Abstract
To examine 143 diabetes risk single nucleotide polymorphisms (SNPs), identified from genome-wide association studies, in association with breast cancer (BC) incidence and subsequent mortality. A population-based sample of Caucasian women with first primary invasive BC (n = 817) and controls (n = 1021) were interviewed to assess diabetes status. Using the National Death Index, women with BC were followed for >18 years during which 340 deaths occurred (139 BC deaths). Genotyping was done using DNA extracted from blood samples. We used unconditional logistic regression to estimate age-adjusted odds ratios and 95% confidence intervals (CIs) for BC incidence, and Cox regression to estimate age-adjusted hazard ratios and CIs for all-cause and BC-specific mortality. Twelve SNPs were associated with BC risk in additive genotype models, at α = 0.05. The top three significant SNPs included SLC30A8- rs4876369 (P = 0.0034), HHEX-rs11187146 (P = 0.0086), and CDKN2A/CDKN2B-rs1333049 (P = 0.0094). Diabetes status modified the associations between rs4876369 and rs2241745 and BC incidence, on the multiplicative interaction scale. Six SNPs were associated with all-cause (CDKAL1-rs981042, P = 0.0032; HHEX-rs1111875, P = 0.0361; and INSR-rs919275, P = 0.0488) or BC-specific (CDKN2A/CDKN2B-rs3218020, P = 0.0225; CDKAL1-rs981042, P = 0.0246; and TCF2/HNF1B-rs3094508, P = 0.0344) mortality in additive genotype models, at α = 0.05. Genetic polymorphisms that increase the risk of developing diabetes may also increase the risk of developing and dying from BC.
Keywords: breast cancer, diabetes, genetics, incidence, mortality, single nucleotide polymorphisms, survival
1 |. INTRODUCTION
Over 11 million women aged ≥18 years were living with diagnosed diabetes in the United Sates (US) in 20151. Diabetes is a known risk factor for heart disease, stroke, nephropathy, and neuropathy and cancers of the liver, pancreas, endometrium, and colon/rectum.2 Accumulating epidemiologic evidence suggests that diabetes may also increase the risk of developing breast cancer (BC), the most frequently diagnosed cancer among women in the US.3 A meta-analysis of five case-control and 15 cohort studies reported a 20% increased risk of developing BC among women with diabetes, compared to those without diabetes.4
Diabetes and BC are both complex multifactorial diseases that share several common predisposing risk factors, including age, overweight/obesity, alcohol consumption, and physical inactivity.1,5,6 A number of biological mechanisms that focus on the underlying pathologic characteristics of diabetes have been proposed to explain a potential causal association between diabetes and carcinogenesis.5 High circulating levels of glucose (ie, hyperglycemia), for example, may facilitate neoplastic proliferation due to high rates of glucose uptake required and adopted by many cancers.5,7 Furthermore, high circulating levels of insulin (ie, hyperinsulinemia) may promote carcinogenesis directly through insulin receptor-mediated mitogenesis,5,8,9 and indirectly through the reduced hepatic transcription of the insulin- like growth factor binding protein-1 (IGFBP-1) gene,10 which results in increased circulating levels of free bioactive IGF-1, a potent mitogen of human breast MCF-7 cells.11 Insulin resistance is also associated with reduced levels of sex hormone binding globulin (SHBG) and thus increases in bioavailable estrogen, although the directionality of association has not been established.12
Given the proliferative effects of insulin on BC cells, hyperinsulinemia is also hypothesized to increase risk of mortality following cancer.5 To date, few studies13 have investigated diabetes status in relation to mortality following BC. While studies consistently report a 40–50% increase in risk of all-cause mortality following BC,13,14 results of BC-specific mortality have been mixed.15–17 However, diabetes has also been associated with more advanced stage at BC presentation,18,19 and for those with diabetes, BC treatment tends to be less aggressive and causes more adverse effects.13,19
Despite observational studies linking diabetes to BC, a few studies20–22 have examined a small number (≤40) of diabetes risk single nucleotide polymorphisms (SNPs) in association with risk of incident BC and subsequent mortality. Given that diabetes is strongly influenced by genetic factors,23 we examined the associations between 143 SNPs in or near 29 genes identified from genome-wide association studies of diabetes risk, and BC incidence and subsequent mortality among participants of a population-based study of BC.
2 |. MATERIALS AND METHODS
2.1 |. Study population
This study included 1838 Caucasian women from the Long Island Breast Cancer Study Project (LIBCSP), a population-based study initiated as a case-control study and then continued as a follow-up study. The LIBCSP study protocol was approved by the Institutional Review Boards of all participating institutions and written informed consent was obtained from participants prior to data collection.
2.2 |. Case-control design
Details of the LIBCSP case-control design have been previously reported.24 In brief, adult women with a first diagnosis of in situ or invasive BC during August 1, 1996 and July 31, 1997 were identified using a rapid reporting system established for the LIBCSP. Approximately 82.1% (n = 1,508) of eligible women with BC completed a comprehensive 100-min interviewer-administered questionnaire, on average within 3 months of diagnosis (25th percentile = 1.2 months, 75th percentile = 4.0 months). Seventy-three percent of BC participants provided blood samples, of which the majority were collected prior to chemotherapy (77.2%). Medical records were abstracted to obtain information on estrogen and progesterone receptor (ER and PR, respectively) status and first course of treatment. The mean age at BC diagnosis among the women included in this study was 59 (range = 25.1–91.9). The majority of women with BC were post-menopausal at diagnosis (68.1%). Most women were diagnosed with ER-positive (76.5%) or PR-positive (66.6%) BC, or both (61.9%).
Women without BC were residents of the same two Long Island counties who were frequency-matched to the expected distribution of women with BC in 5-year age groups in 1996–1997. Women without BC who were 65 years of age and older were identified by Health Care Finance Administration (HCFA) rosters and those under 65 years of age were identified by random digit dialing. Approximately 62.7% (n = 1556) of eligible women without breast cancer completed the questionnaire and, of these, 73.3% provided blood samples. The majority of LIBCSP participants were Caucasian (93%) and ranged in age from 20 to 98.
2.3 |. Follow-up design
Details of the LIBCSP follow-up design and ascertainment of vital status have also been previously reported.25 In brief, the National Death Index (NDI), a centralized database of death record information maintained by the National Center for Health Statistics,26 was used to ascertain date and cause of death for the women with BC. International Statistical Classification of Diseases codes 9/10 174.9 and C-50.9 listed on the death certificate were used to identify BC-related deaths. Follow-up for mortality occurred from the date of diagnosis in 1996 or 1997 until December 31, 2014. The median follow-up was 17.6 years (range = 0.39–18.41 years) during which 340 deaths occurred, 139 of which were from BC.
2.4 |. Diabetes status
Diabetes status was assessed by self-report as part of the case–control interview. Participants were asked whether they had ever been told by a physician that they had diabetes, sugar diabetes, or high blood sugar.15 If participants responded in the affirmative, they were also asked the year in which the doctor first told them they had diabetes and whether they had been prescribed medication for their diabetes. Five women (three with BC and two without BC) were missing information on diabetes status. The prevalence of diabetes was 6.9% among women with BC, and 7.0% among women without BC.
2.5 |. SNP selection and genotyping
We selected 158 polymorphisms in or near 29 genes for genotyping (Supplemental Table S1). These diabetes-related genes and SNPs were selected based on meta-analyses of genome wide association studies (GWAS),27–30 which reported statistically significant associations between each SNP and diabetes risk. Genotyping was done in 2011 at the University of North Carolina at Chapel Hill using the Illumina GoldenGate assay (Illumina, Inc., San Diego, CA) on genomic DNA extracted from mononuclear cells in whole blood. Genome Studio software v. 2011.1 was used to review assay intensity data and genotype cluster images for all SNPs. Blind duplicates of 56 samples were genotyped to verify the reproducibility of genotype calls; concordance between duplicates was greater than 99.4% for all pairs.
2.6 |. Statistical analysis
This study was restricted to the 1838 LIBCSP participants (817 women with invasive BC and 1021 women without BC) who self-identified as Caucasian and for whom genotyping data were available. We first tested all 158 SNPs for Hardy-Weinberg equilibrium (HWE) among the women without BC using Proc Allele in SAS/Genetics version 9.4 (SAS Institute Inc., Cary, NC) at α = 0.05. Seven SNPs (rs1333040, rs10505312, rs11196199, rs7084875, rs6749108, rs7605725, rs4689382) exhibited significant departure from HWE and were not considered further. We next examined the minor allele frequencies of the remaining 151 SNPs in both women with and without BC; rs11708719 had a minor allele frequency <5% (4.55% in women with BC and 4.80% in women without BC). We examined the remaining 150 SNPs for linkage disequilibrium (LD) using the SNAP database based on HapMap.31 The following seven SNPs were excluded due to LD: rs10010131, rs2046916, rs4712524, rs6744642, rs6769511, rs7748720, rs7923837. QC procedures resulted in 143 SNPs available for statistical analysis.
We used unconditional logistic regression in SAS software version 9.4 (SAS Institute, Inc., Cary, NC) to estimate age-adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for the associations between the 143 SNPs and BC incidence. We used multivariable Cox regression in SAS to estimate age-adjusted hazard ratios (HRs) and 95% CIs of the associations between the 143 SNPs and all-cause and BC-specific mortality. For analyses of mortality, observations were censored at the end of follow-up in 2014, if alive. For analyses using BC-specific mortality as the outcome, non-BC deaths were censored at the time of death. For both BC incidence and mortality, we first examined each SNP using additive genotype models (ie, each variant copy is assumed to add to the expression of the phenotype). SNPs significantly associated with BC incidence or mortality at α = 0.05 were subsequently examined using co-dominant genotype models (ie, each variant copy is assumed to have an effect on the phonotype) with common homozygous genotypes defined among the women without breast cancer as the referent group. Although we present results for SNPs associated with BC incidence or mortality at α = 0.05, we also compared our results to a Bonferroni corrected α of 0.0003, to account for multiple comparisons given that we examined 143 SNPs.32 SNPs associated with all-cause or breast cancer-specific mortality were also assessed for the proportional hazards assumption using Kaplan-Meier survival curves and Schoenfeld residuals;33 there were no violations of the proportional hazards assumption. All logistic and Cox regression models were adjusted for age to account for frequency matching of women without BC to women with BC, but no other covariates, given that few factors are causally associated with genetic variants.34
For BC incidence, all-cause mortality, and BC-specific mortality, we created three “risk scores” by summing the “at-risk” alleles significantly associated with each outcome at α = 0.05. The “at-risk” allele was defined as the less common (variant) allele, unless the variant was inversely associated with breast cancer in this population, in which case the more common allele was defined as the “at-risk” allele. We categorized the continuous summary scores into tertiles based on the distributions in women without BC.
We examined diabetes status (yes vs no) as an effect modifier of the associations between SNPs found to be significantly associated and BC incidence or mortality. Multiplicative interactions were evaluated using likelihood ratio tests, comparing additive genotype models with diabetes-by-SNP cross-product terms against reduced models without the interaction terms. We did not examine interactions on the additive scale as we did not consider dominant genotype models.
3 |. RESULTS
3.1 |. Diabetes-related gene SNPs and breast cancer incidence
Twelve SNPs were significantly associated with the risk of developing BC in additive genotype models at α = 0.05 (Figure 1), but none were statistically significant at the Bonferroni-corrected alpha of 0.0003. The most significant SNPs included rs4876369 (P = 0.0034), for which the heterozygous (vs common homozygous) genotype was associated with an OR of BC of 1.41 (95% CI = 1.12–1.76), and rs11187146 (P = 0.0086) and rs1333049 (P = 0.0094), for which the variant (vs common) homozygous genotypes were associated with ORs of BC of 2.06 (95% CI = 1.03–4.10) and 1.40 (95% CI = 1.08–1.82), respectively, in co-dominant genotype models (Table 1). Variant (vs common) homozygous genotypes of rs10830963 (P = 0.0125), rs6794209 (P = 0.0201), rs290494 (P = 0.0412), and rs2241745 (P = 0.0458) were inversely associated with BC incidence (Table 1). The highest (vs lowest) category of the summary score of “at-risk” alleles comprised of the sum of the 12 statistically significant SNPs was associated with a 109% increase in risk of BC (OR = 2.09, 95% CI = 1.64–2.66, P < 0.0001).
FIGURE 1.
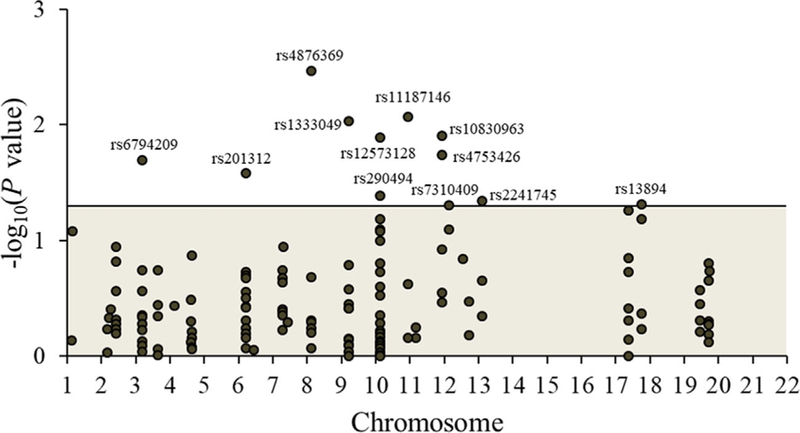
Manhattan plot showing the significance of additive association between all 143 diabetes-related SNPs and incident breast cancer among 817 women with invasive breast cancer cases and 1021 age-matched women without breast cancer in the Long Island Breast Cancer Study Project. The 12 SNPs that reached significance at alpha of 0.05 (those above the horizontal line) are listed in Table 1. [Color figure can be viewed at wileyonlinelibrary.com]
TABLE 1.
Co-dominant genotype model age-adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for diabetes-related SNPs and breast cancer (BC) risk in the LIBCSP (n = 1838)
| Gene | rs ID location | Genotype | 817 BC/1021 no BC | OR (95% CI) | Pa |
|---|---|---|---|---|---|
| SLC30A8 | rs4876369 | AA | 588/802 | 1.00 | |
| Intron | AG | 206/198 | 1.41 (1.12–1.76) | ||
| GG | 16/16 | 1.37 (0.68–2.77) | 0.0034 | ||
| HHEX-EXOC6 | rs11187146 | GG | 582/777 | 1.00 | |
| Intergene | GC | 210/229 | 1.24 (1.00–1.54) | ||
| CC | 21/14 | 2.06 (1.03–4.10) | 0.0086 | ||
| CDKN2A-CDKN2B | rs1333049 | CC | 209/319 | 1.00 | |
| Intergene | CG | 404/483 | 1.28 (1.03–1.59) | ||
| GG | 201/216 | 1.40 (1.08–1.82) | 0.0094 | ||
| MTNR1B | rs10830963 | CC | 458/507 | 1.00 | |
| Intron | CG | 301/426 | 0.78 (0.64–0.95) | ||
| GG | 52/81 | 0.75 (0.52–1.09) | 0.0125 | ||
| TCF7L2 | rs12573128 | AA | 537/725 | 1.00 | |
| Intron | AG | 236/263 | 1.19 (0.97–1.47) | ||
| GG | 35/28 | 1.74 (1.04–2.91) | 0.0129 | ||
| MTNR1B | rs4753426 | CC | 173/249 | 1.00 | |
| Upstream 2KB | CT | 407/528 | 1.09 (0.86–1.38) | ||
| TT | 231/239 | 1.37 (1.05–1.79) | 0.0184 | ||
| IGF2BP2 | rs6794209 | CC | 576/683 | 1.00 | |
| Intron | CT | 221/290 | 0.91 (0.74–1.12) | ||
| TT | 17/43 | 0.44 (0.25–0.79) | 0.0201 | ||
| CDKAL1 | rs201312 | AA | 509/681 | 1.00 | |
| Intron | AG | 262/303 | 1.17 (0.96–1.43) | ||
| GG | 39/32 | 1.60 (0.98–2.59) | 0.0263 | ||
| TCF7L2 | rs290494 | TT | 601/714 | 1.00 | |
| Intron | TG | 192/269 | 0.86 (0.69–1.07) | ||
| GG | 18/34 | 0.60 (0.33–1.07) | 0.0412 | ||
| IRS2 | rs2241745 | AA | 635/755 | 1.00 | |
| Intron | AG | 168/246 | 0.81 (0.64–1.01) | ||
| GG | 9/14 | 0.72 (0.31–1.67) | 0.0458 | ||
| SAT2, SHBG | rs13894 | CC | 700/912 | 1.00 | |
| Intron | CT | 111/105 | 1.36 (1.02–1.81) | ||
| TT | <5/<5 | – | 0.0492 | ||
| HNF1A | rs7310409 | GG | 258/349 | 1.00 | |
| Intron | GA | 378/489 | 1.05 (0.85–1.30) | ||
| AA | 175/177 | 1.33 (1.02–1.73) | 0.0496 | ||
| Sum of “at-risk” allelesb | 0–9 | 172/312 | 1.00 | ||
| 10–11 | 258/375 | 1.22 (0.96–1.56) | |||
| 12–24 | 361/314 | 2.09 (1.64–2.66) | <0.0001 |
Long Island Breast Cancer Study Project (LIBCSP) women without breast cancer were age-matched to women diagnosed with breast cancer between August 1, 1996 and July 31, 1997.
Additive genotype model P values.
The “at-risk” allele was defined as follows: rs4876369: G allele; rs11187146: C allele; rs1333049: G allele; rs10830963: C allele; rs12573128: G allele; rs4753426: T allele; rs6794209: C allele; rs201312: G allele; rs290494: T allele; rs2241745: A allele; rs13894: A allele; rs7310409: A allele.
Diabetes status modified two SNP-BC incidence associations: rs4876369 was associated with an OR of BC of 1.25 (95% CI = 1.02– 1.53) per allele increase among women without diabetes, and with an OR of BC of 4.30 (95% CI = 1.66–11.17) per allele increase among women with diabetes (P for multiplicative interaction = 0.0150) (Table 2). rs2241745 was associated with an OR of BC of 0.76 (95% CI = 0.61–0.94) per allele increase among women without diabetes, and with an OR of BC of 1.76 (95% CI = 0.86–3.58) per allele increase among women with diabetes (P for multiplicative interaction = 0.0283).
TABLE 2.
Additive genotype model age-adjusted odds ratios (ORs) and 95% confidence intervals (CIs) for diabetes-related SNP genotypes and breast cancer risk, overall and stratified by diabetes status in the LIBCSP (n = 1,833).
| Gene | rs ID | Alleles | Overall |
Diabetes status |
PInteractiona | |
|---|---|---|---|---|---|---|
| No (n = 1706) |
Yes (n = 127) |
|||||
| OR (95% CI) | OR (95% CI) | OR (95% CI) | ||||
| SLC30A8 | rs4876369 | A/G | 1.34 (1.10–1.63) | 1.25 (1.02–1.53) | 4.30 (1.66–11.17) | 0.0150 |
| HHEX-EXOC6 | rs11187146 | G/C | 1.29 (1.07–1.56) | 1.36 (1.12–1.66) | 0.72 (0.35–1.46) | 0.0925 |
| CDKN2A-CDKN2B | rs1333049 | C/G | 1.19 (1.04–1.35) | 1.17 (1.03–1.34) | 1.24 (0.75–2.03) | 0.7644 |
| MTNR1B | rs10830963 | C/G | 0.83 (0.71–0.96) | 0.80 (0.69–0.94) | 1.35 (0.76–2.41) | 0.1008 |
| TCF7L2 | rs12573128 | A/G | 1.24 (1.05–1.47) | 1.23 (1.03–1.47) | 1.28 (0.66–2.48) | 0.9562 |
| MTNR1B | rs4753426 | C/T | 1.17 (1.03–1.34) | 1.21 (1.05–1.39) | 0.79 (0.48–1.31) | 0.1194 |
| IGF2BP2 | rs6794209 | C/T | 0.81 (0.68–0.97) | 0.80 (0.66–0.95) | 1.00 (0.53–1.89) | 0.3237 |
| CDKAL1 | rs201312 | A/G | 1.21 (1.02–1.42) | 1.20 (1.01–1.42) | 1.19 (0.61–2.30) | 0.9777 |
| TCF7L2 | rs290494 | T/G | 0.83 (0.69–0.99) | 0.82 (0.68–0.99) | 0.88 (0.46–1.70) | 0.7418 |
| IRS2 | rs2241745 | A/G | 0.81 (0.66–1.00) | 0.76 (0.61–0.94) | 1.76 (0.86–3.58) | 0.0283 |
| SAT2, SHBG | rs13894 | C/T | 1.32 (1.00–1.75) | 1.32 (0.99–1.76) | 1.23 (0.40–3.81) | 0.9506 |
| HNF1A | rs7310409 | G/A | 1.14 (1.00–1.30) | 1.14 (0.99–1.31) | 1.17 (0.73–1.85) | 0.9377 |
| Sum of “at-risk” allelesb | 1.19 (1.13–1.24) | 1.19 (1.14–1.25) | 1.06 (0.88–1.28) | 0.2253 | ||
Long Island Breast Cancer Study Project (LIBCSP) women without breast cancer were age-matched to women diagnosed with breast cancer between August 1, 1996 and July 31, 1997.
Multiplicative interaction P values.
The “at-risk” allele was defined as follows: rs4876369: G allele; rs11187146: C allele; rs1333049: G allele; rs10830963: C allele; rs12573128: G allele; rs4753426: T allele; rs6794209: C allele; rs201312: G allele; rs290494: T allele; rs2241745: A allele; rs13894: A allele; rs7310409: A allele.
3.2 |. Diabetes-related gene SNPs and all-cause and breast cancer-specific mortality
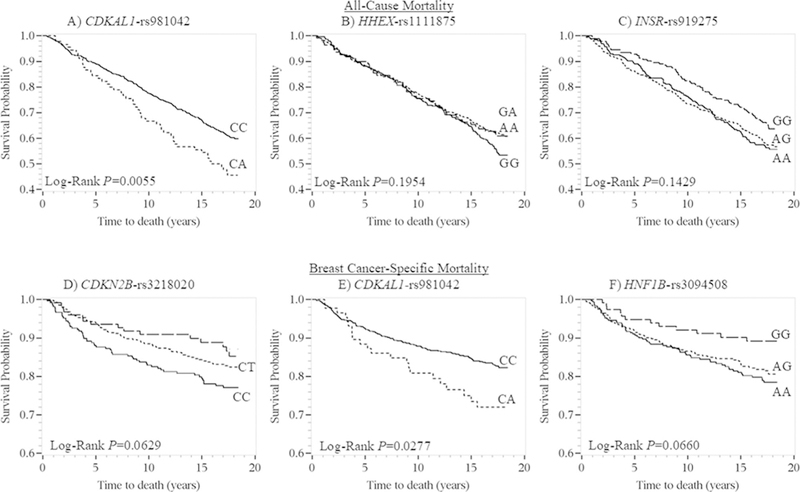
Three SNPs (rs981042, P = 0.0032; rs1111875, P = 0.0361; and rs919275, P = 0.0488) were significantly associated with all-cause mortality in additive genotype models at α = 0.05 (Figure 2), but none were statistically significant at the Bonferroni-corrected alpha of 0.0003. In co-dominant genotype models, the rs981042-CA (vs CC) genotype was associated with worse overall survival in the Kaplan-Meier survival curves (Figure 3) and with an all-cause mortality Cox model HR of 1.48 (95% CI = 1.09–2.01) (Table 3). The variant (vs common) homozygous genotypes of rs1111875 and rs919275 were associated with all-cause mortality Cox model HRs of 0.75 (95% CI = 0.53–1.06) and 0.73 (95% CI = 0.54–0.99), respectively (Table 3); however, in the Kaplan-Meier survival curves, the inverse association was only apparent for rs919275 (Figure 3). The highest (vs lowest) category of summary score of “at-risk” alleles comprised of the sum of the three statistically significant SNPs, was associated with a Cox model HR of all-cause mortality of 1.52 (95% CI = 1.14–2.04, P = 0.0004).
FIGURE 2.
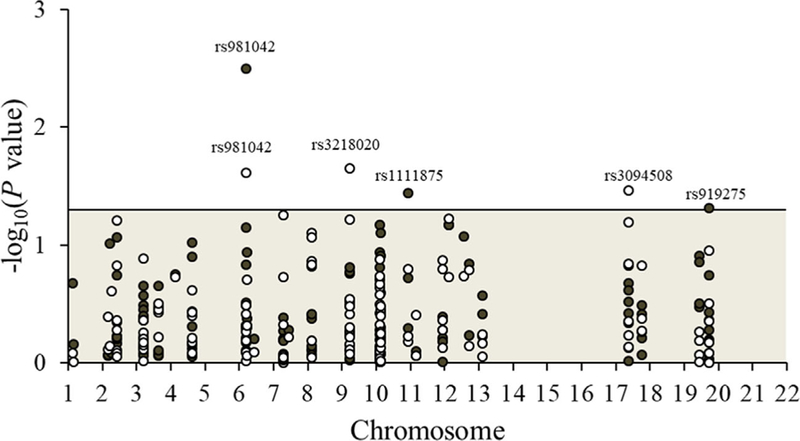
Manhattan plot showing the significance of additive association between all 143 diabetes-related SNPs and all-cause (black circles) and breast cancer-specific (white circles) mortality among 817 women with invasive breast cancer in the Long Island Breast Cancer Study Project. The six SNPs that reached significance at alpha of 0.05 (those above the horizontal line) are listed in Table 3. [Color figure can be viewed at wileyonlinelibrary.com]
FIGURE 3.
Kaplan-Meier survival plots for all-cause for (A) CDKAL1-rs981042, (B) HHEX-rs1111875, and (C) INSR-rs919275; and breast cancer-specific mortality for (D) CDKN2B-rs3218020, (E) CDKAL1-rs981042, and (F) HNF1B-rs3094508 stratified by genotype among LIBCSP women diagnosed with invasive breast cancer in 1996–1997 (n = 817). The x-axis shows times to death in years; the y-axis shows proportion of participants alive
TABLE 3.
Co-dominant genotype model age-adjusted hazard ratios (HRs) with 95% confidence intervals (CIs) for diabetes-related SNPs and mortality following breast cancer in the LIBCSP (n = 817).
| Gene | rs ID |
Genotype | All-cause mortality (n deaths = 340) |
|||
|---|---|---|---|---|---|---|
| Deaths | Censored | HR (95% CI) | Pa | |||
| Location | ||||||
| CDKAL1 | rs981042 | CC | 286 | 434 | 1.00 | |
| Intron | CA | 49 | 41 | 1.48 (1.09–2.01) | ||
| AA | <5 | <5 | – | 0.0032 | ||
| HHEX-EXOC6 | rs1111875 | GG | 137 | 158 | 1.00 | |
| Intergene | GA | 157 | 249 | 0.79 (0.62–0.99) | ||
| AA | 43 | 67 | 0.75 (0.53–1.06) | 0.0361 | ||
| INSR | rs919275 | AA | 112 | 142 | 1.00 | |
| Intron | AG | 159 | 212 | 0.92 (0.72–1.18) | ||
| GG | 66 | 117 | 0.73 (0.54–0.99) | 0.0488 | ||
| Sum of “at-risk” allelesb | 0–2 | 159 | 266 | 1.00 | ||
| 3 | 112 | 140 | 1.27 (1.00–1.62) | |||
| 4–6 | 63 | 65 | 1.52 (1.14–2.04) | 0.0004 | ||
| Breast cancer-specific mortality (n deaths = 139) | ||||||
| Gene | rs ID | Genotype | Deaths | Censored | HR (95% CI) | Pa |
| CDKN2A, CDKN2B | rs3218020 | CC | 58 | 211 | 1.00 | |
| Intron | CT | 65 | 344 | 0.72 (0.51–1.03) | ||
| TT | 16 | 110 | 0.57 (0.33–1.00) | 0.0225 | ||
| CDKAL1 | rs981042 | CC | 116 | 600 | 1.00 | |
| Intron | CA | 22 | 66 | 1.66 (1.05–2.61) | ||
| AA | <5 | <5 | – | 0.0246 | ||
| TCF2, HNF1B | rs3094508 | AA | 62 | 254 | 1.00 | |
| Intron | AG | 65 | 306 | 0.89 (0.63–1.27) | ||
| GG | 12 | 106 | 0.49 (0.26–0.90) | 0.0344 | ||
| Sum of “at-risk” allelesc | 0–2 | 52 | 326 | 1.00 | ||
| 3 | 47 | 236 | 1.25 (0.84–1.85) | |||
| 4–6 | 40 | 99 | 2.26 (1.50–3.42) | 0.0002 | ||
Long Island Breast Cancer Study Project (LIBCSP) women diagnosed with breast cancer between August 1, 1996 and July 31, 1997 and followed-up for vital status through December 31, 2014.
Additive genotype model P values.
The “at-risk” allele was defined as follows: rs981042: A allele; rs1111875: G allele; rs919275: A allele.
The “at-risk” allele was defined as follows: rs3218020: C allele; rs981042: A allele; rs3094508: A allele.
Three SNPs (rs3218020, P = 0.0225; rs981042, P = 0.0246; and rs3094508, P = 0.0344) were significantly associated with BC-specific mortality in additive genotype models (Figure 2). In co-dominant genotype models, the variant (vs common) homozygous genotypes of rs3218020 and rs3094508 were associated with improved BC- specific survival in the Kaplan-Meier survival curves (Figure 3) and with BC-specific mortality Cox model HRs of 0.57 (95% CI = 0.33–1.00) and 0.49 (95% CI = 0.26–0.90), respectively (Table 3). The rs981042- CA (vs CC) genotype was associated with worse BC-specific mortality in the Kaplan-Meier survival curves (Figure 3) and with a BC-specific mortality Cox model HR of 1.66 (95% CI = 1.05–2.61) (Table 3). The highest (vs lowest) category of the summary score of “at-risk” alleles comprised of the sum of the three statistically significant SNPs was associated with a Cox model HR of BC-specific mortality of 2.26 (95% CI = 1.50–3.42, P = 0.0002).
Diabetes status did not modify the associations between rs981042, rs1111875, and rs919275 and all-cause mortality or between rs3218020, rs981042, and rs3094508 and breast cancer- specific mortality, on the multiplicative scale (Table 4).
TABLE 4.
Additive genotype model age-adjusted hazard ratios (HRs) and 95% confidence intervals (CIs) for diabetes-related SNP genotypes and mortality following breast cancer, overall and stratified by diabetes status in the LIBCSP (n = 817)
| Gene | rs ID | Alleles | Overall |
All-Cause Mortality |
PInteractiona | |
|---|---|---|---|---|---|---|
| Diabetes Status | ||||||
| No (n = 758) |
Yes (n = 56) |
|||||
| HR (95% CI) | HR (95% CI) | HR (95% CI) | ||||
| CDKAL1 | rs981042 | C/A | 1.49 (1.14–1.94) | 1.46 (1.08–1.96) | 1.33 (0.75–2.35) | 0.9699 |
| HHEX-EXOC6 | rs1111875 | G/A | 0.84 (0.72–0.99) | 0.85 (0.72–1.01) | 0.90 (0.56–1.44) | 0.7751 |
| INSR | rs919275 | A/G | 0.86 (0.75–1.00) | 0.89 (0.76–1.04) | 0.61 (0.39–0.97) | 0.1618 |
| Sum of “at-risk” allelesb | 1.21 (1.09–1.34) | 1.17 (1.05–1.31) | 1.33 (1.01–1.76) | 0.3962 | ||
| Breast cancer-specific mortality | ||||||
| Diabetes Status | ||||||
| Overall | No (n = 758) | Yes (n = 56) | ||||
| Alleles | HR (95% CI) | HR (95% CI) | HR (95% CI) | PInteractiona | ||
| CDKN2A, CDKN2B | rs3218020 | C/T | 0.74 (0.58–0.96) | 0.76 (0.58–0.99) | 0.58 (0.25–1.39) | 0.4963 |
| CDKAL1 | rs981042 | C/A | 1.61 (1.06–2.44) | 1.58 (1.00–2.48) | 1.61 (0.60–4.28) | 0.8581 |
| TCF2, HNF1B | rs3094508 | A/G | 0.77 (0.60–0.98) | 0.76 (0.59–0.99) | 0.90 (0.41–1.96) | 0.6659 |
| Sum of “at-risk” allelesc | 1.49 (1.21–1.85) | 1.37 (1.15–1.64) | 1.42 (0.82–2.47) | 0.8127 | ||
Long Island Breast Cancer Study Project (LIBCSP) women diagnosed with breast cancer between August 1, 1996 and July 31, 1997 and followed-up for vital status through December 31, 2014
Multiplicative interaction P values.
The “at-risk” allele was defined as follows: rs981042: A allele; rs1111875: G allele; rs919275: A allele.
The “at-risk” allele was defined as follows: rs3218020: C allele; rs981042: A allele; rs3094508: A allele.
4 |. DISCUSSION
In this population-based sample, of the 143 diabetes risk variants genotyped, 12 SNPs were associated with the risk of developing BC, three with all-cause mortality, and three with BC-specific mortality, in additive genotype models at an alpha of 0.05, but none were statistically significant at the Bonferroni-corrected alpha of 0.0003. The top three most significant SNPs associated with BC risk included: rs4876369, an intron variant of SLC30A8 (solute carrier family 30 member 8), which encodes a zinc efflux transporter involved in the accumulation of zinc in intracellular vesicles;35 rs11187146, an intergene variant in the IDE-KIF11-HHEX gene cluster at 10q23.3336; and rs1333049, an intergene variant in the cyclin-dependent kinase inhibitor 2A/B (CDKN2A/B) locus at 9p21.3, hypothesized to play a pivotal role in the development of cardiovascular disease by altering the dynamics of vascular cell proliferation.37,38 ORs per allele increase for the 12 statistically significant SNPs ranged from 1.14 to 1.34 for those positively associated with BC and from 0.81 to 0.83 for those inversely associated with BC risk. Furthermore, diabetes status modified the association between two SNPs (SLC30A- rs4876369 and IRS2-rs2241745) and BC risk. In co-dominant genotype models, the variant homozygous genotypes were associated with ORs ranging from 1.33 to 2.06 for those positively associated and from 0.44 to 0.75 for those inversely associated with breast cancer risk, relative to the common homozygous genotypes. Among the four SNPs inversely associated with breast cancer was rs10830963, an intron of MTNR1B (melatonin receptor 1B), which encodes one of two high affinity forms of a receptor for melatonin, a pineal gland hormone that regulates glucose metabolism by affecting circadian insulin secretion.39 Given that these 12 SNPs consist of intron and intergene variants, their functional impact on BC risk is not clear. These SNPs may be in LD with functionally relevant SNPs not included in our study or they may affect gene function by altering the stability, splicing, or localization of the mRNA.40
In a previous large study of type 2 diabetes susceptibility variants and the risk of developing BC in women of European ancestry, one SNP (TCF7L2-rs7903146) was positively associated and two (FTO- rs9939609 and PRC1-rs8042680) were inversely associated with BC risk.20 In our study, we included rs7903146, an intron variant of TCF7L2 (transcription factor 7 like 2); however, we found no association between this SNP and either risk of developing BC or mortality after BC. It is possible that our study lacked adequate power to detect this association, given that the study by Zhao et al. reported an OR estimate for the risk of developing BC of 1.04 (95% CI = 1.02– 1.06). A second study of type 2 diabetes risk alleles and BC incidence in Caucasian women reported positive associations for rs5945326 and rs1251809 and inverse associations for rs1111875 and rs10923931.21 In contrast to the study by Hou et al., which reported an OR of 0.88 (95% CI = 0.78–0.99) for the C allele of HHEX (hematopoietically expressed homeobox),41 rs1111875 was not associated with risk of developing BC in our study. The rs1111875- A (vs G) allele was, however, inversely associated with all-cause mortality (HR = 0.84, 95% CI = 0.72–0.99) in our study.
For all-cause mortality, we observed that rs981042, an intron variant of CDKAL1 (CDK5 regulatory subunit associated protein 1 like 1), a gene of unknown function,42 was associated with a HR per allele increase of 1.49. rs1111875, an intergene variant located near HHEX,41 and rs919275, an intron variant of INSR (insulin receptor),43 were associated with HRs of 0.84 and 0.86, respectively. For breast cancer-specific mortality, rs981042 and rs3218020, intron variants of CDKAL144 and HNF1B,45 respectively, were associated with HRs per allele of 0.74 and 0.77. One previous study examined type 2 diabetes genetic variants in association with BC survival among 6000 Chinese women.46 The study by Bao et al. examined a gene risk score based on 33 GWAS-identified diabetes risk variants. In their study, there was no association between the gene risk score and subsequent survival among women with breast cancer; however, among women with ER- negative breast cancer, a higher gene risk score was associated with worse overall survival and this association was modified by a history of diabetes.46 In our study, the majority of women with BC were diagnosed with ER-positive BC, which limited our ability to examine effect modification by ER status. Individually, rs7403531 and rs391300 were positively associated with all-cause mortality in their study.46 In contrast to our study, in their study rs4430796 was inversely associated with all-cause mortality. rs2028299 and rs1359790, SNPs not included in our study, were also significantly associated with BC recurrence/mortality.46
Our study had several strengths, including a larger number of SNPs examined than studies published to date, a genetically homogenous population, and the use of existing resources from a population-based study of BC. Additionally, Mendelian randomization, the random assortment of alleles at the time of gamete formation, minimizes the potential for the association between diabetes risk variants and BC to be confounded by environmental factors.34 While several larger studies have examined diabetes risk variants in association with breast cancer risk, to our knowledge, ours is the first study to examine diabetes risk variants in association with BC-specific mortality among US Caucasian women. However, our study was limited by the comparatively smaller sample than studies published to date, which may have limited our ability to detect associations between the SNPs examined here and BC risk/mortality. Second, the large number of statistical tests could have resulted in spurious results; however, we used a targeted approach in selecting diabetes variants of interest and also interpreted our results using a Bonferroni corrected threshold. Third, we relied on GWAS published before November 2007, and thus potentially interesting variants identified since then may have been missed. Furthermore, many of the diabetes SNPs we found to be significantly related to BC risk are intron variants; their role in BC carcinogenesis remains to be clarified. Last, in examining effect modification by diabetes status, we relied on self-reported diabetes and were not able to distinguish between type 1 and type 2 diabetes; however, in the LIBCSP, the majority of women who reported taking diabetes medications (85%) listed medications that are used to treat type 2 diabetes15 and type 2 diabetes is the most common type, accounting for 95% of prevalent cases.1 Furthermore, both type 1 and type 2 diabetes share common predisposing genetic factors as well as the metabolic sequalae hypothesized to influence breast carcinogenesis and progression.47
5 |. CONCLUSION
In summary, genetic polymorphisms that increase the risk of developing diabetes may also increase the risk of developing and dying from BC. Our study helps further clarify the association between diabetes and BC risk and may highlight important biological mechanisms of breast carcinogenesis and progression. The prevalence of diabetes is expected to increase by 54% to more than 54.9 million Americans between 2015 and 2030. Thus, a better understanding of how diabetes impacts breast cancer risk may be important for reducing the high burden of BC in the US.3
Supplementary Material
ACKNOWLEDGMENTS
The authors gratefully acknowledge grant support from the National Cancer Institute and the National Institute of Environmental Health Sciences (UO1 CA/ES66572, UO1 CA66572, and T32 ES007018, and L30 CA220767), and from the Susan G. Komen Foundation (Career Catalyst Award).
Funding information
National Cancer Institute, Grant numbers: L30 CA220767, UO1 CA/ES66572, UO1 CA66572; National Institute of Environmental Health Sciences, Grant number: T32 ES007018; Susan G. Komen Foundation (Career Catalyst Award)
Abbreviations:
- BC
breast cancer
- CDKAL1
CDK5 regulatory subunit associated protein 1 like 1
- CDKN2A/B
cyclin-dependent kinase inhibitor 2A/B
- CI
confidence interval
- ER
estrogen receptor
- GWAS
genome wide association study
- HCFA
health care finance administration
- HHEX
hematopoietically expressed homeobox
- HR
hazard ratio
- HWE
Hardy-Weinberg equilibrium
- IGFBP-1
insulin-like growth factor binding protein-1
- INSR
insulin receptor
- LIBCSP
Long Island Breast Cancer Study Project
- MTNR1B
melatonin receptor 1B
- NDI
National Death Index
- OR
odds ratio
- PR
progesterone receptor
- SHBG
sex hormone binding globulin
- SLC30A8
solute carrier family 30 member 8
- SNP
single nucleotide polymorphism
- TCF7L2
transcription factor 7 like 2
- US
United States
Footnotes
SUPPORTING INFORMATION
Additional supporting information may be found online in the Supporting Information section at the end of the article.
CONFLICTS OF INTEREST
None declared.
REFERENCES
- 1.Centers for Disease Control and Prevention. National Diabetes Statistics Report, 2017 2017. https://www.cdc.gov/diabetes/pdfs/data/statistics/national-diabetes-statistics-report.pdf.
- 2.Vigneri P, Frasca F, Sciacca L, Pandini G, Vigneri R. Diabetes and cancer. Endocr Relat Cancer 2009;16:1103–1123. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018;68:7–30. [DOI] [PubMed] [Google Scholar]
- 4.Larsson SC, Mantzoros CS, Wolk A. Diabetes mellitus and risk of breast cancer: a meta-analysis. Int J Cancer 2007;121:856–862. [DOI] [PubMed] [Google Scholar]
- 5.Giovannucci E, Harlan DM, Archer MC, et al. Diabetes and cancer: a consensus report. Diabetes Care 2010;33:1674–1685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mcpherson K, Steel C, Dixon J. ABC of breast diseases: breast cancerepidemiology, risk factors, and genetics. BMJ 2000;321:624–628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fadaka A, Ajiboye B, Ojo O, Adewale O, Olayide I, Emuowhochere R. Biology of glucose metabolization in cancer cells. J Oncol Sci 2017;3:45–51. [Google Scholar]
- 8.Ish-Shalom D, Christoffersen CT, Vorwerk P, et al. Mitogenic properties of insulin and insulin analogues mediated by the insulin receptor. Diabetologia 1997;40:S25–S31. [DOI] [PubMed] [Google Scholar]
- 9.van der Burg B, Rutteman GR, Blankenstein MA, de Laat SW, van Zoelen EJ. Mitogenic stimulation of human breast cancer cells in a growth factor-defined medium: synergistic action of insulin and estrogen. J Cell Physiol 1988;134:101–108. [DOI] [PubMed] [Google Scholar]
- 10.Ooi GT, Tseng LY, Tran MQ, Rechler MM. Insulin rapidly decreases insulin-like growth factor-binding protein-1 gene transcription in streptozotocin-diabetic rats. Mol Endocrinol 1992;6:2219–2228. [DOI] [PubMed] [Google Scholar]
- 11.Weinstein D, Simon M, Yehezkel E, Laron Z, Werner H. Insulin analogues display IGF-I-like mitogenic and anti-apoptotic activities in cultured cancer cells. Diabetes Metab Res Rev 2009;25:41–49. [DOI] [PubMed] [Google Scholar]
- 12.Wallace IR, McKinley MC, Bell PM, Hunter SJ. Sex hormone binding globulin and insulin resistance. Clin Endocrinol (Oxf) 2013;78: 321–329. [DOI] [PubMed] [Google Scholar]
- 13.Peairs KS, Barone BB, Snyder CF, et al. Diabetes mellitus and breast cancer outcomes: a systematic review and meta-analysis. J Clin Oncol 2011;29:40–46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Barone BB, Yeh H-C, Snyder CF, et al. Long-term all-cause mortality in cancer patients with preexisting diabetes mellitus. JAMA 2008;300: 2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleveland RJ, North KE, Stevens J, Teitelbaum SL, Neugut AI, Gammon MD. The association of diabetes with breast cancer incidence and mortality in the Long Island Breast Cancer Study Project. Cancer Causes Control 2012;23:1193–1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Verlato G, Zoppini G, Bonora E, Muggeo M. Mortality from sitespecific malignancies in type 2 diabetic patients from Verona. Diabetes Care 2003;26:1047–1051. [DOI] [PubMed] [Google Scholar]
- 17.Du W, Simon MS. Racial disparities in treatment and survival of women with stage I-III breast cancer at a large academic medical center in metropolitan Detroit. Breast Cancer Res Treat 2005;91: 243–248. [DOI] [PubMed] [Google Scholar]
- 18.Fleming ST, Pursley HG, Newman B, Pavlov D, Chen K. Comorbidity as a predictor of stage of illness for patients with breast cancer. Med Care 2005;43:132–140. [DOI] [PubMed] [Google Scholar]
- 19.an de Poll-Franse LV, Houterman S, Janssen-Heijnen MLG, Dercksen MW, Coebergh JWW, Haak HR Less aggressive treatment and worse overall survival in cancer patients with diabetes: a large population based analysis. Int J Cancer 2007;120:1986–1992. [DOI] [PubMed] [Google Scholar]
- 20.Zhao Z, Wen W, Michailidou K, et al. Association of genetic susceptibility variants for type 2 diabetes with breast cancer risk in women of European ancestry. Cancer Causes Control 2016;27: 679–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hou N, Zheng Y, Gamazon ER, et al. Genetic susceptibility to type 2 diabetes and breast cancer risk in women of European and African ancestry. Cancer Epidemiol Biomarkers Prev 2012;21:552–556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chen F, Wilkens LR, Monroe KR, et al. No association of risk variants for diabetes and obesity with breast cancer: the Multiethnic Cohort and PAGE studies. Cancer Epidemiol Biomarkers Prev 2011;20: 1039–1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature 2016;536:41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gammon MD, Neugut AI, Santella RM, et al. The Long Island Breast Cancer Study Project: description of a multi-institutional collaboration to identify environmental risk factors for breast cancer. Breast Cancer Res Treat 2002;74:235–254. [DOI] [PubMed] [Google Scholar]
- 25.Parada H, Wolff MS, Engel LS, et al. Organochlorine insecticides DDT and chlordane in relation to survival following breast cancer. Int J Cancer 2016;138:565–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Centers for Disease Control and Prevention. National Death Index http://www.cdc.gov/nchs/ndi.htm. Published 2017. Accessed June 26, 2018.
- 27.Perry JRB, Frayling TM. New gene variants alter type 2 diabetes risk predominantly through reduced beta-cell function. Curr Opin Clin Nutr Metab Care 2008;11:371–377. [DOI] [PubMed] [Google Scholar]
- 28.Zeggini E, Scott LJ, Saxena R, et al. Meta-analysis of genomewide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet 2008;40:638–645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parra EJ, Below JE, Krithika S, et al. Genome-wide association study of type 2 diabetes in a sample from Mexico City and a meta-analysis of a Mexican-American sample from Starr County, Texas. Diabetologia 2011;54:2038–2046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saxena R, Voight BF, Lyssenko V, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 2007;316:1331–1336. [DOI] [PubMed] [Google Scholar]
- 31.Johnson AD, Handsaker RE, Pulit SL, Nizzari MM, O’Donnell CJ, de Bakker PIW. SNAP: a web-based tool for identification and annotation of proxy SNPs using HapMap. Bioinformatics 2008;24:2938–2939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Rothman KJ, Greenland S, Lash TL. Modern Epidemiology 3rd ed Philadelphia, PA: Lippincott, Williams & Wilkins; 2008. [Google Scholar]
- 33.Allison P Survival Analysis Using SAS: A Practical Guide. 2nd ed Cary, NC: 2010. [Google Scholar]
- 34.Smith GD, Ebrahim S. Mendelian randomization: prospects, potentials, and limitations. Int J Epidemiol 2004;30–42. [DOI] [PubMed] [Google Scholar]
- 35.NCBI. SLC30A8 solute carrier family 30 member 8 [Homo sapiens (human)] https://www.ncbi.nlm.nih.gov/gene/169026. Published 2018. Accessed May 17, 2018.
- 36.Liu S, Qian Y, Lu F, et al. Genetic variants at 10q23.33 are associated with plasma lipid levels in a Chinese population. J Biomed Res 2014;28: 53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhao J, Wu X, Nie S, et al. Association of CDKN2B-AS1 rs1333049 with brain diseases: a case-control study and a meta-Analysis. Clin Psychopharmacol Neurosci 2017;15:53–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hannou SA, Wouters K, Paumelle R, Staels B. Functional genomics of the CDKN2A/B locus in cardiovascular and metabolic disease: what have we learned from GWASs? Trends Endocrinol Metab 2015;26: 176–184. [DOI] [PubMed] [Google Scholar]
- 39.Rosta K, Al-Aissa Z, Hadarits O, et al. Association study with 77 SNPs confirms the robust role for the rs10830963/G of MTNR1B variant and identifies two novel associations in gestational diabetes mellitus development. PLoS ONE 2017;12:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tabor HK, Risch NJ, Myers RM. Candidate-gene approaches for studying complex genetic traits: practical considerations. Nat Rev Genet 2002;3:391–397. [DOI] [PubMed] [Google Scholar]
- 41.US National Library of Medicine. Reference SNP (rs) Report: rs1111875 https://www.ncbi.nlm.nih.gov/snp/rs1111875. Published 2018. Accessed May 19, 2018.
- 42.NCBI. CDKAL1 CDK5 regulatory subunit associated protein 1 like 1 [Homo sapiens (human)] 2018. [Google Scholar]
- 43.US National Library of Medicine. Reference SNP (rs) Report: rs 9192 75 [Google Scholar]
- 44.US National Library of Medicine. Reference SNP (rs) Report: rs981042 https://www.ncbi.nlm.nih.gov/snp/rs981042. Published 2018. Accessed May 19, 2018.
- 45.US National Library of Medicine. Reference SNP (rs) Report: rs3218020 https://www.ncbi.nlm.nih.gov/snp/rs3218020. Published 2018. Accessed May 19, 2018.
- 46.Bao P-P, Zhao Z-G, Gao Y-T, et al. Association of type 2 diabetes genetic variants with breast cancer survival among Chinese women. PLoS ONE 2015;10:e0117419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tuomi T Type 1 and type 2 diabetes: what do they have in common? Diabetes 2005;54:S40–S45. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.