Abstract
Deep learning has dramatically improved object recognition, speech recognition, medical image analysis and many other fields. Optical coherence tomography (OCT) has become a standard of care imaging modality for ophthalmology. We asked whether deep learning could be used to segment cornea OCT images. Using a custom-built ultrahigh-resolution OCT system, we scanned 72 healthy eyes and 70 keratoconic eyes. In total, 20,160 images were labeled and used for the training in a supervised learning approach. A custom neural network architecture called CorneaNet was designed and trained. Our results show that CorneaNet is able to segment both healthy and keratoconus images with high accuracy (validation accuracy: 99.56%). Thickness maps of the three main corneal layers (epithelium, Bowman’s layer and stroma) were generated both in healthy subjects and subjects suffering from keratoconus. CorneaNet is more than 50 times faster than our previous algorithm. Our results show that deep learning algorithms can be used for OCT image segmentation and could be applied in various clinical settings. In particular, CorneaNet could be used for early detection of keratoconus and more generally to study other diseases altering corneal morphology.
1. Introduction
Optical coherence tomography (OCT) is a medical imaging technology based on low-coherence interferometry [1–3]. Using OCT, high-resolution volumetric imaging of tissues can be performed in vivo non-invasively and both morphological and functional information about the tissues can be provided. Over the years, OCT has revolutionized ophthalmology and is now considered to be a standard of care [4].
In the medical imaging field, segmentation aims to determine the boundary of different types of tissue. Using segmentation, tissue morphology can be quantified. Tissue thickness or volume can be used as a biomarker for the diagnosis of various diseases. In opthalmology, retinal nerve fiber layer (RNFL) thickness is used as a diagnostic parameter for characterizing structural damage in glaucoma [5, 6]. Tear film thickness can be used as a biomarker for studying dry eye syndrome [7–9]. In the cornea, several diseases cause morphological changes [10] and anterior segment OCT has become an important tool for characterizing them [11, 12]. Keratoconus, a common corneal dystrophy, modifies corneal morphology, particularly that of the epithelial layer [13, 14].
Several algorithms have been developed for the segmentation of medical images. These algorithms can be classified in two categories: those entirely designed by humans and those using machine learning. Various non-machine-learning algorithms for the segmentation of healthy cornea OCT images have been reported: approaches using graph theory [15–18], fast active contour and polynomial fitting [19], Canny edge detection [20], Gaussian mixture models [21] and Hough transform combined with Kalman filtering [22] have been published.
In machine learning approaches, the computer learns how to perform a task after being trained on a given set of correct examples (supervised learning). Until recently, machine learning required a handcrafted feature extractor which demands considerable expertise to develop [23]. The key aspect of deep learning is that the feature extractors are not designed by humans but are learned from the data [23].
Segmentation can be viewed as a classification problem for each pixel of the input image. In a deep learning approach, the neural network maps each pixel to its corresponding class. This process corresponds to a succession of linear and nonlinear transformations applied to the input image. These transformations are learned from the training data.
Deep learning algorithms applied to retina OCT images have been reported recently [24–29]. Roy et al. reported RelayNet for retinal layer and fluid segmentation of macula OCT images [24]. Lee et al. used deep learning for age-related macular degeneration (AMD) detection [25] and for the segmentation of macular edema [26]. Fang et al. used deep learning to automatically segment nine retinal layer boundaries in OCT images of non-exudative AMD [27]. Devalla et al. reported a deep learning approach to digitally stain optic nerve head images [28].At the time of writing, however, to our best knowledge no deep learning approach employed for the segmentation of cornea OCT images has been reported.
Here we report CorneaNet, a deep fully-convolutional neural network for the segmentation of OCT images of healthy and keratoconic corneas. Several neural networks are tested for the segmentation task. The training and the final performance of each model are analyzed. A comparison of the performance with a human-designed algorithm is shown. Finally, thickness maps of the corneal layers are shown in healthy and keratoconus cases.
2. Material and methods
The learning process of a neural network corresponds to an optimization process in which an error is minimized. For deep learning applications, the optimization is mainly done using stochastic gradient descent (SGD), which is described in appendix A.1. The error is represented by a loss function that depends on the model prediction and the true value.
2.1. Dataset
We acquired volumetric OCT data consisting of voxels (slow × fast × depth axis) corresponding to , respectively. The measurements were performed using an ultra-high resolution OCT (UHR-OCT) system previously published [12]. Measurements were performed in 72 eyes of 36 healthy subjects and 70 eyes of 57 patients with keratoconus. Some of these measurements were used for a clinical study previously published [13]. The study protocol was approved by the Ethics Committee of the Medical University of Vienna and was performed in adherence to the Declaration of Helsinki and the Good Clinical Practice (GCP) guidelines of the European Union. Subjects gave written informed consent before they entered the study. The OCT volume acquisition time was approximately 5 s.
Our initial dataset is composed of 140 OCT volumes of keratoconic eyes and 140 OCT volumes of healthy eyes, leading to 143,360 B-scans in total. Our telecentric scanning configuration of the cornea entails that the signal-to-noise ratio (SNR) decreases strongly as the distance from the apex increases [7]. Therefore, the border area of each B-scan and the B-scans far from the apex are not usable. From each OCT volume, 72 images centered around the corneal apex and with a size pixels () were generated leading to a total of 20,160 images. The training images' width corresponds to 2.81 mm and their maximum distance to apex along the slow scanning axis is 0.89 mm. Later in this paper, it will be seen that the network is able to predict beyond this region.
To generate the label images, we used an algorithm implemented in Matlab R2017b (The MathWorks Inc., Natick, MA, USA) based on iterative robust fitting (IRF) that we have previously described [7]. Briefly, the periodogram maxima points belonging to each boundary are selected using an iterative algorithm and are robustly fitted. For each image, the segmentation boundaries were checked and corrected manually if necessary. From the boundary curves, a label image containing the class of each pixel was generated. The 4 classes are epithelium, Bowman’s layer, stroma and background. The background class corresponds both to the aqueous humor and the air. The label image was finally converted to a categorical array containing the class of each pixel (size : ) for the model fitting within the deep learning library Keras [30].
2.2. Loss function and metrics
The learning process of neural networks corresponds to the minimization of the loss function. At each step, the gradient of the loss function is calculated with respect to all parameters of the model (cf. appendix A.1). As such, the loss function must be differentiable and thus smooth. Here, cross-entropy is used as a loss function. Other useful metrics to quantify the performance of the model are presented below.
2.2.1. Cross-entropy
In the following paragraphs, the rationale of using cross-entropy as a loss function is explained.
The true labels and the output of the network can be seen as probability distributions for all pixels. Let x be the class index, be the probability distribution provided by the network (for a given pixel) and be the true probability distribution (i.e. if the pixel belongs to the class x and else).
The entropy of a probability distribution p is defined by
| (1) |
The entropy is a measure of the uncertainty of the probability distribution. is equal to the number of bits on average required to describe the underlying random variable (the unit is bits with base 2 logarithms) [31].
The relative entropy or Kullback-Leibler distance between two probability distributions p and q is defined as
| (2) |
is a measure of the distance between the probability distributions p and q. is non negative and is zero if and only if .
The cross-entropy is defined as
| (3) |
Using Eqs. (1), (2) and (3), the Kullback-Leibler distance can be rewritten as
| (4) |
Using the definition of and Eq. (1), it follows that , the entropy of the true probability distribution is zero. In this case, , the cross-entropy is equal to the Kullback-Leibler distance. Therefore, the cross-entropy is non negative and is zero if and only if and can be used to measure the "distance" between p0 and . The average value of the cross-entropy over all pixels is the definition of categorical cross-entropy used in Keras and TensorFlow.
The pixel class is finally estimated using the following estimator .
2.2.2. Accuracy
The accuracy is defined as the fraction of correctly labeled pixels. More formally, the accuracy is the average over all pixels, of either value 1 if the pixel is correctly labeled, or 0 if not. The accuracy is not a smooth function, because of the discontinuity or "jump" when one pixel value changes. Thus, the accuracy cannot be used as a loss function. However, the accuracy is useful as metric to compare the performances of different models.
2.2.3. Additional metrics
Several other metrics were used to analyze the performances of the tested models. The recall r is the fraction of the true labels that were correctly identified, for each class. The precision p is the fraction of the predicted labels that are correct, for each class. The Dice coefficient and the Jaccard index are standard metrics for segmentation problems. The Dice coefficient is defined as , where X is the predicted set and Y is the true set, for each class. The Dice coefficient is identical to the harmonic mean of the recall and precision . The Jaccard index, also known as intersection over union is defined as . The Jaccard index can be related to the Dice coefficient using the identity . The support is defined as the fraction of the pixels belonging to each class over all images of the dataset. The average precision AP is the support-weighted average of the precision and the accuracy is the support-weighted average of the recall. AD is the support-weighted average of the Dice coefficient and AJ is the support-weighted average of the Jaccard index. These metrics are used to present the results in Table 2.
Table 2.
Results of the different models on validation data obtained using six-fold cross-validation (average ± standard deviation). The support is the fraction of the images covered by each class in the dataset. The sensitivity is the fraction of the true labels that were correctly identified for each class. The precision is the fraction of the predicted labels that are correct for each class. The accuracy is the fraction of correctly identified pixels. All metrics are defined in section 2.2. The background class corresponds to air or aqueous humor.
| Epithelium | Stroma | Bowman’s | Background | Total | |
|---|---|---|---|---|---|
| Support (%) | |||||
| 5.50 | 47.10 | 1.58 | 45.82 | 100.00 | |
|
| |||||
| Recall / Sensitivity (%) | Accuracy (%) | ||||
| CUnet1 | 99.33 ± 0.03 | 99.74 ± 0.02 | 93.99 ± 0.52 | 99.60 ± 0.03 | 99.57 ± 0.02 |
| CorneaNet | 99.35 ± 0.07 | 99.74 ± 0.01 | 94.60 ± 0.25 | 99.57 ± 0.04 | 99.56 ± 0.02 |
| CUnet3 | 99.23 ± 0.02 | 99.58 ± 0.02 | 94.13 ± 0.50 | 99.53 ± 0.03 | 99.45 ± 0.02 |
| CUnet4 | 99.28 ± 0.01 | 99.58 ± 0.02 | 93.62 ± 0.45 | 99.54 ± 0.07 | 99.45 ± 0.03 |
| CUnet5 | 99.10 ± 0.01 | 99.60 ± 0.03 | 96.41 ± 0.02 | 99.55 ± 0.03 | 99.50 ± 0.02 |
|
| |||||
| Precision (%) | AP (%) | ||||
| CUnet1 | 99.20 ± 0.04 | 99.56 ± 0.03 | 95.28 ± 0.28 | 99.76 ± 0.03 | 99.56 ± 0.02 |
| CorneaNet | 99.24 ± 0.01 | 99.55 ± 0.03 | 95.56 ± 0.20 | 99.77 ± 0.02 | 99.57 ± 0.02 |
| CUnet3 | 99.13 ± 0.04 | 99.51 ± 0.02 | 94.23 ± 0.33 | 99.62 ± 0.01 | 99.46 ± 0.01 |
| CUnet4 | 99.11 ± 0.03 | 99.51 ± 0.05 | 94.83 ± 0.17 | 99.60 ± 0.03 | 99.46 ± 0.03 |
| CUnet5 | 99.28 ± 0.02 | 99.58 ± 0.02 | 93.06 ± 0.10 | 99.68 ± 0.03 | 99.51 ± 0.02 |
|
| |||||
| Dice coefficient / F1-score (%) | AD (%) | ||||
| CUnet1 | 99.26 ± 0.04 | 99.65 ± 0.01 | 94.63 ± 0.40 | 99.69 ± 0.02 | 99.57 ± 0.01 |
| CorneaNet | 99.30 ± 0.04 | 99.64 ± 0.01 | 95.07 ± 0.23 | 99.67 ± 0.01 | 99.56 ± 0.01 |
| CUnet3 | 99.18 ± 0.01 | 99.54 ± 0.02 | 94.18 ± 0.09 | 99.57 ± 0.02 | 99.45 ± 0.01 |
| CUnet4 | 99.20 ± 0.01 | 99.54 ± 0.02 | 94.22 ± 0.14 | 99.57 ± 0.02 | 99.45 ± 0.01 |
| CUnet5 | 99.19 ± 0.01 | 99.59 ± 0.03 | 94.71 ± 0.04 | 99.62 ± 0.03 | 99.50 ± 0.02 |
|
| |||||
| Jaccard index / IoU (%) | AJ (%) | ||||
| CUnet1 | 98.53 ± 0.08 | 99.30 ± 0.03 | 89.81 ± 0.73 | 99.38 ± 0.04 | 99.15 ± 0.02 |
| CorneaNet | 98.61 ± 0.07 | 99.28 ± 0.01 | 90.60 ± 0.41 | 99.34 ± 0.02 | 99.14 ± 0.01 |
| CUnet3 | 98.37 ± 0.01 | 99.08 ± 0.03 | 89.00 ± 0.16 | 99.14 ± 0.04 | 98.91 ± 0.03 |
| CUnet4 | 98.41 ± 0.03 | 99.08 ± 0.04 | 89.07 ± 0.25 | 99.14 ± 0.04 | 98.92 ± 0.03 |
| CUnet5 | 98.39 ± 0.03 | 99.18 ± 0.05 | 89.95 ± 0.08 | 99.24 ± 0.06 | 99.02 ± 0.04 |
2.3. Network architecture
A deep neural network can be seen as a directed graph in which each node corresponds to a module performing a certain type of trainable transformation. Examples of these modules include: convolution, pooling, up-sampling, activation and concatenation. These modules are standard in various deep learning libraries like TensorFlow, Keras, CNTK and Torch.
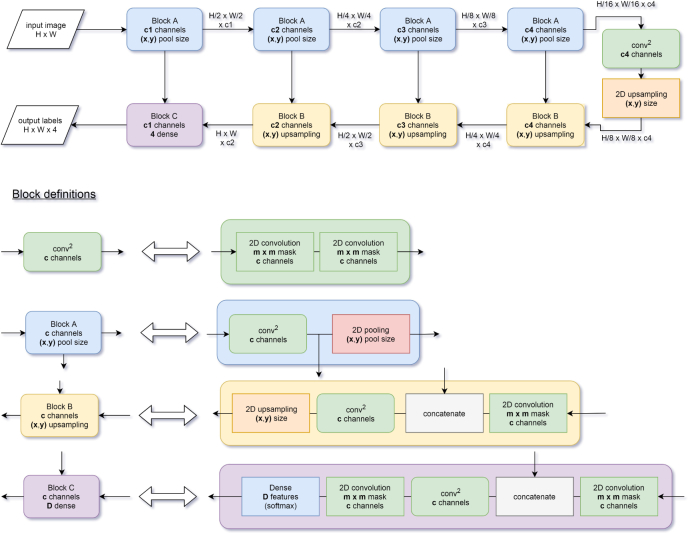
Several network architectures have previously been used for image segmentation, e.g. FCN [32], Unet [33], Segnet [34], Mask R-CNN [35], or Deeplab [36]. U-shape networks have been commonly used for various biomedical segmentation problems [24, 33, 37]. U-net [33] has pioneered the use U-shape architecture for biomedical image segmentation and is inspired by FCN [32] that was used for semantic segmentation [33]. RelayNet [24], a network for multiple retinal layers and delineation of fluid pockets in eye OCT images is inspired by U-net. Figure 1 shows a schematic of a U-shape network architecture. The process sequence in a U-shape network can be described as follows: first, the number of pixels of the representation decreases while the number of features increases. Second, the representation progressively returns to the original size while concatenating with the previous representation to avoid losing resolution. The last layer classifies the category based on the extracted features using a soft-max classifier.
Fig. 1.
Diagram of a typical U-net architecture. After passing through all the blocks, the input image is transformed into a label image. The building blocks A, B and C are defined in terms of basic building blocks (convolution, pooling, concatenation, up-sampling and fully-connected (dense) layer). The last layer of the network is a fully-connected layer with four channels followed by a soft-max activation (the probability for each class). The number of channels of this layer corresponds to the number of classes of the labels (i.e. 4). The activation and batch normalization layers are omitted for clarity.
We compared the performances of five models for cornea OCT image segmentation. Batch normalization helps to accelerate the training [38]. The original U-net model does not contain batch normalization and its optimization did not converge in all cases with our data and optimizers. We wanted to investigate if the use of average pooling instead of maximum pooling could lead to better performances. OCT images can contain a significant amount of noise, and average pooling could be more robust than maximum pooling with respect to the handling of this noise. The original Unet network contains the following amount of features at different depths: . This amount of features leads to more than million parameters. We wanted to investigate whether a lighter model with less parameters could perform equally well or better for our application. More than 30 different models with various architectures were tested. For clarity, we selected five models and present them here. Four of these models, with U-shape architecture, were selected because of their accuracy and time performance. CUnet 5 was selected in order to investigate unconventional WW-shape architecture. Table 1 shows a summary of the essential characteristics of the tested models. The detailed network architecture of the different models is provided in the Appendix A.3.
Table 1.
Characteristics of the architectures of the studied models.
| Model name | Depth | Type of pooling | Mask size | # channels of the conv. layers | # Layers | # Parameters |
|---|---|---|---|---|---|---|
| CUnet1 | 4 | maximum | 3 × 3 | [16, 32, 64, 128, 256] | 82 | 2,167,636 |
| CorneaNet | 4 | average | 3 × 3 | [16, 32, 64, 128, 256] | 82 | 2,167,636 |
| CUnet3 | 3 | maximum | 3 × 3 | [8, 16, 32, 64] | 64 | 136,980 |
| CUnet4 | 3 | average | 3 × 3 | [8, 16, 32, 64] | 64 | 136,980 |
| CUnet5 | 3 | average | 5 × 5 | [32, 64, 128, 256] | 247 | 23,871,732 |
2.4. Training
Each model of Table 1 was trained on our dataset. Six-fold cross-validation was performed. Categorical cross-entropy was used as a loss function. The optimization was performed using SGD [39]. The used optimizer was RMSprop with initial learning rate [40]. A learning rate scheduler that automatically decreases the learning rate by a factor of five if the loss didn’t improve for 15 epochs was employed. Each of the 600 epochs was composed of 100 training steps [30]. The batch size was selected to saturate the GPU memory for each model. The training was performed on images with a size of pixels (depth×width). Because our networks are fully-convolutional, they can be used with various input image sizes. The images from our dataset were pixels from which several pixels images were extracted. This can be viewed as a form of data augmentation. From each image, having a width of 384 pixels, 81 different training images with a width of 64 pixels were extracted (minimum lateral spacing of 4 pixels). During training, images of keratoconic or healthy corneas were provided to the network with equal probability. No preprocessing nor filtering was applied to the input images. An example of the training loss curves of one fold is shown in Fig. 2 (log. scale). Cross-validation, also called rotation estimation is a technique to avoid overfitting and selection bias. It ensures that the model testing is done on first-seen data. Measurements in the same eye can be correlated and were therefore always put in the same set (either training set or test set depending on the rotation index of the cross-validation).
Fig. 2.
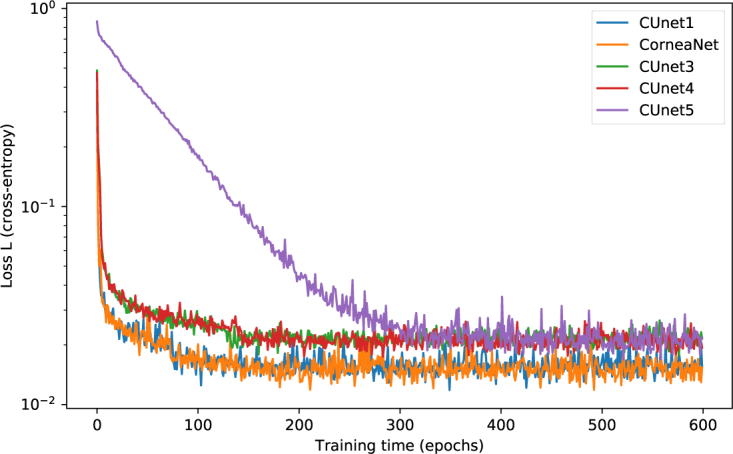
Training loss as a function of the epoch for the five tested models for one fold (logarithmic scale). During the learning process of each model, the loss, representing the error, decreases. CUnet1 and CorneaNet learn faster and achieve finally lower loss than other models.
2.5. Computer hardware and software
The deep learning computations presented here were performed on a personal computer with an Intel Core i7-6850K CPU @ 3.60GHz and with two Nvidia Geforce GTX 1080 TI GPUs. Modern GPUs are much faster than CPUs in terms of number of floating point operations per second (FLOPS): Our CPU provides 0.060 TFLOPS while each GPU provides 11.5 TFLOPS. This difference can partly be explained by the much larger number of cores of the GPU in comparison to the CPU (3584 cores vs. 6 cores). Deep neural network computation is highly parallelizabel and therefore benefits from the large number of GPU cores. The deep neural network was implemented in the Python programming language using Keras 2.2.4 and TensorFlow 1.12.0 libraries [30, 41]. The monitoring of the training was performed using TensorBoard. The operating system of the PC is Linux Ubuntu 16.04 LTS. The tensor computation is using CUDA 9.2 and CUDNN 7.0.5. The GPU driver version is 396.54.
3. Results
3.1. Automated segmentation of healthy and keratoconic corneas
In Fig. 3, representative segmentations of a healthy and a keratoconic cornea are depicted. In the left panel, the results of the segmentation using our Matlab algorithm are shown, while on the right, the results of CorneaNet are provided. As can be seen, CorneaNet is capable of segmenting both healthy and keratoconic corneas.
Fig. 3.
CorneaNet automatically segments cornea OCT images with high accuracy. Segmentation of healthy and keratoconic corneas using the Matlab algorithm and CorneaNet. Keratoconic corneas exhibit usually at least one layer with non-uniform thickness. Scale bar: 250 μm
3.2. Performance analysis
Table 2 shows the performance metrics of the tested models obtained using six-fold cross-validation. All models revealed very similar performances and have a validation accuracy ranging from 99.45 % to 99.57 %. CorneaNet has a slighlty better performance than other tested models in terms of average precision. Globally, the sensitivity and precision is slightly inferior for the Bowman’s layer. This finding can be explained by the fact that the Bowman’s-stroma boundary is not always well defined in keratoconus and some healthy cases, due to a smaller SNR in the OCT data. Therefore the training data itself might contain errors for this layer.
The accuracy is fundamentally limited by the pixelation of the image. By comparing the true segmentation and the one obtained using the neural network, it can be seen that the error always occurs at the boundary between two layers. A manual segmentation would also perform errors at the border. If a pixel is exactly between two segments, it can be classified into either one of the segments causing the segmentation error. Improvement of the pixel resolution of the image can reduce this problem. Concretely, one pixel error at each of the four interfaces for the whole image width corresponds to an accuracy of (). A single pixel itself contains little information or features. As a result of this, one can understand that the obtained accuracy is not far from the optimum.
Table 3 shows the time and memory performances of the studied models. The model memory ranges from 0.5 MB to 91 MB. The duration to predict one image ranges from 13 ms to 337 ms. Models with more parameters are slower.
Table 3.
Time and memory characteristics of the studied models. The image memory is the memory required to store all the feature images data of hidden layers in the GPU memory. B is the batch size we used for the training.1 The values are obtained with an input image size of pixels.
| Model name | # Trainable parameters | Model memory (MB) | Images memory (MB)1 | Batch size B1 | Duration for one prediction1 (ms) |
|---|---|---|---|---|---|
| CUnet1 | 2,164,212 | 8.3 | 887.2 | 4 | 25.3 |
| CorneaNet | 2,164,212 | 8.3 | 887.2 | 4 | 24.9 |
| CUnet3 | 136,164 | 0.5 | 425.2 | 9 | 12.8 |
| CUnet4 | 136,164 | 0.5 | 425.2 | 9 | 12.8 |
| CUnet5 | 23,858,676 | 91.1 | 6646.5 | 1 | 337.6 |
3.2.1. Segmentation speed of CorneaNet and previous algorithms
We compared the segmentation speed of CorneaNet to the previously used iterative robust-fitting (IRF) algorithm implemented in Matlab. Table 4 summarizes the time required for various task by the two algorithms on the same computer system. The IRF algorithm runs on the CPU and was designed for robust segmentation on low SNR images, prioritizing accuracy over speed. Clearly, the deep learning segmentation algorithm performs several orders of magnitude faster. The speed of CorneaNet is fast enough for video-rate B-scan segmentation. This evaluation does not take into account disk reading and writing, but includes transfers between the computer RAM and the GPU memory. One full volume is approximately 3619 MB and, thus, can be fully stored in the RAM.
Table 4.
Comparison of the durations of various tasks for several algorithms .
| Algorithm | 1 image (384×1024 pixels) | 1 volume (512 images) | full dataset (280 volumes) |
|---|---|---|---|
| CorneaNet | 24.9 ms | 12.7 s | 59 min |
| IRF | 9 346 ms | 1 h 19 min 45 s | 15 days 12 h 11 min |
We observed that the deep learning algorithm works robustly and can even predict layer boundaries in partly saturated images, in regions where the IRF algorithm does not work. The saturation is caused by a too high optical power reaching the CMOS camera (central corneal reflex). The speed of the Matlab-based IRF algorithm is mainly limited by the robust fitting steps that are performed several times for each interface. Furthermore, this algorithm, initially designed for the determination of tear film thickness [7] allows for subpixel accuracy which is not essential for the segmentation task presented here. The speed difference can be also explained by the fact that CorneaNet runs on the GPU, unlike the Matlab algorithm that runs on the CPU. Nevertheless, the observed speed difference is very large and could be useful for practical clinical applications.
Several previous cornea segmentation studies have reported the segmentation time per B-scan: 1.13 s [15], 0.512 s [22], 3.09 s [17] (the B-scan sizes were 1000×1024 [15, 22] and 256×1024 [17], respectively). By assuming a proportionality between the time and the number of A-scans per B-scan, we can convert to our image size and obtain processing time of 434 ms/image, 197 ms/image and 4635 ms/image, respectively.
3.3. Thickness maps
Keratoconus causes structural changes in the cornea. We investigated if these structural changes can be visualized on 2D thickness maps obtained using CorneaNet.
The thickness maps were generated by segmenting each B-scan of the volumetric data set using CorneaNet. These thickness maps are the direct output of the network without any further processing and no averaging nor filtering was used to produce them. We observe a good agreement between adjacent B-scans.
In Fig. 4 thickness maps of the three major corneal layers are shown in both a healthy and a keratoconus eye. These measurements belong to the test set of the model, and were therefore not used for training. The size of the maps () is larger than the size of the region used for training (). This indicates that the features detected by the network for segmentation are consistent over a larger region of the cornea. The thickness scale bar is shared by the maps horizontally. The A-scan corresponding to Fig. 4 is indicated as a vertical line in Fig. 4 (g-i).
Fig. 4.
Using CorneaNet, the thicknesses of the epithelium, stroma and Bowman’s layer were computed in a healthy and a keratoconus case. The healthy case shows close to uniform thicknesses for all three layers, while for the keratoconus case, in a specific region of the cornea, the epithelium and stroma are thinner and the Bowman’s layer is thicker. (a-c) Thickness calculation in one tomogram. (a) UHR-OCT tomogram of a keratoconus patient, (b) corresponding labels map computed using CorneaNet. (c) Thicknesses of the three corneal layers computed using the label maps. (d-f) Thickness maps in a healthy subject case. (g-i) Thickness maps in a keratoconus case. The thickness scale bar is shared by the maps horizontally. Scale bar: 1 mm.
Comparison of the thickness maps of the keratoconus patient with that of the healthy subject revealed irregularities in all three layers (Fig. 4 (d-i)). While the healthy cornea shows homogeneous thickness maps of epithelium, Bowman’s layer and stroma, the cornea of the patient suffering from keratoconus reveals non uniform thicknesses for each layer. Despite the limited scan range, in the superior region of the map, part of a torus profile as a sign of the compensatory thickening in the area surrounding the thinnest zone can be observed.
3.4. Comparison with Pentacam
The thickness map obtained using UHR-OCT and CorneaNet was compared with the map of corneal refractive power as extracted by Scheimpflug tomography (Pentacam HR, Oculus Pentacam, Wetzlar, Germany). In Fig. 5, exemplary maps obtained from a patient suffering from keratoconus are depicted. As seen in Fig. 5, the full corneal thickness map is non uniform. The thinnest region of the cornea as measured with UHR-OCT was found in the inferotemporal part of the cornea and corresponds well to the steepest corneal zone as measured with Scheimpflug tomography. While UHR-OCT provides thickness measurements with a precision as high as 1 μm, images might contain motion artifacts and the area of imaging is fundamentally limited by the SNR that decreases as the distance to apex increases.
Fig. 5.
Comparison between (a) full corneal thickness map obtained using UHR-OCT and CorneaNet; and (b) an Oculus Pentacam total power image. Both measurements were done in the same eye suffering from keratoconus.
4. Discussion
Our results show that deep learning can be used to segment OCT images of both healthy and keratoconic corneas. We report CorneaNet, a deep fully-convolutional neural network designed for fast and robust segmentation of cornea OCT images with a validation accuracy of over 99.5% and a segmentation time of less than 25 ms. To our best knowledge, this is the first time that the segmentation of cornea OCT images is performed using deep learning.
Thickness maps of the epithelium, Bowman’s layer and stroma as well as of the full cornea were presented for measurements obtained from healthy and keratoconic corneas. In the inferotemporal part of the keratoconic cornea, a thinner zone compared to the surrounding area could be detected. This finding indicates that theses maps could be used to study the formation of keratoconus and might aid in its early detection. Full corneal thickness maps were compared with the measurement of a commercial device (Oculus Pentacam) and a good agreement was found.
The most promising feature of deep learning-based segmentation is probably the speed. For example, CUnet3 provided a segmentation rate of 66 frames per second (FPS) for an image with pixels. The FPS values will increase in the future with hardware and software improvements. We predict that real time live volumetric segmentation will soon become possible.
Our results are consistent with previous reports about the application of deep learning to OCT images. Several applications to retina OCT images have been published [24–28]. Depending on the application, the reported Dice coefficient ranged from 71% to 99%. CorneaNet provides an average Dice coefficient larger than 99.5%. Additionally, several studies for cornea OCT image segmentation with non-machine-learning approaches were reported [15, 16, 18–20, 22]. Some studies did not provide segmentation for all corneal layers. The segmentation of the Bowman’s layer requires high axial resolution and SNR. Several algorithms allowed for subpixel segmentation accuracy at each interface with typical values ranging from 0.6–1.7 px [15, 17, 22]. Williams et al. reported a Dice coefficient of 96.7 % [16]. As a pixel classification approach, deep learning segmentation is always limited by the pixel resolution of the image.
The segmentation accuracy might be limited by the network architecture itself, by the noise of the image, by the lack of training data and by inaccuracies in the training data. We observed that all models provided very similar validation accuracy values; and for all models, the sensitivity and precision were slightly inferior for the Bowman’s layer. This suggests that for this layer, the accuracy is not limited by the model architecture itself, but by the accuracy of the training data or the SNR of the images. Further work is needed to determine what factor limits the sensitivity and precision of the segmentation for each layer.
Overall, deep learning is suitable for extracting the features in OCT images and using these features to classify each pixel. We observed that the network is able to predict over a larger area than the one used for training. The network was able to predict correctly at a distance beyond 1.6 mm from the corneal apex, while it was trained on images with a maximum distance of 0.89 mm. This suggests that the learned feature extractor is applicable to a larger area of the cornea, to various subjects and to various SNR levels. Furthermore, we observed that the segmentation is robust in the sense that it also performs well in non-regular cases. For example, the network can predict the layer boundaries in partly saturated image (central corneal reflex) contrary to the IRF algorithm that is not working correctly in this region [7].
Concerning the network architecture, we found that batch normalization accelerated training, which is consistent with previous work [38]. Furthermore, it was shown that the use of average pooling instead of maximum pooling works also well for this segmentation problem. The performances are slightly different. We hypothesize that average pooling is more robust against noise which could a be a reason for the better performance at the Bowman’s layer (Table 2). Further work is needed to confirm this hypothesis. For deep learning models, there is a trade-off between the number of parameters and the time performance. We found that by diminishing the number of features of the convolutional layers, leading to a parameter decrease from over 2 million to 0.14 million, the validation accuracy decreased only by 0.1 points. Increasing the number of parameters to over 23 million did not lead to a performance increase, but to a performance decrease, possibly caused by overfitting (cf. Table 2). Thus, depending on the application, a lighter architecture might be more suitable [42]. CorneaNet provides the highest average precision among the tested models.
5. Conclusion
In summary, we reported CorneaNet, a fully-convolutional neural network for the segmentation of cornea OCT images with high accuracy. The segmentation speed of this network is much higher than the one of our previous algorithm. Deep learning algorithms could be used for OCT image segmentation in various clinical settings. In particular, CorneaNet could be used for early detection of keratoconus and more generally to study other diseases altering corneal morphology.
Acknowledgments
We thank Dr. Niklas Pircher of the Department of Ophthalmology of the Medical University of Vienna for his help for the recruitment and measurement of patients with keratoconus.
A. Appendix
A.1. Stochastic gradient descent
The learning process of a neural network corresponds to an optimization process. In most deep learning applications, the optimization algorithm is stochastic gradient descent (SGD), which is described in the next paragraphs.
Consider a deep learning model parameterized by a vector parameter θ. Let be a training example and N be the total number of training examples.
The model parameters θ are estimated by minimizing the global loss function L
| (5) |
computed on the full set of training examples.
The idea of SGD is to minimize the loss function iteratively using a gradient estimate, computed on a small batch of examples (instead of the whole training dataset ). Let G be the gradient estimate on a batch
| (6) |
The parameter update rule is:
| (7) |
which corresponds to the steepest descent update rule where k is the step index and η is a parameter called the learning rate.
SGD is not the only method that can be used for model optimization. Full-batch methods like L-BFGS or non-linear conjugate gradient use the full dataset to compute the gradient [40]. SGD has several advantages over these approaches. It requires less memory, which is particularly useful if the dataset is large and is faster [39]. Nowadays SGD has become the standard optimization method in the deep learning field [23, 43].
A.2. Statistical analysis of the dataset
We performed a basic statistical analysis of the dataset. The results are summarized in Table 5. The group refractive index of the cornea was assumed to be uniform and equal to 1.385 [44]. For each OCT volume, the thickness of each layer was computed for each A-scan, where x corresponds to the fast scanning axis and y to the slow scanning axis. The average thickness along the slow and fast axis was computed for each volume, i.e. . To analyze the uniformity of the thicknesses, we defined a quantity called non-uniformity (NU) as the difference between the maximum thickness and the minimum thickness along the fast axis: . We observe a significant difference of the non-uniformity between both groups using a two-sample t-test (all ). This indicates that non-uniformity could be used as a simple indicator for studying the formation of keratoconus.
Table 5.
Statistical analysis of the dataset. The thicknesses of different layers in both groups are shown. The reported value is the mean ± standard deviation.
| Group | Epithelium | Bowman’s | Stroma | Unit | |
|---|---|---|---|---|---|
| Thickness T | Healthy | 52.91 ± 4.3 | 14.44 ± 1.7 | 461.54 ± 30.0 | μm |
| Thickness T | Keratoconus | 49.36 ± 6.4 | 14.12 ± 3.8 | 414.28 ± 36.0 | μm |
| Non-uniformity NU | Healthy | 0.46 ± 0.1 | 0.48 ± 0.1 | 24.25 ± 12.2 | μm |
| Non-uniformity NU | Keratoconus | 13.15 ± 6.9 | 8.14 ± 7.3 | 49.20 ± 21.8 | μm |
A.3. Definition of the tested models
In Tables 6, 7 and 8, the detailed definition of the architecture of the tested networks is presented. All used layers are standard in any deep learning library like TensorFlow, Keras, CNTK and Torch. In each of the 2D convolutional modules, we perform batch normalization before activation to help accelerate the learning [38]. The activation is a standard ReLU function [42, 45]. The Conv2D mask values are defined in Table 1. Pooling and up-sampling size is always . The dropout layer – helping to reduce overfitting – was used with a factor equal to 0.5. As defined in Table 1, the only difference between CUnet1 and CorneaNet (respectively CUnet3 and CUnet4) is that all maximum pooling layers are replaced by average pooling. The CUnet5 architecture is a WW-shape (i.e. composed of four repetitions of the U-shape structure). The analysis of the time and memory performances of these networks is shown in Table 3. The results of these models on our dataset is shown in Table 2.
Table 6.
Architecture of CUNet 1 and CorneaNet
| ID | Type | Output size | # parameters | Connected with (ID) |
|---|---|---|---|---|
| 1 | InputLayer | 0 | ||
| 2 | Conv2D + BatchNorm. + Activation | 224 | 1 | |
| 3 | Conv2D + BatchNorm. + Activation | 2,384 | 2 | |
| 4 | MaxPooling2D | 0 | 3 | |
| 5 | Conv2D + BatchNorm. + Activation | 4,768 | 4 | |
| 6 | Conv2D + BatchNorm. + Activation | 9,376 | 5 | |
| 7 | MaxPooling2D | 0 | 6 | |
| 8 | Conv2D + BatchNorm. + Activation | 18,752 | 7 | |
| 9 | Conv2D + BatchNorm. + Activation | 37,184 | 8 | |
| 10 | MaxPooling2D | 0 | 9 | |
| 11 | Conv2D + BatchNorm. + Activation | 74,368 | 10 | |
| 12 | Conv2D + BatchNorm. + Activation | 148,096 | 11 | |
| 13 | MaxPooling2D | 0 | 12 | |
| 14 | Conv2D + BatchNorm. + Activation | 296,192 | 13 | |
| 15 | Conv2D + BatchNorm. + Activation | 591,104 | 14 | |
| 16 | Dropout | 0 | 15 | |
| 17 | UpSampling2D | 0 | 16 | |
| 18 | Conv2D + BatchNorm. + Activation | 295,552 | 17 | |
| 19 | Concatenate | 0 | 18 & 12 | |
| 20 | Conv2D + BatchNorm. + Activation | 295,552 | 19 | |
| 21 | Conv2D + BatchNorm. + Activation | 148,096 | 20 | |
| 22 | UpSampling2D | 0 | 21 | |
| 23 | Conv2D + BatchNorm. + Activation | 74,048 | 22 | |
| 24 | Concatenate | 0 | 23 & 9 | |
| 25 | Conv2D + BatchNorm. + Activation | 74,048 | 24 | |
| 26 | Conv2D + BatchNorm. + Activation | 37,184 | 25 | |
| 27 | UpSampling2D | 0 | 26 | |
| 28 | Conv2D + BatchNorm. + Activation | 18,592 | 27 | |
| 29 | Concatenate | 0 | 28 & 6 | |
| 30 | Conv2D + BatchNorm. + Activation | 18,592 | 29 | |
| 31 | Conv2D + BatchNorm. + Activation | 9,376 | 30 | |
| 32 | UpSampling2D | 0 | 31 | |
| 33 | Conv2D + BatchNorm. + Activation | 4,688 | 32 | |
| 34 | Concatenate | 0 | 33 & 3 | |
| 35 | Conv2D + BatchNorm. + Activation | 4,688 | 34 | |
| 36 | Conv2D + BatchNorm. + Activation | 2,384 | 35 | |
| 37 | Conv2D + BatchNorm. + Activation | 2,384 | 36 | |
| 38 | Dense | 68 | 37 | |
|
| ||||
| Total # parameters | 2,167,636 | |||
Table 7.
Architecture of CUNet3
| ID | Type | Output size | # parameters | Connected with (ID) |
|---|---|---|---|---|
| 1 | InputLayer | 0 | ||
| 2 | Conv2D + BatchNorm. + Activation | 112 | 1 | |
| 3 | Conv2D + BatchNorm. + Activation | 616 | 2 | |
| 4 | MaxPooling2D | 0 | 3 | |
| 5 | Conv2D + BatchNorm. + Activation | 1,232 | 4 | |
| 6 | Conv2D + BatchNorm. + Activation | 2,384 | 5 | |
| 7 | MaxPooling2D | 0 | 6 | |
| 8 | Conv2D + BatchNorm. + Activation | 4,768 | 7 | |
| 9 | Conv2D + BatchNorm. + Activation | 9,376 | 8 | |
| 10 | MaxPooling2D | 0 | 9 | |
| 11 | Conv2D + BatchNorm. + Activation | 18,752 | 10 | |
| 12 | Conv2D + BatchNorm. + Activation | 37,184 | 11 | |
| 13 | Dropout | 0 | 12 | |
| 14 | UpSampling2D | 0 | 13 | |
| 15 | Conv2D + BatchNorm. + Activation | 18,592 | 14 | |
| 16 | Concatenate | 0 | 15 & 9 | |
| 17 | Conv2D + BatchNorm. + Activation | 18,592 | 16 | |
| 18 | Conv2D + BatchNorm. + Activation | 9,376 | 17 | |
| 19 | UpSampling2D | 0 | 18 | |
| 20 | Conv2D + BatchNorm. + Activation | 4,688 | 19 | |
| 21 | Concatenate | 0 | 20 & 6 | |
| 22 | Conv2D + BatchNorm. + Activation | 4,688 | 21 | |
| 23 | Conv2D + BatchNorm. + Activation | 2,384 | 22 | |
| 24 | UpSampling2D | 0 | 23 | |
| 25 | Conv2D + BatchNorm. + Activation | 1,192 | 24 | |
| 26 | Concatenate | 0 | 25 & 3 | |
| 27 | Conv2D + BatchNorm. + Activation | 1,192 | 26 | |
| 28 | Conv2D + BatchNorm. + Activation | 616 | 27 | |
| 29 | Conv2D + BatchNorm. + Activation | 1,232 | 28 | |
| 30 | Dense | 68 | 29 | |
|
| ||||
| Total # parameters | 136,980 | |||
Table 8.
Architecture of CUNet5
| ID | Type | Output size | # parameters | Connected with (ID) |
|---|---|---|---|---|
| 1 | InputLayer | 0 | ||
| 2 | Conv2D + BatchNorm. + Activation | 960 | 1 | |
| 3 | Conv2D + BatchNorm. + Activation | 25,760 | 2 | |
| 4 | AveragePooling2D | 0 | 3 | |
| 5 | Conv2D + BatchNorm. + Activation | 51,520 | 4 | |
| 6 | Conv2D + BatchNorm. + Activation | 102,720 | 5 | |
| 7 | AveragePooling2D | 0 | 6 | |
| 8 | Conv2D + BatchNorm. + Activation | 205,440 | 7 | |
| 9 | Conv2D + BatchNorm. + Activation | 410,240 | 8 | |
| 10 | AveragePooling2D | 0 | 9 | |
| 11 | Conv2D + BatchNorm. + Activation | 820,480 | 10 | |
| 12 | Conv2D + BatchNorm. + Activation | 1,639,680 | 11 | |
| 13 | Dropout | 0 | 12 | |
| 14 | UpSampling2D | 0 | 13 | |
| 15 | Conv2D + BatchNorm. + Activation | 819,840 | 14 | |
| 16 | Concatenate | 0 | 15 & 9 | |
| 17 | Conv2D + BatchNorm. + Activation | 819,840 | 16 | |
| 18 | Conv2D + BatchNorm. + Activation | 410,240 | 17 | |
| 19 | UpSampling2D | 0 | 18 | |
| 20 | Conv2D + BatchNorm. + Activation | 205,120 | 19 | |
| 21 | Concatenate | 0 | 20 & 6 | |
| 22 | Conv2D + BatchNorm. + Activation | 205,120 | 21 | |
| 23 | Conv2D + BatchNorm. + Activation | 102,720 | 22 | |
| 24 | UpSampling2D | 0 | 23 | |
| 25 | Conv2D + BatchNorm. + Activation | 51,360 | 24 | |
| 26 | Concatenate | 0 | 25 & 3 | |
| 27 | Conv2D + BatchNorm. + Activation | 51,360 | 26 | |
| 28 | Conv2D + BatchNorm. + Activation | 25,760 | 27 | |
|
| ||||
| 3 × repetition | ID 2 ID 28 (U-architecture) | |||
|
| ||||
| 110 | Conv2D + BatchNorm. + Activation | 4,688 | 109 | |
| 111 | Dense | 68 | 110 | |
|
| ||||
| Total # parameters | 23,845,620 | |||
Typographical corrections were made to the title, abstract, appendix, and acknowedgments.
Funding
Vienna Science and Technology Fund (WWTF) (LS14- 067); Christian Doppler Research Association; Austrian Federal Ministry of Digital and Economic Affairs; National Foundation of Research, Technology and Development.
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Fercher A. F., Mengedoht K., Werner W., “Eye-length measurement by interferometry with partially coherent light,” Opt. Lett. 13, 186–188 (1988). 10.1364/OL.13.000186 [DOI] [PubMed] [Google Scholar]
- 2.Hitzenberger C. K., “Optical measurement of the axial eye length by laser Doppler interferometry,” Investig. Ophthalmol. & Vis. Sci. 32, 616–624 (1991). [PubMed] [Google Scholar]
- 3.Huang D., Swanson E. A., Lin C. P., Schuman J. S., Stinson W. G., Chang W., Hee M. R., Flotte T., Gregory K., Puliafito C. A., Et A., “Optical coherence tomography,” Science 254, 1178–1181 (1991). 10.1126/science.1957169 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Drexler W., Fujimoto J. G., Optical Coherence Tomography: Technology and Applications (Springer, 2015), 2nd ed. 10.1007/978-3-319-06419-2 [DOI] [Google Scholar]
- 5.Bowd C., Zangwill L. M., Berry C. C., Blumenthal E. Z., Vasile C., Sanchez-Galeana C., Bosworth C. F., Sample P. A., Weinreb R. N., “Detecting Early Glaucoma by Assessment of Retinal Nerve Fiber Layer Thickness and Visual Function,” Investig. Ophthalmol. & Vis. Sci. 42, 1993–2003 (2001). [PubMed] [Google Scholar]
- 6.Tatham A. J., Medeiros F. A., “Detecting Structural Progression in Glaucoma with Optical Coherence Tomography,” Ophthalmol. 124, S57–S65 (2017). 10.1016/j.ophtha.2017.07.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Aranha dos Santos V., Schmetterer L., Gröschl M., Garhofer G., Schmidl D., Kucera M., Unterhuber A., Hermand J.-P., Werkmeister R. M., “In vivo tear film thickness measurement and tear film dynamics visualization using spectral domain optical coherence tomography,” Opt. Express 23, 21043 (2015). 10.1364/OE.23.021043 [DOI] [PubMed] [Google Scholar]
- 8.Aranha dos Santos V., Schmetterer L., Triggs G. J., Leitgeb R. A., Gröschl M., Messner A., Schmidl D., Garhofer G., Aschinger G., Werkmeister R. M., “Super-resolved thickness maps of thin film phantoms and in vivo visualization of tear film lipid layer using OCT,” Biomed. Opt. Express 7, 2650 (2016). 10.1364/BOE.7.002650 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schmidl D., Witkowska K. J., Kaya S., Baar C., Faatz H., Nepp J., Unterhuber A., Werkmeister R. M., Garhofer G., Schmetterer L., “The Association Between Subjective and Objective Parameters for the Assessment of Dry-Eye Syndrome,” Investig. Ophthalmol. & Vis. Sci. 56, 1467–1472 (2015). 10.1167/iovs.14-15814 [DOI] [PubMed] [Google Scholar]
- 10.Adler F. H., Kaufman P. L., Levin L. A., Alm A., Adler’s Physiology of the Eye (Elsevier; Health Sciences, 2011). [Google Scholar]
- 11.Ang M., Baskaran M., Werkmeister R. M., Chua J., Schmidl D., Aranha dos Santos V., Garhöfer G., Mehta J. S., Schmetterer L., “Anterior segment optical coherence tomography,” Prog. Retin. Eye Res. (2018). 10.1016/j.preteyeres.2018.04.002 [DOI] [PubMed]
- 12.Werkmeister R. M., Sapeta S., Schmidl D., Garhöfer G., Schmidinger G., Santos V. A. D., Aschinger G. C., Baumgartner I., Pircher N., Schwarzhans F., Pantalon A., Dua H., Schmetterer L., “Ultrahigh-resolution OCT imaging of the human cornea,” Biomed. Opt. Express 8, 1221–1239 (2017). 10.1364/BOE.8.001221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pircher N., Schwarzhans F., Holzer S., Lammer J., Schmidl D., Bata A. M., Werkmeister R. M., Seidel G., Garhöfer G., Gschließer A., Schmetterer L., Schmidinger G., “Distinguishing Keratoconic Eyes and Healthy Eyes Using Ultrahigh-Resolution Optical Coherence Tomography–Based Corneal Epithelium Thickness Mapping,” Am. J. Ophthalmol. 189, 47–54 (2018). 10.1016/j.ajo.2018.02.006 [DOI] [PubMed] [Google Scholar]
- 14.McMonnies C. W., “Corneal endothelial assessment with special references to keratoconus,” Optom. Vis. Sci. Off. Publ. Am. Acad. Optom. 91, e124–e134 (2014). 10.1097/OPX.0000000000000271 [DOI] [PubMed] [Google Scholar]
- 15.LaRocca F., Chiu S. J., McNabb R. P., Kuo A. N., Izatt J. A., Farsiu S., “Robust automatic segmentation of corneal layer boundaries in SDOCT images using graph theory and dynamic programming,” Biomed. Opt. Express 2, 1524–1538 (2011). 10.1364/BOE.2.001524 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Williams D., Zheng Y., Davey P. G., Bao F., Shen M., Elsheikh A., “Reconstruction of 3d surface maps from anterior segment optical coherence tomography images using graph theory and genetic algorithms,” Biomed. Signal Process. Control. 25, 91–98 (2016). 10.1016/j.bspc.2015.11.004 [DOI] [Google Scholar]
- 17.Williams D., Zheng Y., Bao F., Elsheikh A., “Automatic segmentation of anterior segment optical coherence tomography images,” J. Biomed. Opt. 18, 056003 (2013). 10.1117/1.JBO.18.5.056003 [DOI] [PubMed] [Google Scholar]
- 18.Keller B., Draelos M., Tang G., Farsiu S., Kuo A. N., Hauser K., Izatt J. A., “Real-time corneal segmentation and 3d needle tracking in intrasurgical OCT,” Biomed. Opt. Express 9, 2716–2732 (2018). 10.1364/BOE.9.002716 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li Y., Shekhar R., Huang D., “Corneal Pachymetry Mapping with High-speed Optical Coherence Tomography,” Ophthalmol. 113, 792 (2006). 10.1016/j.ophtha.2006.01.048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Schmoll T., Unterhuber A., Kolbitsch C., Le T., Stingl A., Leitgeb R., “Precise thickness measurements of Bowman’s layer, epithelium, and tear film,” Optom. & Vis. Sci. 89, E795–E802 (2012). 10.1097/OPX.0b013e3182504346 [DOI] [PubMed] [Google Scholar]
- 21.Jahromi M. K., Kafieh R., Rabbani H., Dehnavi A. M., Peyman A., Hajizadeh F., Ommani M., “An Automatic Algorithm for Segmentation of the Boundaries of Corneal Layers in Optical Coherence Tomography Images using Gaussian Mixture Model,” J. Med. Signals Sensors 4, 171–180 (2014). [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang T., Elazab A., Wang X., Jia F., Wu J., Li G., Hu Q., “A Novel Technique for Robust and Fast Segmentation of Corneal Layer Interfaces Based on Spectral-Domain Optical Coherence Tomography Imaging,” IEEE Access 5, 10352–10363 (2017). 10.1109/ACCESS.2017.2712767 [DOI] [Google Scholar]
- 23.LeCun Y., Bengio Y., Hinton G., “Deep learning,” Nature 521, 436–444 (2015). 10.1038/nature14539 [DOI] [PubMed] [Google Scholar]
- 24.Roy A. G., Conjeti S., Karri S. P. K., Sheet D., Katouzian A., Wachinger C., Navab N., “ReLayNet: retinal layer and fluid segmentation of macular optical coherence tomography using fully convolutional networks,” Biomed. Opt. Express 8, 3627–3642 (2017). 10.1364/BOE.8.003627 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lee C. S., Baughman D. M., Lee A. Y., “Deep Learning Is Effective for Classifying Normal versus Age-Related Macular Degeneration OCT Images,” Ophthalmol. Retin. 1, 322–327 (2017). 10.1016/j.oret.2016.12.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee C. S., Tyring A. J., Deruyter N. P., Wu Y., Rokem A., Lee A. Y., “Deep-learning based, automated segmentation of macular edema in optical coherence tomography,” Biomed. Opt. Express 8, 3440–3448 (2017). 10.1364/BOE.8.003440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Fang L., Cunefare D., Wang C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8, 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Devalla S. K., Chin K. S., Mari J.-M., Tun T. A., Strouthidis N. G., Aung T., Thiéry A. H., Girard M. J. A., “A Deep Learning Approach to Digitally Stain Optical Coherence Tomography Images of the Optic Nerve Head,” Investig. Ophthalmol. & Vis. Sci. 59, 63–74 (2018). 10.1167/iovs.17-22617 [DOI] [PubMed] [Google Scholar]
- 29.Venhuizen F. G., Ginneken B. V., Liefers B., Asten F. V., Schreur V., Fauser S., Hoyng C., Theelen T., Sánchez C. I., “Deep learning approach for the detection and quantification of intraretinal cystoid fluid in multivendor optical coherence tomography,” Biomed. Opt. Express 9, 1545–1569 (2018). 10.1364/BOE.9.001545 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chollet F., Keras (2015).
- 31.Cover T. M., Thomas J. A., Elements of information theory(John Wiley & Sons, 2012). [Google Scholar]
- 32.Long J., Shelhamer E., Darrell T., “Fully convolutional networks for semantic segmentation,” in Proceedings of the IEEE conference on computer vision and pattern recognition, (2015), pp. 3431–3440. [DOI] [PubMed] [Google Scholar]
- 33.Ronneberger O., Fischer P., Brox T., “U-net: Convolutional networks for biomedical image segmentation,” in International Conference on Medical image computing and computer-assisted intervention, (Springer, 2015), pp. 234–241. [Google Scholar]
- 34.Badrinarayanan V., Kendall A., Cipolla R., “Segnet: A deep convolutional encoder-decoder architecture for image segmentation,” arXiv preprint arXiv:1511.00561 (2015). [DOI] [PubMed]
- 35.He K., Gkioxari G., Dollár P., Girshick R., “Mask r-cnn,” in Computer Vision (ICCV), 2017 IEEE International Conference on, (IEEE, 2017), pp. 2980–2988. [Google Scholar]
- 36.Chen L.-C., Papandreou G., Kokkinos I., Murphy K., Yuille A. L., “Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs,” IEEE transactions on pattern analysis and machine intelligence 40, 834–848 (2018). 10.1109/TPAMI.2017.2699184 [DOI] [PubMed] [Google Scholar]
- 37.Apostolopoulos S., Zanet S. D., Ciller C., Wolf S., Sznitman R., “Pathological OCT Retinal Layer Segmentation Using Branch Residual U-Shape Networks,” in Medical Image Computing and Computer-Assisted Intervention - MICCAI 2017, (Springer, Cham, 2017), Lecture Notes in Computer Science, pp. 294–301. 10.1007/978-3-319-66179-7_34 [DOI] [Google Scholar]
- 38.Ioffe S., Szegedy C., “Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift,” arXiv:1502.03167 [cs] (2015). ArXiv: 150203167. [Google Scholar]
- 39.LeCun Y., Bottou L., Orr G. B., Müller K.-R., “Efficient BackProp,” in Neural Networks: Tricks of the Trade, This Book is an Outgrowth of a 1996 NIPS Workshop, (Springer-Verlag, London, UK, UK, 1998), pp. 9–50. [Google Scholar]
- 40.Ruder S., “An overview of gradient descent optimization algorithms,” arXiv:1609.04747 [cs] (2016). ArXiv: 160904747. [Google Scholar]
- 41.Abadi M., Barham P., Chen J., Chen Z., Davis A., Dean J., Devin M., Ghemawat S., Irving G., Isard M., “Tensorflow: a system for large-scale machine learning,” in OSDI, vol. 16 (2016), pp. 265–283. [Google Scholar]
- 42.Goodfellow I., Bengio Y., Courville A., Bengio Y., Deep learning, vol. 1 (MIT press; Cambridge, 2016). [Google Scholar]
- 43.Jastrzębski S., Kenton Z., Arpit D., Ballas N., Fischer A., Bengio Y., Storkey A., “Three Factors Influencing Minima in SGD,” arXiv:1711.04623 [cs, stat] (2017). ArXiv: 171104623. [Google Scholar]
- 44.Drexler W., Hitzenberger C. K., Baumgartner A., Findl O., Sattmann H., Fercher A. F., “Investigation of dispersion effects in ocular media by multiple wavelength partial coherence interferometry,” Experimental Eye Research 66, 25–33 (1998). 10.1006/exer.1997.0401 [DOI] [PubMed] [Google Scholar]
- 45.Glorot X., Bordes A., Bengio Y., “Deep sparse rectifier neural networks,” in Proceedings of the fourteenth international conference on artificial intelligence and statistics, (2011), pp. 315–323. [Google Scholar]