Abstract
Purpose
Despite substantial progress in sequencing, current strategies can genetically solve only about 55–60% of inherited retinal degeneration (IRD) cases. This can partially be attributed to elusive mutations in the known IRD genes, which are not easily identified by the targeted next-generation sequencing (NGS) or Sanger sequencing approaches. We hypothesized that copy number variations (CNVs) are a major contributor to the elusive genetic causality of IRDs.
Methods
Twenty-eight patients, previously unsolved with a targeted NGS, were investigated with whole-genome SNP and CGH arrays.
Results
Deletions in the IRD genes were detected in five of twenty-eight families, including a de novo deletion. We suggest that the de novo deletion occurred through non-allelic homologous recombination (NAHR) and we constructed a genomic map of NAHR-prone regions with overlapping IRD genes. In this study we also report an unusual case of recessive retinitis pigmentosa due to compound heterozygous mutations in SNRNP200, a gene that is typically associated with the dominant form of this disease.
Conclusions
CNV mapping substantially increased the genetic diagnostic rate of IRDs, detecting genetic causality in 18% of previously unsolved cases. Extending the search to other structural variations (SVs) will likely demonstrate an even higher contribution to genetic causality of IRDs.
Keywords: Inherited retinal degenerations, retinitis pigmentosa, rod-cone dystrophy, cone dystrophy, Jalili syndrome, blue cone monochromacy, copy number variation
INTRODUCTION
Inherited retinal degenerations (IRDs) are important causes of blindness affecting more than 2 million people worldwide1. IRDs are a family of blinding diseases characterized by progressive death and dysfunction of rod and cone photoreceptor cells 1,2. There are several major clinical subtypes of IRDs, where the most common is retinitis pigmentosa (RP) also called rod-cone dystrophy, which accounts for roughly 25% of vision loss in adults2. Other subtypes of IRDs have been reported, e.g. affecting predominantly cones and/or the macula, or pan-retinal degenerations such a Leber Congenital Amaurosis1. Retinal degeneration is also one of the clinical manifestations of syndromic disorders such as Usher or Bardet-Biedl syndromes1,2. Even though distinct IRD subtypes have been identified, there is considerable phenotypic overlap between the different types of IRD, which becomes even more apparent upon genetic testing1.
IRDs are mostly monogenic, with over 200 genes associated with the isolated or syndromic forms3. However, despite substantial progress in sequencing and new disease gene discovery, current strategies can genetically solve only about 55–60% of IRD cases4–7. The high number of unsolved cases can be attributed to as yet-unidentified IRD genes or elusive mutations in the known disease genes. The latter may include silent mutations leading to aberrant splicing, deep intronic mutations, or structural variations (SVs), examples of which were previously identified in IRD disease genes8–11.
SVs include large insertions and deletions also called copy number variations (CNVs), inversions, translocations and other complex genomic rearrangements12. They are not readily detected by targeted next-generation sequencing (NGS) or Sanger sequencing approaches. Therefore, we hypothesized that the presence of SVs is one of the reasons why the molecular cause of disease in many IRD patients remains elusive. We investigated CNVs in 28 genetically unsolved families using a dense SNP-array (Illumina Human Omni 2.5/5.0) and/or a genome-wide Comparative Genomic Hybridization (CGH) array. We detected large deletions in known IRD disease genes in five families, demonstrating that a combined approach of targeted NGS and deletion mapping can lead to an estimated diagnostic rate of 67% of IRD cases. In two families we detected an essentially the same 1.1 Mb deletion, spanning two IRD genes (SNRNP200 and CNNM4). The recurrent deletion likely occurred through the mechanism of non-allelic homologous recombination (NAHR), where the rearranged genomic regions are flanked by paralogous repeat sequences12. We have therefore created a genome-wide map of NAHR-prone regions and superimposed known IRD genes. Overall 35 IRD genes and four genes with deletions reported in this study (SNRNP200, CNNM4, OPN1LW and OPN1MW) matched these regions.
MATERIALS AND METHODS
Patient cohort
The study protocol adhered to the tenets of the Declaration of Helsinki and was approved by the Institutional Review Boards of Massachusetts Eye and Ear Infirmary (MEEI, Harvard Medical School) and The Children’s Hospital of Philadelphia (CHOP, University of Pennsylvania Perelman School of Medicine). The patients included in the study were recruited and clinically examined at MEEI and/or CHOP. Patients underwent a full ophthalmic examination which included best-corrected Snellen visual acuity, dynamic Goldmann visual field testing with the I-4e and V-4e objects of Goldmann, dark adaptation testing performed after 45 minutes of dark adaptation with an 11° white test light in the Goldmann-Weekers dark adaptometer and full-field electroretinographic (ERG) testing with assessment of 0.5 Hz ERG amplitude and 30 Hz ERG amplitudes (MEEI) and an ISCEV-standard ERG testing (CHOP). After patients signed informed consent, blood samples were collected from patients and available family members for DNA extraction and genetic analysis.
Selective exon capture and whole exome sequencing (WES)
For custom selective exon capture, paired-end SureSelect targeted enrichment capture libraries (Agilent Technologies, Santa Clara, CA) were generated and sequenced on a MiSeq NGS platform (Illumina, San Diego, CA) as previously described4. Targeted enrichment included 258 known monogenic inherited retinal degeneration genes3,4. For WES, targeted enrichment capture libraries were generated using the SureSelect Human All Exon targeted enrichment kit (V4+UTR, Agilent Technologies) and sequenced using a HiSeq 2000 NGS instrument (Illumina) as before13. The NGS data was analyzed as previously described4,13 with annotations taken from dbSNP (http://www.ncbi.nlm.nih.gov/SNP/), Exome Aggregation Consortium (ExAC), 1000 Genomes Project, UK10K project data (http://www.uk10k.org/), Genomic Evolutionary Rate Profiling (GERP), SIFT, PolyPhen2 and retinal expression14. Rare variants were selected based on the minor allele frequency (MAF) in public databases of less than 0.5%.
Mutations were annotated based on the following transcripts: CNNM4 (NM_020184), DMD (NM_004007), EYS (NM_001142800), GPM6B (NM_001001995), KRT84 (NM_033045), MMGT1 (NM_173470), PDE6C (NM_006204), PNPLA4 (NM_004650), SNRNP200 (NM_014014), STARD9 (NM_020759) and ZNF449 (NM_152695).
Copy Number Variation (CNV) Analysis
Genomic DNA (gDNA) samples from probands and family members were analyzed with dense whole-genome SNP microarrays (Human Omni 2.5 or Human Omni 5.0, Illumina) according to the manufacturer’s instructions. The hybridized SNP arrays were analyzed using an array reader (iScan array scanner, Illumina) and the SNP calls were made with the genotyping module of the data analysis software (GenomeStudio, Illumina). A custom SNP genotyping report containing 1) SNP chromosome coordinates, 2) genotyping calls, 3) logR ratios, and 4) B-allele frequencies (BAF) was generated for each patient, followed by analysis using the CNV Workshop Suite15.
A genome-wide CGH array enriched with high density probes against the IRD disease genes was designed with the eArray online tool (4 × 180K microarray, Agilent Technologies), as described previously4. The high probe density enables detection of single exon deletions if more than one probe targets the exon. Samples were prepared using sex-matched control gDNA provided with the kit according to standard methods (SureTag Complete DNA Labeling Kit, Agilent Technologies). CGH analysis was done using CytoGenomics software (Agilent Technologies).
Quantitative real time PCR (qRT-PCR)
Deletions were validated and fine mapped using qRT-PCR on gDNA with primers specific to sequences flanking the presumed deletion breakpoint and normalized to two reference genes ZNF80 and GPR15 (Table S1). For each qRT-PCR reaction 5 ng of gDNA, 200 nM of each primer and 10 µl of Fast SYBR Green master mix (Life Technologies, Grand Island, NY) were used. The amplification was performed in a qPCR system (Stratagene Mx3000P®, Agilent Technologies) using the standard thermo-cycling program: 95°C for 3 min, 40 cycles of 95°C for 20 s, 60°C for 1 min followed by a melting curve. Each sample was assayed in triplicate, with two reference genes at each run (ZNF80 and GPR15). The standard deviation reflecting normalization to each of the reference genes was calculated and presented as the error bars.
PCR and Sanger sequencing
The validation of deletion breakpoints in families OGI-086 and OGI-036 was performed by PCR amplification using Herculase II fusion DNA Polymerase (Agilent Technologies) with the following primers: 5’-GACAGAGAGAAACTGGTCTC-3’ (OGI-086-Forward), 5’-CTTGTAGGTCAGGTGACTAG-3’ (OGI-086-Reverse), 5’-TAATTACAAAGGATTTGCAGGGAACAAG-3’ (OGI-036-Forward), 5’-CAGACGCAGTACGCAAAGAT-3’ (OGI-036-Reverse). The amplification conditions were the following: 92°C for 2 minutes; 36 cycles of 92°C for 20 seconds, 62°C for 30 seconds and 68°C for 1 minute (OGI-086) and 92°C for 2 minutes; 36 cycles of 92°C for 20 seconds, 58°C for 30 seconds and 68°C for 6 minutes (OGI-036). Sanger sequencing was performed after PCR cleanup (ExoSap-IT, Affymetrix, Santa Clara, CA, USA) and sequenced (BigDye Terminator v3.1, ABI 3730xl, Life Technologies) using the PCR primers.
Generating non-allelic homologous recombination (NAHR) prone regions
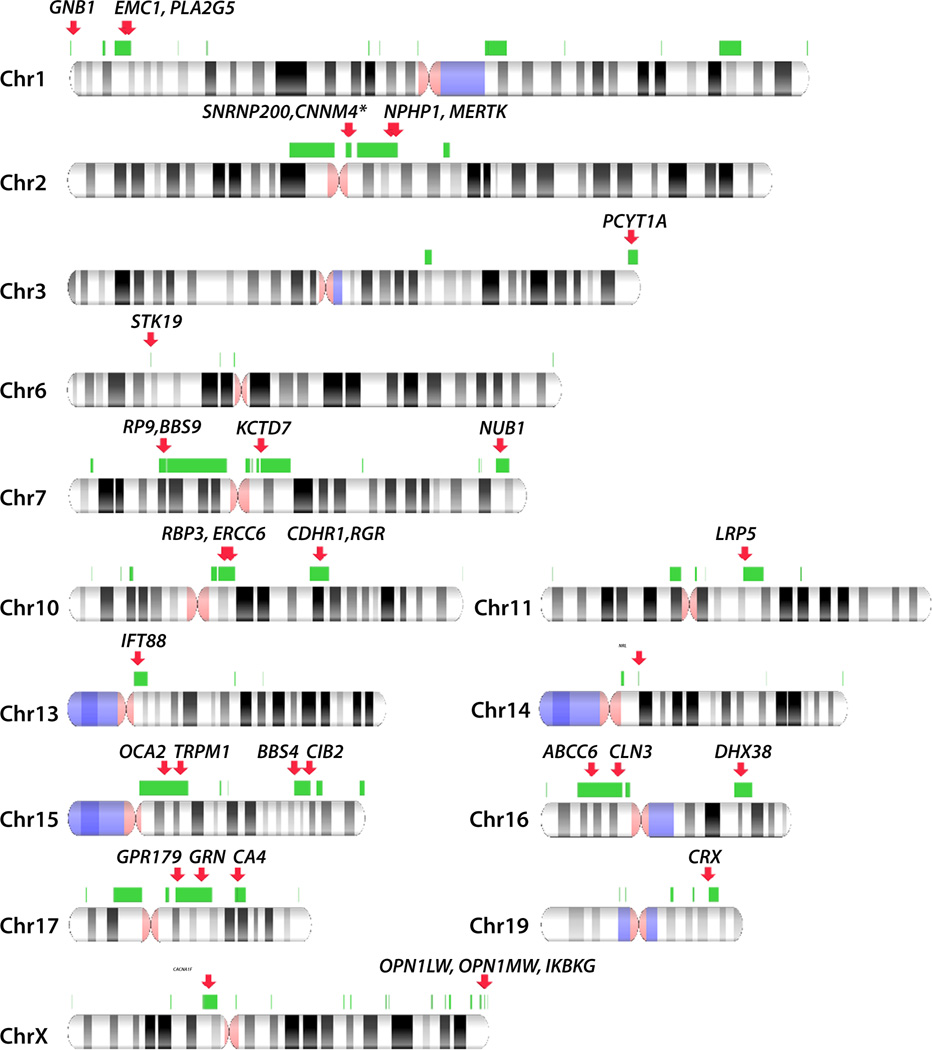
Segmental duplication track was obtained from the USCS genome browser (http://genome.ucsc.edu/) using the GRCh37/hg19 genome assembly. The regions were filtered to contain repeat pairs according to the following criteria: >10kb long regions, >95% identity, with intervening sequence between 50 kb and 10 Mb and not spanning the centromere. Direct and inverted repeats are included. The overlapping regions were subsequently merged to obtain 156 regions spanning ≈300Mb of genomic sequence (Table S2). Similar methodology was applied by Liu and colleagues12. IRD genes were superimposed unto the NAHR regions (Figure 4) using NCBI Genome Decoration Page (http://www.ncbi.nlm.nih.gov/genome/tools/gdp)
Figure 4. Non-allelic homologous recombination prone regions and IRD genes.
NAHR-prone regions (green bars) were computationally predicted using the following criteria: >10kb long regions of >95% identity, with intervening sequence between 50 kb and 10 Mb and not spanning the centromere. Direct and inverted repeats are included. IRD genes overlapping the NAHR-prone regions are indicated as red arrows. Only the chromosomes, where NAHR-prone regions and IRD genes overlap are presented. The full genome-wide map of NAHR-prone regions is presented in Table S2. The asterisk denotes IRD whole-gene CNVs reported in this manuscript for the first time.
RESULTS
Twenty-six families previously unsolved with whole-exome sequencing (WES) and two families with apparently homozygous mutations in known IRD genes were studied for deletions with a dense whole-genome SNP-array (Illumina Human Omni 2.5/5.0) and/or a CGH array. Using these techniques we detected large deletions in IRD genes in five families (Table 1). These copy number variations (CNVs) were further validated using PCR and quantitative real-time qRT-PCR.
Table 1.
Deletions detected in the IRD families
| Family | Deletion from SNP chip | Final deletion | IRD Gene affected | |||
|---|---|---|---|---|---|---|
| Position | Size | No. SNPs |
Position | Size | ||
| OGI-046 | chr2: 96741757- 98188430 |
1.4 Mb | 558 | ≈chr2:96755045-97823903 | ≈1.1 Mb | SNRNP200, CNNM4 |
| OGI-023 | chr2: 96688990- 97765355 |
1.1 Mb | 1099* | ≈chr2:96755045-97823903 | ≈1.1 Mb | SNRNP200, CNNM4 |
| OGI-086 | chr19: 54604319- 54632673 |
28.3 kb | 34 | chr19: 54602436-54636367 | 33,932 bp | PRPF31 |
| OGI-014 | chr6: 65604204- 65655039 |
50.8 kb | 36 | chr6:(65659138-65658256)- (65602857-65602460) |
≈55.4–56.7 kb | EYS (ex15-18) |
| OGI-036 | chrX: 153404608- 153420176 |
15.6 kb | 3 | chrX: 153400587- 153453251 | 52,664 bp | OPN1LW, OPN1MW |
This sample was analyzed using the Omni 5.0 SNP array
Patient OGI-046-116 presented with retinitis pigmentosa (RP) at age 9. Fundus examination showed early bone spicule pigmentation in the retinal peripheries of both eyes. The patient had constricted visual fields and elevated dark adaptation thresholds (Table S3). OCT imaging showed thinning of the peripheral retina and cystic changes in the macula. The proband and his parents were studied with WES, which revealed a hemizygous missense variant (c.3133C>A, p.Pro1045Thr) in SNRNP200 [MIM #601664], a gene typically associated with a dominant form of RP16. This variant was previously reported in a Chinese cohort of LCA patients; however no details of phenotype, family segregation or functional analysis of the variant were available17. The p.Pro1045Thr change is rare (2/104,468 alleles in the ExAC database) and the Pro1045 residue is conserved throughout all vertebrates available in the UCSC genome browser (Figure S1), therefore this variant was considered to be likely pathogenic as a recessive allele. The p.Pro1045Thr change was heterozygous in the father (OGI-046-114) but absent in the mother, which suggested the possibility of a deletion of SNRNP200 in trans with the missense mutation.
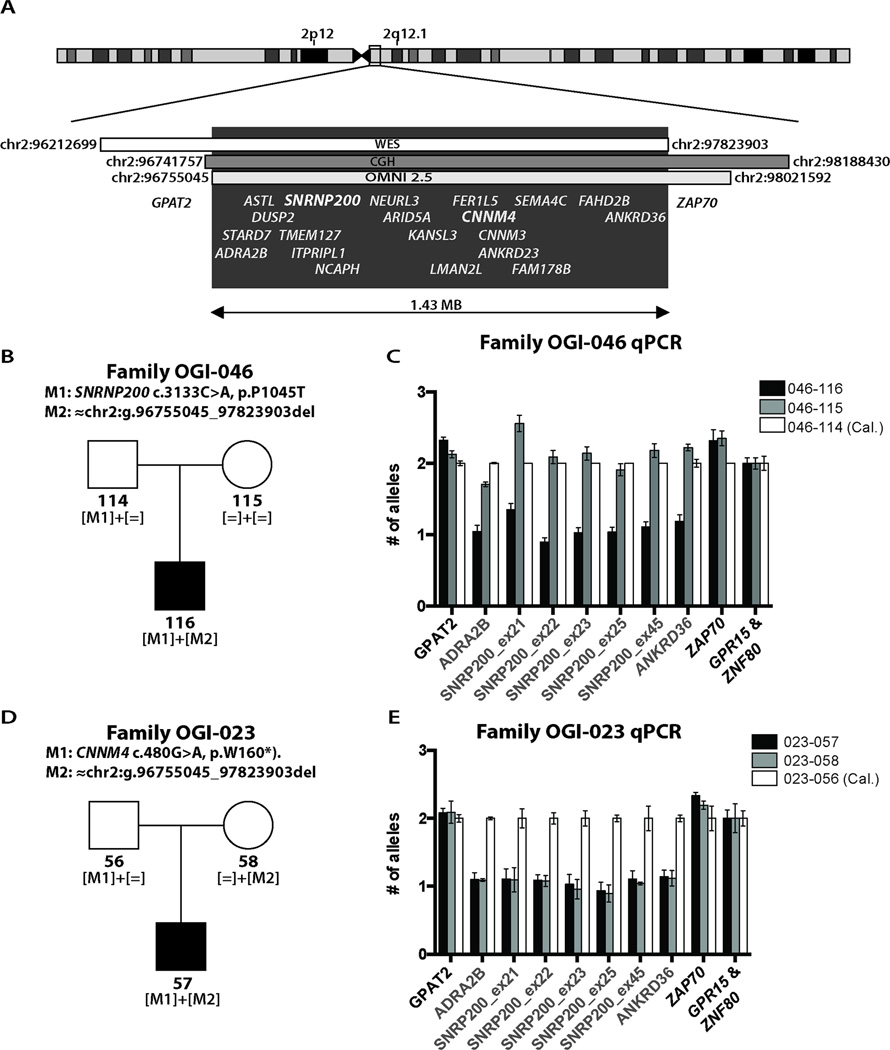
Results from a dense SNP chip indicated a deletion of 1.4 Mb (chr2: 96741757-98188430) in the proband. CGH array analysis and estimation of the homozygosity stretch identified by WES narrowed down the interval to 1.1 Mb (chr2:96755045-97823903) (Figure 1A). This region encompasses 20 genes, including SNRNP200, and CNNM4 [MIM #607805], a gene associated with a cone-rod dystrophy and amelogenesis imperfecta18,19, known as Jalili syndrome. Further qPCR validation in all family members indicated that this is a de novo deletion (Figure 1C). The absence of the deletion in the mother was further confirmed by 4 heterozygous SNPs in ADRA2B (rs34667759), ASTL (rs183967306 and chr2:g.96803404C>T) and in TMEM127 (rs13022177), all within the deleted region in the proband. In addition, whole genome SNP analysis of the proband and his mother confirmed the family relationship.
Figure 1. Deletion mapping in families OGI-046 and OGI-023.
A) A heterozygous deletion of 20 genes on chr2q12.1 found in family OGI-046 by CGH and SNP arrays, and validated by SNPs found in WES B) Pedigree of family OGI-046 with mutant allele segregation indicated. C) Fine mapping of the deletion by qPCR in family OGI-046. D) Pedigree of family OGI-023 with mutant allele segregation indicated. E) Fine mapping of the deletion by qPCR in family OGI-023. Each primer pair in the qPCR experiments was normalized to two reference genes GPR15 and ZNF80, where the error bars represent standard deviation between these results.
Interestingly, we found another family with a deletion of the same region (Family OGI-023, Figure 1D). In this family, the proband presented with the combination of cone dystrophy and amelogenesis imperfecta, typical of Jalili syndrome, at age 11 (Table S3) 18,19. Targeted genetic testing showed a homozygous nonsense mutation (c.480G>A, p.Trp160*) in CNNM4. Segregation analysis revealed that while the subject’s father carried the mutation (Figure 1D), his mother’s sequence was wild type at this location. To determine if this result was due to a maternally inherited deletion involving CNNM4, we analyzed the family using a dense SNP chip and a CGH array. These analyses identified a ~1.1Mb multigenic deletion involving CNNM4 and SNRNP200 in both the proband and the mother, which was confirmed and fine-mapped by qPCR analysis (Figure 1E). The qPCR analyses demonstrated that the deletion in both families occurs between the GPAT2 and ANDRA2B genes on the 5’ side and ANKRD36 and ZAP70 on the 3’ end. Unfortunately, we were not able to map the exact breakpoints in this family due to highly repetitive regions near the centromere. These highly repetitive regions flanking the deletion were at the same time informative about the possible mechanism of this CNV, since recurrent genomic rearrangements often occur through NAHR, between long highly homologous sequences on the same chromosome12. Indeed, we found that the deleted region is flanked by ≈16kb repeats bearing 98.5% sequence identity (chr2: 96598520- 96614694 and chr2: 97820062- 97836456).
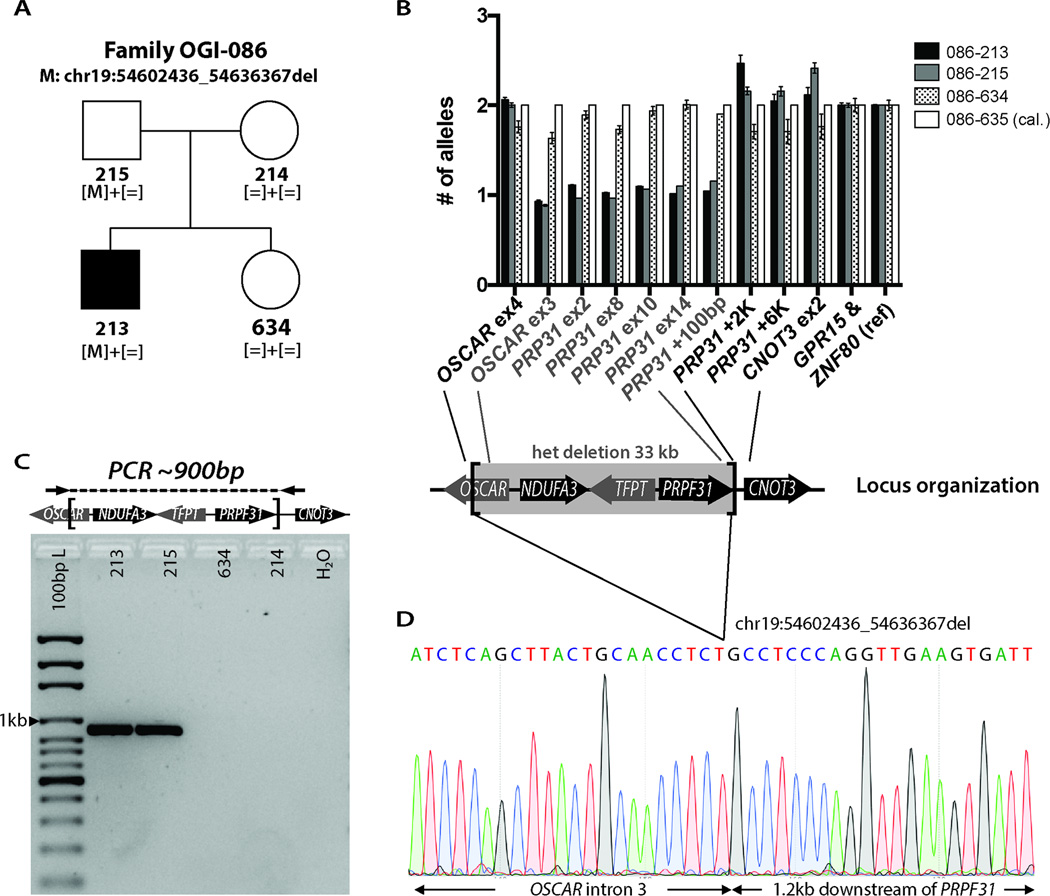
Patient OGI-086-213 was diagnosed with RP at age 13. During his last visit at age 18, he was found to have bilateral constricted visual fields and reduced ERG responses, bone spicule pigmentation in the retinal periphery and granularity in the macula (Table S3). In addition, examination of the anterior chamber of the eye showed bilateral anterior polar cataracts. Similar cataracts were also present in the father, who did not have any symptoms of visual impairment and whose retina showed no clinical signs of degeneration upon fundus examination. WES analysis showed rare variants in 7 genes (Table S4), none of which were previously associated with IRD. CNV analysis performed using the SNP array revealed a heterozygous 28 kb deletion on chromosome 19 in the proband and the father, which affected four genes: OSCAR, NDUFA3, TFPT and PRPF31 (Table 1). Mutations in PRPF31 are known to cause a dominant form of RP [MIM #600138], where the disease is caused by haploinsufficiency and large deletions have been documented to be pathogenic11. In addition, partial penetrance is a common feature in families with mutations in this gene, where the wild-type allele can have a disease-rescuing effect 20,21. Therefore, the unaffected status of the father was not surprising. The large deletion, which included PRPF31, was considered as the most likely cause of disease in the proband. This deletion was subsequently confirmed by qPCR and the breakpoints were PCR amplified and Sanger sequenced, which showed the deletion to be exactly 33,932 bp (chr19:54602436-54636367) (Figure 2).
Figure 2. Deletion mapping in family OGI-086.
A) Pedigree of family OGI-086 with mutant allele segregation. B) Fine mapping of the deletion by qPCR with deleted genes indicated (error bars represent standard deviation between two normalization genes GPR15 and ZNF80). C) PCR amplification across the breakpoints. D) Sanger sequencing electropherogram showing the precise position of the deletion.
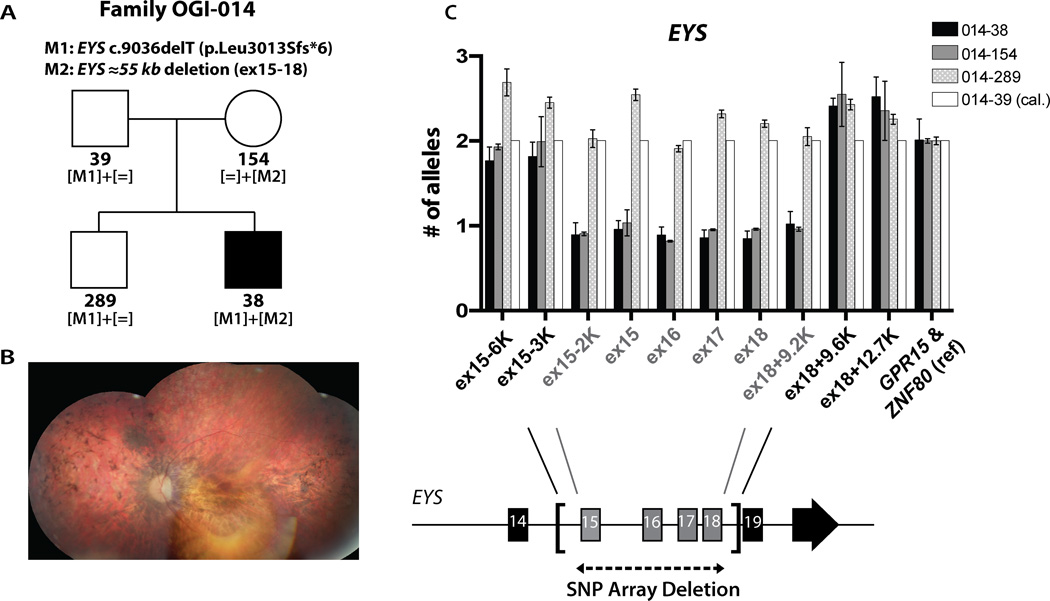
Patient OGI-014-038 had decreased vision and difficulty with night vision since age 15 and he was diagnosed with RP at age 33 (Table S3, Figure 3A and B). The proband and his unaffected family members were genetically investigated with WES and with the dense SNP array. WES analysis revealed rare coding changes indicative of recessive disease in 3 genes: PNPLA4 (homozygous c.575C>T, p.Pro192Leu); KRT84 (heterozygous c.1327C>T, p.Arg443Trp and heterozygous c.364T>G, p.Phe122Val) and STARD9 (heterozygous c.5584_5589del, p.Ser1862_Thr1863del and heterozygous c.8803G>C, p.Glu2935Gln), none of which were previously associated with IRD. Additional rare coding changes in trans with non-coding variants in 5 genes were identified (Table S4). Two heterozygous variants in known autosomal recessive IRD genes were found: a paternally inherited mutation in EYS (c.9036delT, p.Leu3013Sfs*6) and a maternally inherited variant in PDE6C (c.413T>C, p.Leu138Ser). These sequence alterations were not sufficient to explain the disease, however the subsequent CNV analysis revealed a heterozygous maternally inherited deletion of approximately 51 kb in EYS, encompassing exons 15–18, leading to a frameshift (c.2260_2846del, p.Ser754Ifs*3). The above data suggested that EYS is the most likely cause of the disease in this patient. The EYS deletion was further mapped by qPCR, demonstrating that the deletion is between 55.4 and 56.7 kb in size, where the 5’ breakpoint is 2.5–3.4 kb upstream of exon 15 (chr6:65659138-65658256) and the 3’ breakpoint is 9.2–9.6 kb downstream of exon 18 (chr6:65602857-65602460) (Figure 3C). Unfortunately, despite numerous attempts we were not able to PCR amplify across the breakpoints. This could be due to a more complex genomic rearrangement, such as translocation.
Figure 3. Deletion mapping in family OGI-014.
A) Pedigree of family OGI-014 with mutant allele segregation. B) Composite fundus photograph of the left eye of patient OGI-014-038. C) Fine mapping of the deletion by qPCR with four deleted exons of EYS indicated (error bars represent standard deviation between two normalization genes GPR15 and ZNF80).
Patient OGI-036-091 and his half-brother OGI-036-336 presented with decreased visual acuity, reduced cone responses on full field ERG testing and decreased color vision (Table S3, Figure S2A). Dark adaptation was within normal limits. Fundus examination showed pigment granularity in the macula and OCT imaging showed normal retinal architecture and thickness in the periphery with a diminished photoreceptor layer centrally, indicative of cone dystrophy. Family history was consistent with an X-linked inheritance pattern, since maternal grandfather was reported to have a retinal dystrophy (no clinical data available) (Figure S2B). The family was studied with whole-exome sequencing to detect causative variants on chrX. With the frequency criteria described in the methods section we detected only four variants shared by the three affected males (OGI-036-091, 336 and 753), heterozygous in the obligate carrier (OGI-036-333) and absent in the father (OGI-036-334). These were three 3’UTR variants (GPM6B c.*525T>C, ZNF449 c.*1661A>G and MMGT1 c.*2599T>C) and one intronic change (DMD c.7173+13A>G) (Table S4). None of these variants seemed likely pathogenic, we therefore performed SNP array analysis to detect deletions. Using this technique we detected a 15.6 kb deletion affecting the promoter region and exons 1–4 of OPN1LW (Table 1, Figure S2C), a gene known to be implicated in blue cone monochromacy22. PCR amplification of selected regions (Figure S2D), as well as amplification and Sanger sequencing across the breakpoints (Figure S2E) revealed a much larger deletion of 52,664 bp, which affects the entire OPN1LW gene with its promoter and the first exon of the adjacent OPN1MW (Figure S2F). In this family, the genetic diagnosis helped to refine the clinical diagnosis, which is likely blue cone monochromacy.
DISCUSSION
The results reported here indicate that CNVs contribute significantly to the genetic causality of IRDs. Of the 28 patients whose genetic cause of disease was not identified by panel based NGS testing or WES, likely disease-causing deletions were identified in five or 18% of cases. Four of the patients with deletions came from an original WES cohort of 56 patients; therefore we estimate that ≈7% of IRD patients carry large deletions in known IRD genes. This result has important implications for genetic diagnostic testing of patients with IRDs, as well as for studies seeking to identify novel IRD disease genes. The finding of the same deletion in two families with distinct causes of disease suggests that previously identified mechanisms that create SVs in the genome, such as NAHR, can create pathogenic alleles in IRD genes12,23. This suggests that methods capable of detecting more complex forms of genomic rearrangement, such as WGS or single-molecule long read sequencing, will help identify novel genetic causes of IRD8,24,25. SVs can also be detected by other techniques such as fluorescence in situ hybridization (FISH), however this technique is more useful to test a candidate genomic rearrangement or validate results from other methods rather than unbiased detection of genome wide SVs26.
Patients from two unrelated families carried essentially the same deletion, which spanned two known IRD disease genes: SNRNP20016 and CNNM418,19. The deletion in patient OGI-023–057 was maternally inherited and in addition he carried a paternally inherited stop mutation in CNNM4, which correlated with his phenotype of cone dysfunction and amelogenesis imperfecta as part of Jalili syndrome and the recessive inheritance of CNNM4-associated retinal degeneration18,19. The large deletion in patient OGI-046-116 occurred de novo and was accompanied by a likely pathogenic missense change in SNRNP200 (p.Pro1045Thr) inherited from his father. The proband showed typical signs of non-syndromic rod-cone dystrophy (Table S3)1,2. This genotype is unusual, since previously SNRNP200 was associated with dominantly inherited retinal degeneration16,27 and there is only one report of recessive disease due to mutations in SNRNP20017. SNRNP200 codes for an RNA helicase hBRR2, a component of the pre-mRNA spliceosome28. Mutations in other splicing genes (PRPF3, PRPF4, PRPF6, PRPF8, PRPF31, RP9) were also associated with dominant RP29–34, where disease mechanism in patients with PRPF31 mutations was shown to be haploinsufficiency 20,21. Therefore, originally, we hypothesized that the disease in patient OGI-046-116 is due to the haploinsufficiency of SNRNP200 and that the p.Pro1045Thr change has no functional effect, even though the amino acid residue is highly conserved (Figure S1). However, since we discovered another family with a large deletion encompassing SNRNP200, where the deletion carrier is unaffected (OGI-023–058), we concluded that missing one copy of SNRNP200 is not sufficient to cause the disease. We believe that the p.Pro1045Thr mutation likely creates a hypomorphic allele, and the combination of hemizygosity for SNRNP200 with the p.Pro1045Thr allele is compatible with life, but insufficient to maintain a healthy retina. Alternatively, the disease mechanism in SNRNP200-associated IRD is due to haploinsufficiency and the partial penetrance is due to other genetic factors, e.g. SNRNP200 expression level from the wild-type allele, as was demonstrated for PRPF3120,21. To test these hypotheses, however, a larger cohort of patients with SNRNP200 deletions is needed and so far the only reported mutations were missense changes and one splice-site variant27.
Another unusual finding is that both families OGI-023 and OGI-046 carried the same deletion that occurred de novo in patient OGI-046-116. Such recurrent genomic rearrangements are thought to result from NAHR, where the rearranged sequence is flanked by paralogous repeat sequences12,35. Indeed, the deleted interval of approximately 1.1 Mb, is flanked by ≈16kb repeats bearing 98.5% sequence identity. This region was also predicted to be prone to NAHR before12. Therefore, even though we are the first to report this 1.1Mb deletion encompassing SNRNP200 and CNNM4 associated with inherited retinal degenerations, we expect that this is a more common event. Deletion of this large region carries also additional implications, since other deleted genes may lead to phenotypes unrelated to IRDs, e.g. heterozygous loss-of-function mutations in TMEM127 have been associated with susceptibility to pheochromocytoma36. None of the subjects from families OGI-023 and OGI-046 with a heterozygous deletion in TMEM127 were reported to have pheochromocytoma or any other type of cancer.
Encouraged by the above example we hypothesized that other IRD genes may fall into the NAHR-prone regions and have recurrent CNVs. We therefore constructed a genome-wide map of NAHR regions and superimposed known IRD genes, implicated in isolated and syndromic disease (Figure 4). We found that 35 IRD genes are likely to be affected by NAHR, 13 of which were never associated with a CNV before, 9 had large indels affecting only some exons of the gene and the remaining 13 had CNVs affecting the entire gene reported (Figure 4, Table S2)27. Among the genes likely affected by NAHR were OPN1LW and OPN1MW, deletion in which was associated with blue cone monochromacy in this study (family OGI-036). Other researchers have also reported deletions in this region37,38, which validates our predictions of regions prone to recombination. We believe that identifying such recombination hotspots will facilitate searching for the missing genetic causality in IRD patients, and perhaps in patients with other Mendelian disorders.
The disease in the remaining two families was due to deletions in PRPF31 and EYS. Even though new deleted regions were found in this study, structural variations in these genes were reported previously11,39,40. Deletions in PRPF31 and EYS were also found to be major contributors to IRD pathology by other researchers, accounting for 2.5% of adRP and 4% of previously unsolved arRP cases respectively 11,39. These genes do not lie in the proximity of the NAHR regions and therefore other mechanisms of structural variations are likely implicated35. Before ascertaining that the disease in proband OGI-014-038 is due to disruption of EYS, there were three potential candidates for new genetic types of IRD. Variants in these three genes (PNPLA4, KRT84 and STARD9) were rare, affecting conserved residues and the genes were expressed in the retina (Table S4). It seems therefore imperative that CNVs in the known disease genes be investigated before new gene-disease associations are reported.
In summary, we detected deletions that contribute to the disease in five out of 28 IRD patients. The high rate of CNVs (≈18%) in the families that were not previously solved by the targeted NGS approaches indicates that these types of mutations are the frequent cause of the missing inheritance in IRDs and CNV detection should be included in genetic diagnostic testing. We expect that the integrated approach of the targeted NGS and CNV mapping could provide a genetic diagnosis for up to 67% of IRD cases (60% by NGS4–7 and additional 7% from CNV mapping). The techniques used in the present study (SNP and CGH arrays) can only detect large deletions or duplications, which is inherent to these techniques that rely on the color or strength of the fluorescence signal after hybridization of the gDNA to specific probes complementary to the target regions of the genome. Smaller CNVs, which are not targeted by the genome-wide probes in the arrays, as well as translocations, inversions or other complex structural rearrangements, which will not alter the intensity of the fluorescence signal, cannot be detected by SNP and CGH arrays. Consequently, it is possible that SVs in known IRD genes are a more important contributor to disease than detected in this study. Other researchers proposed using the NGS read-depth to infer CNVs, which also increased the genetic diagnostic rate of IRDs40. The read-depth analysis of NGS reads has similar limitations, therefore whole-genome-sequencing may be necessary to map certain SVs, as shown before8.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Eye Institute [EY012910 (EAP) and P30EY014104 (MEEI core support)], the Foundation Fighting Blindness (USA, EAP), the Fleming Family Foundation (KMB), BPL is a Senior Clinical Investigator of the Research Foundation Flanders (FWO), Belgium. The authors would like to thank the patients and their family members for their participation in this study and the Ocular Genomics Core members for their experimental assistance. The authors would like to thank the Exome Aggregation Consortium and the groups that provided exome variant data for comparison. A full list of contributing groups can be found at http://exac.broadinstitute.org/about.
Footnotes
The authors declare no conflict of interest related to the work presented in this manuscript.
Supplementary information is available at the Genetics in Medicine website.
References
- 1.Berger W, Kloeckener-Gruissem B, Neidhardt J. The molecular basis of human retinal and vitreoretinal diseases. Prog Retin Eye Res. 2010;29(5):335–375. doi: 10.1016/j.preteyeres.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 2.Hartong DT, Berson EL, Dryja TP. Retinitis pigmentosa. Lancet. 2006;368(9549):1795–1809. doi: 10.1016/S0140-6736(06)69740-7. [DOI] [PubMed] [Google Scholar]
- 3. [Accessed May 2016];Retinal Information Network (RetNet) available at https://sph.uth.edu/retnet/home.htm.
- 4.Consugar MB, Navarro-Gomez D, Place EM, et al. Panel-based genetic diagnostic testing for inherited eye diseases is highly accurate and reproducible, and more sensitive for variant detection, than exome sequencing. Genet Med. 2014;17(4):253–261. doi: 10.1038/gim.2014.172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Audo I, Bujakowska KM, Léveillard T, et al. Development and application of a next-generation-sequencing (NGS) approach to detect known and novel gene defects underlying retinal diseases. Orphanet J Rare Dis. 2012;7:8. doi: 10.1186/1750-1172-7-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neveling K, Collin RWJ, Gilissen C, et al. Next Generation Genetic Testing for Retinitis Pigmentosa. Hum Mutat. 2013;34(8):1181–1181. doi: 10.1002/humu.22045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang X-F, Huang F, Wu K-C, et al. Genotype-phenotype correlation and mutation spectrum in a large cohort of patients with inherited retinal dystrophy revealed by next-generation sequencing. Genet Med. 2015;17(4):271–278. doi: 10.1038/gim.2014.138. [DOI] [PubMed] [Google Scholar]
- 8.Nishiguchi KM, Tearle RG, Liu YP, et al. Whole genome sequencing in patients with retinitis pigmentosa reveals pathogenic DNA structural changes and NEK2 as a new disease gene. Proc Natl Acad Sci U S A. 2013;110(40):16139–16144. doi: 10.1073/pnas.1308243110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bitner-Glindzicz M, Lindley KJ, Rutland P, et al. A recessive contiguous gene deletion causing infantile hyperinsulinism, enteropathy and deafness identifies the Usher type 1C gene. Nat Genet. 2000;26:56–60. doi: 10.1038/79178. [DOI] [PubMed] [Google Scholar]
- 10.den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79(3):556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sullivan LS, Bowne SJ, Seaman CR, et al. Genomic rearrangements of the PRPF31 gene account for 2.5% of autosomal dominant retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2006;47(10):4579–4588. doi: 10.1167/iovs.06-0440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Liu P, Carvalho CMB, Hastings PJ, Lupski JR. Mechanisms for recurrent and complex human genomic rearrangements. Curr Opin Genet Dev. 2012;22(3):211–220. doi: 10.1016/j.gde.2012.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bujakowska KM, Zhang Q, Siemiatkowska AM, et al. Mutations in IFT172 cause isolated retinal degeneration and Bardet-Biedl syndrome. Hum Mol Genet. 2014;24(1):230–242. doi: 10.1093/hmg/ddu441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Farkas M, Grant G, White J, Sousa M, Consugar M, Pierce E. Transcriptome analyses of the human retina identify unprecedented transcript diversity and 3.5 Mb of novel transcribed sequence via significant alternative splicing and novel genes. BMC Genomics. 2013;14(486) doi: 10.1186/1471-2164-14-486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gai X, Perin JC, Murphy K, et al. CNV Workshop: an integrated platform for high-throughput copy number variation discovery and clinical diagnostics. BMC Bioinformatics. 2010;11:74. doi: 10.1186/1471-2105-11-74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao C, Bellur DL, Lu S, et al. Autosomal-dominant retinitis pigmentosa caused by a mutation in SNRNP200, a gene required for unwinding of U4/U6 snRNAs. Am J Hum Genet. 2009;85(5):617–627. doi: 10.1016/j.ajhg.2009.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang X, Wang H, Sun V, et al. Comprehensive molecular diagnosis of 179 Leber congenital amaurosis and juvenile retinitis pigmentosa patients by targeted next generation sequencing. J Med Genet. 2013;50(10):674–88. doi: 10.1136/jmedgenet-2013-101558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Polok B, Escher P, Ambresin A, et al. Mutations in CNNM4 cause recessive cone-rod dystrophy with amelogenesis imperfecta. Am J Hum Genet. 2009;84(2):259–65. doi: 10.1016/j.ajhg.2009.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Parry DA, Mighell AJ, El-Sayed W, et al. Mutations in CNNM4 cause Jalili syndrome, consisting of autosomal-recessive cone-rod dystrophy and amelogenesis imperfecta. Am J Hum Genet. 2009;84(2):266–73. doi: 10.1016/j.ajhg.2009.01.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rivolta C, McGee TL, Rio Frio T, Jensen RV, Berson EL, Dryja TP. Variation in Retinitis Pigmentosa-11 (PRPF31 or RP11) Gene Expression Between Symptomatic and Asymptomatic Patients With Dominant RP11 Mutations. Hum Mutat. 2006;27(7):644–653. doi: 10.1002/humu.20325. [DOI] [PubMed] [Google Scholar]
- 21.Rose AM, Shah AZ, Venturini G, et al. Transcriptional regulation of PRPF31 gene expression by MSR1 repeat elements causes incomplete penetrance in retinitis pigmentosa. Sci Rep. 2016 Jan;6:19450. doi: 10.1038/srep19450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nathans J, Merbs S, Sung C-H, Weitz C, Wang Y. Molecular Genetics of Human Visual Pigments. Annu Rev Genet. 1992;26:403–424. doi: 10.1146/annurev.ge.26.120192.002155. [DOI] [PubMed] [Google Scholar]
- 23.Lindstrand A, Davis EE, Carvalho CMB, et al. Recurrent CNVs and SNVs at the NPHP1 locus contribute pathogenic alleles to Bardet-Biedl syndrome. Am J Hum Genet. 2014;94(5):745–754. doi: 10.1016/j.ajhg.2014.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fromer M, Moran JL, Chambert K, et al. Discovery and statistical genotyping of copy-number variation from whole-exome sequencing depth. Am J Hum Genet. 2012;91(4):597–607. doi: 10.1016/j.ajhg.2012.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang DW, Raley C, Jiang MK, et al. Towards Better Precision Medicine: PacBio Single-Molecule Long Reads Resolve the Interpretation of HIV Drug Resistant Mutation Profiles at Explicit Quasispecies (Haplotype) Level. J Data Mining Genomics Proteomics. 2016;7(1) doi: 10.4172/2153-0602.1000182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bejjani BA, Shaffer LG. Clinical Utility of Contemporary Molecular Cytogenetics. Annu Rev Genomics Hum Genet. 2008;9(1):71–86. doi: 10.1146/annurev.genom.9.081307.164207. [DOI] [PubMed] [Google Scholar]
- 27.Human Gene Mutation Database (HGMD) available at: http://www.biobase-international.com/product/hgmd.
- 28.Liu S, Rauhut R, Vornlocher H-PP, Luhrmann R, Lührmann R. The network of protein-protein interactions within the human U4/U6.U5 tri-snRNP. Rna. 2006;12(7):1418–1430. doi: 10.1261/rna.55406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vithana EN, Abu-Safieh L, Allen MJ, et al. A human homolog of yeast pre-mRNA splicing gene, PRP31, underlies autosomal dominant retinitis pigmentosa on chromosome 19q13.4 (RP11) Mol Cell. 2001;8(2):375–381. doi: 10.1016/s1097-2765(01)00305-7. [DOI] [PubMed] [Google Scholar]
- 30.Chakarova CF, Hims MM, Bolz H, et al. Mutations in HPRP3, a third member of pre-mRNA splicing factor genes, implicated in autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2002;11(1):87–92. doi: 10.1093/hmg/11.1.87. [DOI] [PubMed] [Google Scholar]
- 31.Chen X, Liu Y, Sheng X, et al. PRPF4 mutations cause autosomal dominant retinitis pigmentosa. Hum Mol Genet. 2014;23:2926–2939. doi: 10.1093/hmg/ddu005. [DOI] [PubMed] [Google Scholar]
- 32.Tanackovic G, Ransijn A, Ayuso C, Harper S, Berson EL, Rivolta C. A missense mutation in PRPF6 causes impairment of pre-mRNA splicing and autosomal-dominant retinitis pigmentosa. Am J Hum Genet. 2011;88:643–649. doi: 10.1016/j.ajhg.2011.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.McKie AB, McHale JC, Keen TJ, et al. Mutations in the pre-mRNA splicing factor gene PRPC8 in autosomal dominant retinitis pigmentosa (RP13) Hum Mol Genet. 2001;10(15):1555–1562. doi: 10.1093/hmg/10.15.1555. [DOI] [PubMed] [Google Scholar]
- 34.Keen TJ, Hims MM, McKie AB, et al. Mutations in a protein target of the Pim-1 kinase associated with the RP9 form of autosomal dominant retinitis pigmentosa. Eur J Hum Genet. 2002;10:245–249. doi: 10.1038/sj.ejhg.5200797. [DOI] [PubMed] [Google Scholar]
- 35.Malhotra D, Sebat J. CNVs: Harbingers of a rare variant revolution in psychiatric genetics. Cell. 2012;148(6):1223–1241. doi: 10.1016/j.cell.2012.02.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Qin Y, Yao L, King EE, et al. Germline mutations in TMEM127 confer susceptibility to pheochromocytoma. Nat Publ Gr. 2010;42(3):229–233. doi: 10.1038/ng.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Carroll J, Rossi Ea, Porter J, et al. Deletion of the X-linked opsin gene array locus control region (LCR) results in disruption of the cone mosaic. Vision Res. 2010;50(19):1989–1999. doi: 10.1016/j.visres.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ayyagari R, Kakuk LE, Coats CL, et al. Bilateral macular atrophy in blue cone monochromacy (BCM) with loss of the locus control region (LCR) and part of the red pigment gene. Mol Vis. 1999 Jul;5:13. [PubMed] [Google Scholar]
- 39.Pieras JI, Barragán I, Borrego S, et al. Copy-number variations in EYS: A significant event in the appearance of arRP. Investig Ophthalmol Vis Sci. 2011;52(8):5625–5631. doi: 10.1167/iovs.11-7292. [DOI] [PubMed] [Google Scholar]
- 40.Eisenberger T, Neuhaus C, Khan AO, et al. Increasing the yield in targeted next-generation sequencing by implicating CNV analysis, non-coding exons and the overall variant load: the example of retinal dystrophies. PLoS One. 2013;8(11):e78496. doi: 10.1371/journal.pone.0078496. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.