Abstract
Genome editing technologies such as zinc finger nucleases, transcription activator-like effector nucleases, and the clustered regularly interspaced short palindromic repeat/CRISPR-associated protein system have revolutionized biological research. Each biotechnology consists of a DNA binding protein that can be programmed to recognize and initiate double stranded breaks for sitespecific gene modification. These technologies have the potential to be harnessed to cure diseases caused by aberrant gene expression. In order to be successful therapeutically, their functionality depends on their safe and efficient delivery into the cell nucleus. This review discusses the challenges in the delivery of genome editing tools and highlights recent innovations in non-viral delivery that have potential to overcome these limitations and advance the translation of genome editing towards patient care.
Keywords: ZFN, TALEN, CRISPR/Cas, Non-viral, Gene Editing ZFNs, TALENs, CRISPR/Cas
Zinc finger nucleases (ZFNs) (see Glossary) and transcription activator-like effector nucleases (TALENs) are hybrid restriction enzymes composed of a DNA binding domain and a DNA cleavage domain based on the FokI endonuclease [1]. In contrast to ZFNs and TALENs, both of which use protein structures to recognize DNA sequences, the clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated protein (Cas) system enables DNA recognition through RNA-defined specificity. This simplifies the design of a targetspecific gene editing complex to a short guide RNA (sgRNA) with a 20 bp complementarity with the target DNA sequence, as opposed to a new protein array, though the target sequence must be upstream of a protospacer adjacent motif (PAM) sequence [2, 3]. Table 1 summarizes the characteristics of each gene editing system. Gene editing technologies continue to advance rapidly and recent reviews have provided coverage of many physical and chemical strategies for intracellular delivery of macromolecules that can potentially be adopted for gene editing applications as well as strategies for specific modification of genome editing proteins [4, 5]. In this review, we provide in-depth discussion of the most recent studies on non-viral gene editing, most of which were published in the last year, with an emphasis on the utilization of nanoparticle-based delivery vehicles.
Table 1.
Comparison of ZFN, TALEN, and CRISPR/Cas gene editing systems.
| DNA binding domain | DNA cleavage domain | Mechanism for target specificity | Challenges and restrictions | |
|---|---|---|---|---|
| ZFN | Zinc finger protein repeats | FokI endonuclease | Each zinc finger protein recognizes 3 bp DNA | Target sequence length must be multiple of 3; longer target sequence requires larger protein |
| TALEN | Central repeat domains of transcription activator-like effector proteins | FokI endonuclease | Each TALE protein unit recognizes 1 bp DNA | Cloning of TALE repeat arrays can be technically challenging; longer target sequence requires larger protein |
| CRISPR/Cas9 | Cas9 endonuclease in complex with sgRNA | Cas9 endonuclease | 20 bp targeting region of sgRNA confers specificity through DNA-RNA complementarity | Target sequence must be upstream of PAM site |
All three gene editing systems result in sequence-specific DNA cleavage, at which point the cell’s own DNA repair machinery can be harnessed to achieve gene modification. In the absence of a donor DNA template, double stranded breaks (DSBs) are repaired through error-prone non-homologous end joining (NHEJ), which introduces small indels that can shift the reading frame and result in gene knockout. When a donor DNA template is present, entire genes can be knocked in at the cut site. This is commonly achieved through homology directed repair (HDR) [6, 7], although some studies have shown higher integration rates through a process termed homology-independent targeted integration (HITI) [8]. The DNA binding capabilities of these genome editing enzymes have been further harnessed to enable site-specific single base pair editing [9, 10] and epigenetic [11, 12] or transcriptional [13, 14] modulation of gene expression.
In order for gene editing complexes to be functionally active, they must first be delivered into the cell nucleus, necessitating delivery across both the plasma and nuclear membranes. Historically, the most common way to achieve this was to use viral vectors, in which nucleic acids coding for the enzyme complexes are packaged into viruses and delivered to a target cell. While viral vectors are often highly effective, the complexity and challenge associated with scale-up of virus production, potential for insertional mutagenesis, and possible immune responses against the viral vector limit their use in a therapeutic capacity [15]. Viral vector cargo size limitations additionally limit efficacy particularly in AAV systems, which can necessitate editing enzymes and donor templates to be packaged into separate viral particles [16, 17]. Non-viral delivery methods have emerged as a viable alternative as they can be engineered to largely avoid these problems, but they require substantial improvements in order to reach the efficacy of their viral counterparts. Challenges to non-viral delivery include protection of nucleic acid or protein cargo from degradation, opsonization, and immune avoidance, as well as delivery to specific cell targets and cellular compartments. Recent research has further highlighted risks of host humoral and cell mediated immunity to the Cas9 protein [18] as well as double-stranded break P53 responses associated with genome editing that risk selection of P53 deficient edited cells [19, 20] that have yet to be overcome with any delivery strategy.
Cargo selection: DNA, mRNA, or Protein
Plasmid DNA
Genome editing complexes can be delivered by non-viral vectors in the form of plasmid DNA, a format that offers flexibility in design as DNA sequences can be easily incorporated into plasmids using simple molecular cloning techniques. However, gene editing efficiency is often limited by the efficiency of nuclear delivery and gene expression that is required to generate the final gene editing protein complexes. Upon systemic injection, DNA nanoparticles face multiple barriers before successful delivery to target tissues (Figure 1, Key Figure). Nanoparticles with highly positive surface charges are prone to serum protein adsorption, aggregation, premature cargo release [21], and nanoparticle clearance by immune cells [22]. Coating nanoparticles with hydrophilic molecules such as poly(ethylene glycol) (PEG) is a common anti-fouling strategy that has been shown to reduce immune stimulation and increase circulation time [23], but anti-PEG responses resulting in accelerated clearance upon repeated administration may limit the applicability of PEGylation in the clinic for some applications [24]. Nanoparticle size is another important feature as molecules smaller than 5.5 nm in diameter have been shown to experience rapid clearance from the kidneys [25]. Complexation into nanoparticles decreases renal clearance of DNA by increasing its size beyond the renal filtration limit, and nanoparticles effective for gene delivery have been found to fall in the range of 100–250 nm in diameter [26].
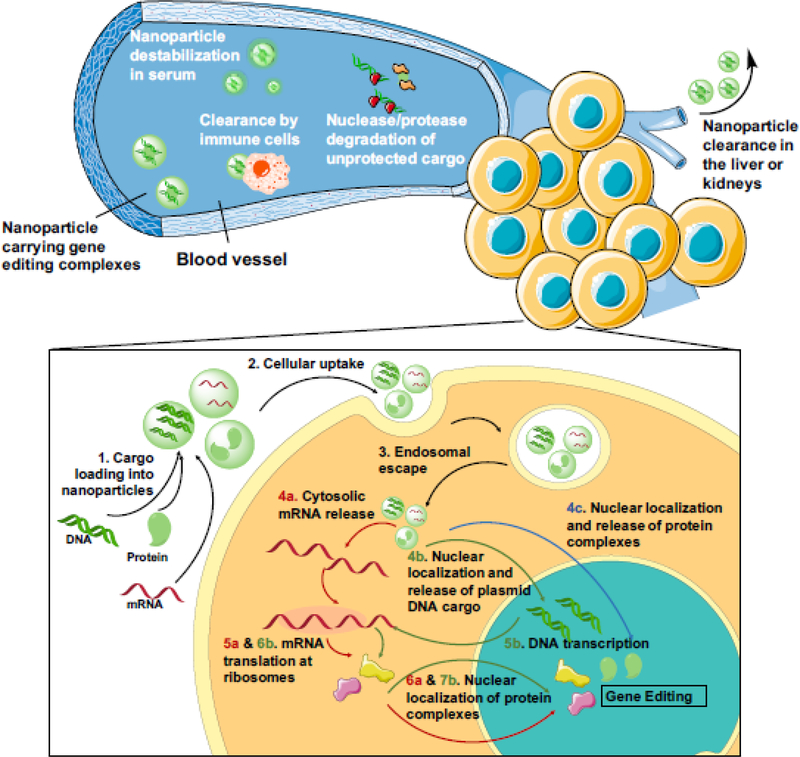
Figure 1. Key Figure. Extracellular and intracellular obstacles for non-viral delivery of gene editing tools.
Systemically injected nanoparticles carrying gene editing complexes must protect their cargo from destabilizing serum components, enzyme degradation, and clearance by immune cells or in the liver and kidneys. Upon reaching the target tissue, they face further intracellular delivery barriers. Protein or nucleic acid cargo must be (1) encapsulated in nano-carriers for (2) efficient intracellular uptake to occur. Upon endocytosis into the cell, nanoparticles need to (3) escape from degradative endo-lysosomal compartments. For mRNA delivery, cargo release should occur in the cytosol (4a) to enable mRNA translation at the ribosomes (5a). For DNA delivery, plasmid DNA needs to traffic to the nucleus (4b) where DNA can be transcribed into mRNA (5b); the mRNA transcript must then enter the cytosol to be translated into protein (6b). Protein complexes, whether synthesized in the cell (6a and 7b) or delivered directly by the nanocarrier (4c), must localize to the nucleus for gene editing to occur. Intracellular delivery steps common to all cargo formats are shown in black arrows and those specific to mRNA, plasmid DNA, and protein complexes are shown in red, green, and blue arrows, respectively.
Upon arrival at target cells, nanoparticles must first cross the cell membrane. Cellular uptake can be enabled by attaching cell-penetrating peptides on nanoparticle surfaces to promote direct internalization [27] or through non-specific interactions between cationic nanoparticles and the anionic cell surface [28, 29]. If internalized via endocytosis, nanoparticles must also escape from degradative endo-lysosomal compartments. Many materials have been proposed to escape the endosome via the debated proton sponge mechanism, where nanomaterials containing uncharged amines at neutral pH acquire charge during endosome acidification, leading to polymer swelling and membrane destabilization for transient endosomal escape [30]. Several recent studies suggest that upon protonation in the acidifying endosome, polycations such as polyethylenimine (PEI) cause endosomal deformation and increase the permeability of the endosomal membrane, allowing nanoparticles to leak out through small pores [31, 32]. Finally, plasmid DNA must localize to the nucleus, which can be facilitated by nanomaterials that expedite cytosolic and nuclear trafficking [33]. In rapidly dividing cells, nuclear entry of plasmids generally occurs following nuclear membrane breakdown during cell division but in post-mitotic cells, import to the nucleus generally occurs through nuclear pores, which can be facilitated by transcription factor binding sequences in the plasmid that bind to importins [33]. Gene editing complexes face an additional trafficking step as proteins synthesized at cytosolic ribosomal sites must again localize to the nucleus to bind to and cleave genomic DNA. This could be especially challenging for the CRISPR/Cas system as Cas9 endonucleases must also bind sgRNA in the nucleus before they can form functional gene editing units. Utilization of minicircle DNA, which is more efficient on a per-mass basis and less immunogenic than plasmid DNA due to elimination of bacterial expression sequences, offers an alternative to some of these challenges and has been utilized for delivery of CRISPR [34], ZFN and TALEN [35] systems. The reduced size of minicircles compared to plasmids facilitates their cytosolic trafficking and nuclear import while the removal of sequences of bacterial origin reduces transcriptional silencing associated with plasmid DNA sequences [36].
Delivery of genome editing factors as DNA sequences also carries a substantial risk of unintentional genomic integration, which can induce insertional mutagenesis due to incorporation of highly active promoter elements in chromosomal DNA or disruption of tumor suppressor genes [37]; while the risk of insertional mutagenesis with non-viral delivery of plasmid DNA is generally much lower than with DNA viral vectors, this risk must be taken into account for translationally relevant therapies. Finally, plasmid DNA delivery is generally not feasible for cell types that are refractory to plasmid DNA transfection. This is especially problematic in immune cells, where it has been demonstrated that the same delivery system that enabled up to 50% editing in human embryonic kidney cells achieved less than 4% editing in CD4+ T cells [38]. This could potentially be due to the fact that T cells can sense the intracellular presence of foreign nucleic acids [39], leading to an innate immune response and prompting the need for other delivery methods.
mRNA
Another nucleic acid delivery cargo to enable genome editing is mRNA, which can be synthesized from a DNA template using in vitro transcription. mRNA delivery circumvents the need for nuclear localization of the nucleic acid cargo as protein expression can occur following cytosolic delivery, and protein expression is detectable as short as 4–6 hours post-transfection [40, 41]. This virtually eliminates the risk of insertional mutagenesis and also reduces the probability of off-target effects as the duration of protein expression is much shorter for mRNA compared to DNA. Cas9 protein expression can be effectively undetectable 72 hours post-transfection with mRNA in vitro [41] and 24 hours post-injection in vivo [40]. One major challenge for mRNA delivery is that in CRISPR applications, the delivery of Cas9 mRNA and sgRNA may require specialized materials as the two have very different lengths and different kinetics of expression. One study showed that the optimal condition for an RNA-mediated CRISPR/Cas9 editing system required the Cas9 mRNA to be delivered 24 hours before sgRNA delivery [42]. For gene insertion applications, the need for a donor template DNA may also necessitate an alternative delivery mechanism as Wang and colleagues demonstrated when the authors used electroporation to deliver mRNA coding for ZFNs and a viral vector to deliver donor DNA [43].
Proteins
Delivery of gene editing protein complexes synthesized outside the cell eliminates the need for intracellular transcription and translation, and gene editing can occur immediately following intranuclear delivery. This could potentially boost gene editing efficacy in post-mitotic or hard-totransfect cells, in which limits to the cell’s transcriptional or translational capacity could result in lower gene editing efficacy when using plasmid DNA or mRNA. On the other hand, this method also reduces the cargo’s ability to enable cell type specificity as transcriptional targeting cannot be used. In the case of CRISPR/Cas9, there is the additional concern that the Cas9 protein variant from the bacterial species S. pyogenes and S. aureus have both been shown to be recognized by antibodies in greater than 60% of human patients [18], which could result in rapid clearance of these proteins upon systemic delivery. Furthermore, Cas9 and TALENs generally cannot cross the cell membrane on their own, though ZFNs have been shown to have some inherent cell membrane permeability [44]. Due to the nucleic acid binding nature of the Cas9 protein, unmodified SpCas9 possesses a net charge of +20, which becomes further positively charged with addition of a nuclear localization signal (NLS) peptide [45]. This net positive charge of the Cas9 protein can be neutralized with protein engineering, by inclusion of a glutamate tag up to 20 amino acids long, which enabled direct cytosolic delivery when Cas9/sgRNA complexes were assembled with arginine-gold nanoparticles [45]. An alternative strategy to increase the plasma membrane permeability of genome editing enzymes is to fuse multiple viral SV40 nuclear localization signal (NLS) domains onto these proteins, which was reported to enable editing without an additional vector material both in vitro and in vivo [46, 47]. Similarly, cell-penetrating peptides have been conjugated to Cas9 and gRNAs directly to facilitate uptake and endosomal escape as selfcondensed cationic nanoparticles, although these particles mediated <10% knockout efficiency in vitro in HEK293T cells [48].
Physical Delivery Methods
Delivery methods that physically disrupt the cell and nuclear membranes have been used extensively for delivery of ZFNs, TALENs and CRISPR but are generally limited to ex vivo delivery. Gene editing proteins or their DNA or RNA precursors have been directly injected into cells or embryos through the process of microinjection and have been successfully used to generate disease models in rodents, but this technique is highly limited in the number of cells that can be edited in a timely manner [49, 50]. Other physical methods include electroporation, in which an electric field causes small pores to temporarily form in the cell membrane to allow nucleic acids and proteins to pass through, and nucleofection, which combines electroporation with chemicallyenhanced delivery. Many groups have utilized electroporation to deliver nucleic acids or proteins for ZFNs, TALENs or CRISPR ribonucleoproteins (RNPs) directly in manners that have been extensively reviewed, with many on-going clinical trials utilizing electroporation for delivery to T-cells [4]. Overall, physical delivery methods have a high potential to be used for ex vivo editing of isolated circulating lymphocytes ex vivo, which could potentially then be adoptively transferred into a patient for cellular therapy.
Additionally, hydrodynamic injection has been used in vivo to induce expression of Cas9 and gRNA from plasmid DNA in the liver of rodents, although this method is not regarded as translatable to humans [51]. Other physical delivery methods include direct mechanical disruption of cellular membranes via micro-constriction [52] or induction of cell uptake and endosomal escape of Cas9 RNP complexes via manipulation of osmotic potential [53]. D’Astolfo and colleagues utilized hypertonic solutions to trigger macropinocytosis in vitro followed by endosomal disruption mediated by zwitterionic propanebetaines to deliver Cas9 RNP complexes, providing an alternative to electroporation or physical membrane disruption [53].
Chemical material-based delivery approaches for genome editing
Lipids and lipid-like materials
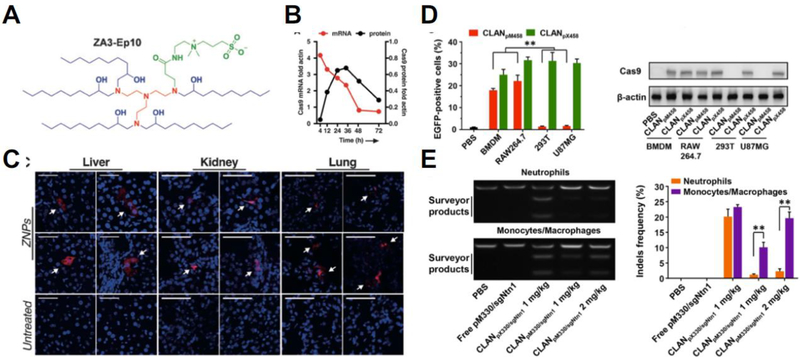
Chemical approaches to non-viral delivery utilize biomaterials to form nanostructures that encapsulate genome-editing cargos and then shuttle them into cells (see Figure 2 for chemical structures of these materials). Many lipids and lipid-like materials have been developed for the intracellular delivery of nucleic acids such as oligonucleotides [54], mRNA [55], and plasmid DNA [56]. For genome editing applications, these materials have largely been used to deliver genome editing tools in the form of nucleic acids (see Table 2 for summary). Plasmid DNA encoding a Cas9-sgRNA complex targeting VEGF was delivered using a PEG-PEI-cholesterol lipopolymer and achieved ~50% gene knockout in osteosarcoma cells in vitro and in vivo [57] while a CRISPR DNA construct with a CD68 promoter was delivered using lipid-containing PEGPLGA nanoparticles to enable macrophage-specific gene editing, resulting in 30% gene knockout in vitro and 20% in vivo [58]. Cas9 mRNA and in vitro transcribed sgRNAs were delivered by Miller and colleagues using a zwitterionic amino lipid library [42] and by Jiang and colleagues using N1,N3,N5-tris(2-aminoethyl)benzene-1,3,5-tricarboxamide (TT) based lipid-like nanoparticles [40] for gene editing in vitro and in vivo (Figure 3). Noting the ability to deliver the RNAs despite their vastly different sized (100 nt for sgRNA and >4500 nt for mRNA), the lipidoid nanoparticles engineered by Miller and colleagues achieved 95% knockout in vitro and detectable editing in the liver, lungs and kidney.
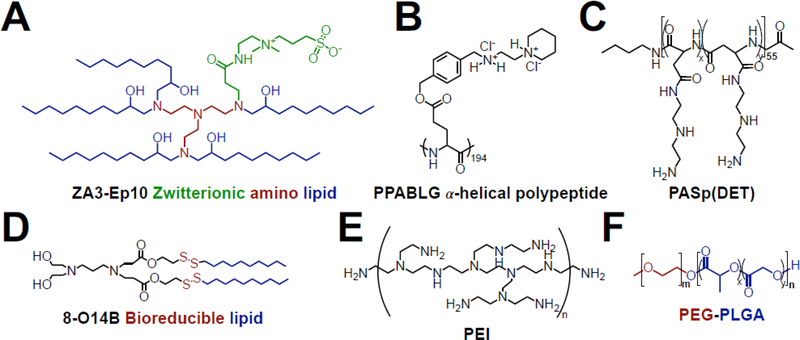
Figure 2. Chemical structures of materials that have been used to deliver non-viral gene editing complexes.
(A) Zwitterionic amino lipid were used to deliver Cas9 mRNA and sgRNA [42]; (B) PPABLG polypeptides condensed plasmid DNA into nanoparticles for CRISPR editing [70]; (C) PASp(DET) coating enabled endosomal escape of CRISPR-gold constructs [67]; (D) bioreducible lipids enabled intracellular delivery of RNP complexes [60]; (E) PEI was used in hybrid nanoparticle systems to deliver the CRISPR system as RNPs [69] and plasmid DNA [57]; (F) PEG-PLGA nanoparticles enabled CRISPR editing in macrophages in vivo [58].
Table 2.
Examples of non-viral nanoparticle-mediated CRISPR delivery.
| Nanoparticle Cargo format Main findings material | Refs | ||
|---|---|---|---|
| PEG-PEI- cholesterol lipopolymer |
Plasmid DNA | 50% knockout of VEGF gene in osteosarcoma cells in vitro and in vivo |
[57] |
| Lipid-assisted PEG-PLGA |
Plasmid DNA | Macrophage-specific knockout of netrin1; 30% in vitro and 20% in vivo | [58] |
| Zwitterionic amino lipid | Cas9 mRNA and in vitro transcribed sgRNA | Achieved gene deletion-mediated turning on of mCherry expression in Ai9 mice in vivo | [42] |
| Lipofectamine 2000® | Ribonucleoprotein complex | 24% knockout of eGFP in vitro in neuron-derived mouse embryonic stem cells and 13% knockout in vivo in mouse cochlea hair cells | [59] |
| PEG-lipid | Cas9 mRNA and in vitro transcribed sgRNA | 97% knockout of mouse transthyretin protein in liver cells for; sustained for at least 12 months following single systemic injection |
[61] |
| Polymer and gold nanoparticle | Ribonucleoprotein complex | 5.4% homology-directed repair in mouse mdx Duchenne muscular dystrophy model after intramuscular injection |
[67] |
| PEI coated on DNA nanoparticle |
Ribonucleoprotein complex | ~40% GFP knockout in vitro and 25% knockout in vivo in U2OS cells following intratumoral injection | [69] |
| Cationic cell- penetrating polypeptide |
Plasmid DNA | 35% gene deletion, 67% protein knockdown, and reduced tumor growth by >71% in HeLa cells after intratumoral injections in vivo | [70] |
Figure 3. Lipid-containing delivery systems enable gene editing after systemic injections in vivo.
(A) Structure of the zwitterionic amino lipid used by Miller and colleagues [42] to formulate nanoparticles delivering the CRISPR/Cas9 system in the form of Cas9 mRNA and sgRNA. (B) Expression profile of Cas9 after mRNA delivery. (C) Systemic injection of these zwitterionic lipid nanoparticles achieved gene editing in the liver, kidney, and lung (as indicated by red fluorescence). Reproduced with permission from [42]. (D) Cationic lipid-assisted PEG-b-PLGA nanoparticles (CLAN) developed by Luo and colleagues [58] delivering the CRISPR/Cas9 system in the form of plasmid DNA showed preferential expression in macrophages (BMDM and RAW264.7) when using a macrophage-specific promoter (pM458). (E) Systemic injection of these nanoparticles enabled macrophage-specific gene editing in mice. Reproduced with permission from [58].
Lipid materials have also been explored as vehicles for genome editing proteins. Zuris and colleagues engineered Cas9 and TALENs fused to anionic GFP proteins to increase the negative surface charge of these proteins and complexed them with the commercially-available cationic lipid transfection reagent Lipofectamine 2000™ and demonstrated 24% gene knockout in neuronderived mouse embryonic stem cells in vitro and 13% gene knockout in mouse cochlea hair cells in vivo [59]. Likewise, Wang and colleagues utilized more effective bioreducible lipid nanoparticles to deliver charge neutralized Cas9 RNP complexes for up to 70% knockout in vitro [60]. Lipid materials have also shown impressive levels of gene editing after systemic delivery, primarily in the liver. Finn and colleagues reported that lipid nanoparticles formulated with PEGlipids showed excellent serum stability, and when used to deliver Cas9 mRNA and sgRNAs targeting the mouse transthyretin gene in liver cells resulted in >97% reduction in serum protein levels that persisted for at least 12 months after a single systemic injection [61]. Yin and colleagues utilized the highly effective lipid cKK-E12 lipid nanoparticle formulation for co-delivery to the liver of Cas9 mRNA and sgRNA highly modified for enhanced efficacy, demonstrating 80% gene knockout primarily in hepatocytes [62]. Importantly, lipid nanoparticles also have the ability to complement viral delivery strategies for genome editing to improve tissue specificity; Yin and colleagues reported using lipid nanoparticles to deliver Cas9 mRNA non-virally to the liver, while using AAV encoding a sgRNA and HDR DNA template to achieve 6% editing correction of hepatocytes [63]. Using biologically derived materials, Kim and colleagues used cancer-derived exosomes as the delivery vehicle for plasmid DNA encoding a CRISPR/Cas system and showed efficient and targeted editing in an ovarian cancer model [64]. An exosome-liposome hybrid vector developed by Lin and colleagues enabled CRISPR-interference in mesenchymal stem cells, which could not be transfected using liposomes alone [65]. These results showcase the great potential of lipid formulations for in vivo delivery of genome editing tools to treat genetic diseases, rivaling that of viral mediated delivery for some tissues such as the liver.
Polymeric materials
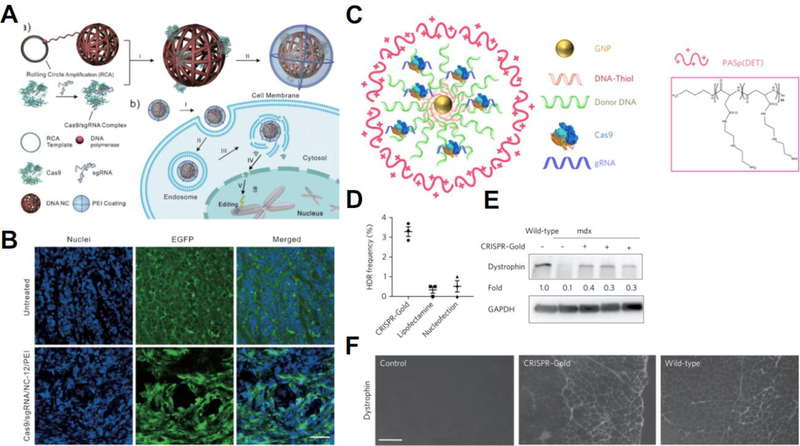
Polymeric materials have been used to deliver DNA, mRNA, and oligonucleotides [66]. For delivering genome editing tools, polymers have often been used in multi-component delivery systems to promote endosomal escape (Figure 4). Lee and colleagues used the endosomal disruptive polymer poly(N-(N-(2-aminoethyl)-2-aminoethyl) aspartamide) to coat Cas9-sgRNA RNPs adsorbed onto gold nanoparticles and used this vehicle, which they termed CRISPR-Gold, to correct the dystrophin gene in a mouse model of Duchenne muscular dystrophy in vivo; despite an in vivo HDR frequency of only 5% following intramuscular injection, these nanoparticles mediated a statistically measurable increase in muscle strength compared to scramble nanoparticles [67]. CRISPR-Gold has been utilized in follow-up work to mediate genome editing in rodent brains following local injection using both Cas9 and Cpf1, managing to reduce mRNA and protein levels of the target gene by up to 50% [68]. In an alternative approach, Sun and colleagues used rolling circle amplification to create DNA nanoclews that enabled encapsulation of RNP complexes in a DNA based particle core that were then coated with the cationic polymer polyethylenimine (PEI) and achieved CRISPR-mediated gene editing in vitro of up to 28% knockout compared to <2% knockout with PEI only as well as an estimated 25% knockout of eGFP in vivo following intratumoral injection of the nanoparticles [69]. Recent work by the labs of Cheng and Leong have demonstrated the promise of a cationic α-helical polypeptide to deliver Cas9 and sgRNA plasmids for enhanced efficiency at gene editing in vitro and in vivo [70], leading to 67% targeted protein knockdown in HeLa cells in vivo following repeated intratumoral injections and reducing tumor growth by >71%, consequently significantly extending survival in the HeLa xenograft mouse model [70]. These strategies, using cationic polymers to deliver either nucleic acids, nucleic acid neutralized RNP complexes, or anionic modified RNP complexes, have all relied upon local delivery due to the cationic nature of the particles being utilized presenting potential systemic delivery challenges. While this relatively high efficacy of cationic polymer-based materials for local delivery is promising in mice, for certain applications, it may face challenges in scale-up to patients due to the larger length scales required for sufficient transport and therapeutic coverage.
Figure 4. Hybrid delivery systems for CRISPR/Cas9 using polymers to facilitate cellular uptake and endosomal escape.
(A) Assembly schematic for the DNA nanoclew system in which Cas9-sgRNA RNPs are assembled into nanocomplexes with DNA nanoparticles coated with the cationic polymer PEI. (B) DNA nanoclews enabled knockout of an eGFP gene in vivo. Reproduced with permission from [69]. (C) Assembly schematic for CRISPR-gold, which complexed RNPs with gold nanoparticles and a PASp(DET) polymer. CRISPR-gold corrected the dystrophin gene in primary myoblasts from the mdx mouse model for Duchenne muscular dystrophy in vitro (D&E), and enabled restoration of the dystrophin gene in vivo after intramuscular injection (F). Reproduced with permission from [67].
Advances in systemic delivery strategies could be utilized to give polymeric gene editing delivery vehicles greater reach. The most commonly used strategy is PEGylation, in which the hydrophilic molecule polyethylene glycol (PEG) is used to reduce fouling of the nanoparticle surface and increase circulation time [23]. More recently, the use of zwitterionic materials have gained interest as another strategy for systemic delivery. Recent advances in the field of systemic siRNA delivery are applicable to genome editing machinery nucleic acids or proteins; particularly, use of zwitterionic materials including cationic quaternary ammonium sulfonamide amino lipids [71] and zwitterionic phosphorylcholine-based polymer corona in a diblock polymer have been shown to improve systemic delivery and offer more effective alternatives to PEGylation [72].
Another avenue to explore is the use of polymeric materials to deliver gene editing proteins. In contrast to nucleic acid delivery, where cationic polymers are typically used to condense anionic nucleic acids into nanoparticles via electrostatic interactions, protein delivery platforms must be more flexible to accommodate protein molecules of various surface charges. Chang and colleagues reported a protein delivery system composed of a dendrimer end-capped with guanidyl groups to facilitate protein binding through hydrogen bonding and salt bridges as well as phenyl groups to promote endocytosis and endosomal escape [73]. This polymer successfully encapsulated proteins of different sizes and surface charges and promoted the efficient intracellular delivery of functionally-intact proteins in vitro. Yan and colleagues employed a different strategy in which thin polymer shells were synthesized around individual protein molecules in situ, resulting in a core-shell architecture in which polymer shells were covalently linked to the protein cores [74]. The authors showed that this system protected proteins against protease degradation and enabled efficient cellular uptake in vitro and in vivo of proteins that retained their efficacy following intracellular delivery without relying on electrostatic charge based interactions, which is beneficial for gene editing protein complexes and Cas9 RNP complexes which possess regions of varying charge. Recently, several groups have reported the use of zinc/imidazole-based metal-organic frameworks (MOF) for intracellular protein delivery [75, 76]. These MOF nanoparticles have been demonstrated to effectively protect protein cargo from protease digestion and were used by Alsaiari and colleagues to enable intracellular delivery of CRISPR RNPs, though the editing efficiency was quite low (30% gene knockout in CHO cells in vitro) and further optimization of this system is required [77]. Overall, these strategies could be adapted to delivering gene editing protein complexes, which are mostly impermeable to cell membranes on their own and unsuitable for encapsulation using cationic polymers as they have slightly cationic surface charges.
Concluding Remarks and Future Perspectives
In August 2017, the FDA approved the first therapeutic that involved a gene therapy step as a medicine in the United States [78]. Tisagenlecleucel, a cell-based cancer immunotherapy for children and young adults with B-cell lymphoblastic leukemia, uses viruses to insert a gene encoding chimeric antigen receptors (CARs) into patient-derived T cells and infuses these CAR T cells back into the patient. This drug has achieved remission in >80% of patients who had been refractory to traditional radiation therapy [79]. Subsequently, the first gene therapy in the United States for use in vivo to directly treat a genetic disease was approved by the FDA in December 2017 [80]. The therapy, voretigene neparvovec-rzyl, uses an adeno-associated virus (AAV2) to deliver RPE65 cDNA to patients with biallelic RPE65 mutation-associated retinal dystrophy and demonstrated improved vision without product-related serious adverse events [81]. The approval of these first gene therapy-based medicines in the United States marks the beginning of an exciting new era of precision medicine in which gene-based therapeutics, such as gene editing technologies, are being brought to market.
While these impressive steps forward bode well for the future clinical translation of gene editing technologies, their efficacy largely depends on the efficiency of intracellular delivery and the suitability of the delivery system for the genetic cargo being delivered. Viral delivery systems can be effective but can also have significant limitations to cargo size as well as potential adverse effects such as insertional mutagenesis and immunogenicity, which may limit many applications to modifying cells ex vivo for re-infusion into patients as in the case with CAR T cells [82]. The field is moving quickly as clinical trials have begun in China using CRISPR to engineer CAR T cells targeting lung cancer [83] and in the UK using TALENs to engineer CAR T cells targeting pediatric leukemia [84], and the first CRISPR-engineered CAR T cell clinical trials in the US were recently approved [85]. Non-viral platforms could expand the scope of gene editing therapies by facilitating safe and effective direct delivery to native cells in vivo. Recent studies using a nonviral polymer and gold nanoparticle hybrid system resulted in effective correction of disease phenotypes in muscle cells after intramuscular injection [67]. Clinically-relevant levels of gene editing in the liver were achieved from systemic injections of lipid nanoparticles that could be promising in treating a host of inherited liver diseases such as primary hyperoxaluria type 1, transthyretin amyloidosis, and hepatitis B [61]. Other advances in achieving more targeted delivery of gene editing tools could be used to decrease the chances of off-target editing. Even with these advances, much work still needs to be done in order to adapt gene editing technologies to treat various disease types in a safe and effective way (see Outstanding Questions). Innovating genome editing complexes as well as nanostructured delivery vehicles with higher specificity, enhanced efficiency, and lower toxicity brings these promising technologies closer to the clinic, where they have the potential to precisely cure genetic diseases instead of merely treating disease symptoms.
Highlights.
ZFNs, TALENs, and CRISPR/Cas are programmable nucleases that can be designed to target virtually any gene in a highly site-specific manner.
Once bound to a target sequence, nucleases can initiate DSBs that enable gene knock-out via NHEJ or gene knock-in via HDR or HITI.
Successful intracellular delivery is crucial to functional genome editing.
Gene editing complexes can be delivered to cells as plasmid DNA, mRNA, or proteins.
Non-viral nanoparticulate delivery using lipid-like and polymeric materials can overcome systemic delivery hurdles and bring gene editing closer to the clinic.
Outstanding Questions.
What cargo type should gene editing molecules be delivered as to maximize editing efficiency while minimizing off-target effects and immunogenicity in the most cell types?
What type of non-viral delivery vehicle would best enable clinically relevant levels of gene editing in humans?
Are gene editing technologies safe to use as a systemically delivered drug in light of P53 damage responses to double stranded DNA breaks?
What is the best way to develop and implement gene editing technologies to treat diseases in which the same disease phenotype can be caused by multiple different genotypes in different patients?
Acknowledgements
Y.R. and D.W. thank the NSF for graduate fellowships. The authors would like to thank the Bloomberg~Kimmel Institute for Cancer Immunotherapy, Research to Prevent Blindness / Dr. H. James and Carole Free Catalyst Award, and the NIH (R01EB022148) for support.
Glossary
- Clustered regularly interspaced short palindromic repeats (CRISPR)
the RNA targeted, DNA cleaving system originating in bacteria that has been re-appropriated for RNA guided DNA cleavage in mammalian cells
- CRISPR associated protein (Cas)
a protein associated with the CRISPR system, frequently Cas9 from S. aureus or S. pyogenes
- Double stranded break (DSB)
occurs when both strands of DNA are cleaved, triggering DNA repair pathways
- Homology directed repair (HDR)
occurs when a donor template containing a gene insert flanked by homology arms that are complementary to DNA sequences flanking the cut site is present after double-stranded breaks
- Homology-independent target integration (HITI)
introduces gene inserts via donor templates that do not contain homology arms
- Non-homologous end-joining (NHEJ)
the primary, non-specific repair pathway of doublestranded break ligation in mammalian cells
- Protospacer adjacent motif (PAM)
a short (2–6 bp) region of DNA recognized by the CRISPR associated proteins as non-bacterial in origin that is required to mediate CRISPR cleavage at RNA targeted. The canonical PAM sequence is 5’-NGG-3’ but other Cas variants with different PAM sequences have been discovered or engineered.
- Ribonucleoprotein (RNP)
in the context of CRISPR, the assembled complex of gRNA and Cas protein that can actively induce DNA cleavage
- Transcription activator-like effector nuclease (TALEN)
a FokI based nuclease that is targeted to specific DNA sequences by central repeat domains in which 33–35 amino acids specify a single target DNA base
- Zinc finger nuclease (ZFN)
a FokI based, targeted DNA cleaving protein that can introduce double stranded breaks targeted by triplet DNA bp recognizing zinc finger motifs
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Mani M et al. (2005) Binding of two zinc finger nuclease monomers to two specific sites is required for effective double-strand DNA cleavage. Biochemical and Biophysical Research Communications 334 (4), 1191–1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mojica FJM et al. (2009) Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology 155 (3), 733–740. [DOI] [PubMed] [Google Scholar]
- 3.Deltcheva E et al. (2011) CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature 471 (7340), 602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yin H et al. (2017) Delivery technologies for genome editing. Nature reviews Drug discovery 16 (6), 387. [DOI] [PubMed] [Google Scholar]
- 5.Wang H-X et al. (2017) CRISPR/Cas9-based genome editing for disease modeling and therapy: challenges and opportunities for nonviral delivery. Chem Rev 117 (15), 9874–9906. [DOI] [PubMed] [Google Scholar]
- 6.Moehle EA et al. (2007) Targeted gene addition into a specified location in the human genome using designed zinc finger nucleases. Proceedings of the National Academy of Sciences 104 (9), 3055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yoshimi K et al. (2016) ssODN-mediated knock-in with CRISPR-Cas for large genomic regions in zygotes. Nature communications 7, 10431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Suzuki K et al. (2016) In vivo genome editing via CRISPR/Cas9 mediated homology-independent targeted integration. Nature 540, 144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kim YB et al. (2017) Increasing the genome-targeting scope and precision of base editing with engineered Cas9-cytidine deaminase fusions. Nat Biotechnol 35 (4), 371376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Rees HA et al. (2017) Improving the DNA specificity and applicability of base editing through protein engineering and protein delivery. Nature Communications 8, ncomms15790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hilton IB et al. (2015) Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology 33 (5), 510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amabile A et al. (2016) Inheritable silencing of endogenous genes by hit-and-run-targeted epigenetic editing. Cell 167 (1), 219–232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Polstein LR and Gersbach CA (2012) Light-Inducible Spatiotemporal Control of Gene Activation by Customizable Zinc Finger Transcription Factors. Journal of the American Chemical Society 134 (40), 16480–16483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konermann S et al. (2014) Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature 517, 583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kotterman MA et al. (2015) Viral vectors for gene therapy: translational and clinical outlook. Annual review of biomedical engineering 17, 63–89. [DOI] [PubMed] [Google Scholar]
- 16.Yang Y et al. (2016) A dual AAV system enables the Cas9-mediated correction of a metabolic liver disease in newborn mice. Nat Biotechnol 34 (3), 334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yu W et al. (2017) Nrl knockdown by AAV-delivered CRISPR/Cas9 prevents retinal degeneration in mice. Nature Communications 8, 14716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Charlesworth CT et al. (2018) Identification of Pre-Existing Adaptive Immunity to Cas9 Proteins in Humans. bioRxiv, 243–345. [Google Scholar]
- 19.Ihry RJ et al. (2018) p53 inhibits CRISPR–Cas9 engineering in human pluripotent stem cells. Nat Med, 1. [DOI] [PubMed] [Google Scholar]
- 20.Haapaniemi E et al. (2018) CRISPR–Cas9 genome editing induces a p53-mediated DNA damage response. Nat Med, 1. [DOI] [PubMed] [Google Scholar]
- 21.Liu J et al. (2005) Influence of serum protein on polycarbonate-based copoly mermicelles as a delivery system for a hydrophobic anti-cancer agent. Journal of Controlled Release 103 (2), 481–497. [DOI] [PubMed] [Google Scholar]
- 22.Jones SW et al. (2013) Nanoparticle clearance is governed by Th1/Th2 immunity and strain background. The Journal of Clinical Investigation 123 (7), 3061–3073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Suk JS et al. (2016) PEGylation as a strategy for improving nanoparticle-based drug and gene delivery. Advanced drug delivery reviews 99, 28–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ando H et al. (2018) Reactivity of IgM antibodies elicited by PEGylated liposomes or PEGylated lipoplexes against auto and foreign antigens. J Control Release 270, 114119. [DOI] [PubMed] [Google Scholar]
- 25.Choi HS et al. (2007) Renal Clearance of Nanoparticles. Nature biotechnology 25 (10), 1165–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sunshine JC et al. (2012) Uptake and transfection with polymeric nanoparticles are dependent on polymer end-group structure, but largely independent of nanoparticle physical and chemical properties. Mol Pharm 9 (11), 3375–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Freimann K et al. (2018) Formulation of Stable and Homogeneous Cell-Penetrating Peptide NF55 Nanoparticles for Efficient Gene Delivery In Vivo. Molecular Therapy - Nucleic Acids 10, 28–35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Verma A and Stellacci F (2010) Effect of surface properties on nanoparticle–cell interactions. Small 6 (1), 12–21. [DOI] [PubMed] [Google Scholar]
- 29.Lin J and Alexander-Katz A (2013) Cell Membranes Open “Doors” for Cationic Nanoparticles/Biomolecules: Insights into Uptake Kinetics. ACS Nano 7 (12), 10799–10808. [DOI] [PubMed] [Google Scholar]
- 30.Stewart MP et al. (2016) Challenges in carrier-mediated intracellular delivery: moving beyond endosomal barriers. Wiley Interdisciplinary Reviews: Nanomedicine and Nanobiotechnology 8 (3), 465–478. [DOI] [PubMed] [Google Scholar]
- 31.Vermeulen LMP et al. (2018) Endosomal Size and Membrane Leakiness Influence Proton Sponge-Based Rupture of Endosomal Vesicles. ACS Nano 12 (3), 2332–2345. [DOI] [PubMed] [Google Scholar]
- 32.Clark SR et al. (2018) Determining the effects of PEI adsorption on the permeability of 1,2-dipalmitoylphosphatidylcholine/bis(monoacylglycero)phosphate membranes under osmotic stress. Acta Biomaterialia 65, 317–326. [DOI] [PubMed] [Google Scholar]
- 33.Vaughan EE et al. (2006) Intracellular trafficking of plasmids for gene therapy: mechanisms of cytoplasmic movement and nuclear import. Curr Gene Ther 6 (6), 671–681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen Z et al. (2017) Targeted Delivery of CRISPR/Cas9-Mediated Cancer Gene Therapy via Liposome-Templated Hydrogel Nanoparticles. Advanced Functional Materials 27 (46). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Dad AB et al. (2014) Enhanced gene disruption by programmable nucleases delivered by a minicircle vector. Gene Ther 21 (11), 921. [DOI] [PubMed] [Google Scholar]
- 36.Chen Z-Y et al. (2003) Minicircle DNA vectors devoid of bacterial DNA result in persistent and high-level transgene expression in vivo. Mol Ther 8 (3), 495–500. [DOI] [PubMed] [Google Scholar]
- 37.Yin H et al. (2014) Non-viral vectors for gene-based therapy. Nat Rev Genet 15 (8), 541–555. [DOI] [PubMed] [Google Scholar]
- 38.Mandal Pankaj K. et al. (2014) Efficient Ablation of Genes in Human Hematopoietic Stem and Effector Cells using CRISPR/Cas9. Cell Stem Cell 15 (5), 643–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Monroe KM et al. (2014) IFI16 DNA Sensor Is Required for Death of Lymphoid CD4 T Cells Abortively Infected with HIV. Science 343 (6169), 428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Jiang C et al. (2017) A non-viral CRISPR/Cas9 delivery system for therapeutically targeting HBV DNA and pcsk9 in vivo. Cell Research 27, 440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Liang X et al. (2015) Rapid and highly efficient mammalian cell engineering via Cas9 protein transfection. J Biotechnol 208, 44–53. [DOI] [PubMed] [Google Scholar]
- 42.Miller JB et al. (2017) Non-Viral CRISPR/Cas Gene Editing In Vitro and In Vivo Enabled by Synthetic Nanoparticle Co-Delivery of Cas9 mRNA and sgRNA. Angewandte Chemie International Edition 56 (4), 1059–1063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang J et al. (2016) Highly efficient homology-driven genome editing in human T cells by combining zinc-finger nuclease mRNA and AAV6 donor delivery. Nucleic Acids Research 44 (3), e30–e30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gaj T et al. (2012) Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nature Methods 9, 805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mout R et al. (2017) Direct cytosolic delivery of CRISPR/Cas9-ribonucleoprotein for efficient gene editing. ACS nano 11 (3), 2452–2458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Liu J et al. (2015) Improved Cell-Penetrating Zinc-Finger Nuclease Proteins for Precision Genome Engineering. Molecular Therapy. Nucleic Acids 4 (3), e232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Staahl BT et al. (2017) Efficient genome editing in the mouse brain by local delivery of engineered Cas9 ribonucleoprotein complexes. Nat Biotechnol 35 (5), 431–434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ramakrishna S et al. (2014) Gene disruption by cell-penetrating peptide-mediated delivery of Cas9 protein and guide RNA. Genome Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wefers B et al. (2013) Direct production of mouse disease models by embryo microinjection of TALENs and oligodeoxynucleotides. Proceedings of the National Academy of Sciences 110 (10), 3782. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Geurts AM et al. (2009) Knockout Rats via Embryo Microinjection of Zinc-Finger Nucleases. Science 325 (5939), 433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yin H et al. (2014) Genome editing with Cas9 in adult mice corrects a disease mutation and phenotype. Nat Biotechnol 32 (6), 551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Han X et al. (2015) CRISPR-Cas9 delivery to hard-to-transfect cells via membrane deformation. Science advances 1 (7), e1500454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.D’Astolfo DS et al. (2015) Efficient intracellular delivery of native proteins. Cell 161 (3), 674–690. [DOI] [PubMed] [Google Scholar]
- 54.Wang Y et al. (2015) Delivery of oligonucleotides with lipid nanoparticles. Advanced drug delivery reviews 87, 68–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hajj KA and Whitehead KA (2017) Tools for translation: non-viral materials for therapeutic mRNA delivery. Nature Reviews Materials 2, 17056. [Google Scholar]
- 56.de Lima MCP et al. (2001) Cationic lipid–DNA complexes in gene delivery: from biophysics to biological applications. Advanced drug delivery reviews 47 (2–3), 277–294. [DOI] [PubMed] [Google Scholar]
- 57.Liang C et al. (2017) Tumor cell-targeted delivery of CRISPR/Cas9 by aptamer-functionalized lipopolymer for therapeutic genome editing of VEGFA in osteosarcoma. Biomaterials 147, 68–85. [DOI] [PubMed] [Google Scholar]
- 58.Luo Y-L et al. (2018) Macrophage-Specific in Vivo Gene Editing Using Cationic Lipid-Assisted Polymeric Nanoparticles. ACS Nano. [DOI] [PubMed] [Google Scholar]
- 59.Zuris JA et al. (2015) Cationic lipid-mediated delivery of proteins enables efficient protein-based genome editing in vitro and in vivo. Nat Biotechnol 33 (1), 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Wang M et al. (2016) Efficient delivery of genome-editing proteins using bioreducible lipid nanoparticles. Proceedings of the National Academy of Sciences 113 (11), 2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Finn JD et al. (2018) A Single Administration of CRISPR/Cas9 Lipid Nanoparticles Achieves Robust and Persistent In Vivo Genome Editing. Cell Reports 22 (9), 2227–2235. [DOI] [PubMed] [Google Scholar]
- 62.Yin H et al. (2017) Structure-guided chemical modification of guide RNA enables potent non-viral in vivo genome editing. Nature Biotechnology 35, 1179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Yin H et al. (2016) Therapeutic genome editing by combined viral and non-viral delivery of CRISPR system components in vivo. Nat Biotechnol 34 (3), 328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim SM et al. (2017) Cancer-derived exosomes as a delivery platform of CRISPR/Cas9 confer cancer cell tropism-dependent targeting. J Control Release 266, 816. [DOI] [PubMed] [Google Scholar]
- 65.Lin Y et al. (2018) Exosome–Liposome Hybrid Nanoparticles Deliver CRISPR/Cas9 System in MSCs. Advanced Science 5 (4), 1700611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Yin H et al. (2014) Non-viral vectors for gene-based therapy. Nature Reviews Genetics 15, 541. [DOI] [PubMed] [Google Scholar]
- 67.Lee K et al. (2017) Nanoparticle delivery of Cas9 ribonucleoprotein and donor DNA in vivo induces homology-directed DNA repair. Nature Biomedical Engineering 1 (11), 889–901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Lee B et al. (2018) Nanoparticle delivery of CRISPR into the brain rescues a mouse model of fragile X syndrome from exaggerated repetitive behaviours. Nature Biomedical Engineering 2 (7), 497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sun W et al. (2015) Self assembled DNA nanoclews for the efficient delivery of CRISPR–Cas9 for genome editing. Angewandte Chemie International Edition 54 (41), 12029–12033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Wang H-X et al. (2018) Nonviral gene editing via CRISPR/Cas9 delivery by membrane-disruptive and endosomolytic helical polypeptide. Proceedings of the National Academy of Sciences. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Miller JB et al. (2018) Development of Cationic Quaternary Ammonium Sulfonamide Amino Lipids for Nucleic Acid Delivery. ACS Applied Materials & Interfaces 10 (3), 23022311. [DOI] [PubMed] [Google Scholar]
- 72.Jackson MA et al. (2017) Zwitterionic Nanocarrier Surface Chemistry Improves siRNA Tumor Delivery and Silencing Activity Relative to Polyethylene Glycol. ACS Nano 11 (6), 5680–5696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Chang H et al. (2017) Rational Design of a Polymer with Robust Efficacy for Intracellular Protein and Peptide Delivery. Nano letters 17 (3), 1678–1684. [DOI] [PubMed] [Google Scholar]
- 74.Yan M et al. (2009) A novel intracellular protein delivery platform based on single protein nanocapsules. Nature Nanotechnology 5, 48. [DOI] [PubMed] [Google Scholar]
- 75.Chen T-T et al. (2018) Biomineralized Metal-Organic Framework Nanoparticles Enable Intracellular Delivery and Endo-Lysosomal Release of Native Active Proteins. Journal of the American Chemical Society. [DOI] [PubMed] [Google Scholar]
- 76.Cheng G et al. (2018) Self-Assembly of Extracellular Vesicle-like Metal–Organic Framework Nanoparticles for Protection and Intracellular Delivery of Biofunctional Proteins. Journal of the American Chemical Society. [DOI] [PubMed] [Google Scholar]
- 77.Alsaiari SK et al. (2017) Endosomal Escape and Delivery of CRISPR/Cas9 Genome Editing Machinery Enabled by Nanoscale Zeolitic Imidazolate Framework. Journal of the American Chemical Society 140 (1), 143–146. [DOI] [PubMed] [Google Scholar]
- 78.Ledford H (2017) Engineered cell therapy for cancer gets thumbs up from FDA advisers. Nature 547 (7663), 270. [DOI] [PubMed] [Google Scholar]
- 79.Bach PB et al. (2017) FDA Approval of Tisagenlecleucel: Promise and Complexities of a $475 000 Cancer Drug. Jama 318 (19), 1861–1862. [DOI] [PubMed] [Google Scholar]
- 80.Fischer A, FDA approves novel gene therapy to treat patients with a rare form of inherited vision loss, FDA, FDA.gov, 2017. [Google Scholar]
- 81.Russell S et al. (2017) Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: a randomised, controlled, open-label, phase 3 trial. The Lancet 390 (10097), 849–860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Eyquem J et al. (2017) Targeting a CAR to the TRAC locus with CRISPR/Cas9 enhances tumour rejection. Nature 543 (7643), 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Cyranoski D (2016) Chinese scientists to pioneer first human CRISPR trial. Nature News 535 (7613), 476. [DOI] [PubMed] [Google Scholar]
- 84.Qasim W et al. (2017) Molecular remission of infant B-ALL after infusion of universalTALEN gene-edited CAR T cells. Science Translational Medicine 9 (374). [DOI] [PubMed] [Google Scholar]
- 85.Reardon S (2016) First CRISPR clinical trial gets green light from US panel. Nature News. [Google Scholar]