Abstract
Almost 55% to 80% of patients with breast cancer have an unfavorable pathological complete response to chemotherapy. MicroRNAs are small noncoding RNAs involved in cancer progression; however, their utility as predictors of pathological complete response to neoadjuvant chemotherapy is unclear. Here, we investigated if miR-143 could discriminate between pathological complete response and no-polymerase chain reaction of patients with locally advanced triple negative breast cancer that have received a fluorouracil-cisplatin/paclitaxel-based neoadjuvant treatment. Data showed that miR-143 exhibited a significant low expression (P < .0006) in patients that achieved pathological complete response in comparison to nonresponder group. Receiver operating characteristic curve analysis suggested that miR-143 could be a good predictor of pathological complete response (area under curve = 0.849, P < .0006). Moreover, Kaplan-Meier analysis indicated that before neoadjuvant therapy low levels of miR-143 were associated to increased disease free survival. To gain insights into cellular functions of miR-143, we firstly showed that miR-143 was severely repressed in breast cancer cell lines and tumors in comparison to normal mammary cells and tissues. Ectopic restoration of miR-143 using RNA mimics inhibited both cell proliferation and migration and sensitized breast cancer cells to cisplatin therapy in vitro. To decipher the signaling networks regulated by miR-143, we used a high-throughput enzyme-linked immunosorbent assay-based phosphorylation antibody array. Phospho-proteomic profiling revealed that miR-143 coordinately reduced the protein levels and phosphorylation status of multiple oncoproteins involved in AKT, WNT/β-catenin, SAPK/JNK, FAK, and JAK/STAT signaling pathways. Moreover, low miR-143 and high GSK3-β, RAF1, paxillin, and p21CIP1 expression levels in a large cohort of patients with breast cancer were associated with worst outcome. In summary, miR-143 could be a potential predictor of response to neoadjuvant therapy and it may function as a divergent regulator of diverse signaling networks to suppress cell proliferation and migration in breast cancer.
Keywords: breast cancer, pathological complete response, neoadjuvant therapy, miR-143, cell proliferation, cell migration, signaling pathways
Introduction
Breast cancer is the most frequently diagnosed malignancy and the leading cause of cancer death among women worldwide.1 Triple-negative breast cancer is the disease subtype that occurs in nearly 15% of total cases and is associated with highly aggressiveness and poor prognosis. Neoadjuvant chemotherapy (also called preoperative or primary chemotherapy) constitutes an important standard approach for treatment of locally advanced breast cancer. Neoadjuvant treatment takes place before surgical extraction of tumors with the objective of reducing high tumor size to breast size ratio in order to not only facilitate the removal of tumors but also to go ahead with breast conservation and improve the postoperative recovery and long-term outcome for patients. Pathological complete response (pCR) is defined by the absence of residual disease after neoadjuvant chemotherapy.2 Patients with breast cancer who better benefit from neoadjuvant chemotherapy are those who achieve a successful pCR which have been associated with both improved disease-free survival (DFS) and overall survival (OS).3-8 The CTNeoBC study in a large cohort of women with breast cancer had definitively established a good association between pCR and DFS/OS.9 Molecular and clinical factors inherent to tumor biology are known to influence the degree of response to neoadjuvant chemotherapy. Unfortunately, no highly specific tumor biomarkers that can predict the clinical response to neoadjuvant chemotherapy and outcome of patients with breast cancer have been clearly defined yet.
MicroRNAs (miRNAs) are evolutionary conserved small noncoding RNAs of 25 nucleotides length that function as negative regulators of gene expression by either inhibiting translation or inducing deadenylation-dependent degradation of target messenger RNAs in cytoplasmic P-bodies.10 Transcriptional and epigenetic mechanisms leading to aberrant expression of miRNAs and its target genes have been involved in the initiation and progression of breast cancer. Importantly, miRNAs have also been recently investigated as potential predictors of clinical response to chemo-radiotherapy in diverse types of cancers.11-17 Previously, we reported that miRNA expression signatures could be associated to pCR in patients with triple negative breast cancer.18 Here, we focused in the clinical and molecular analysis of miR-143. Our data showed that miR-143 could be a predictor of pCR to neoadjuvant chemotherapy in patients with breast cancer. Also, we provided experimental findings suggesting a potential function for miR-143 as regulator of diverse genes involved in signaling networks involved in carcinogenesis.
Materials and Methods
Ethic Statement
All procedures involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki Declaration and its later amendments or comparable ethical standards. An informed consent was obtained from all individual participants included in this study.
Tissues Samples
Tumors from patients with triple negative breast cancer with or without pCR were collected from National Institute of Cancerology, Mexico. The institutional review board of the Institute (http://www.incan.salud.gob.mx/) approved the protocol and the informed consent to perform the research with human samples (number 011/013/TMI; CB659). All recruited patients received neoadjuvant chemotherapy based in 5-fluorouracil, adriamycin, cyclophosphamide-cisplatin/paclitaxel regimen. After tumor resection specimens were formalin-fixed paraffin-embedded. An expert pathologist confirmed the triple negative status of tumors by immunohistochemistry using specific antibodies against the receptors of estrogen, progesterone, and HER2/neu in clinical specimens (Supplementary Table 1). Only tissues containing at least 80% of tumor cells were used in this study.
Cell Lines
Human triple negative MDA-MB-231 and estrogen positive MCF-7 breast cancer cell lines were obtained from the American Type Culture Collection and routinely grown in Dulbecco’s modification of Eagle’s minimal medium supplemented with 10% fetal bovine serum and penicillin-streptomycin (50 U/mL; Invitrogen, Carlsbad, California) in 5% CO2 atmosphere at 37°C.
Reverse Transcription and Real-Time Polymerase Chain Reaction
Total RNA was isolated using the RNeasy FFPE kit (Qiagen Inc, Valencia, California) following the manufacture’s protocol. Briefly, 10 sections of 10 µm paraffin embedded samples were incubated in xylene for 1 hour at 63°C for deparaffinization. Total RNA was extracted using Trizol protocol (Ambion, Austin, Texas). RNA concentration and purity were evaluated for spectrophotometry (Nano-Drop Technologies, Wilmington, Delaware) and integrity was analyzed by 1% agarose gel electrophoresis. Quantitative reverse transcription polymerase chain reaction (RT-PCR) analysis of individual miR-143 was performed using microRNA assays (4427975; Thermo-Fisher, Waltham, Massachusetts). Briefly, 100 ng total RNA were reverse transcribed using a stem looped-RT specific primer, 0.15 μL dNTPs (100 mM), 1.0 μL reverse transcriptase MultiScribe (50 U/μL), 1.5 μL 10× buffer, 0.19 μL RNase inhibitor (20 U/μL), and 4.16 μL RNase-free water. Then, retrotranscription reaction (1:15 dilution) was mixed with 10 μL master mix TaqMan Universal PCR Master Mix, No AmpErase UNG 2×, 7.67 μL RNase free water, and 1.0 μL PCR probe. Polymerase chain reaction was performed in a GeneAmp System 9700 (Applied Biosystems, Foster, California) as follows: 95°C for 10 minutes, and 40 cycles at 95°C for 15 seconds, and 60°C for 1 minute. Tests were normalized using RNU44 as internal control. Experiments were performed 3 times by triplicate and results were expressed as mean (SD). P < .05 was considered as statistically significant.
MicroRNAs-143 Restoration in Breast Cancer Cells
The precursor of miR-143 (60 nM, MC12540; ThermoFisher) and scramble (60 nM) sequence (AM17110; ThermoFisher) used as negative control were individually transfected into MDA-MB-231 and MCF-7 breast cancer cells using siPORT amine transfection agent (Ambion). Briefly, miR-143 mimics and scramble were added to wells containing 1 × 107 cells and incubated for 48 hours. Then, total RNA was extracted using Trizol and efficacy of RNA mimics treatment was evaluated by qRT-PCR using specific stem-looped RT oligonucleotide and TaqMan probe (4427975; ThermoFisher) as implemented in the TaqMan MicroRNA Assays protocol. Experiments were performed 3 times by triplicate and results were expressed as mean (SD). P < .05 was considered as statistically significant.
Cell Proliferation Assays
For cell proliferation analysis, the MTT reagent ([3-[4,5-dimethylthiazol-2-yl]-2,5 diphenyltetrazolium bromide] was added to MDA-MB-231 and MCF-7 cells (1 × 105) and incubated for 3.5 hours at 37°C. Then, dissolution buffer (99% isopropanol, 0.3% HCl, 0.7% NP-40) was added to cells and incubated for additional 15 minutes. Absorbance was recorded at different time points using a spectrophotometer (570-630 nm). Data were analyzed using the BioStat software. For cisplatin sensitization studies, MDA-MB-231 and MCF-7 cells (1 × 105) transfected with miR-143 mimics (60 nM) or scramble (60 nM) were treated with cisplatin (55 μM) during 24 hours and cell proliferation analysis was performed by MTT assays as described. Experiments were performed 3 times by triplicate and results were expressed as mean (SD). P < .05 was considered as statistically significant.
Cell Migration Assays
Both MDA-MB-231 and MCF-7 breast cancer cells (1 × 105) treated with miR-143 mimics (60 nM) or scramble sequence (60 nM) were seeded in a 6-well plate and grown to 80% confluence. Twenty-four hours postransfection, a vertical wound was traced in the cell monolayer. After 12 and 24 hours, cells were fixed with 4% paraformaldehyde and the scratched area was quantified. Experiments were performed in triplicate and results were expressed as mean (SD). P < .05 was considered as statistically significant.
Phosphorylation Antibody Array
The MDA-MB-231 cells were transfected with the miR-143 (60 nM) mimics and scramble (60 nM) as control and incubated during 48 hours. Then, whole protein extracts (100 μg) were obtained in the presence of phosphatase and protease inhibitors (complete protease/phosphatase inhibitor cocktail, Sigma-Aldrich, St. Louis Missouri), and treated following the manufacturer protocol (PAA137; Full Moon BioSystems, California). This assay is designed as a high-throughput enzyme-linked immunosorbent assay (ELISA)-based antibody array for qualitative protein phosphorylation profiling. It contains 137 antibodies against 36 signaling proteins and 6 replicates printed on standard-size three-dimensional polymer coated glass slides. Briefly, phosphorylation antibody arrays were blocked for 45 minutes followed by incubation with biotin-labeled whole protein extracts for 1 hour at room temperature. After washing, the biotin-labeled proteins bound to signaling antibodies in the arrays were detected using Cy3-conjugated streptavidin (Amersham Biosciences, Little Chalfont, UK), and then slides were documented at 530 nm in a GenePix 4100 scanner[Please provide manufacturer name and location (city and state [if USA] or city and country [if other than USA]) for “GenePix 4100 scanner.”]. Phosphorylation ratio was computed as follows: phosphorylation ratio = (phopho experiment/unphospho experiment)/(phopho control/unphospho control). Changes in protein levels and phosphorylation status were taken as significant if the signal was above the background represented on the array relative to values in control cells (fold change ratio <1.0 as the cutoff criteria for decrease), as recommended by the manufacturer. Cutoff values range from 0 to 2.0 and values less than 1.0 indicated a decrease in proteins phosphorylation.
Bioinformatic Prediction of miR-143 Gene Targets
The miR-143 targets were predicted using TargetScan 7.2 and PicTar 2007 software. Only targets predicted by the 2 algorithms were included in downstream analysis.
Kaplan-Meier Analysis
Kaplan-Meier curves for miR-143 and GSK3-β, RAF1, paxillin, and p21CIP1 genes were determined as described.19,20 Briefly, gene expression data and OS information were downloaded together from GEO (Affymetrix microarrays, Santa Clara, California), EGA and TCGA databases. To evaluate the prognostic value of a particular gene, the patient’s data were split into 2 groups according to various quantile expressions of miR-143 (miRpower tool 2178 patients with breast cancer) and GSK3-β, RAF1, paxillin, and p21CIP1 genes (Start KM plotter, 3951 patients). The 2 patient cohorts were compared by a Kaplan-Meier survival plot, and the hazard ratio with 95% confidence intervals (CIs) and log rank P value were calculated.
Results
Low miR-143 Levels Predicted Response to Chemotherapy and Were Associated With Higher DFS
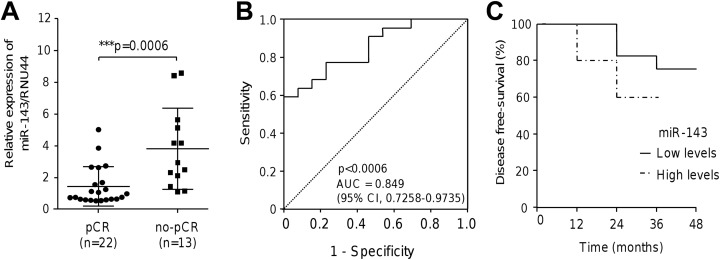
In order to define if miR-143 expression could distinguish between patients with breast cancer that achieved pCR (n = 22) from nonresponder (n = 13) patients, we performed RT-PCR assays. Data showed that miR-143 was differentially expressed between both groups (Figure 1A). The miR-143 exhibited a significant low expression (P < .0006) in responders in comparison to nonresponder group. Moreover, receiver operating characteristic curve analysis indicated that miR-143 could be a good predictor of pCR. The area under the ROC curve for miR-145 in pCR patients was 0.849 (95% CI: 0.7258-0.9735; Figure 1B). Kaplan-Meier survival analysis based on miR-143 expression was performed to estimate the DFS in the same cohort of triple negative patients with breast cancer. Patients were dichotomized into 2 groups with low- and high-miR-143 expression. Data showed that patients with breast cancer with low levels of miR-143 had a higher DFS in comparison to patients with relative high levels of miR-143 (Figure 1C).
Figure 1.
The miR-143-5p expression, ROC, and Kaplan-Meier survival analysis. A, Stem-loop qRT-PCR assays for miR-143 expression in locally advanced patients with triple negative breast cancer that achieved pCR and no-pCR. Data were normalized using endogenous RNU44 as control. Experiments were performed in triplicate, P = .0006. B, ROC curve using miR-143 expression levels to discriminate pCR from no-pCR in patients with breast cancer. C, DFS analysis based in miR-143 high and low expression levels. Continuous line denotes the behavior for patients with higher DFS. AUC indicates area under the ROC curve; CI, confidence interval; DFS, disease free survival; pCR, pathological complete response; ROC, receiver operating characteristic.
MicroRNAs-143 was Repressed in Breast Cancer Cells Lines and Tumors and Inhibited cell Proliferation and Migration
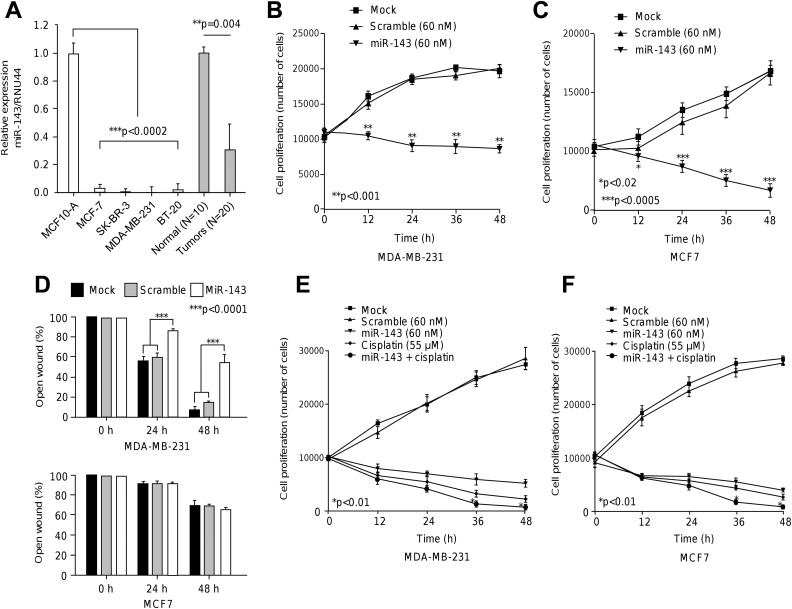
To study the functional relevance of miR-143, we firstly analyzed its expression in breast cancer cells. Data showed that miR-143 expression was significantly (P < .0002) repressed in MDA-MB-231, MCF-7, SK-BR-3, and BT-20 breast cancer cell lines in comparison to MCF10-A nontumorigenic cells (Figure 2A). Likewise, miR-143 was severely downregulated in locally advanced breast tumors relative to normal mammary tissues (P = .004). We next investigated whether the ectopic restoration of miR-145 has effects in cell proliferation. Data from MTT assays showed that the growth rate of triple negative MDA-MB-231 cells transfected with miR-145 precursor (60 nM) was significantly (P < .01) decreased in comparison with mock and scramble (60 nM) transfected control cells after 48 hours (Figure 2B). To extend these findings to other breast cancer subtypes, we also analyzed the estrogen-positive MCF-7 breast cancer cells. Similar results in cell proliferation were found in MCF-7 cells (Figure 2C). Then, we performed scratch/wound-healing assays to evaluate the role of miR-143 overexpression in cell migration. Data showed that cell migration ability was significantly (P < .0001) impaired by miR-143 in MDA-MB-231 cells compared to mock and scramble controls. In contrast, we did not found significantly changes on cell migration of poor invasive MCF-7 cells (Figure 2D).
Figure 2.
The miR-143 suppressed cell proliferation, migration, and sensitized breast cancer cells to therapy. A, Stem-loop quantitative RT-PCR assays for miR-143 expression in breast cancer and normal (MCF-10A) cell lines and locally advanced triple negative breast tumors and normal tissues. Data were normalized using RNU44 as control. Experiments were performed in triplicate. B and C, MTT assays for cell proliferation in MDA-MB-231 and MCF-7 cells transfected with miR-143 or scramble control. Experiments were performed in triplicate. Data are presented as the mean (SD). D, Scratch/wound healing assays for MDA-MB-231 and MCF-7 cells transfected with miR-143 or scramble control. Percentage of open wound after 24 hours and 48 hours was quantified and graphed. Experiments were performed in triplicate and data represented as the mean (SD). E and F, Sensitization assays in MDA-MB-231 and MCF-7 cells transfected with miR-143 or scramble control and combinations treated with cisplatin.
MicroRNAs-143 Sensitized Breast Cancer Cells to Cisplatin Therapy
We next analyzed the effects of miR-143 restoration on the cytotoxicity of cisplatin. MDA-MB-231 cells transfected and nontransfected with miR-143 precursor were submitted to cisplatin (IC50 55 μM) intervention during 48 hours. Results showed that cisplatin alone induced a significant decrease in cell proliferation at 12 hours and 48 hours in comparison with mock and scramble controls (Figure 2E). Combined cisplatin and miR-143 therapy produced a modest but significant (P < .01) and synergistic effect in cell death at 36 hours and 48 hours in comparison with cisplatin and miR-143 monotherapies (Figure 2E). No differences between treatments were found at 12 hours. To extend these findings to other breast cancer subtypes, we also analyzed the estrogen positive MCF-7 cells. Our results indicate that MCF-7 cells treated with cisplatin alone exhibit a significant (P < .01) reduction in cell viability at 12 hours and 48 hours in comparison to controls (Figure 2F). Combined cisplatin plus miR-143 therapy induced a significant (P < .01) cytotoxic effect at 36 hours and 48 hours relative to cisplatin and miR-143 alone. In contrast, no significant differences between treatments were found at 12 hours and 24 hours.
MicroRNAs-143 Targets Multiple Signaling Pathways involved in Cell Proliferation, Migration, and Response to Chemotherapy
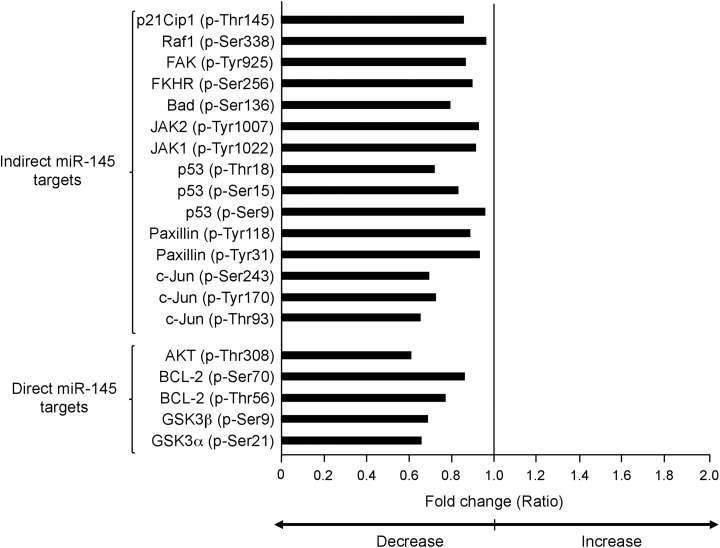
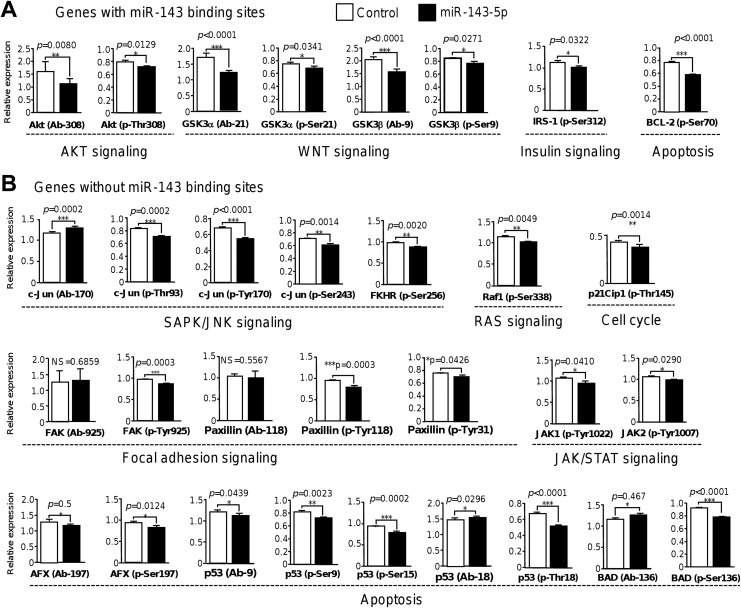
We next wondered if miR-143 modulates signaling pathways related to cancer. For this purpose, we used a high-throughput ELISA-based phospho antibody array (PAA137; Full Moon BioSystems) in which the protein levels and phosphorylation status of 36 signaling proteins were simultaneously screened. Phosphoproteomic profiling data showed that restoration of miR-143 resulted in a significant decrease in protein levels and phosphorylation status (ratio < 1.0) of 14 of 39 proteins evaluated (Figure 3). Proteins were grouped into 4 major intracellular signaling networks including AKT, WNT, SAPK/JNK, JAK/STAT, and FAK pathways (Figure 4). Relative expression analysis indicated that treatment of MDA-MB-231 cells with miR-143 led to a significant reduction of total AKT (P = .080) and phosphorylated AKT (p-Thr308) kinases (P = .012). Likewise, activation of WNT/β-catenin signaling was also impacted as the levels of total GSK3α and GSK3β were decreased by miR-143. Moreover, phosphorylated GSK3α(p-Ser21) and GSK3β(p-Ser9) were slightly but significantly diminished (P = .034 and P = .027, respectively). Also, the level of insulin signaling-related IRS-1(p-Ser312), a protein involved in chemotherapy resistance, was suppressed (P = .032). On the other hand, we found a set of modulated genes involved in SAPK/JNK, JAK/STAT, and FAK signaling, as well as in cell cycle and apoptosis processes. The c-Jun transcription factor is a downstream effector of SAPK/JUNK signaling pathway. Although no significant changes in protein levels of total c-Jun were found, phosphorylated c-Jun(p-Thr93), c-Jun(p-Tyr170), and c-Jun(Ser243) phosphorylated proteins showed a significant decrease after miR-143 transfection (Figure 4B). In addition, levels of phosphorylated FKHR(p-Ser256) transcription factor were also decreased (P = .002). Evidence for alterations in FAK signaling activation represented by a slight but significantly reduced phosphorylation status of paxillin at p-Tyr118 and p-Tyr31 amino acid residues was found. In contrast, no significant changes in protein levels of paxillin were detected. Activation of RAS signaling was also affected as we observed a low phosphorylation status of RAS signaling-related Raf1(p-Ser338) and the cell cycle p21Clp1(p-Thr145) proteins. The JAK/STAT signaling activation was also hampered as phosphorylation levels of JAK1(p-Tyr1022) and JAK2(p-Tyr1007) declined after miR-143 treatment. Finally, phosphorylation levels of cell cycle and apoptosis regulators p53(pSer9), p53(pSer15), p53(pThr18), and AFX(pSer97; also known as FOXO4) were lower in miR-143-expressing cells.
Figure 3.
Protein levels and phosphorylation status of 12 signaling proteins in MDA-MB-231 cells expressing miR-143. Graph indicates the decrease (<1; P < .05 vs control) in phosphorylation status of signaling transducers in MDA-MB-231 cells transfected with miR-143 mimics (60 nM) by 48 hours in comparison to nontransfected control cells. Direct and indirect targets of miR-143 are indicated with brackets.
Figure 4.
The miR-143 decreases the protein levels and phosphorylation status of multiple cell signaling proteins. Phosphorylation Antibody Arrays were used to detect the changes in protein levels and phosphorylation status of signaling proteins in MDA-MB-231 cells transfected with pre-miR-143 (60 nM) and nontransfected as control. A and B, Relative expression analysis showed the changes in protein levels and phosphorylation status. Data were grouped as protein-encoding genes containing (A) miR-143 binding sites, and (B) without miR-143 binding sites as well as in signaling pathways they are involved. Black bars indicate protein levels and phosphorylation levels in breast cancer cells transfected with miR-143 relative to nontransfected control cells (white bars).
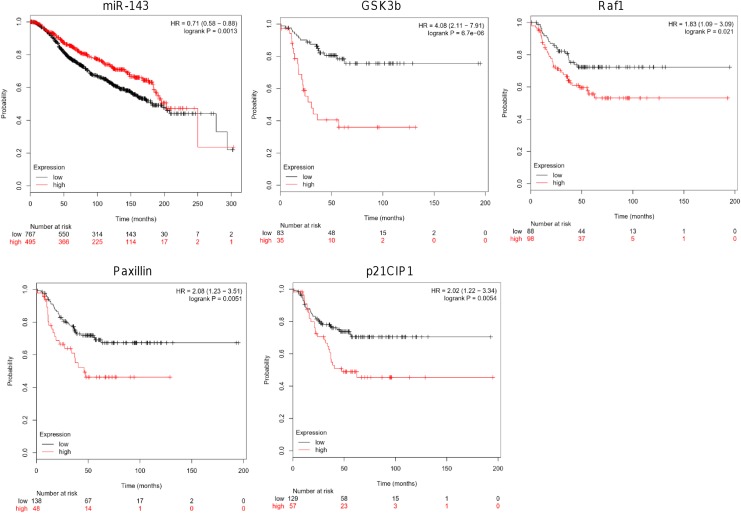
Expression Levels of miR-143 and Target Genes Correlate With Poor Patient’s Outcome
To obtain insights about the clinical implications of miR-143, we performed Kaplan-Meier OS analysis using a large cohort of patients as describe in the Kaplan-Meier plotter database (http://kmplot.com/analysis/). Results showed that low expression of miR-143 (hazard ratio [HR] = 0.71, logrank P = .0013) and higher expression of its targets GSK3-β (HR = 4.08, logrank P = 6.7e-06), RAF1 (HR = 1.83, logrank P = .021), paxillin (HR = 2.08, logrank P = .0051), and p21CIP1 (HR = 2.02, logrank P = .0054) correlate with worst outcomes of patients with breast cancer (Figure 5).
Figure 5.
Overall survival analysis. Kaplan-Meier curves for miR-143 and GSK3-β, RAF1, paxillin and p21CIP1 genes. Patients were split into 2 groups according to various quantile expressions of miR-143 (miRpower tool 2178 patients with breast cancer) and GSK3-β, RAF1, paxillin and p21CIP1 genes (start KM plotter, 3951 patients). The 2 patient cohorts were compared by a Kaplan-Meier survival plot, and the hazard ratio with 95% confidence intervals and log rank P value were calculated.
Discussion
Neoadjuvant chemotherapy is used as a model for research because it provides opportunities to test novel drugs that may lead to pCR and better outcomes. However, still there is a lack of effective methods to select patients with breast cancer who would have a benefit from pCR, thus there is a need to identify novel biomarkers that may predict response to neoadjuvant therapy. Previously, several studies reported that miRNA expression signatures could be associated with pCR after neoadjuvant treatment in different types of human cancers.13-18 Here, we showed that patients with triple negative breast cancer with low miR-143 levels achieved pCR, which has a prognostic impact on DFS and OS (Figure 1). Before neoadjuvant chemotherapy, we distinguish 2 groups of patients: (1) a group with very low miR-143 (low miR-143) and (2) a group with relative high expression (high miR-143). Both groups have lower miR-143 expression in comparison to normal breast tissues and cell lines (Figure 2A). In this cohort of patients, relative high expression of miR-143 before the therapy was associated with worst response to chemotherapy (no-pCR) and low DFS in comparison to miR-143 low group (Figure 1A). These data highlight the role of miR-143 as a potential predictor of pCR in triple negative breast cancer. Similar data were reported in patients with locally advanced rectal cancer in which low serum levels of miR-143 predicted pCR to neoadjuvant chemoradiotherapy.21 We also found that miR-143 expression was significantly repressed in breast tumors and in 4 breast cancer cell lines relative to normal mammary tissues and nontumorigenic cells, respectively (Figure 2A). In addition, ectopic restoration of miR-143 inhibited cell proliferation and migration and sensitized tumor cells to cisplatin treatment (Figure 2). Our data are in agreement with previous reports in other types of human cancer were miR-143 was downregulated and its ectopic overexpression inhibited cell proliferation, migration and sensitized cells to therapy in gastric,22 endometrial,23 gallbladder,24 breast,25 and colorectal cancers,26 as well as in glioma,27 reinforcing the idea that miR-143 is a bona fide tumor suppressor associated with therapy response. In this scenario, we hypothesize that higher levels of miR-143 may be suppressing apoptotic proteins resulting in the inhibition of tumor cell apoptosis and cell proliferation (Figure 1A), thus explaining in part, why these patients did not achieved a good response to therapy.
Besides, here we provide novel data on the cellular signaling cascades modulated by miR-143 in MDA-MB-231 cells. Phosphoproteomic profiling indicated that miR-143 decreased the protein levels and phosphorylation status of 14 of 39 proteins tested (Figure 3 and 4). Phosphorylation levels of AKT (p-Thr308), GSK3-α(pSer21) and GSK3-βpSer9), IRS-1(p-312) and BCL2(pSer70) were decreased in miR-143-expressing cancer cells. Markedly, AKT, GSK3-α, GSK3-β, IRS-1, and BCL2 protein-encoding genes contain canonical binding sites for miR-143 indicating that they could be direct posttranscriptional targets. Our findings indicated that activation of AKT signaling was hampered as AKT and AKT(p-Thr308) protein levels were significantly decreased by miR-143 which maybe related to suppression of cell proliferation and migration, as well as sensitization to cisplatin therapy (Figures 2 and 4A). It was previously reported that increased AKT(p-Thr308) phosphorylation was associated with Gli1 upregulation which promoted the progression of cell cycle in breast tumors with a stem cell phenotype.28 In addition, the combination of everolimus plus trastuzumab treatment reduced the levels of AKT(p-Thr308), affecting cell growth and tumorigenicity through induction of cell cycle arrest and promotion of early apoptosis in breast cancer stem cells.29 Moreover, cell migration, adhesion, and invasion were suppressed through downregulation of AKT(p-Thr308) levels after knockdown of cPLA2γ in breast cancer cells.30
Likewise, we found that activation of WNT pathway was also impaired because phosphorylation levels of GSK3-α(pSer21) and GSK3-β(pSer9) were decreased by miR-143. The consequences of reduction in phosphorylation of the aforementioned proteins have been well documented. For instance, suppression of GSK3-β(pSer9) levels inhibited cell migration and invasion, and promoted cell death of triple negative breast cancer cells. 31,32 In addition, knockdown of GSK3-α(pSer21) and GSK3-β(pSer9) using CG0009 inhibitor enhanced apoptosis and inhibited cell growth in breast cancer cell lines.33 Similarly, a reduction in GSK3-α(pSer21) and GSK3-β(pSer9) levels resulted in decreased cell proliferation and cell cycle arrest (G1) in prostate cancer cells34 and apoptosis in colon cancer cells.35
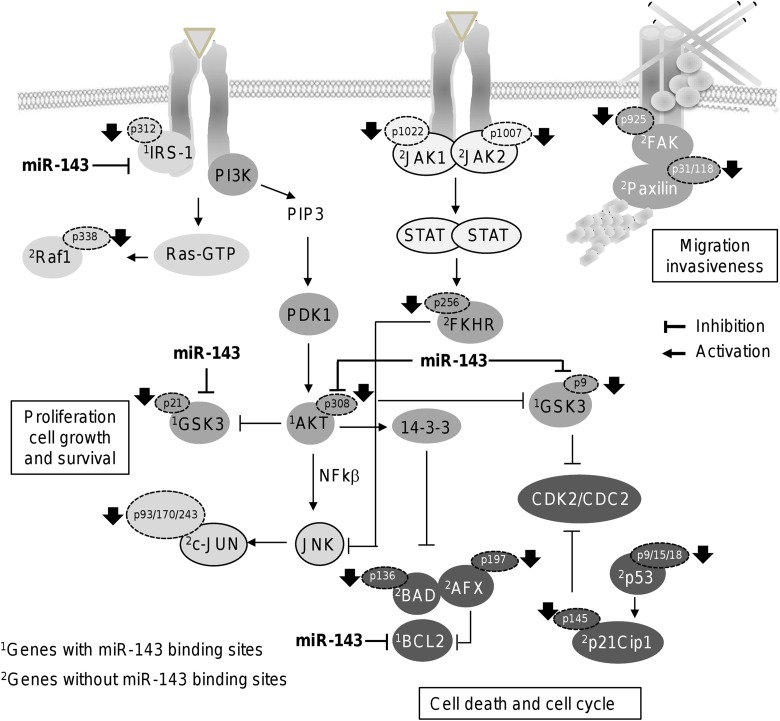
Phosphoproteomic profiling also leads us to the identification of a set of modulated proteins involved in SAPK/JNK, focal adhesion, and apoptosis signaling (Figure 4B). These protein-encoding genes did not contain putative miR-143 binding sites at their 3′-untranslated region indicating that they could be regulated by an indirect mechanism. The activation of the JNK/SAPK pathway culminates in the phosphorylation of c-Jun, activating transcription factor-2 and p53. The JNK/SAPKs play a central role in cell proliferation, apoptosis, motility, metabolism, and DNA repair in cancer cells. We found no changes in c-Jun protein levels in miR-143 expressing MDA-MB-231 cells. In contrast, phosphorylated levels of c-Jun(pThr93), c-Jun(pTyr170), and c-Jun(pSer243) significantly dropped after restoration of miR-143 (Figure 4B). In addition, FKHR (pSer256) was also significantly downregulated. It was previously reported that epidermal growth factor treatment induced the phosphorylation of AKT which in turn activates FKHR which resulted in the extrusion of FKHR (pSer256) from nucleus to cytoplasm,36 impairing its ability to activate nuclear target genes involved in cell-cycle progression and apoptosis in MDA-MB-231 cells. Taken altogether, we suggested a working model for miR-143 regulation of signaling networks (Figure 6). We propose that cell migration could be suppressed by indirect targeting of FAK signaling as FAK(pTyr925) and paxillin(pTyr31) and paxillin (pTyr118) were declined by miR-143 which may impact assembly of focal adhesions and cytoskeleton reorganization required for proper cell migration and invasion. In addition, an inhibitory effect of miR-143 in the crosstalk of RAS and AKT pathways may also occur as it directly targets IRS-1 and AKT kinase and indirectly RAF1(pSer338). Diminished levels of AKT(pThr308) could lead to the impairment of JNK and c-Jun phosphorylation impacting cell proliferation. The WNT/β-catenin signaling also could be severely affected as the protein levels and phosphorylation status of GSK3-α(pSer21) and GSK3-β(pSer9) were decreased thus impacting cell death and survival. Proteins with a pivotal role in cell death and cell cycle were also suppressed. All these changes in proteins levels and phosphorylation may converge resulting in the inhibition of breast cancer hallmarks. In summary, our data suggested that miR-143 could be a potential predictor of response to neoadjuvant therapy, and also provide clues in miR-143 molecular functions in signaling networks related to carcinogenesis in breast cancer.
Figure 6.
Working model for miR-143 functions. Model for regulation of cancer-related signaling pathways by miR-143. Gray circles indicate the direct miR-204 targets. Protein-encoding genes with and without miR-143 binding sites are denoted by numbers 1 and 2, respectively.
Statistical Analysis
Experiments were performed in triplicate and results were represented as mean (SD). One-way analysis of variance followed by Tukey test were used to compare the differences between means. A P < .05 was considered as statistically significant.
Supplemental Material
Supplementary_table_1 for MicroRNA-143 is Associated With Pathological Complete Response and Regulates Multiple Signaling Proteins in Breast Cancer by Raúl García-Vázquez, Laurence A. Marchat, Erika Ruíz-García, Horacio Astudillo-de la Vega, Abelardo Meneses-García, Claudia Arce-Salinas, Enrique Bargallo-Rocha, Ángeles Carlos-Reyes, José Sullivan López-González, Carlos Pérez-Plasencia, Rosalío Ramos-Payán, Maribel Aguilar-Medina, and César López-Camarillo in Technology in Cancer Research & Treatment
Acknowledgments
We acknowledge Universidad Autónoma de la Ciudad de México for support. Raul García Vázquez was recipient of a fellowship from CONACYT (No. 441111).
Abbreviations
- DFS
disease-free survival
- ELISA
enzyme-linked immunosorbent assay
- miRNAs
microRNAs
- OS
overall survival
- pCR
pathological complete response
- ROC
receiver operating characteristic
- RT-PCR
reverse transcription polymerase chain reaction.
Footnotes
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This study was funded by Consejo Nacional de Ciencia y Tecnología CONACyT México, Fondo SSA/IMSS/ISSSTE (grant 233370).
ORCID iD: César López-Camarillo, PhD  https://orcid.org/0000-0002-9417-2609
https://orcid.org/0000-0002-9417-2609
Supplemental Material: Supplemental material for this article is available online.
References
- 1. Torre LA, Siegel RL, Ward EM, Jemal A. Global cancer incidence and mortality rates and trends—an update. Cancer Epidemiol Biomarkers Prev. 2016;25(1):16–27. [DOI] [PubMed] [Google Scholar]
- 2. von Minckwitz G, Untch M, Blohmer JU, et al. Definition and impact of pathologic complete response on prognosis after neoadjuvant chemotherapy in various intrinsic breast cancer subtypes. J Clin Oncol. 2012;30(15):1796–1804. [DOI] [PubMed] [Google Scholar]
- 3. Liedtke C, Mazouni C, Hess KR, et al. Response to neoadjuvant therapy and long-term survival in patients with triple negative breast cancer. J Clin Oncol. 2008;26(8):1275–1281. [DOI] [PubMed] [Google Scholar]
- 4. Ring AE, Smith IE, Ashley S, et al. Oestrogen receptor status, pathological complete response and prognosis in patients receiving neoadjuvant chemotherapy for early breast cancer. Br J Cancer. 2004;91(12):2012–2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Tanioka M, Sasaki M, Shimomura A, et al. Pathologic complete response after neoadjuvant chemotherapy in HER2-overexpressing breast cancer according to hormonal receptor status. Breast. 2014;23(4):466–472. [DOI] [PubMed] [Google Scholar]
- 6. Zhang P, Yin Y, Mo H, et al. Better pathologic complete response and relapse-free survival after carboplatin plus paclitaxel compared with epirubicin plus paclitaxel as neoadjuvant chemotherapy for locally advanced triple-negative breast cancer: a randomized phase 2 trial. Oncotarget. 2016;7(37):60647–60656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kong X, Moran MS, Zhang N, et al. Meta-analysis confirms achieving pathological complete response after neoadjuvant chemotherapy predicts favourable prognosis for breast cancer patients. Eur J Cancer. 2011;47(14):2084–2090. [DOI] [PubMed] [Google Scholar]
- 8. Luangdilok S, Samarnthai N, Korphaisarn K. Association between pathological complete response and outcome following neoadjuvant chemotherapy in locally advanced breast cancer patients. J Breast Cancer. 2014;17(4):376–385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cortazar P, Zhang L, Untch M, et al. Pathological complete response and long-term clinical benefit in breast cancer: the CTNeoBC pooled analysis. Lancet. 2014;384(9938):164–172. [DOI] [PubMed] [Google Scholar]
- 10. Esquela-Kerscher A, Slack FJ. Oncomirs—microRNAs with a role in cancer. Nat Rev Cancer. 2006;6(4):259–269. [DOI] [PubMed] [Google Scholar]
- 11. Kolacinska A, Morawiec J, Fendler W, et al. Association of microRNAs and pathologic response to preoperative chemotherapy in triple negative breast cancer: preliminary report. Mol Biol Rep. 2014;41(5):2851–2857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hummel R, Hussej DJ, Haier J. MicroRNAs: predictors and modifiers of chemo- and radiotherapy in different tumor types. Eur J Cancer. 2010;46(2):298–311. [DOI] [PubMed] [Google Scholar]
- 13. Gasparini P, Cascione L, Fassan M, et al. MicroRNA expression profiling identifies a four microRNA signature as a novel diagnostic and prognostic biomarker in triple negative breast cancers. Oncotarget. 2014;5(5):1174–1184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ohzawa H, Miki A, Teratani T, et al. Usefulness of miRNA profiles for predicting pathological responses to neoadjuvant chemotherapy in patients with human epidermal growth factor receptor 2-positive breast cancer. Oncol Lett. 2017;13(3):1731–1740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Raychaudhuri M, Bronger H, Buchner T, Kiechle M, Weichert W, Avril S. MicroRNAs miR-7 and miR-340 predict response to neoadjuvant chemotherapy in breast cancer. Breast Cancer Res Treat. 2017;162(3):511–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Pedroza-Torres A, Fernández-Retana J, Peralta-Zaragoza O, et al. A microRNA expression signature for clinical response in locally advanced cervical cancer. Gynecol Oncol. 2016;142(3):557–565. [DOI] [PubMed] [Google Scholar]
- 17. Petrillo M, Zannoni GF, Beltrame L, et al. Identification of high-grade serous ovarian cancer miRNA species associated with survival and drug response in patients receiving neoadjuvant chemotherapy: a retrospective longitudinal analysis using matched tumor biopsies. Ann Oncol. 2016;27(4):625–634. [DOI] [PubMed] [Google Scholar]
- 18. García-Vazquez R, Ruiz-García E, Meneses García A, et al. A microRNA signature associated with pathological complete response to novel neoadjuvant therapy regimen in triple-negative breast cancer. Tumour Biol. 2017;39(6):1010428317702899. [DOI] [PubMed] [Google Scholar]
- 19. Lánczky Á, Nagy G, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients. Breast Cancer Res Treat. 2016;160(3):439–446. [DOI] [PubMed] [Google Scholar]
- 20. Gyorffy B, Lanczky A, Eklund AC, et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treatment. 2010;123(3):725–731. [DOI] [PubMed] [Google Scholar]
- 21. Hiyoshi Y, Akiyoshi T, Inoue R, et al. Serum miR-143 levels predict the pathological response to neoadjuvant chemoradiotherapy in patients with locally advanced rectal cancer. Oncotarget. 2017;8(45):79201–79211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Guoping M, Ran L, Yanru Q. miR-143 inhibits cell proliferation of gastric cancer cells through targeting GATA6. Oncol Res. 2018;26(7);1023–1029. doi:10.3727/096504018X15151515028670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Chang L, Zhang D, Shi H, Bian Y, Guo R. MiR-143 inhibits endometrial cancer cell proliferation and metastasis by targeting MAPK1. Oncotarget. 2017;8(48):84384–84395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. He M, Zhan M, Chen W, et al. MiR-143-5p deficiency triggers EMT and metastasis by targeting HIF-1α in gallbladder cancer. Cell Physiol Biochem. 2017;42(5):2078–2092. [DOI] [PubMed] [Google Scholar]
- 25. Johannessen C, Moi L, Kiselev Y, et al. Expression and function of the miR-143/145 cluster in vitro and in vivo in human breast cancer. PLoS One. 2017;12(10):e0186658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Qian X, Yu J, Yin Y, et al. MicroRNA-143 inhibits tumor growth and angiogenesis and sensitizes chemosensitivity to oxaliplatin in colorectal cancers. Cell Cycle. 2013;12(9):1385–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Wang L, Shi ZM, Jiang CF, et al. MiR-143 acts as a tumor suppressor by targeting N-RAS and enhances temozolomide-induced apoptosis in glioma. Oncotarget. 2014;5(14):5416–5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ni W, Yang Z, Qi W, Cui C, Cui Y, Xuan Y. Gli1 is a potential cancer stem cell marker and predicts poor prognosis in ductal breast carcinoma. Hum Pathol. 2017;69:38–45. [DOI] [PubMed] [Google Scholar]
- 29. Zhu Y, Zhang X, Liu Y, et al. Antitumor effect of the mTOR inhibitor everolimus in combination with trastuzumab on human breast cancer stem cells in vitro and in vivo. Tumour Biol. 2012;33(5):1349–1362. [DOI] [PubMed] [Google Scholar]
- 30. Tian G, Wang X, Zhang F, et al. Downregulation of cPLA2γ expression inhibits EGF-induced chemotaxis of human breast cancer cells through Akt pathway. Biochem Biophys Res Commun. 2011;409(3):506–512. [DOI] [PubMed] [Google Scholar]
- 31. Choi CH, Lee BH, Ahn SG, Oh SH. Proteasome inhibition-induced p38 MAPK/ERK signaling regulates autophagy and apoptosis through the dual phosphorylation of glycogen synthase kinase 3β. Biochem Biophys Res Commun. 2012;418(4):759–764. [DOI] [PubMed] [Google Scholar]
- 32. Gollavilli PN, Kanugula AK, Koyyada R, Karnewar S, Neeli PK, Kotamraju S. AMPK inhibits MTDH expression via GSK3β and SIRT1 activation: potential role in triple negative breast cancer cell proliferation. FEBS J. 2015;282(20):3971–3985. [DOI] [PubMed] [Google Scholar]
- 33. Kim HM, Kim CS, Lee JH, et al. CG0009, a novel glycogen synthase kinase 3 inhibitor, induces cell death through cyclin D1 depletion in breast cancer cells. Wagner B, ed. PLoS ONE. 2013;8(4):e60383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Hsieh T, Yang CJ, Lin CY, Lee YS, Wu JM. Control of stability of cyclin D1 by quinone reductase 2 in CWR22Rv1 prostate cancer cells. Carcinogenesis. 2012;33(3):670–677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Hu C, Dong T, Li R, Lu J, Wei X, Liu P. Emodin inhibits epithelial to mesenchymal transition in epithelial ovarian cancer cells by regulation of GSK-3β/β-catenin/ZEB1 signaling pathway. Oncol Rep. 2016;35(4):2027–2034. [DOI] [PubMed] [Google Scholar]
- 36. Jackson JG, Kreisberg JI, Koterba AP, Yee D, Brattain MG. Phosphorylation and nuclear exclusion of the forkhead transcription factor FKHR after epidermal growth factor treatment in human breast cancer cells. Oncogene. 2000;19(40):4574–4581. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary_table_1 for MicroRNA-143 is Associated With Pathological Complete Response and Regulates Multiple Signaling Proteins in Breast Cancer by Raúl García-Vázquez, Laurence A. Marchat, Erika Ruíz-García, Horacio Astudillo-de la Vega, Abelardo Meneses-García, Claudia Arce-Salinas, Enrique Bargallo-Rocha, Ángeles Carlos-Reyes, José Sullivan López-González, Carlos Pérez-Plasencia, Rosalío Ramos-Payán, Maribel Aguilar-Medina, and César López-Camarillo in Technology in Cancer Research & Treatment