Summary
Many questions regarding proteins involved in microbial sulfur metabolism remain unsolved. For sulfur respiration at low pH, the terminal electron acceptor is still unclear. Desulfurella amilsii is a sulfur‐reducing bacterium that respires elemental sulfur (S0) or thiosulfate, and grows by S0 disproportionation. Due to its versatility, comparative studies on D. amilsii may shed light on microbial sulfur metabolism. Requirement of physical contact between cells and S0 was analyzed. Sulfide production decreased by around 50% when S0 was trapped in dialysis membranes, suggesting that contact between cells and S0 is beneficial, but not strictly needed. Proteome analysis was performed under the aforementioned conditions. A Mo‐oxidoreductase suggested from genome analysis to act as sulfur reductase was not detected in any growth condition. Thiosulfate and sulfite reductases showed increased abundance in thiosulfate‐reducing cultures, while rhodanese‐like sulfurtransferases were highly abundant in all conditions. DsrE and DsrL were abundantly detected during thiosulfate reduction, suggesting a modified mechanism of sulfite reduction. Proteogenomics suggest a different disproportionation pathway from what has been reported. This work points to an important role of rhodaneses in sulfur processes and these proteins should be considered in searches for sulfur metabolism in broader fields like meta‐omics.
Introduction
Elemental sulfur is one of the most ubiquitous sulfur species in sediments and geological deposits, formed by biological and chemical oxidation of H2S (Rabus et al., 2013). Elemental sulfur has a low solubility in water [5 μg l−1 at 20°C (Boulegue, 1978)] hampering fast growth of sulfur‐utilizing prokaryotes (Blumentals et al., 1990; Schauder and Müller, 1993). However, microorganisms able to utilize elemental sulfur are great in number and widespread over the tree of life (Dahl et al., 2001; Friedrich et al., 2001; Rabus et al., 2013; Florentino et al., 2016a,b).
Two mechanisms have been postulated in sulfur‐reducers to cope with the low solubility of elemental sulfur: (a) direct physical attachment of the cells to the solid S0 phase (Laska et al., 2003), or (b) a soluble intermediate form of sulfur terminally accepts electron. Regarding the latter, when sulfide is available in the environment, it cleaves the S0‐ring of elemental sulfur by a nucleophilic attack, which generates polysulfide (Eq. 1), enabling an enhancement of sulfur respiration (Blumentals et al., 1990, Schauder and Müller, 1993, Hedderich et al., 1999).
| (1) |
The concentration and chain length of polysulfide depend on several parameters, such as pH, elemental sulfur and sulfide concentrations, redox potential, and temperature. The maximum polysulfide concentration in solution increases with increasing pH (Schauder and Müller, 1993). Some binding proteins produced by sulfur‐reducing microorganisms, such as the polysulfide sulfur transferase showed high affinity for polysulfide at low concentrations in Wolinella succinogenes (Klimmek et al., 1999, Lin et al., 2004).
At low pH, however, polysulfide is unstable and is readily converted to elemental sulfur (Steudel, 2003). While at 20°C and pH 6.5, the maximum concentration of polysulfide in solution is around 0.1 mM, at pH 3 (in the presence of elemental sulfur in excess and 1 mM of hydrogen sulfide) it is just around 10−12 M at 80°C (Schauder and Müller, 1993).
Therefore, depending on the environmental conditions, the terminal electron acceptor for sulfur‐reducing enzymes might differ. The enzymes possibly involved in the respiration of those compounds were only isolated and characterized from a few microorganisms, such as Wolinella succinogenes (Klimmek et al., 1991), Pyrococcus furiosus (Ma and Adams, 1994) and Acidianus ambivalens (Laska et al., 2003). Unfortunately, a well‐defined activity of those enzymes in the metabolism of sulfur utilizers is not yet available.
Sulfur disproportionation, a process involving oxidative and reductive routes, is also a poorly characterized part of the sulfur metabolism. Although those reactions have been described for several sulfur reducers from the Proteobacteria, Firmicutes and Thermodesulfobacteria phyla (Finster et al., 1998, Finster, 2008, Hardisty et al., 2013, Florentino et al., 2016a,b), the formation and oxidation of sulfite as a key intermediate is not yet understood (Finster, 2008, Hardisty et al., 2013). In a similar manner, sulfite is postulated to be an intermediate in thiosulfate reduction (Surkov et al., 2000, Stoffels et al., 2012), but the complete mechanism of thiosulfate respiration leading to sulfide is still unclear.
Desulfurella amilsii is an acidotolerant sulfur‐reducing bacterium isolated from sediments of the acidic Tinto river in Spain. It can grow in a broad range of pH (from 3 to 7) and it is able to grow by respiring elemental sulfur or thiosulfate, and by disproportionating elemental sulfur (Florentino et al., 2016a,b). Based on its genome sequence, D. amilsii encodes an NADH‐dependent ferredoxin:NADP+ oxidoreductase (NfnAB; DESAMIL20_1852–1853) that in the past was reported to act as sulfide dehydrogenase (Ma and Adams, 1994); and a Mo‐oxidorreductase with no confident functional assignment (DESAMIL20_1358–1361), but likely involved in sulfur metabolism, possibly playing a role as sulfur reductase (Laska et al., 2003) or tetrathionate reductase. The multiple enzymes open possibilities for both polysulfide and elemental sulfur serving as terminal electron acceptors (Florentino et al., 2017). The reduction of thiosulfate is likely catalyzed by another Mo‐oxidoreductase serving as thiosulfate reductase (PhsAB; DESAMIL20_9–10) and the sulfite reductase (DsrAB; DESAMIL20_1434–1435) encoded in the genome of D. amilsii. A number of rhodanese‐like sulfurtransferase genes are also encoded in the genome of D. amilsii without a clear function.
Performing important conversions of the reductive sulfur cycle, D. amilsii was used to combine strategies for sulfur metabolism research. Analyses at the protein level can help to shed some light on the pathways involved in the metabolism of this microorganism. In this respect, this study aimed to: (i) investigate the requirement for cell‐sulfur interaction in sulfur respiration by trapping sulfur in dialysis membranes; and through comparative proteomics (ii) elucidate enzymes involved in sulfur/thiosulfate respiration and sulfur disproportionation, and (iii) compare autotrophic and organotrophic growth to identify the enzymes involved. This combined strategy allowed us to obtain information about energy and carbon metabolism of D. amilsii, as well as to reveal the importance of some enzymes – rhodanese‐like sulfurtransferases – in sulfur metabolism. This study is the first proteomic study performed under different sulfur metabolizing conditions in a comparative way, helping to elucidate some aspects of this important metabolism.
Material and methods
Growth medium and cultivation
D. amilsii cells were grown in 1 L bottles containing 500‐ml anoxic medium (Florentino et al., 2015). To adjust the pH of the medium, bicarbonate‐buffer was omitted as described by Sánchez‐Andrea et al. (2013) and pH was adjusted to 3.5 or 6.5 with HCl. In the set analyzed for disproportionation of elemental sulfur, elemental sulfur was added at a concentration of 25 mM with dissolved carbon dioxide or bicarbonate likely used as carbon source. For the cultures incubated under sulfur‐respiring conditions, either elemental sulfur or thiosulfate (25 mM) were added as electron acceptors; in the chemolithotrophic cultures subset H2/CO2 (1.5 atm, 80:20, v/v) was added to provide H2 as electron donor and CO2 as carbon source, while the heterotrophic cultures were supplied with acetate (5 mM) as electron donor and carbon source. Bottles with medium were sealed with butyl rubber stoppers (Rubber BV, Hilversum, The Netherlands) and autoclaved for 30 min at 105°C. Except otherwise indicated, cultures were incubated statically at 50°C and in the dark.
Chemically synthesized elemental sulfur was the only form of elemental sulfur utilized for all experiments in this study. Sample identification stands for electron donor, electron acceptor and pH of cultivation: e−donor_e−acceptor_pH. When there was no electron donor added, S_ stands for disproportionation.
Dialysis membranes experiment
Sulfur‐ and heavy metals‐free Spectra/Por (Spectrum, Rancho Dominguez, CA) cellulose membranes with a molecular weight cut‐off of 6–8 kDa were used. Prior to the utilization, they were rinsed and dipped in demineralized water to remove preservatives. The membranes were filled with chemically synthesized elemental sulfur (0.4 g) and 4 ml of the cultivation medium, sealed with standard Spectra/Por closures (Spectrum, CA), placed in culture vials containing 500 ml of anoxic medium with H2/CO2 (1.5 atm, 80:20, v/v). An active culture incubated at 50°C with H2/CO2 (1.5 atm, 80:20, v/v) was used as inoculum (1% v/v). Incubations were set in triplicate, with and without membrane, and at pH 3.5 and 6.5 and incubated statically at 50°C. Uninoculated controls were also included.
Growth and sulfur reductive activity were followed weekly via cell counts and sulfide production respectively. The number of cells was determined by using a Petroff‐Hausser counting chamber with a cell‐depth of 0.02 mm and ruling pattern 1:400 mm2 (Hausser Scientific, Horsham, PA). Sulfide production was determined colorimetrically using the methylene blue method described by Cline (1969). The cellular elemental sulfide production rates (cESPR) were calculated from the formation of hydrogen sulfide and the cell numbers as described by Surkov et al. (2000) with modifications (Eq. 2):
| (2) |
where, S, C and t refer to the amount of hydrogen sulfide produced (μmol), the averaged cell number and the reaction time (day), respectively, at time intervals i and i − 1.
Integrity of the dialysis membranes was checked after each experiment by visual inspection and scanning electron microscopy (SEM). For SEM, a 3% (v/v) solution of glutaraldehyde (Sigma Aldrich, St. Louis, MI) in phosphate‐buffered saline solution was used to fix the cells for 1 h at room temperature. Afterward samples were dehydrated in increasing concentrations of ethanol (10%, 30%, 50%, 70%, 80%, 90%, 96% and 100%) and air‐dried. Cells were analyzed using a JEOL JSM‐6480LV microscope (JEOL, Peabody, MA).
Proteomics
Proteins were extracted with an average protein concentration of 5 μg ml−1, during the late exponential phase from biological triplicates of cultures grown as above‐mentioned by disproportionating, sulfur‐ and thiosulfate‐reducing conditions at pH 6.5. The sulfur‐reducing condition was additionally tested at pH 3.5. Growth curves of the cells cultivated under the studied conditions are presented in Supporting Information Fig. S1.
Cultures were centrifuged (10 min, 4°C, 10 000g) and the cell pellets were re‐suspended in 0.5 ml SDT‐lysis buffer [50 mM dithiothreitol +4% (w/v) sodium dodecyl sulfate in 100 mM Tris/HCl pH 7.6] with 100 μM of phenylmethylsulfonyl fluoride. The suspension was sonicated six times using a Branson sonifier SFX150 equipped with a 3 mm tip (Branson, Carouge, CH), in cycles of 30 s pulse and 30 s rest intervals on ice. Unbroken cells and cell debris were removed by centrifugation at 13 000 rpm for 10 min and the protein concentration in the supernatant was measured with the Pierce BCA Protein Assay Kit (Thermo Scientific, Waltham, MA).
Proteins (15 μg) were loaded to Precise™ 12% Tris‐HEPES Gels, 58 mm × 80 mm × 1 mm, 10‐Well (ThermoFisher Scientific, Waltham, MA) and run for 30 min at 120 V. The gels were stained for 3 h with the Colloidal Blue Staining Kit (ThermoFisher Scientific, Waltham, MA) and de‐stained for 15 h in demineralized water. Gels were washed with demineralized water three times. Disulfide bonds of the proteins were dissociated by incubating the gels in 10 mM dithiothreitol in 50 mM ammonium bicarbonate at 60°C for 1 h. Gels were washed with demineralized water three times. Carboxamidomethylation of the reduced cysteines was performed with 20 mM iodoacetamide in 100 mM Tris (pH 8) at room temperature in dark conditions for 1 h. Gels were washed with demineralized water three times. Each lane was cut into three slices of 1 cm height which were individually cut into pieces of about 1 mm2 and transferred to 1.5 ml‐Eppendorf Protein LoBind tubes (Eppendorf, Hamburg, Germany). Enzymatic digestion was performed by adding 250 ng of trypsin in 100 μl of ammonium bicarbonate (50 mM). The tryptic digestion was incubated at room temperature shaking at 50 rpm for 18 h. In order to stop the trypsin digestion, 10% trifluoroacetic acid in H2O was added to the samples until the pH dropped to 3, followed by sonication for 15 s. The trypsin‐digested peptide sample was loaded onto a clean‐up column prepared in‐house, which contained a stage tip with 2 mg extra 0.10 × 32 mm Magic C18AQ 200A (5 μm bead size) material (Bruker Nederland B.V., Leiderdorp, NL), as described previously (Rappsilber et al., 2007, Lu et al., 2011). Gel pieces were washed with 100 μl formic acid (23.6 mM) and added to the column. The content of the column was eluted, and the column was washed once more with 100 μl formic acid (23.6 mM). Peptides were eluted manually with 50 μl 1:1 acetonitrile (ACN):formic acid (23.6 mM). The ACN content was reduced by concentrating the samples at 45°C for 1 h in an Eppendorf™ Vacufuge™ Concentrator (Eppendorf, Hamburg, Germany). The peptide sample volumes were brought to 50 μl with 23.6 mM formic acid.
The peptide samples were analyzed by injecting 18 μl sample over a 0.10 × 32 mm Magic C18AQ 200A 5 μm beads (Bruker Nederland B.V.) pre‐concentration column (prepared in‐house, Lu et al., 2011) at a constant pressure of 270 bar (normally resulting in a flow of ca., 7 μl min−1). Peptides were eluted from the pre‐concentration column onto a 0.10 × 250 mm Magic C18AQ 200A (3 μm bead size) analytical column (prepared in‐house, Lu et al., 2011) with an ACN gradient at a flow of 0.5 μl min−1 with a Proxeon EASY nanoLC. The gradient increased from 8% to 33% acetonitrile in water with 23.6 mM formic acid in 50 min, followed by a fast increase in the percentage of ACN to 80% (with 20% water and 23.6 mM formic acid in both the acetonitrile and the water) in 3 min as a column cleaning step.
A P‐777 Upchurch Microcross column was positioned between the pre‐concentration and analytical column. An electrospray potential of 3.5 kV was applied directly to the eluent via a stainless‐steel needle fitted into the waste line of the microcross. Full scan positive mode FTMS spectra were measured between m/z 380 and 1400 on a LTQ‐Orbitrap XL (Thermo electron, San José, CA) in the Orbitrap at high resolution (60 000). CID fragmented MSMS scans of the four most abundant 2+ and 3+ charged peaks in the FTMS scan were recorded in data dependent mode in the linear trap (MSMS threshold = 5000, 45 s exclusion duration for the selected m/z ± 25 ppm).
LCMS data with all MSMS spectra were analyzed with MaxQuant 1.5.2.8 (Cox and Mann, 2008) using default settings for the Andromeda search engine (Cox et al., 2011), except that extra variable modifications were set for deamidation of N and Q.
The D. amilsii protein sequence database (Florentino et al., 2017) with 2045 protein sequences (NCBI accession number MDSU01) was used together with a database which contains sequences of common contaminants such as BSA (P02769, bovine serum albumin precursor), trypsin (P00760, bovine), trypsin (P00761, porcine), keratin K22E (P35908, human), keratin K1C9 (P35527, human), keratin K2C1 (P04264, human) and keratin K1CI (P35527, human). De‐amidated peptides were allowed to be used for protein quantification. The MaxQuant MaxLFQ algorithm (Cox et al., 2014), a MS1 peak intensity‐based label‐free quantification (LFQ) method (Cox and Mann, 2008; Hubner et al., 2010), was used to combine the normalized peptide intensities of the fractions (in relation to the total amount of protein and all of its identified peptides) to make possible statistical comparisons of normalized intensities of protein groups between conditions.
Filtering and further analysis of the MaxQuant/Andromeda workflow output and the analysis of the abundances of the identified proteins were performed with the Perseus 1.5.5.3 module (available at the MaxQuant suite). Peptides were accepted for further analysis when they had a false discovery rate (FDR) of less than 1% and proteins with at least two identified peptides of which at least one was unique and one unmodified. Reversed hits and contaminants were deleted from the MaxQuant output. The nLC‐MSMS system quality was checked with PTXQC (Bielow et al., 2016) using the MaxQuant result files. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD008496.
Comparative analyses of the proteins sequences with matches were performed on the Rapid Annotation using Subsystem technology (RAST) database (http://rast.nmpdr.org/). The BLAST tool of the Joint Genome Institute (JGI) website was used to check the presence of homologous genes in selected metagenomes (https://img.jgi.doe.gov/).
Quantitative reverse transcription PCR (qRT‐PCR)
Cultures grown at pH 3.5 and 6.5 (500 ml) were harvested in the exponential phase and centrifuged at 4000g for 20 min. The pellet was re‐suspended in 1 ml of Tris‐EDTA buffer; 20 U of lysozyme were added, and the mixture was incubated for 10 min at room temperature. Pure β‐mercaptoethanol (3 μl) and proteinase‐K (0.8 U) were added to the samples with 150 μl of cell lysis solution (Epicentre, Madison, WI). The mixture was incubated at 60°C for 10 min, following the instructions of the manufacturer and cooled down on ice for 5 min. A volume of 175 μl of MPC Protein Precipitation Reagent (Epicentre, Madison, WI, USA) was added to the samples and centrifuged for 10 min at 4°C prior to removal of pellet debris. The supernatant was used for RNA extraction by the Maxwell® 16S RNA Purification System (Promega, Madison, WI). cDNA was synthesized by the GoScript™ Reverse Transcription System (Promega, Madison, WI), followed by the amplification of the subsequent transcripts.
Primers targeting sulfur reductase genes were designed using the Primer3plus tool available at http://www.bioinformatics.nl/cgi‐bin/primer3plus/primer3plus.cgi, considering the target length average between 18 and 27 nucleotides, melting temperature (T m) between 52 and 60°C and the GC content between 40% and 60%. The forward sequence of the primer designed to target SreA was 5′TGCTCTACCGCATGTTTCTG3′, and the reverse 5′CTGCGTGAGCGATTTGATTA3′. For the primer targeting SreC, the forward sequence was 5′CTGGGGTGGAGCATTATCTG3′ and the reverse 5′ATCAAAGCCTAGCCATGGTG3′. The primer pairs were tested for specificity at its optimal annealing temperature (58°C).
PCR amplification was performed in triplicate in a BioRad CFX96 system (Bio‐Rad Laboratories, Hercules, CA) in a total volume of 25 μl using iTaq Universal SYBR Green Supermix (Bio‐Rad Laboratories), template cDNA at a final concentration of 0.2–0.4 ng μl−1 and 0.4 μM of forward and reverse primers, according to the manufacture's recommendations. Relative quantification was used to determine the changes in steady‐state mRNA levels of sreA gene and to express it relative to the levels of 16S rRNA gene. Results were based on the comparison of the distinct quantification cycle values (Cq) at a constant level of fluorescence. Cq refers to the cycle in which fluorescence can be detected, and lower Cq values mean higher initial copy numbers of the target.
Results
Dialysis membranes experiment
The permeable membrane used ensured that cells had no direct contact with elemental sulfur but allowed soluble molecules smaller than 6–8 kDa to diffuse through the pores. Sulfide production levels – as marker for metabolic activity – ranged from 0 to 5.0 mmol per culture with the corresponding consumption of hydrogen (data not shown). Starting levels of sulfide in all the cultures were around 0.4 mmol, due to its release during medium autoclavation from the cysteine used as reducing agent. Besides, the addition of an active culture as inoculum contributed with 0.4 mmol of sulfide in the cultures, resulting in 0.8 mmol of sulfide at time zero. The uninoculated control group, with starting sulfide level of 0.4 mmol, did not show any increase in sulfide along the 42 days of experiment. The sulfide production data differed more by the presence or absence of the membrane than by pH, showing a decrease of 47% and 51% when S0 was trapped in dialysis bags at pH 6.5 and 3.5 respectively (Fig. 1A). In the same way, the planktonic cells number decreased when S0 was trapped in dialysis bags (Fig. 1B), by 78% and 97% at pH 6.5 and 3.5 respectively.
Figure 1.
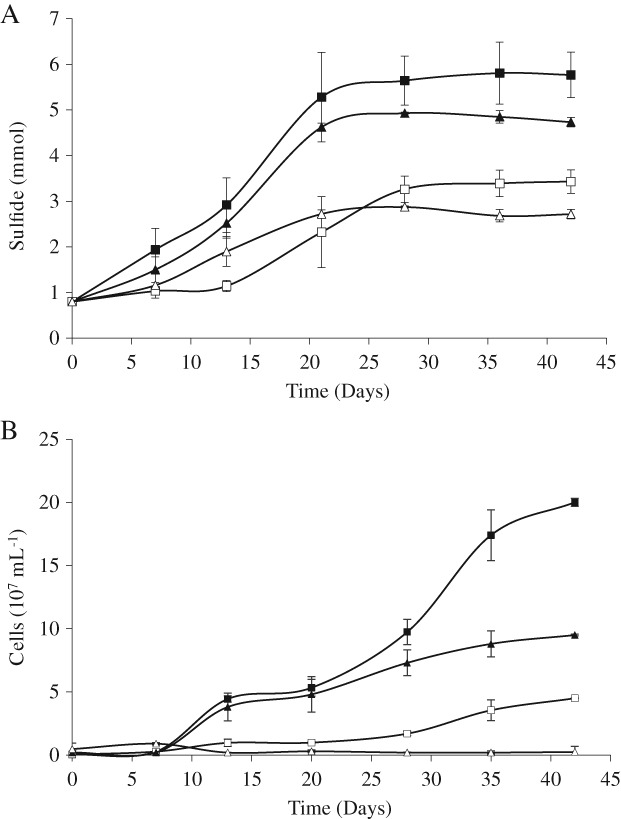
Hydrogen sulfide production (A) and number of planktonic cells (B) of D. amilsii at pH 6.5 and 3.5 with sulfur dispersed in the medium and sulfur trapped in dialysis bags of 6–8 kDa molecular weight cut‐off. The results were averaged from biological triplicate measurements and the standard deviation is shown. The squares represent cultures grown at pH 6.5 and the triangles represent pH 3.5. Filled symbols represent cultures with sulfur dispersed in the medium and open symbols represent cultures with sulfur trapped in dialysis bags.
Cultures incubated with sulfur dispersed in the medium at pH 6.5 and 3.5 showed sulfide production rates per cell of 18.9 and 10.7 fmol cell−1 day−1 respectively. For the cultures with sulfur trapped in dialysis bags, the rates obtained were equal at pH 6.5 (18.9 fmol cell−1 day−1), while at pH 3.5 the rate per cell would give an unrealistic value of 730 fmol cell−1 day−1 due to the low number of planktonic cells detected. Since the sulfide production levels were similar with or without membrane, likely the low number of planktonic cells detected was an underestimation of the real cell number due to a large number of cells being attached to the membrane.
To test this hypothesis, SEM was performed at the surface of the dialysis membrane at both pH conditions. At pH 3.5, where planktonic cells were hardly detected, a large number of cells were attached to the surface of the membrane (Fig. 2A). Although some cells could be visualized on the surface of the membrane at pH 6.5 (Fig. 2B), cells were mostly present in suspension (2 × 108 cells ml−1). Extracellular polymeric substance (EPS) was visible in both conditions, but at low pH, significant cell aggregation was observed (Fig. 2C), while at high pH cells appeared mostly single or in pairs (Fig. 2B and D).
Figure 2.
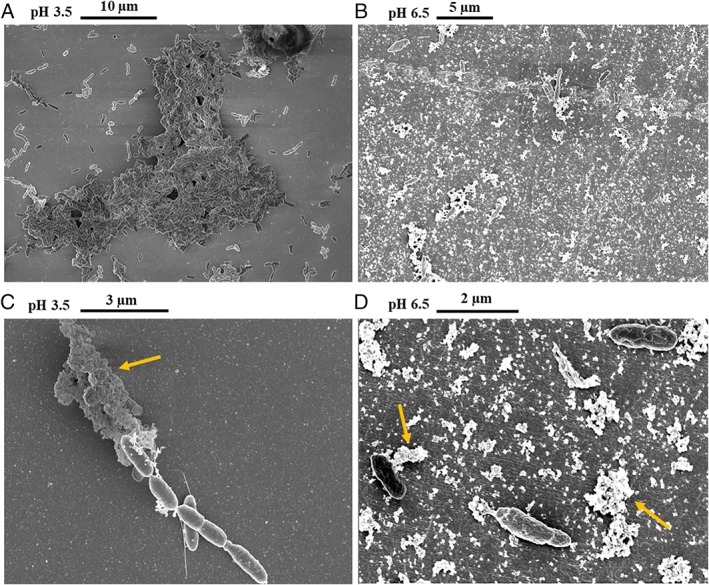
Scanning electron micrographs of the dialysis bags surface. A. Aggregate of cells of D. amilsii grown at pH 3.5, depicted from a larger field. B. Larger field depicted revealing few cells attached to surface of the membrane at pH 6.5. C. Presence of EPS (indicated by yellow arrow) and appendages in the cells attached to the surface of the membrane at pH 3.5. D. Presence of EPS, indicated by yellow arrows, covering and attaching the cells to the membrane at pH 6.5. [Color figure can be viewed at wileyonlinelibrary.com]
Comparative proteomic analysis
Global and differential proteomic results
Five growth conditions were studied by proteomic analysis: two respiratory conditions for thiosulfate or sulfur reduction with acetate as electron donor (pH 6.5), and three autotrophic conditions with sulfur reduction and hydrogen as electron donor (pH 6.5 and pH 3.5) and sulfur disproportionation (pH 6.5). From a total of 2088 protein coding sequences identified in the genome of D. amilsii (Florentino et al., 2017), 698 proteins were identified in the five conditions studied. A core of 492 proteins was identified in all the analyzed conditions. The highest number of total proteins was identified in the culture growing chemolithotrophically by sulfur respiration at circumneutral pH 6.5 (698), while thiosulfate‐respiring cultures showed the lowest number of total proteins identified (582). In Fig. 3, the number of common proteins identified at the different conditions is displayed. Overall, 10–24 proteins were exclusively present in one of the growth conditions (Supporting Information Table S1).
Figure 3.
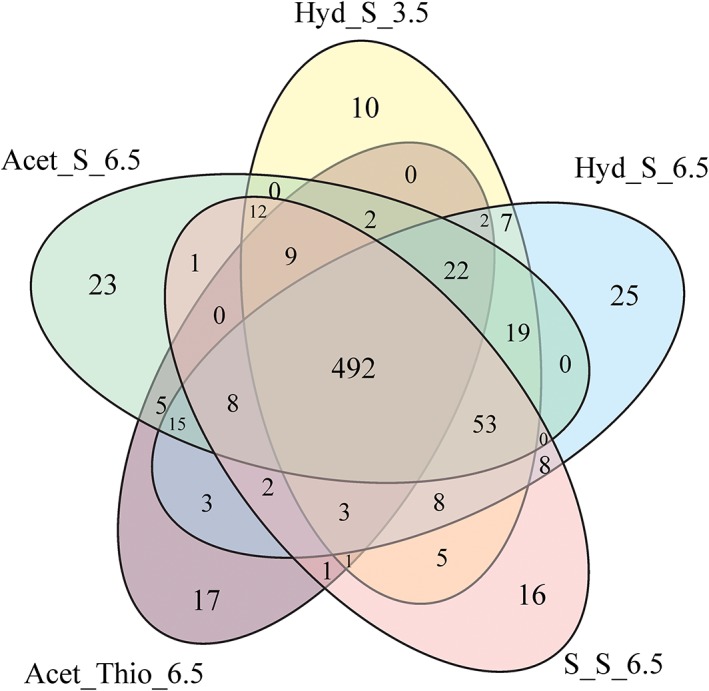
Overview of common and unique proteins identified among different conditions in D. amilsii of a total of 698 proteins. [Color figure can be viewed at wileyonlinelibrary.com]
A total of 524 proteins were significantly abundant in cultures grown on acetate by sulfur respiration, 293 in cultures grown by respiration of thiosulfate, and 454 in sulfur‐disproportionating cultures. In a comparative analysis, the majority of the abundant proteins was related to reactions involved in the tricarboxylic acid (TCA) cycle, oxidoreductases and proteins of unknown function.
In a pairwise comparison, 112 proteins showed significantly different intensities between sulfur‐reducing and sulfur‐disproportionating cultures; 92 between sulfur and thiosulfate‐reducing cultures, 137 between thiosulfate‐reducing and sulfur‐disproportionating cultures and 47 proteins showed significantly different intensities within pH 3.5 and 6.5 for chemolithotrophic cultures (p value <0.05 and S0 = 1). The complete lists of proteins with significantly different abundances in pairwise comparisons are given in Supporting Information Tables S2–S5. In Supporting Information Tables S6 and S7, raw values for normalized protein abundance (LFQ intensity) of the proteins involved in the analyzed metabolisms.
Sulfur metabolism
Cell–sulfur interaction and sulfur respiration
Flagellar proteins were abundant in all the conditions with sulfur as electron acceptor as shown in Fig. 4, such as FliD FlgK, FlgL, FlaG and FlaA, as well as proteins involved in the biosynthesis of pili, such as TraB, PilQ, PilT, PilB, PliM and PilC, which are reported to be involved in attachment and biofilm formation of microorganisms (O'Toole and Kolter, 1998, Klausen et al., 2003).
Figure 4.
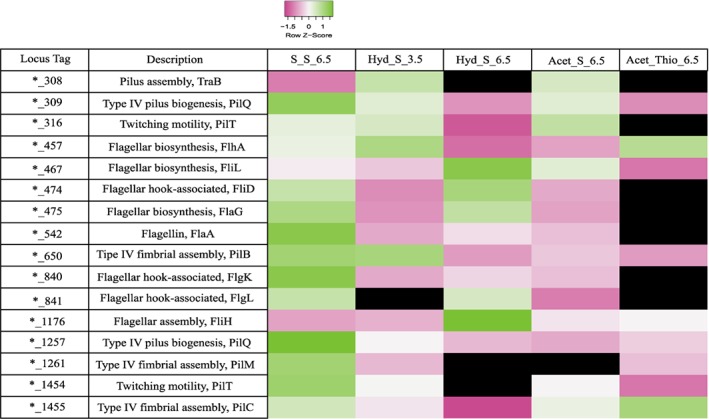
Relative abundance levels of proteins possibly involved in motility, cell‐sulfur attachment and/or uptake processes in D. amilsii TR1 in the 5 conditions analyzed. LFQ intensities of the biological triplicates were averaged and are shown after Z‐score normalization. The colours gradient indicates the degree of intensity, in which low relative intensity is indicated in pink and high relative intensity in green. Black colour indicates protein not detected in the respective condition. Raw values of LFQ intensities are shown in Supplementary Table S6. [Color figure can be viewed at wileyonlinelibrary.com]
A thiol:disulfide interchange protein (DESAMIL20_693) was revealed in equal abundance in cultures grown by sulfur respiration (Fig. 5) and was absent in the thiosulfate condition. Besides, three rhodanese‐like sulfurtransferases (DESAMIL20_1100, DESAMIL20_270 and DESAMIL20_1419) were found to be highly abundant in all the conditions, while the DESAMIL20_1987 was only abundant in cultures growing at pH 3.5 and the DESAMIL20_2007 was only abundant also in the cultures respiring sulfur at pH 3.5 and disproportionating ones. In the proteome of D. amilsii, the subunits of the Mo‐oxidorreductase possibly acting as sulfur reductase or tetrathionate reductase (DESAMIL20_1360) (Florentino et al., 2017), were not detected in any condition, while the putative NfnAB ferredoxin:NADP+ oxidoreductase (DESAMIL20_1852 and DESAMIL20_1853), previously annotated as sulfide dehydrogenase, was highly abundant at both low and high pH. Moreover, from the five rhodanese‐like sulfurtransferases encoded in the genome of D. amilsii (DESAMIL20_270, DESAMIL20_1100 and DESAMIL20_1419, DESAMIL20_2007, DESAMIL20_1987), the first three are highly abundant in cultures growing by sulfur and thiosulfate respiration (Fig. 5). In parallel with the abundance of the electron bifurcating NfnAB NADH‐dependent ferredoxin:NADP+ oxidoreductase (Wang et al., 2010; Buckel and Thauer, 2018), ferredoxins (DESAMIL20_472) were present in the cultures, with 2.6‐fold abundance at pH 6.5 in comparison to pH 3.5.
Figure 5.
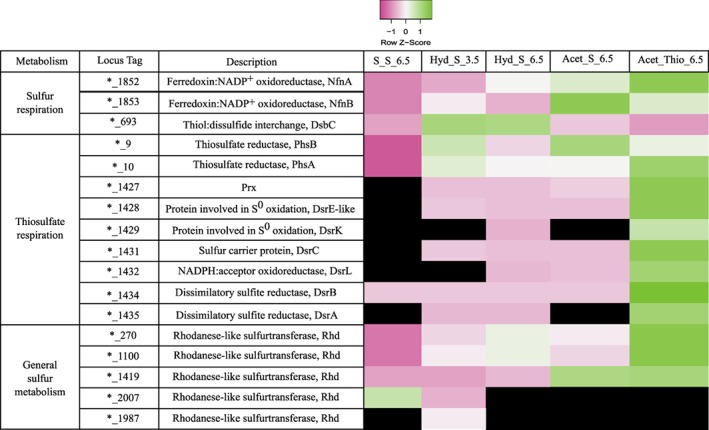
Relative abundance levels of proteins possibly involved in sulfur and thiosulfate reduction in D. amilsii TR1. LFQ intensities of the biological triplicates were averaged and are shown after Z‐score normalization. The colours gradient indicates the degree of intensity, in which low relative intensity is indicated in pink and high relative intensity in green. Black colour indicates protein not detected in the related condition. Raw values of LFQ intensities are shown in Supporting Information Table S7. The locus tag for the genes encoded in D. amilsii are represented in the table by the specific identifier preceded *_, in which * stands for DESAMIL20_. [Color figure can be viewed at wileyonlinelibrary.com]
Although membrane proteins constitute about 30% of the proteome in an organism, proteomics protocols normally lead to low recovery of such proteins (Bill et al., 2011). The major challenges for membrane proteomics are related to: (1) low abundance due to their hydrophobicity and therefore limited solubility, rendering them poorly soluble, aggregating and precipitating in aqueous environments (Rawlings, 2016; Chandramouli and Qian, 2009); and (2) restricted enzyme accessibility for hydrolysis of peptide bonds by trypsin, since this has as target residues lysine and arginine, which are mostly absent in transmembrane helices (Chandramouli and Qian, 2009). Therefore, due to the absence of the membrane‐bound Mo‐oxidorreductase (DESAMIL20_1360) in the proteomics data and to thrust aside possible technical limitations of the proteomics protocol, a qRT‐PCR targeting the catalytic and membrane‐bound subunits of this Mo oxidoreductase was performed. Cultures grown on hydrogen and sulfur at pH 6.5 and 3.5 (Hyd_S_6.5 and Hyd_S_3.5) were used to determine whether this transcript was present in D. amilsii. A difference of circa 10 cycles in the reaction can be observed between D. amilsii cultures and the negative control, indicating expression of the genes DESAMIL20_1360 and DESAMIL20_1359 in the studied microorganism. However, no difference in expression was observed at both pH values studied. Besides, high numbers for quantification cycles reflect their low expression levels (Table 1), suggesting that this enzyme might instead play a tetrathionate reductase role in D. amilsii.
Table 1.
Quantification cycles of the primers specific for the genes of interest in D. amilsii grown at pH 3.5 and 6.5.
| Gene | Condition | ||
|---|---|---|---|
| D. amilsii (pH 3.5) | D. amilsii (pH 6.5) | NC | |
| 16S rRNA | 27.2 (±0.4) | 26.7 (±0.1) | 44.3 (±0.3) |
| DESAMIL20_1360 | 36.5 (±0.8) | 35.8 (±0.4) | 45.8 (±0.6) |
| DESAMIL20_1359 | 32.5 (±0.3) | 32.8 (±0.1) | 45.4 (±0.5) |
NC stands for negative control.
Thiosulfate respiration
Thiosulfate respiration is typically reported to involve a thiosulfate reductase which catalyzes the reaction displayed in Eq. 3 (Stoffels et al., 2012). The sulfite produced is reduced to sulfide by the dissimilatory sulfite reductase DsrAB (Eq. 4), together with DsrC and with the involvement of DsrMKJOP complex (Eq. 5), which is overall an energy‐yielding reaction (Santos et al., 2015).
The catalytic subunit of thiosulfate reductase is quite similar to the catalytic subunits of polysulfide reductase and sulfur reductase, all of which are Mo‐oxidoreductases. The very high abundance of the Mo‐oxidorreductase (DESAMIL20_10) in thiosulfate‐respiring cultures likely indicates its role as a thiosulfate‐reducing enzyme in D. amilsii. DESAMIL20_10 (PhsA) was 8.3 times more abundant in thiosulfate‐respiring cultures than in sulfur‐reducing cultures and 31.4 times more abundant than in sulfur‐disproportionating cultures (Fig. 5). The beta subunit of the thiosulfate reductase (DESAMIL20_9) was 2.5 times more abundant in thiosulfate respiration than in sulfur‐disproportionating cultures but was not differentially produced in comparison to the sulfur‐respiring cultures. The operon encoding this enzyme lacks a membrane subunit, so the thiosulfate reductase of D. amilsii is apparently a dimeric periplasmic enzyme, as there is a Tat signal peptide in DESAMIL20_10. The alpha and beta subunits of the dissimilatory sulfite reductase (DsrAB, DESAMIL20_1434‐DESAMIL20_1435), together with DsrC (DESAMIL20_1431) and the DsrK subunit (DESAMIL20_1429) of the DsrMKJOP complex were also more abundant in thiosulfate‐reducing cultures.
Only one peptide of the DsrM subunit (DESAMIL20_1430) of the DsrMK complex was identified and therefore the DsrM subunit was not included in the strictly filtered proteome dataset. On the contrary, the DsrK subunit was identified with 15 peptides.
Interestingly, a single cysteine is present at the N‐terminus of the D. amilsii DsrC protein, which hampers the formation of a protein‐based trisulfide, with a sulfite‐derived sulfur connecting the two conserved cysteines of DsrC, as described by Santos et al., (2015). The DsrJOP subunits typically present in the Gram‐negative bacteria are surprisingly not encoded in the genome of D. amilsii, which is an interesting anomaly. It is recognized that DsrMK are the minimum subunits needed for electron transfer from the quinone pool to the sulfite reduction process as it has been shown for some Gram‐positive Firmicutes (Junier et al., 2010, Pereira et al., 2011) However, all the Deltaproteobacteria sulfite reducers analyzed so far possess the complete DsrMKJOP complex (Grein et al., 2013).
| (3) |
| (4) |
| (5) |
A protein related to DsrE (DESAMIL20_1428), which is a sulfurtransferase in sulfur oxidizers (Dahl, 2015) was highly abundant exclusively in D. amilsii cultures growing by thiosulfate respiration (Fig. 5), supporting its involvement in sulfite reduction. Another typically sulfur oxidation‐associated protein, DsrL (DESAMIL20_1432), was also detected in high abundance in cultures grown by thiosulfate respiration, as well as by sulfur respiration at pH 6.5.
Moreover, three rhodanese‐like proteins were detected in high abundance in thiosulfate‐respiring cultures (DESAMIL20_270, DESAMIL20_1100 and DESAMIL20_1419). The abundance of those proteins was not significantly different among the conditions analyzed in this study, and therefore they may be relevant for all routes of sulfur metabolism in D. amilsii.
Disproportionation of elemental sulfur
Proteomic analysis of D. amilsii growing by disproportionation revealed the largest number of unique proteins differentially expressed among the pool of samples studied. D. amilsii is not able to use sulfate as electron acceptor and accordingly, its genome does not encode the ATP sulfurylase and adenylyl‐sulfate reductase (Florentino et al., 2017). Although, as aforementioned, the dissimilatory sulfite reductase is encoded in its genome, sulfite was never detected in the cultures growing by sulfur disproportionation and a high abundance of the dissimilatory sulfite reductase DsrAB, the DsrC and the DsrMKJOP complex were only detected in cultures growing by thiosulfate reduction (Fig. 5).
In the sulfur‐disproportionating condition, a rhodanese‐like sulfurtransferase (DESAMIL20_2007) is present in high abundance (Fig. 5). Interestingly, this protein is only present in sulfur‐disproportionating and low pH cultures. In addition, a monomeric protein related to sulfide: quinone reductase (DESAMIL20_2009) was measured in two samples of the triplicates, with 3 peptides. Unfortunately, the rather low concentration in this condition resulted in poor statistics (−log p‐value = 0.93); and therefore, its role in sulfur disproportionation could not be demonstrated.
Several ribosomal proteins like SSU ribosomal proteins (S4, S5, S8, S10 and S12) and LSU ribosomal proteins (L1, L2, L3, L13, L16, L17, L22 and L31) are highly produced in D. amilsii cultures grown by sulfur disproportionation when compared with the thiosulfate‐respiring cultures (Supporting Information Table S4); but only the L31p protein is highly produced when compared with sulfur‐respiring cultures, while the SSU proteins S7, S9 and S13, and the LSU proteins L14, L15 and L23 are highly produced under respiration of elemental sulfur.
Hydrogen metabolism and carbon fixation
The [Ni‐Fe] membrane‐bound hydrogenase – HybABC (HybA – DESAMIL20_503; HybB – DESAMIL20_502; HybC – DESAMIL20_504) and its maturation complex HypABCDEF (HypA – DESAMIL20_500; HypB – DESAMIL20_499; HypC – DESAMIL20_506; HypD – DESAMIL20_507; HypE – DESAMIL20_508; HypF – DESAMIL20_505) are encoded in the genome of D. amilsii TR1. HybAC was highly abundant in the proteome of the cultures grown with hydrogen as electron donor (Hyd_S_3.5 and Hyd_S_6.5). However, the membrane‐bound cytochrome b subunit HybB could not be identified, likely due to the proteomics preparation process.
All the enzymes involved in the reverse TCA cycle, linked with CO2 fixation, are found in the proteome when D. amilsii is grown chemolithotrophically. ATP citrate lyase (DESAMIL20_1597), fumarate reductases (DESAMIL20_1536 and DESAMIL20_1537) and the catalytic subunit of 2‐oxoglutarate ferredoxin oxidoreductases (DESAMIL20_178) are abundant. Besides, the enzyme citrate synthase (DESAMIL20_1709) was detected in high abundance in all analyzed conditions. Moreover, three subunits of the tetrameric pyruvate synthase/pyruvate: ferredoxin oxidoreductase (DESAMIL20_1626, DESAMIL20_1627 and DESAMIL20_1628) were found in high abundance in chemolithotrophic cultures (Hyd_S_3.5, Hyd_S_6.5 and S_S_6.5). This enzyme is likely to convert the acetyl‐CoA generated into pyruvate that might enter the classical gluconeogenesis route, for which all component enzymes could be detected.
A carbon starvation protein A (DESAMIL20_1476) involved in the peptide utilization during carbon starvation is only produced under disproportionation condition, as well as some amino acid transport ATP‐binding proteins (DESAMIL20_576, DESAMIL20_572 and DESAMIL20_2006).
The identification and intensities of all abundant proteins in the proteome analysis potentially involved in hydrogen and acetate metabolism are presented in Supporting Information Table S8.
Acetate metabolism
According to the genome analysis, D. amilsii could activate acetate to acetyl‐CoA via the enzyme acetyl‐CoA synthetase (ACS, DESAMIL20_135) or the combination of acetate kinase (AK, DESAMIL20_1989) and the phosphate acetyltransferase (PAT, DESAMIL20_1988) (Florentino et al., 2017). However, only ACS was detected in the proteome when acetate was used as electron donor (Acet_S_6.5 and Acet_Thio_6.5). The enzymes required for the oxidation of acetate via the operation of the TCA cycle were also abundant in the cultures grown with acetate, such as the succinate dehydrogenase (DESAMIL20_1536–DESAMIL20_1539), the citrate synthase (DESAMIL20_1709) and the 2‐oxoglutarate oxidoreductase (DESAMIL20_178–180). In comparison to chemolithotrophic cultures, heterotrophic cultures of D. amilsii showed higher abundance of the putative sulfur transferase DsrE‐like protein (DESAMIL20_1428), a flavoprotein‐quinone oxidoreductase (DESAMIL20_1711) and an uncharacterized ferredoxin oxidoreductase (DESAMIL20_1499), while NADPH dehydrogenase (DESAMIL20_251) was not detected in any culture (Supporting Information Table S8).
Discussion
In this study, results for growth and sulfide production in cultures with sulfur trapped in dialysis membrane showed that sulfur reduction by D. amilsii is favoured by its contact with sulfur particles. Nevertheless, growth is possible even in the presence of a dialysis membrane.
Considering the conditions analyzed in this study, with polysulfide expected to be present in concentrations of circa of 0.1 mM at pH 6.5, and the presence of planktonic cells despite the dialysis membrane, it is reasonable to hypothesize that polysulfide can act as an electron acceptor for D. amilsii at pH 6.5. In the pH 3.5 culture, polysulfide is expected at very low concentration (in the order of pM), but nanosulfur particles less than 6–8 kDa could have been formed from elemental sulfur as it was shown for A. sulfurireducens (Boyd and Druschel, 2013). Nanosulfur particles would be able to diffuse through the dialysis membrane and become available for respiration by D. amilsii. The formation and diffusion through the pores of the dialysis membrane seem to be a bottleneck for the process, lowering the rates and activity. Microscopic observations suggested that, especially at low pH, D. amilsii tends to grow in proximity to the sulfur particles (attached to the membrane) and the presence of pili and flagella proteins in the proteome analysis evidences their involvement in the process (Fig. 4).
Four strategies for interaction between the cells and the solid S0 can be hypothesized (Fig. 6): (1) the nucleophilic attack of sulfur by sulfide in the circumneutral surroundings of the cell, generating small polysulfide molecules as soluble intermediates that may cross the cell membrane through channels or other dedicated proteins and react with the cytoplasmic rhodanese sulfur transferases; (2) the interaction between —SH or —S—S— groups on the surface of sulfur and thiol groups present in the outer‐membrane, generating linear soluble polysulfanes (Rohwerder and Sand, 2003), likely via the thiol:disulfide interchange protein (DsbC); (3) the formation of pili leading to extracellular electron transport or in a non‐conductive way just to the establishment of biofilm and (4) the direct uptake of the polymeric fraction of elemental sulfur (Franz et al., 2007, 2009) and/or its immobilization by binding the thiol groups present on the flagellar protein – FliD (Ohmura et al., 1996).
Figure 6.
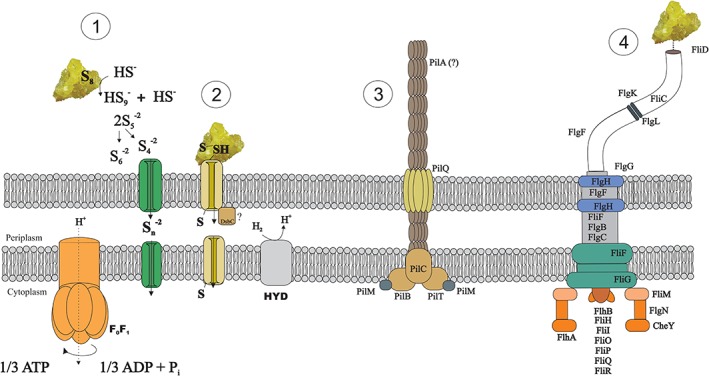
Summary scheme of the possible strategies adopted by D. amilsii to uptake and reduce element sulfur or polysulfide/persulfide sulfanes. 1 – Nucleophilic attack of sulfur by sulfide, generating polysulfide; 2 – Attachment cell‐sulfur and interaction between sulfur and thiol groups via the DsbC, generating soluble polysulfanes; 3 – Pili formation; 4 ‐ Chemical bond between polymeric elemental sulfur and flagellum via FliD. [Color figure can be viewed at wileyonlinelibrary.com]
Pili‐related proteins were abundant in thiosulfate‐respiring cultures, in which there is no cell‐sulfur attachment, dismissing their involvement. The abundance of different cytoplasmic rhodanese‐like proteins indicates their role in sulfur respiration, suggesting that sulfur molecules somehow reach the cytoplasm, by a still unidentified mechanism, and making the electrical conductance of pili in the process seem unlikely. Although further investigation is required to elucidate the role of pili and flagella in reductive processes of D. amilsii, it seems that such appendages might have a significant contribution by motility toward insoluble particles and/or adhesion to the bulk solid substrate.
Surprisingly, proteomic analyses did not reveal abundance of the Mo‐oxidorreductase related to sulfur metabolism (DESAMIL20_1360) in any studied condition. This enzyme is strongly associated to the membrane, as it includes nine transmembrane helices in the membrane subunit. The proteomics preparation process might have led to an underrepresentation of this enzyme in the dataset, as happened for the DrsM subunit of the DsrMK complex or the membrane‐bound HybB subunit of the HybABC hydrogenase. Nevertheless, the soluble subunits could have been detected. The qPCR analysis also did not reveal an abundant presence of its subunits' transcripts, corroborating the hypothesis that this protein may not be a key player in these sulfur conditions. DESAMIL20_1360 shows highest similarity to tetrathionate reductases, so maybe it will play a role in the presence of this substrate. The high abundance of the putative NfnAB ferredoxin:NADP+ oxidoreductase (DESAMIL20_1852, DESAMIL20_1853) in the cultures, with 2.6‐fold abundance at pH 6.5, is in agreement with a transhydrogenase function. Furthermore, ferredoxin (DESAMIL20_472) showed up in high abundance in all the conditions in which sulfur was added as electron acceptor, further supporting that this enzyme acts physiologically as an NADH‐dependent ferredoxin:NADP+ oxidoreductase as in other anaerobes (Wang et al., 2010).
The abundance of rhodanese‐like sulfurtransferases in D. amilsii cultures is remarkable. It has been shown that rhodanese‐like sulfurtransferases work as a sulfur supplier for key sulfur‐reducing enzymes in Aquifex aeolicus (Giuliani et al., 2010, Aussignargues et al., 2012). The role of those proteins, however, has only been deeply investigated for endogenous sulfidogenic metabolism studies in human gastrointestinal tract, in which sulfur might be provided to the enzyme by 3‐mercaptopyruvate (Mikami et al., 2011). This study leads to the hypothesis that rhodanese‐like sulfurtransferases may play a crucial role in the sulfur/polysulfide respiration process. Further studies are required to evidence the role of the rhodanese‐like sulfur transferases and other sulfur‐related enzymes in the process of sulfur respiration, and the link between the carbon and sulfur metabolism.
During thiosulfate respiration, normally sulfite is generated in the first step via the PhsABC complex (thiosulfate reductase) and reduced to sulfide via the dissimilatory sulfite reductase (DsrAB), its substrate protein DsrC and the membrane‐associated electron transport complex DsrMKJOP. Generally, as shown by Santos et al. (2015), the DsrAB reduces the sulfur in sulfite and transfers it to DsrC that has two cysteine residues, forming, a trisulfide species. The DsrK subunit of the DsrMKJOP complex is then proposed to act on the DsrC trisulfide, releasing hydrogen sulfide and restoring DsrC to its reduced state. The DsrC protein in D. amilsii is atypical and presents only one cysteine residue. Santos et al. (2015) showed that DsrC mutants lacking this penultimate Cys were still able to reduce sulfite, but with slower growth and lower cell yields. In the case of D. amilsii, although only one Cys‐residue is present, reduction of thiosulfate occurred efficiently (in comparison with the other metabolisms present), without an apparent interference in the cell yields and growth of the microorganism (Florentino et al., 2016a,b). It is noteworthy that dsrAB, dsrC and dsrMK genes are found in a putative operon (Fig. 7) that is distinct from the dsr operons commonly found in sulfate reducers (Pereira et al., 2011) and containing additional genes. These include genes for two small proteins (DESAMIL20_1427 and DESAMIL20_1428) related to DsrE, a sulfur‐transfer protein commonly involved in sulfur oxidation processes (Dahl, 2015). They both contain a conserved cysteine that may be involved in the reaction of sulfite with DsrAB/DsrC, and thus compensate for the absence of the second conserved Cys in DsrC (DESAMIL20_1431). In the acidothermophilic sulfur‐oxidizing archaeon Metallosphaera cuprina, two DsrE proteins were recently shown to be involved in thiosulfate transfer (Liu et al., 2014). Conspicuously, a third gene in this operon encodes also another sulfur oxidation‐associated protein DsrL (DESAMIL20_1432). The presence of three dsr genes associated with sulfur oxidation may indicate that the D. amilsii is capable of sulfur oxidation, which has to be tested. Nevertheless, their up‐regulation during thiosulfate reduction is a strong indication that they can also function in a reductive mode, most likely involved in sulfite reduction. Overall, the new dsr operon arrangement of D. amilsii suggests a slightly modified sulfite reduction mechanism.
Figure 7.
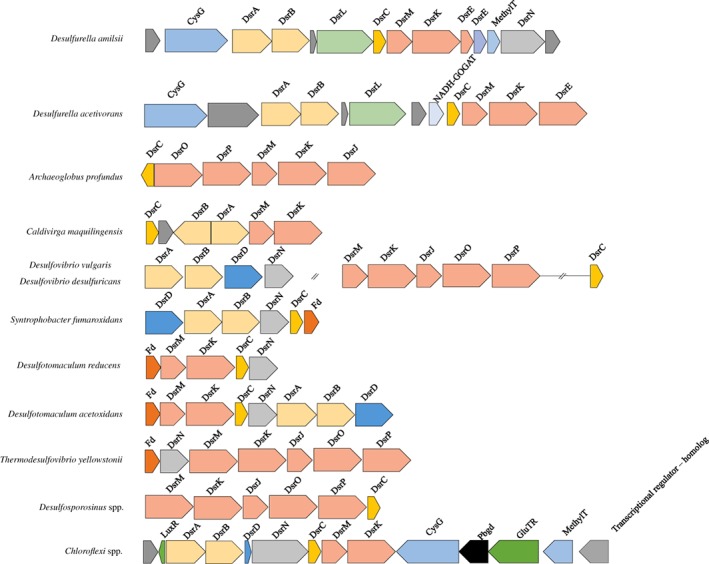
Neighbourhood organization at the dsr locus in D. amilsii in comparison to other sulfur utilizers. Fd, ferredoxin; DsrN, cobyrinic acid a,c‐diamide synthase. Operons of sulfur utilizers were adapted from Pereira, Ramos et al. (2011). [Color figure can be viewed at wileyonlinelibrary.com]
During disproportionation, elemental sulfur acts simultaneously as electron donor and electron acceptor. Disproportionation of elemental sulfur by D. amilsii yielded 0.6 mM of sulfate and 1.9 mM of sulfide, in accordance with the expected stoichiometry (ratio 1:3 sulfate/sulfide) of Eq. 6 (Florentino et al., 2015).
| (6) |
The absence of sulfite in sulfur‐disproportionating cultures, and the exclusive abundance of the DsrAB, DsrC and the Dsr membrane complex subunits in cultures growing by thiosulfate reduction suggest that sulfite is not an intermediate in sulfur disproportionation in D. amilsii. Besides, the unique presence of rhodanese‐like proteins (DESAMIL20_1987 and DESAMIL20_2007) in sulfur‐disproportionating and acidotolerant sulfur‐respiring cultures is a remarkable output in this study, raising the hypothesis that they may not only act as sulfur‐respiring enzymes, but as direct sulfur‐disproportionating enzymes in D. amilsii cultures. Therefore, proteome data from D. amilsii suggests a different disproportionation pathway from the one reported by others (Finster, 2008, Hardisty et al., 2013).
A major finding of this study is the involvement of rhodanese‐like sulfurtransferases as key players in reductive routes of sulfur metabolism, indicating that the current view on sulfur cycle needs to be broadened as novel pathways are likely to soon be discovered. Moreover, ecological studies, such as metagenomics data mining, might benefit from the utilization of those key rhodanese‐like sulfurtransferases to detect sulfur metabolism in different environments, as existing metagenome datasets contain untapped potential for understanding metabolic pathways and their biological impact.
This study also confirms the pathway for hydrogen utilization as electron donor by the uniquely abundant Ni‐Fe hydrogenase HybABC during chemolithotrophic growth. D. amilsii is capable of synthesizing cell carbon from C1‐compounds likely via the reductive TCA cycle, in a similar way as reported for other Proteobacteria (Hugler et al., 2007, Berg, 2011), green sulfur bacteria (Tang and Blankenship, 2010) and microaerophilic bacteria of the phylum Aquificae (Hugler et al., 2007). Moreover, a citrate synthase was detected in very high abundance in all the analyzed conditions, which can indicate a similar metabolism of carbon fixation to the recently discovered for its closest relative Desulfurella acetivorans (Mall et al., 2018). In the reported study, citrate synthase is shown to cleave citrate adenosine triphosphate independently into acetyl coenzyme A and oxaloacetate, in a so‐called reversed oxidative TCA cycle (roTCA cycle), previously considered impossible under physiological conditions (Mall et al., 2018). The operation of the roTCA cycle requires reduced ferredoxin, which is probably synthesized through electron bifurcation by the NADH‐dependent ferredoxin:NADP+ oxidoreductase (NfnAB–DESAMIL20_1852; DESAMIL20_1853) (Mall et al., 2018), also detected in high abundance in the D. amilsii proteome. Furthermore, the high abundance of carbon starvation protein A and some amino acid transport ATP‐binding proteins in cultures growing by sulfur disproportionation indicates that D. amilsii might utilize peptides from the yeast extract available in the medium as carbon source to help in the CO2 fixation, as it is a costly process for the cell, especially under harsh conditions.
Our results indicate the involvement of ACS in acetate activation by D. amilsii, with its oxidation happening via TCA cycle. Acetate activation, therefore, happens in a different way from the two sulfur‐reducing bacteria previously investigated: D. acetooxidans which uses succinyl‐CoA:acetate CoA‐transferase (Gebhardt et al., 1985) via and D. acetivorans (Schmitz et al.,1990), which used AK, PAT and succinyl‐CoA synthetase.
In Fig. 8, a proposed metabolic reconstruction of D. amilsii is graphically displayed, summarizing the most relevant aspects of the sulfur metabolism addressed in this manuscript.
Figure 8.
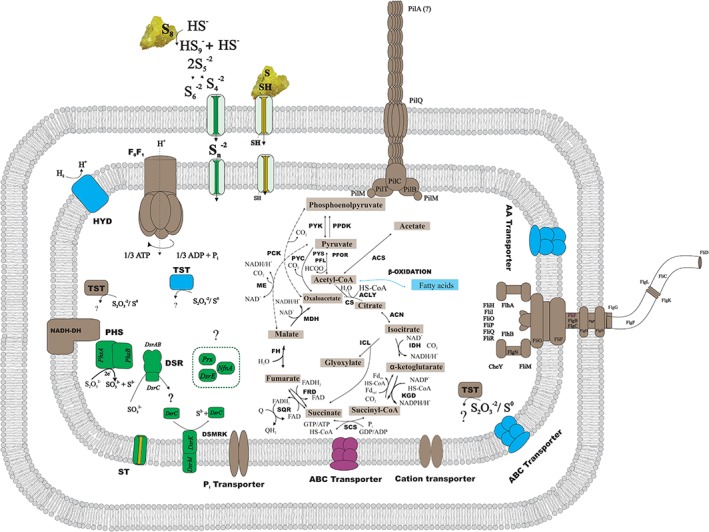
Metabolic reconstruction of D. amilsii. Relative abundance of proteins during heterotrophic growth by sulfur or thiosulfate reduction and sulfur disproportionation is shown. Proteins highlighted in brown are found in similar levels in all the analyzed conditions, proteins highlighted in green are found exclusively during thiosulfate respiration; proteins highlighted in blue are found more abundantly present in sulfurdisproportionating cultures and proteins highlighted in purple are found exclusively during sulfur reduction. ACLY, ATP citrate lyase; ACS, acetyl‐CoA synthetase; CS, citrate synthase; DSR, Dissimilatory sulfite reductase; FH, fumarate hydratase; FR, fumarate reductase; HYD, hydrogenase; ICL, Isocitrate lyase; IDH, Isocitrate dehydrogenase; KGD, α‐ketoglutarate dehydrogenase; MDH, malate dehydrogenase; ME, malic enzymes; PCK, phosphoenolpyruvate carboxinase; PFL, pyruvate:formate lyase; PFOR, pyruvate:ferredoxin oxidoreductase; PHS, thiosulfate reductase; PPDK, pyruvate phosphate dikinase; PTA, phosphotransacetylase; PYC, pyruvate carboxylase; PYK, pyruvate kinase; PYS, Pyruvate synthase; SCS, Succinyl‐CoA synthetase; SQR, Succinate‐coenzyme Q reductase; NADH‐DH, NADH dehydrogenase; MK, menaquinone; ST, sulfite transporter; TST, rhodanese‐like sulfurtransferase. The rhodanese‐like sulfurtransferase represented in blue has the locus tag DESAMIL20_2007; while the locus tags for the ones represented in black are DESAMIL20_270, DESAMIL20_1100. [Color figure can be viewed at wileyonlinelibrary.com]
Concluding remarks
This study reports a comparative proteomic analysis of a sulfur‐reducing microorganism grown under different sulfur reductive conditions. The apparent low expression level of a Mo‐oxidoreductase, previously thought to be a sulfur reductase, and the high abundance of different sulfurtransferases raise the hypothesis that rhodaneses are key players in sulfur/polysulfide reduction in D. amilsii. Further analyses need to be performed to confirm the role of the rhodanese‐like proteins in sulfur respiration at different pH values. Moreover, our results suggest the Mo‐oxidorreductase might in fact play a role as tetrathionate reductase and not a sulfur reductase. In cultures growing by thiosulfate respiration, proteomic and genomic analyses showed the presence of the atypical DsrMK complex (instead of the complete DsrMKJOP complex), which thus far was only present in some Firmicutes and Archaeoglobus spp. In addition, although the DsrC in D. amilsii contains only one cysteine residue, sulfite reduction to sulfide could still be successfully performed in this microorganism possibly due to the detected high levels of supporting proteins like DsrE, which implies a novel sulfite reduction mechanism in D. amilsii. DsrL and DsrE‐like proteins, well known for their involvement in the oxidation of reduced sulfur compounds, were abundantly detected during thiosulfate reduction by D. amilsii. Encoded enzymes previously described to be involved in disproportionation of elemental sulfur were not detected in D. amilsii cultures, and so rhodanese‐like sulfurtransferases are thought to play a major role in this metabolic process, representing again a novel mechanism. As a first comparative study on the protein level of sulfur metabolism routes, this study helps in the elucidation of the roles of sulfur‐reducing enzymes, which will improve the search for sulfur metabolism in broader fields, such as metaomics.
Conflict of interest
The authors declare no competing financial interest.
Supporting information
Figure S1. Sulfide production levels of Desulfurella amilsii cultures incubated under different reductive conditions.
Table S1. Unique proteins detected per condition analyzed. The locus tag for the genes encoded in D. amilsii is DESAMIL20_*. To avoid repetition of the prefix in the tables, the locus tags are represented only by the specific identifier.
Table S2. Differentially expressed proteins in a pairwise comparison Acet_S_6.5 vs Acet_thio_6.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant in sulfur respiration, while the negative values (highlighted orange) represent the proteins highly abundant during thiosulfate respiration. ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S3 – Differentially expressed proteins in a pairwise comparison Acet_S_6.5 vs S_S_6.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant in sulfur respiration, while the negative values (highlighted orange) represent the proteins highly abundant during sulfur disproportionation. ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S4 Differentially expressed proteins in a pairwise comparison Acet_thio_6.5 vs S_S_6.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant in thiosulfate respiration, while the negative values (highlighted orange) represent the proteins highly abundant during sulfur disproportionation. ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S5. Differentially expressed proteins in a pairwise comparison Hyd_S_6.5 vs Hyd_S_3.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant at circumneutral pH, while the negative values (highlighted orange) represent the proteins highly abundant in acidotolerant cultures (pH 3.5). ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S6. Normalized protein abundance (LFQ intensity) of the biological triplicates for proteins possibly involved in motility, cell‐sulfur attachment and/or uptake processes in D. amilsii TR1 in the 5 conditions analyzed. The locus tag for the genes encoded in D. amilsii are represented in the figure by the specific identifier preceded DESAMIL20_, in which * stands for DESAMIL20*.
Table S7. Normalized protein abundance (LFQ intensity) of the biological triplicates for proteins possibly involved in sulfur and thiosulfate reduction in D. amilsii TR1. The locus tag for the genes encoded in D. amilsii are represented in the figure by the specific identifier preceded DESAMIL20_, in which * stands for DESAMIL20*.
Table S8. Normalized protein abundance (LFQ intensity) of the biological triplicates for proteins possibly involved in hydrogen and acetate metabolisms by D. amilsii TR1 in the 5 conditions analyzed. The locus tag for the genes encoded in D. amilsii are represented in the figure by the specific identifier preceded DESAMIL20_, in which * stands for DESAMIL20*.
Acknowledgements
This research was supported by the organization of the Brazilian Government for the development of Science and Technology CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico), by ERC project 323009 and a Gravitation project from the Netherlands Ministry of Education, Culture and Science (024.002.002). The authors thank Monika Jarzembowska (WUR, Wageningen, The Netherlands) for the support with the scanning electron microscopy analysis.
References
- Aussignargues, C. , Giuliani, M. C. , Infossi, P. , Lojou, E. , Guiral, M. , Giudici‐Orticoni, M. T. , and Ilbert, M. (2012) Rhodanese functions as sulfur supplier for key enzymes in sulfur energy metabolism. J Biol Chem 287: 19936–19948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berg, I. A. (2011) Ecological aspects of the distribution of different autotrophic CO2 fixation pathways. Appl Environ Microbiol 77: 1925–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bielow, C. , Mastrobuoni, G. , and Kempa, S. (2016) Proteomics quality control: quality control software for MaxQuant results. J Proteome Res 15: 777–787. [DOI] [PubMed] [Google Scholar]
- Bill, R. M. , Henderson, P. J. , Iwata, S. , Kunji, E. R. , Michel, H. , Neutze, R. , et al (2011) Overcoming barriers to membrane protein structure determination. Nat Biotechnol 29: 335–340. [DOI] [PubMed] [Google Scholar]
- Blumentals, I. , Itoh, M. , Kelly, R. M. , and Olson, G. J. (1990) Role of polysulfides in reduction of elemental sulfur by the hyperthermophilic archaebacterium Pyrococcus furiosus . Appl Environ Microbiol 56: 1255–1262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulegue, J. (1978) Solubility of elemental sulfur in water at 298 K. Phosphorus Sulfur Silicon Relat Elem 5: 127–128. [Google Scholar]
- Boyd, E. S. , and Druschel, G. K. (2013) Involvement of intermediate sulfur species in biological reduction of elemental sulfur under acidic, hydrothermal conditions. Appl Environ Microbiol 79: 2061–2068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buckel, W. , and Thauer, R.K. (2018) Flavin‐based electron bifurcation, ferredoxin, flavodoxin, and anaerobic respiration with protons (Ech) or NAD+ (Rnf) as electron acceptors: A historical review. Front Microbiol 14: 401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chandramouli, K. , and Qian, P. (2009) Proteomics: challenges, techniques and possibilities to overcome biological sample complexity. Hum Genomics Proteomics 2009: 239204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cline, J. D. (1969) Spectrophotometric determination of hydrogen sulfide in natural waters. Limnol Oceanogr 14: 454–458. [Google Scholar]
- Cox, J. , Hein, M. Y. , Luber, C. A. , Paron, I. , Nagaraj, N. , and Mann, M. (2014) Accurate proteome‐wide label‐free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol Cell Proteomics 13: 2513–2526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox, J. , and Mann, M. (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.‐range mass accuracies and proteome‐wide protein quantification. Nat Biotechnol 26: 1367–1372. [DOI] [PubMed] [Google Scholar]
- Cox, J. , Neuhauser, N. , Michalski, A. , Scheltema, R. A. , Olsen, J. V. , and Mann, M. (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10: 1794–1805. [DOI] [PubMed] [Google Scholar]
- Dahl, C. (2015) Cytoplasmic sulfur trafficking in sulfur‐oxidizing prokaryotes. IUBMB Life 67: 268–274. [DOI] [PubMed] [Google Scholar]
- Dahl, C. , Friedrich, C. , and Kletzin, A. (2001) Sulfur oxidation in prokaryotes In Encyclopedia of Life Sciences (ELS). Chichester: John Wiley & Sons, Ltd. [Google Scholar]
- Finster, K. (2008) Microbiological disproportionation of inorganic sulfur compounds. J Sulfur Chem 29: 281–292. [Google Scholar]
- Finster, K. , Leiesack, W. , and Thamdrup, B. O. (1998) Elemental sulfur and thiosulfate disproportionation by Desulfocapsa sulfoexigens sp. nov., a new anaerobic bacterium isolated from marine surface sediment. Appl Environ Microbiol 64: 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino, A. P. , Brienza, C. , Stams, A. J. M. , and Sánchez‐Andrea, I. (2016a) Desulfurella amilsii sp. nov., a novel acidotolerant sulfur‐respiring bacterium isolated from acidic river sediments. Int J Syst Evol Microbiol 66: 1249–1253. [DOI] [PubMed] [Google Scholar]
- Florentino, A. P. , Stams, A. J. M. , and Sánchez‐Andrea, I. (2017) Genome sequence of Desulfurella amilsii strain TR1 and comparative genomics of Desulfurellaceae family. Front Microbiol 8:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Florentino, A. P. , Weijma, J. , Stams, A. J. M. , and Sánchez‐Andrea, I. (2015) Sulfur reduction in acid rock drainage environments. Environ Sci Technol 49: 11746–11755. [DOI] [PubMed] [Google Scholar]
- Florentino, A. P. , Weijma, J. , Stams, A. J. M. , and Sánchez‐Andrea, I. (2016b) Ecophysiology and application of acidophilic sulfur‐reducing microorganisms In Biotechnology of Extremophiles: Advances and Challenges, Rampelotto H. P. (ed). Cham: Springer International Publishing, pp. 141–175. [Google Scholar]
- Franz, B. , Gehrke, T. , Lichtenberg, H. , Hormes, J. , Dahl, C. , and Prange, A. (2009) Unexpected extracellular and intracellular sulfur species during growth of Allochromatium vinosum with reduced sulfur compounds. Microbiology 155: 2766–2774. [DOI] [PubMed] [Google Scholar]
- Franz, B. , Lichtenberg, H. , Hormes, J. , Modrow, H. , Dahl, C. , and Prange, A. (2007) Utilization of solid "elemental" sulfur by the phototrophic purple sulfur bacterium Allochromatium vinosum: a sulfur K‐edge X‐ray absorption spectroscopy study. Microbiology 153: 1268–1274. [DOI] [PubMed] [Google Scholar]
- Friedrich, C. G. , Rother, D. , Bardischewsky, F. , Quentmeier, A. , and Fischer, J. (2001) Oxidation of reduced inorganic sulfur compounds by bacteria: emergence of a common mechanism? Appl Environ Microbiol 67: 2873–2882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebhardt, N. A. , Thauer, R. K. , Linder, D. , Kaulfers, P. , and Pfennig, N. (1985) Mechanism of acetate oxidation to CO2 with elemental sulfur in Desulfuromonas acetoxidans . Arch Microbiol 141: 7. [Google Scholar]
- Giuliani, M.‐C. , Jourlin‐Castelli, C. , Leroy, G. , Hachani, A. , and Giudici‐Orticoni, M. T. (2010) Characterization of a new periplasmic single‐domain rhodanese encoded by a sulfur‐regulated gene in a hyperthermophilic bacterium Aquifex aeolicus . Biochimie 92: 388–397. [DOI] [PubMed] [Google Scholar]
- Grein, F. , Ramos, A. R. , Venceslau, S. S. , and Pereira, I. A. C. (2013) Unifying concepts in anaerobic respiration: insights from dissimilatory sulfur metabolism. Biochim Biophys Acta 1827: 145–160. [DOI] [PubMed] [Google Scholar]
- Hardisty, D. S. , Olyphant, G. A. , Bell, J. B. , Johnson, A. P. , and Pratt, L. M. (2013) Acidophilic sulfur disproportionation. Geochim Cosmochim Acta 113: 136–151. [Google Scholar]
- Hedderich, R. , Klimmek, O. , Kröger, A. , Dirmeier, R. , Keller, M. , and Stetter, K. O. (1999) Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev 22: 29. [Google Scholar]
- Hubner, N. C. , Bird, A. W. , Cox, J. , Splettstoesser, B. , Bandilla, P. , Poser, I. , et al (2010) Quantitative proteomics combined with BAC TransgeneOmics reveals in vivo protein interactions. J Cell Biol 189: 739–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugler, M. , Huber, H. , Molyneaux, S. J. , Vetriani, C. , and Sievert, S. M. (2007) Autotrophic CO2 fixation via the reductive tricarboxylic acid cycle in different lineages within the phylum Aquificae: evidence for two ways of citrate cleavage. Environ Microbiol 9: 81–92. [DOI] [PubMed] [Google Scholar]
- Junier, P. , Junier, T. , Podell, S. , Sims, D. R. , Detter, J. C. , Lykidis, A. , et al (2010) The genome of the gram‐positive metal‐ and sulfate‐reducing bacterium Desulfotomaculum reducens strain MI‐1. Environ Microbiol 12: 2738–2754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klausen, M. , Heydorn, A. , Ragas, P. , Lambertsen, L. , Aaes‐Jorgensen, A. , Molin, S. , and Tolker‐Nielsen, T. (2003) Biofilm formation by Pseudomonas aeruginosa wild type, flagella and type IV pili mutants. Mol Microbiol 48: 1511–1524. [DOI] [PubMed] [Google Scholar]
- Klimmek, O. , Kröger, A. , Steudel, R. , and Holdt, G. (1991) Growth of Wolinella succinogenes with polysulphide as terminal acceptor of phosphorylative electron transport. Arch Microbiol 155: 177–182. [Google Scholar]
- Klimmek, O. , Stein, T. , Pisa, R. , Simon, J. , and Kroger, A. (1999) The single cysteine residue of the Sud protein is required for its function as a polysulfide‐sulfur transferase in Wolinella succinogenes . Eur J Biochem 263: 79–84. [DOI] [PubMed] [Google Scholar]
- Laska, S. , Lottspeich, F. , and Kletzin, A. (2003) Membrane‐bound hydrogenase and sulfur reductase of the hyperthermophilic and acidophilic archaeon Acidianus ambivalens . Microbiology 149: 2357–2371. [DOI] [PubMed] [Google Scholar]
- Lin, Y. J. , Dancea, F. , Lohr, F. , Klimmek, O. , Pfeiffer‐Marek, S. , Nilges, M. , et al (2004) Solution structure of the 30 kDa polysulfide‐sulfur transferase homodimer from Wolinella succinogenes . Biochemistry 43: 1418–1424. [DOI] [PubMed] [Google Scholar]
- Liu, L. J. , Stockdreher, Y. , Koch, T. , Sun, S. T. , Fan, Z. , Josten, M. , et al (2014) Thiosulfate transfer mediated by DsrE/TusA homologs from acidothermophilic sulfur‐oxidizing archaeon Metallosphaera cuprina . J Biol Chem 289: 26949–26959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu, J. , Boeren, S. , de Vries, S. C. , van Valenberg, H. J. F. , Vervoort, J. , and Hettinga, K. (2011) Filter‐aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J Proteomics 75: 34–43. [DOI] [PubMed] [Google Scholar]
- Ma, K. , and Adams, M. W. (1994) Sulfide dehydrogenase from the hyperthermophilic archaeon Pyrococcus furiosus: a new multifunctional enzyme involved in the reduction of elemental sulfur. J Bacteriol 176: 6509–6517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mall, A. , Sobotta, J. , Huber, C. , Tschirner, C. , Kowarschik, S. , Bacnik, K. , et al (2018) Reversibility of citrate synthase allows autotrophic growth of a thermophilic bacterium. Science 359: 563–567. [DOI] [PubMed] [Google Scholar]
- Mikami, Y. , Shibuya, N. , Kimura, Y. , Nagahara, N. , Ogasawara, Y. , and Kimura, H. (2011) Thioredoxin and dihydrolipoic acid are required for 3‐mercaptopyruvate sulfurtransferase to produce hydrogen sulfide. Biochem J 439: 479–485. [DOI] [PubMed] [Google Scholar]
- O'Toole, G. A. , and Kolter, R. (1998) Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol Microbiol 30: 295–304. [DOI] [PubMed] [Google Scholar]
- Ohmura, N. , Tsugita, K. , Koizumi, J. , and Saika, H. (1996) Sulfur‐binding protein of flagella of Thiobacillus ferrooxidans . J Bacteriol 178: 5776–5780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira, I. A. C. , Ramos, A. , Grein, F. , Marques, M. , Da Silva, S. , and Venceslau, S. (2011) A comparative genomic analysis of energy metabolism in sulfate‐reducing bacteria and archaea. Front Microbiol 2: 1–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabus, R. , Hansen, T. A. , and Widdel, F. (2013). In Dissimilatory Sulfate‐ and Sulfur‐Reducing Prokaryotes. The Prokaryotes: Prokaryotic Physiology and Biochemistry, Rosenberg E., DeLong E. F., Lory S., Stackebrandt E., and Thompson F. (eds). Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 309–404. [Google Scholar]
- Rappsilber, J. , Mann, M. , and Ishihama, Y. (2007) Protocol for micro‐purification, enrichment, pre‐fractionation and storage of peptides for proteomics using StageTips. Nat Protoc 2: 1896–1906. [DOI] [PubMed] [Google Scholar]
- Rawlings, A. E. (2016) Membrane proteins: always an insoluble problem? Biochem Soc Trans 44: 790–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rohwerder, T. , and Sand, W. (2003) The sulfane sulfur of persulfides is the actual substrate of the sulfur‐oxidizing enzymes from Acidithiobacillus and Acidiphilium spp. Microbiology 149: 1699–1710. [DOI] [PubMed] [Google Scholar]
- Sánchez‐Andrea, I. , Stams, A. J. M. , Amils, R. , and Sanz, J. L. (2013) Enrichment and isolation of acidophilic sulfate‐reducing bacteria from Tinto River sediments. Environ Microbiol Rep 5: 672–678. [DOI] [PubMed] [Google Scholar]
- Santos, A. A. , Venceslau, S. S. , Grein, F. , Leavitt, W. D. , Dahl, C. , Johnston, D. T. , and Pereira, I. A. (2015) A protein trisulfide couples dissimilatory sulfate reduction to energy conservation. Science 350: 1541–1545. [DOI] [PubMed] [Google Scholar]
- Schauder, R. , and Müller, E. (1993) Polysulfide as a possible substrate for sulfur‐reducing bacteria. Arch Microbiol 160: 377–382. [Google Scholar]
- Schmitz, R. A. , Bonch‐Osmolovskaya, E. A. , and Thauer, R. K. (1990) Different mechanisms of acetate activation in Desulfurella acetivorans and Desulfuromonas acetoxidans . Arch Microbiol 154: 274–279. [Google Scholar]
- Steudel, R. (2003) Inorganic polysulfides Sn2− and radical anions Sn− In Elemental Sulfur Und Sulfur‐Rich Compounds II, Steudel R. (ed). Berlin, Heidelberg: Springer Berlin Heidelberg, pp. 127–152. [Google Scholar]
- Stoffels, L. , Krehenbrink, M. , Berks, B. C. , and Unden, G. (2012) Thiosulfate reduction in Salmonella enterica is driven by the proton motive force. J Bacteriol 194: 475–485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Surkov, A. V. , Bottcher, M. E. , and Kuever, J. (2000) Stable sulfur isotope fractionation during the reduction of thiosulfate by Dethiosulfovibrio russensis . Arch Microbiol 174: 448–451. [DOI] [PubMed] [Google Scholar]
- Tang, K. H. , and Blankenship, R. E. (2010) Both forward and reverse TCA cycles operate in green sulfur bacteria. J Biol Chem 285: 35848–35854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizcaíno, J. A. , Csordas, A. , del‐Toro, N. , Dianes, J. A. , Griss, J. , Lavidas, I. , et al (2016) 2016 update of the PRIDE database and related tools. Nucleic Acids Res 44: D447–D456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, S. , Huang, H. , Moll, J. , and Thauer, R. K. (2010) NADP+ reduction with reduced ferredoxin and NADP+ reduction with NADH are coupled via an electron‐bifurcating enzyme complex in Clostridium kluyveri . J Bacteriol 192: 5115–5123. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Sulfide production levels of Desulfurella amilsii cultures incubated under different reductive conditions.
Table S1. Unique proteins detected per condition analyzed. The locus tag for the genes encoded in D. amilsii is DESAMIL20_*. To avoid repetition of the prefix in the tables, the locus tags are represented only by the specific identifier.
Table S2. Differentially expressed proteins in a pairwise comparison Acet_S_6.5 vs Acet_thio_6.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant in sulfur respiration, while the negative values (highlighted orange) represent the proteins highly abundant during thiosulfate respiration. ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S3 – Differentially expressed proteins in a pairwise comparison Acet_S_6.5 vs S_S_6.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant in sulfur respiration, while the negative values (highlighted orange) represent the proteins highly abundant during sulfur disproportionation. ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S4 Differentially expressed proteins in a pairwise comparison Acet_thio_6.5 vs S_S_6.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant in thiosulfate respiration, while the negative values (highlighted orange) represent the proteins highly abundant during sulfur disproportionation. ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S5. Differentially expressed proteins in a pairwise comparison Hyd_S_6.5 vs Hyd_S_3.5. Positive values of the t‐test difference (highlighted blue) represent the proteins highly abundant at circumneutral pH, while the negative values (highlighted orange) represent the proteins highly abundant in acidotolerant cultures (pH 3.5). ‐Log p value represents ‐log10 transformed p‐value from a pairwise two‐tailed T test comparing the two groups.
Table S6. Normalized protein abundance (LFQ intensity) of the biological triplicates for proteins possibly involved in motility, cell‐sulfur attachment and/or uptake processes in D. amilsii TR1 in the 5 conditions analyzed. The locus tag for the genes encoded in D. amilsii are represented in the figure by the specific identifier preceded DESAMIL20_, in which * stands for DESAMIL20*.
Table S7. Normalized protein abundance (LFQ intensity) of the biological triplicates for proteins possibly involved in sulfur and thiosulfate reduction in D. amilsii TR1. The locus tag for the genes encoded in D. amilsii are represented in the figure by the specific identifier preceded DESAMIL20_, in which * stands for DESAMIL20*.
Table S8. Normalized protein abundance (LFQ intensity) of the biological triplicates for proteins possibly involved in hydrogen and acetate metabolisms by D. amilsii TR1 in the 5 conditions analyzed. The locus tag for the genes encoded in D. amilsii are represented in the figure by the specific identifier preceded DESAMIL20_, in which * stands for DESAMIL20*.


