Abstract
Transmembrane voltage and intracellular calcium concentration are coupled parameters essential to the function of neurons, cardiomyocytes, and other excitable cells. Here we introduce the Firefly-HR microscope for simultaneous optogenetic stimulation and voltage and calcium imaging with fluorescent proteins using three spectrally distinct visible color bands. Firefly-HR combines patterned stimulation, near-total internal reflection laser excitation through a prism located between the sample and a water-immersion objective, and concurrent imaging of three color channels. The microscope has efficient light collection, low fluorescent background, and a large field of view (0.24 x 1.2 mm @ 1000 frames/sec). We characterize optical crosstalk and demonstrate capabilities with three applications: (1) probing synaptically connected neuronal microcircuits, (2) examining the coupling between neuronal action potentials and calcium influx, and (3) studying the pharmacology of paced human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs) via simultaneous recordings of voltage, calcium, and contraction.
Introduction
Recent decades have seen a rapid expansion in the number of protein-based optical tools for exploring cellular systems, both optogenetic actuators [1,2] and fluorescent sensors [3–6]. One research focus has been the development of constructs for optical electrophysiology, both manipulation of transmembrane potential by light-gated ion channels and light driven pumps [7–9], and the recording of transmembrane potential by fluorescent voltage sensors [10]. Combining tools yielded the all-optical electrophysiology platform Optopatch, which pairs the blue-light gated channelrhodopsin CheRiff with the red-light excited voltage sensor QuasAr [11]. Optopatch can recapitulate many traditional patch clamp measurements, but at a much higher throughput.
In parallel with developments in voltage sensors and actuators, there have been huge strides in fluorescent calcium-sensing proteins, which have been a workhorse for in vivo imaging in neuroscience. In addition to the canonical GFP-based calcium sensors [12], there have been numerous developments making red-shifted calcium reporters [13–16]. Here, we pair the Optopatch platform with orange cytosolic calcium sensor jRGECO1a [15] (Fig. 1(A)). Optopatch uses blue for CheRiff stimulation and the red and near-infrared for voltage readout, leaving a window in the yellow-orange portion of the visible spectrum for jRGECO1a (Fig. 1(F)). It is natural to pair calcium and voltage recordings, as calcium often converts electrical signaling between cells into a local, intracellular signal. Recording both in tandem opens a window in to key cellular processes such as synaptic plasticity [17], activity-dependent regulation of transcription in neurons [18], and excitation-contraction coupling in myocytes [19].
Fig. 1.
Firefly-HR design. (A) The proteins involved in the measurement system: Membrane bound, light-gated ion channel CheRiff is opened by 470 nm light. Cytosolic calcium sensor jRGECO1a is excited by 560 nm light. Membrane bound voltage sensor QuasAr is excited by 638 nm light. (B) An optical diagram of the microscope. There are four key light sources: 1) Patterned blue light reflected off the digital micromirror device (DMD) for e.g. targeted stimulation of individual neurons. 2-3) 638 & 560 nm lasers for at near-total internal reflection (TIR) for fluorescence excitation of QuasAr and jRGECO1a. 4) Green light in trans-illumination for measuring cardiac contractions. Two synchronized cameras record the three light sources simultaneously. The top camera is designated as a brightfield collector, while the bottom camera is designated as a fluorescent collector. (C) An expanded view of prism. The water-immersion objective is optically coupled to the bottom of the prism with water, and the prism is optically coupled to the glass-bottomed dish with immersion oil. This geometry enables TIR or near TIR illumination over a large field of view, does not rout intense lasers through the objective, and maintains the high numerical aperture of 1.0. “1” and “2”, which are reproduced in (E), are the objective access and laser access apertures, respectively. (D) The prism design. (E) The design of the prism plate that prevents oil and water from mixing or obscuring the laser path. (F) Spectra. JRGECO1A spectrum is mApple, and filters are from Semrock. Optopatch component spectra from [11].
Customized optics are required to implement Optopatch because of the fast frame rate necessary for voltage imaging and because the voltage-sensing protein QuasAr is ~200-fold dimmer than GFP [11,20]. We build on optical design element from the Firefly microscope, designed for high-throughput screening applications with Optopatch, which can simultaneously stimulate and record action potentials in hundreds of neurons [21]. We dub the new microscope Firefly-HR (high resolution). Firefly-HR pairs patterned optogenetic stimulation with simultaneous voltage and calcium recordings from tens of neurons in parallel at a 1 kHz frame rate with a high signal-to-noise ratio (SNR).
Here, we first show the optical and mechanical design of Firefly-HR (Fig. 1) and characterize its optical performance (Fig. 2). We then demonstrate capabilities in several different systems. The first demonstration highlights SNR and patterned stimulation in measurements of synaptic transmission (Fig. 3). Next, we consider optical crosstalk when adding calcium imaging with jRGECO1a (Fig. 4). We then show optogentetic stimulation paired with voltage and calcium imaging in neurons (Fig. 5) and examine how simultaneous voltage and calcium signals can be used to access biological information unavailable with either recording alone. We finish with optical pacing and simultaneous imaging of motion, voltage, and calcium in cardiomyocytes (Fig. 6), and show how distinct facets of cardiac function are targeted by different pharmacological modulators.
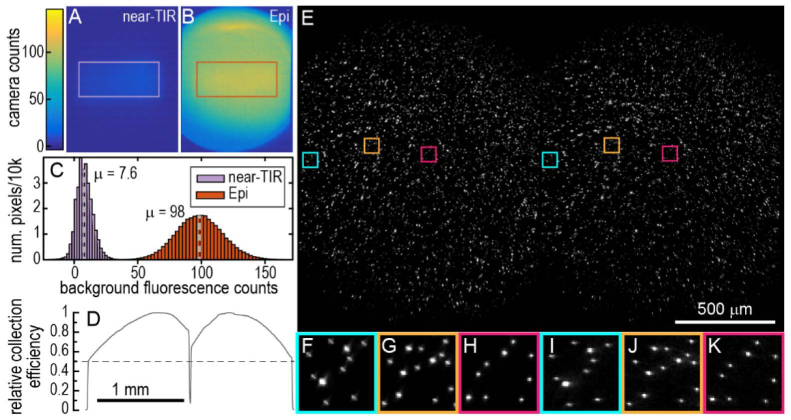
Fig. 2.
Optical characterization. Images in the QuasAr fluorescence channel of a pristine, empty cyclic olefin copolymer (COC) substrate illuminated by a 638 nm laser routed (A) through the prism at near-TIR and (B) through the objective in traditional Epi illumination. For both light paths, the 638 nm laser was apertured to illuminate exactly the same area, and the laser power in both paths was tuned to match illumination intensities. Counts in the Epi sample were scaled so that the median of measured bead intensities in a separate sample (after background subtraction) matched between Epi and near-TIR illumination. In near-TIR illumination, there is a small amount of background from the immersion oil and substrate. In Epi illumination, there is a much larger amount of out-of-focus autofluorescence from glass in the objective, still present when the sample is removed entirely. (C) The histogram of pixel intensities after background subtraction from the rectangles shown in A and B show 13x lower autofluorescence in the near-TIR illumination geometry. (D) The light collection efficiency of the 2 fluorescence imaging pathways [QuasAr left, GFP right]. A relatively homogeneous fluorescent orange paper sample was illuminated by an oversized, homogeneous LED source from above. Images were captured at many locations within the paper and averaged to minimize effects of sample inhomogeneity. The lineout reveals some light loss from the objective where the colors are still colinear and some light loss in the dual-view path after the colors have been separated. (E) A dualview image of 1 um Tetraspeck beads emitting multiple fluorescent colors is captured on the bottom camera (of Fig. 1(B)). The image spans the full camera width and shows the 1.2 mm wide FOV for both the QuasAr channel (left) and the GFP channel (right). The dichroic reflecting green light to the first camera was removed. Each half of the image has been independently background subtracted and contrasted. Expanded views of indicated sub-regions in (E) for red (F - H) and green (I - K) fluorescence channels. Expanded images from each fluorescence channel are shown with the same brightness and contrast values.
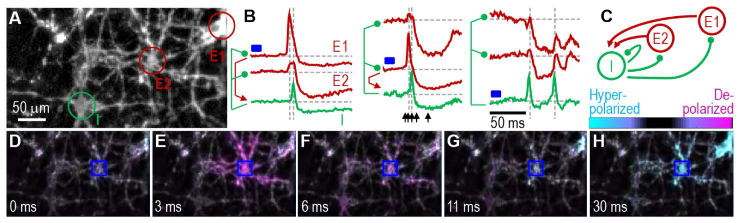
Fig. 3.
Synaptic microcircuit. Neurons were collected from an E18 rat hippocampus and cultured for 14 days. (A) QuasAr fluorescence image of excitatory cells E1 and E2 connected in a microcircuit with inhibitory cell I. (B) The digital micromirror device (DMD) is used to stimulate single cell individually, indicated by the blue rectangle, while recording from the other 2. The left of each panel shows inferred connectivity for excitatory (red) and inhibitory (green) synaptic coupling. (C) A diagram of microcircuit connectivity determined from the functional recordings in B. (D) – (H) Frames from the video where E2 is stimulated (blue square), at times indicated by black arrows in B. (D) Before cell E2 fires. (E) The stimulated cell E2 fires, which triggers (F) cell I to fire. (G) & (H) Cell I induces inhibitory post synaptic potentials in cells E1 and E2.
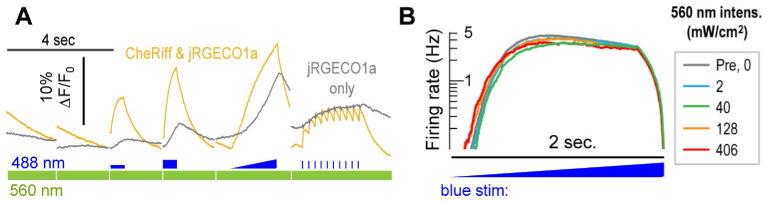
Fig. 4.
Optical Crosstalk. (A) The average fluorescence from 19 FOV’s of jRGECO1a-expressing neurons with (gold) and without (gray) CheRiff expression. Blue light photoconverts jRGECO1a into a brighter fluorescent state as shown in the blue stimulated portions of the gray trace. 560nm laser intensity: 80 mW/cm2. 488nm illumination intensities: 10 – 100 mW/cm2. At higher blue stimulation intensities, the optical crosstalk can exceed the true calcium signal, where the true calcium signal is calculated by the difference between the yellow trace and the gray trace. Direct blue-light induced fluorescence, captured with the 560nm laser off, is negligible. (B) 560 nm laser-induced stimulation of the CheRiff. The average firing rate from at least 780 neurons/condition is shown before any intervention (gray) and after illuminating the cells with different intensities of yellow light in response to a slow ramped blue stimulus. The highest 560 nm intensity of 406 mW/cm2 is the largest we use in any experiment. There may be a slight change in the blue light intensity to induce the first action potential at the stronger yellow illumination intensities, but it does not rise to statistical significance and typically one can ignore this effect.
Fig. 5.
Simultaneous voltage and calcium imaging in neurons. (A) QuasAr fluorescence from an example field of view. (B) jRGECO1a fluorescence from the same field, imaged on the other half of the camera. (C) A color overlay after image registration. (D) The pixel weights used to calculate the time traces for 5 neurons in this FOV. (E) Synchronized voltage and calcium recordings from the cells labeled in D. The blue light stimulus of CheRiff is shown below. (F) A raster plot from 47 FOVs in 3 dishes with no pharmacological modulation. Each row in 1 cell and each point is one action potential. (G) The calcium traces from the same cells. (H) The average spike rate for all cells treated with 30 μM 4-AP (green), 10 μM isradipine (purple), or vehicle control (blue). (I) The corresponding average calcium time traces. The voltage action potential waveform averaged over all spikes during (J) spontaneous recording and (K) all epochs excluding the comb. (L) – (N) For each cell, the integrated calcium signal is plotted against the total number of spikes. Although the change is not statistically significant, the changes in slope suggest that the calcium flux per action potential may change. (O) The number of spiking neurons per FOV, demonstrating throughput.
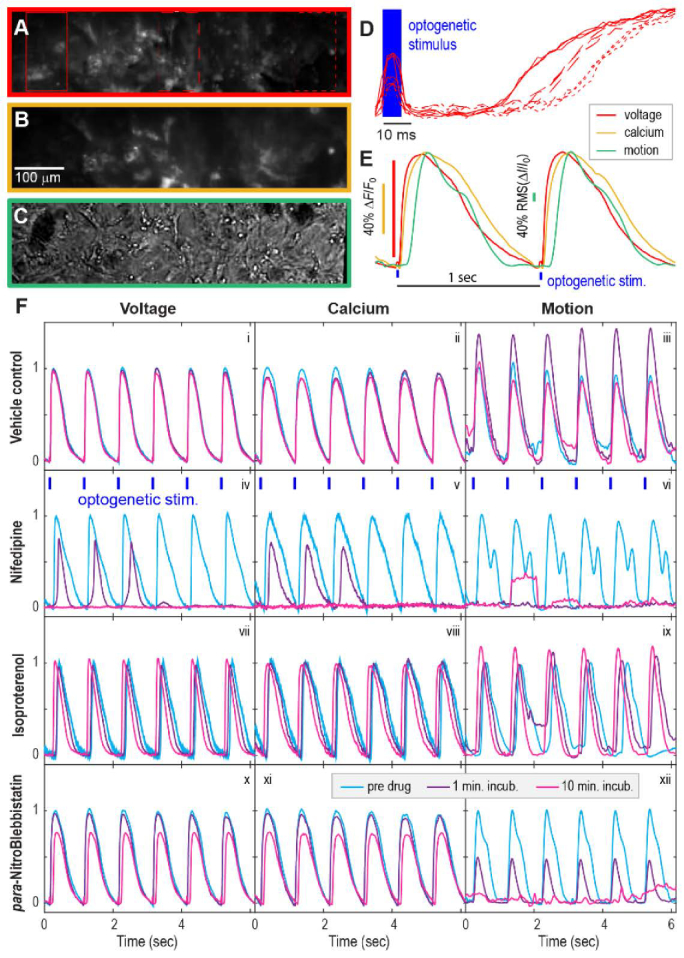
Fig. 6.
Tri-functional recordings in human iPS-derived cardiomyocytes. (A) Voltage and (B) calcium fluorescent recordings were captured on one camera, and (C) motion was captured on a second, synchronized camera. The optogenetic stimulus was delivered from above with a 470 nm LED offset to stimulate left of the FOV (not shown in Fig. 1(B)). The action potential propagated from left to right with a velocity of ~20 μm/ms. (D) Multiple beats from the regions in A are aligned in time to the optogenetic stimulus. The good overlap shows the highly reproducible conduction velocity. (E) Simultaneous voltage, calcium, and motion recordings. Motion was calculated as the root-mean-square deviation from the average baseline image. As an action potential enters the FOV, the voltage deflection precedes the calcium transient, which precedes motion, as expected. (Fi - Fxii) Pharmacology in the tri-view system. Each FOV was measured three times: before drug application (blue), 1 min after drug application (purple), and 10 min. after drug application (magenta). Columns show functional readouts and rows correspond to different drugs. After application of a vehicle control (0.1% DMSO), voltage and calcium changed minimally upon re-imaging while motion showed an unusual, temporary change. Calcium channel blocker nifedipine (0.1 μM) drastically changed AP and CT shapes after 1 min treatment and eliminated firing part way through the recording. Motion was stopped before electrical activity ceased. β-adrenergic receptor agonist isoproterenol (0.1 μM) increased conduction velocity and contraction strength after 10 min. It also increased spontaneous beat rate (data not shown). para-Nitroblebbistatin (20 μM), a photostable myosin II inhibitor [30], had relatively small effects on AP’s and CT’s but drastically reduced, and then eliminated, contraction.
Protein constructs
For Optogenetic stimulation, we use the channelrhodopsin CheRiff, with peak photocurrent at ~460 nm and a larger photocurrent than the widely used ChR2 [11]. We use the voltage sensor QuasAr3 [22], which is similar to QuasAr2 [11] but with additional amino acid sequences that improve trafficking to the neuronal membrane. We tested three orange calcium sensors: R-CaMP2 [14], jRGECO1a, and jRCaMP1a [15]. Consistent with literature results, we found that jRGECO1a was clearly the most sensitive for detecting the calcium transients from individual action potentials, so used it in all measurements below.
Firefly-HR design and characterization
For all-optical electrophysiology measurements using Optopatch, we had previously worked with a more traditional, high-magnification microscope system [23] (Table 1, Col. 1) and the low-magnification Firefly microscope [21] (Table 1, Col. 3). The high-magnification system makes high signal-to-noise ratio (SNR) recordings due to the high light collection efficiency and can record from two fluorescence channels simultaneously, but it can only record from a few neurons simultaneously because of the relatively small field of view (FOV). The Firefly microscope can record from ~100 neurons simultaneously with sufficient SNR for reliable action potential (AP) detection in primary rat pyramidal neurons, and so is the preferred instrument for applications where throughput is paramount, such as phenotypic drug screening. However, the SNR is marginal for detecting APs in some small, human induced pluripotent stem (iPS) cell derived inhibitory neurons and for recording post-synaptic potentials (PSPs) resulting from synaptic signaling between pairs of neurons. On the Firefly, it is also difficult to implement multicolor fluorescence recordings because of the large etendue of the fluorescence light and the requirement for very large optics. To address these limitations, we introduce the intermediate magnification Firefly-HR (Table 1, Col. 2), which enables multicolor fluorescence recordings for simultaneously tracking voltage and calcium during optogenetic stimulation, has an FOV large enough to explore neuronal circuits in vitro, and has excellent SNR. In addition to QuasAr and jRGECO1a fluorescence channels, it can record trans-illumination in the green channel for tracking contraction in myocytes. Figure 1 shows optical and mechanical designs.
Table 1. Microscope Specifications.
| Spec. | “high-mag.” microscope | Firefly-HR | Firefly |
|---|---|---|---|
| Objective Mag. | 60x | 20x | 2x |
| Microscope Mag. | 15x | 5x | 2x |
| Immersion fluid | Oil | Water | Air |
| NA | 1.5 | 1.0 | 0.5 |
| Illumination geometry | Through-obj. near-TIR | Through-prism near-TIR | Through-prism near-TIR |
| FOV @ 1 kHza | 0.08 x 0.36 mm | 0.24 x 1.2 mm | 0.6 x 6 mm |
| Resolutionb | 0.4 μm | 1.3 μm | 3.3 μm |
| Multicolor readout | Potentially | Yes | No |
sCMOS can record ~200 rows of pixels at a 1 kHz frame rate.
Camera pixel size (6.5 μm) divided by magnification.
Through-prism, near TIR red and yellow illumination
One of the unique design features of Firefly-HR is laser illumination through a prism to implement near total internal reflection (TIR) shown in Fig. 1(B)-1(E), which is essential to minimize background autofluorescence. Background fluorescence is particularly important in our application for 3 reasons: 1) QuasAr2 is ~200x dimmer than EGFP [11,20], so the background autofluorescence is much larger relative to the signal than for typical fluorescent protein imaging. 2) We image a large area, so more laser power is required, which can produce significant out-of-focus autofluorescence from glass in the objective. 3) We have short, 1 millisecond exposure times to capture voltage dynamics, so high laser powers are required, again causing out-of-focus background autofluorescence. The prism is made from low-autofluorescence fused silica, and the sample is grown on cyclic-olefin copolymer (COC) coverslip-bottomed dishes, whose background autofluorescence is many times lower than typical glass coverslips (data not shown). The prism is optically coupled to the coverslip by an index-matched, low-autofluorescence oil (Olympus Type F). At the near-TIR angle of incidence, the beam refracts at the COC/buffer interface and propagates nearly horizontally (Fig. 1(C)), parallel to the COC surface. This refraction compresses the beam, and in spite of reflection losses near the critical angle, the beam intensity is increased roughly 4-fold [21], reducing the total laser power required. The beam rapidly propagates out of the FOV, minimizing out-of-focus fluorescence from the imaging buffer. This excitation geometry reduces background autofluorescence by ~13-fold (Fig. 2(A)-2(C)).
To have the advantages of the through-prism near-TIR illumination, but also to leverage the high numerical aperture of the water immersion objective, we built a prism holder that enabled oil coupling above and water coupling below the prism (Fig. 1(C) and 1(E)). The 45°-angled facets of the prism through which the laser entered was isolated from immersion fluids above and below the prism and kept dry. An 8 W, 638 nm laser (DILAS/Coherent, M1B-638.3-8C-SS4.3-T3) for QuasAr excitation and a 300 mW, 560 nm laser (MPB, VFL-P-300-560-OEM1) for jRGECO1a excitation were re-shaped and combined. Each laser could be modulated by a fast shutter (Uniblitz, DSS10B1ZM0L). Lasers were routed through an orthogonal pair of slits to create a homogeneous illumination rectangle with tunable width and height, which was imaged onto the sample, and tuned in size to match the FOV. The final mirror before the sample was located on a vertical translation stage in the Fourier plane of the final lens, which decoupled degrees of freedom: position in the sample plane was set by angular adjustment and the incidence angle was tuned around TIR with the translation stage.
Imaging path
The full path is diagrammed in Fig. 1(B). Starting from the sample plane, all three imaging wavelengths (brightfield, jRGECO1a, and QuasAr) are transmitted through the fused silica prism (Fig. 1(D)) to the 20x water immersion objective (Zeiss, 421452-9900), which is positioned so the image of the sample is formed at infinity in the absence of additional lenses. Immersion oil between the prism and sample and water between the objective and prism ensure that the objective’s full numerical aperture is utilized. A tube lens forms an intermediate image plane at the slit; the slit imposes sharp edges on the image, so QuasAr and jRGECO1a images can be nearly touching on the camera. After a lens located 1 focal length from the slit, a dichroic mirror (Chroma, ZT488/640rpc) reflects the collimated green brightfield light while transmitting JRGECO1A and QuasAr fluorescence. The brightfield light passes through a bandpass filter (Semrock, FF01-542/20) and is imaged onto a Hamamatsu ORCA-Flash4.0 sCMOS camera. A second dichroic mirror (Semrock, FF640-FDi01-25x36) separates jRGECO1a and QuasAr fluorescence, each of which is filtered (Semrock, FF01-600/37; FF02-736/128). The fluorescence is recombined by a second dichroic identical to the splitter and imaged onto the camera with a 50 mm photography lens (Zeiss, Zeiss Makro-Planar T* 50mm f/2 ZF.2 Lens). Optics in the infinity space with separated QuasAr and jRGECO1a fluorescence were carefully positioned and aligned to minimize clipping; because of the large spatial extent of the FOV and high angular collection efficiency of the objective, fluorescent light has a large spatial and angular content. In particular, QuasAr and jRGECO1a fluorescence were overlapped at the final lens pupil and incident with equal and opposite angles. QuasAr and jRGECO1a fluorescence were mapped to camera left and right (e.g Fig. 2(E)). The total magnification of the imaging system was 5x, which enabled us to map essentially the full, 1.2 mm wide voltage and calcium images from the objective side-by-side on the camera. The low magnification also fits more area into the central 200 rows of pixels that can be read out from the sCMOS chip at 1 kHz. The magnification cannot be reduced further without sacrificing detection efficiency because the sCMOS chip and microlens array in front of it start to lose light beyond and incidence angle of ± 15°.
The two cameras can be synchronized by triggering each frame in the movie, “synchronous” mode on the camera, which locks the acquisition time of each frame. Camera synchronization and modulation of light sources was controlled by a data acquisition card (National Instruments, NI PCIe-6323).
Light collection efficiency across the FOV was characterized with a homogeneous fluorescent sample (Fig. 2(D)). The resulting efficiency readout results from collection efficiency of the objective (narrower peaked curves within each color) multiplied with the efficiency of optics after fluorescence colors are separated, predominantly the final lens. The ~2x reduction in light collection at the image edges results in a degradation in SNR due to shot noise, but strong signals can still easily be detected. The effects of optical aberration are shown in Fig. 2(E)-2(K). 1 μm Tetraspeck beads, which absorb and emit across the visible spectrum, were imaged. Pixelation-limited resolution was achieved at the center of each color channel (Fig. 2(H) and 2(K)). Although resolutions degraded slightly towards the image edge (Fig. 2(F) and 2(I)), it was still sufficient for functional fluorescence imaging. The optical system provides high-sensitivity, multi-color imaging over a large FOV.
Patterned stimulation
A 488 nm laser (Coherent OBIS 488 LX 50 mW) was routed through a poly chromatic acousto optic modulator (Gooch & Housego, 48062-1.0-0.55-1W) for rapid analog modulation, expanded, and collimated before reflecting of a digital micromirror device (DMD; Texas Instrument, DLP3000). The beam overfilled the DMD so blue light intensity was flat across the pixel array. The DMD was imaged onto the sample, so any image displayed on the DMD was projected onto the cells. The maximum laser intensity at the sample was 600 mW/cm2, which opens >90% of CheRiff channels and achieves near-maximal photocurrent [11].
Methods
Rat hippocampal neuronal culture
Cells were plated on either Ø10 mm glass bottom dishes (MatTek, # P35G-1.5-10-C), or cyclic olefin copolymer (COC) bottomed dishes in a single well (Ibidi, # 81156) or 8-well (Ibidi, # 80826) format. The COC dishes had lower autofluorescence and slightly improved optical performance. Prior to cell plating, the imaging area of each dish was coated with Poly-D-Lysine (Sigma Aldrich, # P6407-10X5MG; 100 μg/mL, overnight incubation at 4°C), rinsed twice with PBS, and coated with Laminin (Thermofisher Scientific, #23017-015; 20 μg/mL, overnight at 4°C or 2 hours at room temperature). E18 rat hippocampal tissue, maintained at 4°C between dissection and dissociation (Brain Bits, # SDEHP), were dissociated with papain following the manufacturer protocol for neuron production, pelleted (200g, 1 min), and re-suspended in rat neuron feeding media [475 mL Neurobasal-A (Thermofisher, #10888022), 5 mL 100x Glutamax (Thermofisher, #35050061), 5 mL 5,000 IU Penicillin/ 5,000 µg/mL Streptomycin (Corning, #30-001-CI), 10 mL 50x B27 supplement (Thermofisher, #17504044), 5 mL 100x N2 supplement (Thermofisher, #17502048), 1 μg/mL Laminin (Thermofisher, #23017015), 10 ng/mL BDNF (R&D systems, #248-BD), 10 ng/mL GDNF (R&D systems, #212-GD)]. Neurons were plated at a density of 25k/cm2. For glass bottomed dishes, 70 μL of cell suspension was added to the recessed region dropwise. The following day, the media volume in each dish was increased to 1.5 mL with rat neuron feeding media. Cells were transduced on DIV6 with lentiviral vectors encoding optogenetic constructs. For dual view Ca/V measurements, 2 μL of hSyn-CheRiff- EBFP2, 10 μL of CaMKII-jRGECO1a, and 10 μL of CaMKII-QuasAr2-darkCitrine were used. For excitability measurements without the calcium sensor, we used the voltage sensor construct hSyn-QuasAr-Citrine for easy readout of expression levels and trafficking during culture. The virus mixture was diluted with rat neuron feeding media to a final volume of 70 μL/dish and added carefully to the recessed region of the dish after aspirating existing media. The virus was removed after 16 hours and the dish volume was returned to 1.5 mL feeding media. 40,000 rat cortical glia were added on DIV8 to each dish, which increased neuron viability, increased neurite outgrowth, and reduced cell clumping. 70 μL of glial suspension was added carefully to the recessed region of the dish after aspirating existing media, and the media volume was increased to 1.5 mL the following day. 48 hours before imaging, cell media was exchanged and supplemented with 100 nM retinal, an essential cofactor for rhodopsins, including CheRiff and QuasAr. Dishes were imaged on DIV13-15.
Rat cortical glial culture
Rat cortex tissues are obtained from Brain Bits (#PRcx Rat) and dissociated as per the manual provided with the tissue. The tissue is digested in Papain enzyme (Brain Bits, #PAP/HE), titurated, and plated in Glia media [500 mL NbAstro media (Brain Bits, #NbASTRO), 5 mL 5,000 IU Penicillin/ 5,000 µg/mL Streptomycin (Corning, #30-001-CI)] in T25 PDL coated flasks (VWR, #12777-090). They are cultured for 2 weeks, transitioning from the initial sparse plating to a confluent monolayer, with 90% media exchange every 4 days. After 2 weeks, the glial cells are trypsinized (VWR, #45000-660), aliquoted, frozen in freezing media [20% DMSO (Sigma Aldrich, # D2650-100ML) and 80% FBS (GE Healthcare, #SH30070.03HI)] and stored in liquid nitrogen for future use.
Lentivirus production
To prepare lentivirus with consistently high-quality batches, we expanded HEK293T cells (ATCC, #CRL-3216), and produce a large stock of cryovials (2.5M cells/vial in 40% DMEM, 10% DMSO, and 50% FBS). For each round of virus production, HEK cells were thawed and expand into 15 cm plates, passaging at 80-90% confluence. All culture was performed in DMEM10 [440 mL high-glucose DMEM (Life Technologies, #11995-073), 5 mL 100x Glutamax (Thermofisher, # 35050061), 50 mL fetal bovine serum (HyClone, #SH30071.02HI), and 5 mL 100x non-essential amino acids (Life Technologies, # 11140-050)]. Once cells reached 70-80% confluence they were transfected with second generation lentiviral plasmids. 20 μg of the plasmid containing the fluorescent construct, 25 μg of the mixed packaging and envelope plasmids PSPax2/pMD2.G (Cellecta, #CPCP-K2A), 500 μL of Opti-MEM reduced serum media (Thermofisher, #31985-070), and 180 μL of 1 mg/mL PEI transfection reagent (Polysciences, #24765-2) were mixed and incubated for 10 min. The solution was added to 25 mL DMEM10 and added to the 15 cm plate after aspiration of the old media. The virus-laden media was harvested 48-60 hours after transfection and centrifuged at 1500g for 5 min to pellet out any cell debris. The viral supernatant was sterile filtered using a 0.45 µm steriflip, and then concentrated 10x following the Lenti-X concentrator protocol (Clontech, #PT4421-2). The lentivirus was then aliquoted and stored at −80°C. The lentiviral stocks are qualitatively titered with Lenti-X GoStix from Clontech and quantitatively by qRT-PCR protocols. Viral genome titers of 109 to 1010 copies/mL are achieved routinely with functional titers ~50 times lower at ~108 IFU/mL.
iPSC-derived cardiomyocyte cell culture
Human induced pluripotent stem cell derived iCell cardiomyocytes were purchased from Cellular Dynamics (CDI no. CMC-100-110-001) and cultured following manufacturer recommendations. 35 mm MatTek dishes with 10 mm glass bottoms (MatTek no. P35G-1.5-10-C) were coated with 150 μL of 0.1% w/v gelatin and incubated at 37°C for one hour. The dishes were kept at room temperature for at least one hour prior to use. After thawing, cells were resuspended in plating medium (CDI no. CMM-100-110-001) and seeded at a density of 1.27 x 105 cells/cm2, calculated using the lot-specific plating efficiency. 72 μL of cell suspension was added to the central recessed region of the glass-bottomed dish dropwise. After 16-24 hours, 1.5 mL of plating medium was added to each dish. 48 hours post-plating, the dish was gently agitated to dislodge non-adherent cells, and the plating media was replaced with 1.5 mL of maintenance medium (CDI no. CMM-100-120-001). Media was exchanged every 2-3 days. On DIV 7, the cardiomyocytes were transduced with Optopatch vectors: 4μL of CMV-CheRiff-GFP (viral titer: 9 x 108 genome copies/mL), 4 μL of CMV-jRGECO1a (4 x 109 genome copies/mL), and 8 μL of CMV-QuasAr2-dark-mOrange2 (3 x 109 copies/mL). The three lentiviruses were combined and diluted with maintenance medium for a final volume of 70 μL/dish and added to the recessed region of the dish following complete dish aspiration. The virus was removed after 16 hours and the cells fed with 1.5 mL of fresh maintenance medium. 48 hours before imaging, the cell medium was supplemented with 100 nM retinal, a required co-factor for Optopatch rhodopsins. Cardiomyocytes were imaged on DIV14.
Neuronal imaging
Neurons were imaged in a buffer with minimal autofluorescence containing salts and sugars in identical concentration to the BrainPhys culture medium [24]. The formulation was (in mM): 121 NaCl, 4.2 KCl, 1.1 CaCl2, 1 MgSO4, 26.5 sodium gluconate, 0.5 Na2HPO4, 0.45 NaH2PO4, 10 HEPES, 2.5 glucose, 0.5 sodium pyruvate. Buffer was exchanged 15 minutes before imaging to allow equilibration, and imaging was performed at room temperature. For non-synaptic measurements, synaptic blockers were added to uncouple the cells and to obtain cell-autonomous excitability: 25 μM D-AP5 (Tocris, #0106; blocks NMDA channel), 10 μM NBQX (Tocris, #0373; blocks AMPA channel), 20 μM gabazine (Alomone Labs, #G-216; blocks GABA channel). Typical light illumination intensities were: 300 W/cm2 at 638 nm for QuasAr excitation, 80 mW/cm2 at 560 nm for jRGECO1a excitation, and 0 – 100 mW/cm2 at 488 nm for CheRiff stimulation.
Cardiomyocyte imaging
Cardiomyocytes were imaged in a buffer with minimal autofluorescence containing salts and sugars in concentration matched to the CDI culture medium. The formulation was (in mM): 129 NaCl, 5.3 KCl, 1.8 CaCl2, 0.8 MgSO4, 0.9 NaH2PO4, 25 HEPES, 10 D-( + )-Galactose, 1 sodium pyruvate. Buffer was exchanged 15 minutes before imaging to allow equilibration, and imaging was performed at 37°C. Typical light illumination intensities were: 5 W/cm2 at 638 nm for QuasAr excitation, 25 mW/cm2 at 560 nm for jRGECO1a excitation, 25 mW/cm2 at 488 nm for pacing with CheRiff, and 1 mW/cm2 at 530nm for brightfield imaging.
Optopatch recordings of synaptic signaling
As a first demonstration of microscope performance and SNR, we probe synaptic signaling using targeted stimulation with the DMD. Figure 3(A) shows QuasAr fluorescence image of excitatory cells E1 and E2 connected in a microcircuit with inhibitory cell I. Visualization 1 (438.7KB, avi) , Visualization 2 (601KB, avi) , and Visualization 3 (193KB, avi) are the corresponding movies. Recordings were made in imaging media without synaptic blockers. To probe microcircuit connections, we stimulated each individual neuron with the digital micromirror device (DMD) and observed the voltage responses of all 3 cells, as shown in the three panels of Fig. 3(B). In the left panel, cell E1 is stimulated, which in turn triggers cell I, which inhibits all three neurons. The potentials of all three neurons drop well below baseline after each action potential, and the duration of the voltage drop is longer than would be expected for an action potential afterhyperpolarization, suggesting inhibitory synaptic drive. Neuron I appears to form an autapse and inhibits itself. In the middle panel, cell E2 is stimulated, which triggers cell I, which again inhibits all three neurons. In the right panel, cell I is stimulated to fire 2 action potentials that lead directly to inhibition. Figure 3(C) summarizes the microcircuit connectivity based on the observations. Figure 3(D)-3(H) are selected frames (corresponding to black arrows in middle panel of Fig. 3(B)) from Visualization 2 (601KB, avi) . The stimulated neuron E2 first fires an action potential (Fig. 3(E)), which triggers an action potential in neuron I (Fig. 3(F)), which inhibits all three neurons (Fig. 3(H)).
Simultaneous optogenetic stimulation and voltage and calcium imaging
Characterization of optical crosstalk
Optical crosstalk must be carefully characterized when using optogenetic tools in concert because action spectra of actuators and the absorption and fluorescence spectra of sensors are broad and partially overlapped. There are 6 types of potential crosstalk: each engineered protein can interact with either of the other two. Crosstalk between CheRiff and QuasAr is small, and carefully quantified in the original Optopatch publication [11], so we will not discuss further here. A third crosstalk, QuasAr fluorescence in the jRGECO1a channel, is undetectable in our system. When excited by intense 638 nm laser light, QuasAr fluorescence, which must be lower in energy than the excitation light, is fully blocked by the jRGECO1a emission filter. QuasAr leakage in the jRGECO1a channel from excitation with the 560 nm laser is also negligible because of poor spectral overlap with the jRGECO1a emission filter and because QuasAr is much dimmer than jRGECO1a.
The three remaining types of crosstalk are non-negligible. Overlap of jRGECO1a fluorescence with the QuasAr channel is shown in Fig. 1(F). The jRGECO1a emission filter (Semrock, FF01-600/37) captures 48% of jRGECO1a’s emission spectrum while the QuasAr filter collects 12% of this emission spectrum (Semrock, FF02-736/128). To address this spectral leakage, we subtract 0.25x the jRGECO1a signal from the QuasAr signal. QuasAr raw counts are typically ~10x higher than jRGECO1a raw counts because we drive the QuasAr with intense 638 nm illumination to capture fast voltage dynamics, so even without subtraction the effect is generally small.
The most difficult of the 6 types of crosstalk is the known change in basal jRGECO1a fluorescence induced by blue light [15]. Quasi-instantaneous changes in jRGECO1a fluorescence from direct jRGECO1a excitation by the blue light are small relative to 560 nm-excited fluorescence and can be corrected because they directly follow the blue light intensity. However, long-lasting changes in jRGECO1a fluorescence levels, presumably caused a change in the quantum mechanical state, are comparable in magnitude to fluorescent changes from real calcium dynamics (Fig. 4(A)), particularly at high blue stimulus intensities. It is difficult to separate this optical cross-talk from calcium signaling, as they occur on similar timescales. During experiment design, care must be taken to avoid this confound. Strategies we have used include 1) Keeping the blue stimulus intensity low (e.g. Figure 4(A), first blue step). 2) Looking for drug-induced changes in behavior. Although crosstalk is present, any observed changes in jRGECO1a fluorescence will result from drug-induced changes in calcium levels (e.g. Figure 5(I)). 3) Spatially separating optogenetic stimulation and fluorescence recording. Spatial separation is possible because a remote, optogenetically triggered electrical signal can propagate through the electrically connected cardiomyocyte syncytium (e.g. Figure 6) or along neuronal axons and induce a response in the detection region.
Finally, we examined the possibility that the 560 nm laser light used to excite jRGECO1a could directly stimulate CheRiff through the long-wavelength tail of the action spectrum. The CheRiff photocurrent is at least 20x smaller at 560 nm than at its spectral peak at 470 nm [11], but blue stimuli can drastically perturb neuronal behaviors significantly lower than typical 560 nm imaging intensities. To probe whether the yellow light stimulates CheRiff and perturbs neuronal behavior, we examined the neuronal firing rate during ramped blue stimulus while cells were bathed in different intensities of 560 nm light (Fig. 4(B)). Cells were transduced with QuasAr and CheRiff only, and the average neuronal firing rate was calculated as shown in Fig. 4(B). Cells were illuminated with sequentially increasing intensities of 560 nm laser light. There may be a small shift in behavior at 128 and 406 mW/cm2, but the effect is small, indicating at most a modest stimulation of CheRiff by the jRGECO1a excitation light. For experiments below, yellow laser intensities were kept low to minimize perturbation: 80 mW/cm2 in neurons and 25 mW/cm2 in cardiomyocytes.
Simultaneous voltage and calcium imaging in neurons
Visualization 4 (10MB, avi) and Fig. 5 show synchronized voltage and calcium recording during optogenetic stimulation to probe neuronal excitability and calcium signaling. Rat hippocampal neurons were imaged after 14 days in culture. They were lentivirally transduced at day 7 with CheRiff, jRGECO1a, and QuasAr; expression of all three constructs was driven by the neuron-specific human synapsin promoter (see methods). As in Fig. 2(E), QuasAr and jRGECO1a fluorescence were imaged side by side onto Fig. 1(B)’s bottom camera (Fig. 5(A) and 5(B)). Image registration (Fig. 5(C)) was performed using an affine transformation; one transformation matrix was applied to the whole data set. The transformation was determined by manual control point selection.
To analyze the movies, the voltage image was first segmented into smaller spatial regions with a watershed algorithm, and then pixel weight maps and voltage time-traces were extracted from each region using principal component analysis/independent component analysis (PCA/ICA) [23]. This produces a pixel weight mask (e.g. colored blobs in Fig. 5(D)) and a voltage time trace (e.g. red traces in Fig. 5(E)) for each cell. Calcium time traces for each cell (e.g. orange traces in Fig. 5(E)) were calculated by multiplying each frame in the movie by the (affine transformed) pixel weight mask and averaging over space. Passing a single movie through these analytical steps generates Figs. 5(D) and 5(E), where 5 electrically active cells are detected within the FOV. In Fig. 5(E), each voltage spike (red) is synchronized to a blip in calcium signal (orange). Individual cytosolic calcium transients can be observed for lone action potentials, but faster spike trains result in larger calcium deflections where the contributions from individual action potentials are difficult to identify. Figure 5(F) displays the spike raster for data from ~250 neurons – each row corresponds to one cell and each point represents one action potential. Figure 5(G) shows the calcium “raster” with each row representing a single cell with yellow high calcium and blue low calcium. Action potentials are mostly confined to stimulus epochs, while the jRGECO1a signal extends long after firing has ceased.
Figures 5(H) and 5(I) show the average rate of action potentials and the average calcium signal, respectively. In response to a 500 ms step stimulus, vehicle control neurons (blue) initially fire rapidly and then the firing rate slows as neurons adapt. The corresponding average calcium trace shows matched, but slower dynamics.
The 3 colors correspond to different drug treatments: vehicle control (blue), potassium channel blocker 4-aminopyradine (4-AP, 30 μM, green), and calcium channel blocker isradipine (10 μM, purple). 4-AP is low potency (high μM), non-selective blocker of voltage gated potassium channels. These Kv channels are responsible for the fast downstroke after an action potential, returning the neuron voltage to baseline [25]. Here, 4-AP increases both the amplitude and width of recorded action potentials (Fig. 5(J)-5(K)). As a result, although 4-AP reduces the spike rate (Fig. 5(H)), the calcium signal is increased (Fig. 5(I)), presumably because of increased calcium influx during each action potential. Isradipine is a potent (low nM) and selective blocker of L-type (Cav1.x) voltage-gated calcium channels, which in neurons is responsible for much of the calcium influx during an action potential [26]. Isradipine reduced the spike rate (Fig. 5(H)), reduced the calcium influx (Fig. 5(I)), shrank the AP area, and reduced spike afterhyperpolarization (Fig. 5(K)), presumably due to an indirect effect on calcium-activated potassium channels.
We then examined whether we could detect changes in the calcium influx per spike, an important parameter in calcium-mediated intracellular signaling that cannot be detected from either voltage or calcium imaging alone. Figures 5(L)-5(N) show the number action potentials vs integrated jRGECO1a signal for each neuron (both signals exclude the final 10 pulse stimulus epoch). The integrated calcium signal is normalized to the average non-spiking vehicle control cell. Each scatter of points is fit with a line, the slope of which is a measure of calcium influx per action potential. 4-AP has a slope greater than vehicle, suggesting increased calcium flux/AP, while isradipine has a slope less than vehicle, suggesting decreased calcium flux/AP. The trends align with expectations for an action potential widener and calcium blocker, suggesting that simultaneous voltage and calcium imaging gives access to new physiological information.
Simultaneous voltage, calcium, and contraction recordings in cardiomyocytes
As a final application, we examined human induced pluripotent stem cell-derived cardiomyocytes (hiPSC-CMs). In addition to voltage and calcium, we tracked motion (green path in Fig. 1(B)), a third critical readout to assay excitation contraction coupling and cardiomyocyte function. Having all three readout modalities gives critical information for evaluating efficacy of cardioprotective drug candidates and toxicity of any candidate therapeutic.
To track motion, we positioned a green LED over the sample and recorded a phase image in the transmitted light. Contrast was created by slightly defocusing the camera. The green much weaker than fluorescence excitation light, so it did not appreciably stimulate CheRiff or excite the fluorescent sensors. To calculate the motion trace from the phase movie, we first average all phase images when the calcium recording is at baseline to calculate the at-rest, uncontracted image. Next, each frame in the phase movie was normalized by this image to get the fractional change in light intensity for each pixel. Finally, the root-mean-square (RMS) deviation of the fractional change was calculated for each frame, yielding a measure of average displacement from the resting position across the FOV (Fig. 6).
Cardiomyocytes were cultured for 14 days and transduced with lentivirus with CheRiff, jRGECO1a, and QuasAr (see methods). Cells were paced with 3 x 3 mm extended blue light illumination to the left of the FOV. From one FOV, we simultaneously captured voltage (Fig. 6(A)) and calcium (Fig. 6(B)) fluorescence images and a phase image (Fig. 6(C)) (Visualization 5 (5.7MB, avi) ). The action potential propagated from the stimulus region through the electrotonically coupled cardiomyocyte syncytium into and across the record region. The voltage propagation is visible as temporal lag from different spatial locations in the FOV (Fig. 6(D)). Figure 6(E) shows the voltage (red), calcium (orange), and motion (green) responses of the cardiomyocytes during optogenetic stimulation with a 1 Hz train of 6 ms wide blue pulses. The expected sequence of signals, voltage followed by calcium followed by contraction, was observed.
To demonstrate the value of this three-modality recording, we tested cardiomyocyte response to vehicle control, calcium channel blocker nifedipine, β-adrenergic agonist isoproterenol, and myosin II blocker para-Nitroblebbistatin during 1 Hz optogenetic pacing (Fig. 6(F), panels i – xii). Recordings were made before compound addition, 1 minute after addition, and 10 minutes after addition. In our vehicle control, DMSO, there is little to no change in voltage (i) and calcium (ii) activity, although there was an unexplained change in motion that we occasionally observe in these iPSC-CM systems. (iii) Nifedipine, which blocks the L-type calcium current responsible for the sustained cardiac action potential [27], induced large changes in AP (iv) and calcium transient shape (v) by 1 min; part-way through the recording, beating terminated (vi) Contraction stopped before voltage and calcium transients ceased completely, presumably because the calcium transient is too small to trigger calcium-induced-calcium release through ryanodine channels in the sarcoplasmic reticulum [28]. The non-selective β-adrenergic agonist isoproterenol, which in vivo triggers fight-or-flight signaling to make the heart beat faster (positive chronotropic) and harder (positive inotropic) [29], had dramatic effects in vitro. Although un-paced cardiomyocytes beat faster in response to isoproterenol, here cells were locked to the optogenetic pacing rate of 1 Hz. The conduction velocity was increased by 10 min (vii), and contraction strength increased, as reported by cell displacement (ix) Para-Nitroblebbistatin, a photostable and non-cytotoxic version of the traditional myosin II inhibitor Blebbistatin [30], should block calcium-induced contraction downstream of voltage and calcium signaling. We saw a dramatic reduction in contraction strength by 1 min (xii) with minimal effect on voltage (x) and calcium (xi), and a complete lack of coordinated contraction by 10 min (xii). The diverse functional responses highlight the value of tri-functional readout and optogenetic pacing for understanding compound effects.
Discussion
Because of its versatility and unique capabilities, the Firefly-HR microscope could be of interest to a variety of labs. It is designed to be a replicable system for the biological research community. Except for the customized fused silica prism, and its corresponding holder (Fig. 1(D)-(E)), all other components are commercially available. The microscope can be built fully equipped for a little over $100k. It is compatible with a variety of coverslip-bottomed dishes, although fused silica and cyclic olefin copolymer coverslips offer the lowest background for demanding voltage-sensing applications.
Prior to the Firefly-HR platform, simultaneous voltage and calcium neural recordings during stimulation generally had to be implemented with manual patch clamp paired with a calcium-sensing protein or dye. Recording of synaptically mediated electrical signaling between neurons had to be implemented with a dual patch-clamp system. Although patch-clamp data has superb SNR, temporal resolution, and control over the cells electrical state, the throughput is very low, typically no more than a couple cells per hour for a skilled operator. Attempts have been made to automate the patch-clamp process to achieve throughputs of 10’s of cells an hour [31], but simultaneous calcium recording is challenging, no more than 1 cell can be recorded at a time, and extensive human intervention is require to swap out patch pipettes. All-optical tools, in contrast, lend themselves to high-throughput recordings. The large FOV enables us to routinely record from ~13 neurons at once (Fig. 5(O)). When paired with a motorized xy stage and motorized focus control paired with a standard autofocus algorithm, we can record from 2 FOV’s/min, for a throughput exceeding 1000 cells/hour. We have used 8-well plates (Ibidi, µ-Slide) with the microscope to speed pharmacology measurements, and plan to make the microscope compatible with 96-well plates for high-throughput applications.
In the past, other researchers have demonstrated simultaneous voltage and calcium optical recordings, but they did not have the versatility of the current system. Work using dyes enabled promising voltage and calcium optical readouts, but the blue-shifted dyes Fura Ca2+ and JWP1114 [32] are spectrally incompatibility with channelrhodopsin stimulation. Other work imaged voltage and calcium using engineered proteins using an Arch-based near-infrared voltage sensor, from the same family as QuasAr used in this study, and the GFP-based calcium sensor GCaMP6F [33,34]. However, GCaMP6F is also spectrally incompatible with direct channelrhodopsin stimulation. We developed Firefly-HR to enable voltage recording, calcium recording, and optogenetic stimulation all at the same time.
There are many applications that can benefit from the multi-modal readout of the Firefly-HR platform. One example is in vitro disease modeling using either rodent or human iPS cell-derived neurons or cardiomyocytes. It is becoming ever faster and easier to generate disease-model iPS cell lines from patients or with genome editing [35], and applications in drug discovery and personalized medicine are multiplying. The richer the assay readout, the more likely one is to discover a robust, disease-relevant phenotype with these cellular tools, which plays to Firefly-HR’s strengths. Another application is for drug discovery centered around calcium channels, which have been implicated in numerous indications including chronic pain [36], familial hemiplegic migraine [37], schizophrenia [38], epilepsy [39], autism spectrum disorder [40], and cardiac arrhythmias [41]. Simultaneous recoding of voltage and calcium will give much deeper insight into a candidate therapeutic’s mechanism and performance. A final example is in cardiotoxicity measurements: cardiotox accounts for 7% of attrition during drug development [42,43], and simultaneous recordings of voltage, calcium, and motion in cardiomyocytes would be sensitive to a broad set of toxicity mechanisms.
Finally, the biggest current complication with the platform is optical crosstalk, specifically blue-light induced changes in jRGECO1a baseline fluorescence levels which has similar temporal dynamics to calcium signals (Fig. 4(A)). Currently, care must be taken in designing experiments to avoid this confound. Fortunately, there is a strong motivation in the field to develop improved red calcium sensors as there is high demand for pairing calcium imaging with optogenetics or other GFP-based sensors. Hopefully this issue will soon be resolved. Although we have not yet tested it, K-GECO1 [16] looks very promising.
Conclusion
The Firefly-HR is a custom microscope optimized for simultaneously recording of calcium, voltage, and motion at frame rates up to 1 kHz during optogenetic stimulation. High SNR, a wide FOV, and patterned stimulation enable a variety of measurements, including neuronal excitability and calcium signaling, synaptic transmission and microcircuits, and cardiomyocyte tri-functional recordings. The versatility, rich functional readout, and throughput make it a powerful tool for basic science and drug discovery.
Acknowledgements
The authors would like to thank Adam Cohen, Sami Farhi, and Yoav Adam for helpful discussions regarding the optogenetic constructs.
Funding
The National Institute of Health (NIH) (SBIR grants R44MH112474 and R43MH112273).
Disclosures
CAW: Q-State Biosciences (I,E,P), All other authors: Q-State Biosciences (I,E).
References
- 1.Dugué G. P., Akemann W., Knöpfel T., “A comprehensive concept of optogenetics,” in Progress in Brain Research, Knöpfel T., Boyden E. S., eds. (Elsevier B.V., 2012), Vol. 196, pp. 1–28. [DOI] [PubMed] [Google Scholar]
- 2.Zhang K., Cui B., “Optogenetic control of intracellular signaling pathways,” Trends Biotechnol. 33(2), 92–100 (2015). 10.1016/j.tibtech.2014.11.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Newman R. H., Fosbrink M. D., Zhang J., “Genetically encodable Fluorescent Biosensors for Tracking Signaling Dynamics in Living Cells,” Chem. Rev. 111(5), 3614–3666 (2011). 10.1021/cr100002u [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mehta S., Zhang J., “Reporting from the Field: Genetically Encoded Fluorescent Reporters Uncover Signaling Dynamics in Living Biological Systems,” Annu. Rev. Biochem. 80(1), 375–401 (2011). 10.1146/annurev-biochem-060409-093259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tantama M., Hung Y. P., Yellen G., Optogenetic Reporters: Fluorescent Protein-Based Genetically Encoded Indicators of Signaling and Metabolism in the Brain., 1st ed. (Elsevier B.V., 2012), Vol. 196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lin M. Z., Schnitzer M. J., “Genetically encoded indicators of neuronal activity,” Nat. Neurosci. 19(9), 1142–1153 (2016). 10.1038/nn.4359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fenno L., Yizhar O., Deisseroth K., “The development and application of optogenetics,” Annu. Rev. Neurosci. 34(1), 389–412 (2011). 10.1146/annurev-neuro-061010-113817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lin J. Y., “A user’s guide to channelrhodopsin variants: features, limitations and future developments,” Exp. Physiol. 96(1), 19–25 (2011). 10.1113/expphysiol.2009.051961 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Klapoetke N. C., Murata Y., Kim S. S., Pulver S. R., Birdsey-Benson A., Cho Y. K., Morimoto T. K., Chuong A. S., Carpenter E. J., Tian Z., Wang J., Xie Y., Yan Z., Zhang Y., Chow B. Y., Surek B., Melkonian M., Jayaraman V., Constantine-Paton M., Wong G. K.-S., Boyden E. S., “Independent optical excitation of distinct neural populations,” Nat. Methods 11(3), 338–346 (2014). 10.1038/nmeth.2836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu Y., Zou P., Cohen A. E., “Voltage imaging with genetically encoded indicators,” Curr. Opin. Chem. Biol. 39, 1–10 (2017). 10.1016/j.cbpa.2017.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hochbaum D. R., Zhao Y., Farhi S. L., Klapoetke N., Werley C. A., Kapoor V., Zou P., Kralj J. M., Maclaurin D., Smedemark-Margulies N., Saulnier J. L., Boulting G. L., Straub C., Cho Y. K., Melkonian M., Wong G. K.-S., Harrison D. J., Murthy V. N., Sabatini B. L., Boyden E. S., Campbell R. E., Cohen A. E., “All-optical electrophysiology in mammalian neurons using engineered microbial rhodopsins,” Nat. Methods 11(8), 825–833 (2014). 10.1038/nmeth.3000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chen T.-W., Wardill T. J., Sun Y., Pulver S. R., Renninger S. L., Baohan A., Schreiter E. R., Kerr R. A., Orger M. B., Jayaraman V., Looger L. L., Svoboda K., Kim D. S., “Ultrasensitive fluorescent proteins for imaging neuronal activity,” Nature 499(7458), 295–300 (2013). 10.1038/nature12354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhao Y., Araki S., Wu J., Teramoto T., Chang Y.-F., Nakano M., Abdelfattah A. S., Fujiwara M., Ishihara T., Nagai T., Campbell R. E., “An expanded palette of genetically encoded Ca2+ indicators,” Science 333(6051), 1888–1891 (2011). 10.1126/science.1208592 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Inoue M., Takeuchi A., Horigane S., Ohkura M., Gengyo-Ando K., Fujii H., Kamijo S., Takemoto-Kimura S., Kano M., Nakai J., Kitamura K., Bito H., “Rational design of a high-affinity, fast, red calcium indicator R-CaMP2,” Nat. Methods 12(1), 64–70 (2015). 10.1038/nmeth.3185 [DOI] [PubMed] [Google Scholar]
- 15.Dana H., Mohar B., Sun Y., Narayan S., Gordus A., Hasseman J. P., Tsegaye G., Holt G. T., Hu A., Walpita D., Patel R., Macklin J. J., Bargmann C. I., Ahrens M. B., Schreiter E. R., Jayaraman V., Looger L. L., Svoboda K., Kim D. S., “Sensitive red protein calcium indicators for imaging neural activity,” eLife 5, 1–24 (2016). 10.7554/eLife.12727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shen Y., Dana H., Abdelfattah A. S., Patel R., Shea J., Molina R. S., Rawal B., Rancic V., Chang Y. F., Wu L., Chen Y., Qian Y., Wiens M. D., Hambleton N., Ballanyi K., Hughes T. E., Drobizhev M., Kim D. S., Koyama M., Schreiter E. R., Campbell R. E., “A genetically encoded Ca2+ indicator based on circularly permutated sea anemone red fluorescent protein eqFP578,” BMC Biol. 16(1), 9 (2018). 10.1186/s12915-018-0480-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zucker R. S., “Calcium- and activity-dependent synaptic plasticity,” Curr. Opin. Neurobiol. 9(3), 305–313 (1999). 10.1016/S0959-4388(99)80045-2 [DOI] [PubMed] [Google Scholar]
- 18.Flavell S. W., Greenberg M. E., “Signaling Mechanisms Linking Neuronal Activity to Gene Expression and Plasticity of the Nervous System,” Annu. Rev. Neurosci. 31(1), 563–590 (2008). 10.1146/annurev.neuro.31.060407.125631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bers D. M., “Cardiac Excitation-Contraction Coupling,” Nature 415(6868), 198–205 (2002). 10.1038/415198a [DOI] [PubMed] [Google Scholar]
- 20.Maclaurin D., Venkatachalam V., Lee H., Cohen A. E., “Mechanism of voltage-sensitive fluorescence in a microbial rhodopsin,” Proc. Natl. Acad. Sci. U.S.A. 110(15), 5939–5944 (2013). 10.1073/pnas.1215595110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Werley C. A., Chien M.-P., Cohen A. E., “Ultrawidefield microscope for high-speed fluorescence imaging and targeted optogenetic stimulation,” Biomed. Opt. Express 8(12), 5794–5813 (2017). 10.1364/BOE.8.005794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Adam Y., Kim J. J., Lou S., Zhao Y., Brinks D., Wu H., Mostajo-Radji M. A., Kheifets S., Parot V., Chettih S., Williams K. J., Farhi S. L., Madisen L., Harvey C. D., Zeng H., Arlotta P., Campbell R. E., Cohen A. E., “All-optical electrophysiology reveals brain-state dependent changes in hippocampal subthreshold dynamics and excitability,” bioRxiv (2018).
- 23.Werley C. A., Brookings T., Upadhyay H., Williams L. A., McManus O. B., Dempsey G. T., “All-optical electrophysiology for disease modeling and pharmacological characterization of neurons,” Curr. Protocols Pharmacol. 78(1), 1 (2017). 10.1002/cpph.25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bardy C., van den Hurk M., Eames T., Marchand C., Hernandez R. V., Kellogg M., Gorris M., Galet B., Palomares V., Brown J., Bang A. G., Mertens J., Böhnke L., Boyer L., Simon S., Gage F. H., “Neuronal medium that supports basic synaptic functions and activity of human neurons in vitro,” Proc. Natl. Acad. Sci. U.S.A. 112(20), E2725–E2734 (2015). 10.1073/pnas.1504393112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bean B. P., “The action potential in mammalian central neurons,” Nat. Rev. Neurosci. 8(6), 451–465 (2007). 10.1038/nrn2148 [DOI] [PubMed] [Google Scholar]
- 26.Catterall W. A., “Voltage-gated calcium channels,” Cold Spring Harb. Perspect. Biol. 3(8), a003947 (2011). 10.1101/cshperspect.a003947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hensley J., Billman G. E., Johnson J. D., Hohl C. M., Altschuld R. A., “Effects of Calcium Channel Antagonists on Ca2+ Transients in Rat and Canine Cardiomyocytes,” J. Mol. Cell. Cardiol. 29(3), 1037–1043 (1997). 10.1006/jmcc.1996.0348 [DOI] [PubMed] [Google Scholar]
- 28.Fill M., Copello J. A., “Ryanodine receptor calcium release channels,” Physiol. Rev. 82(4), 893–922 (2002). 10.1152/physrev.00013.2002 [DOI] [PubMed] [Google Scholar]
- 29.Sniecinski R. M., Wright S., Levy J. H., “Cardiovascular Pharmacology,” in Cardiothoracic Critical Care, Sidebotham D., Mckee A., Gillham M., Levy J. H., eds. (Elsevier Inc., 2007), pp. 33–52. [Google Scholar]
- 30.Képiró M., Várkuti B. H., Végner L., Vörös G., Hegyi G., Varga M., Málnási-Csizmadia A., “para-Nitroblebbistatin, the non-cytotoxic and photostable myosin II inhibitor,” Angew. Chem. Int. Ed. Engl. 53(31), 8211–8215 (2014). 10.1002/anie.201403540 [DOI] [PubMed] [Google Scholar]
- 31.Kodandaramaiah S. B., Franzesi G. T., Chow B. Y., Boyden E. S., Forest C. R., “Automated whole-cell patch-clamp electrophysiology of neurons in vivo,” Nat. Methods 9(6), 585–587 (2012). 10.1038/nmeth.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vogt K. E., Gerharz S., Graham J., Canepari M., “High-resolution simultaneous voltage and Ca2+ imaging,” J. Physiol. 589(Pt 3), 489–494 (2011). 10.1113/jphysiol.2010.200220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hou J. H., Venkatachalam V., Cohen A. E., “Temporal dynamics of microbial rhodopsin fluorescence reports absolute membrane voltage,” Biophys. J. 106(3), 639–648 (2014). 10.1016/j.bpj.2013.11.4493 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dempsey G. T., Chaudhary K. W., Atwater N., Nguyen C., Brown B. S., McNeish J. D., Cohen A. E., Kralj J. M., “Cardiotoxicity screening with simultaneous optogenetic pacing, voltage imaging and calcium imaging,” J. Pharmacol. Toxicol. Methods 81, 240–250 (2016). 10.1016/j.vascn.2016.05.003 [DOI] [PubMed] [Google Scholar]
- 35.Avior Y., Sagi I., Benvenisty N., “Pluripotent stem cells in disease modelling and drug discovery,” Nat. Rev. Mol. Cell Biol. 17(3), 170–182 (2016). 10.1038/nrm.2015.27 [DOI] [PubMed] [Google Scholar]
- 36.Cao Y. Q., “Voltage-gated calcium channels and pain,” Pain 126(1-3), 5–9 (2006). 10.1016/j.pain.2006.10.019 [DOI] [PubMed] [Google Scholar]
- 37.Ophoff R. A., Terwindt G. M., Vergouwe M. N., van Eijk R., Oefner P. J., Hoffman S. M. G., Lamerdin J. E., Mohrenweiser H. W., Bulman D. E., Ferrari M., Haan J., Lindhout D., van Ommen G. J. B., Hofker M. H., Ferrari M. D., Frants R. R., “Familial hemiplegic migraine and episodic ataxia type-2 are caused by mutations in the Ca2+ channel gene CACNL1A4,” Cell 87(3), 543–552 (1996). 10.1016/S0092-8674(00)81373-2 [DOI] [PubMed] [Google Scholar]
- 38.Purcell S. M., Moran J. L., Fromer M., Ruderfer D., Solovieff N., Roussos P., O’Dushlaine C., Chambert K., Bergen S. E., Kähler A., Duncan L., Stahl E., Genovese G., Fernández E., Collins M. O., Komiyama N. H., Choudhary J. S., Magnusson P. K. E., Banks E., Shakir K., Garimella K., Fennell T., DePristo M., Grant S. G. N., Haggarty S. J., Gabriel S., Scolnick E. M., Lander E. S., Hultman C. M., Sullivan P. F., McCarroll S. A., Sklar P., “A polygenic burden of rare disruptive mutations in schizophrenia,” Nature 506(7487), 185–190 (2014). 10.1038/nature12975 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zamponi G. W., Lory P., Perez-Reyes E., “Role of voltage-gated calcium channels in epilepsy,” Pflugers Arch. 460(2), 395–403 (2010). 10.1007/s00424-009-0772-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rubeis S., He X., Goldberg A. P., Poultney C. S., Samocha K., Cicek A. E., Kou Y., Liu L., Fromer M., Walker S., Singh T., Klei L., Kosmicki J., Shih-Chen F., Aleksic B., Biscaldi M., Bolton P. F., Brownfeld J. M., Cai J., Campbell N. G., Carracedo A., Chahrour M. H., Chiocchetti A. G., Coon H., Crawford E. L., Curran S. R., Dawson G., Duketis E., Fernandez B. A., Gallagher L., Geller E., Guter S. J., Hill R. S., Ionita-Laza J., Jimenz Gonzalez P., Kilpinen H., Klauck S. M., Kolevzon A., Lee I., Lei I., Lei J., Lehtimäki T., Lin C.-F., Ma’ayan A., Marshall C. R., McInnes A. L., Neale B., Owen M. J., Ozaki N., Parellada M., Parr J. R., Purcell S., Puura K., Rajagopalan D., Rehnström K., Reichenberg A., Sabo A., Sachse M., Sanders S. J., Schafer C., Schulte-Rüther M., Skuse D., Stevens C., Szatmari P., Tammimies K., Valladares O., Voran A., Li-San W., Weiss L. A., Willsey A. J., Yu T. W., Yuen R. K. C., Cook E. H., Freitag C. M., Gill M., Hultman C. M., Lehner T., Palotie A., Schellenberg G. D., Sklar P., State M. W., Sutcliffe J. S., Walsh C. A., Scherer S. W., Zwick M. E., Barett J. C., Cutler D. J., Roeder K., Devlin B., Daly M. J., Buxbaum J. D., Roeder K., Devlin B., Daly M. J., Buxbaum J. D., “Synaptic, transcriptional and chromatin genes disrupted in autism,” Nature 515(7526), 209–215 (2014). 10.1038/nature13772 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Splawski I., Timothy K. W., Decher N., Kumar P., Sachse F. B., Beggs A. H., Sanguinetti M. C., Keating M. T., “Severe arrhythmia disorder caused by cardiac L-type calcium channel mutations,” Proc. Natl. Acad. Sci. U.S.A. 102(23), 8089–8096, discussion 8086–8088 (2005). 10.1073/pnas.0502506102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Laverty H., Benson C., Cartwright E., Cross M., Garland C., Hammond T., Holloway C., McMahon N., Milligan J., Park B., Pirmohamed M., Pollard C., Radford J., Roome N., Sager P., Singh S., Suter T., Suter W., Trafford A., Volders P., Wallis R., Weaver R., York M., Valentin J., “How can we improve our understanding of cardiovascular safety liabilities to develop safer medicines?” Br. J. Pharmacol. 163(4), 675–693 (2011). 10.1111/j.1476-5381.2011.01255.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guengerich F. P., Review Mechanisms of Drug Toxicity and Relevance to Pharmaceutical Development 26, 3–14 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]