Abstract
Recent insights supporting the fallopian tube epithelium (FTE) and serous tubal intraepithelial carcinomas (STIC) as the tissue of origin and the precursor lesion, respectively, for the majority of high-grade serous ovarian carcinomas (HGSOC) provide the necessary context to study the mechanisms that drive the development and progression of HGSOC. Here we investigate the role of the E3 ubiquitin ligase RNF20 and histone H2B monoubiquitylation (H2Bub1) in serous tumorigenesis and report that heterozygous loss of RNF20 defines the majority of HGSOC tumors. At the protein level, H2Bub1 was lost or downregulated in a large proportion of STIC and invasive HGSOC tumors, implicating RNF20/H2Bub1 loss as an early event in the development of serous ovarian carcinoma. Knockdown of RNF20, with concomitant loss of H2Bub1, was sufficient to enhance cell migration and clonogenic growth of FTE cells. To investigate the mechanisms underlying these effects, we performed ATAC-seq and RNA-seq in RNF20 knockdown FTE cell lines. Loss of RNF20 and H2Bub1 was associated with a more open chromatin conformation leading to upregulation of immune signaling pathways, including interleukin 6 (IL6). IL6 was one of the key cytokines significantly upregulated in RNF20- and H2Bub1-depleted FTE cells and imparted upon these cells an enhanced migratory phenotype. These studies provide mechanistic insight into the observed oncogenic phenotypes triggered by the early loss of H2Bub1.
Keywords: ovarian cancer, fallopian tube, H2Bub1, RNF20, chromatin, cytokines
INTRODUCTION
In the United States, the American Cancer Society predicts approximately 22,240 new cases of ovarian cancer and 14,070 related deaths in 2018 (1). Sixty percent of cases are diagnosed at an advanced clinical stage, with a 5-year overall survival rate of less than 30% (1). High-grade serous ovarian carcinoma (HGSOC) is the most common form of ovarian cancer, accounting for over 70% of cases and the majority of deaths. Standard treatment for HGSOC is surgical debulking followed by platinum-based chemotherapy (2). While chemotherapy is initially effective in controlling the disease, the vast majority of patients subsequently progress and succumb to recurrent, chemotherapy-resistant disease (2). Thus, there is an unmet medical need to develop more effective screening and therapeutic approaches for HGSOC. Recent advances in elucidating the pathogenesis of HGSOC have greatly aided this process. A growing body of evidence now points to the fallopian tube (FT) as the primary site of origin for HGSOCs (3–5). The leading argument for tubal origin of HGSOC is the presence of occult, non-invasive carcinoma in the distal fallopian tube, termed serous tubal intraepithelial carcinoma (STIC). STIC is considered an early manifestation of HGSOC and shares a high degree of morphologic and genetic similarity with the concomitant invasive tubal, ovarian, or peritoneal carcinomas (4–9). As an early form of HGSOC, STIC has allowed us to appreciate the genomic, molecular, and phenotypic changes associated with progression of the disease from localized tumors to a disseminated state. Despite progress in this area, the molecular pathogenesis of HGSOC remains poorly understood and its oncogenic drivers must be identified to guide novel therapeutic approaches (4–9).
Epigenetic mechanisms of gene regulation are defined by their capacity to change gene expression and phenotype without altering the DNA sequence (10). Aberrant epigenetic changes play a prominent role in many types of cancer, including ovarian cancer. This has spurred an interest in the development of epigenetic therapies for this disease (10). Given the genome-wide reach of epigenetic regulation, epigenetic therapies have the capacity to suppress aberrant expression of multiple oncogenic drivers through widespread reprogramming of the cancer cell genome. Early studies have shown that epigenetic therapies may be able to re-sensitize chemoresistant ovarian cancer (11–13). However, more research is needed to better define potential actionable epigenetic vulnerabilities in HGSOC.
The majority of epigenetic events involve post-translational modifications of core histone proteins (14). Permutations of these modifications have been hypothesized to make up a histone code that controls cell fate by directing essentially all DNA-associated nuclear processes (14). In addition to the more common histone modifications such as methylation, acetylation, and phosphorylation, histones H2A and H2B can also be monoubiquitylated (14). Unlike polyubiquitylation, which mainly marks proteins for degradation, monoubiquitylation of histone H2B is an epigenetic mark that regulates gene expression, DNA damage response and repair, DNA replication, and chromatin segregation (15). H2B monoubiquitylation (abbreviated H2Bub1) in mammalian cells is mediated primarily by a heteromeric E3 ubiquitin ligase consisting of ring finger proteins RNF20 and RNF40 (16). RNF20 is the key catalytic unit of this E3 ligase and its depletion leads to downregulation or loss of H2Bub1 (17,18).
Global levels of H2Bub1 are often low to absent in a variety of advanced neoplasms including breast, colorectal, lung, and parathyroid (15,19). These observations suggest that H2Bub1 may act as tumor suppressor. Mechanistically, loss of H2Bub1 is thought to contribute to tumor progression through a combination of events involving changes in protooncogene expression, increased cell survival signaling, increased cell migration, decreased DNA double-strand break repair, and increased chromosomal instability, events that are heavily implicated in HGSOC tumorigenesis (20).
In this study, we examined the status and potential oncogenic role of RNF20/H2Bub1 loss in the development and progression of HGSOC. We found that RNF20 expression is reduced in more than 50% of HGSOC cases, and that H2Bub1 is downregulated or lost early in the pathogenesis of HGSOC from the FT. We address the impact of loss of H2Bub1 on chromatin accessibility and identify key pathways that contribute to the oncogenic behavior of H2Bub1-depleted cells.
MATERIALS AND METHODS
This study was approved by the Institutional Review Boards at the Cedars-Sinai Medical Center (CSMC), Brigham and Women’s Hospital (BWH), Dana-Farber Cancer Institute (DFCI), Yale University, and the University of Pennsylvania.
Case Selection
The cases for this study were obtained from the Departments of Pathology at CSMC, BWH, and Yale University. Formalin-fixed paraffin embedded blocks of fallopian tube tissues were cut from 25 cases whose original pathology reports indicated the presence of STIC and/or invasive HGSOC. These H&E slides were reviewed by three pathologists (VP, MSH, RD) to confirm the presence of STICs and possibly invasive carcinoma in the deeper tissue sections, based on criteria described in the Supplementary Materials and Methods.
Evaluation of H2Bub1 immunohistochemistry (IHC)
The H2Bub1 immunostains were scored semi-quantitatively for intensity and distribution of immunoreactivity (% positive cells). In brief, the distribution of immunoreactivity was scored as follows: 0 (negative or occasional positive cells), 1+ (<10% cells positive), 2+ (10%−75% cells positive), 3+ (76%−100% cells positive). IHC stain intensity was assessed as follows: 0 (negative), 1 (weak), 2 (moderate), 3 (strong). Ultimately, a composite score for each lesion or normal FTE was calculated by multiplying the distribution of immunoreactivity score by the corresponding intensity score.
Cell culture and gene silencing
Immortalized fallopian tube secretory epithelial cells (FTSEC): FT190, FT194, and FT246 were previously described (21,22) and grown in fallopian tube medium (FTM) consisting of DMEM/F12 supplemented with Ultroser G serum substitute (22) and 25 mM HEPES buffer (pH 7.2 – 7.5). Human HGSOC cell lines OVKATE (Japanese Collection of Research Bioresources Cell Bank) and SKOV3 (ATCC) were grown in RPMI1640, 10% FBS and 1% penicillin/streptomycin. HGSOC cell line Kuramochi (Japanese Collection of Research Bioresources Cell Bank) was cultured in RPMI1640 supplemented with 10% FBS, 1% MEM Non-essential amino acids (Gibco), 0.25 U/ml Insulin and 1% penicillin/streptomycin. All cell lines were authenticated using Short Tandem Repeat (STR) profiling and tested to be free of Mycoplasma using the Cambrex MycoAlert assay at the University of Pennsylvania Perelman School of Medicine Cell Center (Philadelphia, PA) in May 2018.
To stably silence RNF20 in FT190 and FT194, cells were transduced with lentiviral vectors (Mission, Sigma-Aldrich) encoding two separate shRNAs: shRNF20_692 (TRCN00000692) or shRNF20_890 (TRCN00000890), or a non-targeting control shRNA: shNTC (SHC002V). The cells were transduced at MOI = 40 followed by antibiotic selection with puromycin. For siRNA-mediated silencing of RNF20 in Kuramochi, OVKATE, SKOV3, FT190, FT194 and FT246 the cells were plated and 24 hr later transfected with pooled siRNAs targeting RNF20, or with non-targeting control pool, using Lipofectamine RNAiMAX (Life Technologies). The siRNAs, SMARTpool ON-TARGET Plus RNF20 siRNA (Cat# J-007027 (05–08), and Control pool siRNA (cat# D-001810–10-05), were purchased from Dharmacon (Lafayette, USA).
In vitro cell assays
For the clonogenic assay, cells were seeded in 6-well plates at 100 – 500 cells per well in triplicate wells. Three to four weeks later, cell were fixed with 4% paraformaldehyde in PBS, stained with 0.5% crystal violet, and colonies >1mm were counted using ImageJ.
The transwell migration assay was performed as previously described (18). Briefly, 2.5×104 cells in 100μl of serum free medium was dispensed into the upper compartment of a Boyden chamber with 8μm pore size filter and 650 μl of complete medium with or without EGF (10ng/mL) was dispensed into the lower compartment. The cells were allowed to migrate overnight, followed by removal of cells from the upper chamber. Cells that migrated to the bottom of the Boyden chamber were fixed in 100% methanol, and stained with crystal violet for 30 min. The underside of each chamber was imaged at 10X. The filter was cut out and the crystal violet staining was dissolved in 10% acetic acid. Absorbance was measured at 570nm for quantification. Transwell migration assay was performed in the presence or absence of human recombinant IL6 (hIL6) (Cell signaling Cat# 8904, Danvers, USA). The IL6 neutralization was performed by using anti-IL6 antibody (R&D Cat# MAB206–500, Minneapolis, USA) 100x the concentration of IL6 for dose dependent experiments with control cells and 25 – 50 μg/ml for experiments with knockdown cells.
Cell proliferation assay was performed using CellTiter-Glo Luminescent reagent (Promega) following the manufacturer’s guidelines. Briefly, 2000 – 5000 cells were seeded in 96 well plates in 100μl of the media and allowed to grow for various time intervals. At the end of the experiment, equal volume of CellTiter-Glo reagent was added, contents were mixed and incubated for 10 min and luminescence was recorded.
All assays were repeated for reproducibility with n ≥ 3 for each condition.
Preparation of conditioned media
Cells were plated in 10cm dish and were allowed to grow at 37°C and 5% CO2. When the cells reached 60–70% confluency media was removed, cells were washed with PBS twice, 8ml of regular FTM media was added to the dish and cells were placed back into the incubator. After 72 hrs, the conditioned media was collected, centrifuged at 2000g at 4°C for 5 min to remove cell debris and filtered through a 0.45μm filter.
Human cytokine array profile and ELISA assays
Levels of cytokines were analyzed using the proteome profiler antibody arrays from R&D systems (cat# ARY005B). Levels of individual cytokines, IL6 (Cat# D6050), TNFα (Cat# DTA00C) and IL1α (Cat# DLA50) were analyzed using Quantitative ELISA assay kits from R&D systems. Conditioned media from cells was used to perform the above assays as per the manufacturer’s recommendations. Media not exposed to cells was used as the negative control. Data for cytokine array profile was acquired using Bio-rad clarity western ECL substrate on Bio-rad ChemiDoc and images were analyzed using image lab 5.0 software. Data for individual cytokines was acquired and analyzed using SoftMax pro 7 software (Molecular Devices) version 7.0.2.
RNA sample processing and sequencing
RNA was isolated using the RNeasy Plus Mini Kit from Qiagen as per the manufacturer’s protocol (Cat# 74134). The extracted RNA samples where checked for overall quality and only samples with high quality (RIN ≥ 8.0) and high purity (OD 260/280 = 1.8–2.0) were used to generate libraries with the Bioo Scientific NEXTflex Rapid Directional RNA-seq Library Prep Kit (Bioo Scientific, Austin TX). The resulting mRNA libraries were then sequenced at the JWCI Sequencing Center on an Illumina HiSeq 2500 in Rapid Mode using 101 bp paired-end reads. See supplementary materials and methods for data processing and downstream analysis.
Assay for Transposase-accessible chromatin sequencing (ATAC-seq) sample processing and sequencing
ATAC-seq was performed as previously described (23). Briefly, 50,000 cells were washed in ice cold 1X PBS and re-suspended in 50 μl ice cold lysis buffer. Then nuclei were pelleted at 500 x g for 10 min at 4°C. The pellet was then incubated with transposition reaction mix for 30 min at 37°C and purified using the MinElute PCR Purification kit (Qiagen). As with the RNA-seq libraries above, the ATAC-seq libraries were sequenced on an Illumina HiSeq 2500 in Rapid Mode using 101bp paired-end reads at the JWCI Sequencing Center. See supplementary materials and methods for data processing and downstream analysis.
The RNA sequencing and the ATAC-seq data presented in this study were submitted to the Gene Expression Omnibus and can be accessed by the accession number GSE122238.
Statistical Analysis
Statistical analysis was performed using GraphPad Prism 7 (GraphPad Software Inc.). Differences in H2Bub1 immunoreactivity or composite scores among morphologically normal FTE, STIC, and invasive HGSOC were examined using the Kruskal-Wallis test, followed by Mann-Whitney test for pair-wise comparison. Data for Trans-well migration, clonogenic assays, and other in-vitro cell assays was analyzed using GraphPad Prism. One-way ANOVA followed by Student’s t-test for pair-wise comparisons was used to determine the statistical significance. In all analyses, p < 0.05 was considered statistically significant. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001; no asterisks, p not calculated.
Antibodies and Reagents
Immunohistochemical staining, Western blots, Hanging drop method, Scratch or wound healing assay
RESULTS
Heterozygous loss of RNF20 is present in the majority of HGSOCs and correlates with mRNA expression levels
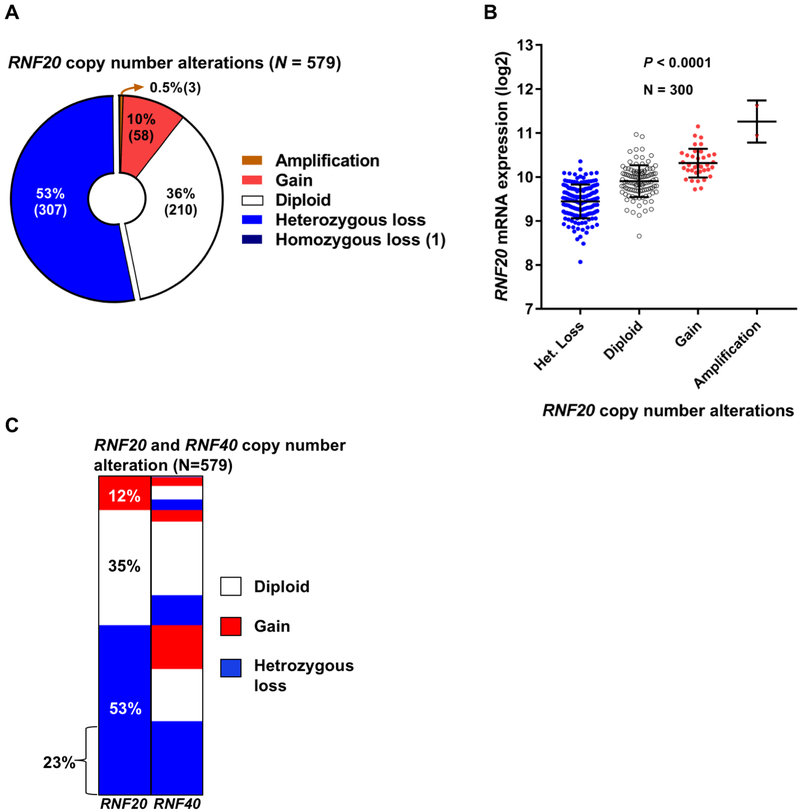
To explore the possibility that alterations in RNF20 are involved in ovarian cancer, we queried The Cancer Genome Atlas (TCGA) database (24,25). We analyzed the provisional dataset containing 579 cases of HGSOC with mRNA (RNAseq V2 RSEM) data and putative somatic copy number calls determined by GISTIC 2.0 (26). Of the 579 cases, more than half (53%) were characterized by heterozygous loss of RNF20, while 36% were diploid and 10% exhibited copy number gain (Fig. 1A). RNF20 amplification and homozygous deletion events were rare, present in only four cases. In line with expected changes in transcript levels due to somatic copy number loss, RNF20 mRNA levels varied according to the gene copy number, with decreased RNF20 mRNA in tumors with heterozygous loss of the RNF20 allele (P<0.0001, one-way ANOVA test, Fig. 1B). We also analyzed the data set to explore the copy number alteration for RNF20 binding partner RNF40. Of 579 cases, 36% of the cases were characterized by heterozygous loss of RNF40, while 44% were diploid and 19% were gains (Supplementary Fig. 1). Additionally, we explored the data set to characterize the combined copy number alteration for both RNF20 and RNF40. Of 579 cases, 23% of the cases exhibited heterozygous loss of both RNF20 and RNF40 allele (Fig. 1C). The analysis of TCGA dataset suggests that a majority of HGSOC cases exhibit a heterozygous loss of RNF20 or RNF40.
Figure 1.
Heterozygous loss of RNF20 is present in the majority of HGSOCs. (A) RNF20 somatic copy number alterations in a cohort of 579 HGSOC cases from The Cancer Genome Atlas (TCGA). (B) RNF20 mRNA expression profile in the TCGA cases, RNF20 mRNA is downregulated in tumors with heterozygous loss of the allele. (C) Distribution of copy number alterations of the RNF20 and RNF40 genes in a TCGA Ovarian cancer cohort (n=579).
Loss of H2Bub1 is an early event in serous tumorigenesis
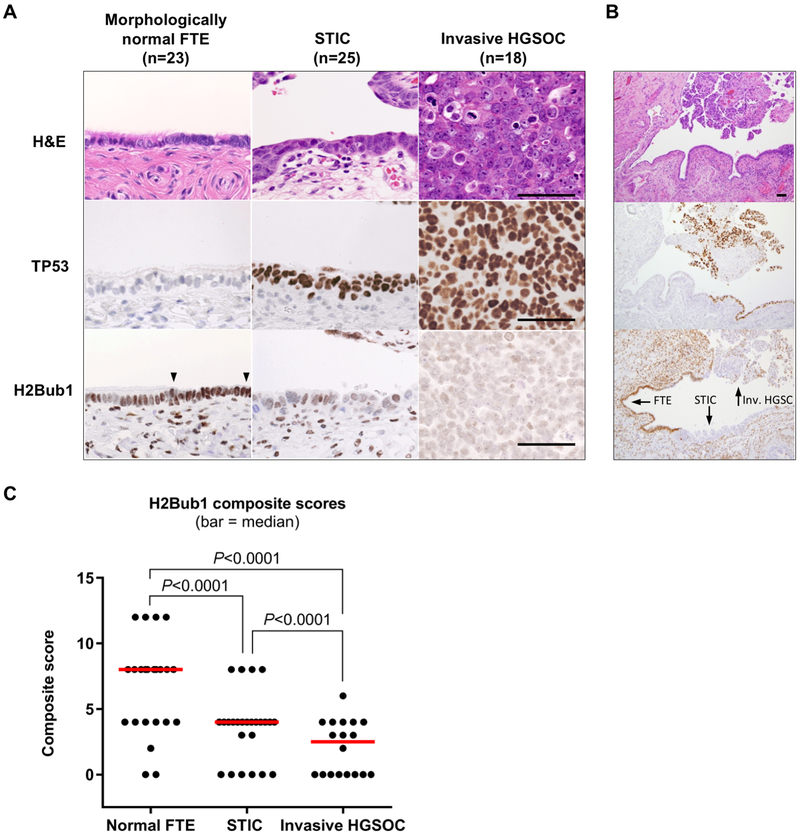
RNF20 is the obligate component of the E3 ubiquitin ligase responsible for H2Bub1 (14,16). Multiple reports (reviewed in (14,15)) have shown that decrease or loss of RNF20 expression leads to concomitant downregulation or loss of H2Bub1. We examined whether the decrease in RNF20 mRNA seen in the TCGA dataset translates to an appreciable reduction of H2Bub1 in HGSOC, and whether this is an early or late event in HGSOC tumorigenesis. To this end, we performed immunohistochemical (IHC) analysis of H2Bub1 in fallopian tube specimens from 25 patients diagnosed with non-invasive high-grade serous tumors (termed STICs) and/or with invasive HGSOC. We identified one or more STICs in all 25 cases, while concurrent invasive HGSOCs were found in 18 cases, and morphologically normal FTE was present in 23 cases. Figures 2A and 2B shows relative H2Bub1 levels in normal FTE, STIC, and in invasive HGSOC, respectively. All stains in Figure 2 were performed on one case to capture the continuity of tumor progression and H2Bub1 abundance. H2Bub1 was scored semi-quantitatively for intensity and distribution of immunoreactivity (% positive cells), the product of which is a composite score. Table 1 shows distribution of the semi-quantitative scores for stain intensity and median composite scores. Distribution of the composite scores is depicted graphically in Figure 2C. In morphologically normal FTE (Fig. 2A) we observed strong, diffuse, nuclear H2Bub1 immunoreactivity (median score = 6; Fig 2C) with a similar pattern and stain intensity noted for ciliated and secretory cells. In contrast to morphologically normal FTE, H2Bub1 immunoreactivity in STIC was focal with varying, but markedly reduced stain intensity overall (Fig. 2B), resulting in lower median composite score (median score = 3; Fig 2C). H2Bub1 immunoreactivity was further reduced in invasive HGSOC (Fig. 2C) (median score = 2, Fig. 2C). While the IHC stain intensity was not significantly different between STIC and invasive HGSOC, the percentage of positive cells was lower in invasive HGSOC, decreasing the composite score (Supplementary Fig. S2A and S2B). Importantly, total H2B protein levels were unaltered in specimens that exhibited decrease or loss of H2Bub1 (Supplementary Fig. S2C). STICs are considered early manifestations of HGSOC and are thought to progress to invasive HGSOC (4,7–9). Therefore, gradual decrease of H2Bub1 immunoreactivity in STICs and further decrease in late-stage invasive tumors suggests that H2Bub1 loss occurs early in, and progresses throughout, the evolution of HGSOC.
Figure 2.
Loss of H2bub1 is an early event in serous tumorigenesis. (A) Immunohistochemical analysis of H2Bub1 expression in morphologically normal fallopian tube epithelium (FTE), serous tubal intraepithelial carcinoma (STIC), and invasive high-grade serous cancer (HGSOC), with p53 staining marking the carcinoma cells. Abundant H2Bub1 is expressed in both secretory cells (between arrowheads) and ciliated cells of morphologically normal FTE, but it is strongly downregulated or lost in STIC and invasive HGSOC. All micrographs (40x, Scale bar = 20 μm) were imaged from one representative case. (B) Low magnification (10x) images of (A) displaying the lesions (arrows) in continuity with normal FTE. (C) Composite H1Bub1 immunostaining scores (range - 0–12), obtained by multiplying semiquantitative scores for stain intensity and distribution (each scored as: 0+, 1+, 2+, 3+;) (horizontal bars – median; P - Mann-Whitney test).
Table 1.
H2Bub1 expression in in-situ and invasive HGSOC
| H2Bub1 stain intensity |
Composite Score |
||||
|---|---|---|---|---|---|
| Negative (%) | Weak (%) | Moderate (%) | Strong (%) | (median; 95% CI) | |
| Normal FTE (n=23) a | 2/23 (9) | 6/23 (26) | 11/23 (48) | 4/23 (17) | 6 (3-6) d |
| STIC (n=25) b | 6/25 (24) | 14/25 (56) | 5/25 (20) | 0/25 (0) | 3 (2-3) e |
| Invasive HGSOC (n=18) c | 8/18 (44) | 9/18 (50) | 1/18 (6) | 0/18 (0) | 2 (0-3) f |
| a vs. b; (p=0.0014)* | d vs. e; (p=0.0007)* | ||||
| a vs. c; (p<0.0001)* | d vs. f; (p<0.0001)* | ||||
| b vs. c; (p=0.0995)* | e vs. f; (p=0.0323)* | ||||
Mann Whitney test
Abbreviations: FTE, fallopian tube epithelium; STIC, serous tubal intraepithelial carcinoma; HGSOC, high-grade serous ovarian carcinoma
RNF20 knockdown in immortalized FTSEC results in downregulation of H2Bub1 and promotes complex oncogenic phenotypes
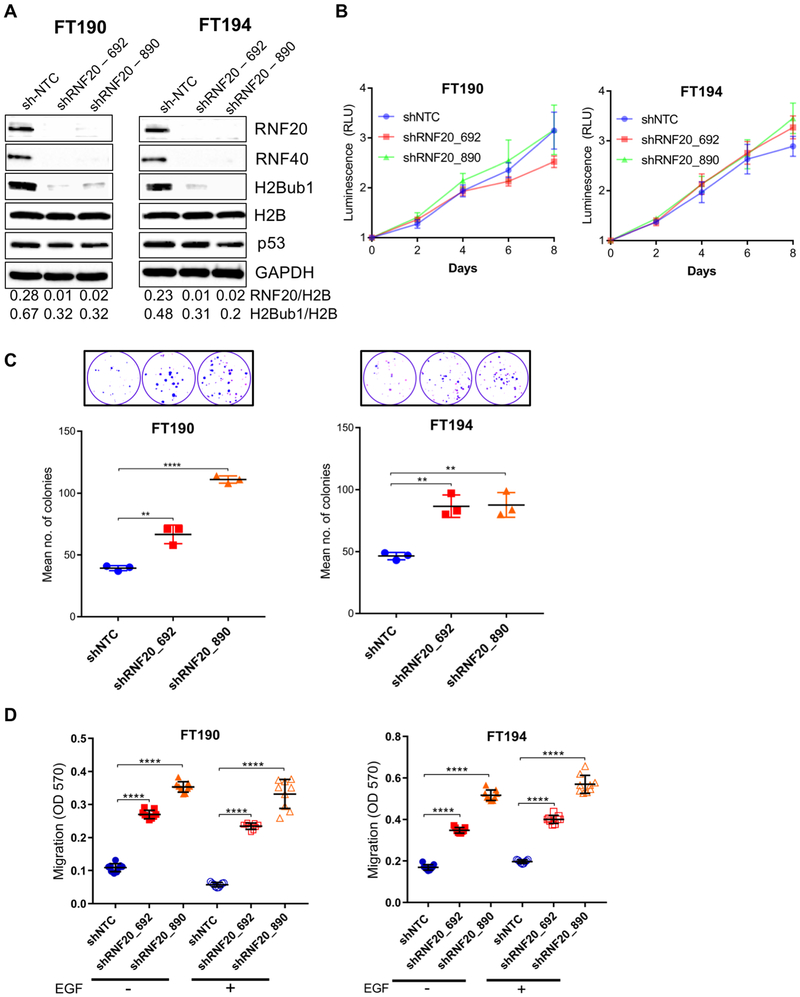
The decrease in H2Bub1 observed in both early and late-stage HGSOC lesions prompted us to examine whether experimental downregulation of RNF20, and consequently of its product H2Bub1, is sufficient to elicit cancer-associated features in non-transformed FTSECs. We used two different shRNA constructs targeting the RNF20 transcript to knockdown RNF20 gene expression in two cell lines, FT190 and FT194. These cell lines have been established as a model platform to study high-grade serous ovarian carcinogenesis from the fallopian tube (21,27). Both cell lines were immortalized with hTERT, the catalytic subunit of telomerase, and SV40 large T plus small t antigens (27,28). Stable expression of RNF20 shRNAs effectively knocked down RNF20 protein levels in both cell lines (Fig. 3A). Depletion of RNF20 resulted in the expected reduction of RNF40, the binding partner of RNF20 in the E3 heterocomplex that mediates monoubiquitination of Lys-120 on histone H2B, which is in line with a previous report (17); the stability of either RNF20 or RNF40 is dependent upon the presence of the other partner, such that depletion of either component results in a substantial decrease in the entire complex (14). As expected, RNF20 knockdown led to pronounced reduction of global ubiquitylated H2B in both cell lines, while total H2B levels were not affected (Fig 3A). p53, whose expression is positively regulated by H2Bub1 (18) did not decrease along with H2B ubiquitylation. However, both FT190 and FT194 express the SV40 large T antigen, which inactivates p53 and stabilizes it (27).
Figure 3.
RNF20 knockdown in immortalized FTSEC results in downregulation of H2Bub1 and promotes complex oncogenic phenotypes. (A) Immortalized human FTSEC cell lines FT190 and FT194 were stably transduced with two RNF20 shRNA variants. Western blot analysis was used to validate knockdown of RNF20 and the resulting downregulation of H2Bub1 (note parallel downregulation of RNF40), p53 protein levels were not affected by manipulating RNF20/H2Bub1. (B) RNF20 knockdown had no significant effect on the proliferation rate. (C) Stable knockdown of RNF20 promoted clonogenic growth in both immortalized FTSEC cell lines (top shows fixed and stained colonies). (D) RNF20 knockdown also promoted trans-well migration in both cell lines. Addition of EGF chemoattractant showed no significant increase in motility in RNF20 knockdown cells compared to RNF20 knockdown cells without EGF. RLU, Relative Light Units. **, p ≤ 0.01; ****, p ≤ 0.0001.
Next, we evaluated the functional impact of RNF20 and H2Bub1 downregulation on proliferation, clonogenic and anchorage-independent growth, and cell migration. To determine the effect of RNF20 knockdown on proliferation, we used the CellTiter-Glo viability assay. As shown in Figure 3B, RNF20 depleted cells and control (shNTC) cells exhibited similar growth kinetics at the six-day endpoint (Fig. 3B). These observations indicate that RNF20 knockdown in FTSECs does not lead to a substantial increase in cellular proliferation. Next, we assessed whether RNF20 knockdown promotes clonogenic growth. The cell lines were plated as single cells at a density of 100 cells per well and the number of colonies (>1mm) were counted after 3–4 weeks. Both FT190 and FT194 cells expressing the control non-targeting shRNA were modestly clonogenic giving rise on average to 39.3 and 46.3 colonies per 100 cells, respectively. However, RNF20 depletion in both cell lines significantly increased their clonogenicity (Fig. 3C). As shown in Figure 3C, in both FT190 and FT194, RNF20 and H2Bub1 downregulated cells formed on average twice the number of colonies compared to control cells.
Shema and colleagues previously reported that stable knockdown of RNF20 in NIH3T3 mouse fibroblasts increased anchorage-independent proliferation, a key indicator of cell transformation (18). Given these observations, we tested whether RNF20 knockdown in FT190 and FT194 cells provides sufficient growth advantage in 3D culture. Using the hanging drop method, cells were seeded and grown for 4 weeks and then examined for sphere formation. As shown in supplementary Figure S3A, RNF20 knockdown led to a modest yet statistically significant increase in sphere forming ability in FT190 and FT194 cells. We used the Boyden chamber system to evaluate trans-well migration in control and RNF20-depleted FT190 and FT194, with and without the EGF attractant. In the absence of EGF, FT190 and FT194 control cells displayed a low level of migration, which was significantly enhanced in all RNF20-depleted cell lines (Fig. 3D). Enhancement of cell motility by RNF20 knockdown was observed also in the presence of EGF in FT190 and FT194 cells. The presence of EGF did not further enhance the RNF20 knockdown mediated increased migration ability. We observed similar results using a scratch assay (Supplementary Fig. S3B). To confirm the shRNA results, we used a siRNA pool to transiently knockdown RNF20 in FT190, FT194 and FT246 cell lines. In all cases, knockdown of RNF20 led to decreased levels of H2Bub1 (Supplementary Fig. S4A) with a concomitant increase in cell migration (Supplementary Fig. S4B).
RNF20 and H2Bub1 depletion escalates oncogenic behavior in cells derived from late-stage HGSOC
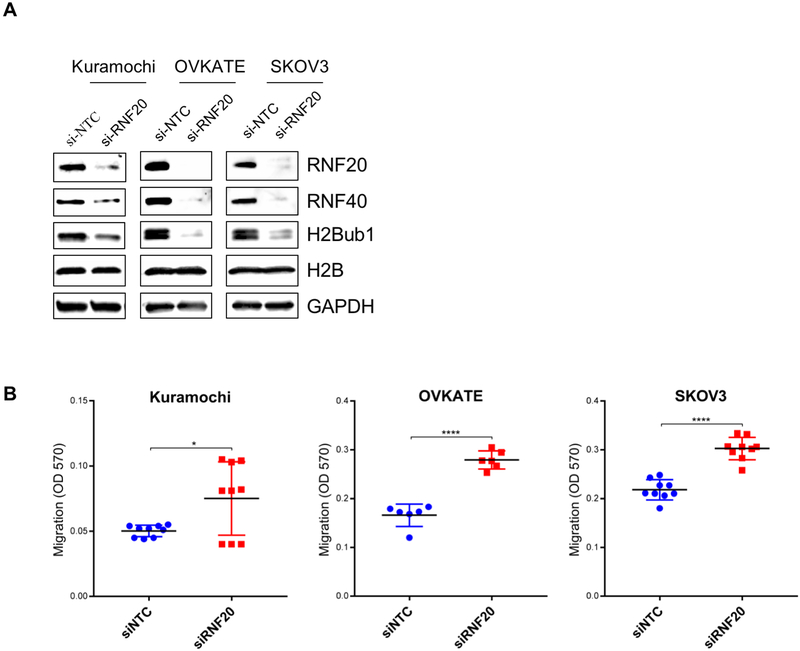
The results described thus far indicate that RNF20 depletion is sufficient to enhance clonogenicity and motility in two immortalized, tubal-derived cell lines. Clonogenic growth and enhanced cell migration are distinctive features of advanced tumors, suggesting that decreased RNF20 expression in tubal neoplasms may potentially facilitate cancer progression. As noted in the H2Bub1 IHC analysis, H2Bub1 staining in tubal neoplasms was focal, often with variable staining intensity among individual tumor cells. This observation suggests that H2Bub1 is depleted gradually during tumor progression. To address this hypothesis, we examined whether RNF20 and H2Bub1 depletion facilitates tumor progression in cells derived from late-stage carcinoma. To this end, we screened by Western blot a number of ovarian cancer cell lines to select those that retained relatively high RNF20 and H2Bub1 expression (Supplemental Figure S4C). We selected Kuramochi, OVKATE and SKOV3 cell lines and used siRNA pools targeting RNF20 to deplete RNF20 and H2Bub1 in these cell lines. The siRNA treatment led to strong downregulation of RNF20 in all cell lines at 72 hrs following transfection (Fig. 4A). The degree of RNF20 depletion in these established cancer lines was comparable to that seen in FTSEC lines (Supplemental Figure S4A) and led to effective downregulation of H2Bub1 and markedly increased cell migration in all three cell lines (Figure 4B).
Figure 4.
RNF20 and H2Bub1 depletion escalates oncogenic behavior in late-stage HGSOC and SKOV3 cells. Cell lines which retained H2Bub1 were selected and RNF20 was silenced by pooled siRNAs. (A) Western blot was used to validate RNF20 knockdown and the resulting H2Bub1 downregulation. (B) RNF20/H2Bub1 downregulation promoted trans-well migration in HGSOC cell lines Kuramochi, OVKATE and SKOV3. *, p ≤ 0.05; ****, p ≤ 0.0001.
RNF20 and H2Bub1 depletion re-distributes open chromatin regions and alters immune signaling pathways
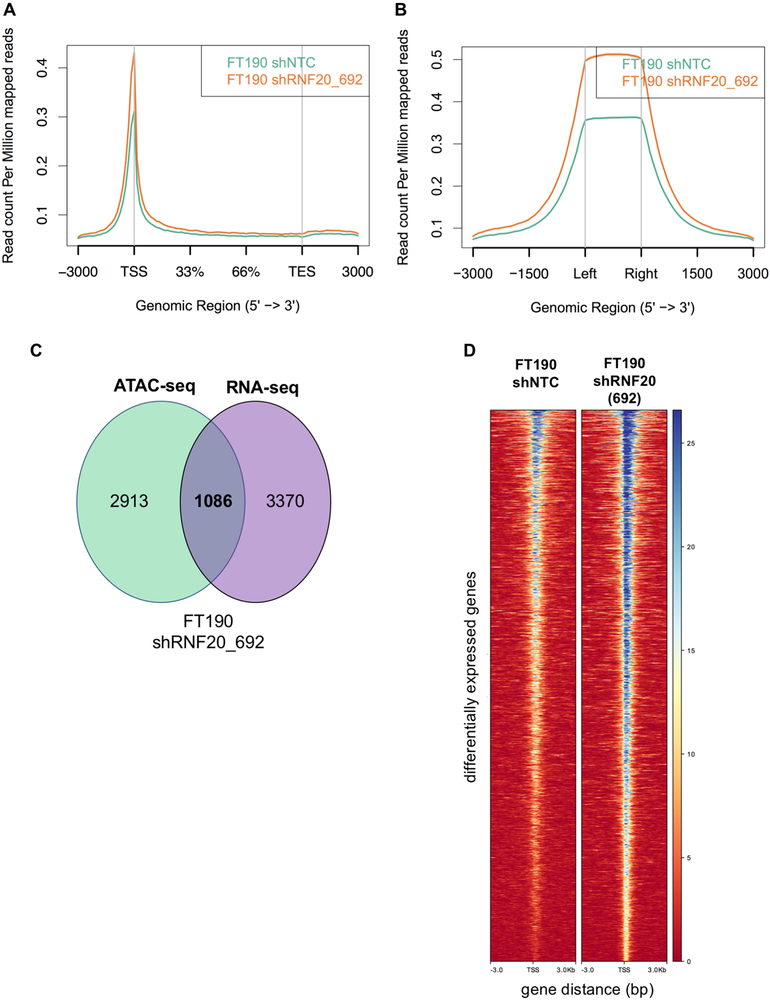
Cell migration and clonogenic growth represent complex cell behaviors. To explore how RNF20 knockdown and the resulting H2Bub1 downregulation promote these phenotypes we analyzed genome-wide changes in chromatin state using Assay for Transposase-Accessible Chromatin using sequencing (ATAC-seq) and gene expression using RNA-seq. Several models have been proposed for the function of H2Bub1, including a role in nucleosome stabilization, leading to a more closed chromatin conformation (29), and conversely, interference with chromatin compaction leading to an open conformation (30). Surprisingly, in our genome-wide ATAC-seq analysis we found that the ATAC-seq read counts in RNF20-depleted cells showed enrichment in promoter regions (Fig. 5A) and known DNase I hypersensitivity regions (Fig. 5B) as compared to respective control cells. This suggests that the loss of H2Bub1 could lead to a more open chromatin conformation in FTSEC cells. To our knowledge, this is among the first observations to show a direct relationship between the loss of H2Bub1 and open chromatin conformation in mammalian cells. Next, we identified the set of differentially expressed (DE) genes from the RNA-seq data that overlapped with significant ATAC-seq peaks in RNF20-depleted cells. We identified 1,086 genes that overlapped between the RNA-seq and ATAC-seq data in FT190 shRNF20_692 cells (Fig. 5C). Consistent with the genome-wide patterns observed in Figure 5A and 5B, the 1,086 genes were found to be located in genomic regions that show a more open chromatin state in RNF20-depleted cells as compared to controls (Figure 5D). Furthermore, the majority (675) of the overlapped genes represented in Figure 5C and 5D showed increased expression. Similar results were obtained using the FT194 cell line (Supplementary Fig. S5A – S5J).
Figure 5.
Changes in chromatin accessibility and gene expression between RNF20-depleted and control cells. (A) The distribution of ATAC-seq reads in FT190 shRNF20_692 cells show enrichment around transcription start site and (B) known DNase I hypersensitivity sites as compared to control cells. (C) Overlap between ATAC-seq peaks and RNA-seq significantly differentially expressed genes. All overlap genes were upregulated in RNA-seq. (D) Heatmaps for the set of significantly differentially expressed genes within ATAC-seq peaks for control and RNF20-depleted FT190 cells. ATAC-seq reads are centered on transcription start sites (TSS) with 3KB of flanking sequence. The color scale indicates an increase in read counts from red to blue, with darker blue representing a more open chromatin state.
To identify the processes and pathways altered due to the loss of H2Bub1, we performed an enrichment analysis using the Molecular Signatures Database (MSigDB) (see supplemental methods). Interestingly, enrichment analysis of the set of differentially expressed genes within ATAC-seq peaks identified significant enrichment for immune function related pathways (Supplementary Figure S6A – S6D). Within these immune signaling pathways, IL6 was identified as one of the significantly altered immune signaling genes in our ATAC-seq and RNA-seq analysis. An IGV browser view of the IL6 gene region showed enrichment for ATAC-seq reads in the RNF20-depleted cells as compared to the control cells (Supplementary Figure S7A-S7D), again indicating a more open chromatin state in the IL6 gene region in RNF20-depleted cells. Overall, our ATAC-seq and RNA-seq data analysis identified that loss of H2Bub1 changes the chromatin landscape to a more open conformation thereby altering various key pathways including immune signaling pathways.
Loss of RNF20 and H2Bub1 upregulates the levels of cytokine IL6 leading to enhanced migration in FTSECs
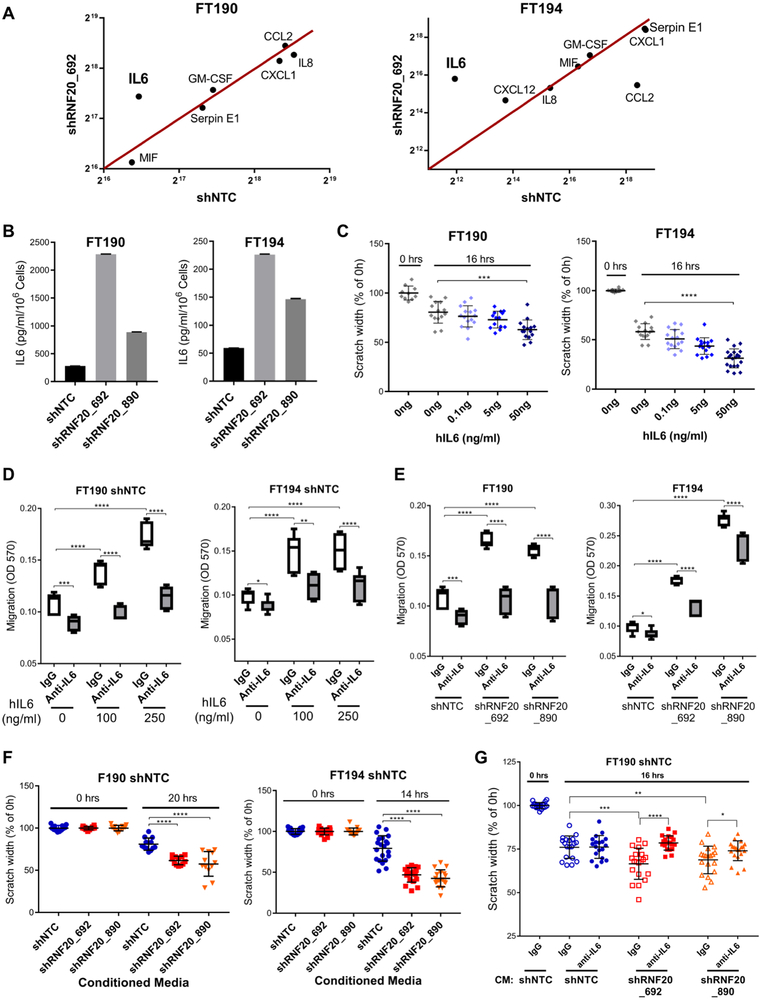
To further explore the impact of H2Bub1 downregulation on cytokine production, we performed a cytokine array profile of the cell supernatants from control and RNF20-depleted cells. Of all the detected cytokines in the array, IL6 was found to be significantly upregulated in both FT190 and FT194 RNF20-depleted cells (Fig. 6A and Supplementary Fig. S8A), consistent with our pathway enrichment analyses. To quantitatively measure the levels of IL6 in cell supernatant we performed an ELISA assay. Figure 6B shows that IL6 levels are highly upregulated due to the loss of RNF20 and H2Bub1. Similarly, we found that the levels of both TNFα and IL1α were also significantly upregulated in RNF20-depleted cells (Supplementary Fig. S8B).
Figure 6.
RNF20/H2Bub1 depletion upregulates the levels of IL6 cytokine promoting oncogenic behavior in FTSECs. (A) Cytokine array profiling identifies IL6 as the key altered cytokine in RNF20/H2Bub1 deficient cells. (B) ELISA assay shows that the levels of IL6 are elevated in the conditioned media of RNF20/H2Bub1 deficient cells. (C) Scratch assay showing that hIL6 added to the media promotes the rate of migration in control cells (shNTC) in a dose-dependent manner. (D) Trans-well migration assay showing that hIL6 induces migration in control cells which can be neutralized by using the anti-IL6 antibody. (E) Trans-well migration assay showing that the anti-IL6 antibody could reverse the RNF20/H2Bub1 mediated enhanced migration to the basal levels equivalent to that of control cells in FT190 cells, while FT194 cells showed partial, but significant, recovery of the oncogenic phenotype. (F) Conditioned media from control cells and RNF20/H2Bub1 deficient cells was transferred onto the control cells in a scratch assay. Conditioned media from RNF20/H2Bub1 deficient cells significantly enhanced the migration/scratch closing ability of control cells. (G) Conditioned media from control and RNF20-depleted FT190 cells was pre-incubated with control IgG or anti-IL6 IgG for 20 minutes and transferred on to the control cells (shNTC) in a scratch assay. Scratch width was measured at various time points. Anti-IL6 antibody could neutralize the migratory phenotype induces by conditioned media from RNF20-depleted cells. Control IgG and anti-IL6 antibody concentrations were 25 μg/ml for FT190 and 50 μg/ml for FT194. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001; ****, p ≤ 0.0001. For scratch assay in figure 6C and 6G only one data set is shown for reference at 0 hrs because the scratch width at 0 hrs was 100 percent for all conditions.
To assess the effect of IL6 on cell migration ability we performed a scratch assay and trans-well migration assay in the presence or absence of recombinant human-IL6 (hIL6). FT190 and FT194 cells were allowed to migrate in the scratch assay in the presence of increasing concentrations of hIL6. Both FT190 and FT194 showed a dose-dependent increase in migration (Fig. 6C). Similar results were observed with the trans-well migration assay (Fig 6D). However, when we co-incubated hIL6 with an anti-IL6 specific antibody versus the respective control IgG1, we could neutralize this effect (Fig. 6D). The above data shows that IL6 alone can enhance the migration of FTSECs thereby mimicking the enhanced migration seen in RNF20-depleted cells. Therefore, we asked whether the increased levels of IL6 in RNF20-depleted cells are responsible for the enhanced migratory phenotype of the cells. To this end, we performed trans-well migration assay with control and RNF20-depleted cells in the presence of control IgG or anti-IL6 antibody. Importantly, the anti-IL6 antibody was able to inhibit the enhanced migration of RNF20-depleted cells almost completely (FT190 cells), or at least partially (FT194 cells) (Fig. 6E).
Our data suggests that RNF20 and H2Bub1 depleted cells secrete increased amounts of IL6 (Fig. 6B), which can account, in part, for the enhanced migratory potential of the cells (Fig. 6D and 6E). These observations prompted us to ask whether the conditioned media from RNF20-depleted cells could impact the migratory ability of control cells in trans. To test this possibility, we collected the conditioned media from control and RNF20-depleted cells and transferred the conditioned media onto the control cells in a scratch assay. Reassuringly, the conditioned media from RNF20-depleted cells was able to enhance the migration of control cells (Fig. 6F), and the results were consistent in both cell lines, FT190 and FT194 cell lines. To assess whether the effect of the conditioned media was due to the presence of IL6, we repeated the experiment in the presence or absence of anti-IL6 antibody. We pre-incubated the conditioned media with control (IgG1) or anti-IL6 antibody and then transferred it to the control cells. While FT190 control cells exposed to RNF20-depleted conditioned media containing control antibody retained enhanced migration, this migration was blocked by the presence of anti-IL6 antibody in the conditioned media (Fig. 6G). Thus, RNF20 and H2Bub1 depleted cells produce higher levels of IL6, which may stimulate autocrine and/or paracrine signaling, leading to enhanced migration of neighboring cells.
Finally, to validate the correlation between the level of H2Bub1 and the expression of IL6 at the tissue level we performed RNAscope in situ hybridization for IL6 on FFPE samples of HGSOC and controls. HGSOC tumors that were H2Bub1 negative by IHC, exhibited the highest levels of IL6, while HGSOC tumors that displayed low levels of H2Bub1 has modest expression of IL6. Importantly, control ovary and uterine tissues were negative for IL6 (Supplementary Fig. S9).
DISCUSSION
Here we report on the role of RNF20 and H2Bub1 in ovarian cancer pathogenesis. We use orthogonal approaches to make three novel observations. First, we used immunohistochemistry to show that H2Bub1 is lost in STIC precursors of HGSOC. Second, we show that loss of H2Bub1 leads to the acquisition of a malignant phenotype in FT cells. Finally, ATAC-Seq and RNA seq reveal that loss of H2Bub1 leads to a more accessible chromatin state that causes upregulation of immune modulators that stimulate growth and migration of early HGSOC precursors.
At the genomic level we show that over half of HGSOC cases are marked by heterozygous loss of the RNF20 allele, an event that is strongly associated with decreased RNF20 transcript levels. It is noteworthy that among the HGSOC cases in the TCGA dataset, RNF20 amplification was present in only three cases and homozygous deletion in just one case, while three cases with RNF20 mutations were noted. The relative rarity of these genomic events in a large cohort indicates that gaining more copies of the RNF20 allele is unlikely to provide selective advantage. Complete loss or mutational inactivation of RNF20 may be strongly resisted by selective pressure, which is in line with reported embryonic lethality of RNF20 knockout in mice (15,31).
Given that RNF20 is depleted in HGSOCs yet is responsible for carrying out H2B monoubiquitylation, we used IHC to examine H2Bub1 levels in early non-invasive human HGSOC lesions (STICs) and in the concomitant invasive HGSOC tumors. The IHC analysis showed that H2Bub1 is attenuated early and continues to decrease throughout HGSOC progression. These findings are in agreement with previous reports showing decreased RNF20 transcript levels in metastatic prostate cancer cells relative to benign disease cells (32) and H2Bub1 decrease or loss by IHC in malignant breast tumors and even further decrease in metastatic lesions (33). In addition, a recent study reported a global loss of H2Bub1 in 77% of the tumors in a cohort of 407 HGSOC samples (34). However, none of these studies examine the status of H2Bub1 in precursor lesions. The output of our IHC analysis was a composite immunoreactivity score, the product of stain intensity and the percentage of positive cells. It is important to point out that while STIC and invasive HGSOC displayed similar stain intensity, we observed a lower percentage of immunoreactive cells in invasive HGSOC. Therefore, the decrease in composite H2Bub1 immunoreactivity between normal FTE and STIC appears to be driven by downregulation of H2Bub1 expression (lower stain intensity), while further decrease in H2Bub1 reactivity between STIC and invasive HGSOC is driven by complete loss of H2Bub1 expression in individual cells. It is unlikely that the complete loss of H2Bub1 in a subset of tumor cells within invasive HGSOC is a consequence of RNF20 depletion due to homozygous loss of both RNF20 alleles; according to our analysis of the TCGA dataset, at least one intact RNF20 allele was present in all but one HGSOC case. However, it is possible that expression of the remaining allele may be silenced later by promoter hypermethylation (18), which could act as the second hit. It is also plausible that one RNF20 allele may be silenced asymmetrically (7) through allele-specific methylation in STIC, resulting in lower expression of RNF20 and H2Bub1, while the transcribed allele is subject to heterozygous loss during tumor progression. Either scenario with a genetically intact but epigenetically repressed RNF20 allele in late-stage HGSOC would present a potential therapeutic opportunity. It is important to note that our purpose for using RNF20 silencing was not to study RNF20 per se, but rather as a tool to deplete H2Bub1. We believe that much of the loss of H2Bub1 may indeed be accounted for by elevated activity of one or more of the many deubiquitinating enzymes (DUBs) that have been reported to deubiquitinate H2B (15).
We showed that stable depletion of RNF20 and the consequent loss of H2Bub1 promotes clonogenic growth and motility in immortalized FTSECs with little to no effect on cell proliferation. The observed effect on clonogenic growth is in agreement with a previous report by Prenzel and colleagues (35), who reported increased clonogenic growth in the luminal breast cancer cell line MCF7. Likewise, the increase in cell motility following RNF20 depletion reported here is in line with the analogous effect observed in mammary MCF10A, which, similar to FT190 and FT194, is also a non-tumorigenic epithelial cell line. The effect of RNF20 knockdown on cell proliferation was previously evaluated in several mouse cell lines where RNF20 was stably depleted by shRNA (33). In those studies, RNF20 depletion led to impaired cell growth and increased apoptosis. It is possible that any anti-proliferative or pro-apoptotic tendencies resulting from RNF20 knockdown may have been offset in our FT cell lines by the proliferative and anti-apoptotic effects of the SV40 TAg (27). Additionally, we showed that knockdown of RNF20 and H2Bub1 enhances migration in HGSOC cell lines that retained H2Bub1 mark. Together, our observations show that loss of RNF20 and H2Bub1 promotes clonogenic growth and enhances migration ability of FT cells.
It is plausible that RNF20/H2Bub1 depletion drives oncogenic phenotypes through changes in chromatin state and alterations in signaling pathways. Previous studies have reported conflicting roles for H2Bub1. For example, some studies suggest that H2Bub1 regulates chromatin dynamics by enhancing nucleosome stability and a more closed conformation (29), while other studies suggest that H2Bub1 disrupts local and higher-order compaction leading to a more open conformation (30). Our ATAC-seq data shows that the loss of RNF20 and H2Bub1 leads to an overall more open chromatin conformation at promoter and DNase I hypersensitive regions in FT190 and FT194 cells. Interestingly, analysis of our RNA-seq and ATAC-seq data identified enrichment in key immune signaling pathways in RNF20-depleted cells. The enrichment of immune/inflammatory signaling pathways, due to the loss of RNF20 and H2Bub1, observed in our study is consistent with mouse studies showing that RNF20 heterozygosity leads to chronic colitis and inflammation-associated colorectal cancer in mice (31).
Our data shows that the levels of IL6 protein were significantly upregulated, along with other cytokines, in RNF20-depleted FT190 and FT194 cells. The observed increase in the levels of IL6 in RNF20-depleted cells is in line with a previous study in colorectal cancer which reported elevated transcript levels of IL6 and other inflammatory cytokines upon downregulation of RNF20 and H2Bub1 (31). In addition, we showed that exogenously added IL6 is sufficient to enhance the migration in FT190 and FT194 cells. This finding led us to ask whether the conditioned media from RNF20-depleted cells would be enriched in inflammatory cytokines, including IL6, which could induce oncogenic phenotypes in FT190 and FT194 cells. Indeed, when we transferred the cytokine enriched conditioned media from RNF20-depleted cells to FT190 and FT194 control cells it significantly enhanced their migration. This suggests that IL6 secretion is enhanced in RNF20-depleted cells which could stimulate autocrine and/or paracrine signaling to modulate the tumor microenvironment and promoting tumor progression. In line with our data, previous studies have shown that high IL6 in HGSOC is correlated with therapy resistance and poor prognosis (36,37). Mechanistically, it is likely that IL6 is operating through the Janus Activated Kinase/Signal Transducer and Activator of Transcription (JAK/STAT) pathway in FT cells, but further studies are needed to define the precise intracellular response to IL6 in FT cells (38). Moreover, these studies highlight the ability of epigenetic changes to influence the tumor microenvironment through immune autocrine and paracrine changes.
Targeting the epigenome, in particular the epigenetic changes involving histone modifications and the expression of histone modifying enzymes, is highly relevant in HGSOC. This strategy, through its global reach over the entire genome, has the potential to overcome limitations created by focusing on selective reactivation or suppression of single genes (39). RNF20 expression was previously shown to be suppressed by promoter hypermethylation, making its reactivation and therefore restoration of H2Bub1 possible through the use of DNA methylation inhibitors (40). The potent oncogenic effect of RNF20/H2Bub1 loss and the potential amenability of this axis to pharmacologic reactivation make it a particularly attractive therapeutic target that could be exploited in HGSOC.
Supplementary Material
Statement of Significance:
Loss of RNF20 and H2Bub1 contributes to transformation of the fallopian tube epithelium and plays a role in the initiation and progression of high-grade serous ovarian cancer.
ACKNOWLEDGMENTS
This work was supported by the National Cancer Institute P50-CA083636 (R. Drapkin), NIH U01 CA152990 (R. Drapkin); the Honorable Tina Brozman `Tina’s Wish’ Foundation (R. Drapkin), the Dr. Miriam and Sheldon G. Adelson Medical Research Foundation (R. Drapkin, M. Oren, D. Hoon), the European Research Council grant 293438 (RUBICAN) (M. Oren), the Basser Center for BRCA (R. Drapkin), the Department of Obstetrics and Gynecology at the University of Pennsylvania Perelman School of Medicine (R. Drapkin), the American Cancer Society Early Detection Professorship (SIOP-06–258-01-COUN) (B. Karlan), the Foundation for Women’s Wellness (R. Drapkin), the Claneil Foundation (R. Drapkin), the Run & Walk for Family & Friends with Cancer Foundation (R. Drapkin), and the Ann and Sol Schreiber Mentored Investigator Award from the Ovarian Cancer Research Alliance (J. Hooda). The authors thank members of the Drapkin lab for fruitful discussions. We thank Mei Zhang for the immunohistochemistry and Michael Cooper for the graphic abstract. We dedicate this work to the memory of Dr. Ellen Fitzgibbon, an ardent supporter of the Penn Ovarian Cancer Research Center and an inspiration to all that knew her.
Footnotes
Conflict of interest statement: RD serves on the scientific advisory board of Repare Therapeutics and Siamab Therapeutics and advises to Mersana Therapeutics and nVision Medical. The authors declare no other conflicts of interest.
REFERENCES
- 1.Torre LA, Trabert B, DeSantis CE, Miller KD, Samimi G, Runowicz CD, et al. Ovarian cancer statistics, 2018. CA: a cancer journal for clinicians 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bowtell DD, Bohm S, Ahmed AA, Aspuria PJ, Bast RC Jr., Beral V, et al. Rethinking ovarian cancer II: reducing mortality from high-grade serous ovarian cancer. Nature reviews Cancer 2015;15:668–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ducie J, Dao F, Considine M, Olvera N, Shaw PA, Kurman RJ, et al. Molecular analysis of high-grade serous ovarian carcinoma with and without associated serous tubal intra-epithelial carcinoma. Nat Commun 2017;8:990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Labidi-Galy SI, Papp E, Hallberg D, Niknafs N, Adleff V, Noe M, et al. High grade serous ovarian carcinomas originate in the fallopian tube. Nat Commun 2017;8:1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eckert MA, Pan S, Hernandez KM, Loth RM, Andrade J, Volchenboum SL, et al. Genomics of Ovarian Cancer Progression Reveals Diverse Metastatic Trajectories Including Intraepithelial Metastasis to the Fallopian Tube. Cancer Discov 2016;6:1342–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Piek JM, van Diest PJ, Zweemer RP, Jansen JW, Poort-Keesom RJ, Menko FH, et al. Dysplastic changes in prophylactically removed Fallopian tubes of women predisposed to developing ovarian cancer. The Journal of pathology 2001;195:451–6 [DOI] [PubMed] [Google Scholar]
- 7.Kindelberger DW, Lee Y, Miron A, Hirsch MS, Feltmate C, Medeiros F, et al. Intraepithelial carcinoma of the fimbria and pelvic serous carcinoma: Evidence for a causal relationship. The American journal of surgical pathology 2007;31:161–9 [DOI] [PubMed] [Google Scholar]
- 8.Kuhn E, Kurman RJ, Vang R, Sehdev AS, Han G, Soslow R, et al. TP53 mutations in serous tubal intraepithelial carcinoma and concurrent pelvic high-grade serous carcinoma--evidence supporting the clonal relationship of the two lesions. The Journal of pathology 2012;226:421–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McDaniel AS, Stall JN, Hovelson DH, Cani AK, Liu CJ, Tomlins SA, et al. Next-Generation Sequencing of Tubal Intraepithelial Carcinomas. JAMA oncology 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Murphy SK. Targeting the epigenome in ovarian cancer. Future oncology 2012;8:151–64 [DOI] [PubMed] [Google Scholar]
- 11.Fu S, Hu W, Iyer R, Kavanagh JJ, Coleman RL, Levenback CF, et al. Phase 1b-2a study to reverse platinum resistance through use of a hypomethylating agent, azacitidine, in patients with platinum-resistant or platinum-refractory epithelial ovarian cancer. Cancer 2011;117:1661–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matei D, Ghamande S, Roman L, Alvarez Secord A, Nemunaitis J, Markham MJ, et al. A Phase I Clinical Trial of Guadecitabine and Carboplatin in Platinum-Resistant, Recurrent Ovarian Cancer: Clinical, Pharmacokinetic, and Pharmacodynamic Analyses. Clin Cancer Res 2018;24:2285–93 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pulliam N, Fang F, Ozes AR, Tang J, Adewuyi A, Keer H, et al. An Effective Epigenetic-PARP Inhibitor Combination Therapy for Breast and Ovarian Cancers Independent of BRCA Mutations. Clin Cancer Res 2018;24:3163–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Johnsen SA. The enigmatic role of H2Bub1 in cancer. FEBS letters 2012;586:1592–601 [DOI] [PubMed] [Google Scholar]
- 15.Fuchs G, Oren M. Writing and reading H2B monoubiquitylation. Biochimica et biophysica acta 2014;1839:694–701 [DOI] [PubMed] [Google Scholar]
- 16.Kim J, Hake SB, Roeder RG. The human homolog of yeast BRE1 functions as a transcriptional coactivator through direct activator interactions. Molecular cell 2005;20:759–70 [DOI] [PubMed] [Google Scholar]
- 17.Kim J, Guermah M, McGinty RK, Lee JS, Tang Z, Milne TA, et al. RAD6-Mediated transcription-coupled H2B ubiquitylation directly stimulates H3K4 methylation in human cells. Cell 2009;137:459–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shema E, Tirosh I, Aylon Y, Huang J, Ye C, Moskovits N, et al. The histone H2B-specific ubiquitin ligase RNF20/hBRE1 acts as a putative tumor suppressor through selective regulation of gene expression. Genes & development 2008;22:2664–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Urasaki Y, Heath L, Xu CW. Coupling of glucose deprivation with impaired histone H2B monoubiquitination in tumors. PloS one 2012;7:e36775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shiloh Y, Shema E, Moyal L, Oren M. RNF20-RNF40: A ubiquitin-driven link between gene expression and the DNA damage response. FEBS letters 2011;585:2795–802 [DOI] [PubMed] [Google Scholar]
- 21.Perets R, Wyant GA, Muto KW, Bijron JG, Poole BB, Chin KT, et al. Transformation of the fallopian tube secretory epithelium leads to high-grade serous ovarian cancer in Brca;Tp53;Pten models. Cancer cell 2013;24:751–65 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Karst AM, Drapkin R. Primary culture and immortalization of human fallopian tube secretory epithelial cells. Nature protocols 2012;7:1755–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bustos MA, Salomon MP, Nelson N, Hsu SC, DiNome ML, Hoon DS, et al. Genome-wide chromatin accessibility, DNA methylation and gene expression analysis of histone deacetylase inhibition in triple-negative breast cancer. Genom Data 2017;12:14–6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Integrated genomic analyses of ovarian carcinoma. Nature 2011;474:609–15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO, et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Science signaling 2013;6:pl1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mermel CH, Schumacher SE, Hill B, Meyerson ML, Beroukhim R, Getz G. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome biology 2011;12:R41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Karst AM, Levanon K, Drapkin R. Modeling high-grade serous ovarian carcinogenesis from the fallopian tube. Proceedings of the National Academy of Sciences of the United States of America 2011;108:7547–52 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boichuk S, Hu L, Hein J, Gjoerup OV. Multiple DNA damage signaling and repair pathways deregulated by simian virus 40 large T antigen. Journal of virology 2010;84:8007–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chandrasekharan MB, Huang F, Sun ZW. Ubiquitination of histone H2B regulates chromatin dynamics by enhancing nucleosome stability. Proceedings of the National Academy of Sciences of the United States of America 2009;106:16686–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fierz B, Chatterjee C, McGinty RK, Bar-Dagan M, Raleigh DP, Muir TW. Histone H2B ubiquitylation disrupts local and higher-order chromatin compaction. Nat Chem Biol 2011;7:113–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tarcic O, Pateras IS, Cooks T, Shema E, Kanterman J, Ashkenazi H, et al. RNF20 Links Histone H2B Ubiquitylation with Inflammation and Inflammation-Associated Cancer. Cell reports 2016;14:1462–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Varambally S, Yu J, Laxman B, Rhodes DR, Mehra R, Tomlins SA, et al. Integrative genomic and proteomic analysis of prostate cancer reveals signatures of metastatic progression. Cancer cell 2005;8:393–406 [DOI] [PubMed] [Google Scholar]
- 33.Chernikova SB, Razorenova OV, Higgins JP, Sishc BJ, Nicolau M, Dorth JA, et al. Deficiency in mammalian histone H2B ubiquitin ligase Bre1 (Rnf20/Rnf40) leads to replication stress and chromosomal instability. Cancer research 2012;72:2111–9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dickson KA, Cole AJ, Gill AJ, Clarkson A, Gard GB, Chou A, et al. The RING finger domain E3 ubiquitin ligases BRCA1 and the RNF20/RNF40 complex in global loss of the chromatin mark histone H2B monoubiquitination (H2Bub1) in cell line models and primary high-grade serous ovarian cancer. Hum Mol Genet 2016;25:5460–71 [DOI] [PubMed] [Google Scholar]
- 35.Prenzel T, Begus-Nahrmann Y, Kramer F, Hennion M, Hsu C, Gorsler T, et al. Estrogen-dependent gene transcription in human breast cancer cells relies upon proteasome-dependent monoubiquitination of histone H2B. Cancer research 2011;71:5739–53 [DOI] [PubMed] [Google Scholar]
- 36.Sanguinete MMM, Oliveira PH, Martins-Filho A, Micheli DC, Tavares-Murta BM, Murta EFC, et al. Serum IL-6 and IL-8 Correlate with Prognostic Factors in Ovarian Cancer. Immunol Invest 2017;46:677–88 [DOI] [PubMed] [Google Scholar]
- 37.Worzfeld T, Pogge von Strandmann E, Huber M, Adhikary T, Wagner U, Reinartz S, et al. The Unique Molecular and Cellular Microenvironment of Ovarian Cancer. Frontiers in oncology 2017;7:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roxburgh CS, McMillan DC. Therapeutics targeting innate immune/inflammatory responses through the interleukin-6/JAK/STAT signal transduction pathway in patients with cancer. Transl Res 2016;167:61–6 [DOI] [PubMed] [Google Scholar]
- 39.Marsh DJ, Shah JS, Cole AJ. Histones and their modifications in ovarian cancer - drivers of disease and therapeutic targets. Frontiers in oncology 2014;4:144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wang Y, Cardenas H, Fang F, Condello S, Taverna P, Segar M, et al. Epigenetic targeting of ovarian cancer stem cells. Cancer research 2014;74:4922–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.