for the multiple tested parameters [95]. The Bonferroni correction, having type I error rate α/m for each of m tests, is the most conservative and commonly used method to correct the multiple comparison issue. Sometimes, the Bonferroni’s correction is too conservative to find the clinical significance within given samples. Various methods have been suggested, such as less conservative post-hoc analyses, e.g., the Holm’s procedure and false-discovery rate, which use sequentially adjusted P values [96, 97]. In addition, multivariable analysis is recommended to evaluate additional usefulness and independent significance of texture features compared with SUVmax and MTV, on which the parameters have dependence, as well as already established clinical parameters.
Machine Learning
Machine learning methods used in radiomics researches are classified as supervised learning and unsupervised learning [98]. Supervised learning is used to solve problems of regression predicting continuous variables, and classification predicting categorical variables. Supervised learning trains a function fw based on a labeled training data set consisting of a paired input (x) and desired output (y). There are various types of supervised learning algorithms including support vector machines, linear regression, logistic regression, linear discriminant analysis, decision trees, k-nearest neighbor algorithms, and neural networks. The typical data structure of supervised learning is composed of three subsets: the training set fitting models, the validation set estimating prediction error for model selection, and the test set assessing the generalization error of the final chosen model [99]. The validation set is also regarded as a part of model training and a test set should be separated from training parts. Enough samples are needed for each set to get the best learning and testing results. If the data set is not enough to split into training and testing parts, the cross-validation method could be more efficient for use in order to validate the established model. In k-fold cross-validation, the data set D is randomly partitioned into k mutually exclusive subsets D1, D2, …, Dk of equal size. In each of learning experiments, one of those subsets is picked as a test set and the remaining subsets then as a training set. Learning experiments and testing performances are performed k separate times and the performance of k-fold cross-validation is obtained by averaging the test results from the k different learning experiments [100]. Unsupervised learning is used for unlabeled data to find hidden structure of data set. Typical algorithms include k means clustering, hierarchical clustering, etc. To perform clustering, it is important to determine how represent a cluster of more than one point and how calculate the distance between clusters. A cluster center called centroid is often used to represent each cluster. Most commonly used distance functions are Euclidean distance and Manhattan distance.
Issues: High Dimensionality in Machine Learning with Radiomics Data
One thing should be considered in radiomics research using machine learning is the high-dimensionality of radiomics feature space. Although hundreds of radiomics features could be extracted from cases, most radiomics studies usually have a restricted number of cases for analysis usually no more than 200. The small sample size but high dimensionality causes lower information density in radiomics research, which results in the higher risk of an overfitted model. This is related to one of well-known issues in the machine learning field, namely “The Curse of Dimensionality” [101]. As the number of features or dimensions grows, the amount of data needed to generalize accurately grows exponentially. As the dimensionality grows over the optimal number of features without increasing the number of training samples, the classifier’s performance decreases. To avoid the curse of dimensionality, training machine with carefully selected features is the most commonly used strategies in radiomics field [87, 102]. Further, highly correlated features provide redundant rather than complementary information; it can be used as a basis for selecting features in radiomics analysis [28, 103]. Non-linear classifiers are more vulnerable to overfitting due to high dimensionality than linear and simple classifiers [104]; therefore, non-linear classifiers are more restricted in feature number. Another approach like dimensionality reduction such as the principal component analysis, by combining the original feature values to have the largest variance, would help to manage the problems [105]. However, combined features with the largest variance do not guarantee having the most discriminative information.
Conclusions
Radiomics studies have been conducted in two perspectives: ITH assessment and high-throughput extraction of information. Radiomics feature extraction is less intuitive than conventional PET parameters and served as an entry barrier to this field of study. All the steps including PET/CT imaging, preprocessing, matrix design and feature extraction, and statistics/modeling affect the quality of radiomics analysis. Therefore, it is important to understand the issues of metabolic radiomics including tumor segmentation, quantization, robustness/repeatability, and dependence on SUVmax/MTV. Further, statistical validity should be ensured with cares in analyzing hundreds of parameters. Based on the successful validation of the clinical role of radiomics by individual studies, multicenter research or meta-analysis is expected to be actively pursued in a near future. In this regard, standardization of methodology and harmonization of radiomics results will become more important in later studies. Despite the pending issues of methodology, radiomics is expected to be clinically useful in precision medicine for oncology.
Abbreviations
- AQ
Absolute quantization
- AUC-CSH
Area under curve of cumulative SUV-volume histogram
- CSH
Cumulative SUV-volume histogram
- CV
Coefficient of variation
- FBN
Fixed bin number
- FBS
Fixed bin size
- FCM
Fuzzy C-means
- FDR
False-discovery rate
- 18F-FDG
18F-fluorodeoxyglucose
- FLAB
Fuzzy locally adaptive Bayesian
- GLCM
Gray-level co-occurrence matrix
- GLRLM
Gray-level run-length matrix
- GLSZM
Gray-level size zone matrix
- IFH
Intensity frequency histogram
- ITH
Intratumoral heterogeneity
- IVH
Intensity volume histogram
- MTV
Metabolic tumor volume
- NGTDM
Neighborhood gray-tone difference matrix
- PET/CT
Positron emission tomography/computed tomography
- PVC
Partial volume correction
- RQ
Relative quantization
- SAM
Second angular moment
- SGLDM
Spatial gray-level dependence matrix
- SUV
Standardized uptake value
- TLG
Total lesion glycolysis
Conflict of Interest
Seunggyun Ha, Hongyoon Choi, Jin Chul Paeng, and Gi Jeong Cheon declare no conflict of interest.
Ethical Statement
This work does not contain any studies with human participants or animals performed by any of the authors. For this type of study, formal consent is not required.
Informed Consent
Not applicable.
Footnotes
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
References
- 1.Shackney SE, Shankey TV. Genetic and phenotypic heterogeneity of human malignancies: finding order in chaos. Cytometry Part A. 1995;21(1):2–5. doi: 10.1002/cyto.990210103. [DOI] [PubMed] [Google Scholar]
- 2.Gerlinger M, Swanton C. How Darwinian models inform therapeutic failure initiated by clonal heterogeneity in cancer medicine. Br J Cancer. 2010;103(8):1139. doi: 10.1038/sj.bjc.6605912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sankin A, Hakimi AA, Mikkilineni N, Ostrovnaya I, Silk MT, Liang Y, et al. The impact of genetic heterogeneity on biomarker development in kidney cancer assessed by multiregional sampling. Cancer Med. 2014;3(6):1485–1492. doi: 10.1002/cam4.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim L, Tsao MS. Tumour tissue sampling for lung cancer management in the era of personalised therapy: what is good enough for molecular testing? Eur Respir J. 2014;44:erj01970–2013. doi: 10.1183/09031936.00197013. [DOI] [PubMed] [Google Scholar]
- 6.Fletcher JW, Djulbegovic B, Soares HP, Siegel BA, Lowe VJ, Lyman GH, et al. Recommendations on the use of 18F-FDG PET in oncology. J Nucl Med. 2008;49(3):480–508. doi: 10.2967/jnumed.107.047787. [DOI] [PubMed] [Google Scholar]
- 7.Hoppe RT, Advani RH, Ai WZ, Ambinder RF, Aoun P, Armand P, et al. NCCN guidelines insights: Hodgkin lymphoma, version 1.2018. J Natl Compr Cancer Netw: JNCCN. 2018;16(3):245–254. doi: 10.6004/jnccn.2018.0013. [DOI] [PubMed] [Google Scholar]
- 8.Gallamini A, Barrington SF, Biggi A, Chauvie S, Kostakoglu L, Gregianin M, et al. The predictive role of interim positron emission tomography for Hodgkin lymphoma treatment outcome is confirmed using the interpretation criteria of the Deauville five-point scale. Haematologica. 2014;99(6):1107–1113. doi: 10.3324/haematol.2013.103218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arimoto MK, Nakamoto Y, Higashi T, Ishimori T, Ishibashi M, Togashi K. Intra- and inter-observer agreement in the visual interpretation of interim 18F-FDG PET/CT in malignant lymphoma: influence of clinical information. Acta Radiol (Stockholm, Sweden : 1987) 2018;59:1218–1224. doi: 10.1177/0284185117751279. [DOI] [PubMed] [Google Scholar]
- 10.Duncan JS, Ayache N. Medical image analysis: Progress over two decades and the challenges ahead. IEEE Trans Pattern Anal Mach Intell. 2000;22(1):85–106. [Google Scholar]
- 11.Berghmans T, Dusart M, Paesmans M, Hossein-Foucher C, Buvat I, Castaigne C, et al. Primary tumor standardized uptake value (SUVmax) measured on fluorodeoxyglucose positron emission tomography (FDG-PET) is of prognostic value for survival in non-small cell lung cancer (NSCLC): a systematic review and meta-analysis (MA) by the European Lung Cancer Working Party for the IASLC Lung Cancer Staging Project. J Thorac Oncol. 2008;3(1):6–12. doi: 10.1097/JTO.0b013e31815e6d6b. [DOI] [PubMed] [Google Scholar]
- 12.Casasnovas R-O, Meignan M, Berriolo-Riedinger A, Bardet S, Julian A, Thieblemont C, et al. SUVmax reduction improves early prognosis value of interim positron emission tomography scans in diffuse large B-cell lymphoma. Blood. 2011;118:37–43. doi: 10.1182/blood-2010-12-327767. [DOI] [PubMed] [Google Scholar]
- 13.Sher A, Lacoeuille F, Fosse P, Vervueren L, Cahouet-Vannier A, Dabli D, et al. For avid glucose tumors, the SUV peak is the most reliable parameter for [18 F] FDG-PET/CT quantification, regardless of acquisition time. EJNMMI Res. 2016;6(1):21. doi: 10.1186/s13550-016-0177-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kajáry K, Tokés T, Dank M, Kulka J, Szakáll S, Jr, Lengyel Z. Correlation of the value of 18F-FDG uptake, described by SUVmax, SUVavg, metabolic tumour volume and total lesion glycolysis, to clinicopathological prognostic factors and biological subtypes in breast cancer. Nucl Med Commun. 2015;36(1):28–37. doi: 10.1097/MNM.0000000000000217. [DOI] [PubMed] [Google Scholar]
- 15.Costelloe CM, Macapinlac HA, Madewell JE, Fitzgerald NE, Mawlawi OR, Rohren EM, et al. 18F-FDG PET/CT as an indicator of progression-free and overall survival in osteosarcoma. J Nucl Med. 2009;50(3):340–347. doi: 10.2967/jnumed.108.058461. [DOI] [PubMed] [Google Scholar]
- 16.Choi ES, Ha SG, Kim HS, Ha JH, Paeng JC, Han I. Total lesion glycolysis by 18F-FDG PET/CT is a reliable predictor of prognosis in soft-tissue sarcoma. Eur J Nucl Med Mol Imaging. 2013;40(12):1836–1842. doi: 10.1007/s00259-013-2511-y. [DOI] [PubMed] [Google Scholar]
- 17.Moon SH, Hyun SH, Choi JY. Prognostic significance of volume-based PET parameters in cancer patients. Korean J Radiol. 2013;14(1):1–12. doi: 10.3348/kjr.2013.14.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haralick RM, Shanmugam K. Textural features for image classification. IEEE Trans Syst Man Cybern. 1973;3(6):610–621. [Google Scholar]
- 19.Gillies RJ, Kinahan PE, Hricak H. Radiomics: images are more than pictures, they are data. Radiology. 2015;278(2):563–577. doi: 10.1148/radiol.2015151169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lambin P, Rios-Velazquez E, Leijenaar R, Carvalho S, van Stiphout RG, Granton P, et al. Radiomics: extracting more information from medical images using advanced feature analysis. Eur J Cancer. 2012;48(4):441–446. doi: 10.1016/j.ejca.2011.11.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fang Y-HD, Lin C-Y, Shih M-J, Wang H-M, Ho T-Y, Liao C-T, et al. Development and evaluation of an open-source software package CGITA for quantifying tumor heterogeneity with molecular images. Biomed Res Int. 2014;2014:9. doi: 10.1155/2014/248505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L, Fried DV, Fave XJ, Hunter LA, Yang J, Court LE. IBEX: an open infrastructure software platform to facilitate collaborative work in radiomics. Med Phys. 2015;42(3):1341–1353. doi: 10.1118/1.4908210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nioche C, Orlhac F, Boughdad S, Reuze S, Goya-Outi J, Robert C, et al. LIFEx: a freeware for radiomic feature calculation in multimodality imaging to accelerate advances in the characterization of tumor heterogeneity. Cancer Res. 2018. 10.1158/0008-5472.can-18-0125. [DOI] [PubMed]
- 24.Cid YD, Castelli J, Schaer R, Scher N, Pomoni A, Prior JO, et al. QuantImage: an online tool for high-throughput 3D radiomics feature extraction in PET-CT. Biomedical texture analysis. Amsterdam: Elsevier; 2018. pp. 349–377. [Google Scholar]
- 25.Folkert MR, Setton J, Apte AP, Grkovski M, Young RJ, Schöder H, et al. Predictive modeling of outcomes following definitive chemoradiotherapy for oropharyngeal cancer based on FDG-PET image characteristics. Phys Med Biol. 2017;62(13):5327. doi: 10.1088/1361-6560/aa73cc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kirienko M, Cozzi L, Antunovic L, Lozza L, Fogliata A, Voulaz E, et al. Prediction of disease-free survival by the PET/CT radiomic signature in non-small cell lung cancer patients undergoing surgery. Eur J Nucl Med Mol Imaging. 2018;45(2):207–217. doi: 10.1007/s00259-017-3837-7. [DOI] [PubMed] [Google Scholar]
- 27.Lovat E, Siddique M, Goh V, Ferner RE, Cook GJR, Warbey VS. The effect of post-injection (18)F-FDG PET scanning time on texture analysis of peripheral nerve sheath tumours in neurofibromatosis-1. EJNMMI Res. 2017;7(1):35. doi: 10.1186/s13550-017-0282-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yip S, McCall K, Aristophanous M, Chen AB, Aerts HJ, Berbeco R. Comparison of texture features derived from static and respiratory-gated PET images in non-small cell lung cancer. PLoS One. 2014;9(12):e115510. doi: 10.1371/journal.pone.0115510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vaidya M, Creach KM, Frye J, Dehdashti F, Bradley JD, El Naqa I. Combined PET/CT image characteristics for radiotherapy tumor response in lung cancer. Radiother Oncol. 2012;102(2):239–245. doi: 10.1016/j.radonc.2011.10.014. [DOI] [PubMed] [Google Scholar]
- 30.Grootjans W, Tixier F, van der Vos CS, Vriens D, Le Rest CC, Bussink J, et al. The impact of optimal respiratory gating and image noise on evaluation of intratumor heterogeneity on 18F-FDG PET imaging of lung cancer. J Nucl Med. 2016;57(11):1692–1698. doi: 10.2967/jnumed.116.173112. [DOI] [PubMed] [Google Scholar]
- 31.Oliver JA, Budzevich M, Zhang GG, Dilling TJ, Latifi K, Moros EG. Variability of image features computed from conventional and respiratory-gated PET/CT images of lung cancer. Transl Oncol. 2015;8(6):524–534. doi: 10.1016/j.tranon.2015.11.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Galavis PE, Hollensen C, Jallow N, Paliwal B, Jeraj R. Variability of textural features in FDG PET images due to different acquisition modes and reconstruction parameters. Acta Oncol (Stockholm, Sweden) 2010;49(7):1012–1016. doi: 10.3109/0284186X.2010.498437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yan J, Chu-Shern JL, Loi HY, Khor LK, Sinha AK, Quek ST, et al. Impact of image reconstruction settings on texture features in 18F-FDG PET. J Nucl Med. 2015;56(11):1667–1673. doi: 10.2967/jnumed.115.156927. [DOI] [PubMed] [Google Scholar]
- 34.Shiri I, Rahmim A, Ghaffarian P, Geramifar P, Abdollahi H, Bitarafan-Rajabi A. The impact of image reconstruction settings on 18F-FDG PET radiomic features: multi-scanner phantom and patient studies. Eur Radiol. 2017;27(11):4498–4509. doi: 10.1007/s00330-017-4859-z. [DOI] [PubMed] [Google Scholar]
- 35.Hatt M, Tixier F, Cheze Le Rest C, Pradier O, Visvikis D. Robustness of intratumour (1)(8)F-FDG PET uptake heterogeneity quantification for therapy response prediction in oesophageal carcinoma. Eur J Nucl Med Mol Imaging. 2013;40(11):1662–1671. doi: 10.1007/s00259-013-2486-8. [DOI] [PubMed] [Google Scholar]
- 36.van Velden FH, Cheebsumon P, Yaqub M, Smit EF, Hoekstra OS, Lammertsma AA, et al. Evaluation of a cumulative SUV-volume histogram method for parameterizing heterogeneous intratumoural FDG uptake in non-small cell lung cancer PET studies. Eur J Nucl Med Mol Imaging. 2011;38(9):1636–1647. doi: 10.1007/s00259-011-1845-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lasnon C, Majdoub M, Lavigne B, Do P, Madelaine J, Visvikis D, et al. (18)F-FDG PET/CT heterogeneity quantification through textural features in the era of harmonisation programs: a focus on lung cancer. Eur J Nucl Med Mol Imaging. 2016;43(13):2324–2335. doi: 10.1007/s00259-016-3441-2. [DOI] [PubMed] [Google Scholar]
- 38.Orlhac F, Boughdad S, Philippe C, Stalla-Bourdillon H, Nioche C, Champion L, et al. A postreconstruction harmonization method for multicenter radiomic studies in PET. J Nucl Med. 2018;59(8):1321–1328. doi: 10.2967/jnumed.117.199935. [DOI] [PubMed] [Google Scholar]
- 39.Bagci U, Chen X, Udupa JK. Hierarchical scale-based multiobject recognition of 3-D anatomical structures. IEEE Trans Med Imaging. 2012;31(3):777–789. doi: 10.1109/TMI.2011.2180920. [DOI] [PubMed] [Google Scholar]
- 40.Foster B, Bagci U, Mansoor A, Xu Z, Mollura DJ. A review on segmentation of positron emission tomography images. Comput Biol Med. 2014;50:76–96. doi: 10.1016/j.compbiomed.2014.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bagci U, Foster B, Miller-Jaster K, Luna B, Dey B, Bishai WR, et al. A computational pipeline for quantification of pulmonary infections in small animal models using serial PET-CT imaging. EJNMMI Res. 2013;3(1):55. doi: 10.1186/2191-219X-3-55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nestle U, Walter K, Schmidt S, Licht N, Nieder C, Motaref B, et al. 18F-deoxyglucose positron emission tomography (FDG-PET) for the planning of radiotherapy in lung cancer: high impact in patients with atelectasis. Int J Radiat Oncol Biol Phys. 1999;44(3):593–597. doi: 10.1016/s0360-3016(99)00061-9. [DOI] [PubMed] [Google Scholar]
- 43.Fiorino C, Reni M, Bolognesi A, Cattaneo GM, Calandrino R. Intra- and inter-observer variability in contouring prostate and seminal vesicles: implications for conformal treatment planning. Radiother Oncol. 1998;47(3):285–292. doi: 10.1016/s0167-8140(98)00021-8. [DOI] [PubMed] [Google Scholar]
- 44.Erasmus JJ, Gladish GW, Broemeling L, Sabloff BS, Truong MT, Herbst RS, et al. Interobserver and intraobserver variability in measurement of non-small-cell carcinoma lung lesions: implications for assessment of tumor response. J Clin Oncol. 2003;21(13):2574–2582. doi: 10.1200/JCO.2003.01.144. [DOI] [PubMed] [Google Scholar]
- 45.Fox JL, Rengan R, O'Meara W, Yorke E, Erdi Y, Nehmeh S, et al. Does registration of PET and planning CT images decrease interobserver and intraobserver variation in delineating tumor volumes for non-small-cell lung cancer? Int J Radiat Oncol Biol Phys. 2005;62(1):70–75. doi: 10.1016/j.ijrobp.2004.09.020. [DOI] [PubMed] [Google Scholar]
- 46.Hatt M, Cheze Le Rest C, Albarghach N, Pradier O, Visvikis D. PET functional volume delineation: a robustness and repeatability study. Eur J Nucl Med Mol Imaging. 2011;38(4):663–672. doi: 10.1007/s00259-010-1688-6. [DOI] [PubMed] [Google Scholar]
- 47.Bagci U, Yao J, Caban J, Turkbey E, Aras O, Mollura DJ. A graph-theoretic approach for segmentation of PET images. Conf Proc IEEE Eng Med Biol Soc. 2011;2011:8479–82. 10.1109/iembs.2011.6092092. [DOI] [PMC free article] [PubMed]
- 48.Nestle U, Kremp S, Schaefer-Schuler A, Sebastian-Welsch C, Hellwig D, Rube C, et al. Comparison of different methods for delineation of 18F-FDG PET-positive tissue for target volume definition in radiotherapy of patients with non-small cell lung cancer. J Nucl Med. 2005;46(8):1342–1348. [PubMed] [Google Scholar]
- 49.Hong R, Halama J, Bova D, Sethi A, Emami B. Correlation of PET standard uptake value and CT window-level thresholds for target delineation in CT-based radiation treatment planning. Int J Radiat Oncol Biol Phys. 2007;67(3):720–726. doi: 10.1016/j.ijrobp.2006.09.039. [DOI] [PubMed] [Google Scholar]
- 50.Ha S, Park S, Bang JI, Kim EK, Lee HY. Metabolic radiomics for pretreatment (18)F-FDG PET/CT to characterize locally advanced breast cancer: histopathologic characteristics, response to neoadjuvant chemotherapy, and prognosis. Sci Rep. 2017;7(1):1556. doi: 10.1038/s41598-017-01524-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Orlhac F, Soussan M, Maisonobe JA, Garcia CA, Vanderlinden B, Buvat I. Tumor texture analysis in 18F-FDG PET: relationships between texture parameters, histogram indices, standardized uptake values, metabolic volumes, and total lesion glycolysis. J Nucl Med. 2014;55(3):414–422. doi: 10.2967/jnumed.113.129858. [DOI] [PubMed] [Google Scholar]
- 52.Erdi YE, Mawlawi O, Larson SM, Imbriaco M, Yeung H, Finn R, et al. Segmentation of lung lesion volume by adaptive positron emission tomography image thresholding. Cancer. 1997;80(12 Suppl):2505–2509. doi: 10.1002/(sici)1097-0142(19971215)80:12+<2505::aid-cncr24>3.3.co;2-b. [DOI] [PubMed] [Google Scholar]
- 53.Schaefer A, Kremp S, Hellwig D, Rube C, Kirsch CM, Nestle U. A contrast-oriented algorithm for FDG-PET-based delineation of tumour volumes for the radiotherapy of lung cancer: derivation from phantom measurements and validation in patient data. Eur J Nucl Med Mol Imaging. 2008;35(11):1989–1999. doi: 10.1007/s00259-008-0875-1. [DOI] [PubMed] [Google Scholar]
- 54.Davis JB, Reiner B, Huser M, Burger C, Szekely G, Ciernik IF. Assessment of 18F PET signals for automatic target volume definition in radiotherapy treatment planning. Radiother Oncol. 2006;80(1):43–50. doi: 10.1016/j.radonc.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 55.Drever L, Robinson DM, McEwan A, Roa W. A local contrast based approach to threshold segmentation for PET target volume delineation. Med Phys. 2006;33(6):1583–1594. doi: 10.1118/1.2198308. [DOI] [PubMed] [Google Scholar]
- 56.Lu L, Lv W, Jiang J, Ma J, Feng Q, Rahmim A, et al. Robustness of radiomic features in [(11)C]choline and [(18)F]FDG PET/CT imaging of nasopharyngeal carcinoma: impact of segmentation and discretization. Mol Imaging Biol. 2016;18(6):935–945. doi: 10.1007/s11307-016-0973-6. [DOI] [PubMed] [Google Scholar]
- 57.Cheng NM, Fang YH, Tsan DL, Hsu CH, Yen TC. Respiration-averaged CT for attenuation correction of PET images—impact on PET texture features in non-small cell lung cancer patients. PLoS One. 2016;11(3):e0150509. doi: 10.1371/journal.pone.0150509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Krak NC, Boellaard R, Hoekstra OS, Twisk JW, Hoekstra CJ, Lammertsma AA. Effects of ROI definition and reconstruction method on quantitative outcome and applicability in a response monitoring trial. Eur J Nucl Med Mol Imaging. 2005;32(3):294–301. doi: 10.1007/s00259-004-1566-1. [DOI] [PubMed] [Google Scholar]
- 59.Hatt M, Cheze-le Rest C, van Baardwijk A, Lambin P, Pradier O, Visvikis D. Impact of tumor size and tracer uptake heterogeneity in (18)F-FDG PET and CT non-small cell lung cancer tumor delineation. J Nucl Med. 2011;52(11):1690–1697. doi: 10.2967/jnumed.111.092767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhu W, Jiang T Automation segmentation of PET image for brain tumors. Nuclear Science Symposium Conference Record. 2003 IEEE; 2003: IEEE.
- 61.Hatt M, Cheze le Rest C, Turzo A, Roux C, Visvikis D. A fuzzy locally adaptive Bayesian segmentation approach for volume determination in PET. IEEE Trans Med Imaging. 2009;28(6):881–893. doi: 10.1109/TMI.2008.2012036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Werner-Wasik M, Nelson AD, Choi W, Arai Y, Faulhaber PF, Kang P, et al. What is the best way to contour lung tumors on PET scans? Multiobserver validation of a gradient-based method using a NSCLC digital PET phantom. Int J Radiat Oncol Biol Phys. 2012;82(3):1164–1171. doi: 10.1016/j.ijrobp.2010.12.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Geets X, Lee JA, Bol A, Lonneux M, Gregoire V. A gradient-based method for segmenting FDG-PET images: methodology and validation. Eur J Nucl Med Mol Imaging. 2007;34(9):1427–1438. doi: 10.1007/s00259-006-0363-4. [DOI] [PubMed] [Google Scholar]
- 64.Hatt M, Tixier F, Pierce L, Kinahan PE, Le Rest CC, Visvikis D. Characterization of PET/CT images using texture analysis: the past, the present... any future? Eur J Nucl Med Mol Imaging. 2017;44(1):151–165. doi: 10.1007/s00259-016-3427-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Hatt M, Cheze le Rest C, Descourt P, Dekker A, De Ruysscher D, Oellers M, et al. Accurate automatic delineation of heterogeneous functional volumes in positron emission tomography for oncology applications. Int J Radiat Oncol Biol Phys. 2010;77(1):301–308. doi: 10.1016/j.ijrobp.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 66.Lambin P, Leijenaar RTH, Deist TM, Peerlings J, de Jong EEC, van Timmeren J, et al. Radiomics: the bridge between medical imaging and personalized medicine. Nat Rev Clin Oncol. 2017;14(12):749–762. doi: 10.1038/nrclinonc.2017.141. [DOI] [PubMed] [Google Scholar]
- 67.Cheng NM, Fang YH, Lee LY, Chang JT, Tsan DL, Ng SH, et al. Zone-size nonuniformity of 18F-FDG PET regional textural features predicts survival in patients with oropharyngeal cancer. Eur J Nucl Med Mol Imaging. 2015;42(3):419–428. doi: 10.1007/s00259-014-2933-1. [DOI] [PubMed] [Google Scholar]
- 68.Tixier F, Le Rest CC, Hatt M, Albarghach N, Pradier O, Metges JP, et al. Intratumor heterogeneity characterized by textural features on baseline 18F-FDG PET images predicts response to concomitant radiochemotherapy in esophageal cancer. J Nucl Med. 2011;52(3):369–378. doi: 10.2967/jnumed.110.082404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Lovinfosse P, Janvary ZL, Coucke P, Jodogne S, Bernard C, Hatt M, et al. FDG PET/CT texture analysis for predicting the outcome of lung cancer treated by stereotactic body radiation therapy. Eur J Nucl Med Mol Imaging. 2016;43(8):1453–1460. doi: 10.1007/s00259-016-3314-8. [DOI] [PubMed] [Google Scholar]
- 70.Leijenaar RT, Carvalho S, Velazquez ER, van Elmpt WJ, Parmar C, Hoekstra OS, et al. Stability of FDG-PET radiomics features: an integrated analysis of test-retest and inter-observer variability. Acta Oncol (Stockholm, Sweden) 2013;52(7):1391–1397. doi: 10.3109/0284186X.2013.812798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Leijenaar RT, Nalbantov G, Carvalho S, van Elmpt WJ, Troost EG, Boellaard R, et al. The effect of SUV discretization in quantitative FDG-PET radiomics: the need for standardized methodology in tumor texture analysis. Sci Rep. 2015;5:11075. doi: 10.1038/srep11075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Orlhac F, Soussan M, Chouahnia K, Martinod E, Buvat I. 18F-FDG PET-derived textural indices reflect tissue-specific uptake pattern in non-small cell lung cancer. PLoS One. 2015;10(12):e0145063. doi: 10.1371/journal.pone.0145063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Orlhac F, Nioche C, Soussan M, Buvat I. Understanding changes in tumor textural indices in PET: a comparison between visual assessment and index values in simulated and patient data. J Nucl Med. 2017;58(3):387–392. doi: 10.2967/jnumed.116.181859. [DOI] [PubMed] [Google Scholar]
- 74.van Velden FH, Kramer GM, Frings V, Nissen IA, Mulder ER, de Langen AJ, et al. Repeatability of radiomic features in non-small-cell lung cancer [(18)F]FDG-PET/CT studies: impact of reconstruction and delineation. Mol Imaging Biol. 2016;18(5):788–795. doi: 10.1007/s11307-016-0940-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Brooks FJ, Grigsby PW. The effect of small tumor volumes on studies of intratumoral heterogeneity of tracer uptake. J Nucl Med. 2014;55(1):37–42. doi: 10.2967/jnumed.112.116715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Hatt M, Majdoub M, Vallieres M, Tixier F, Le Rest CC, Groheux D, et al. 18F-FDG PET uptake characterization through texture analysis: investigating the complementary nature of heterogeneity and functional tumor volume in a multi-cancer site patient cohort. J Nucl Med. 2015;56(1):38–44. doi: 10.2967/jnumed.114.144055. [DOI] [PubMed] [Google Scholar]
- 77.Presotto L, Bettinardi V, De Bernardi E, Belli ML, Cattaneo GM, Broggi S, et al. PET textural features stability and pattern discrimination power for radiomics analysis: an “ad-hoc” phantoms study. Phys Med. 2018;50:66–74. doi: 10.1016/j.ejmp.2018.05.024. [DOI] [PubMed] [Google Scholar]
- 78.Chicklore S, Goh V, Siddique M, Roy A, Marsden PK, Cook GJ. Quantifying tumour heterogeneity in 18F-FDG PET/CT imaging by texture analysis. Eur J Nucl Med Mol Imaging. 2013;40(1):133–140. doi: 10.1007/s00259-012-2247-0. [DOI] [PubMed] [Google Scholar]
- 79.Chung HH, Kang SY. Prognostic value of preoperative intratumoral FDG uptake heterogeneity in early stage uterine cervical cancer. J Gynecol Oncol. 2016;27(2):e15. doi: 10.3802/jgo.2016.27.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.El Naqa I, Grigsby P, Apte A, Kidd E, Donnelly E, Khullar D, et al. Exploring feature-based approaches in PET images for predicting cancer treatment outcomes. Pattern Recogn. 2009;42(6):1162–1171. doi: 10.1016/j.patcog.2008.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Castellano G, Bonilha L, Li LM, Cendes F. Texture analysis of medical images. Clin Radiol. 2004;59(12):1061–1069. doi: 10.1016/j.crad.2004.07.008. [DOI] [PubMed] [Google Scholar]
- 82.Yu H, Caldwell C, Mah K, Mozeg D. Coregistered FDG PET/CT-based textural characterization of head and neck cancer for radiation treatment planning. IEEE Trans Med Imaging. 2009;28(3):374–383. doi: 10.1109/TMI.2008.2004425. [DOI] [PubMed] [Google Scholar]
- 83.Lv W, Yuan Q, Wang Q, Ma J, Jiang J, Yang W, et al. Robustness versus disease differentiation when varying parameter settings in radiomics features: application to nasopharyngeal PET/CT. Eur Radiol. 2018;28(8):3245–3254. doi: 10.1007/s00330-018-5343-0. [DOI] [PubMed] [Google Scholar]
- 84.Amadasun M, King R. Textural features corresponding to textural properties. IEEE Trans Syst Man Cybern. 1989;19(5):1264–1274. [Google Scholar]
- 85.Galloway MM. Texture analysis using gray level run lengths. Comput Graphics Image Process. 1975;4(2):172–179. [Google Scholar]
- 86.Thibault G, Fertil B, Navarro C, Pereira S, Cau P, Levy N, et al. Texture indexes and gray level size zone matrix application to cell nuclei classification. Pattern Recognition and Information Processing. 2009: 140-145.
- 87.Aerts HJ, Velazquez ER, Leijenaar RT, Parmar C, Grossmann P, Carvalho S, et al. Decoding tumour phenotype by noninvasive imaging using a quantitative radiomics approach. Nat Commun. 2014;5:4006. doi: 10.1038/ncomms5006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Vallieres M, Freeman CR, Skamene SR, El Naqa I. A radiomics model from joint FDG-PET and MRI texture features for the prediction of lung metastases in soft-tissue sarcomas of the extremities. Phys Med Biol. 2015;60(14):5471–5496. doi: 10.1088/0031-9155/60/14/5471. [DOI] [PubMed] [Google Scholar]
- 89.Thibault G, Angulo J, Meyer F. Advanced statistical matrices for texture characterization: application to cell classification. IEEE Trans Biomed Eng. 2014;61(3):630–637. doi: 10.1109/TBME.2013.2284600. [DOI] [PubMed] [Google Scholar]
- 90.Thibault G. Advanced statistical matrices for texture characterization: application to DNA chromatin and microtubule network classification. 18th IEEE International Conference on Image Processing. 2011.
- 91.Tixier F, Hatt M, Le Rest CC, Le Pogam A, Corcos L, Visvikis D. Reproducibility of tumor uptake heterogeneity characterization through textural feature analysis in 18F-FDG PET. J Nucl Med. 2012;53(5):693–700. doi: 10.2967/jnumed.111.099127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.van Velden FH, Nissen IA, Jongsma F, Velasquez LM, Hayes W, Lammertsma AA, et al. Test-retest variability of various quantitative measures to characterize tracer uptake and/or tracer uptake heterogeneity in metastasized liver for patients with colorectal carcinoma. Mol Imaging Biol. 2014;16(1):13–18. doi: 10.1007/s11307-013-0660-9. [DOI] [PubMed] [Google Scholar]
- 93.Chung HH, Kang SY, Ha S, Kim J-W, Park N-H, Song YS, et al. Prognostic value of preoperative intratumoral FDG uptake heterogeneity in early stage uterine cervical cancer. J Gynecol Oncol. 2015;27(2):e15. doi: 10.3802/jgo.2016.27.e15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Park S, Ha S, Lee S-H, Paeng JC, Keam B, Kim TM, et al. Intratumoral heterogeneity characterized by pretreatment PET in non-small cell lung cancer patients predicts progression-free survival on EGFR tyrosine kinase inhibitor. PLoS One. 2018;13(1):e0189766. doi: 10.1371/journal.pone.0189766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Bender R, Lange S. Adjusting for multiple testing—when and how? J Clin Epidemiol. 2001;54(4):343–349. doi: 10.1016/s0895-4356(00)00314-0. [DOI] [PubMed] [Google Scholar]
- 96.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 97.Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B Methodol. 1995;57:289–300. [Google Scholar]
- 98.Alpaydin E. Introduction to machine learning. Cambridge: MIT press; 2009. [Google Scholar]
- 99.Hastie T, Friedman J, Tibshirani R. The elements of statistical learning. Berlin: Springer; 2001. Model assessment and selection; pp. 193–224. [Google Scholar]
- 100.Kohavi R, editor. A study of cross-validation and bootstrap for accuracy estimation and model selection. Montreal: Ijcai; 1995. [Google Scholar]
- 101.Köppen M The curse of dimensionality. 5th Online World Conference on Soft Computing in Industrial Applications (WSC5). 2000
- 102.Segal E, Sirlin CB, Ooi C, Adler AS, Gollub J, Chen X, et al. Decoding global gene expression programs in liver cancer by noninvasive imaging. Nat Biotechnol. 2007;25(6):675. doi: 10.1038/nbt1306. [DOI] [PubMed] [Google Scholar]
- 103.Hall MA Correlation-based feature selection for machine learning. 1999
- 104.Misaki M, Kim Y, Bandettini PA, Kriegeskorte N. Comparison of multivariate classifiers and response normalizations for pattern-information fMRI. NeuroImage. 2010;53(1):103–118. doi: 10.1016/j.neuroimage.2010.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Van Der Maaten L, Postma E, Van den Herik J. Dimensionality reduction: a comparative. J Mach Learn Res. 2009;10:66–71. [Google Scholar]