Abstract
Purpose:
MET exon 14 splice site alterations that cause exon skipping at the mRNA level (METex14) are actionable oncogenic drivers amenable to therapy with MET tyrosine kinase inhibitors (TKI); however, secondary resistance eventually arises in most cases while other tumors display primary resistance. Beyond relatively uncommon on-target MET kinase domain mutations, mechanisms underlying primary and acquired resistance remain unclear.
Experimental Design:
We examined clinical and genomic data from 113 lung cancer patients with METex14. MET TKI resistance due to KRAS mutation was functionally evaluated using in vivo and in vitro models.
Results:
Five of 113 patients (4.4%) with METex14 had concurrent KRAS G12 mutations, a rate of KRAS co-occurrence significantly higher than in other major driver-defined lung cancer subsets. In one patient, the KRAS mutation was acquired post-crizotinib, while the remaining 4 METex14 patients harbored the KRAS mutation prior to MET TKI therapy. Gene set enrichment analysis of transcriptomic data from lung cancers with METex14 revealed preferential activation of the KRAS pathway. Moreover, expression of oncogenic KRAS enhanced MET expression. Using isogenic and patient-derived models, we show that KRAS mutation results in constitutive activation of RAS/ERK signaling and resistance to MET inhibition. Dual inhibition of MET or EGFR/ERBB2 and MEK reduced growth of cell line and xenograft models.
Conclusions:
KRAS mutation is a recurrent mechanism of primary and secondary resistance to MET TKIs in METex14 lung cancers. Dual inhibition of MET or EGFR/ERBB2 and MEK may represent a potential therapeutic approach in this molecular cohort. (245/250 words)
Translational Relevance:
METex14 alterations have emerged as one of the most common targetable alterations in non-small cell lung cancers, and are amenable to MET kinase inhibitor therapy; however, inhibitor activity can be limited by primary or acquired genomic aberrations that lead to therapeutic resistance, warranting the investigation of alternative therapeutic approaches. We show that concurrent KRAS mutation is more common in the setting of METex14 than with other major drivers (e.g. sensitizing EGFR and ALK alterations), and confers primary or secondary resistance to MET-directed therapies in METex14-altered lung cancers. Dual inhibition of MET or EGFR/ERBB2 and MEK reduced growth of cell line and xenograft models. These results provide a rationale for clinical evaluation of these combination approaches. (115/120–150 words)
Keywords: MET exon14, KRAS, drug resistance, lung cancer, next-generation sequencing
Introduction
The MET gene encodes the MET receptor tyrosine kinase. A variety of mechanisms activate MET signaling pathways, including MET gene amplification, protein overexpression, activating point mutations, and fusions (1–4). MET exon14 (METex14) skipping alterations were first discovered in lung cancers in 2003 (5), and have been demonstrated to be oncogenic in preclinical models (6). The splice site mutations flanking exon 14 of MET result in an in-frame transcript missing exon 14, and consequently a MET protein lacking the juxtamembrane domain that includes the CBL E3 ubiquitin ligase binding site (Y1003). The MET receptor lacking exon 14 shows decreased ubiquitination and protein turnover, resulting in aberrant MET accumulation, causing activation of downstream signaling, and ultimately oncogenesis (6).
METex14 alterations have been identified in 3–4 % of lung adenocarcinomas (7–10), a frequency comparable to that of ALK fusions, placing them among the top three most common targetable kinase alterations in lung adenocarcinoma, along with EGFR and ALK. Responses to MET inhibitors, such as crizotinib and cabozantinib, have been reported in lung cancer patients with METex14 alterations, including the PROFILE 1001 (NCT00585195) trial which found an objective response rate to crizotinib of 39% with a progression free survival of 8 months in the 28 evaluable patients (8, 11, 12). Based on these data, crizotinib has been incorporated into the National Comprehensive Cancer Network (NCCN) guidelines (Version 1.2018 Non-Small Cell Lung Cancer) as an emerging therapy for the treatment of METex14-altered lung cancers.
Acquired resistance to targeted therapy can occur by a variety of mechanisms, including second-site mutations in the kinase domain, bypass signaling pathway activation, copy number changes, and histological transformation (13–15). Recently, secondary mutations in the MET kinase domain, specifically D1228N/V and Y1230C, have been reported as mechanisms of acquired resistance to crizotinib in patients with METex14 alterations that were treated with MET tyrosine kinase inhibitor (TKI) therapy (16, 17). These point mutations have been shown to alter the binding kinetics of MET TKIs to the ATP-binding pocket (18); however, to date, there have been no reports of target-independent mechanisms of resistance to MET inhibition in METex14-altered lung cancers. In the present study, we show a predilection for METex14 alterations to occur concurrently with mutant KRAS, and demonstrate that activated KRAS represents a novel molecular mechanism mediating either primary or acquired resistance to crizotinib therapy in patients with METex14 skipping alterations.
Materials and methods
Clinical sequencing data.
We performed a retrospective analysis of all patients with METex14 alterations lung cancers seen at Memorial Sloan Kettering Cancer Center (MSKCC) from January 2014 through April 2018. Genomic data were available from the MSK-IMPACT™ large panel sequencing assay, a clinical test designed to detect mutations, copy-number alterations, and select gene fusions involving up to 468 cancer-associated genes (19, 20). A subset of cases were also studied by a custom ArcherDx targeted RNAseq assay based on anchored multiplex PCR (21), designed to detect kinase fusions and METex14 skipping at the mRNA level.
Cell lines.
NIH-3T3, HEK-293T, H1993, and Hs746T cells were purchased from American Type Culture Collection (Manassas, VA). PC9 cells were obtained from RIKEN Cell Bank (Tsukuba, Ibaraki, Japan). The LUAD12C cell line was established under an institutional review board approved biomarker specimen protocol from a pleural effusion sample derived from PT-5 with lung cancer harboring METex14 skipping and KRAS G12S mutation after resistance to crizotinib and a MET bivalent antibody had emerged. Cell lines were routinely tested for mycoplasma. Cell culture media and antibiotics were prepared by the MSKCC Media Preparation Core Facility. LUAD12C cells were cultured in a mixture of Dulbecco’s modified Eagle’s medium (DMEM) and Ham’s F12 medium (1:1 ratio) supplemented with 2% fetal bovine serum (FBS). NIH-3T3 cells were cultured in DMEM supplemented with 10% calf serum. HEK-293T and Hs746T cells were cultured in DMEM media supplemented with 10% FBS. PC9 and H1993 cells were cultured in RPMI-1640 supplemented with 10% FBS.
Gene expression analyses.
Gene expression microarray data for 83 lung adenocarcinomas collected at the British Columbia Cancer Agency (BCCA, Vancouver, BC) were generated using Illumina HumanWG-6 v3.0 arrays and normalized as previously described (GEO Accession Number = GSE75037). Data from Affymetrix SNP6 arrays were used to determine the amplification status of MET for these same cases as previously described while Illumina custom capture was performed to determine METex14 status. In total, five cases had wildtype MET amplification while three had METex14 mutations. RNA-seq data (RSEM) for 230 lung adenocarcinomas profiled by The Cancer Genome Atlas (TCGA, provisional data set) were downloaded from the UCSC Cancer Genomics Browser and MET amplification and METex14 status were determined for these same cases using the MSKCC cBIO Portal. Seven tumors had wildtype MET amplification while seven harbored METex14 mutations. For each dataset, expression of each gene was compared between METex14 and wildtype MET amplified tumors using a T-Test and a fold change between groups determined. Genes were ranked from high to low using the fold change, and the log10 (negative fold change) or –log10 (positive fold change) of the resulting p-value determined to create a ranking metric. Pre-ranked Gene Set Enrichment Analysis (GSEA-P) was then run against the “Oncogenic Pathways” signature database on the resulting gene list using default parameters and weighted enrichment statistic using the above metric, as previously described (22). Resulting FDR values were used to rank METex14 upregulated pathways for both the BCCA and TCGA datasets and enrichment plots determined. Morpheus software (https://software.broadinstitute.org/morpheus) was used to make all heatmaps.
Results
Co-occurrence of KRAS and METex14 alterations
Of the 3,632 patients whose non-small cell lung cancer specimens underwent clinical next-generation sequencing using the MSK-IMPACT™ assay as of April 2018, 113 (3%) were found to harbor METex14 alterations. Of these, 7% (n = 8/113) had concurrent alterations activating KRAS, including 5 with KRAS exon 2 mutations resulting in codon 12 amino acid substitutions (2 of which had concurrent amplification of the mutant allele), and 3 with amplification of wildtype KRAS. In all 8 cases of concurrent METex14 and KRAS altered specimens, the MET alterations were noted on the DNA-based MSK-IMPACT platform and were previously known to be associated with MET exon 14 skipping. Of the remaining 105 METex14 patients identified, 104 underwent MSK-IMPACT testing and 16 had ArcherDx available for review. Of the 16 patients with ArcherDx data, 15 had simultaneous MSK-IMPACT data for review. Of these 15 patients, 1 case was not noted to have the METex14 alteration by MSK-IMPACT. When the prevalence of concurrent KRAS alterations in the METex14 cohort is compared to that in other lung cancer subsets defined by oncogenic drivers in the total lung cancer cohort (n = 3,632), a significant predilection for concurrent KRAS aberrations is evident in the METex14 population (p = 0.0005). While this still represents only a minority of METex14 cases, its statistical significance reflects the fact that, in contrast, concurrent KRAS alterations are absent or rare in cases with EGFR, ALK, RET, or ROS1 alterations (Table 1A).
Table 1:
A: Comparison of concurrent KRAS mutations and amplification by driver status. Evaluation of concurrent KRAS alterations to drivers was evaluated using the Fischer’s exact test. In a pooled analysis of all the above-mentioned drivers for concurrent KRAS mutations (991 patients, 19 of which had concurrent KRAS alterations) compared to the 113 METex14 mutant patients (8 of which had alterations in KRAS) there was a statistically significant increased co-occurrence of KRAS alterations in the METex14cohort (p =0.004); B: Brief description of the 8 patients with METex14 mutant lung cancer with KRAS alterations during treatment course.
| A: | |||
|---|---|---|---|
| Genomic Alteration | Percent of Total Cohort (n=3,632) | Frequency of Concurrent KRAS Alteration1 (n) | P-value2 |
| METex14-altered | 3% (113) | 7% (8: 3M, 3A, 2M/A) | 0.0005 |
| EGFR-mutant3 | 19% (706) | 1.5% (11: 4M, 7A) | 0.47 |
| ALK fusion | 3% (119) | 1% (1A) | 0.71 |
| RET fusion | 2% (62) | 0% (0) | 0.62 |
| ROS1 fusion | 2% (63) | 0% | 0.62 |
| B: | |||
|---|---|---|---|
| Patient ID | MET | KRAS | Targeted Treatment |
| PT-1 | METex14 | G12D | None |
| PT-2 | METex14 | G12A with amplification | Crizotinib |
| PT-3 | Y1003C | G12C | Crizotinib |
| PT-4 | METex14 with amplification | G12C | None |
| PT-5 | METex14 | G12S with amplification | Crizotinib, MET antibody |
| PT-6 | METex14 | Amplification | None |
| PT-7 | METex14 | Amplification | Crizotinib, cabozantinib |
| PT-8 | METex14 | Amplification | None |
KRAS know hotspot mutation (M) or KRAS amplification (A); or both KRAS mutation and amplification
Fisher’s exact test for prevalence of concurrent KRAS alterations in indicated subset compared to aggregate of the other driver subsets in the table.
Defined as known oncogenic alterations in L858R and exon 19 deletion/insertions.
The presence of a concurrent KRAS alteration was associated either with a short period of disease control with MET inhibition, or primary progressive disease in the majority of cases where clinical data were available (Supplemental Table 1A). In 7 of the 8 cases of METex14-altered tumors with concurrent KRAS alterations (mutation or amplification), the KRAS alteration was present de novo (i.e. prior to MET-directed therapy, if administered). Three of these patients received crizotinib (PT-2 and PT-3 with KRAS mutations, and PT-7 with KRAS amplification), and achieved transient disease stabilization with therapy (Table 1B and Supplemental Table 1A).
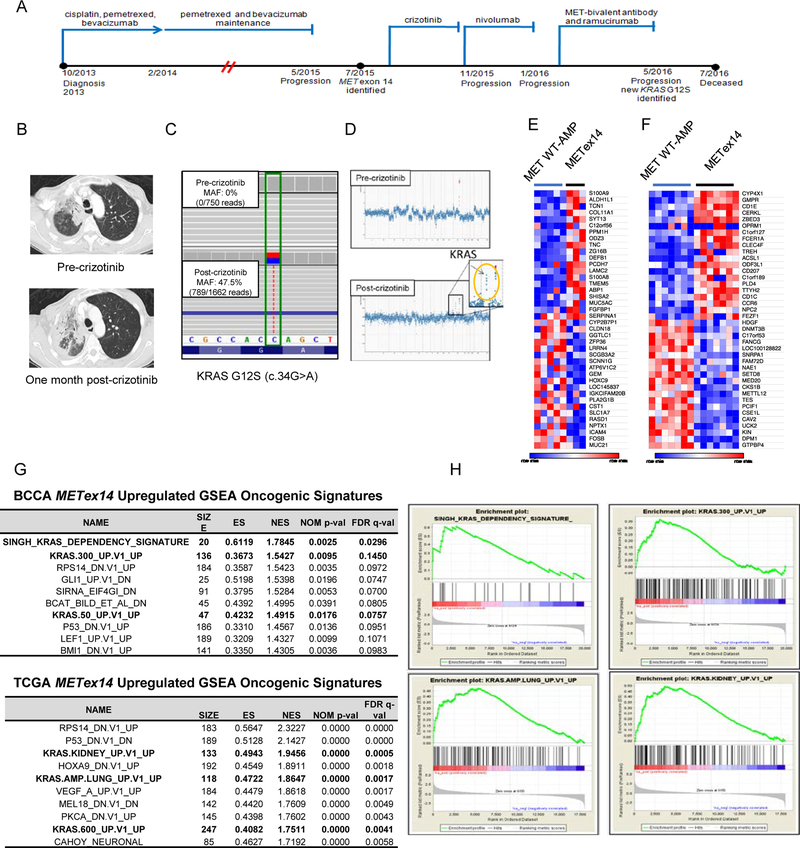
In the remaining patient (PT-5), an acquired KRAS mutation was identified after crizotinib therapy (Table 1B, Supplemental Table 1A). This patient was a 60-year-old female who presented with metastatic lung adenocarcinoma involving the left pleura. Her clinical course is summarized in Figure 1A. She was treated with crizotinib following the identification of a METex14 alteration (c.2888–16_c.2888–2 del). Further imaging after 2 cycles of therapy revealed primary progressive disease (Figure1B). The re-biopsy sample from a dermal metastatic focus revealed acquired KRAS G12S (c.34G>A) mutation present at an elevated allelic fraction (47.5%, 789/1662 reads), reflecting amplification of the mutant allele (Figures 1C and 1D).
Figure 1: A patient with lung adenocarcinoma harboring MET exon14 alteration and acquired KRAS G12S.
A. The clinical course and treatment history of PT-5 with a MET exon 14 alteration (KRAS wild type) at the time of progression on cytotoxic chemotherapy. The next biopsy took place after progression on MET bivalent antibody and showed a newly acquired KRAS G12S mutation. B. Computed tomography scans of the patient before and after progression during crizotinib treatment. C. MSK-IMPACT sequence analysis of paired samples before and after crizotinib resistance. D. The copy number plots for paired samples before and after crizotinib resistance. E. The top and bottom 20 genes differentially expressed in METex14 (n=3) vs MET wildtype amplified (n=5) lung adenocarcinomas from BCCA. F. The same analysis but for TCGA tumors with 7 METex14 and 7 MET wildtype amplified tumors. G. The top GSEA Oncogenic Signatures upregulated in METex14 vs MET wildtype amplified tumors. Gene sets related to KRAS signaling are indicated in bold. H. Representative GSEA enrichment plots for the top KRAS-related signatures in BCCA (top) and TCGA (bottom).
KRAS signaling is upregulated in METex14-driven vs wildtype MET-amplified tumors
To determine factors that might underlie this apparent predilection for KRAS activation, we compared expression profiles of tumors with METex14 to those with wildtype MET amplification in two independent cohorts (Figures 1E and 1F). Gene Set Enrichment Analysis was performed to identify oncogenic pathways preferentially upregulated in the METex14 context. Notably, in two independent datasets (BCCA and TCGA), multiple GSEA signatures related to upregulation of KRAS signaling were among the most enriched in the METex14 tumors (Figures 1G.and 1H). These included signatures specifically related to KRAS activation in the lung (Figures 1G and 1H). Importantly, none of the METex14 cases in these two datasets contained concurrent KRAS mutation suggesting that METex14 drives these effects independently. Thus, METex14 tumors are associated with a higher level of KRAS signaling than tumors with amplification of wild type MET. It may therefore be hypothesized that METex14 tumor cells may be more tolerant of, or may differentially benefit from, further activation of KRAS signaling via KRAS mutations, explaining their increased prevalence of co-occurrence.
Expression of mutant KRAS induces resistance to MET-directed therapies
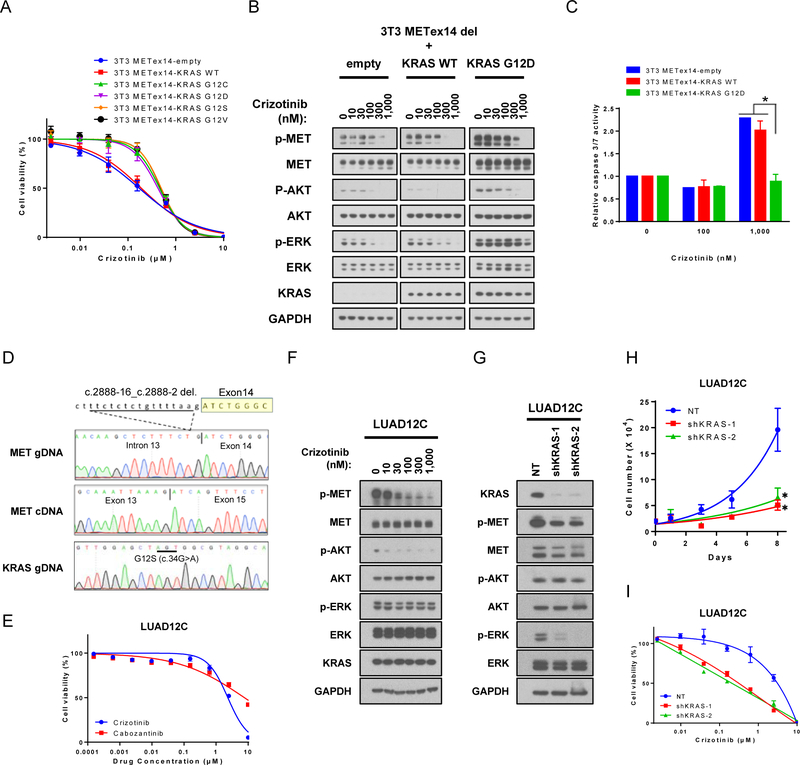
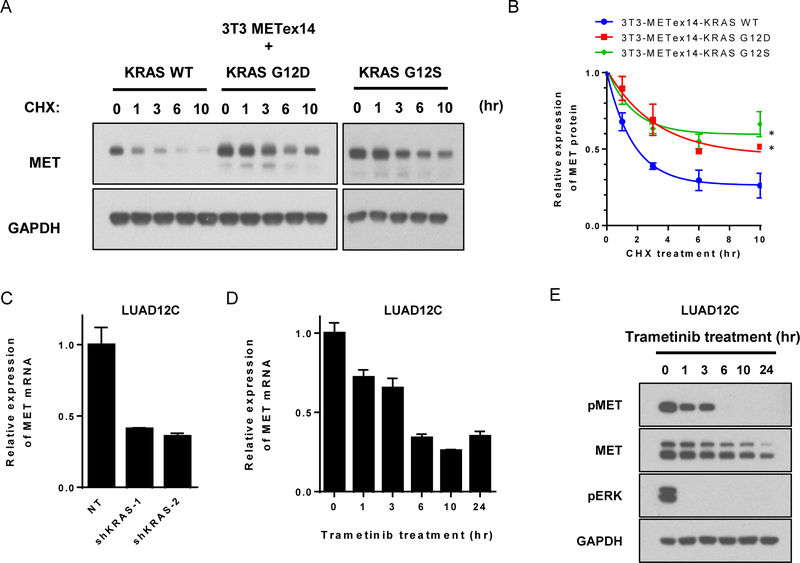
To validate the role of KRAS mutations in resistance to MET-directed therapies, we generated isogenic models. The only publicly available lung cancer cell line with METex14, H596, is insensitive to MET TKI, probably due to a concurrent PIK3CA mutation (23). Therefore, we generated isogenic NIH-3T3 cell line models by transduction with lentiviral vectors driving expression of METex14 or empty vector (3T3-METex14-empty), KRAS wildtype (3T3-METex14-KRAS WT) or KRAS mutants (3T3-METex14-KRAS G12C, G12D, G12S, and G12V). Focus formation assays showed that neither 3T3-METex14-empty nor 3T3-METex14-KRAS WT cells exhibited any transformed foci, but all 3T3-METex14 and KRAS double mutant cells exhibited marked transformation (Figure S1A). Western blotting revealed that KRAS mutants upregulated phosphorylation of AKT and ERK (Figure S1B and S1C). No significant differences were identified between different amino acid substitutions at KRAS codon 12 (Figure S1B and S1C).
Next, to determine the sensitivity of isogenic 3T3 models to MET inhibitors, we compared the growth of cells with METex14 and KRAS-G12 mutations in the presence of crizotinib and cabozantinib, two FDA-approved drugs with potent inhibitory activity against MET, ALK, and ROS1, or MET, VEGFR2, KIT, RET, and AXL, respectively (24). The two agents differ in their mode of binding to the targets: crizotinib is a type I inhibitor that binds the active DFG-in conformation of its targets, whereas cabozantinib is a type II inhibitor which binds the inactive DFG-out conformation of its targets (25). 3T3-METex14-KRAS mutant cells were less sensitive to crizotinib than 3T3-METex14-KRAS WT (Figure 2A). Overexpression of KRAS WT had little effect on sensitivity of 3T3-METex14 cells to crizotinib treatment, even when expressed at the same level as KRAS mutant (Figures 2A and S1D). Cells expressing METex14 and oncogenic KRAS also showed resistance to another MET inhibitor, cabozantinib, to an even greater degree than to crizotinib (Figure S1E). Western blot analysis showed that phosphorylation of METex14 and AKT were inhibited crizotinib in 3T3-METex14-empty, 3T3-METex14-KRAS WT, and 3T3-METex14-KRAS G12D cells. However, phosphorylation of ERK remained largely insensitive to crizotinib treatment in 3T3-METex14-KRAS G12D cells (Figure 2B). Similar results were obtained with the other KRAS mutants (Figure S1F) and with cabozantinib (Figure S1G). The presence of KRAS G12D also resulted in less crizotinib-induced caspase 3/7 activation compared to METex14-empty or METex14-KRAS WT cells (Figure 2C).
Figure 2: KRAS mutations mediates resistance to MET directed therapies in METex14 skipping cells.
A. Isogenic stably transfected 3T3-METex14-KRAS cell lines were treated with crizotinib for 96 hours. Cell viability was determined using AlamarBlue. Each condition was assayed in six-replicate determinations and data are representative of three independent experiments (mean ± SE). B. Isogenic stable 3T3-METex14-KRAS lines were serum starved for 6 hours, and subsequently treated with increasing concentrations of crizotinib for 4 hours and lysates were subjected to immunoblotting. C. Caspase 3/7 activity was analyzed in stable 3T3-METex14-KRAS lines that were treated with crizotinib for 48 hours. Each condition was assayed in triplicate determinations and data are representative of two independent experiments (mean ± SE). *p<0.05, compared to either 3T3-METex14-empty or -KRAS WT group. D. Direct sequencing confirms co-occurrence of METex14 skipping and KRAS G12S mutation in the LUAD12C cell line. E. LUAD12C cells were treated with crizotinib or cabozantinib for 96 hours. Cell viability was determined using AlamarBlue viability dye. Each condition was assayed in six-replicate determinations and data are representative of three independent experiments (mean ± SE). F. LUAD12C cells were treated with increasing concentrations of crizotinib for 4 hours and lysates were subjected to immunoblotting. G. LUAD12C cells were infected with lentivirus expressing shRNAs targeting KRAS or non-targeting shRNA as a control, followed by selection with puromycin for 2 days. Lysates were subjected to immunoblotting. H. A total 2.5 × 104 of KRAS knockdown LUAD12C cells were plated in 6-well plates in HGF supplemented medium, and growth rate determined. Each condition was assayed in duplicate determinations and data are representative of three independent experiments (mean ± SE). *p<0.05, compared to non-targeting shRNA cells. I. KRAS knockdown LUAD12C cells were treated with crizotinib in HGF-supplemented medium for 120 hours. Cell viability was determined using AlamarBlue. Each experiment was assayed in six-replicate determinations and data are representative of three independent experiments (mean ± SE).
To further interrogate the role of oncogenic KRAS in mediating resistance to MET TKI, we used a lung cancer cell line that harbors amplified MET (H1993) and remains dependent on MET expression for survival (26). Consistent with the 3T3-METex14 model, overexpression of KRAS G12D reduced the sensitivity of H1993 cells to both crizotinib and cabozantinib (Figure S2A). As expected, phosphorylation of ERK remained intact in the presence of crizotinib in cells expressing KRAS G12D (Figure S2B). Taken together, these results show that expression of oncogenic KRAS mediates resistance to MET directed therapies in lung cancers with METex14 skipping or MET amplification.
Concurrent expression of mutant KRAS with METex14 mediates resistance to anti-MET therapy in a patient derived cell line model.
A patient-derived cell line, LUAD12C, was developed from a malignant pleural effusion specimen from PT-5 and was utilized to determine whether the presence of oncogenic KRAS with METex14 splice variant correlates with lack of response to MET TKIs. We first confirmed the presence of KRAS G12S and the loss of exon 14 in MET by RT-PCR (Figures 2D and S2C). cDNAs from Hs746T (gastric cancer cell line with METex14) (27) and PC9 (EGFRex19del) were used as RT-PCR controls for METex14 skipping and MET-wildtype, respectively. LUAD12C cells were refractory to crizotinib and cabozantinib, compared to 3T3-METex14-empty cells (Figure 2A and 2E). Western blot analysis demonstrated that phosphorylation of ERK (but not MET and AKT) was largely insensitive to crizotinib in LUAD12C cells (Figure 2F).
Next, we investigated the effect of KRAS mutation on the growth of LUAD12C cells by knocking down KRAS with two independent shRNAs. Expression of KRAS mRNA (Figure S2D) and protein (Figure 2G) were reduced by 80–90% by the shRNAs; this was associated with reduced phosphorylation of ERK (Figure 2G). Notably, a reduction in KRAS also resulted in downregulation of phosphorylated MET, as further explored below. Two days after lentiviral shRNA infection, the growth of KRAS knockdown cells decelerated, and they subsequently died (Figure 2H). Rescuing MET activation by treating cells with HGF (a MET ligand) improved growth and survival of KRAS knockdown cells to a small extent, suggesting that MET signaling can still contribute to growth. Cell culture medium supplemented with HGF (10ng/mL) was therefore used in further experiments using KRAS knockdown LUAD12C cells. KRAS knockdown cells grew slower (doubling time: non-targeting control (NT), 2.1 days; shKRAS-1, 4.5 days; shKRAS-2, 3.9 days, Figure 2H), and exhibited increased sensitivity to crizotinib compared to control NT cells in a viability assay (Figure 2I). These results indicated that LUAD12C cells, that have both METex14 skipping and KRAS mutation, strongly depend on KRAS-ERK signaling, and that KRAS mutation can mediate resistance to crizotinib.
Combined MEK and MET inhibition as a therapeutic strategy for NSCLC with concurrent METex14 and KRAS alterations
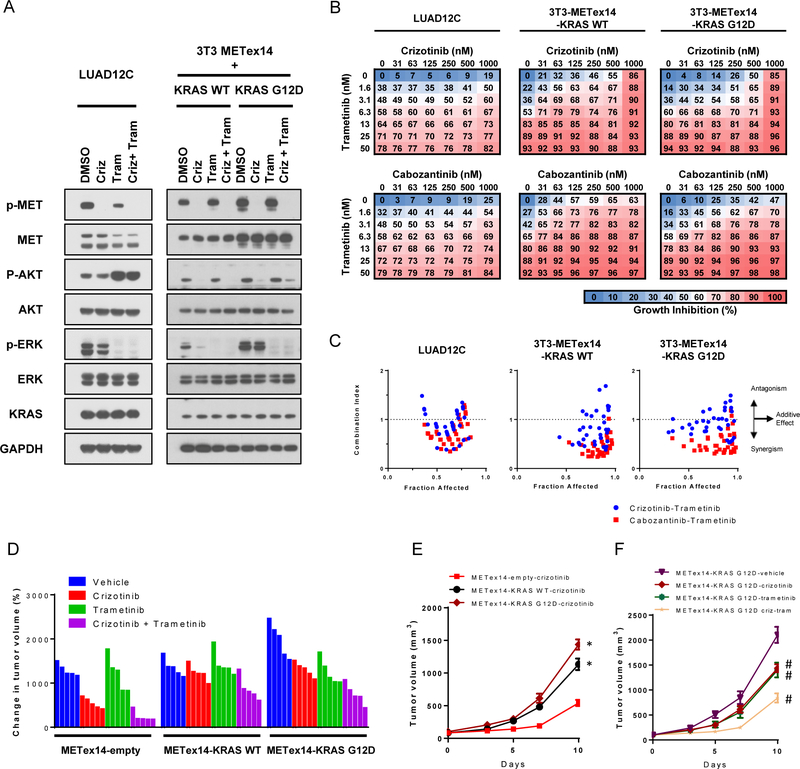
We assessed whether acquired resistance to MET TKI through KRAS mutation could be overcome by combination therapy with MEK and MET inhibitors. Trametinib, a MEK1/2 inhibitor, efficiently inhibited cell proliferation in both LUAD12C and 3T3-METex14-KRAS isogenic cell lines with IC50s below 10 nM (Figure S3A). Expression of KRAS G12D or KRAS WT with METex14 did not alter sensitivity of cells to trametinib and no significant differences in sensitivity to trametinib were found between 3T3-METex14-KRAS WT and 3T3-METex14-KRAS G12D cells. In contrast to their efficient growth inhibition, neither crizotinib nor trametinib alone could inhibit both the ERK and AKT signaling pathways in LUAD12C and 3T3-METex14-KRAS G12D cells (Figure 3A). Identical results were obtained with cabozantinib treatment (Figure S3B). Notably, trametinib, alone or in combination with crizotinib, activated AKT pathway, resulting in robust AKT phosphorylation in LUAD12C cells (Figure 3A).
Figure 3: Trametinib synergizes with MET inhibitors in concurrent METex14 alteration and KRAS mutant cells.
A. LUAD12C, 3T3-METex14-KRAS WT, and -KRAS G12D cells were treated with crizotinib (1 μM), trametinib (25 nM), or a combination of trametinib (25 nM) and crizotinib (1μM) for 4 hours. Lysates were then subjected to immunoblotting. B. LUAD12C, 3T3-METex14-KRAS WT, and -KRAS G12D cells were treated with a combination of trametinib and either crizotinib or cabozantinib for 96 hours. Cell viability was determined by AlamarBlue. Data represented the mean value of growth inhibition ratio at each concentration of the drugs in four independent experiments. C. Dot plot indicates the combination index and fraction affected (inhibition ratio) of various drug concentration. D. 3T3-METex14-empty, -KRAS WT, and -KRAS G12D cells were implanted into a subcutaneous flank of athymic nude mice. When tumors reached approximately 100 mm3, mice were treated with vehicle or 25 mg/kg crizotinib, 1 mg/kg trametinib, or a combination of 25 mg/kg crizotinib and 1 mg/kg trametinib daily for 10 days. The relative change in volume from baseline of Individual tumors are shown in the waterfall plots. E. Tumors volume in animals with crizotinib treatment group are shown stratified by cell lines. *p<0.05, compared to the respective empty group. F. Tumors volume of 3T3-METex14-KRAS G12D xenografts are shown stratified by treatment group. #p<0.05, compared to vehicle-treated group.
We then examined the combinatorial effect of MET TKI and trametinib in cell viability assays. Whereas trametinib alone efficiently inhibited cell proliferation even at low concentrations, the restoration of MET TKI sensitivity upon trametinib treatment was limited (Figure 3B). Therefore, we assessed synergism between MET TKI and trametinib using the Chou-Talalay Method (28) (Figure 3C). The combination index (CI) reflects the extent of synergy or antagonism of two drugs, and generally defines whether drug-drug interactions are synergistic (CI<1), additive (CI=1), or antagonistic (CI>1) (28). In LUAD12C cells and 3T3-METex14 isogenic models, the CI values of most combination points were <1 for crizotinib or cabozantinib when combined with trametinib, representing a synergistic effect.
To define a therapeutic strategy for patients with METex14+KRAS mutation, we performed in vivo experiments testing the efficacy of a combination of crizotinib and trametinib. Mice bearing 3T3-METex14-KRAS xenografts were treated with either crizotinib alone (25 mg/kg), trametinib alone (1 mg/kg), or a combination of the two drugs. The relative change in volume of individual tumors upon treatment is shown in Figures 3D and S3C. For easier viewing, tumor growth data are shown in four separate graphs (Figures 3E and 3F, S3D, and S3E). 3T3-METex14-KRAS G12D xenografts grew significantly faster than 3T3-METex14-empty or KRAS WT xenografts (Figures 3D and S3D). Crizotinib treatment suppressed growth of 3T3-METex14 xenograft tumors, but was less effective at slowing growth of 3T3-METex14-KRAS G12D xenograft tumors (Figure 3E). In contrast to our observations in cell lines, expression of KRAS WT reduced sensitivity to crizotinib in vivo. (Figure 3E). Treatment with trametinib alone showed a small effect on tumor growth in all 3 groups (Figure S3E), however, a combination of trametinib and crizotinib was more effective at showing tumor growth (Figure 3F). The drug combination was well tolerated during the treatment course (Figure S3F). These results show that mutation or amplification of KRAS can confer resistance to crizotinib treatment in 3T3-MET exon14 skipping xenograft tumors in vivo, and that the addition of trametinib can restore tumor inhibition.
Combination of MEK and ERBB inhibitors suppress growth of LUAD12C cells in a synergistic manner
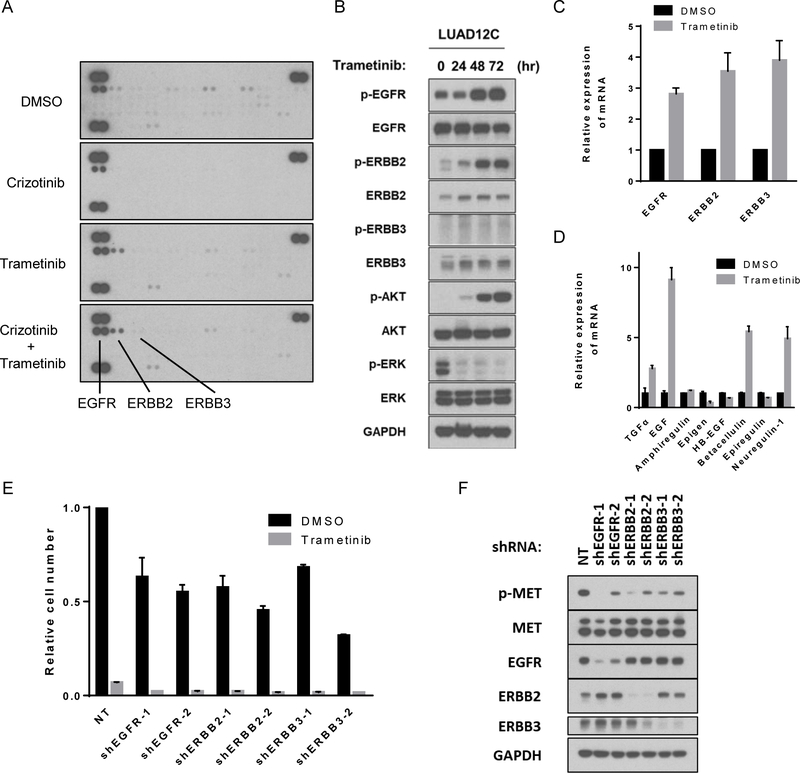
To investigate factors that might hamper response to MET and MEK combination therapy in this setting, we evaluated receptor tyrosine kinase (RTK) activities because MEK inhibition is known to cause feedback activation of RTKs in KRAS-mutant lung cancer by relieving physiological feedback suppression (29–31). Phospho-RTK profiling of LUAD12C treated as indicated in Figure 4A revealed that phosphorylation of EGFR and ERBB2 were upregulated following trametinib treatment (Figure 4A). Time course experiments showed that phosphorylation of EGFR, ERBB2, ERBB3 and AKT increased gradually (Figure 4B). Previous reports have shown that MEK inhibition leads to a decrease in MYC expression, a known negative regulator of ERBB2 and ERBB3 transcription, resulting in upregulation of ERBB2 and ERBB3 (29). Consistent with published reports in other cell lines, trametinib treatment of LUAD12C cells resulted in suppression of MYC protein and mRNA expression (Figures S4A and S4B), and upregulation of ERBB2 and ERBB3 protein and mRNA expression (Figures 4B and 4C). We also examined the expression of ERBB family ligands to evaluate whether MEK inhibition can induce an autocrine activation of RTKs. Using a custom qPCR array designed to profile the ERBB pathway genes, we found increased expression of TGF-α, EGF, BTC, and NRG1 (ligands that activate EGFR and ERBB3) following trametinib treatment (Figure 4D).
Figure 4: Trametinib treatment induces feedback activation of AKT and ERBB family and combination of MEK and ERBB inhibition induces synergetic suppressive effect on LUAD12C cells.
A. LUAD12C cells were treated with crizotinib (1 μM), trametinib (25 nM), or a combination of trametinib (25 nM) and crizotinib (1μM) for 48 hours. Lysates were then applied to phospho-RTK arrays. B. LUAD12C cells were treated with trametinib (25 nM) for indicated time. Lysates were subjected to immunoblotting. C. LUAD12C cells were treated with trametinib (25 nM) for 48 hours and mRNA expression level of ERBB families were determined by RT-qPCR. Experiments were conducted in triplicate and error bars represent mean ± SD. D. LUAD12C cells were treated with trametinib (25 nM) for 48 hours and mRNA expression level of ligands for ERBB families were determined by RT-qPCR. Experiments were conducted in triplicate and error bars represent mean ± SD. E. LUAD12C cells were infected with lentivirus expressing shRNAs targeting genes or non-targeting shRNA as a control, followed by selection with puromycin. Subsequently, a total 1.5 × 105 of knockdown LUAD12C cells were plated in 6-well plates, and treated with DMSO or trametinib (5nM) for 10 days. Relative cell numbers were shown. Each condition was assayed in duplicate determinations in 2 independent experiments and error bar represent mean ± SE. F. Lysates from knockdown LUAD12C cells after selection were subjected to immunoblotting.
To determine whether combined inhibition of both MEK and ERBB family members is more effective at reducing growth, we knocked down each ERBB family member with shRNAs in LUAD12C cells and determined growth in the presence of trametinib. Loss of EGFR, ERBB2 or ERBB3 resulted in decreased cell growth, suggesting that the ERBB family RTKs provide essential growth or survival signals (Figure 4E) and this growth inhibitory effect was further enhanced upon addition of trametinib (Figure 4E).
To gain further insight into the relationship between MET and the ERBB family, we examined MET phosphorylation in LUAD12C cells in which expression of EGFR, ERBB2 or ERBB3 were reduced by shRNAs. As shown in Figure 4F, MET phosphorylation was reduce following downregulation ERBB expression (Figure 4F).
MEK inhibition suppresses MET expression via effects on transcription and protein stability
MET signaling can be affected by downstream MAPK signaling. This is suggested by Figure S1B and S1C, which show that total MET protein expression was increased in 3T3-METex14-KRAS mutant cells compared to 3T3-METex14-KRAS WT cells. 3T3-METex14-KRAS mutants displayed a 2.5-fold increase in phospho-MET and a 1.9-fold increase in total MET expression compared with 3T3-METex14-KRAS WT. Changes in MET expression and activation were also seen upon KRAS knockdown in LUAD12C cells (Figure 2G). We therefore investigated further the regulation of MET expression. Activation of the RAS-ERK pathway is known to upregulate MET expression at the transcription level through activation of the SP-1, AP-1, and ETS-1 transcription factors (32–35). The data in Figures S1B and S1C suggest a posttranscriptional regulation of MET as well, because ectopically expressed METex14 cDNA in the 3T3 cells is under the control of a CMV promoter and is not expected to be regulated by these canonical transcription factors. To compare METex14 protein levels in cells with or without activated KRAS, cells were treated with the protein synthesis inhibitor cycloheximide (CHX), and then METex14 levels at various time points determined by Western blotting. The amount of METex14 was quantified by densitometry and is shown relative to the amount of METex14 expressed in the absence of CHX. METex14 had an extended half-life in the presence of two different KRAS mutations, compared to KRAS WT (Figures 5A and 5B), suggesting that oncogenic KRAS resulted in increased METex14 protein stability. This effect was also seen in 3T3 cells ectopically expressing wildtype MET (Figures S5A and S5B). We next looked at the effect of downregulating the KRAS-MAPK pathway on METex14 mRNA and protein levels in LUAD12C cells. Suppression of KRAS expression with shRNAs or MEK1/2 activity with trametinib reduced METex14 mRNA (Figures 5C and D). Treatment with trametinib also caused a rapid decrease in METex14 phosphorylation, evident within 1 h and completely lost by 6 h (Figure 5E). Although trametinib treatment also caused a reduction in total METex14, this was slower than the loss in phosphorylation (Figure 5E), and therefore, is not the only basis for the reduction of phospho-MET.
Figure 5: MEK inhibition suppresses MET through transcription and protein level.
A. Lysate from stably transfected 3T3-METex14-KRAS WT, G12D, or G12S cells were collected at the indicated time points after addition of cycloheximide (CHX, 100 μg/mL) and subjected to immunoblotting. B. The amount of MET protein was quantified and is shown relative to the amount of MET expressed in absence of CHX. Data are representative of three independent experiments (mean ± SE).*p<0.05, compared to 3T3-METex14-KRAS WT cells. C. MET mRNA expression level of was determined by RT-qPCR in KRAS knockdown LUAD12C cells after selection. Each condition was assayed in triplicate determinations (mean ± SD). D. LUAD12C cells were treated with trametinib (25 nM) for the indicated time and mRNA expression levels of MET were analyzed by RT-qPCR. Experiments were conducted in triplicate and error bars represent mean ± SD. E. LUAD12C cells were treated with trametinib (25 nM) for the indicated time. Lysates were subjected to immunoblotting.
Combined inhibition of MEK and of MET or ERBB suppress growth of LUAD12C cells effectively
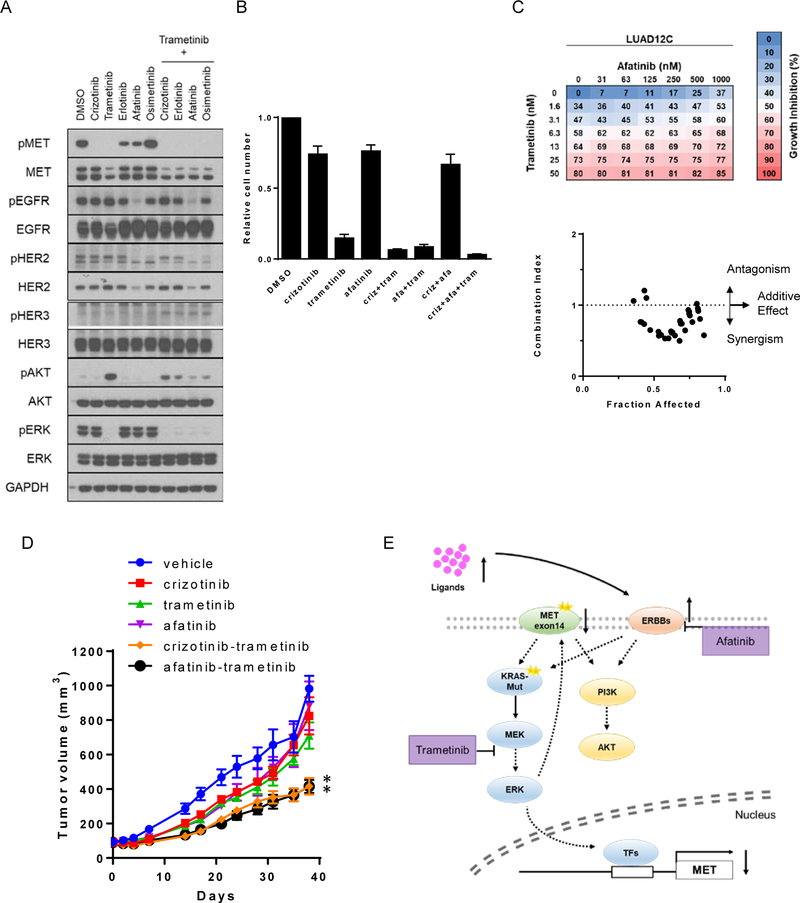
In results described above, we show that inhibition of METex14 and MEK1/2 with crizotinib and trametinib, respectively, was synergistic. In addition, we demonstrated that METex14 activation depends on expression of ERBB family RTKs. Taken together, these findings indicated that combined inhibition of METex14, MEK and ERBB RTKs may be an effective therapeutic strategy for tumors with concurrent METex14 splice variant and KRAS mutation. Therefore, we next assessed which combinational treatment would lead to the most profound growth inhibition in LUAD12C cells. Cells were treated with inhibitors for MET (crizotinib), MEK (trametinib), EGFR (erlotinib and osimertinib) or EGFR/ERBB2 (afatinib) as monotherapy or in combination for 48 hours (Figure 6A). As predicted, trametinib decreased total METex14 protein, with complete inhibition of METex14 phosphorylation. However, as previously reported, and shown in Figure 4B, trametinib treatment reactivated AKT. Addition of either crizotinib, erlotinib, afatinib, or osimertinib prevented the trametinib-induced AKT reactivation (Figure 6A) to varying degree with afatinib exhibiting the most effective suppression. Afatinib (EGFR and ERBB2 inhibitor), but not erlotinib (EGFR only) or osimertinib (mutant EGFR only) was able to suppress phospho-EGFR in the presence of trametinib, indicating that ERBB2 activity may be the predominant mode by which trametinib can stimulate EGFR phosphorylation. Next, we assessed cell growth over a longer duration (12 days). The number of cells were counted and shown relative to the number in the DMSO control group. Growth of LUAD12C cells was not significantly affected by treatment with either crizotinib or afatinib (Figure 6B) whereas trametinib inhibited growth by 90%. Combination of trametinib with crizotinib had an additive effect inhibiting growth by 95% (p = 0.047, compared with trametinb alone). Combination of trametinib and afatinib inhibited growth by 94% (p = 0.14, compared to trametinib alone) and these drugs showed a synergistic effect on growth (Figure 6C). The triple combination of trametinib, crizotinib and afatinib reduced growth by 97% (p = 0.053, compared to trametinib+crizotinib group) (Figure 6B).
Figure 6: Trametinib synergizes with afatinib in concurrent METexon14 alteration and KRAS mutant cells.
A. LUAD12C cells were treated with crizotinib (1 μM), trametinib (25 nM), erlotinib (1 μM), afatinib (1 μM), osimertinib (1 μM), or a combination of them for 48 hours. Lysates were then subjected to immunoblotting. B. A total 3 × 104 of LUAD12C cells were plated in 6-well plates, and treated with DMSO, crizotinib (200nM), trametinib (5nM), afatinib (100 nM), or a combination of them for 11 days. Relative cell numbers are shown. Each condition was assayed in duplicate determinations in 3 independent experiments and error bar represent mean ± SE. C. LUAD12C cells were treated with a combination of trametinib and arfatinib for 96 hours. Cell viability was determined by AlamarBlue. Data represent the mean value of growth inhibition ratio at each concentration of the drugs in three independent experiments. Dot plot indicates the combination index and fraction affected (inhibition ratio) of various drug concentration. D. LUAD12C cells were implanted into a subcutaneous flank of NSG mice. When tumors reached approximately 100 mm3, mice were treated with vehicle or 25 mg/kg crizotinib, 1 mg/kg trametinib, 12.5 mg/kg afatinib, or a combination of them 4 days a week. Tumor volume was determined on the indicated days after the onset of treatment. Data represent mean ± SE (n = 5). *p<0.05, compared to trametinib-treated group. E. A strategy for the treatment of METex14 and KRAS-mutant lung cancer.
To validate these in vitro findings, we tested the efficacy of the combination of trametinib with either crizotinib or afatinib using the patient-derived LUAD12C xenograft model. Mice bearing LUAD12C xenografts were treated with crizotinib (25 mg/kg), trametinib (1 mg/kg), afatinib (12.5 mg/kg), or the indicated combinations on a 4 days-on/3 days-off schedule (Figure 6D and S6A). Consistent with the data obtained with 3T3-MET-KRAS mutant cell line xenograft model, crizotinib monotherapy had minimal effect on growth of LUAD12C xenograft tumor. Similarly, monotherapy with trametinib or afatinib only partially reduced growth of LUAD12C xenograft tumors. A combination of trametinib with either crizotinib or afatinib increased the anti-tumor effect of both drugs. The drug combinations were well tolerated over a 40 day treatment period (Figure S6B).
Discussion
The emergence of acquired resistance to targeted therapy remains an ongoing challenge in patients whose tumors harbor clinically actionable mitogenic drivers. Studies of tumor specimens obtained at time of progression on TKIs have revealed a variety of mechanisms that drive resistance to molecular targeted therapy (13–15) and the elucidation of these TKI resistance mechanisms has led to the nomination of subsequent therapies that are currently in clinical use or being explored in clinical trials (36). Determining mechanisms of resistance of METex14 altered lung cancers to MET therapy is a relatively new area of study with few validated mechanisms of resistance known. To date, only a few second-site mutations in METex14 have been reported as potential mediators of MET TKI resistance (16, 17). In this study, we describe 8 lung cancer patients harboring METex14 alterations and a concurrent KRAS alteration (mutation or amplification) among 113 METex14 patients. The co-occurrence of these two driver oncogenes was statistically significant and similar results were not seen in the setting of other major lung cancer drivers. Significantly, none of these 8 patients harbored a second-site mutations in METex14. For one patient (PT-5), we were able to compare genomic data from paired pre- and post- treatment samples and identified a KRAS G12S mutation in the crizotinib-resistant sample. While an analysis of paired biopsies indicated that the KRAS alteration was not present prior to crizotinib therapy, the presence of primary progressive disease in this patient raises the possibility of a KRAS mutation existing prior to MET inhibition in a subclonal population not detected on clinical sequencing. In addition, the duration of disease control in two other patients with concurrent METex14 and KRAS mutations was relatively short. A literature review found two previous reports of patients with concurrent METex14 and KRAS mutations; in one report, the patient received single agent of crizotinib, resulting in disease progression only in 1 month (37). The other paper mentioned one case with METex14 and KRAS mutations, but further details were not provided (38).
Using isogenic cell line model systems, we found that mutant KRAS alleles but not wildtype KRAS (even when expressed at the same level as the mutant) are able to decrease sensitivity of METex14 cell lines to crizotinib and cabozantinib. However, overexpression of wildtype KRAS was able to blunt the response of METex14 xenografts to crizotinib in vivo, suggesting that amplification of KRAS may also contribute to resistance to MET TKI. Similar findings were observed using cell lines with MET amplification. A combination of trametinib and crizotinib was more effective at reducing tumor growth than either agent alone in vitro and in vivo. Note that in our series, concurrent de-novo KRAS activation in MET exon 14-altered cancers did not preclude response in the clinic. However, it is possible that the duration of disease control on single-agent crizotinib could be affected (recognizing interpatient variability, one patient in our series was on for only 5 months). The use of combination therapy in these cases might represent a more effective therapeutic strategy compared to single-agent TKI use.
Acquired KRAS mutation has been reported not uncommonly in patients with colon cancers that developed resistance to EGFR targeted therapy (39), but in general, concurrent KRAS mutation is an exceedingly rare event in EGFR- and ALK-driven lung cancers (40–43). It has been well documented that strong mitogenic driver mutations (e.g., EGFR, KRAS, NRAS, BRAF, ALK, ROS1, RET alterations) show strong mutual exclusivity (9, 44). Previous studies have shown that co-occurrence of oncogenic KRAS and EGFR mutations will produce synthetic lethal effects due to excessive activation of RAS-ERK-mediated signaling beyond a tolerable threshold, providing a functional basis for the mutually exclusive nature of these two major mitogenic drivers (42). In this regard, we found that transduction of oncogenic KRAS led to a lethal effect in certain MET-driven cell lines (data not shown). These observations imply that not all cancer cells harboring METex14 alteration can tolerate oncogenic KRAS. The high frequency of co-existence of METex14 and KRAS mutation suggests that KRAS may provide a survival advantage to some but not all METex14 tumors. We speculate that this subset of lung cancers likely developed genetic mechanisms that blunt the synthetic lethal effect of having two oncogenes activating MAPK signaling in the same cell. Potentially this can be via upregulation of anti-apoptotic genes, loss of pro-apoptotic genes or changes in autophagy. Identifying the underlying mechanism(s) that make certain cells permissive for co-expression of oncogenic KRAS and METex14 may open up new avenues for better therapy.
MEK inhibitors have been intensely studied in KRAS-driven tumors, even though clinical outcomes with these drugs have been largely unsatisfactory (45). One of the reasons for this lack of efficacy in KRAS-driven tumors is that RTK activation following MEK inhibition can enhance the opportunity for bypass or compensatory signaling. Indeed, the addition of EGFR/ERBB2 dual inhibitor or FGFR1 inhibitor has been shown to enhance tumor cell death in the setting of MEK inhibition in KRAS-mutant cells (29–31). In the LUAD12C model, we found that trametinib treatment activated ERBB family signaling due to upregulation of transcript expression and autocrine ligand stimulation. Reduction of ERBB family expression reduced MET phosphorylation and suppressed growth, suggesting a role for ERBB RTKs in regulating growth of the LUAD12C cell line. Although our study did not investigate the precise nature of the METex14-ERBB interaction, previous studies have shown that MET and ERBB family members can be co-immunoprecipitated (46), EGF-stimulated EGFR phosphorylation leads to transactivation of MET (47, 48); MET and ERBB2 cooperate to enhance cell invasion (49); and MET phosphorylation leads to activation of EGFR or ERBB3 (50, 51). Based on these published data and our experimental findings, we treated mice bearing LUAD12C xenografts with a combination of trametinib and afatinib, and found that this double therapy was more effective at slowing tumor growth than either drug alone. Nevertheless, this combination did not result in tumor regression, suggesting that additional targets for therapy remain to be uncovered in METex14 tumors with oncogenic KRAS alleles.
Importantly, we also found that METex14 tumors have a higher degree of RAS pathway activation than tumors with wildtype MET amplification, suggesting that METex14 tumors may be more tolerant of KRAS activation and perhaps more reliant on this pathway for survival, providing an underlying biological basis for the selection of KRAS mutation in crizotinib resistance. This is further indicative of a complex interplay between these two oncogenes. MET is known to play a prominent role during RAS-mediated tumorigenesis and the activation of the RAS-ERK pathway induces higher MET expression via a transcriptional effect (32, 33, 52). Here we show for the first time that expression of mutant KRAS prolongs the half-life of METex14 protein, perhaps helping to titrate out crizotinib. Conversely, trametinib treatment led to a downregulation of METex14 expression and this was preceded by loss of METex14 phosphorylation. These intriguing findings suggest that activated KRAS is regulating METex14 at multiple levels. Given the interaction between ERBB RTKs and METex14 expression, it is apparent that the signaling complexity of tumors with METex14 and KRAS co-alterations may require new therapeutic strategies.
In summary, our findings confirm KRAS mutation as a recurrent mechanism of primary and secondary resistance to MET-directed therapies in lung cancers harboring METex14 alterations and provide a new potential clinical strategy for patients with concurrent METex14 and KRAS alterations.
Supplementary Material
Acknowledgments
This work was supported by NIH grants P01 CA129243, P30 CA008748, and U54 OD020355.
Footnotes
Conflict of interest:
Charles M. Rudin is a consultant for Bristol-Myers Squibb, Abbvie, Seattle Genetics, Harpoon Therapeutics, Genentech Roche, and AstraZeneca.
Mark G. Kris is a consultant for Ariad, AstraZeneca and Genentech Roche and received research funding from Genentech Roche and Puma Biotechnology.
Alexander Drilon is a consultant for Ignyta, LOXO Oncology, TP Therapeutics, AstraZeneca, Pfizer, Blueprint Medicines, Genentech Roche, Takeda, and has received research funding from Foundation Medicine.
Marc Ladanyi has received advisory board compensation from Boehringer Ingelheim, AstraZeneca, Bristol-Myers Squibb, Takeda, and Bayer, and research support from LOXO Oncology and Helsinn Healthcare.
Romel Somwar has received research support from Helsinn Healthcare
All other authors do not report any relevant conflicts of interest
References
- 1.Trusolino L, Bertotti A, Comoglio PM. MET signalling: principles and functions in development, organ regeneration and cancer. Nat Rev Mol Cell Biol. 2010;11:834–48. [DOI] [PubMed] [Google Scholar]
- 2.Gherardi E, Birchmeier W, Birchmeier C, Vande Woude G. Targeting MET in cancer: rationale and progress. Nat Rev Cancer. 2012;12:89–103. [DOI] [PubMed] [Google Scholar]
- 3.Tovar EA, Graveel CR. MET in human cancer: germline and somatic mutations. Annals of translational medicine. 2017;5:205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Recurrent MET fusion genes represent a drug target in pediatric glioblastoma. Nat Med. 2016;22:1314–20. [DOI] [PubMed] [Google Scholar]
- 5.Ma PC, Kijima T, Maulik G, Fox EA, Sattler M, Griffin JD, et al. c-MET mutational analysis in small cell lung cancer: novel juxtamembrane domain mutations regulating cytoskeletal functions. Cancer Res. 2003;63:6272–81. [PubMed] [Google Scholar]
- 6.Kong-Beltran M, Seshagiri S, Zha J, Zhu W, Bhawe K, Mendoza N, et al. Somatic mutations lead to an oncogenic deletion of met in lung cancer. Cancer Res. 2006;66:283–9. [DOI] [PubMed] [Google Scholar]
- 7.Comprehensive molecular profiling of lung adenocarcinoma. Nature. 2014;511:543–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frampton GM, Ali SM, Rosenzweig M, Chmielecki J, Lu X, Bauer TM, et al. Activation of MET via diverse exon 14 splicing alterations occurs in multiple tumor types and confers clinical sensitivity to MET inhibitors. Cancer Discov. 2015;5:850–9. [DOI] [PubMed] [Google Scholar]
- 9.Jordan EJ, Kim HR, Arcila ME, Barron D, Chakravarty D, Gao J, et al. Prospective Comprehensive Molecular Characterization of Lung Adenocarcinomas for Efficient Patient Matching to Approved and Emerging Therapies. Cancer Discov. 2017;7:596–609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schrock AB, Frampton GM, Suh J, Chalmers ZR, Rosenzweig M, Erlich RL, et al. Characterization of 298 Patients with Lung Cancer Harboring MET Exon 14 Skipping Alterations. J Thorac Oncol. 2016;11:1493–502. [DOI] [PubMed] [Google Scholar]
- 11.Paik PK, Drilon A, Fan PD, Yu H, Rekhtman N, Ginsberg MS, et al. Response to MET inhibitors in patients with stage IV lung adenocarcinomas harboring MET mutations causing exon 14 skipping. Cancer Discov. 2015;5:842–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Drilon A, Ou S-H, Clark J, Camidge DR, Socinski M, Weiss J, et al. Antitumor Activity and Safety of Crizotinib in Patients with MET Exon 14-Altered Advanced Non-Small Cell Lung Cancer. J Thorac Oncol. 2017;12:S438–S9. [Google Scholar]
- 13.Sequist LV, Waltman BA, Dias-Santagata D, Digumarthy S, Turke AB, Fidias P, et al. Genotypic and histological evolution of lung cancers acquiring resistance to EGFR inhibitors. Sci Transl Med. 2011;3:75ra26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ohashi K, Maruvka YE, Michor F, Pao W. Epidermal growth factor receptor tyrosine kinase inhibitor-resistant disease. J Clin Oncol. 2013;31:1070–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lin JJ, Riely GJ, Shaw AT. Targeting ALK: Precision Medicine Takes on Drug Resistance. Cancer Discov. 2017;7:137–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heist RS, Sequist LV, Borger D, Gainor JF, Arellano RS, Le LP, et al. Acquired Resistance to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol. 2016;11:1242–5. [DOI] [PubMed] [Google Scholar]
- 17.Ou SI, Young L, Schrock AB, Johnson A, Klempner SJ, Zhu VW, et al. Emergence of Preexisting MET Y1230C Mutation as a Resistance Mechanism to Crizotinib in NSCLC with MET Exon 14 Skipping. J Thorac Oncol. 2017;12:137–40. [DOI] [PubMed] [Google Scholar]
- 18.Timofeevski SL, McTigue MA, Ryan K, Cui J, Zou HY, Zhu JX, et al. Enzymatic characterization of c-Met receptor tyrosine kinase oncogenic mutants and kinetic studies with aminopyridine and triazolopyrazine inhibitors. Biochemistry. 2009;48:5339–49. [DOI] [PubMed] [Google Scholar]
- 19.Cheng DT, Mitchell TN, Zehir A, Shah RH, Benayed R, Syed A, et al. Memorial Sloan Kettering-Integrated Mutation Profiling of Actionable Cancer Targets (MSK-IMPACT): A Hybridization Capture-Based Next-Generation Sequencing Clinical Assay for Solid Tumor Molecular Oncology. The Journal of molecular diagnostics : JMD. 2015;17:251–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zehir A, Benayed R, Shah RH, Syed A, Middha S, Kim HR, et al. Mutational landscape of metastatic cancer revealed from prospective clinical sequencing of 10,000 patients. Nat Med. 2017;23:703–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng Z, Liebers M, Zhelyazkova B, Cao Y, Panditi D, Lynch KD, et al. Anchored multiplex PCR for targeted next-generation sequencing. Nat Med. 2014;20:1479–84. [DOI] [PubMed] [Google Scholar]
- 22.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102:15545–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Liu X, Jia Y, Stoopler MB, Shen Y, Cheng H, Chen J, et al. Next-Generation Sequencing of Pulmonary Sarcomatoid Carcinoma Reveals High Frequency of Actionable MET Gene Mutations. J Clin Oncol. 2016;34:794–802. [DOI] [PubMed] [Google Scholar]
- 24.Reungwetwattana T, Liang Y, Zhu V, Ou SI. The race to target MET exon 14 skipping alterations in non-small cell lung cancer: The Why, the How, the Who, the Unknown, and the Inevitable. Lung Cancer. 2017;103:27–37. [DOI] [PubMed] [Google Scholar]
- 25.Davare MA, Vellore NA, Wagner JP, Eide CA, Goodman JR, Drilon A, et al. Structural insight into selectivity and resistance profiles of ROS1 tyrosine kinase inhibitors. Proc Natl Acad Sci U S A. 2015;112:E5381–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lutterbach B, Zeng Q, Davis LJ, Hatch H, Hang G, Kohl NE, et al. Lung cancer cell lines harboring MET gene amplification are dependent on Met for growth and survival. Cancer Res. 2007;67:2081–8. [DOI] [PubMed] [Google Scholar]
- 27.Asaoka Y, Tada M, Ikenoue T, Seto M, Imai M, Miyabayashi K, et al. Gastric cancer cell line Hs746T harbors a splice site mutation of c-Met causing juxtamembrane domain deletion. Biochem Biophys Res Commun. 2010;394:1042–6. [DOI] [PubMed] [Google Scholar]
- 28.Chou TC. Drug combination studies and their synergy quantification using the Chou-Talalay method. Cancer Res. 2010;70:440–6. [DOI] [PubMed] [Google Scholar]
- 29.Sun C, Hobor S, Bertotti A, Zecchin D, Huang S, Galimi F, et al. Intrinsic resistance to MEK inhibition in KRAS mutant lung and colon cancer through transcriptional induction of ERBB3. Cell reports. 2014;7:86–93. [DOI] [PubMed] [Google Scholar]
- 30.Manchado E, Weissmueller S, Morris JPt, Chen CC, Wullenkord R, Lujambio A, et al. A combinatorial strategy for treating KRAS-mutant lung cancer. Nature. 2016;534:647–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitai H, Ebi H, Tomida S, Floros KV, Kotani H, Adachi Y, et al. Epithelial-to-Mesenchymal Transition Defines Feedback Activation of Receptor Tyrosine Kinase Signaling Induced by MEK Inhibition in KRAS-Mutant Lung Cancer. Cancer Discov. 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Webb CP, Taylor GA, Jeffers M, Fiscella M, Oskarsson M, Resau JH, et al. Evidence for a role of Met-HGF/SF during Ras-mediated tumorigenesis/metastasis. Oncogene. 1998;17:2019–25. [DOI] [PubMed] [Google Scholar]
- 33.Furge KA, Kiewlich D, Le P, Vo MN, Faure M, Howlett AR, et al. Suppression of Ras-mediated tumorigenicity and metastasis through inhibition of the Met receptor tyrosine kinase. Proc Natl Acad Sci U S A. 2001;98:10722–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gambarotta G, Boccaccio C, Giordano S, Ando M, Stella MC, Comoglio PM. Ets up-regulates MET transcription. Oncogene. 1996;13:1911–7. [PubMed] [Google Scholar]
- 35.Kubic JD, Little EC, Lui JW, Iizuka T, Lang D. PAX3 and ETS1 synergistically activate MET expression in melanoma cells. Oncogene. 2015;34:4964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang JC, Ahn MJ, Kim DW, Ramalingam SS, Sequist LV, Su WC, et al. Osimertinib in Pretreated T790M-Positive Advanced Non-Small-Cell Lung Cancer: AURA Study Phase II Extension Component. J Clin Oncol. 2017;35:1288–96. [DOI] [PubMed] [Google Scholar]
- 37.Liu SY, Gou LY, Li AN, Lou NN, Gao HF, Su J, et al. The Unique Characteristics of MET Exon 14 Mutation in Chinese Patients with NSCLC. J Thorac Oncol. 2016;11:1503–10. [DOI] [PubMed] [Google Scholar]
- 38.Saffroy R, Fallet V, Girard N, Mazieres J, Sibilot DM, Lantuejoul S, et al. MET exon 14 mutations as targets in routine molecular analysis of primary sarcomatoid carcinoma of the lung. Oncotarget. 2017;8:42428–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Misale S, Yaeger R, Hobor S, Scala E, Janakiraman M, Liska D, et al. Emergence of KRAS mutations and acquired resistance to anti-EGFR therapy in colorectal cancer. Nature. 2012;486:532–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yu HA, Arcila ME, Rekhtman N, Sima CS, Zakowski MF, Pao W, et al. Analysis of tumor specimens at the time of acquired resistance to EGFR-TKI therapy in 155 patients with EGFR-mutant lung cancers. Clin Cancer Res. 2013;19:2240–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ohashi K, Sequist LV, Arcila ME, Moran T, Chmielecki J, Lin YL, et al. Lung cancers with acquired resistance to EGFR inhibitors occasionally harbor BRAF gene mutations but lack mutations in KRAS, NRAS, or MEK1. Proc Natl Acad Sci U S A. 2012;109:E2127–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Unni AM, Lockwood WW, Zejnullahu K, Lee-Lin SQ, Varmus H. Evidence that synthetic lethality underlies the mutual exclusivity of oncogenic KRAS and EGFR mutations in lung adenocarcinoma. eLife. 2015;4:e06907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ortiz-Cuaran S, Scheffler M, Plenker D, Dahmen L, Scheel AH, Fernandez-Cuesta L, et al. Heterogeneous Mechanisms of Primary and Acquired Resistance to Third-Generation EGFR Inhibitors. Clin Cancer Res. 2016;22:4837–47. [DOI] [PubMed] [Google Scholar]
- 44.Kris MG, Johnson BE, Berry LD, Kwiatkowski DJ, Iafrate AJ, Wistuba, II, et al. Using multiplexed assays of oncogenic drivers in lung cancers to select targeted drugs. JAMA. 2014;311:1998–2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tomasini P, Walia P, Labbe C, Jao K, Leighl NB. Targeting the KRAS Pathway in Non-Small Cell Lung Cancer. Oncologist. 2016;21:1450–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lai AZ, Abella JV, Park M. Crosstalk in Met receptor oncogenesis. Trends Cell Biol. 2009;19:542–51. [DOI] [PubMed] [Google Scholar]
- 47.Breindel JL, Haskins JW, Cowell EP, Zhao M, Nguyen DX, Stern DF. EGF receptor activates MET through MAPK to enhance non-small cell lung carcinoma invasion and brain metastasis. Cancer Res. 2013;73:5053–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Brevet M, Shimizu S, Bott MJ, Shukla N, Zhou Q, Olshen AB, et al. Coactivation of receptor tyrosine kinases in malignant mesothelioma as a rationale for combination targeted therapy. J Thorac Oncol. 2011;6:864–74. [DOI] [PubMed] [Google Scholar]
- 49.Khoury H, Naujokas MA, Zuo D, Sangwan V, Frigault MM, Petkiewicz S, et al. HGF converts ErbB2/Neu epithelial morphogenesis to cell invasion. Mol Biol Cell. 2005;16:550–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Engelman JA, Zejnullahu K, Mitsudomi T, Song Y, Hyland C, Park JO, et al. MET amplification leads to gefitinib resistance in lung cancer by activating ERBB3 signaling. Science. 2007;316:1039–43. [DOI] [PubMed] [Google Scholar]
- 51.Liu X, Wang Q, Yang G, Marando C, Koblish HK, Hall LM, et al. A novel kinase inhibitor, INCB28060, blocks c-MET-dependent signaling, neoplastic activities, and cross-talk with EGFR and HER-3. Clin Cancer Res. 2011;17:7127–38. [DOI] [PubMed] [Google Scholar]
- 52.Ivan M, Bond JA, Prat M, Comoglio PM, Wynford-Thomas D. Activated ras and ret oncogenes induce over-expression of c-met (hepatocyte growth factor receptor) in human thyroid epithelial cells. Oncogene. 1997;14:2417–23. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.