Abstract
Background
The human leucine-rich, repeat-containing G protein-coupled receptor 5 (LGR5) is a stem cell marker in numerous adult tissues and is overexpressed in a large number of human carcinoma including colon cancer, breast cancer and oral squamous cell carcinomas (OSCC). The role of the full length transcript (LGR5FL) in progression and prognosis of several cancers was reported. However, the biological function of three splice variants of LGR5 (LGR5Δ5, LGR5Δ8 and LGR5Δ5–8) has yet to be thoroughly investigated.
Methods
Seventy-eight frozen tumor samples from adult OSCC patients were studied using quantitative real-time TaqMan™ PCR analysis. The mRNA levels of full length LGR5, the splice variant of LGR5 lacking exon 5 (LGR5Δ5), the splice variant of LGR5 lacking exon 8 (LGR5Δ8) and the mRNA level of all known transcript variants together (LGR5all) were quantified and correlated to overall and disease-specific survival of OSCC patients, clinical parameters and the mRNA level of different tumor-associated markers.
Results
An elevated level of tumoral LGR5Δ5 mRNA, but not LGR5FL, LGR5Δ8 or LGR5all mRNA was significantly associated with a poor prognosis for the overall and disease-specific survival of OSCC patients (hazard ratio (HR) = 2.0; p = 0.02; 95% CI: 1.1–3.7; HR = 3.2; p = 0.01; 95% CI: 1.3–8.0; multivariable Cox regression), respectively. Additionally, a higher tumoral level of LGR5Δ5 mRNA in primary tumors was associated with the occurrence of regional lymph node metastases in OSCC patients (odds ratio (OR) = 3.1; p = 0.022; 95% CI: 1.2–7.9; binary logistic regression). Furthermore, the mRNA levels of all investigated LGR5 transcript variants were significantly correlated with the mRNA expression of Wnt-target genes and markers of epithelial-to-mesenchymal transition (EMT).
Conclusion
The mRNA level of the LGR5 splice variant LGR5Δ5 is an independent negative prognostic marker for overall and disease-specific survival and metastasis in OSCC patients. Additionally, we suggest, all LGR5 transcript variants are involved in the EMT process mainly through activating the Wnt-signalling pathway.
Keywords: Head and neck squamous cell carcinoma, overall survival, stem cell-associated gene; LGR5; Splice variants; EMT
Background
Cancer of the lip and oral cavity represents the 15th most common cancer worldwide with 410,304 new cases and 146,000 deaths in 2015 [1]. Over 90% of all malignancies in the oral cavity are squamous cell carcinomas (SCC) [2]. Although diagnosis at earlier stage improved outcome of the patients in the last decades the 5-years survival rate of OSCC patients has stagnated at approximately 40–50% despite the advances in the therapeutic techniques [3–5]. Therefore, new therapeutic strategies are needed in order to improve the patient’s recurrence rates and the overall survival and therefore independent molecular biomarkers are necessary which help to estimate the prognosis and the efficacy of an individual therapeutic strategy.
As other cancers, OSCCs show a heterogeneity in their cellular morphology [6]. In general, two models have been established in order to explain the underlying mechanisms of tumor heterogeneity: I) the clonal evolution model and II) the cancer stem cell (CSC) hypothesis [7–9]. At this moment a model where the CSC hypothesis is integrated into the clonal evolution model is favoured. This model postulates that genetically distinct tumor subclones harbour subpopulations of different tumor initiating cells (CSC). These CSCs are capable of self-renewal and drive tumor growth, recurrence and metastasis as well as the resistance to therapeutic approaches [10]. Therefore, it is necessary to introduce therapies that target not only the rapidly proliferating tumor cells of the tumor mass, but especially the tumor initiating cells in order to avoid therapeutic failures [11]. New molecular markers which accurately identify CSC cells are essential for those new therapeutic options.
One such candidate molecular marker is the leucine-rich repeat-containing G protein-coupled receptor LGR5, a member of the G-protein-coupled receptor family of proteins and a target of Wnt signalling [12]. It was initially identified as a marker of murine small intestinal and colon stem cells [13]. LGR5 was found to be overexpressed in colorectal cancer [14–18] and several studies indicated that LGR5 expression is associated with colorectal carcinogenesis, tumor growth and metastasis [18–20]. Subsequent studies demonstrated that LGR5 is expressed by a diverse range of adult tissues and organs and acts as a biomarker for adult stem cells in certain tissues including oral tissues [21, 22]. Furthermore, LGR5 was found to be overexpressed in several carcinomas having a close association with initiation and recurrence of different cancer types and correlating with tumor growth, invasion and poor prognosis [18, 20, 23, 24].
Functionally, LGR5 is a part of Wnt signalling complex on the cell membrane, where it appears to be able to enhance the activity of the Wnt/ß-catenin signalling [12]. Thus, LGR5 is a target gene of Wnt signalling, but because of its function also an enhancer of this Wnt signalling in the sense of a positive feedback loop. To our knowledge, three transcript variants of LGR5 have been described until now: one lacking exons 5–8 (LGR5Δ5–8) as introduced by Osawa et al. [25], the second lacking exon 8 (https://www.uniprot.org/uniprot/O75473) and the third lacking exon 5 (LGR5Δ5), which has been previously described by our group [26]. All three variants have a truncated ligand binding domain [27]. In LGR5FL this ligand binding domain interacts with R-Spondins 1–4 resulting in the formation and internalization of a LGR5-RSPO-RNF43 protein complex, leading to a RNF43 membrane clearance, which results in a higher Wnt-activity. However, the functional effects of the truncated ligand binding domain of the LGR5 transcript variants have not been investigated yet.
Recently, the LGR5 protein expression was investigated by immunohistochemistry (IHC) in OSCC, demonstrating an increased LGR5 expression correlating with disease severity but not with patient’s outcome [22]. In a previous work our group demonstrated the mRNA expression of the LGR5 transcript variant LGR5Δ5, but not the expression of full length LGR5 (LGR5FL) being an independent unfavourable prognostic marker for soft tissue sarcoma patients (STS) [26]. Though to date it is not possible to differentiate between the LGR5 isoforms by IHC due to the lack of specific antibodies we were interested whether the transcript variants of LGR5 (LGR5Δ5 or LGR5Δ8) in specific may affect the outcome of OSCC patients.
Therefore, we separately measured the mRNA level of LGR5FL, LGR5Δ5, LGR5Δ8 and of all four known LGR5 variants together (LGR5all) in 78 OSCC samples and correlated them with clinical parameters and the outcome of those patients.
Methods
Tissue samples, histomorphological data and study approval
Frozen primary tumor samples of 78 OSCC patients were analysed using the real-time quantitative PCR (qRT-PCR) analysis. All patients had been treated with surgery at the Department of Oral and Maxillofacial Plastic Surgery, Martin Luther University Halle-Wittenberg, Germany. The tissue samples were cut by a cryocut microtome and the first and the last histologic sections were stained with hematoxylin and eosin. Experienced pathologists (UB, DB) verified the sections. We defined samples as tumor tissue when > 70% of the first and the last histologic sections were tumor tissue. The clinical and histomorphological parameters of OSCC patients are shown in Table 1. The patients’ median age at the time of the diagnosis was 58.5 years (ranging from 25 to 90 years). Forty-nine OSCC patients (63%) died after an average time of 15.4 months (ranging from 0 to 56 months), and 29 OSCC patients (37%) were still alive after an average follow-up time of 44.9 months (ranging from 0 to 81 months). The study was carried out in compliance with the Helsinki Declaration, and it was approved by the Ethics Committee of the Medical Faculty of the University Halle (Ethical registry 210/19.08.09/10). All patients gave written informed consent (Department of Oral and Maxillofacial Plastic Surgery, University of Halle-Wittenberg, Germany).
Table 1.
Clinical, histopathological and survival data
| Parameters | Total (n = 78) | relative LGR5FL mRNA-level ≤134.3 > 134.3 |
relative LGR5Δ5 mRNA-level ≤2,9 > 2,9 |
relative LGR5Δ8 mRNA-level ≤14.9 > 14.9 |
relative LGR5all mRNA-level ≤8449.3 > 8449.3 |
||||
|---|---|---|---|---|---|---|---|---|---|
| 39 | 39 | 39 | 39 | 39 | 39 | 39 | 39 | ||
| Gender | p = 0.15 | p = 0.15 | p = 0.15 | p = 0.15 | |||||
| Male | 63 | 29 | 34 | 29 | 34 | 29 | 34 | 29 | 34 |
| Female | 15 | 10 | 5 | 10 | 5 | 10 | 5 | 10 | 5 |
| Tumor grade | p = 0.54 | p = 0.58 | p = 0.027* | p = 0.20 | |||||
| I | 9 | 6 | 3 | 4 | 5 | 7 | 2 | 4 | 5 |
| II | 57 | 27 | 30 | 30 | 27 | 29 | 28 | 32 | 25 |
| III | 11 | 5 | 6 | 4 | 7 | 2 | 9 | 3 | 8 |
| unknown | 1 | 1 | 0 | 1 | 0 | 1 | 0 | 0 | 1 |
| Tumor stage | p = 0.77 | p = 0.43 | p = 0.36 | p = 0.25 | |||||
| I | 13 | 8 | 5 | 8 | 5 | 6 | 7 | 9 | 4 |
| II | 25 | 13 | 12 | 14 | 11 | 12 | 13 | 11 | 14 |
| III | 9 | 4 | 5 | 5 | 4 | 7 | 2 | 6 | 3 |
| IV | 31 | 14 | 17 | 12 | 19 | 14 | 17 | 13 | 18 |
| Patients at last follow-up | p = 0.48 | p = 0.035* | p = 0.81 | p = 0.48 | |||||
| Alive | 29 | 16 | 13 | 19 | 10 | 14 | 15 | 16 | 13 |
| Dead | 49 | 23 | 26 | 20 | 29 | 25 | 24 | 23 | 26 |
| Recurrence | p = 0.289 | p = 0.933 | p = 0.289 | p = 0.464 | |||||
| Yes | 25 | 15 | 10 | 13 | 12 | 15 | 10 | 14 | 11 |
| No | 51 | 24 | 27 | 26 | 25 | 24 | 27 | 24 | 27 |
| unknown | 2 | 0 | 2 | 0 | 2 | 0 | 2 | 1 | 1 |
| Lymph node status | p = 0.06 | p = 0.02* | p = 0.64 | p = 0.06 | |||||
| N0 | 30 | 19 | 11 | 20 | 10 | 16 | 14 | 19 | 11 |
| N1–3 | 48 | 20 | 28 | 19 | 29 | 23 | 25 | 20 | 28 |
| Distant metastases | p = 0.3 | p = 0.3 | p = 0.3 | p = 1.0 | |||||
| M0 | 74 | 38 | 36 | 38 | 36 | 36 | 38 | 37 | 37 |
| M1 | 4 | 1 | 3 | 1 | 3 | 3 | 1 | 2 | 2 |
| Survival analysis: Overall survival | |||||||||
| Kaplan-Meier analysis | p = 0.21 | p = 0.004* | p = 0.88 | p = 0.063 | |||||
| Median survival (months) | 27 | 17 | 42 | 14 | 25 | 19 | 42 | 15 | |
| 95% CI | 6.0–48.0 | 12.5–21.5 | 22.3–61.7 | 5.6–22.4 | 15.2–34.8 | 8.5–29.5 | 22.4–61.6 | 8–22.0 | |
| Univariable Cox regression | p = 0.22 | p = 0.005* | p = 0.88 | p = 0.069 | |||||
| Hazard ratio | 1.4 | 2.3 | 1.1 | 1.7 | |||||
| 95% CI | 0.8–2.5 | 1.3–4.0 | 0.6–1.8 | 0.9–3.0 | |||||
| Multivariable Cox regression | p = 0.2 | p = 0.02* | p = 0.66 | p = 0.09 | |||||
| Hazard ratio | 1.5 | 2.0 | 1.1 | 1.7 | |||||
| 95% CI | 0.8–2.7 | 1.1–3.7 | 0.6–2.1 | 0.9–3.0 | |||||
| Survival analysis: Disease specific survival | |||||||||
| Kaplan-Meier analysis | p = 0.22 | p = 0.001* | p = 0.40 | p = 0.018 | |||||
| Median survival (months) | n.c. | 17 | n.c. | 14 | n.c. | 19 | n.c. | 15 | |
| 95% CI | 14.0–88 | 0–47 | 7.8–104 | 0.8–111 | |||||
| Univariable Cox regression | p = 0.22 | p = 0.002* | p = 0.4 | p = 0.023* | |||||
| Hazard ratio | 1.6 | 3.7 | 1.38 | 2.5 | |||||
| 95% CI | 0.75–3.4 | 1.6–8.6 | 0.65–2.95 | 1.13–5.5 | |||||
| Multivariable Cox regression | p = 0.31 | p = 0.01* | p = 0.57 | p = 0.071 | |||||
| Hazard ratio | 1.5 | 3.2 | 1.29 | 2.2 | |||||
| 95% CI | 0.67–3.6 | 1.3–8.0 | 0.54–3.1 | 0.94–5.0 | |||||
*p-value ≤0.05 indicates statistical significance. n.c.- not calculable
Quantitative RT-PCR
Total RNA of the frozen tissue samples was extracted using the Trizol reagent (Invitrogen, Karlsruhe, Germany) and 6 μg was used to synthesize cDNA using the RevertAid™ H Minus First Strand cDNA Synthesis Kit (Fermentas, St.Leon-Rot,Germany) according to manufacturer’s instructions. The mean 260/280 value over all samples was determined to be 1.93 (standard deviation SD = 0.06), the mean 230/260 to be 2.04 (SD = 0.16). Real-time quantitative PCR analysis (qRT-PCR) was performed in duplicate on a Rotorgene RG-6000 (LTF, Wasserburg, Germany) using TaqMan™ assays (ABI) for LGR5all (including all known LGR5 transcript variants) OPN, MMP7, TWIST1, NANOG, Oct3/4, SNAI1, P4HA1, ZEB2, TGFβ, CTGF, RSPO1, RNF43, IGF2, Vimentin and RPII, which was used as endogenous control.
qRT-PCR reactions for LGR5FL, LGR5Δ5 and LGR5Δ8 transcript variants were performed using the Biozym Blue Probe qPCR Mix (Biozym) according to manufacturer’s instructions and the primer/probe sets: LGR5FL primer forward 5`-AAACCTCTCCAGCTTGGTAG-3`, primer reverse 5`-CGACCTGATATTGTTGCTATGAAATC-3`, probe 5`-FAM-CCTGGGAAAGAAATGCTTTGATGGGC-BHQ1–3`; LGR5Δ5 primer forward 5`-GCCTTCAATCCCTACATTTC-3`, primer reverse 5`-CGACCTGATATTGTTGCTATGAAATC-3`, probe 5`-FAM-CCTGGGAAAGAAATGCTTTGATGGGC-BHQ1–3`; LGR5Δ8 primer forward 5`-CCAACCTTAAAGAACTACATTTC-3`, primer reverse 5`-AGGTAAATGTTGAAAAGCAG-3`, probe 5`-FAM-TGACAATCCCATCCAGTTTGTTGG-MGB-3`.
The results were normalized to RPII transcripts amount and expressed as ΔΔCt [28]. For the analysis the patients cohort was subdivided in two groups according to the LGR5FL, LGR5Δ5, LGR5Δ8 and LGR5all median mRNA levels. An elevated expression of LGR5FL was determined as a median relative transcript level of > 134.3 LGR5FL mRNA / RPII mRNA, of LGR5Δ5 as a median relative transcript level of > 2.9 LGR5Δ5 mRNA / RPII mRNA, of LGR5Δ8 as a median relative transcript level of > 14.9 LGR5Δ8 mRNA / RPII mRNA and of LGR5all as a median relative transcript level of > 8449 relative LGR5all mRNA level/relative RPII mRNA level.
LGR5 immunohistochemistry
For immunohistochemistry (IHC), the LGR5 mAb LS-C105455 (LifespanBioscience) was used. Tissue samples were deparaffinized with xylol and transferred via alcohol into aqua dest (Elix 5 Filter System, Merck-Millipore). Antigen decloaking was performed by steaming the slides with a preheated T-EDTA buffer (ZUC029–500, 1:10 dissolved, Zytomed Systems) at pH 6.0 and 98 °C for 30 min in an oven (Braun, type 3216). After cooling down for 20 min and rinsing with aqua dest, slides were blocked for 7–10 min with 3% H2O2. Following another rinsing step and application of washing buffer (ZUC202–2500, 1:20 solution, Zytochem Plus HRP Kit / Plus Polymer System, Zytomed) the LGR-5 mAb at a dilution of 1:400 was added dropwise on the tissue area and incubated for 30 min at room temperature (RT). Following a washing step, the slides were incubated with a biotinylated secondary antibody (Broad Spectrum, Zytochem Plus HRP Kit, Zytomed) for 15 min at room temperature, rinsed with washing buffer followed by 15 min of incubation with horse radish peroxidase (HRP; Zytochem Plus HRP, Zytomed). The epitopes were visualized with DAB (10 min of DAB Substrate Kit, Zytomed). After further rinsing steps (aqua dest.), the slides were counterstained with hemalaun (Dr. K. Hollborn & Sons) for 30 s, rinsed in water for 10 min, then transferred via alcohol into xylol and finally cover-slipped (Eukitt, ORSAtec) for bright field analysis.
Statistical analyses
The association between the LGR5FL, LGR5∆5, LGR5∆8 and LGR5all expression level and clinicopathological parameters was analysed by χ2 – test. The association between overall and disease-specific survival and LGR5FL, LGR5Δ5, LGR5∆8 und LGR5all transcript variants mRNA levels was analysed by the log-rank test. Survival statistics were performed employing a multivariable Cox proportional hazard regression adjusted for gender, tumor staging, tumor grading and regional lymph node metastases. Receiver operating characteristic (ROC) curves, area under the curve (AUC) of the ROC and the cut-off point were calculated to determine the impact of tumoral mRNA levels of LGR5FL, LGR5Δ5, LGR5∆8 und LGR5all transcript variants in patients with lymph node metastases versus patients without lymph node metastases. The association between the tumoral mRNA level of LGR5Δ5 and the occurrence of lymph node metastases was tested by binary logistic regression with reporting of odds ratio (OR). Spearman’s correlation was used to assess the association between the mRNA levels of LGR5FL, LGR5Δ5, LGR5∆8 und LGR5all transcript variants and a panel of tumor-associated markers which were analysed from the same RNA sample. Significance was defined by a p value of less than 0.05. For Spearman’s correlation Bonferroni corrected significance level (0.05/15) was used to cater for multiple comparisons. The follow-up time was calculated from the day of diagnosis until the day of last follow-up. The overall survival time and the disease-specific survival time were calculated from the day of diagnosis until the time of death (any reason) or until time of disease specific death of the patients.
Results
Expression of LGR5Δ5 but not LGR5FL or LGR5Δ8 is associated with poor clinical outcome in OSCC patients
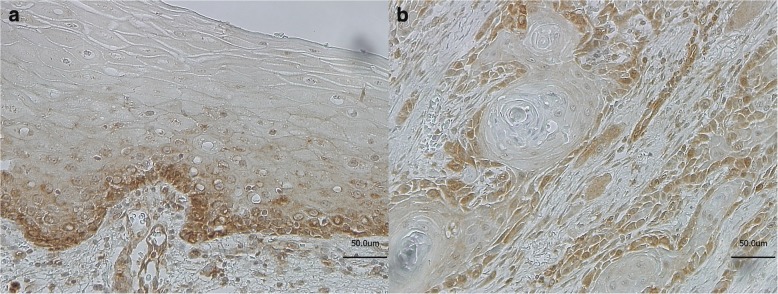
The expression of LGR5 in normal, dysplasia-free oral mucosa (Fig. 1a) and OSCC (Fig. 1b) was analysed by immunohistochemistry. In normal oral mucosa the LGR5 expression was restricted to the stratum basale. In the neoplastic epithelium of the OSCCs the LGR5 expression was diffusely expressed throughout the tumor mass except the keratinized central areas. A differentiation between the LGR5 isoforms originating from the different transcript variants by immunostaining was not possible due to the lack of specific antibodies.
Fig. 1.
Immunostaining showing expression of LGR5 examplary in normal oral mucosa, (morphologically normal, tumor adjacent mucosa) (a) and in OSCC (b)
For the survival analysis, the mRNA levels of LGR5all, LGR5FL and the LGR5 transcript variants LGR5Δ5 and LGR5Δ8 were analysed in 78 OSCC samples and the OSCC patients’ cohort was separated into two cohorts (high and low) based on the median expression level of the LGR5all, LGR5FL, LGR5Δ5 and LGR5Δ8 mRNA. The median relative LGR5FL mRNA level of 78 OSCC samples was 134.3 (ranging from 0.5–5021; mean 391.4); the median relative LGR5Δ5 mRNA level was 2.9 (ranging from 0 to 254.9; mean 15.01); the median relative LGR5Δ8 mRNA level was 14.9 (ranging from 0 to 400; mean 54.3) and the median relative LGR5all mRNA level was 8449 (ranging from 546.4–310,418; mean 30,059), respectively.
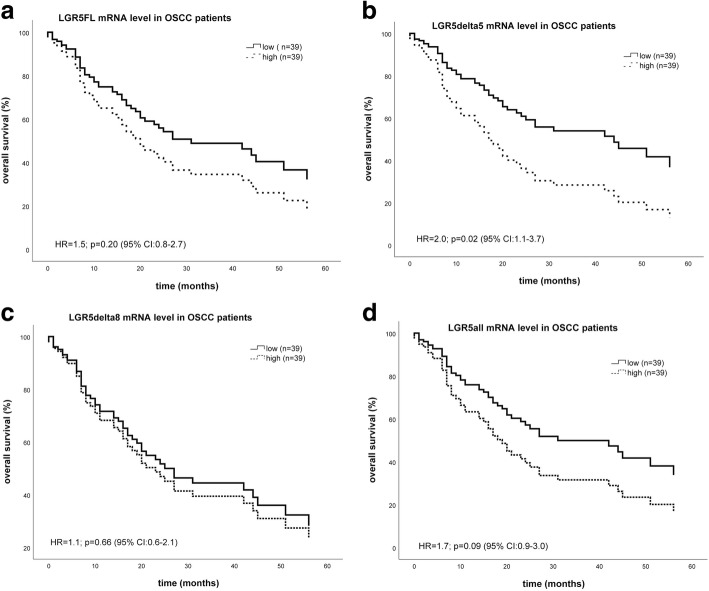
Kaplan-Meier analysis revealed a significant correlation of LGR5∆5 mRNA expression with overall survival. OSCC patients with high tumoral mRNA levels of LGR5Δ5 died on median 28 months earlier (median 14 +/− 8.4 months) as compared to patients with lower tumoral mRNA levels of LGR5Δ5 (median 42 +/− 19.7 months) (p = 0.004) (Table 1). Multivariable Cox proportional hazard regression (confounding factors: gender, staging, grading and regional lymph node metastases) revealed that LGR5Δ5 mRNA level was an independent prognostic factor (p = 0.02) for overall survival with hazard ratio of 2.0 (95% CI: 1.1–3.7) (Table1; Fig. 2b). The mRNA level of LGR5FL, LGR5Δ8 or LGR5all was not associated with overall survival of OSCC patients (Table 1; Fig. 2 a, c, d). The disease-specific survival is associated with the expression of variant LGR5Δ5 in univariable and multivariable Cox analyses and the data regarding the survival analysis are recorded in Table 3.
Fig. 2.
Multivariable Cox hazard regression for LGR5FL (a), LGR5Δ5 (b), LGR5Δ8 (c) and LGR5all (d) mRNA expression level and overall survival in OSCC patients. Relative expression level of LGR5FL, LGR5Δ5, LGR5Δ8 or LGR5all mRNA in 78 OSCC tumor samples was correlated with overall survival. Regarding confounding factors, the Cox model was adjusted to patients’ gender, tumor stage, tumor grading and the occurrence of regional lymph node metastases. The high and low cut-off values for: a. LGR5FL were > 134.3 and ≤ 134.3 LGR5FL mRNA level (HR = 1.5, p = 0.2; CI: 0.8–2.7). b. LGR5Δ5 were > 2.9 and ≤ 2.9 LGR5Δ5 mRNA level (HR = 2.0, p = 0.02; CI: 1.1–3.7). c. LGR5Δ8 were > 14.9 and ≤ 14.9 LGR5Δ8 mRNA level (HR = 1.1, p = 0.66; CI: 0.6–2.1). d. LGR5all were > 8449.3 and ≤ 8449.3 LGR5all mRNA level (HR = 1.7, p = 0.09; CI: 0.9–3.0)
LGR5Δ5 expression in OSCC is correlated with the occurrence of lymph node metastases
The association between LGR5FL, LGR5∆5, LGR5∆8 and LGR5all mRNA levels and clinicopathological parameters was analysed by χ2 – test and results are summarized in Table 1. OSCC cases were subdivided in two groups according to the LGR5FL, LGR5Δ5, LGR5Δ8 and LGR5all median mRNA levels (high and low). High LGR5∆5 mRNA level was found to correlate with the occurrence of lymph node metastases with an odds ratio (OD) of 3.1 (p = 0.022; 95% CI: 1.2–7.9) whereas a high LGR5∆8 mRNA level is associated with a higher tumor grade (p = 0.027) (Table 1). In these cases with lymph node involvement (n = 48) the median level of LGR5∆5 mRNA in the primary lesions was 2-fold higher (4.05 vs. 1.99) when compared with those cases without lymph node metastasis (n = 30). For further analysis of the association between LGR5∆5 mRNA level within the primary tumor lesions and the involvement of regional lymph nodes, ROC curves were constructed by calculating the sensitivities and specificities of the LGR5∆5 mRNA levels to distinguish between the primary tumors with and without lymph node metastasis The cut-off point was 4.2 relative LGR5Δ5 mRNA level (determined by the highest Youden value) at the sensitivity of 50% and a specificity of 83% with a corresponding AUC (area under the curve) of 63.8% (p = 0.041; 95% CI: 51.5–76.2%) (Fig. 3). In contrary to these data, the mRNA levels of LGR5FL, LGR5Δ8 or LGR5all within the primary carcinoma was not associated with the occurrence of lymph node metastases (Table 1).
Fig. 3.
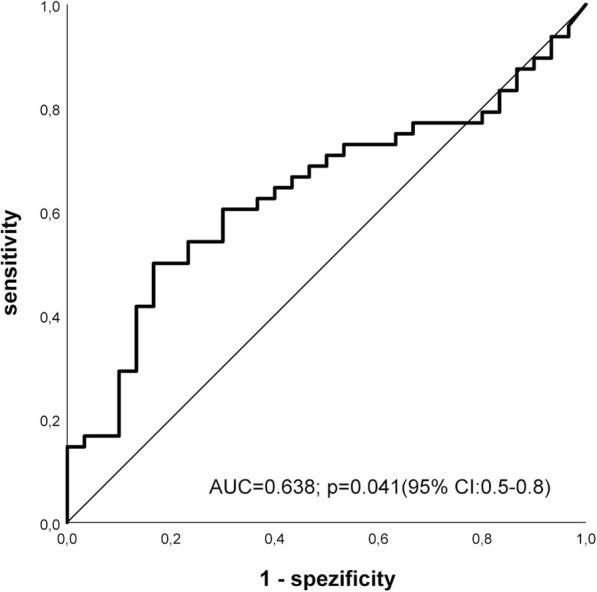
ROC curve demonstrating the sensitivity and specificity of the LGR5∆5 intratumoral mRNA level of patients who developed regional lymph node metastases compared to patients without metastasis. The cut-off point was 4.2 relative LGR5Δ5 mRNA level with a corresponding AUC (area under the curve) = 63.8% (p = 0.041; 95% CI: 0.5–0.8).
LGR5 transcript variants expression correlates with the expression of Wnt-target genes involved in EMT
Analyses according to Spearman-Rho (Table 2) revealed after Bonferroni correction a significant correlation between LGR5 transcript variants mRNA expression and the mRNA expression of tumor-associated markers. The mRNA levels of all LGR5 transcripts (LGR5FL, LGR5Δ5, LGR5Δ8 and LGR5all) were positively associated with the mRNA level of Vimentin. The LGR5FL, LGR5all and LGR5Δ5 mRNA levels were found to be positively correlated with TCF-7 mRNA. Additionally the mRNA level of LGR5FL was positively associated with the TWIST1, ZEB2, MMP7, TGFß, P4HA1, CTGF and IGF2 mRNA. While the LGR5all mRNA correlated with the mRNA level of TWIST1, MMP7 and NANOG. Furthermore, the mRNA level of LGR5Δ5 correlated positively with the ZEB2, CTGF and IGF2 mRNA. Moreover, LGR5Δ8 mRNA level is positively associated with mRNA level of OCT3/4. However, there was no association between the mRNA level of the LGR5 transcript variants and the mRNA levels of RNF43 and RSPO1, which are both directly involved in the LGR5 signalling or the mRNA level of SNAI1, an EMT-`mastermind´ (Table 2).
Table 2.
Correlations between the LGR5 transcripts mRNA levels and the mRNA levels of different biomarkers
| LGR5FL/RPII | LGR5Δ5/RPII | LGR5Δ8/RPII | LGR5all/RPII | ||
|---|---|---|---|---|---|
| EMTmarkers | |||||
| TWIST1 | rs | 0.379 | 0.258 | 0.177 | 0.350 |
| p-value | 0.001* | 0.031 | 0.143 | 0.003 | |
| n | 70 | 70 | 70 | 70 | |
| SNAI1 | rs | 0.320 | 0.330 | 0.228 | 0.158 |
| p-value | 0.007 | 0.005 | 0.057 | 0.191 | |
| n | 70 | 70 | 70 | 70 | |
| ZEB2 | rs | 0.450 | 0.456 | 0.270 | 0.312 |
| p-value | < 0.0001* | < 0.0001* | 0.018 | 0.006 | |
| n | 76 | 76 | 76 | 76 | |
| MMP7 | rs | 0.373 | 0.288 | 0.308 | 0.363 |
| p-value | 0.001* | 0.016 | 0.01 | 0.002* | |
| n | 70 | 70 | 70 | 70 | |
| TCF7 | rs | 0.581 | 0.490 | 0.337 | 0.553 |
| p-value | < 0.0001* | < 0.0001* | 0.004 | < 0.0001* | |
| n | 70 | 70 | 70 | 70 | |
| Vimentin | rs | 0.594 | 0.624 | 0.454 | 0.402 |
| p-value | < 0.0001* | < 0.0001* | < 0.0001* | 0.001* | |
| n | 70 | 70 | 70 | 70 | |
| TGFβ | rs | 0.402 | 0.305 | 0.285 | 0.261 |
| p-value | < 0.0001* | 0.007 | 0.013 | 0,023 | |
| n | 76 | 76 | 76 | 76 | |
| Stem cell markers | |||||
| NANOG | rs | 0.187 | 0.137 | 0.219 | 0.383 |
| p-value | 0.121 | 0.259 | 0.068 | 0.001* | |
| n | 70 | 70 | 70 | 70 | |
| Oct3/4 | rs | 0.251 | 0.216 | 0.377 | 0.207 |
| p-value | 0.036 | 0.073 | 0.001* | 0.086 | |
| n | 70 | 70 | 70 | 70 | |
| Proteins involved in the metastatic process | |||||
| P4HA1 | rs | 0.390 | 0.236 | 0.276 | 0.167 |
| p-value | < 0.0001* | 0.037 | 0.014 | 0.144 | |
| n | 78 | 78 | 78 | 78 | |
| CTGF | rs | 0.492 | 0.469 | 0.302 | 0.301 |
| p-value | < 0.0001* | < 0.0001* | 0.012 | 0.013 | |
| n | 68 | 68 | 68 | 68 | |
| IGF2 | rs | 0.498 | 0.485 | 0.316 | 0.271 |
| p-value | < 0.0001* | < 0.0001* | 0.009 | 0.025 | |
| n | 68 | 68 | 68 | 68 | |
| OPN | rs | 0.281 | 0.240 | 0.301 | 0.079 |
| p-value | 0.020 | 0.049 | 0.013 | 0.522 | |
| n | 68 | 68 | 68 | 68 | |
| Wnt signaling modulating genes | |||||
| RNF43 | rs | −0.129 | −0.171 | −0.204 | −0.123 |
| p-value | 0.287 | 0.158 | 0.090 | 0.311 | |
| n | 70 | 70 | 70 | 70 | |
| RSPO1 | rs | 0.097 | 0.131 | 0.040 | 0.051 |
| p-value | 0.427 | 0.284 | 0.743 | 0.678 | |
| n | 69 | 69 | 69 | 69 | |
Bivariable Spearman’s Rho test. rs: correlation coefficient. The underlined genes have been shown to be Wnt- target genes. P-values were adjusted by the Bonferroni correction.* p-value ≤0.003 indicates statistical significance
Discussion
In this study, we demonstrated that an elevated LGR5Δ5 mRNA level is an independent negative prognostic marker for overall and disease-specific survival and is associated with the occurrence of regional lymph node metastases in OSCC patients while LGR5FL, LGR5Δ8 as well as LGR5all mRNA levels have no prognostic and predictive impact.
An association between an elevated LGR5 expression and unfavourable outcome has been reported for several tumor entities. In glioblastoma the number of the LGR5 expressing cells increased with the tumor staging and correlated with poor outcome [29]. In lung cancer and colon cancer an elevated LGR5 expression was found to correlate with tumor-size, tumor-stage, metastasis and poor outcome [16, 18, 23, 30]. While in gastric carcinoma, a high LGR5 expression correlated with lymphatic invasion but not with the risk of regional lymph node metastasis [31]. In the presented study, we found a significant correlation between LGR5Δ5 mRNA level (p = 0.006) and the occurrence of regional lymph node metastases but not for the other LGR5 transcript variants (LGR5FL and LGR5Δ8) and lymph node involvement in OSCC (Table 1). Furthermore, only the LGR5Δ5 splice variant but not LGR5FL or LGR5Δ8 mRNA level have a prognostic value for OSCC patients. In contrast another study in OSCC showed that the LGR5 expression increased during the process of the malignant transformation but there was no association between the LGR5 protein expression and other clinical parameters [22]. However, these investigators performed LGR5 immunohistochemistry and therefore a discrimination between splice variant products of LGR5 was not possible.
Concerning the LGR5 splice variants only one study analysed the functional difference between LGR5FL and the LGR5 transcript variants LGR5Δ5 and LGR5Δ5–8 in respect to cell proliferation. In that study the scientists observed a higher activation of the Wnt signalling together with a higher proliferative ability upon overexpression of both LGR5 splice variants compared with the cells which overexpressed only LGR5FL in colorectal cancer cells [25].
Several studies indicate an association between LGR5 expression and the expression of other Wnt-target genes, e.g., ß-catenin [29, 32–34]. In our study, we found a positive correlation between the mRNA levels of different LGR5 transcript variants and the mRNA levels of Wnt-target genes MMP7, TCF7, TWIST1, Vimentin, NANOG, OCT3/4 and ZEB2 was seen. Most of these Wnt-target genes are linked to the epithelial-to-mesenchymal transition (EMT). TWIST and ZEB are the ‘mastermind’ genes of the EMT [35] while MMP7, Vimentin, NANOG, OCT3/4 and TGFß are also involved in the EMT. According to this finding, for hepatocellular carcinoma it was reported that LGR5 promotes metastasis through inducting EMT [36]. Moreover, NANOG and OCT3/4 are the key regulators of self-renewal in stem cells [37]. Osawa et al. described that the LGR5FL expression was restricted to stem cells of the crypts while the expression of the LGR5 splice variants (LGR5Δ5 and LGR5Δ5–8) was also seen in the middle and the tips of the villi of the small intestine and was associated with a higher proliferative ability [25]. Furthermore, in our study the mRNA level of the LGR5 transcript variants were positively correlated with the mRNA level of CTGF, P4HA1, and IGF2, as all of these genes are linked to metastasis in cancer [38, 39]. Altogether, our data show that LGR5 is strongly associated with an enhanced Wnt signalling pathway and we suggest that the induction of the EMT program could be mediated by LGR5.
Although the mRNA expression of genes involved in EMT induction and metastasis is associated with the mRNA expression of all investigated LGR5 transcript variants in this study, only OSCC patients with a high tumoral LGR5Δ5 level have a significant higher risk of regional lymph node metastasis. We hypothesize that this finding might be due to the induction of the EMT program combined with a higher proliferative ability [25] of the LGR5Δ5 overexpressing tumor cell, which results in a faster tumor growth and progression and leads to a shorter overall survival of OSCC patients.
Conclusion
The data presented in this study show that an elevated mRNA level of the LGR5 splice variant LGR5Δ5 is an independent negative prognostic factor for OSCC patients as well as correlates with the risk of lymphatic metastasis. Moreover, we suggest that LGR5 is involved in the EMT process and postulate that this happens predominantly through the activation of the Wnt signalling. Thus our results indicate that LGR5 might be involved in tumor progression and metastasis of OSCCs. An elevated expression of the LGR5 splice variant LGR5Δ5 could be used as a potential prognostic biomarker marking an unfavorable prognosis but has to be analyzed in prospective studies for its application as therapeutic biomarker in OSCC patients.
Acknowledgements
We thank our colleagues from the Department of Oral and Maxillofacial Plastic Surgery and the Department of Radiotherapy for contributing to this study and for their continuous support. DB would like to thank the histology laboratory of the institute of pathology and in particular Jana Beer for their comprehensive technical assistance in IHC analysis.
Funding
S.R. and M.K. were supported by the Wilhelm-Roux-Program of BMBF/NBL3 (FKZ: 16/18, 21/25) (purchase of consumables and microarray kits). We acknowledge the financial support of the Open Access Publication Fund of the Martin Luther University Halle-Wittenberg.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AUC
Area under the curve
- CSC
Cancer stem cell, ROC, Receiver operating characteristic
- CTGF
Connective tissue growth factor
- EMT
Epithelial to Mesenchymal Transition
- HR
Hazard ratio; 95% Cl, 95% confidence interval
- IGF2
Insulin like growth factor 2
- LGR5
Leucine-rich repeat-containing G protein-coupled receptor
- MMP7
Matrix metallopeptidase 7
- NANOG
Nanog homeobox
- Oct3/4
POU class 5 homeobox 1
- OPN
Osteopontin
- OSCC
Oral squamous cell carcinoma
- P
Probability
- P4HA1
Prolyl 4-hydroxylase subunit alpha 1
- SNAI1
Snail family transcriptional repressor 1
- TCF7
Transcription factor 7
- TGFβ
Transforming growth factor beta 1
- TWIST1
Twist family bHLH transcription factor 1
- ZEB2
Zinc finger E-box binding homeobox 2
Authors’ contributions
SR, HT and MK designed the study, collected data and performed statistical analysis. MB, TG, BA, CW and DV made substantial contributions regarding the acquisition and interpretation of data. TK, JK, AG, HW performed experimental procedures. DB identified tumor and normal tissue in the frozen sections and performed the immunohistochemistry. UB verified the sections of the tissue samples. AW performed and supervised the statistical analysis. AWE treated the patients, collected material and data and was involved in the statistical interpretation of the data. SR, HT, DB and MK drafted the manuscript. All authors contributed to the critical revision and final approval of the version to be published.
Ethics approval and consent to participate
Ethical registry 210/19.08.09/10 was obtained from the Ethics Committee of the Medical Faculty of the University Halle. All patients gave written informed consent (Department of Oral and Maxillofacial Plastic Surgery, University of Halle-Wittenberg, Germany).
Consent for publication
Not applicable.
Competing interests
All the authors declare that they have no competing interests.
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Contributor Information
Swetlana Rot, Phone: +49 (0) 345 557 7427, Email: swetlana.rot@uk-halle.de, Email: swetlana.rot@medizin.uni-halle.de.
Tom Kaune, Email: tom.kaune@uk-halle.de.
Helge Taubert, Email: Helge.Taubert@uk-erlangen.de.
Thomas Greither, Email: thomas.greither@medizin.uni-halle.de.
Johanna Kotrba, Email: johanna.kotrba@med.ovgu.de.
Antje Güttler, Email: antje.guettler@uk-halle.de.
Henri Wichmann, Email: henri.wichmann@gmx.de.
Udo Bilkenroth, Email: udobilkenroth@gmx.de.
Andreas Wienke, Email: andreas.wienke@uk-halle.de.
Bilal Al-Nawas, Email: al-nawas@uni-mainz.de.
Matthias Bache, Email: matthias.bache@uk-halle.de.
Dirk Vordermark, Email: dirk.vordermark@uk-halle.de.
Claudia Wickenhauser, Email: claudia.wickenhauser@uk-halle.de.
Daniel Bethmann, Email: daniel.bethmann@uk-halle.de.
Alexander W. Eckert, Email: alexander.eckert@uk-halle.de
Matthias Kappler, Email: matthias.kappler@uk-halle.de.
References
- 1.Fitzmaurice C, Allen C, Barber RM, Barregard L, Bhutta ZA, Brenner H, et al. Global, regional, and National Cancer Incidence, mortality, years of life lost, years lived with disability, and disability-adjusted life-years for 32 Cancer groups, 1990 to 2015: a systematic analysis for the global burden of disease study. JAMA Oncol. 2017;3:524–548. doi: 10.1001/jamaoncol.2016.5688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Camargo Cancela M de, Voti L, Guerra-Yi M, Chapuis F, Mazuir M, Curado MP. Oral cavity cancer in developed and in developing countries: population-based incidence. Head Neck 2010;32:357–367. doi:10.1002/hed.21193. [DOI] [PubMed]
- 3.Eckert AW, Wickenhauser C, Salins PC, Kappler M, Bukur J, Seliger B. Clinical relevance of the tumor microenvironment and immune escape of oral squamous cell carcinoma. J Transl Med. 2016;14:85. doi: 10.1186/s12967-016-0828-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pulte D, Brenner H. Changes in survival in head and neck cancers in the late 20th and early 21st century: a period analysis. Oncologist. 2010;15:994–1001. doi: 10.1634/theoncologist.2009-0289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rogers SN, Brown JS, Woolgar JA, Lowe D, Magennis P, Shaw RJ, et al. Survival following primary surgery for oral cancer. Oral Oncol. 2009;45:201–211. doi: 10.1016/j.oraloncology.2008.05.008. [DOI] [PubMed] [Google Scholar]
- 6.Califano J, van der Riet P, Westra W, Nawroz H, Clayman G, Piantadosi S, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 7.Clevers H. The cancer stem cell: premises, promises and challenges. Nat Med. 2011;17:313–319. doi: 10.1038/nm.2304. [DOI] [PubMed] [Google Scholar]
- 8.Dick JE. Stem cell concepts renew cancer research. Blood. 2008;112:4793–4807. doi: 10.1182/blood-2008-08-077941. [DOI] [PubMed] [Google Scholar]
- 9.Greaves M, Maley CC. Clonal evolution in cancer. Nature. 2012;481:306–313. doi: 10.1038/nature10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kreso A, Dick JE. Evolution of the cancer stem cell model. Cell Stem Cell. 2014;14:275–291. doi: 10.1016/j.stem.2014.02.006. [DOI] [PubMed] [Google Scholar]
- 11.Al-Sarraf M. Treatment of locally advanced head and neck cancer: historical and critical review. Cancer Control. 2002;9:387–399. doi: 10.1177/107327480200900504. [DOI] [PubMed] [Google Scholar]
- 12.de LW, Barker N, Low TY, Koo B, Li VSW, Teunissen H, et al. Lgr5 homologues associate with Wnt receptors and mediate R-spondin signalling. Nature. 2011;476:293–297. doi: 10.1038/nature10337. [DOI] [PubMed] [Google Scholar]
- 13.Barker N, van Es JH, Kuipers J, Kujala P, van den Born M, Cozijnsen M, et al. Identification of stem cells in small intestine and colon by marker gene Lgr5. Nature. 2007;449:1003–1007. doi: 10.1038/nature06196. [DOI] [PubMed] [Google Scholar]
- 14.Fan X, Wu H, Yu H, Zhou Q, Zhang Y, Huang Q. Expression of Lgr5 in human colorectal carcinogenesis and its potential correlation with beta-catenin. Int J Color Dis. 2010;25:583–590. doi: 10.1007/s00384-010-0903-z. [DOI] [PubMed] [Google Scholar]
- 15.McClanahan T, Koseoglu S, Smith K, Grein J, Gustafson E, Black S, et al. Identification of overexpression of orphan G protein-coupled receptor GPR49 in human colon and ovarian primary tumors. Cancer Biol Ther. 2006;5:419–426. doi: 10.4161/cbt.5.4.2521. [DOI] [PubMed] [Google Scholar]
- 16.Takahashi H, Ishii H, Nishida N, Takemasa I, Mizushima T, Ikeda M, et al. Significance of Lgr5(+ve) cancer stem cells in the colon and rectum. Ann Surg Oncol. 2011;18:1166–1174. doi: 10.1245/s10434-010-1373-9. [DOI] [PubMed] [Google Scholar]
- 17.Takeda K, Kinoshita I, Shimizu Y, Matsuno Y, Shichinohe T, Dosaka-Akita H. Expression of LGR5, an intestinal stem cell marker, during each stage of colorectal tumorigenesis. Anticancer Res. 2011;31:263–270. [PubMed] [Google Scholar]
- 18.Uchida H, Yamazaki K, Fukuma M, Yamada T, Hayashida T, Hasegawa H, et al. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in colorectal cancer. Cancer Sci. 2010;101:1731–1737. doi: 10.1111/j.1349-7006.2010.01571.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Barker N, Ridgway RA, van Es JH, van de Wetering M, Begthel H, van den Born M, et al. Crypt stem cells as the cells-of-origin of intestinal cancer. Nature. 2009;457:608–611. doi: 10.1038/nature07602. [DOI] [PubMed] [Google Scholar]
- 20.Merlos-Suárez A, Barriga FM, Jung P, Iglesias M, Céspedes MV, Rossell D, et al. The intestinal stem cell signature identifies colorectal cancer stem cells and predicts disease relapse. Cell Stem Cell. 2011;8:511–524. doi: 10.1016/j.stem.2011.02.020. [DOI] [PubMed] [Google Scholar]
- 21.Barker N, Tan S, Clevers H. Lgr proteins in epithelial stem cell biology. Development. 2013;140:2484–2494. doi: 10.1242/dev.083113. [DOI] [PubMed] [Google Scholar]
- 22.Dalley AJ, Abdul Majeed AA, Pitty LP, Major AG, Farah CS. LGR5 expression in oral epithelial dysplasia and oral squamous cell carcinoma. Oral Surg Oral Med Oral Pathol Oral Radiol. 2015;119:436–40.e1. doi: 10.1016/j.oooo.2014.11.014. [DOI] [PubMed] [Google Scholar]
- 23.Ryuge S, Sato Y, Jiang S, Wang G, Kobayashi M, Nagashio R, et al. The clinicopathological significance of Lgr5 expression in lung adenocarcinoma. Lung Cancer. 2013;82:143–148. doi: 10.1016/j.lungcan.2013.06.010. [DOI] [PubMed] [Google Scholar]
- 24.Tanese K, Fukuma M, Yamada T, Mori T, Yoshikawa T, Watanabe W, et al. G-protein-coupled receptor GPR49 is up-regulated in basal cell carcinoma and promotes cell proliferation and tumor formation. Am J Pathol. 2008;173:835–843. doi: 10.2353/ajpath.2008.071091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osawa H, Takahashi H, Nishimura J, Ohta K, Haraguchi N, Hata T, et al. Full-length LGR5-positive cells have chemoresistant characteristics in colorectal cancer. Br J Cancer. 2016;114:1251–1260. doi: 10.1038/bjc.2016.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rot S, Taubert H, Bache M, Greither T, Würl P, Eckert AW, et al. A novel splice variant of the stem cell marker LGR5/GPR49 is correlated with the risk of tumor-related death in soft-tissue sarcoma patients. BMC Cancer. 2011;11:429. doi: 10.1186/1471-2407-11-429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peng WC, de LW, Forneris F, Granneman JCM, Huch M, Clevers H, Gros P. Structure of stem cell growth factor R-spondin 1 in complex with the ectodomain of its receptor LGR5. Cell Rep. 2013;3:1885–1892. doi: 10.1016/j.celrep.2013.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 29.Nakata S, Campos B, Bageritz J, Bermejo JL, Becker N, Engel F, et al. LGR5 is a marker of poor prognosis in glioblastoma and is required for survival of brain cancer stem-like cells. Brain Pathol. 2013;23:60–72. doi: 10.1111/j.1750-3639.2012.00618.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu X, Xi H, Chen L. Lgr5 is a potential marker of colorectal carcinoma stem cells that correlates with patient survival. World J Surg Oncol. 2012;10:244. doi: 10.1186/1477-7819-10-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yamanoi K, Fukuma M, Uchida H, Kushima R, Yamazaki K, Katai H, et al. Overexpression of leucine-rich repeat-containing G protein-coupled receptor 5 in gastric cancer. Pathol Int. 2013;63:13–19. doi: 10.1111/pin.12013. [DOI] [PubMed] [Google Scholar]
- 32.Hirsch D, Barker N, McNeil N, Hu Y, Camps J, McKinnon K, et al. LGR5 positivity defines stem-like cells in colorectal cancer. Carcinogenesis. 2014;35:849–858. doi: 10.1093/carcin/bgt377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Vieira GC, Chockalingam S, Melegh Z, Greenhough A, Malik S, Szemes M, et al. LGR5 regulates pro-survival MEK/ERK and proliferative Wnt/β-catenin signalling in neuroblastoma. Oncotarget. 2015;6:40053–40067. doi: 10.18632/oncotarget.5548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Walker F, Zhang H, Odorizzi A, Burgess AW. LGR5 is a negative regulator of tumourigenicity, antagonizes Wnt signalling and regulates cell adhesion in colorectal cancer cell lines. PLoS One. 2011;6:e22733. doi: 10.1371/journal.pone.0022733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu J, Yu G, Cheng X, Li X, Zeng X, Ren X. LGR5 promotes hepatocellular carcinoma metastasis through inducting epithelial-mesenchymal transition. Oncotarget. 2017;8:50896–50903. doi: 10.18632/oncotarget.15143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Karimabad MN, Mahmoodi M, Jafarzadeh A, Darehkordi A, Hajizadeh MR, Khorramdelazad H, et al. Evaluating of OCT-4 and NANOG was differentially regulated by a new derivative indole in leukemia cell line. Immunol Lett. 2017;190:7–14. doi: 10.1016/j.imlet.2017.06.012. [DOI] [PubMed] [Google Scholar]
- 38.Bache M, Kappler M, Wichmann H, Rot S, Hahnel A, Greither T, et al. Elevated tumor and serum levels of the hypoxia-associated protein osteopontin are associated with prognosis for soft tissue sarcoma patients. BMC Cancer. 2010;10:132. doi: 10.1186/1471-2407-10-132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kappler M, Kotrba J, Kaune T, Bache M, Rot S, Bethmann D, et al. P4HA1: a single-gene surrogate of hypoxia signatures in oral squamous cell carcinoma patients. Clinical and Translational Radiation Oncology. 2017;5:6–11. doi: 10.1016/j.ctro.2017.05.002.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.