Abstract
Purpose:
The lack of a timely and reliable measure of response to cancer immunotherapy has confounded understanding of mechanisms of resistance and subsequent therapeutic advancement. We hypothesized that PET imaging of granzyme B using a targeted peptide, GZP, could be utilized for early response assessment across many checkpoint inhibitor combinations, and that GZP uptake could be compared between therapeutic regimens and dosing schedules as an early biomarker of relative efficacy.
Experimental Design:
Two models, MC38 and CT26, were treated with a series of checkpoint inhibitors. GZP PET imaging was performed to assess tumoral GZP uptake, and tumor volume changes were subsequently monitored to determine response. The average GZP PET uptake and response of each treatment group were correlated to evaluate the utility of GZP PET for comparing therapeutic efficacy.
Results:
In both tumor models, GZP PET imaging was highly accurate for predicting response, with 93% sensitivity and 94% negative predictive value. Mean tumoral GZP signal intensity of treatment groups linearly correlated with percent response across all therapies and schedules. Moreover, GZP PET correctly predicted that sequential dose scheduling of PD-1 and CTLA-4 targeted therapies demonstrates comparative efficacy to concurrent administration.
Conclusion:
Granzyme B quantification is a highly sensitive and specific early measure of therapeutic efficacy for checkpoint inhibitor regimens. This work provides evidence that GZP PET imaging may be useful for rapid assessment of therapeutic efficacy in the context of clinical trials for both novel drugs as well as dosing regimens.
Keywords: Granzyme B, PET, Cancer Immunotherapy, Predictive Biomarker
Introduction
The success of approved immune checkpoint inhibitors targeting PD-1, PD-L1, and CTLA-4 has generated significant interest in the application of these and other therapies across malignancies (1–3). Although many challenges exist in the rapidly progressing field of immuno-oncology, two major concerns include improving the ability to monitor response to both approved and clinically trialed drugs and reducing potentially severe side effects while maintaining maximum anti-tumor efficacy (4,5). Currently, response is monitored by anatomic measurements of tumor volume and overall survival, the latter of which may take years to resolve (4). Adding to the complication of long delays between therapy initiation and outcome is the frequency of severe immune-related adverse events, especially in therapy combinations such as anti-CTLA-4 and anti-PD-1 (5,6). Several pre-clinical and clinical studies have demonstrated that concurrent administration of combination therapies may not be necessary, and that the timing of when each checkpoint inhibitor is delivered may reduce side effects while maintaining efficacy (7).
Although a substantial amount of evidence surrounds the efficacy of PD-1 and CTLA-4 blockade, not all combinations of hypothesized checkpoints under investigation are strongly supported by biological rationale. One such checkpoint of interest, TIM-3, is upregulated in murine and human cancers following treatment with PD-1 checkpoint inhibitors, providing a rational target for combination studies (8). However, the role of TIM-3 in oncologic immunotherapy response has yet to be completely elucidated, as evidence supports its role in both immunogenic and tolerogenic pathways (9,10). The ability to non-invasively compare the effects of therapies targeting both well-understood checkpoint inhibitors, like PD-1 and CTLA-4, and those without consensus, such as TIM-3, could accelerate the clinical development of novel therapies.
Prior to therapy, certain characteristics, such as T cell infiltration, PD-L1 expression, microsatellite instability and mutational burden have been demonstrated to enrich response (11–13). While these are important population-based characteristics, they are not effective for predicting individual tumoricidal immune activity. Recently, we developed a PET imaging agent targeting a marker of effector T cell activation, granzyme B, that permits direct quantification of the anti-tumor response before tumor volume changes (14). Extracellular granzyme B has a biological half-life of 14 days, representing a stable target of immune activation. Using a highly specific peptide PET imaging agent for granzyme B (GZP), we found that GZP PET signal was predictive of eventual response to immunotherapy, with high signal tumors responding to therapy, and low-signal tumors progressing. By designing the GZP imaging agent to only bind to the active, secreted form of granzyme B, we have developed a system in which PET imaging can quantitatively integrate pro- and anti-tumor immune signaling, as well as discriminate between active and exhausted T cells to provide insight beyond T cell infiltration (15,16). Given the initial success of this agent, a more comprehensive analysis of the potential clinical applications of GZP, including as a broad predictive agent and clinical trial assessment tool was warranted. We hypothesized that GZP PET imaging would both be predictive of response to additional immunotherapy regimens across multiple tumor types for an individual tumor and provide a quantitative biomarker that would allow for early comparison of therapeutic regimens efficacy. We hypothesized that it could also be used as an efficacy monitoring tool in an adaptive therapy approach of dose scheduling, permitting a precision approach to immunotherapy.
Materials and Methods
Materials and Cell Lines
Unless otherwise stated, all chemicals were purchased from Sigma-Aldrich (St. Louis, MO). CT26 murine colon carcinoma cells were obtained from ATCC (Manassas, VA) and cultured in Roswell Park Memorial Institute medium with 10% fetal bovine serum (FBS). MC38 murine colon carcinoma cells were obtained from Kerafast (Boston, MA) and cultured in Dulbecco’s modified Eagle medium with 10% FBS. Monthly mycoplasma testing was performed by PCR screening and cells were discarded after 15 passages. All cell-based experiments were done with cells acquired within 6 months in order to ensure fidelity of the cell line identity.
Murine Models
Mice were housed and maintained by the Center for Comparative Medicine following animal protocols approved by the Massachusetts General Hospital Institutional Animal Care and Use Committee. Approximately 1 × 106 cells per xenograft of CT26 (n=5–12) and MC38 (n=5–8) were diluted 1:1 (v:v) in Matrigel (Corning, Tewksbury, MA) and injected into the upper right flank of balb/c or C57BL/6 mice (Charles River Laboratories, Wilmington, MA) respectively. CT26 and MC38 tumors were chosen based on their demonstrated responsiveness to checkpoint inhibitor therapy and whole-exome sequencing confirmed differences in mutation burden. (14,17–19). Anti-mouse PD-1 (clone RPM1–14), anti-mouse CTLA-4 (clone 9H10), and anti-mouse TIM-3 (clone RMT3–23) therapies were obtained from Bio X Cell (West Lebanon, NH). Mice were treated on days 3, 6, and 9 following tumor inoculation by intraperitoneal injection with saline (vehicle), 200 μg anti-PD-l (monotherapy), 200 μg anti-PD-1 and 100 μg anti-CTLA-4 (P+C combination therapy), or 200 μg anti-PD-1 and 250 μg anti-TIM-3 (P+T combination therapy). These doses were chosen based on previous studies that demonstrated efficacy at ranges that approximate the 3–10 mg/kg dosing used for human clinical studies (18,20,21). In subsequent studies, scheduling of immunotherapy doses were modified to 100 μg anti-CTLA-4 on day 3 then 200 μg anti-PD-1 on days 6 and 9 (C+P schedule therapy) or 200 μg anti-PD-1 and 100 μg anti-CTLA-4 on day 3 and PD-1 maintenance on days 6 and 9 (PC+P schedule therapy). A schematic of all treatments can be found in Supplemental Figure 1. Mice were either imaged or sacrificed for ex vivo analysis on day 12 post-tumor inoculation. For survival imaging experiments, tumor dimensions were measured with calipers every 2–3 days beginning on day 5 for MC38 and day 10 for CT26 and ending on day 20. Mice were sacrificed once tumors exceeded a volume of 500 mm3 or developed ulceration.
Biochemical Analysis of Tumors
Tumors were excised from sacrificed mice for ex vivo analysis on day 12 post-tumor inoculation. Tumors designated for Western blot analysis were lysed in 1% SDS solution supplemented with protease inhibitors prior to analysis by Western blotting. Anti-granzyme B (4275S, Cell Signaling Technologies, Danvers, MA), anti-CTLA-4 (ab134090, Abcam, Cambridge, MA), anti-PD-1 (12A7D7, ThermoFisher, Waltham, MA), anti-TIM-3 (ab185703, Abcam), anti-CD4 (sc19643, Santa Cruz Biotechnology, Dallas TX) anti-CD3 (sc-20047, Santa Cruz), anti-CD8 (53–6.7, ThermoFisher) and anti-β-actin (4970S, Cell Signaling Technologies) antibodies were used and subsequently detected by horseradish peroxidase-conjugated goat anti-rabbit polyclonal antibody (ab6721, Abcam) as per manufacturers’ recommendations. Bands were detected using SignalFire™ ECL Reagent (Cell Signaling Technologies) and imaged on an iBright FL1000 Imaging System (ThermoFisher). Images were quantitatively analyzed by ratio of net relative optical intensity of the target of interest to that of β-actin for each sample.
For immunohistochemical staining of patient samples, all specimens were acquired from patients following informed consent and according to a Massachusetts General Hospital institutional review board-approved clinical protocol. Samples were obtained from patients undergoing pembrolizumab treatment between 21–42 days in order to analyze samples for potential markers of response. Immunofluorescent staining was performed on formalin-fixed paraffin embedded sections following antigen retrieval in EDTA buffer using standard heat-based antigen retrieval techniques. Samples were blocked with goat serum before detecting granzyme B expression with an anti-granzyme B antibody (ab5049, Abcam, Cambridge, MA). Bound antibody was detected by AlexaFluor488 goat anti-rabbit (Life Technologies, Carlsbad, CA) for immunofluorescent visualization. Fluorescent microscopy was performed on a BioTek Cytation5 inverted microscope using Gen5 software (BioTek, Winooski, VT).
Flow cytometric analysis was performed on P+C treated and vehicle treated mice by excising murine tumors on day 12 following tumor inoculation and generating a single cell suspension by incubating in RPMI (ThermoFisher) supplemented with 1 mg/ml collagenase type IV for 35 min at 37°C. Cells were filtered into a single cell suspension through a 70 micron cell filter (Greiner Bio-One, Monroe, NC) and stained for live cells by Zombie Violet Fixable Viability Kit (Biolegend, San Diego, CA), and murine anti-CD45 (clone 104; Biolegend), anti-CD8 (clone 53–6.7; Biolegend), anti-PD-1 (clone RMP1–30; Biolegend), anti-EOMES (ab2574227, ThermoFisher), and anti-granzyme B (Grb17, ThermoFisher) according to the manufacturer’s specifications. Flow cytometry was performed on a cytometry cell analyzer BD LSRFortessa X20 (BD Biosciences, San Jose, CA) and gating established using FlowJo software (FlowJo LLC, Ashland, OR).
Peptide Synthesis and Radiolabeling
NOTA–β-Ala–Gly–Gly--Gly–Ile–Glu–Phe–Asp–CHO (NOTA-GZP) was synthesized using standard FMOC chemistry and purity was measured by mass spectroscopy and HPLC (20). 68Ga was eluted from a 68Ge/68Ga generator (Radio Medix, Houston, TX) with 0.1M HCl. The eluent was raised to pH 3.5–4.0 with 2M HEPES before addition of 100 μg of NOTA-GZP. After allowing the labeling reaction to proceed for 10 minutes at room temperature, the peptide was purified on a reverse-phase C18 Sep-Pak mini cartridge (Waters, Milford, MA) and eluted in 200 μL 70% ethanol. Saline was added to create a final formulation of <10% ethanol concentration and approximately 9.5–28 MBq per mouse.
Murine PET Imaging
On day 12 post-inoculation, mice were intravenously injected via tail vein with 68Ga-NOTA-GZP and subsequently imaged after one hour. All imaging was performed on a rodent Triumph PET/CT (GE Healthcare, Wilmington, MA). PET images were acquired for 15 min and followed by CT acquisition. Reconstruction was achieved using 3D-MLEM (4 iterations, 20 subsets) and corrected for scatter and randoms. VivoQuant software (InviCRO, Boston, MA) was used for image processing and analysis. Individual tumors were identified manually by drawing a 3D region of interest using CT-anatomic correlation. Background blood pool radioactivity was measured by identifying the left ventricle of the heart as a region of interest. Specific 68Ga NOTA-GZP uptake was quantified by dividing total tumor uptake by background blood pool to derive a tumor to blood ratio (TBR).
Statistical Analysis
GraphPad Prism Version 7 software (GraphPad Software, Inc., La Jolla, CA) was used for statistical analysis and graphing. Unpaired t tests with Welch’s correction were used for all comparisons between immunotherapy-treated and vehicle-treated tumors. A first-order regression weighted by group size was used to correlate percent response with average tumor to blood ratio TBR for each treatment arm. Statistical inference based on Pearson’s correlation coefficient was used to define 95% confidence bands of the best-fit line. Comparison of therapy schedules was performed using one-way analysis of variance with a Kruskal-Wallis test for multiple comparisons. Survival benefit was calculated by log-rank test for trend.
Results
Western Blot Analysis of Relevant Checkpoint Inhibitor Targets
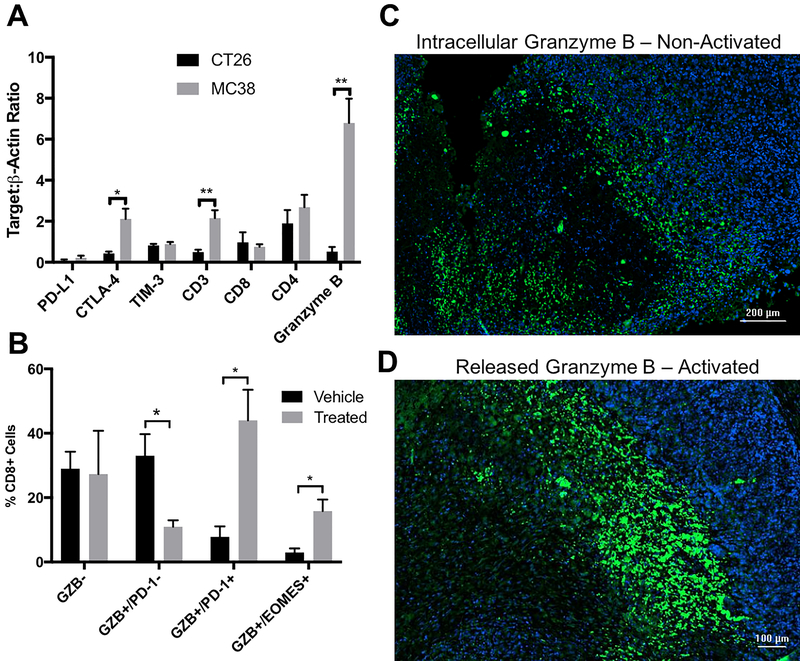
MC38 and CT26 syngeneic tumors treated with concurrent combination anti-PD-1 and CTLA-4 therapy were excised and evaluated by immunoblot on day 12 after three combination doses to determine post-treatment markers that may differentiate response rate prior to therapy. Expression after one cycle of treatment of the checkpoint inhibitor target molecules, PD-1, CTLA-4 and TIM-3, immune cell markers CD3, CD8, CD4 and granzyme B were analyzed (Fig 1A, Supp. Fig. 2). Significant differences were found to exist between moderate and highly responsive tumors for CTLA-4 (MC38 CTLA-4:β-actin= 2.113 versus CT26 0.43, p=0.02) as well as CD3 (MC38 CD3:β-actin= 2.153 versus CT26 0.50, p=0.006). Granzyme B was also evaluated by immunoblot on day 12 and expression was found to be significantly higher in treated MC38 tumors (granzyme B: β-actin = 6.79) than in treated CT26 tumors (0.52 P = 0.002). This 13-fold higher expression in highly responsive MC38 tumors post treatment was the largest magnitude of difference for any of the markers analyzed.
Figure 1-.
A) Western blot quantification of potential predictive proteins associated with anti-tumor response in CT26 (black) and MC38 (gray) tumors treated with anti-PD-1 and anti-CTLA-4 combination therapy. Bars represent the mean of 4 tumors, error bars denote standard error measurement (SEM). B) Flow cytometry quantification of CT26 tumors treated with anti-PD-1 and anti-CTLA-4 combination therapy for granzyme B (GZB), PD-1 and EOMES in CD45+ T cells from vehicle (black) and combination therapy. Bars represent the mean of 4 tumor replicates, with error denoted as SEM C) Tumor specimens from melanoma patients treated with anti-PD-1 therapy stained for nuclei with DAPI (blue) and anti-granzyme B (green). A more diffuse pattern is seen in D) consistent with activated T cells * - P < 0.05, ** - P < 0.01
Despite the significant differences obtained by Western blot, as a destructive technique, it was not possible to determine whether the detected granzyme B was from active, inactive, or exhausted T cells. PD-1 and eomesodermin (EOMES) were chosen as markers of chronic activation and exhaustion, and granzyme B positive T cells found in the tumors were assessed by flow cytometry to determine relative expression levels of these two proteins (Fig 1B). Analysis in anti-PD-1 plus anti-CTLA-4 combination treated and vehicle treated CT26 tumors on day 12 demonstrated that although there were roughly similar levels of granzyme B negative T cells in both treatment groups, the percentage of granzyme B+/PD-1 negative T cells was much higher in vehicle treated tumors, whereas a significantly higher number of granzyme B+/PD-1+ cells were found in treated cells. A similar trend in granzyme B+/EOMES+ CD8+ T cells supports more T cell exhaustion, as previous studies have linked this phenotype to T cell exhaustion, although the active level of granzyme B release cannot be distinguished (22).
Immunofluorescent Microscopy of Human Melanoma
The inability to distinguish active granzyme B release using ex vivo techniques such as flow cytometry and Western blot represents a significant problem for using granzyme B as a protein biomarker, as only actively released granzyme B is capable of initiating apoptosis and generating an anti-tumor response. To demonstrate this concept in immunotherapy patients, immunoflourescent microscopy of human melanoma specimens post treatment of anti-PD-1 illustrates that although similar levels of granzyme B may be present in two separate tumors, the ability to quantify released granzyme B is critical to distinguishing patient non-responders (Figure 1C) from responders (Figure 1D). The biological relevance and magnitude of dynamic range between response and non-response of granzyme B, combined with the necessity to quantify only activated, released granzyme B makes PET imaging with the GZP peptide a logical choice for response monitoring.
Granzyme B PET Imaging
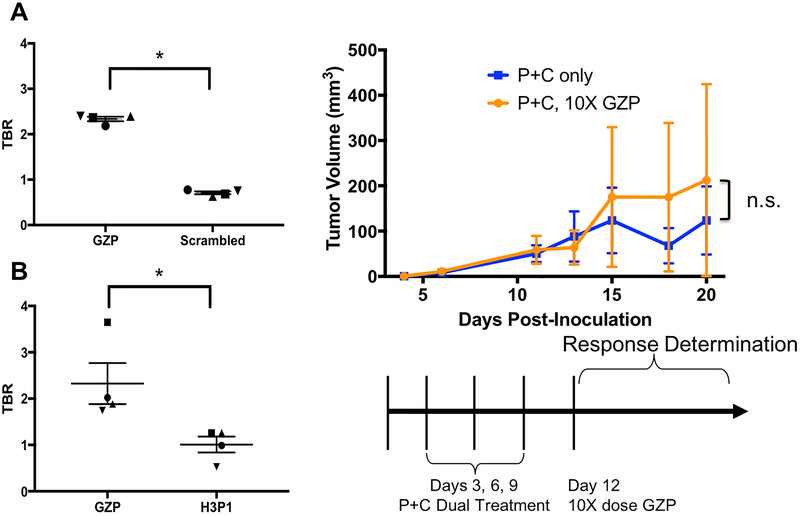
Previously, we demonstrated response prediction in a moderately responsive CT26 model of tumor immunotherapy, however baseline levels of granzyme B were low, which leads to minimal differences between stored granzyme B not contributing to the anti-tumor response and secreted granzyme B that is actively inducing tumoral killing. In order to examine whether the predictive capability of PET imaging could be expanded into a tumor model with higher levels of granzyme B at baseline, both CT26 and MC38 tumor-bearing mice were examined. Prior to the comparison of these models, biodistribution was performed in MC38 mice, which showed that the GZP peptide had similar non-target organ distribution and clearance routes, confirming that comparison across two different mouse strains was appropriate (Supp. Fig. 3). In order to confirm that accumulation of the peptide was due to granzyme B-targeted binding and not perfusional differences between responding and non-responding tumors, imaging was performed in combination anti-PD-1 plus anti-CTLA-4 treated mice with both GZP and one of two non-targeted peptides on the same day. Significantly higher peptide accumulation was observed in responding tumors for GZP as compared to either control peptide, confirming that perfusional differences were not the major driving factor for peptide accumulation (Figure 2A–B). Additionally, the peptide was tested at 10x times the imaging dose in a group of combination treated mice and compared to mice injected with a control peptide. No differences in either tumor growth rate or complete responses were seen between the groups, indicating that the fraction of granzyme B bound by the GZP peptide was not significant enough to alter anti-tumor response (Figure 2C).
Figure 2-.
Tumor to blood ratios of subsequently-responding mice bearing CT26 tumors treated with anti-PD-1 plus anti-CTLA-4 combination therapy and imaged using PET/CT with A) GZP followed 4 hours later by a scrambled peptide or B) GZP followed four hours later with a second peptide targeting a human protein. Background TBR = 1. * - P < 0.05 C) Schematic of treatment with anti-PD-1 and anti-CTLA-4 therapy followed by 10x mass dose injection of unlabeled GZP. Average tumor volume of 10x GZP-treated mice are shown by orange lines and circles, with uninjected tumors shown by blue lines and squares.
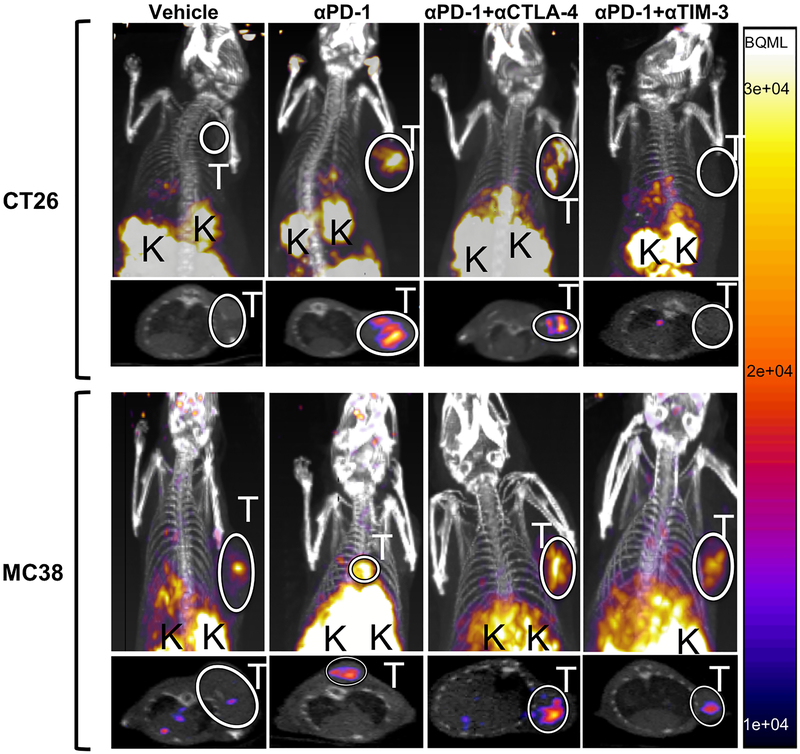
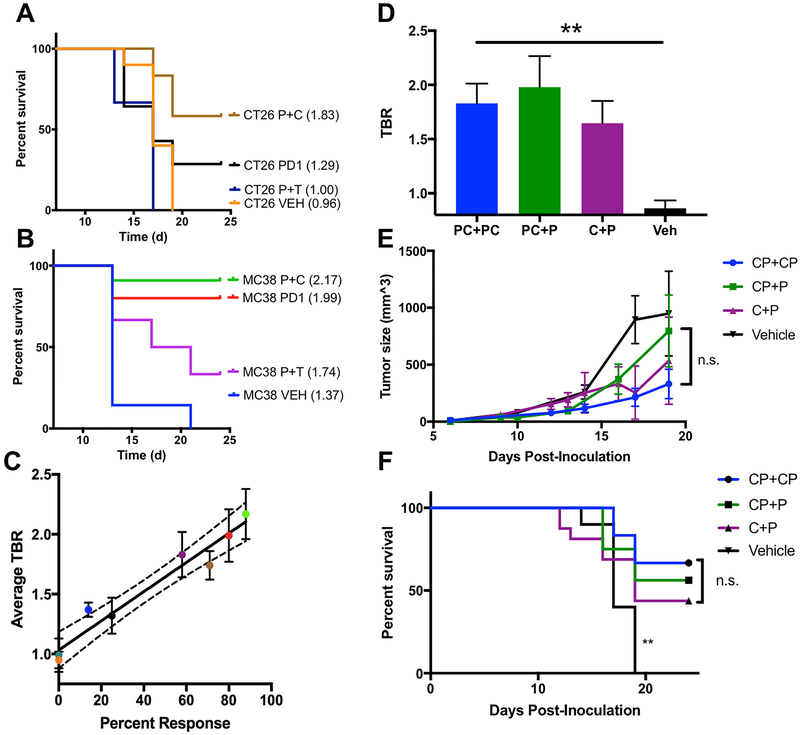
Following these observations, both MC38 and CT26 mice were treated with monotherapy, anti-PD-1+anti-CTLA-4 (P+C) combination therapy, anti-PD-1+anti-TIM-3 (P+T) combination therapy, or saline and imaged using 68Ga-NOTA-GZP on day 12 to non-invasively assess granzyme B levels, and subsequently compared to previous imaging of CT26 P+C (19). PET imaging revealed common organ uptake patterns in all mice analyzed, including kidney and bladder accumulation consistent with renal clearance characteristic of small peptides. Degree of tumor uptake, however, varied based on tumor model and therapy regimen (Fig 3). In the CT26 model, vehicle-treated mice had similar tracer accumulation in the tumor and in the left ventricle of the heart, which was used as a measure of background radioactivity, resulting in a baseline average tumor-to-background ratio (TBR) of 0.97±0.07. In comparison, CT26 tumor-bearing mice receiving PD-1 monotherapy and P+C combination therapy had significantly higher tumor uptake with mean TBRs of 1.32±0.15 (p<0.05) and 1.48±0.19 (p<0.005) respectively (Fig 4A). CT26 xenograft mice treated with P+T combination therapy demonstrated no elevated tumor-specific tracer accumulation in comparison to the vehicle-treated mice (TBR=0.99±0.14).
Figure 3-.
Representative PET/CT coronal and axial images taken from CT26 (top row) or MC38 (bottom row) tumor-bearing mice. Mice were treated with either vehicle, anti-PD-1, anti-PD-1 plus anti-CTLA-4 or anti-PD-1 plus anti-CTLA-4 therapy as denoted in the heading of each row. Tumors are outlined by a white circle and labeled with a white “T” and kidneys, the major route of imaging agent clearance are marked by a black “K”. All PET intensities are displayed in the same intensity, with the scale displayed on the right in Bq/mL.
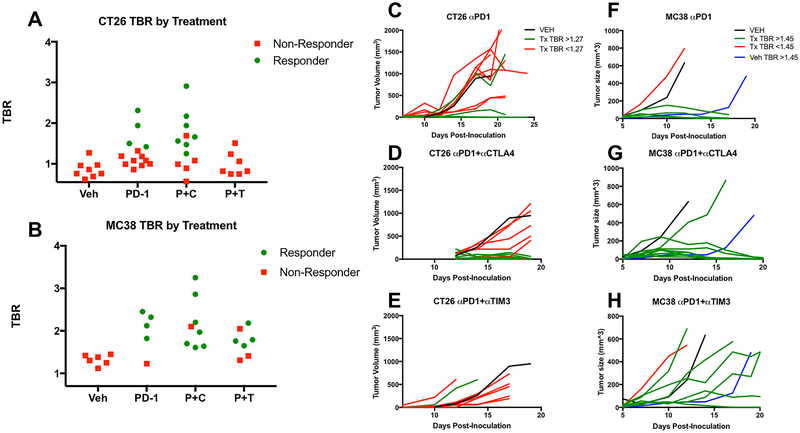
Figure 4-.
Individual tumor to blood (TBR) ratios for mice treated with vehicle (Veh, n=8 CT26, n=7 MC38) anti-PD-1 (PD-1 n=12 CT26, n=5 MC38), anti-PD-1 plus anti-CTLA-4 (P+C, n=12 CT26, n=7 MC38) or anti-PD-1 plus anti-TIM-3 (P+T n=7 CT26, n=7 MC38) therapies for A) CT26 and B) MC38 tumor bearing mice. Green circles denote mice with responses and pink boxes represent non-responsive tumors. Individual tumor growth curves for CT26 (C-E) and MC38 (F-H) tumors treated with the same therapy combinations are shown, with green lines corresponding to high GZP PET TBRs and pink lines to low GZP PET TBRs. The black lines are a representative mean of 8 vehicle mice and blue lines are representative of a single mouse with high GZP PET TBR that exhibited a partial response.
In order to experimentally define a TBR threshold to predict response across the three therapy regimens, the highest TBR for vehicle-treated CT26 tumors, 1.27, was defined as the value above which tumors would be classified as high-uptake, and below which they would be classified as low-uptake. Using this cutoff for the CT26 model, 5 of 12 monotherapy, 7 of 12 P+C combination therapy, and 1 of 6 P+T combination therapy-treated mice were classified as high-uptake.
MC38 tumor-bearing mice also demonstrated differential tumor uptake based on therapy regimen. Saline-treated MC38-bearing mice had an average TBR of 1.37±0.016, significantly higher than that of the saline-treated CT26 tumor-bearing mice (p<0.005) (Figure 4B) and indicative of a comparatively higher baseline granzyme B level in MC38 tumors consistent with Western blot results. As in the CT26 model, MC38 tumor-bearing mice treated with both monotherapy and P+C combination therapy had significantly higher TBRs of 1.99±0.22 (p<0.05) and 2.17±0.21 (p<0.005), respectively, compared to the relevant control mice. However, unlike in the CT26 model, MC38 tumors treated with P+T combination therapy also resulted in higher uptake than control tumors (TBR=1.74±0.12, p<0.05), although this group still had the lowest mean TBR among the three treatments. The threshold for high- and low- uptake for MC38 tumors was defined as 1.45 using the same methodology as for CT26 tumors, with an exception made for a single vehicle-treated tumor with high uptake that later demonstrated delayed growth in comparison to all other vehicle-treated tumors. The number of high-uptake MC38 tumors included 4 of 5 monotherapy, 8 of 8 P+C combination therapy, and 5 of 7 P+T combination therapy-treated mice.
Growth Curve Analysis
Tumor volumes were used as a surrogate response measure of overall survival. CT26 tumor growth was dichotomous, as tumors regressed completely by day 20, or continued to grow until developing ulcerations, reaching maximum volume, or the end of the study. Based on these growth patterns, mice were categorized as responders if the tumor volume decreased to 0 mm3 or non-responders if tumor volume continued to increase. Figure 4C–E illustrates the changes in CT26 tumor volume for each therapy regimen, with an average of vehicle-treated tumors represented in black. None of the vehicle-treated CT26 tumors regressed in size, but 3 of 12 monotherapy (Fig. 4C), 7 of 12 P+C combination therapy (Fig. 4D), and 0 of 6 P+T combination therapy treated mice (Fig 4E) responded to therapy.
In the MC38 tumor model, three groups were identified based on response patterns; complete responders, non-responders, and a third group classified as partial responders. Responding tumors regressed in size by day 20, non-responding tumors grew to 500 mm3 in size on or before day 14, and partially responding tumors reached 500 mm3 at a delayed time point later than day 14. Figure 4 F–H highlights these temporal patterns of tumor volume changes in MC38 by therapy regimen with vehicle-treated tumor growth curves in black. Of note, 1 of the 7 vehicle-treated MC38 tumors showed significantly delayed tumor growth compared to the other vehicle-treated tumors and was thereby classified as a partial responder (Figure 4F). This tumor also had a TBR greater than 1.45 and as such is shown in blue. A higher percentage of MC38 tumors responded to therapy compared to CT26 tumors, with 4 of 5 monotherapy treated and 7 of 8 P+C combination therapy treated tumors completely regressing in size. Unlike CT26, MC38 tumors responded to P+T combination therapy, with 1 of 7 tumors responding completely, and an additional 3 of 7 demonstrating delayed growth in response to therapy, characteristic of partial response.
Analysis of Response Prediction using GZP PET
To compare the predictive value of GZP PET across therapeutic regimens, we defined the threshold for discriminating high from low GZP uptake among treated tumors as the highest TBR seen among vehicle treated tumors, as described above, resulting in TBR thresholds of 1.27 for CT26 tumors and 1.45 for MC38 tumors. Using this classification system, 26 of 28 fully or partially responding mice were categorized as high-uptake by GZP PET imaging, whereas 33 of the 38 non-responding mice were classified as low-uptake (Table 1). This reflects an overall sensitivity and specificity of 93% and 87% respectively for the ability of 68Ga-GZP PET to predict immunotherapy response. A more comprehensive examination as to the reason for the discrepancy between PET and response for the discordant tumors was sought through analysis of the positive and negative predictive values of GZP PET. Most of the non-predictive scans were found in tumors that had high granzyme B PET signal but ultimately did not respond resulting in a positive predictive value of 84% (26/31) but a negative predictive value of 94% (33/35), indicating that low granzyme B PET signal was highly correlated with non-response.
Table 1-.
Summary of all tumor and treatment types analyzed by GZP PET. Data is presented for sensitivity, specificity, positive predictive value (PPV) and negative predictive value (NPV) of GZP PET for each individual tumor and treatment combination, with a global summary in the bottom row.
| Tumor | Therapy | Sensitivity | Specificity | PPV | NPV |
|---|---|---|---|---|---|
| CT26 | Vehiclea | N/A | 8/8 | N/A | 8/8 |
| CT26 | Mono | 4/4 | 7/8 | 4/5 | 7/7 |
| CT26 | PD-1+CTLA-4a | 7/8 | 4/4 | 7/7 | 4/5 |
| CT26 | PD-1+TIM-3 | N/A | 6/7 | 0/1 | 6/6 |
| MC38 | Vehicle | 1/1 | 6/6 | 1/1 | 6/6 |
| MC38 | Mono | 4/4 | 1/1 | 4/4 | 1/1 |
| MC38 | PD-1 +CTLA-4 | 7/7 | 0/1 | 7/8 | N/A |
| MC38 | PD-1+TIM-3 | 3/4 | 1/3 | 3/5 | 1/2 |
| Summary | 93% (26/28) |
87% (33/38) |
84% (26/31) |
94% (33/35) |
Next, the accuracy of granzyme B PET imaging was tested to determine if group-based prediction of efficacy was possible by compiling the mean GZP TBR within each treatment group and correlating with overall percent response. Treatments with the best response survival outcomes also had the highest mean GZP TBR based on a Kaplan Meier plot of survival for each therapy (Figure 5A–B). To determine whether the relationship between percent response and mean tumor GZP TBR was correlated, percent response was plotted against TBR (Fig. 5C). The curve revealed a linear relationship with R2=0.84 and a significantly non-zero slope (p<0.0005), demonstrating a direct relationship between GZP TBR for a treatment group and percent survival for a given treatment group.
Figure 5-.
A) Kaplan-Meier curves for each tumor and therapy combination with average TBR included in parentheses next to each label. B) Linear plot of overall response versus TBR for each tumor/treatment combination, where R2 = 0.84 (p < 0.0001) and dotted lines represent the 95% confidence interval. D) GZP PET tumor to blood ratios (TBR) of concurrent anti-PD-1 and anti-CTLA-4 therapy (PC+PC, blue, n=14), single concurrent anti-PD-1 and anti-CTLA-4 followed by single agent anti-PD-1 therapy (PC+P, green, n=8), single dose anti-CTLA-4 therapy followed by anti-PD-1 monotherapy (C+P, purple, n=14) and vehicle treatment (Veh, black, n=8) are denoted by bars representing the mean of 8–14 mice for each group. Error bars represent SEM E) Average tumor volumes of CP+CP (blue circle), CP+P (green square), C+P (purple triangle), and Vehicle (black triangle) treated tumors, with error bars representing SEM. F) Kaplan-Meier curve of the three treatment schedules and vehicle treated tumors. ** - P < 0.01
Combination Therapy Dose Schedule Assessment
Since GZP PET could successfully predict the efficacy of various combinations of therapy, we next sought to determine if it could also quantify the effectiveness of the order in which a single combination was delivered. The combination of anti-PD-1 and anti-CTLA-4 was given either repeatedly as was done with the previous experiments (PC+PC), as a single concurrent combination followed by single agent anti-PD-1 administration (PC+P), or as a single agent anti-CTLA-4 therapy given once followed by anti-PD-1 therapy (C+P). PET imaging of each of these groups revealed statistically similar TBRs (Figure 5D) (PC+PC = 1.83±0.18, PC+P = 1.98±0.29, C+P = 1.65±0.21). As predicted by GZP PET imaging, neither mean tumor volume (Figure 5E) nor overall survival (Figure 5F) were different between any of the treatment groups. The predictive nature of GZP PET imaging in assessing the timing of anti-PD-1 and anti-CTLA-4 therapy suggests it can also be used to determine when combinations should be given, in addition to which therapies are most effective.
Discussion
In this manuscript, we demonstrate GZP PET imaging to be a promising method for measuring active anti-tumor immune response and for comparing novel checkpoint inhibitors and combination regimens. By classifying tumors as responders based on GZP PET signal intensity, we establish that granzyme B PET imaging is highly sensitive and selective for response across a variety of tumor models and treatments. Even comparison of vehicle-only groups revealed that CT26 tumors had low baseline levels of granzyme B and moderate responsiveness to immunotherapy, whereas MC38 had high baseline levels of granzyme B and high responsiveness to immunotherapy. In fact, one MC38 vehicle-treated tumor that was identified as having levels of granzyme B consistent with response spontaneously regressed, even without therapy. It has been demonstrated that MC38 tumors have a DNA mismatch repair deficiency phenotype, which leads to increased tumor mutational burden and therefore increased probability of immune recognition and high baseline granzyme B as seen in this study (19). Our data support that a more inflamed tumor microenvironment induces high granzyme B secretion even at baseline, and GZB PET could serve as a potential pre-treatment biomarker of response for mismatch repair deficient tumors.
Across both the MC38 and CT26 treatment groups, GZP PET had 94% negative predictive value. The high negative predictive value indicates that the absence of granzyme B is a strong predictor of failure to activate an anti-tumor response. Conversely, high GZP PET may not always signify response, as immune cell activation may not be enough to clear a tumor in cases of rapid tumor growth or a strong tumor evasion. However, an 84% positive predictive value suggests that this outcome occurs in a minority of cases.
In addition to individual response, we also illustrate that subsequent survival for each therapy group correlates with the average PET uptake of that cohort, suggesting that GZP PET imaging could provide a method for early comparison of novel immunotherapy regimens. The most effective therapy of this study, anti-PD-1 and anti-CTLA-4 in combination, yielded higher mean GZP PET uptake in comparison to the other two regimens, and subsequently resulted in higher rates of response. An unexpected diminishment in therapeutic efficacy of PD-1 and TIM-3 combination therapy especially highlights the potential of granzyme B PET imaging to detect and predict combinations that may not be effective. In addition, despite significant differences in the immunogenicity of each of the cell lines monitored, a combined analysis of both tumor types revealed a non-zero slope and a linear correlation between granzyme B levels and percent response, which corroborates the hypothesis that granzyme B PET imaging can be used to determine therapeutic effectiveness on a broad scale.
GZP PET also permitted a rapid evaluation of precise dose timing of anti-PD-1 and anti-CTLA-4 therapy, with GZP PET accurately predicting that a single administration of anti-CTLA-4 followed by anti-PD-1 therapy was as effective as repeated concurrent administration of anti-PD-1 and anti-CTLA-4 therapy. This is concordant with other studies that demonstrate the timing of checkpoint inhibitor therapy is crucial (9,26). GZP PET imaging can be used to gather early evidence regarding proper timing of combination therapy delivery, providing a personalized approach to an adaptive immunotherapy regimen.
Because of the favorable pharmacokinetics and high degree of similarity between the murine and human versions of GZP, our imaging agent provides a readily available route of clinical translation. Previous analysis by our group suggests that differences in the granzyme B expression between responding and non-responding patients can be seen as early as 21 days post-therapy initiation (14). Taken together, these data justify planned clinical trials utilizing the human version of the peptide. This is an important next step, as GZP PET imaging has, to date, only been tested in subcutaneous murine models, which may have different mechanisms of response to immunotherapy. Additionally, the data should be expanded to ensure additional tumor models, immunotherapy regimens, image timing and dosing schedules in larger sample sizes and over longer time frames continue to follow the same linear relationship between GZP uptake and tumor responsiveness. GZP PET holds promise as a significant addition to the evolving immuno-oncology landscape, both for its ability to predict individual response to therapy and for its ability to evaluate novel therapies and combination regimens as they emerge.
Supplementary Material
Statement of Translational Relevance.
As checkpoint inhibitor therapy treatments continue to gain FDA approval across a number of cancers, previous challenges monitoring therapeutic efficacy become amplified. Previously, our group developed a clinically translatable PET imaging agent for granzyme B. Here, we demonstrate the predictive capabilities of granzyme B PET imaging across a range of tumors and treatments, revealing greater than 90% sensitivity and negative predictive value. Granzyme B PET imaging could also be used to compare therapies, with a correlation between mean PET signal and overall response. Finally, we demonstrate that the effectiveness of the order in which CTLA-4 and PD-1 blockade is administered can be ascertained by PET imaging. The data suggest granzyme B PET imaging could provide an early readout of immunotherapy efficacy with benefits to both individual patients and novel therapy evaluation.
Acknowledgements
The authors would like to thank Sengchan Khamhoung and Catharina Dekker for technical assistance in preparing the manuscript. This publication is based on research supported by the Melanoma Research Alliance, and NIH grants R01CA214744, K99CA215604, P50CA090381.
Financial Support: NIH R01CA214744, P50CA090381 and Melanoma Research Alliance to Umar Mahmood; K99CA215604 to Benjamin Larimer
Footnotes
Conflict of Interest: BL, EWK, UM are consultants and co-founders/shareholders of Cytosite Biopharma Inc.
References
- 1.Herbst RS, Baas P, Kim DW, Felip E, Perez-Gracia JL, Han JY, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet 2016;387(10027):1540–50. [DOI] [PubMed] [Google Scholar]
- 2.Hodi FS, O’Day SJ, McDermott DF, Weber RW, Sosman JA, Haanen JB, et al. Improved Survival with Ipilimumab in Patients with Metastatic Melanoma. New England Journal of Medicine 2010;363(8):711–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Massard C, Gordon MS, Sharma S, Rafii S, Wainberg ZA, Luke J, et al. Safety and Efficacy of Durvalumab (MEDI4736), an Anti–Programmed Cell Death Ligand-1 Immune Checkpoint Inhibitor, in Patients With Advanced Urothelial Bladder Cancer. Journal of Clinical Oncology 2016;34(26):3119–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wolchok JD, Hoos A, O’Day S, Weber JS, Hamid O, Lebbé C, et al. Guidelines for the evaluation of immune therapy activity in solid tumors: immune-related response criteria. Clinical Cancer Research 2009;15(23):7412–20. [DOI] [PubMed] [Google Scholar]
- 5.Marrone KA, Ying W, Naidoo J. Immune-Related Adverse Events From Immune Checkpoint Inhibitors. Clinical Pharmacology & Therapeutics 2016:n/a-n/a. [DOI] [PubMed] [Google Scholar]
- 6.Postow MA, Chesney J, Pavlick AC, Robert C, Grossmann K, McDermott D, et al. Nivolumab and ipilimumab versus ipilimumab in untreated melanoma. New England Journal of Medicine 2015;372(21):2006–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Messenheimer DJ, Jensen SM, Afentoulis ME, Wegmann KW, Feng Z, Friedman DJ, et al. Timing of PD-1 blockade is critical to effective combination immunotherapy with anti-OX40. Clinical Cancer Research 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Koyama S, Akbay EA, Li YY, Herter-Sprie GS, Buczkowski KA, Richards WG, et al. Adaptive resistance to therapeutic PD-1 blockade is associated with upregulation of alternative immune checkpoints. 2016;7:10501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Huang Y-H, Zhu C, Kondo Y, Anderson AC, Gandhi A, Russell A, et al. CEACAM1 regulates TIM–3–mediated tolerance and exhaustion. Nature 2015;517(7534):386–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gorman JV, Starbeck-Miller G, Pham N-LL, Traver GL, Rothman PB, Harty JT, et al. Tim-3 Directly Enhances CD8 T Cell Responses to Acute Listeria monocytogenes Infection. The Journal of Immunology 2014;192(7):3133–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Herbst RS, Soria J-C, Kowanetz M, Fine GD, Hamid O, Gordon MS, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature 2014;515(7528):563–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dudley JC, Lin M-T, Le DT, Eshleman JR. Microsatellite instability as a biomarker for PD-1 blockade. Clinical Cancer Research 2016;22(4):813–20. [DOI] [PubMed] [Google Scholar]
- 13.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, et al. Mutational landscape determines sensitivity to PD-1 blockade in non–small cell lung cancer. Science 2015;348(6230):124–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Larimer BM, Wehrenberg-Klee E, Dubois F, Mehta A, Kalomeris T, Flaherty K, et al. Granzyme B PET Imaging as a Predictive Biomarker of Immunotherapy Response. Cancer Research 2017;77(9):2318–27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Larimer BM, Wehrenberg-Klee E, Caraballo A, Mahmood U. Quantitative CD3 PET Imaging Predicts Tumor Growth Response to Anti-CTLA-4 Therapy. Journal of Nuclear Medicine 2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tavaré R, Escuin-Ordinas H, Mok S, McCracken MN, Zettlitz KA, Salazar FB, et al. An Effective Immuno-PET Imaging Method to Monitor CD8-Dependent Responses to Immunotherapy. Cancer Research 2016;76(1):73–82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Selby MJ, Engelhardt JJ, Johnston RJ, Lu L-S, Han M, Thudium K, et al. Preclinical Development of Ipilimumab and Nivolumab Combination Immunotherapy: Mouse Tumor Models, In Vitro Functional Studies, and Cynomolgus Macaque Toxicology. PLoS ONE 2016;11(9):e0161779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lau J, Cheung J, Navarro A, Lianoglou S, Haley B, Totpal K, et al. Tumour and host cell PD-L1 is required to mediate suppression of anti-tumour immunity in mice. Nature Communications 2017;8:14572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Efremova M, Rieder D, Klepsch V, Charoentong P, Finotello F, Hackl H, et al. Targeting immune checkpoints potentiates immunoediting and changes the dynamics of tumor evolution. Nature Communications 2018;9:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kang C-W, Dutta A, Chang L-Y, Mahalingam J, Lin Y-C, Chiang J-M, et al. Apoptosis of tumor infiltrating effector TIM-3+CD8+ T cells in colon cancer. Scientific Reports 2015;5:15659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Holmgaard RB, Zamarin D, Lesokhin A, Merghoub T, Wolchok JD. Targeting myeloid-derived suppressor cells with colony stimulating factor-1 receptor blockade can reverse immune resistance to immunotherapy in indoleamine 2,3-dioxygenase-expressing tumors. EBioMedicine 2016;6:50–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Buggert M, Tauriainen J, Yamamoto T, Frederiksen J, Ivarsson MA, Michaëlsson J, et al. T-bet and Eomes Are Differentially Linked to the Exhausted Phenotype of CD8+ T Cells in HIV Infection. PLOS Pathogens 2014;10(7):e1004251. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.