Abstract
Glaucoma detection in color fundus images is a challenging task that requires expertise and years of practice. In this study we exploited the application of different Convolutional Neural Networks (CNN) schemes to show the influence in the performance of relevant factors like the data set size, the architecture and the use of transfer learning vs newly defined architectures. We also compared the performance of the CNN based system with respect to human evaluators and explored the influence of the integration of images and data collected from the clinical history of the patients. We accomplished the best performance using a transfer learning scheme with VGG19 achieving an AUC of 0.94 with sensitivity and specificity ratios similar to the expert evaluators of the study. The experimental results using three different data sets with 2313 images indicate that this solution can be a valuable option for the design of a computer aid system for the detection of glaucoma in large-scale screening programs.
1. Introduction
Glaucoma is an optic neuropathy characterized by progressive degeneration of retinal ganglion cells [1]. There are many different types of glaucoma, with a variety of etiologies and pathogenic factors, but all have in common typical changes in the structure and la function of the optic nerve. Glaucoma is the leading cause of global irreversible vision loss with a prevalence for population aged 40-80 estimated in 3-4% [2]. The number of people with glaucoma worldwide was estimated in 64.3 million in 2013, increasing to 76.0 million in 2020 and 111.8 million in 2040 [2]. Because glaucoma is an asymptomatic condition until a relatively late stage the diagnosis is frequently delayed. Population-level surveys suggest that only 10-50% of people with glaucoma are aware they suffer the disease [1]. As early diagnosis and treatment of the condition can prevent vision loss, glaucoma screening has been tested in numerous studies worldwide [3–6]. Current studies show that glaucoma screening can be cost-effective in risk population (family history, black ethnicity, age) and can be improved using a test with initial automated classification followed by the expert assessment of a specialist [7].
The standard of care for glaucoma screening consists of routine optometrist visits every 2-3 years, suspicious cases are then referred to an ophthalmologist who performs additional tests and examinations for final confirmation of the diagnosis. A complete glaucoma study usually includes detailed medical history, slit lamp examination, visual field, fundus photography and a tonometry [8,9], and since the 90’s it also includes some optic nerve imaging test such as scanning laser tomography (HRT) [10], optical coherence tomography (OCT) [11] and scanning laser polarimetry with variable corneal compensation (GDx-VCC) [12]. Nowadays, only OCT images and photographs are widely used to assess the structure of the optic nerve in glaucoma.
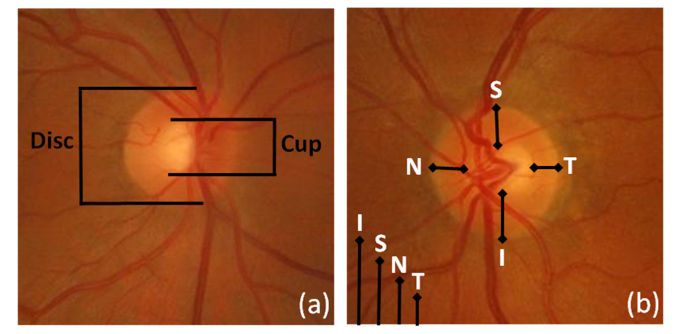
Color fundus imaging has been often used in combination with image processing algorithms to aid in the detection and grading of eye diseases [13,14]. The availability of digital fundus cameras in primary care settings and their extensive use in eye screening programs explains the interest in screening for glaucoma using this image modality. Nevertheless, the subjective interpretation of color fundus images for the identification of glaucomatous signs is a challenging task that requires specific expertise and years of practice. To overcome this difficulty a great effort has been made to develop automatic glaucoma-detection algorithms based on image processing of color fundus images. We can distinguish four main changes in the retinal structures associated with glaucoma: optic nerve head cupping, neuro-retinal rim thinning, retinal nerve fibre layer defects and peripapillary atrophy [15]. Figure 1 shows some of the typical signs assessed to detect glaucoma in color fundus images.
Fig. 1.
Example of findings used to detect glaucoma in color fundus images. (a) Quantification of the optic cup to disc ratio (CDR). The reduction of the optic nerve fibres (typically related with glaucoma) provokes optic disc cupping, central cup becomes larger, with respect to the optic disc (b) The neuroretinal rim usually follows a normal pattern (ISNT rule) where the inferior region is broader than the superior, broader than the nasal, and broader than the temporal region. The alteration of this pattern is a suspicious sign of glaucoma
To aid in the detection of glaucoma numerous image processing algorithms have been proposed. We can find works that focus on the localization and segmentation of the optic disc [16–18] and numerous glaucoma-detection algorithms based on the extraction of features from the image or transformed versions of the image to train different types of classifiers. The extracted features could identify or consider relevant information present in the images, with potential for better representation o case classification than clinical measurements. Among others we can mention glaucoma detection based on a probabilistic combination of previously compressed features extracted from the pixel intensity values, the Fourier Transform (FT) and B-splines coefficients [19], or using higher order spectra analysis and texture-based features extracted from preprocessed images and a Support Vector Machines (SVM) classifier [20], or with a feature extraction based on higher order spectra and discrete wavelet transform and a SVM classifier [21,22], or using an empirical wavelet transform with a least-squares SVM [23] or with an adaptive histogram equalization convolved with several filter banks processed to create local configuration patterns that feed a k-nearest neighbor (kNN) classifier [24]. The previous methods apply the approach of identifying features in the image in order to train a classifier with all the findings extracted directly from the image or from a transformed version of it (using wavelets, FT, high order spectra analysis…). At the end, the different algorithms explore different aspects and transformations of the ONH to determine patterns that are representative and may identify glaucoma. In this work we applied a different approach to the glaucoma detection problem through the use of Convolutional Neural Networks (CNNs).
Convolutional networks, commonly known as one of the most popular deep learning algorithms for image analysis, have become very rapidly a successful alternative for analyzing medical images. These methods could be considered as the evolution of the supervised techniques started at the end of the 1990s, where training data sets of previously classified images are used to develop the system. This strategy supersedes the previous approach based on feature extraction and posterior classification mentioned in previous paragraphs. The new deep learning paradigm implies that computers can perform the feature learning and classification simultaneously. We can usually find in a deep learning algorithm a network (model) formed by many layers that transform an input data (images normally) to outputs (e.g. pathology present/absent). The most successful type of models for medical image analysis is a sub-class of neural networks called convolutional neural networks (CNN) that was introduced in the 1980s [25].
In [26] Litjens et al., provided a thorough review of the current use of these techniques in medical analysis. The study mentions state-of-the art applications of deep learning technology in the main topics of biomedical image processing: classification, object detection, segmentation or registration among others. Shin et al. [27] mentioned three mayor strategies that used CNNs to medical image classification problems: training from scratch, using off-the shell pre-trained CNN features, and conducting unsupervised CNN pre-training with supervised fine-tuning. Training a deep CNN from scratch (or full training) presents relevant limitations. It requires a large amount of labeled data, which in fields like medical imaging could be extremely expensive to collect both in time and budget, especially for images that present pathological findings relevant for diagnosis. Besides, the training of a deep CNN usually requires extensive memory and computational resources and it could be a very time consuming task. Finally, the design of a CNN and the adjustment of the hyper-parameters of the network could be a challenging process that requires dealing with overfitting and other issues that can limit the success of the application of this technology. One alternative to overcome these problems is the use of transfer learning with fine tuning. Transfer learning is a method successfully used in machine learning and data mining for classification, regression and clustering problems. It is generally defined as the capability of the system to utilize the knowledge learned in one domain of interest, to another that shares some common characteristics [28]. The use of state-of-the art performance CNNs on the ImageNet Large-Scale Visual Recognition Challenge (ILSVRC) [29,30] (with millions of labeled images from 1000 different classes) has been successfully tested in several medical image analysis studies [27,31–33]. This last method is the one we have used in this work: the transfer learning of CNN models pre-trained with different image data sets and fine-tuned to solve a specific medical imaging task, the automatic classification of glaucoma in fundus photographs.
CNNs have been successfully applied to color fundus images in the context of Computer-Aided-Detection (CAD) systems and screening programs for eye diseases, achieving state-of-the art performance or outperforming previous implementations. We can find since 2015 several studies applying CNN in retinal vessel segmentation [34], image quality assessment [35], segmentation of the optic disc and the optic disc cup [36,37], diabetic retinopathy detection [38], age-macular degeneration detection [13] or hemorrhage detection [39] among others. The CNN architectures successfully exploit both local and global features present in the images, being a proper tool for the detection of glaucoma. Several studies have already tackled this problem in color fundus images using CNN. In [40] the authors applied a six layers architecture to optic disc patches previously segmented. In [41] the authors used CNN to extract features and train a SVM classifier to detect glaucoma. Recently Fu et. al. [42] presented a novel ensemble network based on the application of different CNNs to the global fundus image and to different versions of optic disc region. The assessment of deep learning algorithms with transfer learning has also been addressed in [43–45] implementing studies with greater number of images than previous works and achieving expert level accuracy and high sensitivity and specificity. Finally, in OCT there are also recent studies applying CNNs for glaucoma detection [46] or segmentation of layers [47,48]. In Table 1 we present a summary of the methods used for glaucoma detection, describing the data sets used and the results reported.
Table 1. Summary of methods for the detection of glaucoma in color fundus images. In the data sets column we indicate the number of normal cases (-) and glaucoma cases ( + ). For the performance we used the reported metric used in the study: AUC, accuracy (Acc) and specificity (Sp) and sensitivity (Sn).
| Authors | Method | Data sets | Performance |
|---|---|---|---|
| Bock et al. (2010) [19] | Pixel values FFT coefficients, B-spline and probabilistic SVM | Private (336 -/239 + ) | AUC – 87% Acc – 80% |
| Krishnan et al. (2013) [21] | HOS, TT, DWT with SVM | Private (30-/30 + ) | Acc – 91.67% |
| Maheshwari et al. (2017) [23] | 2D EWT and LS-SVM | Private (30-/30 + ) RIM-ONE (255-/250 + ) |
Acc – 98.33% (Private) Acc – 81.32% (RIM-ONE) |
| Acharaya et al. (2017) [24] | Texton, LCP features and KNN | Private (143-/559 + ) | Acc – 95,7% |
| Chen etal. (2015) [40] | CNN (6 layers) | ORIGA (168 + 482-) SCES (46+/1676-) |
AUC (83.1%-88.7%) |
| Al-Bander et al. (2017) [41] | CNN 23 layers and SVM | RIM-ONE (200+/255-) | Acc – 88.2%, Sn – 85%, Sp – 89.8% |
| Fu et al. (2018) [42] | Ensemble of 4 CNNs | ORIGA (168+/482-) SCES (1636-/46 + ) |
AUC-91.83%, Sn – 84.78%, Sp – 83.80% |
| Li et al. (2018) [44] | Transfer Learning with Inception Network | Private (48116) | AUC – 98.6%, Sn – 95.6%, Sp – 92.0% |
| Christopher et.al. (2018) [43] | Transfer Learning with ResNet, VGG16 and Inception v3 | Private (5633+/9189-) | AUC-91%, Sn – 88%, Sp – 95% |
| Shibata et.al. (2018) [45] | Transfer Learning with ResNet | Private (1364+/1768-) | AUC – 96.5% |
Our work is aimed at developing tools for prescreening in computer aided diagnosis system for the detection of glaucoma in large-scale screening programs. In this paper we assessed the application of different CNN architectures for the classification of glaucoma with fundus color images. We studied the performance of different architectures as well as some transfer learning schemes from pre-trained CNN models. We have ensured that all the architectures that are the basis of the current state-of-the-art methods are represented in our comparisons including an Inception based network such as GogleLeNet, a RestNet based architecture, concretely ResNet50 and a recently proposed network such as DENet. Finally, we will consider the potential benefit of integrating basic clinical data collected in the screening studies of the patients in the final classification.
The rest of the paper is structured in several sections. In section 2 we describe the data sets, the network architectures, the training process and the performance metrics used in the study. In section 3 we report the results of the different experiments. Finally, section 4 contains the summary of the study with the main conclusions and suggestions for further works.
2. Materials and methods
2.1 Study data sets
We used 2313 retinal fundus images in this work coming from three different data sets: two publicly available (RIM-ONE [49] and DRISHTI-GS [50]) and one private from a screening campaign performed at Hospital de la Esperanza (Parc de Salut Mar) in Barcelona (Spain). We considered two categories, glaucoma and normal. Glaucoma includes the images classified by specialists as suspect of glaucoma or with glaucoma.
The Open Retinal Image Database for Optic Nerve Evaluation (RIM-ONE) is an ophthalmic image group of databases designed in order to be a reference for the design of optic nerve head segmentation algorithms and in development of computer-aided glaucoma diagnosis. The database was created by the collaboration of three Spanish hospitals: Hospital Universitario de Canarias, Hospital Clínico San Carlos and Hospital Universitario Miguel Servet. Our study used the three releases included by the authors until now. The image set was designed in collaboration of 4 glaucoma experts. The camera used to capture the images was a Nidek AFC-210 background camera with a 21.1-megapixel Canon EOS 5D Mark II body. All the images are centered at the optic disc.
The data set DRISHTI-GS consist of 101 retinal fundus images for optic disc segmentation. All images were collected at Aravind Eye Hospital in Madurai (India). Glaucoma patient selection was done by clinical experts based on findings during examination. The retinal images come from Indian patients of 40-80 years old. The images were taken with the eyes dilated, centered on the optic disk, with a field of view of 30-degrees and of dimension 2996x1944 pixels and PNG uncompressed image format.
Finally the ESPERANZA data set consisted of 1446 color fundus images with a field of view 45 degrees, centered on the macula and including the optic disc from patients with age ranging from 55 to 86 years. The retinal images were provided from the glaucoma detection campaign performed to 1006 different patients. During the examination of the patients, a short clinical history was collected (like age, family history of glaucoma, personal record of glaucoma and glaucoma therapy, among others) and besides the color fundus images, other tests (like the measurement of the intraocular pressure (IOP) in both eyes, the visual acuity and Optical Coherence Tomography images) were performed. Special care was taken in order to create a reference gold standard data set. All the images had a double complete glaucoma evaluation performed by six expert (senior) ophthalmologists and nine non-expert (younger) ophthalmologists using a tele-screening tool. The ophthalmologists with more than five years of experience were considered as experts in this work. In case of disagreement between the two evaluations performed, two glaucoma experts decided the final classification of the image by consensus. Each ophthalmologist evaluated a proportional part of all the images inside its category. The assessment of the images included the evaluation of image quality in four categories (good, enough, bad or not evaluable) and the clinical classification in three categories (normal, glaucoma suspect or glaucoma). The selection of images from the campaign to be included in the final data set used in this work corresponds to the images that were labeled by the evaluators with good or enough quality and in the case of glaucoma positive images we included the ones classified as glaucoma suspect or glaucoma.
Table 2 contains information of the retinal images considered in the study from each data set. We considered two classes for the classifications tasks studied in this work because the majority of the data sets used (RIM-ONE r2, r3 and DRISHTI-GS) were defined with only two classes (glaucoma and normal). RIM-ONE r1 is the only one that accounted four classes (normal, early, moderate and deep glaucoma) while ESPERANZA data set defined three classes (normal, glaucoma-suspect and glaucoma). The initial size of the images of the RIM-ONE data sets is very different. In the case of r1 the image sizes vary from 316x342 to 831x869. In the version r2 the sizes vary from 290x290 to 1375x1654. Figure 2 shows examples of images from the ESPERANZA data set included in the glaucoma and normal categories and the findings identified by the clinicians in the evaluation.
Table 2. Data sets used in the study. The term “glaucoma” includes all retinal images in the data sets classified by a specialist as suspect of glaucoma or as suffering the disease in any stage (early, moderate or severe glaucoma).
| Data set | Initial Size | Format | Glaucoma | Normal |
|---|---|---|---|---|
| ESPERANZA | 1024x680 | JPG | 113 | 1333 |
| RIM-ONE r1 | Not fixed | BMP | 40 | 118 |
| RIM-ONE r2 | Not fixed | JPG | 200 | 255 |
| RIM-ONE r3 | 2144x1424 | JPG | 71 | 82 |
| DRISHTI-GS | 2996x1944 | PNG | 70 | 31 |
| TOTAL | 494 | 1819 |
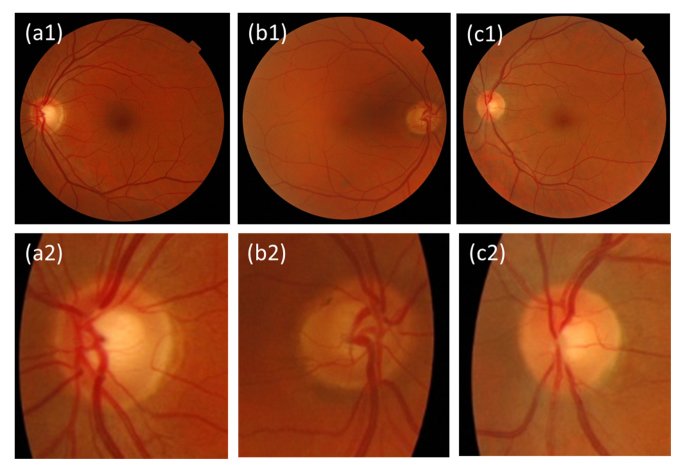
Fig. 2.
Examples of color fundus images from the ESPERANZA data set. All images were labeled with good quality by the evaluators. (a1-2) Left eye of a glaucoma suspect disc with unaccomplished ISNT rule (inferior rim is not wider than superior rim). (b1-2) Right eye of a glaucoma suspect disc due to superior and temporal rim thinning. (c1-2) Left eye with of a normal disc.
As we mentioned before, the ESPERANZA data set had a gold standard classification created by consensus of at least two ophthalmologists. We used the individual evaluation of every ophthalmologist to estimate the performance of human evaluators with respect to the consensus gold-standard taking into account their level of expertise. This performance is compared with the performance of the proposed Deep Learning Architectures. In Table 3 we present a reference of the specificity and sensitivity of the expert and non-expert evaluators with respect to the gold standard. To make a fair comparison for the calculation of these metrics we considered only color fundus images with quality good or enough. The gold standard set included 1735 color fundus images classified as normal and 160 classified with glaucoma or glaucoma suspect.
Table 3. Specificity and sensitivity (defined in section 2.4) reference in the classification of the experts and non-experts evaluators respect to the gold standard in the ESPERANZA data set. The values were calculated considering all the images evaluated by the ophthalmologists during the campaign with quality good or enough.
| Glaucoma Experts | Glaucoma Non-experts | |
|---|---|---|
| Specificity | 0.8914 | 0.8607 |
| Sensitivity | 0.7662 | 0.5875 |
2.2 Preprocessing
In the preprocessing step, we processed the images from the different data sets to a common and standard format in order to train the networks in a homogeneous way. No correction of illumination or contrast enhancement was applied to the images.
We decided to use standard patches centered at the optic disk and the same size for all the data sets, because of the clinical interest for the classification of this region and also to reduce the computational costs in the training step. In the case of the images from the ESPERANZA and DRISHTI-GS data sets we had to localize the optic disk to center the image and scaled them to 256x256 and 224x224 pixels size in order to adapt them to the different networks we used in the study. For the localization of the optic disk we used the same approach previously proposed by [51] with the application of morphological operations of the binary image previously corrected to limit the effect of the blood vessels and small exudates on the image. A region of interest was then obtained to crop the image with the optic disc in the center. The described method localized correctly the optic disc with an accuracy of 84.02% for the images for the ESPERANZA data set and 95.05% for the DRISHTI-GS data set. The images with the optic disk wrongly identified were manually segmented. In the case of the different releases of the RIM-ONE data set, we only had to perform a scaling to the final sizes (256x256 and 224x224 depending of the CNN). Finally we subtracted the mean across every channel, to ensure that all data inputs have the same centered distribution.
2.3 CNNs used and transfer learning
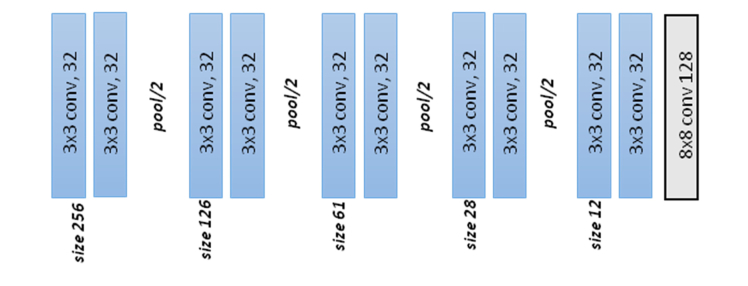
For the selection of the network we tested six different CNNs methods. The first one used the architecture presented in Fig. 3. It consisted of 15 convolutional, pooling, fully-connected and softmax layers and was designed following standard CNN principles. We include batch normalization after each convolutional layer to accelerate the training and to improve the initialization of the network [52]. At the end of the network a softmax layer performed the final binary classification (glaucoma negative and glaucoma positive). The final output of the classifier was a two-element vector. To train the network we used the stochastic gradient descent algorithm and a binary cross-entropy loss function. In Fig. 3 and Table 4, we present the details of the design of the network.
Fig. 3.
Architecture of the standard CNN.
Table 4. Detail of the layers and parameters defined in the standard CNN. The input of the network is an image of size 256x256x3 and the output are two scores of the two classes considered. In the parameters column K indicates the number of filters of the layer. After each convolution layer, batch normalization was applied.
| Layer | Operation | Input size | Parameters | Layer | Operation | Input size | Parameters |
|---|---|---|---|---|---|---|---|
| 1 | Convolution | 256x256 | 3x3, K = 32 | 9 | Max-pooling | 57x57 | 2x2 |
| 2 | Convolution | 254x254 | 3x3, K = 32 | 10 | Convolution | 28x28 | 3x3, K = 32 |
| 3 | Max-pooling | 252x252 | 2x2 | 11 | Convolution | 26x26 | 3x3, K = 32 |
| 4 | Convolution | 126x126 | 3x3, K = 32 | 12 | Max-pooling | 24x24 | 2x2 |
| 5 | Convolution | 124x124 | 3x3, K = 32 | 13 | Convolution | 12x12 | 3x3, K = 32 |
| 6 | Max-pooling | 122x122 | 2x2 | 14 | Convolution | 10x10 | 3x3, K = 32 |
| 7 | Convolution | 61x61 | 3x3, K = 32 | 15 | Convolution | 8x8 | K = 128,Dropout, p = 0.5 |
| 8 | Convolution | 59x59 | 3x3, K = 32 | 16 | Soft-Max | 128x1 | 2 classes |
As we confirmed in this work, the use of these architectures offers two valuable benefits. First, the design and architectural improvements of these CNNs can warranty superior performance ratios even in a training from scratch scheme. Second, the use of pre-trained deep CNNs and the subsequent fine-tune of the weights of the network applying the new labeled images, could lead to even better performance metrics and a potential reduction in training resources in terms of time, memory and computational operations. Besides the STANDARD CNN presented in Fig. 3, we selected other commonly used architectures (VGG19, GoogLeNet, and ResNet50) and the recently presented DENet, specifically designed for glaucoma screening detection. We prepared several experiments to analyze quantitatively the contribution of the selection of the architecture, the training scheme, fine-tuning (VGG19 TL, GOOGLENET TL, RESNET50 TL, DENET DISC TL) versus full training (VGG19, GOOGLENET, RESNET50 and DENET DISC), and the data set used for training.
VGG19 [53] is a publicly available CNN model that includes five stacks, each stack contains between two and four convolutional layer followed by a max-pooling layer, and it ends with three fully connected layers. The main contribution of this architecture was the increasing of the depth of the network (in this work we applied the version with 19 layers) and the use of very small (3x3) convolutional filters. We can find applications of this network in several studies, for the fixed features extraction in CADx for breast cancer detection with different imaging modalities [54] or for classification of 19 different skin diseases [55].
GoogLeNet proposed in [56] accomplished as main contribution the improved utilization of the computing resources inside the network, increasing the depth and width of the network but keeping the computational budget constant. It also proposed a new module called “Inception” which concatenates convolution layers with different kernel sizes and one pooling layer into a single new filter. The complete architecture contains 22 layers including two convolution layers, three pooling layers and nine inception layers. GoogLeNet was successfully used in the detection of lymph node metastases in women with breast cancer [57], in the classification of normal and cancerous lung tissues from CARS (Coherent anti-Stokes Raman scattering) images [58] or retinal pathologies using optical coherence tomography (OCT) images [31].
Residual Networks (ResNet) [59] have been broadly tested in general and medical image classification. The main characteristics of ResNet are the intensive application of batch normalization and the use of “shortcut connections” in order to tackle the low performance issues due to the vanishing divergence and the vanishing gradient problems in deep CNN. ResNet has been successfully adopted in recent works of glaucoma classification [43,45] and it has also been included in the ensemble CNN DENet [42].
The Disc-Aware Ensemble Network (DENet) [42,60] is a glaucoma screening network based on an ensemble of four networks. DENet considers various levels and modules of the fundus images. Two networks exploit global fundus images and are based on ResNet and U-shape convolutional network (U-net). The other two networks are centered on the local optic disc region, previously cropped based on one of the previous networks, and use ResNet to classify the disc region and a polar transformed version of the same disc area. The authors provided the code of the four networks and the trained models using the ORIGA full data set. In this study we used DENet in two different contexts. First, we evaluated the ensemble network using the global fundus images from a subset of our data set (only ESPERANZA and DRISHTI-GS have global images) and the pre-trained models provided by the authors (DENET). Secondly, in order to assess the impact in the performance of the use of transfer learning, fine tuning a pre-trained glaucoma classification model, we used one of the networks of the ensemble, DENet Disc. This model uses segmented optic disc color fundus images and allows us to train the network with all our training data set from scratch (DENET DISC) and with transfer learning initializing it with the pre-trained models provided (DENET DISC TL).
The input in VGG19, GoogleNet, ResNet50 and DENet Disc networks were the preprocessed color fundus images from the data sets of the study with a final size of 224x224x3 and centered at the optic disc. The input images in the case of the standard network were 256x256x3. The region of interest presented to all the networks was the same. We changed the last layer with the softmax classifier in VGG19, GoogleNet and ResNet50 to consider only the two classes of interest in our study (glaucoma and normal).
2.4 Performance metrics
To evaluate the performance of the algorithms Receiver Operating Characteristic (ROC) analysis was performed and sensitivity/specificity ratios were calculated. We defined sensitivity, or true positive rate, as the number of true positives (number of images with glaucoma correctly detected) divided by the sum of the number of true positives and false negatives (images incorrectly classified as normal). Therefore, the sensitivity shows the percentage of glaucoma cases correctly identified by the algorithm. We defined specificity as the number of true negatives (number of images normal correctly detected in our case) divided by sum of the number of true negatives and the false positives (images incorrectly classified as glaucoma). The specificity is a ratio that shows the percentage of normal cases correctly identified. We also considered the Balanced Accuracy (BAcc) as the mean of sensitivity and specificity to take into account the imbalance in the number of positive and negative cases in the testing data set. We used the ROC graph for visualizing the performance of the networks [61]. The ROC graph is a two dimensional representation with the sensitivity in the Y axis and 1-specificity in the X axes. We compared the performance of the algorithms using the area under the receiver operating curve (AUC) generated by ROC curve. For the calculation of the optimum threshold we considered the Youden index, defined as the index where the sum of the specificity −1 and sensitivity is maximum [62].
2.5 Training and testing processes
The complete data set was randomly divided in three different groups: training, validation and test set with the distribution of images presented in Table 5. The validation set was used to monitor the number of epochs of the training process. The number of epochs with better performance in terms of accuracy in both classes (glaucoma and normal) with the validation set is listed in Table 6. After the selection of the hyper-parameters and the cross validation experiments, the rest of the trainings in the study considered in the training set the images from validation set. The final training set contained 370 glaucoma and 1364 normal images. The test sets were the same during all the experiments (124 glaucoma and 455 normal images)
Table 5. Distribution of images in the groups “glaucoma” and “normal”.
| Group | Training | Validation | Test |
|---|---|---|---|
| Glaucoma | 333 | 37 | 124 |
| Normal | 1227 | 137 | 455 |
Table 6. Number of epochs selected after the monitoring of the training process using the validation set.
| Group | Number of epochs |
|---|---|
| VGG19 TL | 20 |
| VGG19 | 100 |
| RESNET TL | 45 |
| RESNET | 80 |
| GOOGLENET TL | 40 |
| GOOGLENET | 100 |
| DENET DISC TL | 25 |
| DENET DISC | 130 |
| STANDARD CNN | 50 |
The overfitting it is a well-known issue in CNNs with limited training data. In order to limit this important problem in the training process we applied different strategies commonly used for this purpose. First, the selection of the number of epochs to complete the process was stopped when the performance on the validation set was reduced. Second, we applied dropout, the technique presented by [63] that consists in temporarily removing units along with all its incoming and outgoing connections in a neural network. We included dropout with p = 0.5 in the standard CNN. In the rest of the tested architectures we maintained the regularization schemes designed by the authors. Finally, we applied data augmentation. This technique consists of training and/or testing on systematically transformed images. The transformations used typically have to maintain the classification of the original image. In our study we first balanced the number of images in each group to 1400 in both groups (glaucoma and normal), with horizontal flips in the case of the normal images and with a random combination of flips and translations of 20 pixels for the glaucoma images. We also used data augmentation during training in all the networks. In each iteration, every image included in the batch could be transformed by a random combination of the operations: random flip, random small rotations between −10 degrees and + 10 degrees and random translation of maximum *-20 pixels in the x or y direction of the image.
2.6 Implementation
The preprocessing steps were implemented using Matlab R2016b 64-bits (Mathworks, Inc.) on a desktop computer equipped with an Intel Xeon CPU ES31245 and the CNN experiments were implemented in Python (version 2.7.12) using the libraries Lasagne (version 0.2) [64] and Theano (version 0.9) [65] in a desktop computer with a NVIDIA GeForce GTX Titan Pascal 12GB GPU.
3. Results and discussion
We defined several experiments to evaluate the best solution in terms of performance and to get more insight of the different alternatives tested.
3.1 Hyper-parameter selection
As described before we used the validation set to select the number of epochs for all the networks and types of training (full training/fine tuning) used during the study. In Table 6 we present the final values selected.
For the STANDARD CNN we used Stochastic Gradient Descent (SDG) updates, a binary cross entropy loss function, a learning rate of 0.005 and a batch size of 64. In the case of the transfer learning networks (VGG19, RESNET50, GOOGLENET and DENET DISC) we selected SDG updates with Nesterov momentum 0.9, a learning rate of 0.0001, the categorical cross entropy loss function and a batch size of 32.
3.2 CNN algorithm comparison
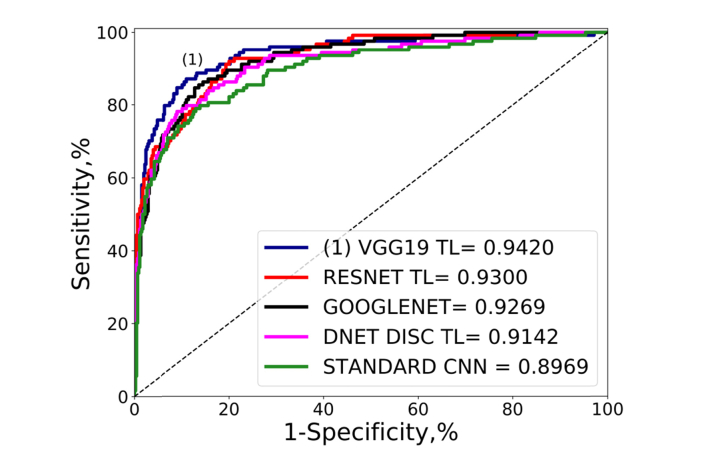
After the selection of the hyper-parameters we evaluated all the architectures under study (STANDARD CNN, VGG19, RESNET, GOOGLENET and two versions of DENET) in terms of the performance metrics, using all the data included in the training and validation subsets for training, and for testing the full test set. In Table 7 and Fig. 4 we present the ROC curves and the performance metrics. The two first options were VGG19 TL and RESNET50 TL with an AUC of 0.942 and 0.930 and in the case of RESNET TL the best sensitivity 91.94%. It is also remarkable that DENET DISC TL (based on ResNet50) and fine tuned from ORIGA data set did not outperform RESNET50 TL fine-tuned from ImageNet. The impact from fine-tuning from a general data set (ImageNet) is clear if we consider the difference between the performance of both trainings (full training and transfer learning) in VGG19 and RESNET50. In the case of DENET DISC, that used fine tuning from a targeted model (ORIGA), we noticed the biggest improvement in AUC (from 0.8371 to 0.9142) compared with the rest of the networks that used ImageNet.
Table 7. Performance ratios of all the CNNs evaluated. The sensitivity and specificity were calculated using the Youden index previously described. The networks were evaluated with the total test set (370 normal and 124 glaucoma). The best option for each metric is highlighted in bold.
| Architecture | AUC | Sensitivity (%) | Specificity (%) | B-Accuracy |
|---|---|---|---|---|
| VGG19 TL | 0.9420 | 87.01 | 89.01 | 88.05 |
| VGG19 | 0.8971 | 82.26 | 86.81 | 84.53 |
| RESNET TL | 0.9300 | 91.94 | 80.00 | 85.97 |
| RESNET | 0.9193 | 83.87 | 86.15 | 85.01 |
| GOOGLENET TL | 0.8994 | 83.96 | 81.76 | 82.41 |
| GOOGLENET | 0.9269 | 84.68 | 87.25 | 85.97 |
| DENET DISC TL [42] | 0.9142 | 78.22 | 90.99 | 84.61 |
| DENET DISC [42] | 0.8371 | 79.03 | 75.16 | 77.09 |
| STANDARD CNN | 0.8969 | 79.03 | 87.03 | 83.03 |
Fig. 4.
Global performance comparison ROC curves and AUC values of the comparison of the networks of the study using the test set described in Table 5. The first option in terms of AUC is labeled as (1).
VGG19 TL performed with the highest AUC, 0.9420, which is in the range of state of art results of the last published studies (AUCs between 0.91 and 0.985) [42–45]. The performance of the VGG19 TL was even higher (AUC 0.9640, sensitivity 94.51%, specificity 90.99% and BAcc 92.75%) if we considered only the glaucoma images at any stage of the disease, without the cases represented as glaucoma-suspect present in the ESPERANZA data set.
As we mentioned before, DENET uses global fundus images. As all the images in RIM-ONE data sets were centered in the optic disc, the number of data sets that we considered to evaluate DENET were limited to ESPERANZA and DRISHTI-GS data sets. Taking into account these two data sets we evaluated DENET directly without further training. The performance ratios (AUC 0.7507, sensitivity 70.45%, specificity 70.26% and BAcc 70.35%) were inferior to the performance of all other networks for this sub-test data set (Table 8).
Table 8. Performance ratios of all the CNNs evaluated considering a subset of the test set including the images from ESPERANZA and DRIHSTI-GS data sets set (343 normal and 45 glaucoma). The sensitivity and specificity were calculated using the Youden index previously described. The best option in each metric is highlighted in bold.
| Architecture | AUC | Sensitivity (%) | Specificity (%) | B-Accuracy |
|---|---|---|---|---|
| VGG19 TL | 0.9270 | 93.33 | 81.63 | 87.48 |
| VGG19 | 0.8491 | 71.11 | 89.21 | 80.16 |
| RESNET TL | 0.8957 | 84.44 | 85.71 | 85.08 |
| RESNET | 0.8778 | 71.11 | 93.00 | 82.06 |
| GOOGLENET TL | 0.8821 | 86.67 | 76.68 | 81.67 |
| GOOGLENET | 0.9276 | 80.00 | 90.96 | 85.48 |
| DENET DISC TL [42] | 0.9272 | 82.22 | 91.55 | 86.88 |
| DENET DISC [42] | 0.8261 | 77.78 | 75.51 | 76.64 |
| DENET [42] | 0.7507 | 70.45 | 70.26 | 70.35 |
| STANDARD CNN | 0.8312 | 75.76 | 78.72 | 77.14 |
These results demonstrate that CNNs can be trained, using large data sets and without having to specify lesion-based features, to identify glaucoma in retinal fundus images with high sensitivity and high specificity. Besides, the best option achieved state-of-the art performance ratios with images coming from multiples sources and different sizes and formats. According to our results this heterogeneity of the data sets could represent a differential value for the training of this type of classification strategies, and suppose a relevant parameter to consider respect to other issues like the complexity of the network or a great amount of more homogeneous images, where the capacity of the network to learn new features could be more limited. In this sense, it is remarkable how recent studies like [43] that reported an AUC of 0.91 or [44] that achieved an AUC of 0.986, required data sets with a significantly higher number of images (Table 1) achieving comparable or worse results in comparison with the proposal of this study.
We additionally investigated the contribution of each learning strategy (full training vs transfer learning) in the final prediction. For that purpose we created ensemble versions of each network considering the mean prediction of the full and the fine tuned trainings. As we present in Table 9 the performance ratios did not outperform the best option of each network in all the performance ratios. This is more significant in VGG19 and DNET were the difference between the performance ratios of both trainings was more relevant. RESNET presented a slight improvement in AUC but worse values in the rest of the ratios. GOOGLENET presented an improvement in AUC and sensitivity but decay in specificity and BAcc. These results suggest that features learned by both trainings for the same network are not complementary and their combination does not represent any clear advantage for the final classification.
Table 9. Performance ratios of the Ensemble of the full training and fine tuned of each CNN.
| Architecture | AUC | Sensitivity (%) | Specificity (%) | B-Accuracy |
|---|---|---|---|---|
| VGG19 | 0.9219 | 83.64 | 89.45 | 86.35 |
| RESNET | 0.9305 | 83.87 | 85.71 | 84.79 |
| GOOGLENET | 0.9277 | 88.71 | 82.85 | 85.78 |
| DENET DISC | 0.9083 | 81.45 | 88.35 | 84.90 |
3.3 Ten-fold cross validation
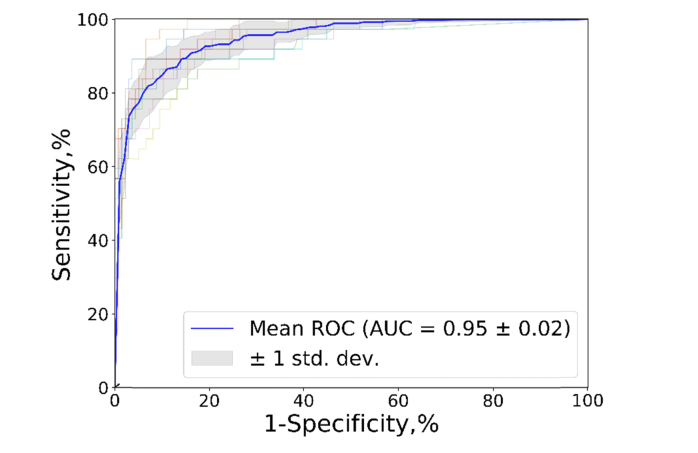
To confirm robustness and stability of the VGG19 TL network, a 10-fold cross validation test was performed considering all the images included in the training and validation subsets (Table 5) to make the different splits of the cross validation and allowing to compute the performance of the network ten times with different training and validation subsets. The results showed an acceptable variability in the performance of the network with a standard deviation of AUC of 0.02 in the Mean ROC. Figure 5 shows the corresponding 10-fold ROC curves and the median of all the cases.
Fig. 5.
ROC curves and AUC values for the 10-fold cross validation experiment on VGG19 TL.
3.4 CNN/human evaluator performance
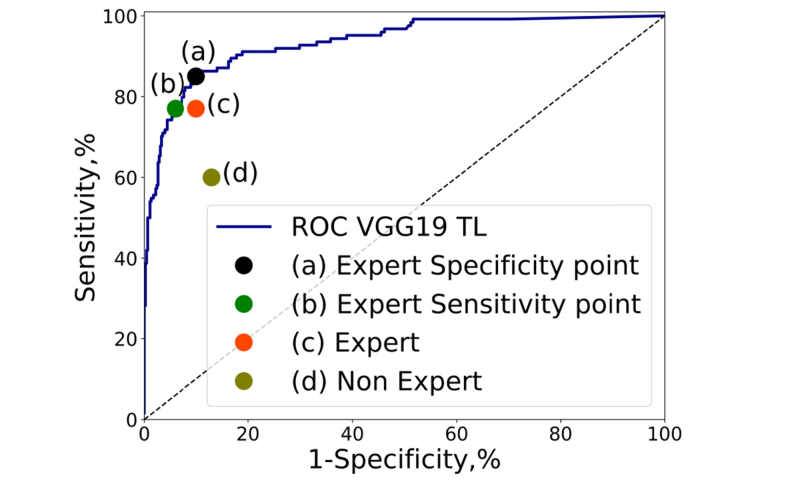
We compared the classification of the best network of previous experiments with the performance of the experts and non-experts evaluators. Like in [66] we selected two operating points. The first operating point approximates the specificity of the expert evaluators (89.14%) and the second operating point corresponded with the sensitivity of the expert evaluators (76.62%). The results are presented in Fig. 6, where we can appreciate the good performance of the solution compared with both groups of evaluators. Assuming the same specificity of an expert evaluator the network achieved higher sensitivity (85.48%) and for the case of the same sensitivity as experts the model scored higher specificity (93.18%). According with these results the proposed method can achieve high sensitivity and specificity, with ratios comparable to an expert ophthalmologist with more than 5 years of experience. Screening populations for a high prevalence disease like glaucoma, require both high sensitivity and high specificity to minimize both false-positive and false-negative results. These conclusions are similar with the ones presented recently by Shibata et. al. [45] that compared the performance of residents grouped in three clusters according to their years of experience and ResNet50.
Fig. 6.
ROC curve of VGG19 with fine tuning (VGG19_TL) over the test set of Table 5 and relevant operating points. (a) Expert specificity operating point of the ROC curve with sensitivity 85.48% and specificity 89.67% (b) Expert sensitivity operating point of the ROC curve with sensitivity 77.41% and specificity 93.18% (c) Reference performance of an ophthalmologist expert in glaucoma considering all the ESPERANZA data set, sensitivity 76.62% and specificity 89.14%. (d) Reference performance of an ophthalmologist non expert in glaucoma considering all the ESPERANZA data set, sensitivity 58.75% and specificity 86.07%.
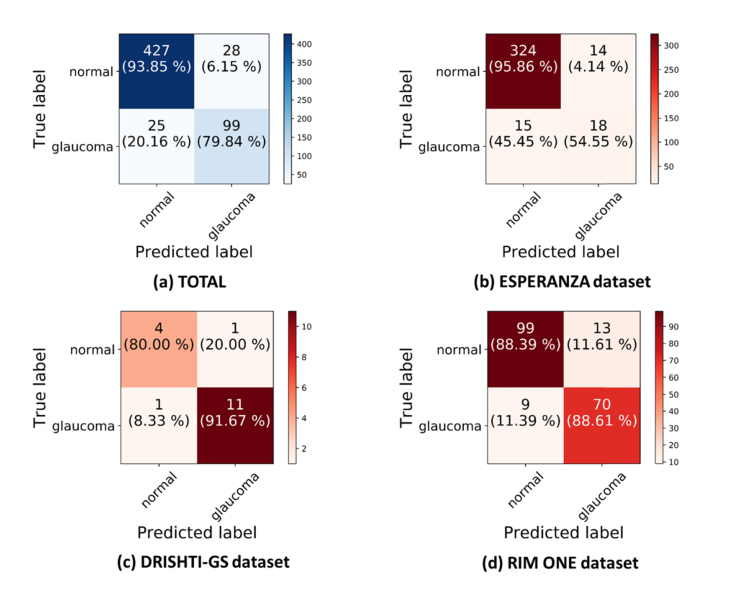
In Fig. 7 we show the different behavior of the network with the different data sets. We can notice the inferior performance in the ESPERANZA data set compared to the other data sets (DRISHTI-GS and RIM-ONE). There are several reasons that could explain this behavior. First, it was obtained in a screening setting so cases were not preselected and all cases with quality images were included. Pre-selection of cases may bias the databases and also the estimation of any classification algorithm, since cases with interpretation doubts are usually excluded although they do exist in the population. Second, although the ESPERANZA data set had a double evaluation with consensus it is a screening labeled data set and further tests are needed to confirm the diagnosis, having considered for this study the positive cases suspected of glaucoma by assessing the fundus images. Third, the position of the optic disc was not centered in the photo since these images were also used for screening of other retinal diseases. The color fundus images acquired for this data set were centered in the macula and the optic disc was at the border of the image. In the other data sets all the images are centered at the optic disc. Finally, the initial image size of the global image in the ESPERANZA data set (1024x680) supposes a possible limitation compared with the images of the other data sets, which were already segmented (RIM-ONE) or had much more resolution (DRISHTI-GS).
Fig. 7.
Confusion matrixes for the test data set considering a training of VGG19 with data augmentation on the fly. (a) All the test data set of the study. (b) Test set from the ESPERANZA database. (c) Test set from DRISHTI-GS database. (d) Test set from RIM-ONE database.
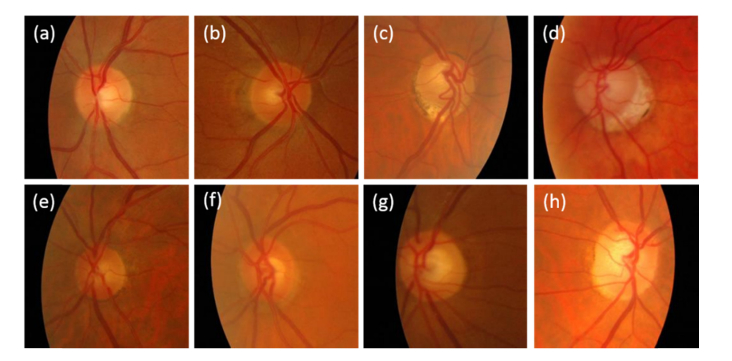
Figure 8 shows examples of the classification of the network in the ESPERANZA data set with true negatives (a, b), true positives (c, d), false negative (e, f) and false positive (g, h) classifications with respect to the consensus gold standard. If we focus on the false negative examples (e and f), it is remarkable that for example in the case (e) the non-expert evaluator labeled the image as normal while the expert evaluator classified it as glaucoma and the final consensus evaluation was glaucoma based on the abnormal ISNT rule. The case (f) is similar, the non-expert classified the disc as normal and the expert as glaucoma, and the consensus labeled the image as being abnormal and showing rim thinning and with an abnormal ISNT rule violation. This is not the case of the false positives (g, h) where the expert and non-expert evaluators labeled the image as normal but the algorithm identified them as glaucoma.
Fig. 8.
Example of the classification of the VGG19_TL network for images from the test set of the ESPERANZA database. (a, b) True negative examples, the human evaluators and the algorithm identified the images as normal. (c, d) True positives examples, the human evaluators and the algorithm identified the images as glaucoma. (e, f) False negatives examples. The human evaluation identified both images as glaucoma but the algorithm labeled them as normal. (g, h) False positives examples. The human evaluators marked the images as normal but the algorithm classified them as glaucoma.
3.5 Data sets performance influence
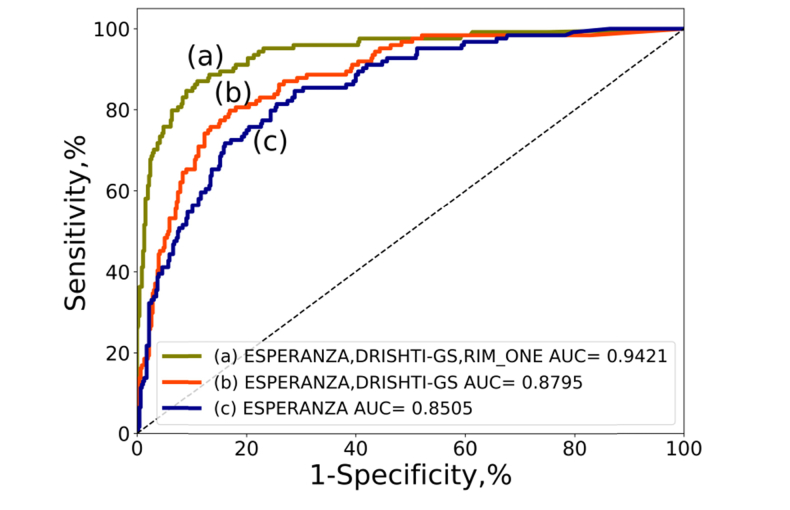
We evaluated the improvement in the performance of VGG19_TL associated with the inclusion of the different data sets for training. For that purpose, we evaluated the network trained with one data set (ESPERANZA), two (ESPERANZA and DRISHTI-GS) and the final data set adding RIM-ONE. To study the performance of each trained model we always used the full test set that includes data of the three data sets. The full test data set was used to verify the performance in the three training configurations. In Fig. 9 we can verify that the evolution in the performance is clear starting in 0.8505 with 113 glaucoma and 1333 normal cases to 0.9436 with the full data set with 494 glaucoma and 1819 normal cases.
Fig. 9.
ROC curves and AUC values using always the test set of Table 5 and training VGG19 with fine tuning and 100 epochs. The red line represents the ROC curve using the network trained only with the EPSERANZA data set. The blue line is the result after training with ESPERANZA and DRISHTI-GS data sets and the green line is the ROC curve after training using all the data sets of the study: ESPERANZA, DRISHTI-GS and RIM-ONE.
3.6 Integration of medical data and CNNs
The final experiment consisted in the integration of clinical history data collected during patient’s screening visit with color fundus images. As we have mentioned, CNNs are especially successful analyzing medical images. But the clinical diagnosis usually involves the assessment of a variety of exams that includes medical images but also data coming from different sources, like the medical history of the patient or from tests where the output could be a simple value. From this perspective, the exclusive use of one test could limit the final diagnosis and represent only a part of the global disease complexity. In order to consider a broader view in the diagnosis, some studies have explored the application of CNN integrating different imaging modalities [67,68] but also combining images with raw data, like in [69] where histological images and genomic data were integrated in a single CNN. Following this approach, we designed one experiment with the ESPERANZA data set integrating color fundus images with medical history data collected from the same patient during the screening campaign. The data selected from the screening visit were: the age, the intraocular pressure (IOP) in both eyes, the family history of glaucoma, the personal record of glaucoma and glaucoma related therapy. The selection of the data was based on some of the major risk factors for glaucoma described in the literature [1,70]. In the next tables we present more information of the clinical data used in the experiment. This data was collected during the examination of the patients in the ophthalmological screening campaign described in section 2.1.
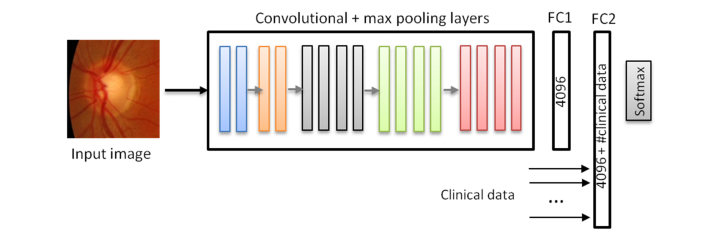
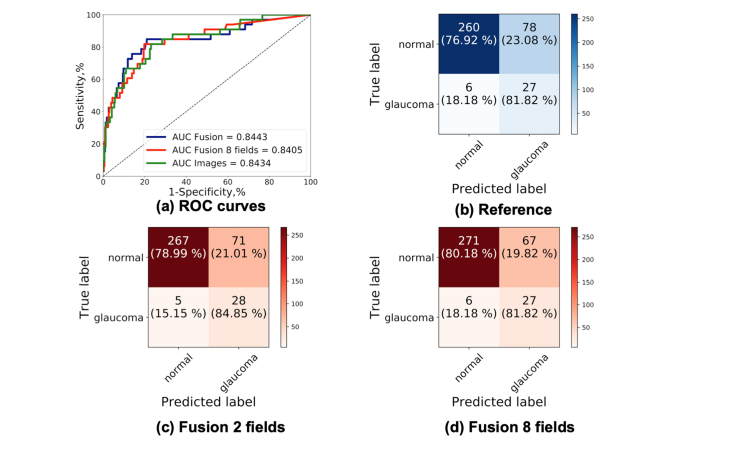
Color fundus images and the examination data were used together during the training by including the raw data into the last fully connected layer of the network. We present more details of the integration in Fig. 10. The clinical data were used in its raw format without post-processing. The value of the “Family record of glaucoma”, “Personal record of glaucoma” and “Personal glaucoma related therapy” had values 0 or 1. All the cases from the training and validation subsets from ESPERANZA were used for training (80 glaucoma and 995 normal) and the ESPERANZA cases from the test data set were used for testing (33 glaucoma and 338 normal). For this experiment we performed a full training and we only used the ESPERANZA data set, because is the only data set that contains information of the examination of the patients. We trained the network for 100 epochs. We performed two different experiments: the first one using all previous data and the second only with the age and the personal record of glaucoma because according with Tables 10 and 11 are the only data which appears different in the glaucoma and normal groups. As we can see in Table 12 and Fig. 11, in the AUC values no significant difference were appreciated by adding the clinical history data, but the sensitivity and specificity had higher values which indicate that this information could be valuable to improve the classification. The results were promising and further tests with other integration architectures and other clinical fields should be considered.
Fig. 10.
CNN Model based on VGG19 integrating the clinical data and color fundus image. The clinical data were incorporated to the model in the last fully connected layer. In the first experiment we included 8 clinical data and in the second the two selected from the analysis of Table 11.
Table 10. Mean IOP and standard deviation and mean age and standard deviation in the training set and test set in the glaucoma and normal classification.
| Data |
Glaucoma Set
Mean ± standard deviation |
Normal set
Mean ± standard deviation |
|---|---|---|
| IOP training set (mmHg) | 13.94 ± 3.16 | 13.97 ± 2.79 |
| IOP test set (mmHg) | 13.64 ± 2.79 | 14.20 ± 2.87 |
| Age training set (years) | 70.99 ± 8.56 | 68.81 ± 7.42 |
| Age test set (years) | 70.81 ± 8.51 | 68.34 ± 6.94 |
Table 11. Percentage of cases with glaucoma family history, personal record of glaucoma and personal related therapy in the training and test set of the glaucoma and normal classification.
| Data | Glaucoma Set | Normal set |
|---|---|---|
| Family history of glaucoma (Training set) | 8.75% | 11.76% |
| Family history of glaucoma (Test set) | 12.12% | 11.83% |
| Personal record of glaucoma (Training set) | 16.25% | 2.61% |
| Personal record of glaucoma (Test set) | 12.12% | 2.66% |
| Personal glaucoma related therapy (Training set) | 0% | 0.80% |
| Personal glaucoma related therapy (Test set) | 0% | 0.59% |
Table 12. AUC, specificity and sensitivity values corresponding with the result of applying the network trained with the ESPERANZA data set considering only the color fundus images (reference), integrating the images with all the data collected from the examination of the patient (fusion all data) and integrating only the age and personal record of glaucoma (fusion age/personal record glaucoma).
| Group | AUC | Sensitivity (%) | Specificity (%) |
|---|---|---|---|
| Reference | 0.8443 | 81.82 | 76.92 |
| Fusion all data | 0.8405 | 81.82 | 80.18 |
| Fusion age/personal record glaucoma | 0.8443 | 84.85 | 78.99 |
Fig. 11.
ROC curves and confusion matrices of the integration of images and clinical data. (a) ROC curves of the three trainings using only the ESPERANZA data set (b) Confusion matrix considering only the color fundus images (reference). (c) Confusion matrix considering the images with the age and personal record of glaucoma. (d) Confusion matrix considering the images with all clinical data collected.
In this study we have concentrated in a direct classification approach but other strategies and tools could be considered in order to aid the clinicians in the final diagnosis. The integration of clinical data from different sources or considering other imaging modalities could improve the performance of the classification, but other techniques that give insight and help to visualize and understand the features that have a relevant role in the final classification represent a valuable option. In this sense we highlight alternatives like the use of occlusion testing to recognize areas of the image with more impact in the classification [43,71] or saliency maps where the clinicians could be informed of the areas of the images with more influence in the prediction [72].
4. Conclusion
In this paper, we exploited and evaluated the application of deep convolutional neural networks for glaucoma detection using color fundus images in the context of large screening campaigns. We studied the influence of the architecture, the data set size, the training strategy and the integration of data collected from the clinical history and the patient examination. We used three different data sets, two publicly available DRISHTI-GS and RIM-ONE, and other created from a glaucoma screening campaign to assess the performance of the alternatives. The five architectures tested, standard CNN, VGG19, ResNet50, DENet and GoogLeNet offered good performance ratios in terms of AUC, sensitivity and specificity. The best option for the data set used was VGG19 with transfer learning and fine tuning, with an AUC of 0.94, a sensitivity of 87.01% and a specificity of 89.01%, which showed a similar performance with respect to the expert evaluators of the screening campaign. We confirmed the great influence of the number of images and data sets using CNNs. The AUC of the VGG19 with fine tuning increased from 0.85 with one data set to 0.94 with all the data sets of the study. Finally, we evaluated the performance of the integration of the data from the clinical history and tonometry tests with the color fundus images. The results show a slight improvement in sensitivity and specificity with similar AUCs. Further tests with more data and new architectural approaches should be developed and assessed to confirm this line of work. The good results presented demonstrated that CNNs are a valuable alternative for CAD systems to assess and classify fundus images for glaucoma detection campaigns.
Funding
Instituto de Salud Carlos III Fondo de Investigaciones Sanitarias (FIS PI15/00412); Spanish Ministry of Science, Innovation and Universities (TEC2015-66978-R).
Disclosures
The authors declare that there are no conflicts of interest related to this article.
References
- 1.Weinreb R. N., Aung T., Medeiros F. A., “The Pathophysiology and Treatment of Glaucoma: A Review,” JAMA 311(18), 1901–1911 (2014). 10.1001/jama.2014.3192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tham Y.-C., Li X., Wong T. Y., Quigley H. A., Aung T., Cheng C.-Y., “Global Prevalence of Glaucoma and Projections of Glaucoma Burden through 2040: A Systematic Review and Meta-Analysis,” Ophthalmology 121(11), 2081–2090 (2014). 10.1016/j.ophtha.2014.05.013 [DOI] [PubMed] [Google Scholar]
- 3.Tielsch J. M., Katz J., Singh K., Quigley H. A., Gottsch J. D., Javitt J., Sommer A., “A Population-based Evaluation of Glaucoma Screening: the Baltimore Eye Survey,” Am. J. Epidemiol. 134(10), 1102–1110 (1991). 10.1093/oxfordjournals.aje.a116013 [DOI] [PubMed] [Google Scholar]
- 4.Fleming C., Whitlock E. P., Beil T., Smit B., Harris R. P., “Screening for Primary Open-Angle Glaucoma in the Primary Care Setting: An Update for the US Preventive Services Task Force,” Ann. Fam. Med. 3(2), 167–170 (2005). 10.1370/afm.293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maul E. A., Jampel H. D., “Glaucoma Screening in the Real World,” Ophthalmology 117(9), 1665–1666 (2010). 10.1016/j.ophtha.2009.11.001 [DOI] [PubMed] [Google Scholar]
- 6.Zhao D., Guallar E., Gajwani P., Swenor B., Crews J., Saaddine J., Mudie L., Varadaraj V., Friedman D. S., Kanwar N., Sosa-Ebert A., Dosto N., Thompson S., Wahl M., Johnson E., Ogega C., “Optimizing Glaucoma Screening in High-Risk Population: Design and 1-Year Findings of the Screening to Prevent (SToP) Glaucoma Study,” Am. J. Ophthalmol. 180, 18–28 (2017). 10.1016/j.ajo.2017.05.017 [DOI] [PubMed] [Google Scholar]
- 7.Burr J. M., Mowatt G., Hernández R., Siddiqui M. A., Cook J., Lourenco T., Ramsay C., Vale L., Fraser C., Azuara-Blanco A., Deeks J., Cairns J., Wormald R., McPherson S., Rabindranath K., Grant A., “The clinical effectiveness and cost-effectiveness of screening for open angle glaucoma: a systematic review and economic evaluation,” Health Technol. Assess. 11(41), 1–190 (2007). 10.3310/hta11410 [DOI] [PubMed] [Google Scholar]
- 8.Einarson T. R., Vicente C., Machado M., Covert D., Trope G. E., Iskedjian M., “Screening for glaucoma in Canada: a systematic review of the literature,” Can. J. Ophthalmol. 41(6), 709–721 (2006). 10.3129/i06-064 [DOI] [PubMed] [Google Scholar]
- 9.Ervin A. M., Boland M. V., Myrowitz E. H., Prince J., Hawkins B., Vollenweider D., Ward D., Suarez-Cuervo C., Robinson K. A., “Screening for Glaucoma: Comparative Effectiveness,” D): Agency for Healthcare Research and Quality (US); 2012 Apr. Report No.: 12-EHC037-EF (2012). [PubMed]
- 10.Healey P. R., Lee A. J., Aung T., Wong T. Y., Mitchell P., “Diagnostic Accuracy of the Heidelberg Retina Tomograph for Glaucoma A Population-Based Assessment,” Ophthalmology 117(9), 1667–1673 (2010). 10.1016/j.ophtha.2010.07.001 [DOI] [PubMed] [Google Scholar]
- 11.Li G., Fansi A. K., Boivin J.-F., Joseph L., Harasymowycz P., “Screening for Glaucoma in High-Risk Populations Using Optical Coherence Tomography,” Ophthalmology 117(3), 453–461 (2010). 10.1016/j.ophtha.2009.07.033 [DOI] [PubMed] [Google Scholar]
- 12.Yamada N., Chen P. P., Mills R. P., Leen M. M., Stamper R. L., Lieberman M. F., Xu L., Stanford D. C., “Glaucoma screening using the scanning laser polarimeter,” J. Glaucoma 9(3), 254–261 (2000). 10.1097/00061198-200006000-00009 [DOI] [PubMed] [Google Scholar]
- 13.Burlina P. M., Joshi N., Pekala M., Pacheco K. D., Freund D. E., Bressler N. M., “Automated Grading of Age-Related Macular Degeneration From Color Fundus Images Using Deep Convolutional Neural Networks,” JAMA Ophthalmol. 135(11), 1170–1176 (2017). 10.1001/jamaophthalmol.2017.3782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Niemeijer M., van Ginneken B., Cree M. J., Mizutani A., Quellec G., Sanchez C. I., Zhang B., Hornero R., Lamard M., Muramatsu C., Wu X., Cazuguel G., You J., Mayo A., Li Q., Hatanaka Y., Cochener B., Roux C., Karray F., Garcia M., Fujita H., Abramoff M. D., “Retinopathy Online Challenge: Automatic Detection of Microaneurysms in Digital Color Fundus Photographs,” IEEE Trans. Med. Imaging 29(1), 185–195 (2010). 10.1109/TMI.2009.2033909 [DOI] [PubMed] [Google Scholar]
- 15.Haleem M. S., Han L., van Hemert J., Li B., “Automatic extraction of retinal features from colour retinal images for glaucoma diagnosis: A review,” Comput. Med. Imaging Graph. 37(7-8), 581–596 (2013). 10.1016/j.compmedimag.2013.09.005 [DOI] [PubMed] [Google Scholar]
- 16.Abràmoff M. D., Alward W. L. M., Greenlee E. C., Shuba L., Kim C. Y., Fingert J. H., Kwon Y. H., “Automated Segmentation of the Optic Disc from Stereo Color Photographs Using Physiologically Plausible Features,” Invest. Ophthalmol. Vis. Sci. 48(4), 1665–1673 (2007). 10.1167/iovs.06-1081 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Walter T., Klein J.-C., “Segmentation of Color Fundus Images of the Human Retina: Detection of the Optic Disc and the Vascular Tree Using Morphological Techniques,” in Medical Data Analysis, Lecture Notes in Computer Science (Springer, Berlin, Heidelberg, 2001), pp. 282–287. [Google Scholar]
- 18.Mookiah M. R. K., Acharya U. R., Chua C. K., Lim C. M., Ng E. Y. K., Laude A., “Computer-aided diagnosis of diabetic retinopathy: A review,” Comput. Biol. Med. 43(12), 2136–2155 (2013). 10.1016/j.compbiomed.2013.10.007 [DOI] [PubMed] [Google Scholar]
- 19.Bock R., Meier J., Nyúl L. G., Hornegger J., Michelson G., “Glaucoma risk index: Automated glaucoma detection from color fundus images,” Med. Image Anal. 14(3), 471–481 (2010). 10.1016/j.media.2009.12.006 [DOI] [PubMed] [Google Scholar]
- 20.Acharya U. R., Dua S., Du X., Sree S V., Chua C. K., “Automated Diagnosis of Glaucoma Using Texture and Higher Order Spectra Features,” IEEE Trans. Inf. Technol. Biomed. 15(3), 449–455 (2011). 10.1109/TITB.2011.2119322 [DOI] [PubMed] [Google Scholar]
- 21.Krishnan M. M. R., Faust O., “Automated glaucoma detection using hybrid feature extraction in retinal fundus images,” J. Mech. Med. Biol. 13(01), 1350011 (2013). 10.1142/S0219519413500115 [DOI] [Google Scholar]
- 22.Mookiah M. R. K., Rajendra Acharya U., Lim C. M., Petznick A., Suri J. S., “Data mining technique for automated diagnosis of glaucoma using higher order spectra and wavelet energy features,” Knowl.- Based Syst. 33, 73–82 (2012). 10.1016/j.knosys.2012.02.010 [DOI] [Google Scholar]
- 23.Maheshwari S., Pachori R. B., Acharya U. R., “Automated Diagnosis of Glaucoma Using Empirical Wavelet Transform and Correntropy Features Extracted From Fundus Images,” IEEE J. Biomed. Health Inform. 21(3), 803–813 (2017). 10.1109/JBHI.2016.2544961 [DOI] [PubMed] [Google Scholar]
- 24.Acharya U. R., Bhat S., Koh J. E. W., Bhandary S. V., Adeli H., “A novel algorithm to detect glaucoma risk using texton and local configuration pattern features extracted from fundus images,” Comput. Biol. Med. 88, 72–83 (2017). 10.1016/j.compbiomed.2017.06.022 [DOI] [PubMed] [Google Scholar]
- 25.Fukushima K., Miyake S., “Neocognitron: A Self-Organizing Neural Network Model for a Mechanism of Visual Pattern Recognition,” in Competition and Cooperation in Neural Nets, Lecture Notes in Biomathematics (Springer, Berlin, Heidelberg, 1982), pp. 267–285. [Google Scholar]
- 26.Litjens G., Kooi T., Bejnordi B. E., Setio A. A. A., Ciompi F., Ghafoorian M., van der Laak J. A. W. M., van Ginneken B., Sánchez C. I., “A survey on deep learning in medical image analysis,” Med. Image Anal. 42, 60–88 (2017). 10.1016/j.media.2017.07.005 [DOI] [PubMed] [Google Scholar]
- 27.Shin H. C., Roth H. R., Gao M., Lu L., Xu Z., Nogues I., Yao J., Mollura D., Summers R. M., “Deep Convolutional Neural Networks for Computer-Aided Detection: CNN Architectures, Dataset Characteristics and Transfer Learning,” IEEE Trans. Med. Imaging 35(5), 1285–1298 (2016). 10.1109/TMI.2016.2528162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pan S. J., Yang Q., “A Survey on Transfer Learning,” IEEE Trans. Knowl. Data Eng. 22(10), 1345–1359 (2010). 10.1109/TKDE.2009.191 [DOI] [Google Scholar]
- 29.Deng J., Dong W., Socher R., Li L. J., Li K., Fei-Fei L., “ImageNet: A large-scale hierarchical image database,” in 2009 IEEE Conference on Computer Vision and Pattern Recognition (2009), pp. 248–255. 10.1109/CVPR.2009.5206848 [DOI] [Google Scholar]
- 30.Russakovsky O., Deng J., Su H., Krause J., Satheesh S., Ma S., Huang Z., Karpathy A., Khosla A., Bernstein M., Berg A. C., Fei-Fei L., “ImageNet Large Scale Visual Recognition Challenge,” Int. J. Comput. Vis. 115(3), 211–252 (2015). 10.1007/s11263-015-0816-y [DOI] [Google Scholar]
- 31.Karri S. P. K., Chakraborty D., Chatterjee J., “Transfer learning based classification of optical coherence tomography images with diabetic macular edema and dry age-related macular degeneration,” Biomed. Opt. Express 8(2), 579–592 (2017). 10.1364/BOE.8.000579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Huynh B. Q., Li H., Giger M. L., “Digital mammographic tumor classification using transfer learning from deep convolutional neural networks,” J. Med. Imaging (Bellingham) 3(3), 034501 (2016). 10.1117/1.JMI.3.3.034501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tajbakhsh N., Shin J. Y., Gurudu S. R., Hurst R. T., Kendall C. B., Gotway M. B., Jianming Liang, “Convolutional Neural Networks for Medical Image Analysis: Full Training or Fine Tuning?” IEEE Trans. Med. Imaging 35(5), 1299–1312 (2016). 10.1109/TMI.2016.2535302 [DOI] [PubMed] [Google Scholar]
- 34.Fu H., Xu Y., Lin S., Wong D. W. K., Liu J., “DeepVessel: Retinal Vessel Segmentation via Deep Learning and Conditional Random Field,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016, Lecture Notes in Computer Science (Springer, Cham, 2016), pp. 132–139. [Google Scholar]
- 35.Mahapatra D., Roy P. K., Sedai S., Garnavi R., “Retinal Image Quality Classification Using Saliency Maps and CNNs,” in Machine Learning in Medical Imaging, Lecture Notes in Computer Science (Springer, Cham, 2016), pp. 172–179. [Google Scholar]
- 36.Zilly J., Buhmann J. M., Mahapatra D., “Glaucoma detection using entropy sampling and ensemble learning for automatic optic cup and disc segmentation,” Comput. Med. Imaging Graph. 55, 28–41 (2017). 10.1016/j.compmedimag.2016.07.012 [DOI] [PubMed] [Google Scholar]
- 37.Sevastopolsky A., “Optic Disc and Cup Segmentation Methods for Glaucoma Detection with Modification of U-Net Convolutional Neural Network,” Pattern Recognit. Image Anal. 27(3), 618–624 (2017). 10.1134/S1054661817030269 [DOI] [Google Scholar]
- 38.Abràmoff M. D., Lou Y., Erginay A., Clarida W., Amelon R., Folk J. C., Niemeijer M., “Improved Automated Detection of Diabetic Retinopathy on a Publicly Available Dataset Through Integration of Deep Learning,” Invest. Ophthalmol. Vis. Sci. 57(13), 5200–5206 (2016). 10.1167/iovs.16-19964 [DOI] [PubMed] [Google Scholar]
- 39.van Grinsven M. J. J. P., van Ginneken B., Hoyng C. B., Theelen T., Sánchez C. I., “Fast Convolutional Neural Network Training Using Selective Data Sampling: Application to Hemorrhage Detection in Color Fundus Images,” IEEE Trans. Med. Imaging 35(5), 1273–1284 (2016). 10.1109/TMI.2016.2526689 [DOI] [PubMed] [Google Scholar]
- 40.X. Chen, Y. Xu, D. W. K. Wong, T. Y. Wong, and J. Liu, “Glaucoma detection based on deep convolutional neural network,” in 2015 37th Annual International Conference of the IEEE Engineering in Medicine and Biology Society (EMBC) (2015), pp. 715–718. 10.1109/EMBC.2015.7318462 [DOI] [PubMed] [Google Scholar]
- 41.Al-Bander B., Al-Nuaimy W., Al-Taee M. A., Zheng Y., “Automated glaucoma diagnosis using deep learning approach,” in 2017 14th International Multi-Conference on Systems, Signals Devices (SSD) (2017), pp. 207–210. [Google Scholar]
- 42.Fu H., Cheng J., Xu Y., Zhang C., Wong D. W. K., Liu J., Cao X., “Disc-Aware Ensemble Network for Glaucoma Screening From Fundus Image,” IEEE Trans. Med. Imaging 37(11), 2493–2501 (2018). 10.1109/TMI.2018.2837012 [DOI] [PubMed] [Google Scholar]
- 43.Christopher M., Belghith A., Bowd C., Proudfoot J. A., Goldbaum M. H., Weinreb R. N., Girkin C. A., Liebmann J. M., Zangwill L. M., “Performance of Deep Learning Architectures and Transfer Learning for Detecting Glaucomatous Optic Neuropathy in Fundus Photographs,” Sci. Rep. 8(1), 16685 (2018). 10.1038/s41598-018-35044-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li Z., He Y., Keel S., Meng W., Chang R. T., He M., “Efficacy of a Deep Learning System for Detecting Glaucomatous Optic Neuropathy Based on Color Fundus Photographs,” Ophthalmology 125(8), 1199–1206 (2018). 10.1016/j.ophtha.2018.01.023 [DOI] [PubMed] [Google Scholar]
- 45.Shibata N., Tanito M., Mitsuhashi K., Fujino Y., Matsuura M., Murata H., Asaoka R., “Development of a deep residual learning algorithm to screen for glaucoma from fundus photography,” Sci. Rep. 8(1), 14665 (2018). 10.1038/s41598-018-33013-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Muhammad H., Fuchs T. J., De Cuir N., De Moraes C. G., Blumberg D. M., Liebmann J. M., Ritch R., Hood D. C., “Hybrid Deep Learning on Single Wide-field Optical Coherence tomography Scans Accurately Classifies Glaucoma Suspects,” https://www.ingentaconnect.com/content/wk/jglau/2017/00000026/00000012/art00008. 10.1097/IJG.0000000000000765 [DOI] [PMC free article] [PubMed]
- 47.Gopinath K., Rangrej S. B., Sivaswamy J., “A deep learning framework for segmentation of retinal layers from OCT images,” ArXiv180608859 Cs (2018).
- 48.Fang L., Cunefare D., Wang C., Guymer R. H., Li S., Farsiu S., “Automatic segmentation of nine retinal layer boundaries in OCT images of non-exudative AMD patients using deep learning and graph search,” Biomed. Opt. Express 8(5), 2732–2744 (2017). 10.1364/BOE.8.002732 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.F. Fumero, S. Alayon, J. L. Sanchez, J. Sigut, and M. Gonzalez-Hernandez, “RIM-ONE: An open retinal image database for optic nerve evaluation,” in 2011 24th International Symposium on Computer-Based Medical Systems (CBMS) (2011), pp. 1–6. 10.1109/CBMS.2011.5999143 [DOI] [Google Scholar]
- 50.J. Sivaswamy, S. R. Krishnadas, G. D. Joshi, M. Jain, and A. U. S. Tabish, “Drishti-GS: Retinal image dataset for optic nerve head(ONH) segmentation,” in 2014 IEEE 11th International Symposium on Biomedical Imaging (ISBI) (2014), pp. 53–56. 10.1109/ISBI.2014.6867807 [DOI] [Google Scholar]
- 51.S. Sekhar, W. Al-Nuaimy, and A. K. Nandi, “Automated localisation of optic disk and fovea in retinal fundus images,” in 2008 16th European Signal Processing Conference (2008), pp. 1–5. [Google Scholar]
- 52.Ioffe S., Szegedy C., “Batch Normalization: Accelerating Deep Network Training by Reducing Internal Covariate Shift,” ArXiv150203167 Cs (2015).
- 53.Simonyan K., Zisserman A., “Very Deep Convolutional Networks for Large-Scale Image Recognition,” ArXiv14091556 Cs (2014).
- 54.Antropova N., Huynh B. Q., Giger M. L., “A deep feature fusion methodology for breast cancer diagnosis demonstrated on three imaging modality datasets,” Med. Phys. 44(10), 5162–5171 (2017). 10.1002/mp.12453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liao H., “A Deep Learning Approach to Universal Skin Disease Classification,” 8 (2016).
- 56.Szegedy C., Liu W., Jia Y., Sermanet P., Reed S., Anguelov D., Erhan D., Vanhoucke V., Rabinovich A., “Going Deeper With Convolutions,” in (2015), pp. 1–9. [Google Scholar]
- 57.Ehteshami Bejnordi B., Veta M., Johannes van Diest P., van Ginneken B., Karssemeijer N., Litjens G., van der Laak J. A. W. M., Hermsen M., Manson Q. F., Balkenhol M., Geessink O., Stathonikos N., van Dijk M. C., Bult P., Beca F., Beck A. H., Wang D., Khosla A., Gargeya R., Irshad H., Zhong A., Dou Q., Li Q., Chen H., Lin H.-J., Heng P.-A., Haß C., Bruni E., Wong Q., Halici U., Öner M. Ü., Cetin-Atalay R., Berseth M., Khvatkov V., Vylegzhanin A., Kraus O., Shaban M., Rajpoot N., Awan R., Sirinukunwattana K., Qaiser T., Tsang Y.-W., Tellez D., Annuscheit J., Hufnagl P., Valkonen M., Kartasalo K., Latonen L., Ruusuvuori P., Liimatainen K., Albarqouni S., Mungal B., George A., Demirci S., Navab N., Watanabe S., Seno S., Takenaka Y., Matsuda H., Ahmady Phoulady H., Kovalev V., Kalinovsky A., Liauchuk V., Bueno G., Fernandez-Carrobles M. M., Serrano I., Deniz O., Racoceanu D., Venâncio R., “Diagnostic Assessment of Deep Learning Algorithms for Detection of Lymph Node Metastases in Women With Breast Cancer,” JAMA 318(22), 2199–2210 (2017). 10.1001/jama.2017.14585 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Weng S., Xu X., Li J., Wong S. T. C., “Combining deep learning and coherent anti-Stokes Raman scattering imaging for automated differential diagnosis of lung cancer,” J. Biomed. Opt. 22(10), 1–10 (2017). 10.1117/1.JBO.22.10.106017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.He K., Zhang X., Ren S., Sun J., “Deep Residual Learning for Image Recognition,” ArXiv151203385 Cs (2015).
- 60.Fu H., Cheng J., Xu Y., Wong D. W. K., Liu J., Cao X., “Joint Optic Disc and Cup Segmentation Based on Multi-Label Deep Network and Polar Transformation,” IEEE Trans. Med. Imaging 37(7), 1597–1605 (2018). 10.1109/TMI.2018.2791488 [DOI] [PubMed] [Google Scholar]
- 61.Fawcett T., “An introduction to ROC analysis,” Pattern Recognit. Lett. 27(8), 861–874 (2006). 10.1016/j.patrec.2005.10.010 [DOI] [Google Scholar]
- 62.Fluss R., Faraggi D., Reiser B., “Estimation of the Youden Index and its Associated Cutoff Point,” Biom. J. 47(4), 458–472 (2005). 10.1002/bimj.200410135 [DOI] [PubMed] [Google Scholar]
- 63.Srivastava N., Hinton G., Krizhevsky A., Sutskever I., Salakhutdinov R., “Dropout: A Simple Way to Prevent Neural Networks from Overfitting,” J. Mach. Learn. Res. 15, 1929–1958 (2014). [Google Scholar]
- 64.Sander Dieleman, Jan Schlüter, Colin Raffel, Eben Olson, Søren Kaae Sønderby, Daniel Nouri, Daniel Maturana, Martin Thoma, Eric Battenberg, Jack Kelly, Jeffrey De Fauw, Michael Heilman, diogo149, Brian McFee, Hendrik Weideman, takacsg84, peterderivaz, Jon, instagibbs, Dr. Kashif Rasul, CongLiu, Britefury, and Jonas Degrave, Lasagne: First Release. (Zenodo, 2015).
- 65.F. Bastien, P. Lamblin, R. Pascanu, J. Bergstra, I. Goodfellow, A. Bergeron, N. Bouchard, D. Warde-Farley, and Y. Bengio, “Theano: new features and speed improvements,” ArXiv12115590 Cs (2012).
- 66.Gulshan V., Peng L., Coram M., Stumpe M. C., Wu D., Narayanaswamy A., Venugopalan S., Widner K., Madams T., Cuadros J., Kim R., Raman R., Nelson P. C., Mega J. L., Webster D. R., “Development and Validation of a Deep Learning Algorithm for Detection of Diabetic Retinopathy in Retinal Fundus Photographs,” JAMA 316(22), 2402–2410 (2016). 10.1001/jama.2016.17216 [DOI] [PubMed] [Google Scholar]
- 67.Xu T., Zhang H., Huang X., Zhang S., Metaxas D. N., “Multimodal Deep Learning for Cervical Dysplasia Diagnosis,” in Medical Image Computing and Computer-Assisted Intervention – MICCAI 2016, Lecture Notes in Computer Science (Springer, Cham, 2016), pp. 115–123. [Google Scholar]
- 68.Liu S., Liu S., Cai W., Che H., Pujol S., Kikinis R., Feng D., Fulham M. J., Adni, ADNI , “Multimodal Neuroimaging Feature Learning for Multiclass Diagnosis of Alzheimer’s Disease,” IEEE Trans. Biomed. Eng. 62(4), 1132–1140 (2015). 10.1109/TBME.2014.2372011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mobadersany P., Yousefi S., Amgad M., Gutman D. A., Barnholtz-Sloan J. S., Velázquez Vega J. E., Brat D. J., Cooper L. A. D., “Predicting cancer outcomes from histology and genomics using convolutional networks,” Proc. Natl. Acad. Sci. U.S.A. 115(13), E2970–E2979 (2018). 10.1073/pnas.1717139115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Coleman A. L., Miglior S., “Risk Factors for Glaucoma Onset and Progression,” Surv. Ophthalmol. 53(6 Suppl1), S3–S10 (2008). 10.1016/j.survophthal.2008.08.006 [DOI] [PubMed] [Google Scholar]
- 71.Zeiler M. D., Fergus R., “Visualizing and Understanding Convolutional Networks,” in Computer Vision – ECCV 2014, Fleet D., Pajdla T., Schiele B., Tuytelaars T., eds., Lecture Notes in Computer Science (Springer International Publishing, 2014), pp. 818–833. [Google Scholar]
- 72.Esteva A., Kuprel B., Novoa R. A., Ko J., Swetter S. M., Blau H. M., Thrun S., “Dermatologist-level classification of skin cancer with deep neural networks,” Nature 542(7639), 115–118 (2017). 10.1038/nature21056 [DOI] [PMC free article] [PubMed] [Google Scholar]