Abstract
Rationale
Familial recurrence studies provide strong evidence for a genetic component to the predisposition to sporadic, non-syndromic Tetralogy of Fallot (TOF), the most common cyanotic congenital heart disease (CHD) phenotype. Rare genetic variants have been identified as important contributors to the risk of CHD, but relatively small numbers of TOF cases have been studied to date.
Objective
We used whole exome sequencing (WES) to assess the prevalence of unique, deleterious variants in the largest cohort of non-syndromic TOF patients reported to date.
Methods and Results
829 TOF patients underwent WES. The presence of unique, deleterious variants was determined; defined by their absence in the Genome Aggregation Database (gnomAD) and a scaled combined annotation-dependent depletion (CADD) score of ≥20. The clustering of variants in two genes, NOTCH1 and FLT4, surpassed thresholds for genome-wide significance (assigned as P<5x10-8) after correction for multiple comparisons. NOTCH1 was most frequently found to harbour unique, deleterious variants. 31 changes were observed in 37 probands (4.5%; 95% confidence interval [CI]:3.2-6.1%) and included seven loss-of-function variants 22 missense variants and two in-frame indels. Sanger-sequencing of the unaffected parents of seven cases identified five de novo variants. Three NOTCH1 variants (p.G200R, p.C607Y and p.N1875S) were subjected to functional evaluation and two showed a reduction in Jagged1-induced NOTCH signalling. FLT4 variants were found in 2.4% (95% CI:1.6-3.8%) of TOF patients, with 21 patients harbouring 22 unique, deleterious variants. The variants identified were distinct to those that cause the congenital lymphoedema syndrome Milroy Disease. In addition to NOTCH1, FLT4 and the well-established TOF gene, TBX1, we identified potential association with variants in several other candidates including RYR1, ZFPM1, CAMTA2, DLX6 and PCM1.
Conclusions
The NOTCH1 locus is the most frequent site of genetic variants predisposing to non-syndromic TOF, followed by FLT4. Together, variants in these genes are found in almost 7% of TOF patients.
Keywords: Congenital heart disease, Tetralogy of Fallot, genetic variation, whole exome sequencing, NOTCH1, FLT4
Subject Terms: Congenital Heart Disease; Genetic, Association Studies; Genetics
Introduction
Congenital heart disease (CHD) is the most common type of birth defect, affecting 8/1000 live births (1). CHD covers a large spectrum of heterogeneous cardiovascular phenotypes that range from single, localised defects to more complex structural abnormalities. Tetralogy of Fallot (TOF) is the most common complex, cyanotic CHD with a prevalence of 1/3000 births (1,2). TOF is considered a malformation of the cardiac outflow tract which comprises four specific structural characteristics postnatally; a ventricular septal defect (VSD), anterocephalad deviation of the outflow septum with resultant overriding of the aorta, variable obstruction of the right ventricular outflow tract (pulmonary stenosis) and consequent hypertrophy of the right ventricle (2,3). Surgical interventions during infancy mean that 85-90% of TOF patients now survive until at least 30 years of age (1,4). However, this is not without consequence; event-free survival is just 25% after 40 years of age (5), since resultant scar tissue from surgery and pulmonary regurgitation cause significant morbidity in adulthood (6,7).
The cause of TOF is elusive and no single candidate gene can be held accountable for the disease phenotype. However, the genetic status of syndromic TOF sufferers has provided valuable insights into causative genes in some patients. Approximately 20% of cases are associated with a recognised syndrome or chromosomal anomaly (2). Most significantly, approximately 15% of TOF patients have 22q11.2 deletion syndrome, wherein the major causal gene is TBX1 (8,9). Approximately 80% of TOF cases are non-syndromic and there is generally no identifiable cause, largely due to their non-Mendelian patterns of inheritance (10–13). Accordingly, a polygenic genetic architecture has been hypothesised and genome-wide approaches have been undertaken to provide insights into the complex genetic alterations responsible for TOF and other CHDs (11,13–18).
Whole exome sequencing (WES) has been used successfully to identify new CHD candidate genes (14,17,19,20). Many lines of evidence indicate a degree of phenotypic specificity of variants in particular genes. For example, the spectrum of phenotypes caused by 22q11.2 deletion or mutations in TBX1 typically involves the outflow tract and great vessels (9,21,22), while Down syndrome or mutations in NKX2-5 typically cause septal defects (23,24). To date, no WES study of CHD has included substantial numbers of any homogeneous phenotype, which should a priori have the highest power to identify causal variants.
Here, we present findings from WES of the largest cohort of non-syndromic TOF patients reported to date. We performed WES in 829 TOF probands and identified the rarest and most deleterious protein-coding variants genome-wide. We sought evidence of pathological relevance for a subset of variants in the most significantly over-represented genes, based on the variants’ de novo occurrence and functional consequences in cellular models.
Methods
Data can be accessed at the European Genome-phenome Archive (https://www.ebi.ac.uk/ega) using accession number EGAS00001003302.
829 TOF probands were subjected to WES and unique (absent in the Genome Aggregation Database [gnomAD]), deleterious (combined annotation-dependent depletion [CADD] score of ≥ 20) variants were identified. Any variants observed in 1252 reference exome samples, that were analysed using the same approach as our case data, were eliminated from further consideration. Clustering analysis within the cases was then used to identify genes in which significantly more variants were observed than expected given background levels of variation across all genes. De novo variants were identified by Sanger sequencing of proband and parent samples where possible. Immunoblotting and luciferase assays were used to assess the expression and signalling activity of selected variants in the most strongly supported candidate gene. Detailed methods can be found in the Supplementary Materials.
Results
Exome-wide analysis of unique, deleterious variants identifies the highest risk loci for non-syndromic TOF
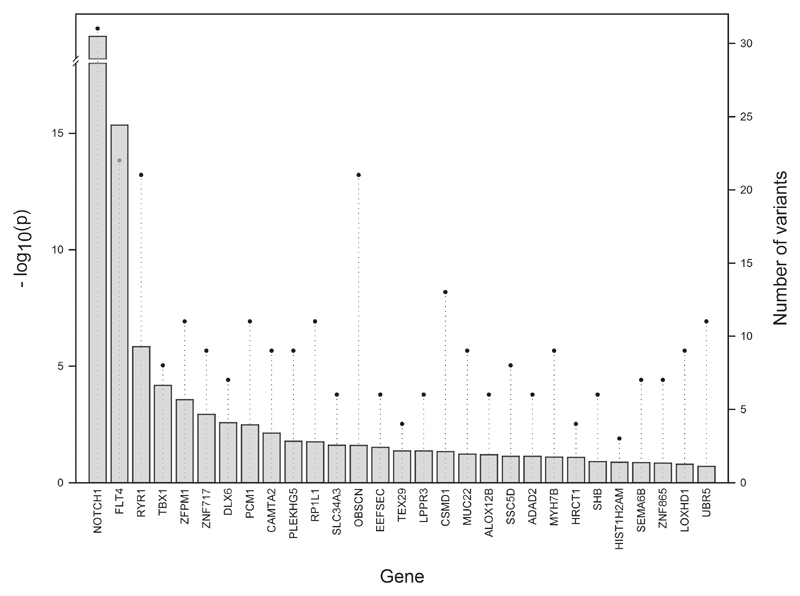
We assessed the incidence of unique, deleterious variants for 829 non-syndromic TOF cases. Any variants observed in 1252 reference exomes were removed from consideration as potential TOF susceptibility variants. The statistical significance of these findings was assessed for each gene using clustering analysis, which corrected for gene size (supplementary table I). Two genes, NOTCH1 and FLT4, surpassed the threshold for genome-wide significance (assessed as P<5 x 10-8) (figure 1) and the unique variants identified in these genes are likely to be contributors to the pathogenesis of TOF. Combined, variants in NOTCH1 and FLT4 account for 6.9% of our TOF cohort, with no overlap between probands with variants in these genes. Additionally, several other genes that harbour an excess of variant clustering are also of interest; including RYR1 and TBX1, which have previously been implicated in CHD (25,26). In particularly, TBX1 is a well-established TOF risk gene which is principally responsible for the cardiac manifestations of 22q11 deletion; additionally, deleterious single nucleotide variants and small functionally significant intragenic deletions in TBX1 have been demonstrated in TOF patients (9,21). A further two genes, ZFPM1/FOG1 and CAMTA2, have roles in heart development and growth, respectively (27). DLX6 is negatively regulated by HAND2, a crucial transcription factor for heart morphogenesis (28) and PCM1 is a regulator of ciliogenesis, a process strongly linked to CHD (29). In addition, we specifically looked at the number of unique, deleterious variants in key cardiac transcription factors including NKX2.5 (30), GATA4 (31), HAND2 (12) and GATA6 (32), since pathogenic variants have previously been identified in TOF cases, typically by targeted candidate gene sequencing. Variants in these genes account for just 1.2% of cases in our cohort. When considering the top nine genes (or a P value cut-off of <0.01), 129 TOF cases had a unique, deleterious variant in one or more genes, accounting for over 16% of our patient cohort (table 1). Just eight samples had variants in more than one of the top nine genes, highlighting the minimal overlap between probands with variants in these genes. Overall, NOTCH1 and FLT4 were found to be by far the most significant contributors to TOF; we therefore explored the variants in these two genes in greater detail.
Figure 1.
The top genes, in order of significance, in which non-syndromic TOF patients carry unique, deleterious variants. Bars indicate the respective significance levels of variant clustering for each gene, represented as –log P values. Circles represent the number of variants. The -log10(p) column for NOTCH1 (P<2.22 x 10-16) goes towards infinity and is shown as arbitrarily high.
Table 1. The top gene candidates, ordered by levels of significance, following the clustering analysis of unique, deleterious variants.
| Gene | Variants | P value | Samples | Cumulative sample count |
|---|---|---|---|---|
| NOTCH1 | 31 | <2.22 x 10-16 | 37 | 37 |
| FLT4 | 22 | 4.44 x 10-16 | 21 | 57 |
| RYR1 | 21 | 1.43 x 10-06 | 22 | 78 |
| TBX1 | 8 | 6.50 x 10-05 | 8 | 86 |
| ZFPM1 | 11 | 0.000266817 | 12 | 98 |
| ZNF717 | 9 | 0.001125519 | 10 | 106 |
| DLX6 | 7 | 0.002583786 | 8 | 114 |
| PCM1 | 11 | 0.003208801 | 11 | 123 |
| CAMTA2 | 9 | 0.007243157 | 9 | 129 |
Variants in NOTCH1 are most commonly present in non-syndromic TOF
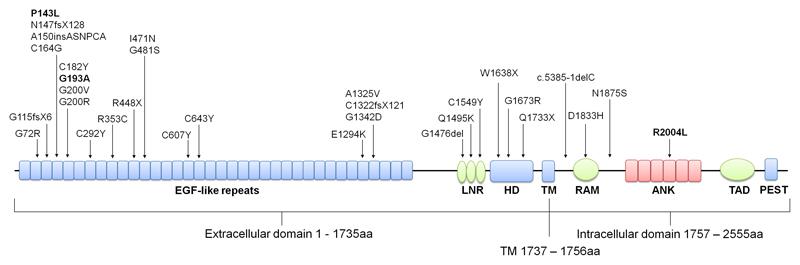
The NOTCH1 locus was most frequently found to harbour a unique, deleterious variant among TOF patients (P<2.22 x 10-16), with 37 probands harbouring 31 NOTCH1 variants (supplementary table II), accounting for 4.5% of our TOF patient cohort (95% CI: 3.2% - 6.1%). Seven of the variants identified were loss-of-function (LOF), including three premature stop codons (p.R448X, p.W1638X and p.Q1733X), three single base pair deletions resulting in frameshifts and eventual premature truncation (p.G115fsX6, p.N147fsX128 and p.C1322fsX121) and a single base pair deletion in a splice site consensus sequence (c.5385-1delC). Of the remaining 24 variants, two were in-frame indels and 22 were missense variants. NOTCH1 is highly intolerant to LOF and missense variation, having a pLI of 1 and a missense z score of 4.48 on the Exome Aggregation Consortium (ExAC). We mapped the distribution of the 31 variants to the various domains of NOTCH1 (figure 2) and found the variants to be located throughout the protein with no significant clusters. The three frameshift mutations were located in the EGF-like repeats in addition to one truncating mutant, p.R448X, whereas the remaining two truncating variants were located in the heterodimerisation domain. Of particular interest, one variant located in EGF-like repeat 5, p.G193A (figure 2, bold), was identified in five unrelated patients and p.P143L (figure 2, bold) located in EGF-like repeat 4 was identified in three unrelated patients. Together, these two variants account for almost 1% of our TOF patient cohort. Interestingly, a further six NOTCH1 variants that map to the EGF-like repeats alter evolutionary conserved cysteine residues that contribute to disulphide bonds essential for maintaining the EGF structure (33). Of the four intracellular domain mutants, a missense variant in the Ankyrin repeats region, p.R2004L is particularly notable (figure 2, bold). R2004 is a surface exposed residue in Ankyrin domain 4 which is located in an interface region with the CSL transcription factor complex (34) and also located at an interface that binds the positive Notch regulator, Deltex (35).
Figure 2.
Unique, deleterious NOTCH1 variants in TOF patients. Diagrammatic representation of the NOTCH1 protein with known protein domains indicated. The location of NOTCH1 variants identified in our TOF cohort is shown. p.P143L, p.G193A and p.R2004L discussed in the main text are indicated (bold). ANK, ankyrin repeats; EGF, epidermal growth factor; HD, heterodimerisation domain; LBR, ligand binding region; LNR, Lin/Notch repeats; PEST, PEST domain; RAM, RBPJ-associated molecule domain; TAD, transactivation domain; TM, transmembrane domain.
Deleterious mutations in other NOTCH pathway genes have been identified in patients with TOF including HEY2 (36) and JAG1 (37,38). For this reason, we compiled a list of NOTCH pathway genes using the MGI Gene Ontology Project and assessed the clustering of variants in these genes. Of 166 genes tested, only NOTCH1 was found to have an excess of unique, deleterious variants (supplementary table III). Hence, variants in other NOTCH pathway genes are not a major cause of TOF in our cohort.
Evidence of pathological consequences for NOTCH1 variants
We investigated the occurrence rate of de novo variants in probands with NOTCH1 variants. Of the 31 probands in our TOF patient cohort that harboured unique, deleterious variants in NOTCH1, samples from both parents were available for seven probands and analysed for variant inheritance. Following Sanger sequencing, five of the seven NOTCH1 variants tested were identified as de novo; two of these were truncating variants, whereas the remaining three de novo variants were missense (table 2). These findings are in keeping with the results of previous WES experiments in CHD, where rare transmitted variants with strong bioinformatic support for functional impact, which are of presumed incomplete penetrance, have been uniformly encountered (14,17,20).
Table 2. Sequencing of parent samples to determine NOTCH1 variant inheritance.
| Amino acid change | Ref | Alt | LOF | Impact | Inheritance status |
|---|---|---|---|---|---|
| p.G200V | C | A | NO | Missense variant | DE NOVO |
| p.C292Y | C | T | NO | Missense variant | FROM UNAFFECTED MOTHER |
| p.R448X | G | A | YES | Stop gained | DE NOVO |
| p.Q1495K | G | T | NO | Missense variant | FROM UNAFFECTED FATHER |
| p.C1549Y | C | T | NO | Missense variant | DE NOVO |
| p.W1638X | C | T | YES | Stop gained | DE NOVO |
| p.N1875S | T | C | NO | Missense variant | DE NOVO |
Ref, reference allele; Alt, alternate allele; loss of function
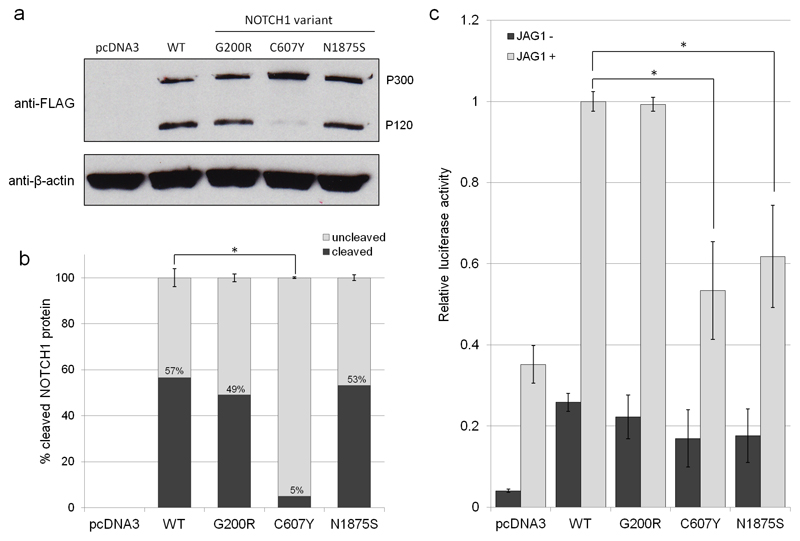
The NOTCH1 gene encodes an evolutionarily conserved transmembrane receptor that mediates cell-cell communication to govern cell fate decisions during development (39). S1 cleavage is an important step in the maturation of the NOTCH1 receptor. During this process, the 300 kDa translation product of NOTCH1 undergoes cleavage in the Golgi by furin-like convertase to generate two polypeptides of 180 and 120 kDa (40). To determine whether NOTCH1 variants affect S1 cleavage, we assessed the expression of three NOTCH1 variants in comparison to wild type (WT) NOTCH1 by immunoblotting. The variants assessed were p.G200R, p.C607Y and p.N1875S (see figure 2); p.G200R is located in a conserved residue located within a β-hairpin turn within EGF5, and p.C607Y, located in EGF16, removes a conserved disulphide bond that normally would be expected to stabilise the EGF-domain conformation. p.N1875S is located in a residue that lies in a linker region between the RAM and Ankyrin repeat regions of the Notch intracellular domain. As expected, we observed two bands at 300 kDa (P300) and 120 kDa (P120), representing full length and cleaved NOTCH1 protein (40); the remaining 180 kDa product was not detectable due to the positioning of our FLAG-tag at the C-terminus (figure 3a). For WT NOTCH1, p.G200R and p.N1875S variants, we observe similar levels of both P300 and P120 (figure 3a). However, the p.C607Y variant exhibited perturbed S1 cleavage. Indeed, quantification confirmed that 5%±0.37% of NOTCH1 p.C607Y underwent cleavage in comparison to 57%±3.96% of WT NOTCH1 (P=0.0002; figure 3b). Hence, the p.C607Y variant affects S1 cleavage of NOTCH1, whereas the receptor is processed normally in the p.G200R and p.N1875S NOTCH1 variants.
Figure 3.
(a) Immunoblot for FLAG to determine the expression and S1 cleavage of NOTCH1 variants p.G200R, p.C607Y and p.N1875S in comparison to WT NOTCH1 following overexpression in HeLa cells. The two bands at 300 kDa (P300) and 120 kDa (P120) represent the full length and the S1-cleaved NOTCH1 protein. β-actin was used as a loading control. (b) Quantification of the percentage of S1 cleaved versus uncleaved NOTCH1 protein for WT NOTCH1 and NOTCH variants p.G200R, p.C607Y and p.N1875S. Error bars: mean ±SEM from three biological replicates and statistical significance was determined using two-tailed paired t-tests. (c) The effect of rare, deleterious NOTCH1 variants on Jagged-induced NOTCH signalling levels. NOTCH signalling activity was measured using a luciferase-based reporter system (RBPJ). HeLa cells were cultured with or without immobilised JAG1 ligand and co-transfected with RBPJ reporter constructs and WT NOTCH1, p.G200R, p.C607Y or p.N1875S. Firefly luciferase readings were normalised to Renilla luciferase readings to control for transfection efficiency and cell number. RBPJ activity was expressed relative to WT NOTCH1 for comparison. Error bars: mean ±SEM from four biological replicates, each with three technical replicates. Statistical significance was assessed using two-tailed paired t-tests and the Hochberg step-up procedure to control for family-wise error rate.
Heterodimeric NOTCH1 is membrane tethered and undergoes further cleavage by γ-secretase which releases the NOTCH intracellular domain (NICD). NICD subsequently translocates to the nucleus where it interacts with transcription factor RBPJ to activate NOTCH target genes (39). To determine whether p.G200R, p.C607Y and p.N1875S variants affect NOTCH1 canonical signalling function, we assessed NOTCH signalling through the RBPJ transcription factor-dependent pathway following stimulation with immobilised Jagged1 (JAG1) ligand. The variants were overexpressed in HeLa cells and NOTCH1 signalling was assessed by RBPJ luciferase activity. Two of the three variants demonstrated reduced NOTCH signalling via RBPJ (figure 3c). The p.C607Y variant, that exhibited perturbed cleavage, significantly reduced NOTCH signalling by 47%±0.12% (P=0.008) compared to WT NOTCH1. Similarly, de novo variant p.N1875S reduced NOTCH signalling by 38%±0.13% (P=0.02). The p.G200R variant exhibited similar canonical NOTCH signalling to WT NOTCH1 (P=0.67) (figure 3c), yet mapping of this variant to the three-dimensional NOTCH1 protein suggests structural implications (supplementary figure II). Furthermore, p.G200R has also been reported in an independent study to segregate with CHD, supporting its pathogenicity (41). No significant differences were observed between WT NOTCH1, p.G200R, p.C607Y and p.N1875S variants in the absence of JAG1 ligand. In each transfection experiment, mRNA expression of WT NOTCH1 and the three NOTCH1 variants was equal (supplementary figure III), thus the differences in NOTCH1 signalling observed were not due to reduced mRNA expression of the variants. Hence, two variants identified in patients that were subjected to functional testing were shown to affect canonical NOTCH1 signalling.
FLT4 variants found in TOF are distinct from those that cause Milroy Disease
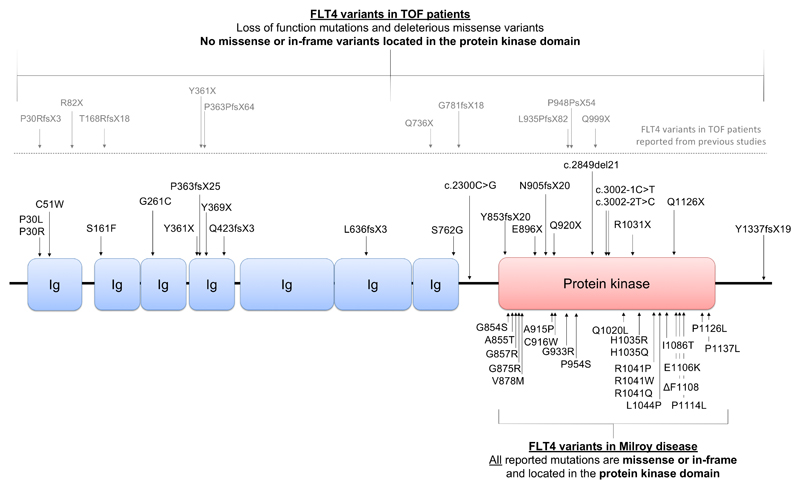
The second most frequent locus of variant clustering in our TOF cohort was FLT4 (P=4.44 x10-16). FLT4 encodes a receptor tyrosine kinase known as vascular endothelial growth factor 3 (VEGFR3). VEGFR3 is indispensable for lymphatic development and FLT4 mutations are a known cause of the hereditary lymphoedema, Milroy disease. Strikingly, all mutations reported for Milroy disease are missense variants or in-frame indels located in the VEGFR3 protein kinase domain (figure 4). In our TOF cohort of 829 probands, we report 22 unique, deleterious FLT4 variants in 21 TOF probands, accounting for 2.4% of cases (supplementary table IV). 16 of the FLT4 variants were LOF, including six premature stop codons (p.Y361X, p.Y369X, p.E896X, p.Q920X, p.R1031X and p.Q1126X) six indels resulting in frameshifts and premature truncation (p.P363fsX25, p.Q423fsX3, p.L636fsX3, p.Y853fsX20, p.N905fsX20 and p.Y1337fsX19) and four splice variants (c.3002-1C>T, c.3002-2T>C, c.2300C>G and c.2849del21). One premature stop codon, p.Y361X, was reported previously in a TOF proband and affected mother (25). The remaining six variants were missense, all of which were located in the immunoglobulin (Ig) domains of VEGFR3. FLT4 is extremely intolerant to both LOF and missense variation, as demonstrated by a pLI of 1 and missense z score of 3.73 on ExAC, respectively. In our 1252 reference exomes, no novel, LOF FLT4 variants were identified. Parent DNA was available for four probands. Three of the variants (p.Q920X, p.Y853fsX20 and c.2300C>G) were inherited from unaffected parents indicating incomplete penetrance, and one missense variant, p.C51W, was de novo (supplementary table V). Frameshift variant Y853fsX20 was identified in two siblings with TOF and was inherited from the mother who was unaffected. Crucially, no missense or in-frame variants were found in the kinase domain, a feature unique to Milroy disease (figure 4). Our findings are in line with a recent publication by Jin et al (2017) that reports LOF variants in FLT4 in 2.3% of 426 TOF probands. Hence, we confirm this finding in the largest TOF cohort reported to date, approximately twice the size of previous studies, endorsing the importance of FLT4 as a major contributor to the incidence of TOF.
Figure 4.
Unique, deleterious FLT4 variants in TOF patients. Schematic representation of FLT4 structure with immunoglobulin (Ig) domains and protein kinase domain, indicated. Top: FLT4 variants identified in our TOF cohort (black) and those previously reported (grey). Bottom: FLT4 missense or in-frame mutations reported in Milroy disease, all located in the protein kinase domain.
Discussion
Despite TOF being the most common, severe cyanotic CHD, variants that could account for the high degree of genetic susceptibility, inferred from familial recurrence risk studies (42), are as yet unidentified. This study represents the largest WES investigation of sporadic, non-syndromic TOF performed to date. Using variant clustering analysis and stringent filtering, we identify two genes that reach genome-wide significance: NOTCH1 and FLT4. As an additional safeguard against false positive results due to systematic methodological differences between our cohort and the studies which contributed to the gnomAD database, we studied a set of over 1000 reference exomes in patients free from CHD; analysed in the same fashion as the case exomes, stringently removing any variant that appeared even once in the reference exome set from consideration as a potential TOF susceptibility variant.
We identify NOTCH1 as the major TOF susceptibility gene; 4.5% of patients carry heterozygous variants in NOTCH1, which based on gnomAD allele frequency, bioinformatic in silico prediction, and functional characterisation, we judged to be likely susceptibility alleles. With the exception of the 22q11 deletion, no single gene locus has been found to account for more TOF cases than NOTCH1. Seven of the variants were LOF, including truncating, frame shift and splice variants, whereas the remaining 24 variants were missense or in-frame indels and anticipated to be pathogenic. Five out of seven variants tested were de novo, adding to the evidence for pathogenicity; the remaining variants were transmitted from unaffected parents indicating incomplete penetrance. Previous sequencing studies of CHD have identified an association of NOTCH1 variants in cardiac malformations including bicuspid aortic valve, aortic valve stenosis, coarctation of the aorta and hypoplastic left heart syndrome and TOF (43–47). However, the extent of NOTCH1 variant contribution to TOF has not been recognised until now. There are no clear distinctions between the type and location of NOTCH1 variants identified in TOF compared to those reported in other isolated cardiovascular abnormalities. We therefore propose that genetic background and/or environmental influences may specify phenotypic expressivity.
A possible role for NOTCH1 in non-syndromic TOF has previously been suggested by copy number variant (CNV) analysis. A study of 34 infants with non-syndromic TOF revealed two patients with CNVs encompassing the NOTCH1 gene (48). Additionally, a microdeletion including the NOTCH1 locus in a patient with TOF was identified in a study of CNVs in 114 TOF patients (49). A recent study that focused primarily on families with left-sided CHD also identified family members with TOF harbouring pathogenic mutations in NOTCH1 (44). Further indirect evidence for NOTCH1 contribution to TOF came from a study that analysed the gene expression patterns in TOF patient right ventricles and found many genes from the NOTCH and WNT signalling pathways were significantly reduced. Interestingly, down-regulation of NOTCH signalling components was also observed in TOF patients with a 22q11.2 deletion (50), highlighting a common transcriptional signature between both syndromic and non-syndromic TOF, initiated by different genetic events. More recently, exome sequencing of 426 TOF patients that focused solely on LOF heterozygous variants did not identify an enrichment of NOTCH1 mutations in TOF patients (25). However, the present study involves, by a substantial margin, the largest TOF cohort studied by WES to date, including both LOF and damaging missense variants, hence providing the most accurate quantification thus far of the contribution of NOTCH1 variants to TOF risk.
Autosomal dominant germ-line mutations in the NOTCH1 gene are also one of the causes of Adams-Oliver syndrome (AOS) which is chiefly characterised by aplasia cutis congenita and terminal transverse limb defects. In addition to these features, around half of patients have congenital cardiac anomalies, including atrial septal defect (ASD), VSD, aortic valve stenosis, pulmonary valve stenosis and TOF (51,52). AOS is an extremely rare syndrome, with a prevalence of approximately 1 in 225,000 (52). No patient in our cohort had diagnostic features of AOS. As with other CHDs associated with NOTCH1 variants, there are no clear distinctions between the NOTCH1 variants we have identified in TOF versus those that cause AOS, though no previously described AOS variant was present in our cases (51,52). Interestingly, the extra-cardiac features of AOS have been suggested to occur due to early embryonic vascular abnormalities (53), raising the possibility that AOS, TOF and other cardiac anomalies that occur due to mutations in NOTCH1 may be a spectrum of disorders. Other examples of syndromic genes that can cause isolated CHD, including TOF, are PTPN11 (Noonan syndrome), (13,54), TBX5 (Holt-Oram syndrome) (55) and JAG1 (Alagille syndrome) (38). Determining the role of genetic background, environmental context and the specific NOTCH1 variants in determining the severity of the cardiac phenotype and the occurrence of extra-cardiac malformations requires further research.
The association of NOTCH1 with a range of cardiac defects is consistent with the reported roles of NOTCH1 during heart development. Active NOTCH1 is observed in the trabecular endocardium and both global and endothelial-specific knockout of Notch1 in mice results in abnormal ventricular trabeculae and abnormal cardiomyocyte patterning (56). Relevant to TOF, Notch1 plays a role in the organisation of the outflow tract, which requires the specification of cells from both the neural crest and secondary heart field (57). Furthermore, Notch1 is important for endocardial epithelial-to-mesenchymal transition, a process that is essential for cardiac valve formation (46,58). It should however be noted that all NOTCH1 variants we report are heterozygous. There are numerous reports of global and tissue specific Notch1 heterozygous mutant mice that appear phenotypically normal, with no obvious cardiovascular pathologies (59,60), although mice lacking endothelial/endocardial Notch1 in various backgrounds do present with TOF-like characteristics including septal defects and abnormal heart valves (61,62). This suggests endothelial NOTCH1 may be partly responsible for the cardiac malformations associated with TOF, and again, emphasising the importance of genetic background. In further support of this, Notch1+/- in a predominantly 129S6 background developed aortic root dilation whereas Notch1+/- in a mixed background did not (63). Altogether, these reports highlight the importance of genetic background in disease expressivity and are consistent with the incomplete penetrance observed.
De novo mutations are a significant cause of early-onset genetic disorders, including CHD. Of the NOTCH1 variants identified in this study where parents were available, five of seven variants were found to be de novo. Similarly, we also found de novo variation in FLT4. For both of our genome-wide significant TOF genes, variants were also found to be inherited from unaffected parents, confirming the role of incompletely penetrant variants observed for other CHD genes and phenotypes (17,20). The incomplete penetrance is in keeping with the complex genetic aetiology of non-syndromic TOF, in which families segregating the condition in a Mendelian fashion are rarely encountered and genetic background, in addition to in utero environmental factors, can be inferred to play significant roles.
For a subset of NOTCH1 variants, we provide evidence of functional impact by assessing canonical NOTCH1 signalling. The p.C607Y missense variant perturbed NOTCH1 receptor S1 cleavage by the calcium-dependent enzyme, furin-like convertase. The S1 cleavage site is located at amino acids 1651 - 1654, some distance away from the variant. A similar observation has been reported by McBride et al (2008) where NOTCH1 variant p.A683T, identified in two patients with left ventricular outflow tract malformations, also perturbed S1 cleavage by similar levels. In both cases, this led to a 50% reduction in RBPJ luciferase activity (64). The mechanism by which such variants alter S1 cleavage to such an extent and reduce signalling by just 50% is unclear and requires further research. Furthermore, de novo variant p.N1875S was shown to have significantly reduced JAG1-induced NOTCH signalling relative to WT NOTCH1, providing further support as to the pathogenicity of de novo variants. p.G200R exhibited signalling levels similar to WT. However, in support of this variants pathogenicity, Blue et al (2014) identified the same NOTCH1 variant in an independent study; p.G200R segregated with disease in two cousins with right-sided CHD, including persistent truncus arteriosus, VSD, pulmonary atresia, and major aorto-pulmonary collateral arteries. Furthermore, a case of TOF was also reported in the preceding generation, although sequencing analysis was not carried out on this relative.
FLT4 was first associated with isolated TOF in a CNV analysis that identified a de novo duplication including FLT4, and a deletion of unknown inheritance upstream of FLT4 (18). Recent WES studies have also identified FLT4 to be a significant contributor to the incidence of TOF. Jin et al (2017) found 2.3% of TOF patients to have LOF FLT4 mutations. Furthermore, Szot et al (2018) also identified a FLT4 variant in a family with TOF (65). Using our larger cohort, we confirm FLT4 variants to be a significant contributor to the incidence of TOF, with 2.4% of our cohort exhibiting deleterious FLT4 variants. In addition to LOF variants, we also identify a small number of pathogenic missense variants, including one variant that is de novo. The encoded product of FLT4, VEGFR3, has a well-established role in lymphatic development and in the adult, VEGFR3 expression is almost entirely restricted to lymphatic vessels (66,67). During embryonic development, VEGFR3 is also expressed in vascular endothelial cells and is crucial for blood vessel development. Loss of VEGFR3 in mice leads to lethality at E9.5 due to defects in blood vessel formation and cardiovascular failure (68–70). This is prior to the emergence of lymphatics, suggesting VEGFR3 plays a unique role in cardiovascular development, independent of lymphangiogenesis. Importantly, patients with VEGFR3 variants causing Milroy disease are not reported to have congenital heart malformations. The distinction between the locations of the mutations in FLT4 that cause Milroy disease in comparison to TOF may shed light on the evidently differing roles of the receptor in lymphatic versus heart development.
In addition to NOTCH1 and FLT4, we also report an excess of clustering in several other genes of interest including RYR1, ZFPM1/FOG1, CAMTA2, DLX6, PCM1 and known TOF gene, TBX1. A summary of in vivo and in vitro functional data currently available for these genes can be found in supplementary table VII. Biallelic heterozygous mutations in RYR1 have previously been linked to CHD, including TOF, in a small number of cases (25,26). In addition, a mouse homozygous for the missense mutation I4895T, displayed notable delays in cardiogenesis including abnormal orientation, improper formation of the outflow tract and an ASD (71), suggesting a role in early heart development. ZFPM1/FOG1 encodes a GATA cofactor previously implicated in heart development. Fog1 null and endothelial lineage knockout mice develop heart malformations including a double outlet right ventricle and abnormal valve formation (27). Morpholino knockdown of fog1 also results in defective cardiac looping in zebrafish (72). While in vivo models suggest a role for FOG1 in heart development, we report a suggestive association of human FOG1 mutations with CHD for the first time. CAMTA2 interacts with NKX2-5, one of the core transcription factors controlling heart development. Together, Camta2 and Nkx2-5 promote cardiac hypertrophy in mice (73). CAMTA2 was also identified as the likely candidate gene from a de novo CNV deletion at 17p13.2 in a patient with congenital pulmonary atresia (74). DLX6 encodes a homeobox protein involved with known role in cranial-facial morphogenesis. Interestingly in mice, Dlx6 is negatively regulated by Hand2 (28), a transcription factor crucial for cardiac morphogenesis. The significance of the relationship between HAND2 and DLX6 in the developing heart is not clear, although the formation of the great vessels and coronary arteries is reported to be independent of Dlx6 in mice (75). PCM1 encodes Pericentriolar Material 1, which is essential for centrosomal proteins and microtubule organisation. PCM1 also positively regulates ciliogenesis (76), a process which has been strongly linked to the development of CHDs (29). Following validation in an independently ascertained cohort, investigations of the role these genes during heart development may be of interest. It should be mentioned that ZNF717 also appears amongst our top TOF-associated genes. ZNF717 is a relatively small gene (less than 4kb) yet of all genes, exhibits the highest frequency of non-synonymous mutations per base pair in our reference exomes. For this reason, we do not consider ZNF717 to be a TOF candidate gene.
In summary, our findings which, in addition to NOTCH1 and FLT4, identified a number of potential novel TOF gene candidates, concur with previous studies regarding the marked locus heterogeneity of the condition. Among the genes that have been implicated in TOF thus far, our large study indicates that NOTCH1 is the most commonly involved. The two most commonly involved genes (NOTCH1 and FLT4) are also both crucial to angiogenesis, suggesting further investigation of common pathways between heart development and angiogenesis may be fruitful. In our top gene candidates, some mutations were de novo, but others were present in apparently asymptomatic individuals, indicating incomplete penetrance. Such incomplete penetrance has been frequently observed, for example, in Mendelian aortopathies, emphasising the importance of genetic background in structural cardiac and vascular diseases. Detailed phenotypic studies of mutation carriers who do not have overt CHD using advanced imaging may be of interest to delineate quantitative phenotypes potentially relevant to CHD.
Supplementary Material
Novelty and Significance.
What Is Known?
Tetralogy of Fallot (TOF) is the most common cyanotic congenital heart disease (CHD).
Non-syndromic TOF is a genetically complex disease, with evidence for contributions from both common and rare variants.
The major causative genes for non-syndromic TOF have yet to be identified.
What New Information Does This Article Contribute?
We performed whole-exome sequencing (WES) in a large cohort of non-syndromic TOF patients and identified very rare deleterious variants in several genes.
Variants in NOTCH1 and FLT4, the most commonly observed genes, were found in 7% of TOF cases, indicating significant contributions from these genes to the population burden of disease.
Functional analysis of NOTCH1 variants found in patients with TOF confirmed a detrimental effect on the NOTCH signalling pathway.
Identification of pathogenic variants in multiple genes in a substantial proportion of non-syndromic TOF points to the utility of the WES approach in discovering the genetic basis of CHD in large cohorts of patients with homogeneous phenotypes.
Congenital heart disease occurs in almost 1% of live births. The most common severe cyanotic form, TOF, is well characterised phenotypically, but the genetic factors associated with non-syndromic cases (80%) are mostly unknown. We performed WES on a large TOF cohort and found variants that were previously unobserved in the general population and were predicted to be highly damaging to protein function in two genes, NOTCH1 and FLT4, in 7% of cases. An in vitro activity assay showed that NOTCH1 variants observed in the patients disrupted NOTCH signalling. Significant (exome-wide p<0.01) excess of very rare deleterious variants were identified in six other genes; such variants were present in 15% of non-syndromic TOF patients.
Acknowledgements
This study makes use of the ICR1000 UK exome series data generated by Professor Nazneen Rahman’s Team at The Institute of Cancer Research, London (77). No other persons besides the authors have made substantial contributions to this manuscript.
Sources of Funding
This work was supported by the British Heart Foundation Programme Grant RG/15/12/31616. BK and SB hold BHF Personal Chairs. SB was supported by the British Heart Foundation funded GOCHD study project grant. BM, CRB and AP were supported by the Netherlands Heart Foundation CVON project CONCOR-genes (CVON 2014-18). The work in Nottingham/Leicester was funded by British Heart Foundation Programme Grant RG/13/10/30376.
Nonstandard Abbreviations and Acronyms
- ASD
atrial septal defect
- CADD
combined annotation-dependent depletion
- CHD
congenital heart disease
- CI
confidence interval
- CNV
copy number variant
- ExAC
Exome Aggregation Consortium
- gnomAD
Genome Aggregation Database
- GWAS
genome wide association study
- HD
heterodimerisation domain
- Ig
immunoglobulin
- JAG1
Jagged1
- LOF
loss-of-function
- MAF
minor allele frequency
- NICD
NOTCH intracellular domain
- SNP
single nucleotide polymorphism
- TOF
Tetralogy of Fallot
- VEGFR3
vascular endothelial growth factor receptor 3
- VSD
ventricular septal defect
- WES
whole exome sequencing
Footnotes
Disclosures
None.
References
- 1.Ferencz C, Rubin JD, McCarter RJ, Brenner JI, Neill CA, Perry LW, Hepner SI, Downing JW. Congenital heart disease: prevalence at livebirth. The Baltimore-Washington Infant Study. Am J Epidemiol. 1985;121:31–36. doi: 10.1093/oxfordjournals.aje.a113979. [DOI] [PubMed] [Google Scholar]
- 2.Bailliard F, Anderson RH. Tetralogy of Fallot. Orphanet J Rare Dis. 2009;4:2. doi: 10.1186/1750-1172-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Shinebourne EA, Babu-Narayan SV, Carvalho JS. Tetralogy of Fallot: from fetus to adult. Heart. 2006;92:1353. doi: 10.1136/hrt.2005.061143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Starr JP. Tetralogy of Fallot: Yesterday and Today. World Journal of Surgery. 2009;34:658. doi: 10.1007/s00268-009-0296-8. [DOI] [PubMed] [Google Scholar]
- 5.Cuypers JA, Menting ME, Konings EE, et al. Unnatural History of Tetralogy of Fallot: Prospective Follow-Up of 40 Years After Surgical Correction. Circulation. 2014;130:1944. doi: 10.1161/CIRCULATIONAHA.114.009454. [DOI] [PubMed] [Google Scholar]
- 6.Folino AF, Daliento L. Arrhythmias after tetralogy of Fallot repair. Indian pacing and electrophysiology journal. 2005;5(4):312–324. [PMC free article] [PubMed] [Google Scholar]
- 7.Fuller S. Tetralogy of Fallot and Pulmonary Valve Replacement: Timing and Techniques in the Asymptomatic Patient. Seminars in Thoracic and Cardiovascular Surgery: Pediatric Cardiac Surgery Annual. 2014;17:30. doi: 10.1053/j.pcsu.2014.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Mercer-Rosa L, Rychik J, Zhao H, Zhang X, Yang W, Shults J, Goldmuntz E. 22q11.2 Deletion Status and Disease Burden in Children and Adolescents With Tetralogy of Fallot. Clinical Perspective. Circulation: Cardiovascular Genetics. 2015;8:74. doi: 10.1161/CIRCGENETICS.114.000819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lindsay EA, Vitelli F, Su H, Morishima M, Huynh T, Pramparo T, Jurecic V, Ogunrinu G, Sutherland HF, Scambler PJ, Bradley A, et al. Tbx1 haploinsufficieny in the DiGeorge syndrome region causes aortic arch defects in mice. Nature. 2001;410:97–101. doi: 10.1038/35065105. [DOI] [PubMed] [Google Scholar]
- 10.Doza JP, Topf A, Bentham J, et al. Low-frequency intermediate penetrance variants in the ROCK1 gene predispose to Tetralogy of Fallot. BMC Genet. 2013;14:57. doi: 10.1186/1471-2156-14-57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Soemedi R, Topf A, Darlay R, et al. Phenotype-specific effect of chromosome 1q21.1 rearrangements and GJA5 duplications in 2436 congenital heart disease patients and 6760 controls. Human Molecular Genetics. 2011;21:1513. doi: 10.1093/hmg/ddr589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Griffin HR, Glen E, Soemedi R, Brown DL, Hall D, Rahman TJ, Eloranta JJ, Jüngst C, Stuart AG, O'Sullivan J, Keavney BD, et al. Functionally significant, rare transcription factor variants in tetralogy of Fallot. PLoS ONE. 2014;9:e95453. doi: 10.1371/journal.pone.0095453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goodship JA, Hall D, Topf A, et al. A Common Variant in the PTPN11 Gene Contributes to the Risk of Tetralogy of Fallot. Circulation: Cardiovascular Genetics. 2012;5:287. doi: 10.1161/CIRCGENETICS.111.962035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zaidi S, Choi M, Wakimoto H, et al. De novo mutations in histone-modifying genes in congenital heart disease. Nature. 2013;498:220. doi: 10.1038/nature12141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordell HJ, Bentham J, Topf A, et al. Genome-wide association study of multiple congenital heart disease phenotypes identifies a susceptibility locus for atrial septal defect at chromosome 4p16. Nature Genetics. 2013;45:822. doi: 10.1038/ng.2637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cordell HJ, Mamasoula C, Postma AV, et al. Genome-wide association study identifies loci on 12q24 and 13q32 associated with tetralogy of Fallot. Human Molecular Genetics. 2013;22:1473–1481. doi: 10.1093/hmg/dds552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Homsy J, Zaidi S, Shen Y, et al. De novo mutations in congenital heart disease with neurodevelopmental and other congenital anomalies. Science. 2015;350:1262. doi: 10.1126/science.aac9396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soemedi R, Bentham J, Darlay R, et al. Contribution of Global Rare Copy-Number Variants to the Risk of Sporadic Congenital Heart Disease. The American Journal of Human Genetics. 2012;91:489. doi: 10.1016/j.ajhg.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Al Turki S, Manickaraj AK, Mercer CL, et al. Rare variants in NR2F2 cause congenital heart defects in humans. The American Journal of Human Genetics. 2014;94:574–585. doi: 10.1016/j.ajhg.2014.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sifrim A, Hitz M-P, Wilsdon A, et al. Distinct genetic architectures for syndromic and nonsyndromic congenital heart defects identified by exome sequencing. Nature Genetics. 2016;48:1060–1065. doi: 10.1038/ng.3627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Griffin HR, Glen E, Zweier C, Stuart AG, Parsons J, Peart I, Deanfield J, O'Sullivan J, Rauch A, Scambler P, Burn J, et al. Systematic survey of variants in TBX1 in non-syndromic tetralogy of Fallot identifies a novel 57 base pair deletion that reduces transcriptional activity but finds no evidence for association with common variants. Heart. 2010;96:1651–1655. doi: 10.1136/hrt.2010.200121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Goldmuntz E, Clark BJ, Mitchell LE, Jawad AF, Cuneo BF, Reed L, McDonald-McGinn D, Chien P, Feuer J, Zackai EH, Emanuel BS, et al. Frequency of 22q11 deletions in patients with conotruncal defects. Journal of the American College of Cardiology. 1998;32:492–498. doi: 10.1016/s0735-1097(98)00259-9. [DOI] [PubMed] [Google Scholar]
- 23.Benhaourech S, Drighil A, El Hammiri A. Congenital heart disease and Down syndrome: various aspects of a confirmed association. Cardiovasc J Afr. 2016;27:287–290. doi: 10.5830/CVJA-2016-019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Benson DW, Silberbach GM, Kavanaugh-McHugh A, Cottrill C, Zhang Y, Riggs S, Smalls O, Johnson MC, Watson MS, Seidman JG, Seidman CE, et al. Mutations in the cardiac transcription factor NKX2.5 affect diverse cardiac developmental pathways. Journal of Clinical Investigation. 1999;104:1567–1573. doi: 10.1172/JCI8154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Jin SC, Homsy J, Zaidi S, et al. Contribution of rare inherited and de novo variants in 2,871 congenital heart disease probands. Nature Genetics. 2017;49:1593–1601. doi: 10.1038/ng.3970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Priest JR, Osoegawa K, Mohammed N, et al. De Novo and Rare Variants at Multiple Loci Support the Oligogenic Origins of Atrioventricular Septal Heart Defects. PLoS Genetics. 2016;12:e1005963. doi: 10.1371/journal.pgen.1005963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katz SG, Williams A, Yang J, Fujiwara Y, Tsang AP, Epstein JA, Orkin SH. Endothelial lineage-mediated loss of the GATA cofactor Friend of GATA 1 impairs cardiac development. Proceedings of the National Academy of Sciences. 2003;100:14030–14035. doi: 10.1073/pnas.1936250100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Barron F, Woods C, Kuhn K, Bishop J, Howard MJ, Clouthier DE. Downregulation of Dlx5 and Dlx6 expression by Hand2 is essential for initiation of tongue morphogenesis. Development. 2011;138:2249. doi: 10.1242/dev.056929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Li Y, Klena NT, Gabriel GC, et al. Global genetic analysis in mice unveils central role for cilia in congenital heart disease. Nature. 2015;521:520. doi: 10.1038/nature14269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Goldmuntz E, Geiger E, Benson DW. NKX2.5 mutations in patients with tetralogy of fallot. Circulation. 2001;104:2565–2568. doi: 10.1161/hc4601.098427. [DOI] [PubMed] [Google Scholar]
- 31.Nemer G, Fadlalah F, Usta J, Nemer M, Dbaibo G, Obeid M, Bitar F. A novel mutation in the GATA4 gene in patients with Tetralogy of Fallot. Hum Mutat. 2006;27:293–294. doi: 10.1002/humu.9410. [DOI] [PubMed] [Google Scholar]
- 32.Lin X, Huo Z, Liu X, Zhang Y, Li L, Zhao H, Yan B, Liu Y, Yang Y, Chen Y-H. A novel GATA6 mutation in patients with tetralogy of Fallot or atrial septal defect. J Hum Genet. 2010;55:662–667. doi: 10.1038/jhg.2010.84. [DOI] [PubMed] [Google Scholar]
- 33.Tien A-C, Rajan A, Bellen HJ. A Notch updated. The Journal of Cell Biology. 2009;184:621. doi: 10.1083/jcb.200811141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Choi SH, Wales TE, Nam Y, O'Donovan DJ, Sliz P, Engen JR, Blacklow SC. Conformational locking upon cooperative assembly of notch transcription complexes. Structure. 2012;20:340–349. doi: 10.1016/j.str.2011.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shimizu H, Woodcock SA, Wilkin MB, Trubenová B, Monk NAM, Baron M. Compensatory flux changes within an endocytic trafficking network maintain thermal robustness of Notch signaling. Cell. 2014;157:1160–1174. doi: 10.1016/j.cell.2014.03.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jordan VK, Rosenfeld JA, Lalani SR, Scott DA. Duplication ofHEY2in cardiac and neurologic development. American Journal of Medical Genetics Part A. 2015;167:2145. doi: 10.1002/ajmg.a.37086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Eldadah ZA, Hamosh A, Biery NJ, Montgomery RA, Duke M, Elkins R, Dietz HC. Familial Tetralogy of Fallot caused by mutation in the jagged1 gene. Human Molecular Genetics. 2001;10:163–169. doi: 10.1093/hmg/10.2.163. [DOI] [PubMed] [Google Scholar]
- 38.Bauer RC, Laney AO, Smith R, Gerfen J, Woyciechowski S, Garbarini J, Loomes KM, Krantz ID, Urban Z, Gelb BD, Goldmuntz E, et al. Jagged1 (JAG1) mutations in patients with tetralogy of fallot or pulmonic stenosis. Hum Mutat. 2010;31:594. doi: 10.1002/humu.21231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersen P, Uosaki H, Shenje LT, Kwon C. Non-canonical Notch signaling: emerging role and mechanism. Trends in Cell Biology. 2012;22:257. doi: 10.1016/j.tcb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Logeat F, Bessia C, Brou C, LeBail O. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proceedings of the National Academy of Sciences. 1998;95(14):8108–12. doi: 10.1073/pnas.95.14.8108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blue GM, Kirk EP, Giannoulatou E, Dunwoodie SL, White SM, Sholler GF, Harvey RP, Winlaw DS. Targeted Next-Generation Sequencing Identifies Pathogenic Variants in Familial Congenital Heart Disease. Journal of the American College of Cardiology. 2014;64:2498. doi: 10.1016/j.jacc.2014.09.048. [DOI] [PubMed] [Google Scholar]
- 42.Burn J, Brennan P, Little J, et al. Recurrence risks in offspring of adults with major heart defects: results from first cohort of British collaborative study. Lancet. 1998;351:311–316. doi: 10.1016/s0140-6736(97)06486-6. [DOI] [PubMed] [Google Scholar]
- 43.Garg V, Muth AN, Ransom JF, Schluterman MK, Barnes R, King IN, Grossfeld PD, Srivastava D. Mutations in NOTCH1 cause aortic valve disease. Nature. 2005;437:270–274. doi: 10.1038/nature03940. [DOI] [PubMed] [Google Scholar]
- 44.Kerstjens-Frederikse WS, van de Laar IM, Vos YJ, et al. Cardiovascular malformations caused by NOTCH1 mutations do not keep left: data on 428 probands with left-sided CHD and their families. Genetics in Medicine. 2016;18:914. doi: 10.1038/gim.2015.193. [DOI] [PubMed] [Google Scholar]
- 45.Yang C, Xu Y, Yu M, et al. Induced pluripotent stem cell modelling of HLHS underlines the contribution of dysfunctional NOTCH signalling to impaired cardiogenesis. Human Molecular Genetics. 2017 doi: 10.1093/hmg/ddx140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McKellar SH, Tester DJ, Yagubyan M, Majumdar R, Ackerman MJ, Sundt TM. Novel NOTCH1 mutations in patients with bicuspid aortic valve disease and thoracic aortic aneurysms. J Thorac Cardiovasc Surg. 2007;134:290–296. doi: 10.1016/j.jtcvs.2007.02.041. [DOI] [PubMed] [Google Scholar]
- 47.Iascone M, Ciccone R, Galletti L, Marchetti D, Seddio F, Lincesso AR, Pezzoli L, Vetro A, Barachetti D, Boni L, Federici D, et al. Identification of de novo mutations and rare variants in hypoplastic left heart syndrome. Clinical Genetics. 2011;81:542. doi: 10.1111/j.1399-0004.2011.01674.x. [DOI] [PubMed] [Google Scholar]
- 48.Bittel DC, Zhou X-G, Kibiryeva N, Fiedler S, O’Brien JE, Marshall J, Yu S, Liu H-Y. Ultra High-Resolution Gene Centric Genomic Structural Analysis of a Non-Syndromic Congenital Heart Defect, Tetralogy of Fallot. PLoS ONE. 2014;9:e87472. doi: 10.1371/journal.pone.0087472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Greenway SC, Pereira AC, Lin JC, et al. De novo copy number variants identify new genes and loci in isolated sporadic tetralogy of Fallot. Nature Genetics. 2009;41:931. doi: 10.1038/ng.415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bittel DC, Butler MG, Kibiryeva N, Marshall JA, Chen J, Lofland GK. Gene expression in cardiac tissues from infants with idiopathic conotruncal defects. BMC Medical Genomics. 2011;4:1. doi: 10.1186/1755-8794-4-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Southgate L, Sukalo M, Karountzos ASV, et al. Haploinsufficiency of the NOTCH1 Receptor as a Cause of Adams-Oliver Syndrome With Variable Cardiac Anomalies. Circulation: Cardiovascular Genetics. 2015;8:572–581. doi: 10.1161/CIRCGENETICS.115.001086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stittrich A-B, Lehman A, Bodian DL, et al. Mutations in NOTCH1 cause Adams-Oliver syndrome. The American Journal of Human Genetics. 2014;95:275–284. doi: 10.1016/j.ajhg.2014.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Swartz EN, Sanatani S, Schreiber RA. Vascular abnormalities in Adams-Oliver syndrome: Cause or effect? American Journal of Medical Genetics. 1999;82:49. doi: 10.1002/(sici)1096-8628(19990101)82:1<49::aid-ajmg10>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 54.Weismann CG, Hager A, Kaemmerer H, Maslen CL, Morris CD, Schranz D, Kreuder J, Gelb BD. PTPN11 mutations play a minor role in isolated congenital heart disease. American Journal of Medical Genetics Part A. 2005;136A:146. doi: 10.1002/ajmg.a.30789. [DOI] [PubMed] [Google Scholar]
- 55.Jia Y, Louw JJ, Breckpot J, Callewaert B, Barrea C, Sznajer Y, Gewillig M, Souche E, Dehaspe L, Vermeesch JR, Lambrechts D, et al. The diagnostic value of next generation sequencing in familial nonsyndromic congenital heart defects. American Journal of Medical Genetics Part A. 2015;167A:1822–1829. doi: 10.1002/ajmg.a.37108. [DOI] [PubMed] [Google Scholar]
- 56.Grego-Bessa J, Luna-Zurita L, del Monte G, et al. Notch Signaling Is Essential for Ventricular Chamber Development. Dev Cell. 2007;12:415. doi: 10.1016/j.devcel.2006.12.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.High FA, Jain R, Stoller JZ, Antonucci NB, Lu MM, Loomes KM, Kaestner KH, Pear WS, Epstein JA. Murine Jagged1/Notch signaling in the second heart field orchestrates Fgf8 expression and tissue-tissue interactions during outflow tract development. Journal of Clinical Investigation. 2009;119(7):1986–96. doi: 10.1172/JCI38922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Timmerman LA. Notch promotes epithelial-mesenchymal transition during cardiac development and oncogenic transformation. Genes Dev. 2004;18:99. doi: 10.1101/gad.276304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Conlon RA, Reaume AG, Rossant J. Notch1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- 60.Swiatek PJ, Lindsell CE, del Amo FF, Weinmaster G, Gridley T. Notch1 is essential for postimplantation development in mice. Genes Dev. 1994;8:707. doi: 10.1101/gad.8.6.707. [DOI] [PubMed] [Google Scholar]
- 61.MacGrogan D, D’Amato G, Travisano S, Martinez-Poveda B, Luxán G, del Monte-Nieto G, Papoutsi T, Sbroggio M, Bou V, Arco PG-D, Gómez MJ, et al. Sequential Ligand-Dependent Notch Signaling Activation Regulates Valve Primordium Formation and MorphogenesisNovelty and Significance. Circulation Research. 2016;118:1480. doi: 10.1161/CIRCRESAHA.115.308077. [DOI] [PubMed] [Google Scholar]
- 62.Koenig SN, Bosse K, Majumdar U, Bonachea EM, Radtke F, Garg V. Endothelial Notch1 Is Required for Proper Development of the Semilunar Valves and Cardiac Outflow Tract. J Am Heart Assoc. 2016;5:4. doi: 10.1161/JAHA.115.003075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Koenig SN, LaHaye S, Feller JD, Rowland P, Hor KN, Trask AJ, Janssen PM, Radtke F, Lilly B, Garg V. Notch1 haploinsufficiency causes ascending aortic aneurysms in mice. JCI Insight. 2017;2:21. doi: 10.1172/jci.insight.91353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.McBride KL, Riley MF, Zender GA, Fitzgerald-Butt SM, Towbin JA, Belmont JW, Cole SE. NOTCH1 mutations in individuals with left ventricular outflow tract malformations reduce ligand-induced signaling. Human Molecular Genetics. 2008;17:2886. doi: 10.1093/hmg/ddn187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Szot JO, Cuny H, Blue GM, Humphreys DT. A Screening Approach to Identify Clinically Actionable Variants Causing Congenital Heart Disease in Exome Data. Am Heart Assoc. 2018;11(3):e001978. doi: 10.1161/CIRCGEN.117.001978. [DOI] [PubMed] [Google Scholar]
- 66.Tammela T, Alitalo K. Lymphangiogenesis: Molecular mechanisms and future promise. Cell. 2010;140:460–76. doi: 10.1016/j.cell.2010.01.045. [DOI] [PubMed] [Google Scholar]
- 67.Partanen TA, Arola J, Saaristo A, Jussila L, Ora A, Miettinen M, Stacker SA, Achen MG, Alitalo K. VEGF-C and VEGF-D expression in neuroendocrine cells and their receptor, VEGFR-3, in fenestrated blood vessels in human tissues. The FASEB Journal. 2000;14:2087. doi: 10.1096/fj.99-1049com. [DOI] [PubMed] [Google Scholar]
- 68.Covassin LD, Villefranc JA, Kacergis MC, Weinstein BM, Lawson ND. Distinct genetic interactions between multiple Vegf receptors are required for development of different blood vessel types in zebrafish. Proceedings of the National Academy of Sciences. 2006;103:6554–9. doi: 10.1073/pnas.0506886103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Tammela T, Zarkada G, Wallgard E, et al. Blocking VEGFR-3 suppresses angiogenic sprouting and vascular network formation. Nature. 2008;454:656. doi: 10.1038/nature07083. [DOI] [PubMed] [Google Scholar]
- 70.Dumont DJ, Jussila L, Taipale J, Lymboussaki A, Mustonen T, Pajusola K, Breitman M, Alitalo K. Cardiovascular failure in mouse embryos deficient in VEGF receptor-3. Science. 1998;282:946–9. doi: 10.1126/science.282.5390.946. [DOI] [PubMed] [Google Scholar]
- 71.Zvaritch E, Depreux F, Kraeva N, Loy RE, Goonasekera SA, Boncompagni S, Kraev A, Gramolini AO, Dirksen RT, Franzini-Armstrong C, Seidman CE, et al. An Ryr1I4895T mutation abolishes Ca2+ release channel function and delays development in homozygous offspring of a mutant mouse line. Proceedings of the National Academy of Sciences. 2007;104:18537. doi: 10.1073/pnas.0709312104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Walton RZ, Olivey HE, Najib K, Johnson V, Earley JU, Ho RK, Svensson EC. Fog1 is required for cardiac looping in zebrafish. Developmental Biology. 2006;289:482. doi: 10.1016/j.ydbio.2005.10.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Song K, Backs J, McAnally J, Qi X, Gerard RD, Richardson JA, Hill JA, Bassel-Duby R, Olson EN. The transcriptional coactivator CAMTA2 stimulates cardiac growth by opposing class II histone deacetylases. Cell. 2006;125:453–466. doi: 10.1016/j.cell.2006.02.048. [DOI] [PubMed] [Google Scholar]
- 74.Xie L, Chen J-L, Zhang W-Z, Wang S-Z, Zhao T-L, Huang C, Wang J, Yang J-F, Yang Y-F, Tan Z-P. Rare de novo copy number variants in patients with congenital pulmonary atresia. PLoS ONE. 2014;9:e96471. doi: 10.1371/journal.pone.0096471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim K-S, Arima Y, Kitazawa T, Nishiyama K, Asai R, Uchijima Y, Sato T, Levi G, Kitanaka S, Igarashi T, Kurihara Y, et al. Endothelin regulates neural crest deployment and fate to form great vessels through Dlx5/Dlx6-independent mechanisms. Mechanisms of Development. 2013;130:553. doi: 10.1016/j.mod.2013.07.005. [DOI] [PubMed] [Google Scholar]
- 76.Wang L, Lee K, Malonis R, Sanchez I, Dynlacht BD. Tethering of an E3 ligase by PCM1 regulates the abundance of centrosomal KIAA0586/Talpid3 and promotes ciliogenesis. eLife. 2016;5 doi: 10.7554/eLife.12950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ruark E, Münz M, Renwick A, Clarke M, Ramsay E, Hanks S, Mahamdallie S, Elliott A, Seal S, Strydom A, Gerton L, et al. The ICR1000 UK exome series: a resource of gene variation in an outbred population. F1000Research. 2015;4:883. doi: 10.12688/f1000research.7049.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.