Abstract
HIV-associated neurocognitive disorders (HAND) represent an important source of neurologic complications in individuals with HIV. The dynamic, often subclinical, course of HAND has rendered diagnosis, which currently depends on neuropsychometric (NP) evaluation, a challenge for clinicians. Here, we present evidence that functional brain connectivity, derived by large-scale Granger causality (lsGC) analysis of resting-state functional MRI (rs-fMRI) time-series, represents a potential biomarker to address this critical diagnostic need. Brain graph properties were used as features in machine learning tasks to 1) classify individuals as HIV+ or HIV− and 2) to predict overall cognitive performance, as assessed by NP scores, in a 22-subject (13 HIV−, 9 HIV+) cohort. Over nearly all seven brain parcellation templates considered, support vector machine (SVM) classifiers based on lsGC-derived brain graph features significantly outperformed those based on conventional Pearson correlation (PC)-derived features (p < 0.05, Bonferroni-corrected). In a second task for which the objective was to predict the overall NP score of each subject, the lsGC-based SVM regressors consistently outperformed the PC-based regressors (p < 0.05, Bonferroni-corrected) on nearly all templates. With the widely used Automated Anatomical Labeling (AAL90) template, it was determined that the brain regions that figured most strongly in the SVM classifiers included those of the default mode network (posterior cingulate cortex, angular gyrus) and basal ganglia (caudate nucleus), dysfunction in both of which have been observed in previous structural and functional analyses of HAND.
Keywords: Resting-state fMRI, functional connectivity, large-scale Granger causality, support vector machine, graph theory, automated diagnosis, HIV-associated neurocognitive disorders (HAND)
1. Introduction
HIV-associated neurocognitive disorders (HAND) are a spectrum of cognitive, motor, and behavioral symptoms that occur in over a quarter of individuals infected with HIV [41] and are associated with significant decreases in health-related quality of life, even among subjects on combined antiretroviral therapy (cART) with well-controlled viral loads [51]. Clinically, HAND present a diagnostic challenge due to the often subtle and dynamic course of cognitive impairment. The Frascati diagnostic consensus [3], based on neuropsychometric (NP) testing, suffers from numerous shortcomings, including lack of sufficient sensitivity and specificity for HAND, as well as susceptibility to learning effects, socioeconomic status, and other confounding factors [54, 36]). Additionally, NP testing-based approaches necessarily cannot detect the disease before the emergence of neuropsychiatric signs. There is thus a substantial need for improved biomarkers of HAND.
Blood oxygen level-dependent (BOLD) functional magnetic resonance imaging (fMRI) has been extensively studied as a potential tool to address such diagnostic needs in a variety of neurologic and psychiatric disorders, including HAND [5, 7, 1]. During an attention task, it was found that HIV+ subjects exhibited greater BOLD activity in the parietal and frontal lobes than HIV− subjects [11]. Hyperactivations in the fronto-striato-parietal network in HIV-infected subjects, particularly in the left inferior frontal gyrus and left caudate nucleus, were also found in a meta-analysis fMRI studies [38]. Resting-state fMRI (rsfMRI) analyses, performed in the absence of a stimulus or task, have gained particular prominence in the past decade, as they can be used to uncover the functional connectivity structure of the brain. Moreover, rsfMRI may have more clinical utility than task-fMRI, since the former is not contingent on a patient’s ability to perform a task, which may be limited due to age or disability. Functional connectivity disruptions have been found in HIV+ subjects, most prominently in the default mode network, but also in the fronto-parietal network and basal ganglia [50, 49, 18, 61]. These group level studies constitute a foundation of evidence on which to pursue the aim of predicting clinically meaningful disease-related parameters at the individual level. In this study, we train and test a machine learning model, using rsfMRI-derived brain graph properties as features, to predict HIV-status and cognitive performance at the level of individual subjects.
The optimal method for extracting connectivity maps from rsfMRI time-series is still an area of active research. We thus compare the predictive ability of machine learning models trained on graphs derived from three time-series analysis techniques, based on either Pearson correlation or the principle of Granger causality, over several brain parcellation templates, which spatially cluster individual rsfMRI voxels into regions-of-interest (ROI). We show that models based on large-scale Granger causality-derived brain graphs are more robustly accurate than those based on conventional Pearson correlation-derived networks, with regard to the prediction of both HIV-status and NP scores.
2. Materials & Methods
2.1. Subjects
This work was performed as part of an NIH sponsored study (R01-DA-034977), for which subjects were recruited from the University of Rochester Medical Center in Rochester, NY, USA. Subjects with any of the following conditions were excluded: severe premorbid or comorbid psychiatric disorders, chronic seizures, stroke, head trauma resulting in loss of consciousness for greater than 30 minutes, non-HIV brain infection, dementia, alcohol or drug abuse, any significant systemic condition that can alter brain function, metallic implants in skull, cardiac devices, or claustrophobia. All subjects provided written informed consent, in accordance with the study protocol reviewed by the institutional review board of the University of Rochester. Cognitive status for all subjects was determined by NP testing. The cohort was comprised of 13 cognitively normal HIV− subjects (mean age SD: 41± 16y; ± 6 females) and 9 cognitively impaired HIV+ subjects (mean age ± SD: 52 ± 10y; 2 females). Of note, one subject in the HIV+ cohort underwent neuroimaging and NP testing in two separate sessions. This second session was considered as a separate subject, thus bringing the total number of subjects in the HIV+ cohort to 10. Among the subjects in the HIV+ cohort, all were diagnosed with asymptomatic neurologic impairment (ANI) with the exception of one subject, who was diagnosed with mild neurocognitive disorder (MND), using the criteria defined in [3].
Viral loads in the HIV+ group were as follows: five subjects had a viral load of less than 50 copies/mL (three of these subjects had undetectable loads), three subjects had a viral load of between 50 and 1000 copies/mL, and one subject had a viral load of nearly 40,000 copies/mL. CD4+ T-cells in this cohort ranged from 253 to 1730 cells/μL. With the exception of one individual, all HIV+ subjects were on a stable cART regimen.
2.2. Neuropsychometric testing
A standard battery of NP tests were performed on all subjects and raw NP scores were obtained for six cognitive domains: executive function (Stroop Interference Task and Trailmaking Test Parts A and B), information processing speed (Symbol Digit Modalities Test and Stroop Color Naming Task), attention (CalCAP(CRT4) and WAIS-III Letter-Number Sequencing Task), learning (Rey Auditory Verbal Learning Test (RAVLT) (trials 1–5) and Rey Complex Figure Immediate Recall Test), memory (RAVLT Delayed Recall Test and Rey Complex Figure Delayed Recall Test), and motor (Grooved Pegboard, left and right hands). An overall score was also generated by combining those of the six domains. All seven scores were then converted to an age- and education-adjusted z-score, as described in [3].
2.3. Neuroimaging data acquisition
Subjects underwent MRI scanning using a 3.0T Siemens Magnetom Trio-Tim system (Siemens Medical Solutions, Erlangen, Germany) housed at the Rochester Center for Brain Imaging (Rochester, NY, USA). A high-resolution structural T1-weighted scan was acquired using a magnetization-prepared rapid gradient echo (MPRAGE) sequence with the following scan parameters: echo time = 3.44ms, repetition time 2530ms, isotropic voxel size = 1mm, flip angle = 7, acquisition time = 3min. BOLD fMRI data were acquired using echo planar imaging with the following scan parameters: echo time = 23ms, repetition time = 1650ms, flip angle = 84, 96 × 96 acquisition matrix. 250 brain volumes with 25 slices separated by 5mm were acquired for each subject.
2.4. fMRI preprocessing
Preprocessing steps were performed using the FMRIB Software Library (FSL, version 5.0, http://fsl.fmrib.ox.ac.uk/) [46]. The first 10 brain volumes for each subject were discarded to eliminate initial saturation effects of the BOLD signal. Subsequently, head motion correction, slice timing correction, brain extraction, and registration to the standard Montreal Neurological Institute (MNI152) atlas space [33, 34] was performed. Removal of the whole brain time-series, as well as correction for head motion and physiologic processes, was done using a nuisance regressor. High-pass filtering was performed to remove signal drifts. Individual voxel time-series were normalized to have a standard deviation of 1 unit and a mean of 0 units in order to prioritize signal dynamics over amplitude [59].
2.5. fMRI spatial parcellation
The preprocessed fMRI scans were spatially subdivided into regions of interest (ROIs) according to seven different parcellation templates: the Automated Anatomical Labeling (AAL90) [52], Harvard-Oxford (HO138) [16], Brainnetome (BN246) [21], and Craddock spectral clustering templates (Craddock100, Crad-dock200, Craddock563, and Craddock831) [14]. These seven templates were chosen because they are widely cited in fMRI literature and because they span a wide range of spatial resolutions (the number in the abbreviated name for each template specifies the number of ROIs). ROI time-series were generated by taking the mean of the time-series from all constituent voxels.
2.6. Time-series analysis and graph generation
For each subject, brain graphs representing the functional connectivity of brain regions were constructed using Pearson correlation (PC) and large-scale Granger causality (lsGC). PC is a bivariate and symmetric method that summarizes the association between two time-series as their covariance normalized by the product of their standard deviations. PC was performed both with and without band-pass filtering (.038Hz .076Hz) the ROI time-series. lsGC [17, 45, 37, 44, 60] is a multivariate and−directional method that is built on the principle of Granger causality (GC) [22], in which time-series A is said to Granger-cause time-series B if the inclusion of information from time-series A in a predictive autoregressive model of time-series B decreases the variance of the residuals between the predicted and actual time-series B. Unlike conventional GC, however, lsGC can be extended to high-dimensional systems due to the embedding of a dimension reduction step using principal components analysis. The two lsGC model parameters, autoregressive order (ρ), i.e., the number of past time-points that are used to predict a future time-point, and the number of principal components used to define the low-dimensional space (c), were optimized using a 10×10 grid search with 5-fold cross-validation on the AAL90 template.
Analysis of the preprocessed fMRI time-series with PC or lsGC generates n×n affinity matrices, where n is the total number of ROIs in the parcellation template. Each entry in the affinity matrices represents the association (detected either by PC or lsGC) of a pair of ROI time-series. Subsequently, the largest 30% of the entries in each affinity matrix were set to 1 and the remainder were set to 0 [31, 43]. This thresholding process allows the matrix to be considered as a binary graph, in which the nodes represent ROIs and the edges connecting the nodes represent functional connectivity between ROIs.
2.7. Graph property computation
The following local properties were calculated for each node of the brain graphs. In-degree: The sum of all incoming edges for a particular node. Out-degree: The sum of all outgoing edges for a particular node. Total-degree: The sum of all edges incident on a particular node, regardless of direction. In-degree and out-degree were calculated for directed lsGC-derived graphs, while Total-degree was calculated for symmetric PC-derived graphs. Clustering coefficient: The fraction of connected triples centered on a node that are also triangles (a connected triple is a set of three nodes for which one node has direct incoming or outgoing edges to the other two; a triangle is a set of three nodes that are all directly connected by either outgoing or incoming edges). Betweenness centrality: The fraction of all shortest paths in a graph that pass through a particular node [35] (the shortest path between nodes i and j is the minimum number of edges that must be traversed to travel from i to j; if such a path does not exist for a given pair of nodes, as might happen when a graph becomes disconnected, then the maximum finite path length for the entire graph is assigned as the shortest path length between that pair of nodes).
The following global properties were also calculated for each brain graph. Degree variance: the variance in the number of incoming, outgoing, or total edges of all nodes in a graph. In-degree variance and out-degree variance were calculated for directed lsGC-derived graphs, while total-degree variance was calculated for symmetric PC-derived graphs, as in-degree and out-degree are identical for symmetric graphs. Modularity: The extent to which a graph can be partitioned into Louvain community detection algorithm-defined modules [10], which have high intramodular connectivity and low intermodular connectivity. Assortativity: The correlation coefficient between the total degree of nodes on opposite ends of all edges in a graph [35]. Small-worldness: The ratio of the random graph-normalized global clustering coefficient and the random graph-normalized shortest path length [56]. Degree distribution-matched random graphs were produced for each empirical brain graph by iteratively rewiring each edge approximately 10 times. Generation of the random graphs and calculation of the graph parameters were performed using the Brain Connectivity Toolbox (brain-connectivity-toolbox.net) [40] in MATLAB (2017b, The MathWorks, Natick, MA).
2.8. Machine learning and feature analysis
Each brain graph property and each NP domain score was normalized across all subjects to have a mean of 0 and a standard deviation of 1. All graph properties for each subject were then concatenated into a single feature vector and used as the input features for a linear support vector machine (SVM) model [13] with ridge regularization [25]. To train and test the model, a 10-fold iterative cross-validation scheme was employed, in which the full data set of brain graphs from all 23 subjects was randomly split into 10 folds. In each iteration, 9 of these folds were used to train the model and the remaining fold was used to test the performance of the learned model. This procedure was then repeated 100 times with redefined data folds.
The outputs of the SVMs for each subject were either the predicted HIV status or NP scores. For the HIV+/− classification task, performance on the testing set was assessed using the area under the receiver operating characteristic curve (AUC). An AUC of 1 indicates perfect classification while an AUC of 0.5 indicates a classification performance that is no better than random chance. For the NP score regression, performance was assessed the coefficient of determination R2. This scheme was iterated 100 times and distributions of the corresponding performance metrics were obtained.
To analyze the relative importance of each brain graph property in the classification task, the mean magnitude of the weights the |βi| in all of the cross-validated models using brain graph features extracted from the AAL 90 template was computed. Subsequently, the weights were ranked and the features i corresponding to the 10 largest |βi| were considered the top features.
To generate surrogate null distributions for the regression SVMs, the NP scores from each cognitive domain were permuted, such that each subject was randomly assigned a set of NP scores drawn from the pool of same-domain NP scores of all subjects. The 10-fold cross validation scheme described above was applied to the permuted samples.
3. Results
3.1. Classification of HIV+ and HIV− subjects
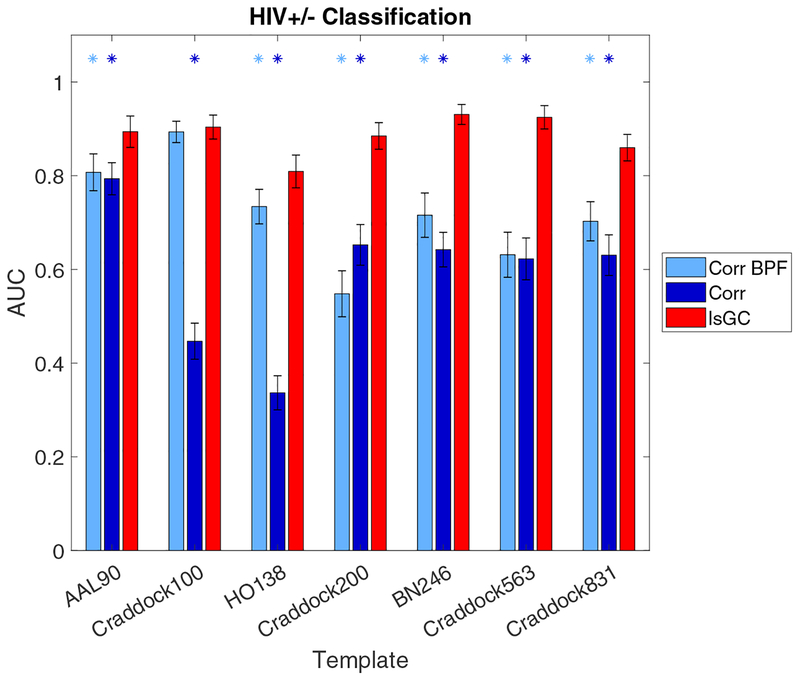
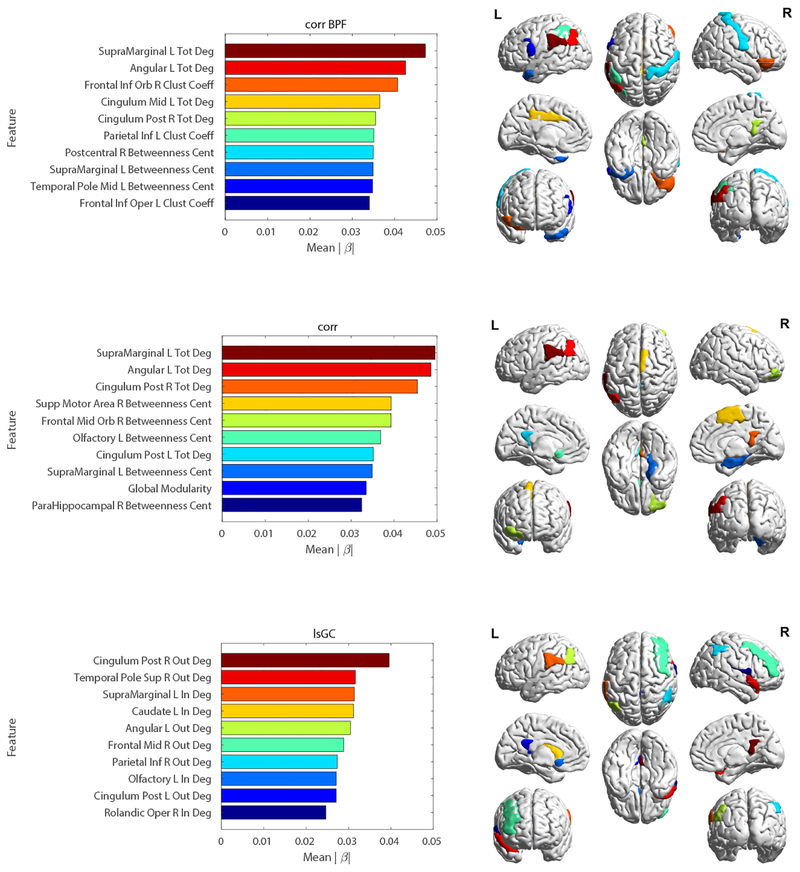
As can be seen in Figure 1, the performance of the lsGC-based SVM classifiers significantly outperform those of both the unfiltered correlation-based and band-pass filtered correlation-based methods across all templates considered, with the exception of the Craddock 100 template, in which the performances of the band-pass filtered correlation model and the lsGC model are similar (p > 0.05). Moreover, while the performances of both correlation-based models vary substantially across templates, that of the lsGC-based model is relatively stable over different templates. The relative SVM model weights |βi| of the top 10 features in the three classification runs on the AAL 90 template-derived brain networks are shown in Figure 2. Note that the top features for the lsGC-based model include features extracted from brain regions in the default mode network (DMN) and basal ganglia. In particular, the right and left posterior cingulate cortex and the left angular gyrus of the DMN are strongly weighted in multiple models. The only basal ganglia region among the top 10 features in any of the three models is the caudate nucleus, which is the fourth most strongly weighted feature in the lsGC model.
Figure 1:
Comparison of time-series analysis methods with regard to HIV+/− classification ability. The vertical axis shows the area under the receiver operating characteristic curve (AUC) while the horizontal axis shows the different parcellation templates. Error bars represent standard deviations. Light blue and dark blue asterisks indicate that the performances of band-pass filtered correlation- and unfiltered correlation-based models, respectively, are significantly different from that of the lsGC-based model (two-sided Wilcoxon rank sum test, p < 0.05, Bonferroni-corrected). Note that over all seven atlases, the classifier based on lsGC-derived features outperforms that based on unfiltered correlation-derived features. The same is true for band-pass filtered correlation, with the exception of the Craddock 100 atlas, in which case the performances of the two models are statistically identical.
Figure 2:
Left: Bar plots depicting the top 10 most strongly weighted features and corresponding weights |βi| for SVM models based on the three methods using the AAL 90 template: correlation with BPF (top), correlation without BPF (middle), and lsGC (bottom). Right: Color-matched brain map of the regions depicted in the corresponding bar plots. Note the prevalence of ROIs previously implicated in HAND, such as the basal ganglia (caudate nucleus) and default mode network (posterior cingulate cortex, angular gyrus) regions. Abbreviations - Tot Deg: Total-Degree; In Deg: In-Degree; Out Deg: Out-Degree; Clust Coeff: Clustering Coefficient; Betweenness Cent: Betweenness Centrality.
3.2. Prediction of cognitive performance
The neuropsychometric (NP) z-scores from each of the seven cognitive domains were compared across the two cohorts. It was found that the mean executive function, attention, learning, memory, motor, and overall z-scores were significantly lower in the HIV+ group than the HIV− group (FDR controlled p < 0.05, one-sided Wilcoxon rank-sum test [8]). Of these, the most commonly affected cognitive domain, assessed by the number of HIV+ subjects who had domain-specific z-scores less than −1.0, was motor function; 5 HIV+ individuals had motor z-scores less than −1.0 and an additional 2 HIV+ individuals had motor z-scores less than 0.
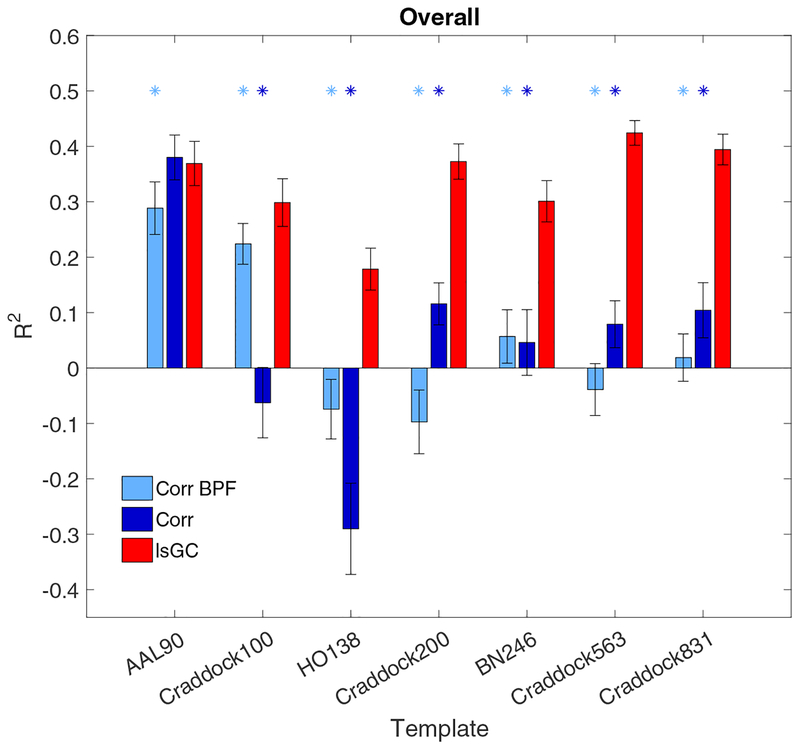
Figure 3 shows that the lsGC-based SVM models perform significantly better in predicting overall cognitive performance, as indicated by larger R2 values, than either of the correlation-based models across nearly all the templates used. The only exception is the AAL 90 template, in which the performance of the lsGC-based model and that of the unfiltered correlation-based model do not exhibit a statistically significant difference. Moreover, over all the templates, the distribution of lsGC performances was substantially different from the corresponding null distribution, generated by randomly permuting the NP score assignments of the subjects.
Figure 3:
The performance of the three SVM models with respect to prediction of cognitive performance, as assessed by overall neuropsychometric (NP) score. The vertical axis shows the coefficient of determination (R2) between the predicted and true NP scores for each model. The horizontal axis shows the seven different parcellation templates used to generate brain graphs, the properties of which served as features in the SVM model. The light blue, dark blue, and red bars represent the R2 value for the band-pass filtered correlation-, unfiltered correlation-, and lsGC-based models, respectively. Light blue asterisks designate significantly different performance between the band-pass filtered correlation model and the lsGC model (Bonferroni-corrected p < 0.05, two-sided Wilcoxon rank-sum test) and the dark blue asterisks designate significantly different performance between the unfiltered correlation model and the lsGC model. Error bars represent two standard deviations.
4. Discussion
The motivation for applying lsGC to obtain brain graphs in addition to traditional correlation-based methods arises from the limitations of bivariate and non-directional methods for the analysis of such a complex and high-dimensional system as the brain. For instance, though Pearson correlation is the most widely used method for performing functional connectivity analyses of the brain, it cannot distinguish indirect from direct connections, as multivariate methods can [47]. Limited multivariate Granger causality formulations have been implemented for functional brain connectivity analysis [29], but the mathematical constraints imposed by the fact that the number of variables, i.e., ROIs, exceeds the number of temporal samples collected prevent the application of such methods to high ROI parcellations or voxel-wise analyses. The dimension reduction step of lsGC allows for this problem to be circumvented. Additionally, though many of the connections in the brain have feedback projections in the opposite direction, asymmetric connectivity in the mammalian brain has also been anatomically verified [27, 26]. The fact that lsGC allows for the inference of directional connectivity may explain the improved performance of lsGC over correlation-based methods in HIV disease state and cognitive score prediction. Indeed, asymmetric functional connectivity has been observed in ROIs of the DMN, including the posterior cingulate cortex and the ventromedial prefrontal cortex [53], which are known to be affected by HAND [49, 61]. Moreover, it was recently shown that global properties of brain graphs generated by another directional connectivity analysis method, mutual connectivity analysis with a generalized radial basis function neural network, were linearly associated with NP scores in two cognitive domains, whereas those generated using PC on the same cohort were not associated with NP scores in any domain [1].
The optimal parcellation template for functional connectivity analyses is not known, as every template has advantages and disadvantages. For instance, although structural parcellations, such as the AAL90 and HO138, are grounded in the anatomical boundaries of brain tissue, there is evidence to suggest that the resultant ROIs do not reflect the functional organization of the underlying tissue [32, 14]. Functional parcellations, such as the Craddock templates, address this issue by generating ROIs on the basis of spatiotemporal clusters in the rsfMRI data. One potential limitation of such data-driven parcellations, however, is their sensitivity to fMRI preprocessing steps, such as spatial smoothing and registration [14]. Other types of parcellations have also been used in the literature, such as randomly defined ROIs and the BN246 atlas, which was generated by subdividing the ROIs of an existing anatomical atlas by incorporating probabilistic tractography data derived from diffusion tensor imaging (DTI) [21]. While most published rs-fMRI network studies use the AAL90 template, it is not clear if the conclusions on brain connectivity and disease prediction made on the basis of one template are intrinsic characteristics of the brain or simply artifacts of that template. Reproduction of the results across other parcellation templates, however, suggests that the findings are truly a feature of the data, rather than of the processing pipeline. In Figures 1 and 3, it can be seen that across all seven templates, lsGC performs as well as or better than, correlation (both band pass-filtered and unfiltered variants) in terms of overall NP score prediction and HIV-status classification.
The plots in Figure 2 show the brain graph features that contributed most strongly to the HIV disease state prediction for each model. It is noteworthy that, for all three models, features extracted from ROIs in the default mode network (DMN) were strongly weighted. The DMN is primarily comprised of nodes in the bilateral posterior cingulate cortex, angular gyrus, precuneus, and medial prefrontal cortex [39] and is believed to play a role in self-referential behavior, including mind-wandering and daydreaming [2]. In the context of HIV, the DMN is an area of interest, as several previous rs-fMRI studies have noted a link between HIV and DMN dysfunction, in terms of both altered intra-network connectivity strength [50, 61] and node centrality [49]. In all three models, features derived from DMN nodes comprise either the top or the second place feature.
Altered functional connectivity in the DMN may also partially underlie the specific memory and attention deficits, as well as the overall cognitive performance deficit, seen in the HIV+ cohort. The DMN is often described as a task-negative network, due to the fact that its activity decreases when subjects are engaged in a goal-directed task [39]. Several studies have found links between the DMN and cognitive performance. For instance, decreased task-induced DMN deactivation was associated with attentional lapses and worsened performance in a selective-attention task [57]. Associations have also been found between the DMN and working memory; in two separate studies, the strength of the functional connectivity between DMN regions was positively correlated with performance in working memory tasks [24, 42]. Additionally, a positive correlation between the strength of DMN functional connectivity at rest and general cognitive performance (as assessed by NP-testing and overall z-scores, as in the present study) was recently found among HIV+ subjects [61]. The disrupted DMN functional connectivity observed here is therefore consistent with the literature that links DMN dysfunction with degraded cognitive performance in multiple domains.
It should be noted, however, that not all fMRI studies of HIV-infected individuals have found alterations in the DMN. For instance, [55] compared functional connectivity between recently-infected HIV+ subjects and HIV− controls and failed to find DMN connectivity differences between the two groups. However, in that study, correlation was used to detect functional connectivity, while in the present study, functional connectivity was determined using lsGC, which has several advantages over correlation, as discussed above. Moreover, given that only subjects who had been infected with HIV in the past year were recruited to the [55] study, DMN pathology may have not yet manifested in those subjects to an extent that could be detected by fMRI. Such results highlight the need for more longitudinal neuroimaging studies of HIV-infected individuals. One such study examining fMRI activations while subjects performed a visual attention task found that several brain regions, including the prefrontal and posterior parietal cortices, exhibited increased activity in HIV+ subjects at the 1-year testing session, relative to baseline [20]. These regions did not include those associated with the DMN. However, it is important to note that [20] examined the altered activation of individual brain regions, rather than connectivity between different regions. Therefore, while activity in the DMN itself may have been unchanged, activity in regions that are connected to the DMN may have been altered in a way that affected the functional connectivity of the DMN. Indeed, changes in connectivity between the DMN and other brain regions has been observed in HIV+ individuals [50].
Unlike the correlation-based models, the lsGC model uniquely included a basal ganglia region (the left caudate nucleus) as a top feature. Histologic abnormalities [4], volumetric atrophy [6], and metabolic dysfunction [58] in the basal ganglia have been previously observed in HIV-infected individuals. Moreover, the caudate is an essential component of the fronto-striato-thalamo-cortical circuit, dysfunction of which is believed to underly many of the cognitive deficits seen in HAND, including in executive function, learning, and memory [19]. Thus, the fact that the lsGC model strongly weighs a basal ganglia region may partially explain its improved performance relative to correlation-based models. Altered basal ganglia functional connectivity may also be the basis of the high prevalence of motor deficits among the HIV+ subjects, as the basal ganglia are known to play a key role in the selection of desired movements and the suppression of unwanted movements [23, 28] and are thus essential for the smooth execution of voluntary movements.
Frequency-specific filtering of rsfMRI time-series prior to correlation-based rsfMRI connectivity analyses is a widespread, but controversial, practice. The rationale behind its use lies in the high signal-to-noise ratio (SNR) of BOLD fMRI and the fact that physiologic noise (e.g., from cardiac and respiratory function) and instrumental noise (e.g., from scanner drift) components are respectively enriched at frequencies above 0.1Hz [12, 30, 48] and below 0.01Hz [9]. There is also evidence, however, that such temporal filtering introduces artificial correlations between pairs of fMRI time-series [15]. Given that the tradeoff between improving SNR and introducing spurious correlations is still a matter of debate, we generated brain graphs based on correlation using both filtered and unfiltered time-series. Results from Figures 1 and 3 show that, in certain contexts, e.g., HIV+/− classification with the Craddock100 and HO138 templates, BPF time-series yield higher predictive ability than unfiltered time-series, while, in other contexts, e.g., overall NP score prediction with the Craddock100 template, the opposite is true, while in still other cases, e.g., most other templates in the HIV+/− classification task, the two models yield comparative predictive abilities.
A few limitations must be kept in mind when interpreting the results of this study. First, because the ground truth connectivity structure of the brain is not known, it cannot be said whether differences in graph properties exploited by the machine learning models to predict disease state and cognitive performance truly represent differences in information transfer between brain regions or some other more amorphous, though still clinically meaningful, aspect of the BOLD signal. Second, the homogeneous and relatively small sample size in this study precluded the rigorous analysis of important disease-related variables such as duration and mode of infection, CD4 levels, and cART status, as well as HIV-related comorbidities, including drug abuse, vascular disease, and depression. Third, because the HIV+ and HIV− subjects were not matched by sex, age, or education level, a direct one-to-one comparison between the two groups could not be performed.
Each of these factors represents a potentially fruitful avenue for future inquiry in the context of functional brain connectivity.
5. Conclusion
It was demonstrated here that machine learning analysis of resting-state functional MRI data with large-scale Granger causality enables the superior prediction of important clinical parameters, including HIV status and cognitive performance, relative to traditional correlation-based methods. Moreover, it was shown that the features of the brain graphs deemed most relevant by the classifiers in distinguishing HIV+ and HIV− subjects were strongly enriched in regions corresponding to the default mode network and basal ganglia regions. The results of these analyses position lsGC as a potential tool for developing biomarkers for diagnosis and staging of HIV-associated neurocognitive disorders.
Large-scale Granger causality (lsGC) was used to derive functional brain networks
Machine learning performed on networks accurately classified subjects as HIV+ or HIV−
Machine learning performed on networks accurately predicted neuropsychometric scores
IsGC-derived networks were more informative than correlation-based networks
Results were robust across several parcellation schemes
Acknowledgments
This work was supported by the National Institutes of Health (R01-DA-034977, R01-MH-099921, T32-GM-007356). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of Health. This work was conducted as a Practice Quality Improvement (PQI) project related to American Board of Radiology (ABR) Maintenance of Certificate (MOC) for Prof. Dr. Axel Wismüller. The study sponsors were not involved in study design, collection and interpretation of data, or in the writing and submission of this manuscript.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: None declared
References
- [1].Abidin AZ, DSouza AM, Nagarajan MB, Wang L, Qiu X, Schifitto G, and A. Wismüller2018. Alteration of brain network topology in HIV-associated neurocognitive disorder: A novel functional connectivity perspective. NeuroImage: Clinical, 17:768–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Andrews-Hanna JR, Reidler JS, Huang C, and Buckner RL 2010. Evidence for the Default Network’s Role in Spontaneous Cognition. Journal of Neurophysiology, 104(1):322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Antinori A, Arendt G, Becker JT, Brew BJ, Byrd DA, Cherner M, Clifford DB, Cinque P, Epstein LG, Goodkin K, Gisslen M, Grant I, Heaton RK, Joseph J, Marder K, Marra CM, McArthur JC, Nunn M, Price RW, Pulliam L, Robertson KR, Sacktor N, Valcour V, and Wojna VE 2007. Updated research nosology for HIV-associated neurocognitive disorders. Neurology, 69(18):1789–1799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Archibald S, Masliah E, Fennema-Notestine C, and Al E 2004. Correlation of in vivo neuroimaging abnormalities with postmortem human immunodeficiency virus encephalitis and dendritic loss. Archives of Neurology, 61(3):369–376. [DOI] [PubMed] [Google Scholar]
- [5].Atluri G, Padmanabhan K, Fang G, Steinbach M, Petrella JR, Lim K, MacDonald A, Samatova NF, Doraiswamy PM, and Kumar V 2013. Complex biomarker discovery in neuroimaging data: Finding a needle in a haystack. NeuroImage: Clinical, 3:123–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Aylward EH, Henderer JD, McArthur JC, Brettschneider PD, Harris GJ, Barta PE, and Pearlson GD 1993. Reduced basal ganglia volume in HIV1associated dementia Results from quantitative neuroimaging. Neurology, 43(10):2099. [DOI] [PubMed] [Google Scholar]
- [7].Balsters JH, Mantini D, Apps MAJ, Eickho SB, and Wenderoth N 2016. Connectivity-based parcellation increases network detection sensitivity in resting state fMRI: An investigation into the cingulate cortex in autism. NeuroImage: Clinical, 11:494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Benjamini Y and Hochberg Y 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the royal statistical society. Series B (Methodological), Pp. 289–300. [Google Scholar]
- [9].Bianciardi M, Fukunaga M, van Gelderen P, Horovitz SG, de Zwart JA, Shmueli K, and Duyn JH 2009. Sources of functional magnetic resonance imaging signal fluctuations in the human brain at rest: a 7 T study. Magnetic resonance imaging, 27(8):1019–1029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Blondel VD, Jean-Loup G, Lambiotte R, and Lefebvre E 2008. Fast unfolding of communities in large networks. Journal of Statistical Mechanics: Theory and Experiment, 2008(10):P10008. [Google Scholar]
- [11].Chang L, Speck O, Miller EN, Braun J, Jovicich J, Koch C, Itti L, and Ernst T 2001. Neural correlates of attention and working memory deficits in HIV patients. Neurology, 57(6):1001–1007. [DOI] [PubMed] [Google Scholar]
- [12].Cordes D, Haughton VM, Arfanakis K, Carew JD, Turski PA, Moritz CH, Quigley MA, and Meyerand ME 2001. Frequencies contributing to functional connectivity in the cerebral cortex in “resting-state” data. AJNR. American journal of neuroradiology, 22(7):1326–1333. [PMC free article] [PubMed] [Google Scholar]
- [13].Cortes C and Vapnik V 1995. Support-vector networks. Machine learning, 20(3):273–297. [Google Scholar]
- [14].Craddock RC, James GA, Holtzheimer PE, Hu XP, and Mayberg HS 2012. A whole brain fMRI atlas generated via spatially constrained spectral clustering. Human Brain Mapping, 33(8):1914–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Davey CE, Grayden DB, Egan GF, and Johnston LA 2013. Filtering induces correlation in fMRI resting state data. NeuroImage, 64(1):728–740. [DOI] [PubMed] [Google Scholar]
- [16].Desikan RS, Ségonne F, Fischl B, Quinn BT, Dickerson BC, Blacker D, Buckner RL, Dale AM, Maguire RP, and Hyman BT 2006. An automated labeling system for subdividing the human cerebral cortex on MRI scans into gyral based regions of interest. Neuroimage, 31(3):968–980. [DOI] [PubMed] [Google Scholar]
- [17].DSouza AM, Abidin AZ, Leistritz L, and Wismüller A 2017. Exploring Connectivity with Large-Scale Granger Causality on Resting-State Functional MRI. Journal of Neuroscience Methods, 287:68–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].du Plessis L, Paul RH, Hoare J, Stein DJ, Taylor PA, Meintjes EM, and Joska JA 2017. Resting-state functional magnetic resonance imaging in clade C HIV: within-group association with neurocognitive function. Journal of NeuroVirology. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Ellis R, Langford D, and Masliah E 2007. HIV and antiretroviral therapy in the brain: neuronal injury and repair. Nature Reviews Neuroscience, 8(1):33–44. [DOI] [PubMed] [Google Scholar]
- [20].Ernst T, Yakupov R, Nakama H, Crocket G, Cole M, Watters M, RicardoDukelow ML, and Chang L 2009. Declined neural e ciency in cognitively stable human immunodeficiency virus patients. Annals of Neurology: Official Journal of the American Neurological Association and the Child Neurology Society, 65(3):316–325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Fan L, Li H, Zhuo J, Zhang Y, Wang J, Chen L, Yang Z, Chu C, Xie S, and Laird AR 2016. The human brainnetome atlas: a new brain atlas based on connectional architecture. Cerebral Cortex, 26(8):3508–3526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Granger CWJ 1969. Investigating Causal Relations by Econometric Models and Cross-spectral Methods. Econometrica, 37(3):424–438. [Google Scholar]
- [23].Graybiel AM, Aosaki T, Flaherty AW, and Kimura M 1994. The basal ganglia and adaptive motor control. Science (New York, N.Y.), 265(5180):1826–1831. [DOI] [PubMed] [Google Scholar]
- [24].Hampson M, Driesen NR, Skudlarski P, Gore JC, and Constable RT 2006. Brain Connectivity Related to Working Memory Performance. The Journal of Neuroscience, 26(51):13338 LP–13343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hoerl AE and Kennard RW 1970. Ridge regression: Biased estimation for nonorthogonal problems. Technometrics, 12(1):55–67. [Google Scholar]
- [26].Honey CJ, Kötter R, Breakspear M, and Sporns O 2007. Network structure of cerebral cortex shapes functional connectivity on multiple time scales. Proceedings of the National Academy of Sciences, 104(24):10240–10245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Kötter R 2004. Online retrieval, processing, and visualization of primate connectivity data from the CoCoMac database. Neuroinformatics, 2(2):127–144. [DOI] [PubMed] [Google Scholar]
- [28].Lanciego JL, Luquin N, and Obeso JA 2012. Functional neuroanatomy of the basal ganglia. Cold Spring Harbor perspectives in medicine, P. a009621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Liao W, Ding J, Marinazzo D, Xu Q, Wang Z, Yuan C, Zhang Z, Lu G, and Chen H 2011. Small-world directed networks in the human brain: Multivariate Granger causality analysis of resting-state fMRI. NeuroImage, 54:2683–2694. [DOI] [PubMed] [Google Scholar]
- [30].Lowe MJ, Mock BJ, and Sorenson JA 1998. Functional Connectivity in Single and Multislice Echoplanar Imaging Using Resting-State Fluctuations. NeuroImage, 7(2):119–132. [DOI] [PubMed] [Google Scholar]
- [31].Lynall M-E, Bassett DS, Kerwin R, McKenna PJ, Kitzbichler M, Muller U, and Bullmore E 2010. Functional Connectivity and Brain Networks in Schizophrenia. The Journal of Neuroscience, 30(28):9477–9487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Margulies DS, Kelly AMC, Uddin LQ, Biswal BB, Castellanos FX, and Milham MP 2007. Mapping the functional connectivity of anterior cingulate cortex. NeuroImage, 37(2):579–588. [DOI] [PubMed] [Google Scholar]
- [33].Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, Mac-Donald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Goualher GL, Boomsma D, Cannon T, Kawashima R, and Mazoyer B 2001a. A probabilistic atlas and reference system for the human brain: International Consortium for Brain Mapping (ICBM). Philosophical Transactions of the Royal Society of London. Series B: Biological Sciences, 356(1412):1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, Mac-Donald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Feidler J, Smith K, Boomsma D, Pol HH, Cannon T, Kawashima R, and Mazoyer B 2001b. A Four-Dimensional Probabilistic Atlas of the Human Brain. Journal of the American Medical Informatics Association, 8(5):401–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Newman MEJ 2003. The structure and function of complex networks. Society for industrial and applied mathematics review. [Google Scholar]
- [36].Overton ET, Kauwe JSK, Paul R, Tashima K, Tate DF, Patel P, Carpenter CCJ, Patty D, Brooks JT, and Clifford DB 2011. Performances on the CogState and standard neuropsychological batteries among HIV patients without dementia. AIDS and Behavior, 15(8):1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Pester B, Lutz L, Herbert W, and Axel W 2013. Exploring Effective Connectivity by a Granger Causality Approach with Embedded Dimension Reduction. Biomedical Engineering, 58:4172. [DOI] [PubMed] [Google Scholar]
- [38].Plessis SD, Vink M, Joska JA, Koutsilieri E, Stein DJ, and Emsley R 2014. HIV infection and the frontostriatal system: a systematic review and meta-analysis of fMRI studies. AIDS, 28(6):803–811. [DOI] [PubMed] [Google Scholar]
- [39].Raichle ME, MacLeod AM, Snyder AZ, Powers WJ, Gusnard DA, and Shulman GL 2001. A default mode of brain function. Proceedings of the National Academy of Sciences, 98(2):676–682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Rubinov M and Sporns O 2010. Complex network measures of brain connectivity: Uses and interpretations. NeuroImage, 52(3):1059–1069. [DOI] [PubMed] [Google Scholar]
- [41].Sacktor N, Skolasky RL, Seaberg E, Munro C, Becker JT, Martin E, Ragin A, Levine A, and Miller E 2016. Prevalence of HIV-associated neurocognitive disorders in the Multicenter AIDS Cohort Study. Neurology, 86(4):334–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Sambataro F, Murty VP, Callicott JH, Tan H-Y, Das S, Weinberger DR, and Mattay VS 2010. Age-related alterations in default mode network: Impact on working memory performance. Neurobiology of Aging, 31(5):839–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Sanz-Arigita EJ, Schoonheim MM, Damoiseaux JS, Rombouts SARB, Maris E, Barkhof F, Scheltens P, and Stam CJ 2010. Loss of Small-World Networks in Alzheimer’s Disease: Graph Analysis of fMRI Resting-State Functional Connectivity. PLOS ONE, 5(11):e13788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Schmidt C, Pester B, Nagarajan M, Witte H, Leistritz L, and Wismueller A 2014. Impact of multivariate granger causality analyses with embedded dimension reduction on network modules. In 2014 36th Annual International Conference of the IEEE Engineering in Medicine and Biology Society, Pp. 2797–2800. [DOI] [PubMed] [Google Scholar]
- [45].Schmidt C, Pester B, Schmid-Hertel N, Witte H, Wismüller A, and Leistritz L 2016. A Multivariate Granger Causality Concept towards Full Brain Functional Connectivity. PLOS ONE, 11(4):e0153105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Smith SM, Jenkinson M, Woolrich MW, Beckmann CF, Behrens TEJ, Johansen-Berg H, Bannister PR, De Luca M, Drobnjak I, Flitney DE, Niazy RK, Saunders J, Vickers J, Zhang Y, De Stefano N, Brady JM, and Matthews PM 2004. Advances in functional and structural MR image analysis and implementation as FSL. NeuroImage, 23:S208–S219. [DOI] [PubMed] [Google Scholar]
- [47].Stephan KE and Friston KJ 2010. Analyzing effective connectivity with functional magnetic resonance imaging. Wiley Interdisciplinary Reviews: Cognitive Science, 1(3):446–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Thomas CG, Harshman RA, and Menon RS 2002. Noise reduction in BOLD-based fMRI using component analysis. NeuroImage, 17(3):1521–1537. [DOI] [PubMed] [Google Scholar]
- [49].Thomas JB, Brier MR, Ortega M, Benzinger TL, and Ances BM 2015. Weighted brain networks in disease: Centrality and entropy in human immunodeficiency virus and aging. Neurobiology of Aging, 36(1):401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Thomas JB, Brier MR, Snyder AZ, Vaida FF, and Ances BM 2013. Pathways to neurodegeneration: effects of HIV and aging on resting-state functional connectivity. Neurology, 80(13):1186–1193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Tozzi V, Balestra P, Murri R, Galgani S, Bellagamba R, Narciso P, Antinori A, Giulianelli M, Tosi G, Fantoni M, Sampaolesi A, Noto P, Ippolito G, and Wu AW 2004. Neurocognitive impairment influences quality of life in HIV-infected patients receiving HAART. International journal of STD & AIDS, 15(4):254–259. [DOI] [PubMed] [Google Scholar]
- [52].Tzourio-Mazoyer N, Landeau B, Papathanassiou D, Crivello F, Etard O, Delcroix N, Mazoyer B, and Joliot M 2002. Automated anatomical labeling of activations in SPM using a macroscopic anatomical parcellation of the MNI MRI single-subject brain. Neuroimage, 15(1):273–289. [DOI] [PubMed] [Google Scholar]
- [53].Uddin LQ, Clare Kelly AM, Biswal BB, Xavier Castellanos F, and Milham MP 2009. Functional connectivity of default mode network components: Correlation, anticorrelation, and causality. Human Brain Mapping, 30(2):625–637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Valcour VG 2011. Evaluating cognitive impairment in the clinical setting: practical screening and assessment tools. Topics in antiviral medicine, 19(5):175–180. [PMC free article] [PubMed] [Google Scholar]
- [55].Wang X, Foryt P, Ochs R, Chung J-H, Wu Y, Parrish T, and Ragin AB 2011. Abnormalities in Resting-State Functional Connectivity in Early Human Immunodeficiency Virus Infection. Brain Connectivity, 1(3):207–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Watts DJ and Strogatz SH 1998. Collective dynamics of’small-world’ networks. Nature, 393(6684):440–442. [DOI] [PubMed] [Google Scholar]
- [57].Weissman DH, Roberts KC, Visscher KM, and Woldor MG 2006. The neural bases of momentary lapses in attention. Nature Neuro-science, 9:971. [DOI] [PubMed] [Google Scholar]
- [58].Wenserski F, von Giesen H-J, Wittsack H-J, Aulich A, and Arendt G 2003. Human Immmunodeficiency Virus 1associated Minor Motor Disorders: Perfusion-weighted MR Imaging and 1H MR Spectroscopy. Radiology, 228(1):185–192. [DOI] [PubMed] [Google Scholar]
- [59].Wismüller A, Lange O, Dersch DR, Leinsinger GL, Hahn K, Pütz B, and Auer D 2002. Cluster Analysis of Biomedical Image Time-Series. International Journal of Computer Vision, 46(2):103–128. [Google Scholar]
- [60].Wismüller A, Nagarajan MB, Witte H, Pester B, and Leistritz L 2014. Pair-wise clustering of large scale Granger causality index matrices for revealing communities. In Proceedings of SPIE, volume 9038, Pp. 90381R–90381R–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Zhuang Y, Qiu X, Wang L, Ma Q, Mapstone M, Luque A, Weber M, Tivarus M, Miller E, Arduino R, Zhong J, and Schifitto G 2017. Combination antiretroviral therapy improves cognitive performance and functional connectivity in treatment-naïve HIV-infected individuals. Journal of NeuroVirology, Pp. 704–712. [DOI] [PMC free article] [PubMed] [Google Scholar]