Graphical abstract
Highlights
-
•
A ‘second green revolution’ using crops with improved below-ground traits is needed.
-
•
Phenotyping plant roots poses practical and data challenges.
-
•
Technologies for 2D root phenotyping include rhizotrons, paper pouches, and plates.
-
•
MRI, PET and X-ray CT allow 3D and 4D root phenotyping in soil.
-
•
Non-invasive techniques for field phenotyping have recently advanced.
-
•
Deep machine learning techniques are transforming the root phenotyping landscape.
Abstract
Major increases in crop yield are required to keep pace with population growth and climate change. Improvements to the architecture of crop roots promise to deliver increases in water and nutrient use efficiency but profiling the root phenome (i.e. its structure and function) represents a major bottleneck. We describe how advances in imaging and sensor technologies are making root phenomic studies possible. However, methodological advances in acquisition, handling and processing of the resulting ‘big-data’ is becoming increasingly important. Advances in automated image analysis approaches such as Deep Learning promise to transform the root phenotyping landscape. Collectively, these innovations are helping drive the selection of the next-generation of crops to deliver real world impact for ongoing global food security efforts.
Current Opinion in Biotechnology 2019, 55:1–8
This review comes from a themed issue on Analytical biotechnology
Edited by Saulius Klimasauskas and Linas Mazutis
For a complete overview see the Issue and the Editorial
Available online 19th July 2018
https://doi.org/10.1016/j.copbio.2018.06.002
0958-1669/© 2018 The Authors. Published by Elsevier Ltd. This is an open access article under the CC BY-NC-ND license (http://creativecommons.org/licenses/by-nc-nd/4.0/).
Introduction
Crop production has to double by 2050 to keep pace with global population growth. This target is even more challenging given the impact of climate change on water availability and the drive to reduce fertilizer inputs to make agriculture environmentally sustainable. Developing crops with improved water and nutrient uptake efficiency would provide a solution. As root architecture influences nutrient and water uptake efficiency, a ‘second green revolution’ has been proposed that deploys crops with improved below ground traits [1]. However, selecting crops based on root system architecture (RSA) poses practical challenges.
This review discusses recent advances in root phenotyping. To date, classical non-destructive 2D techniques such as agar plates or rhizotrons have been integral to our understanding of root development (Figure 1). Non-destructive analysis of 3D root growth is also possible using transparent gels [2,3•,4] but results are often difficult to extrapolate to field conditions. To non-invasively study 3D root growth in soil, more sophisticated approaches are needed. This review explores several promising new approaches to uncover the ‘hidden half’ of plants grown under either lab or field conditions (Box 1). We discuss the ‘big data’ challenges associated with root phenotyping and describe promising solutions being developed by other disciplines, then conclude with a forward look.
Figure 1.
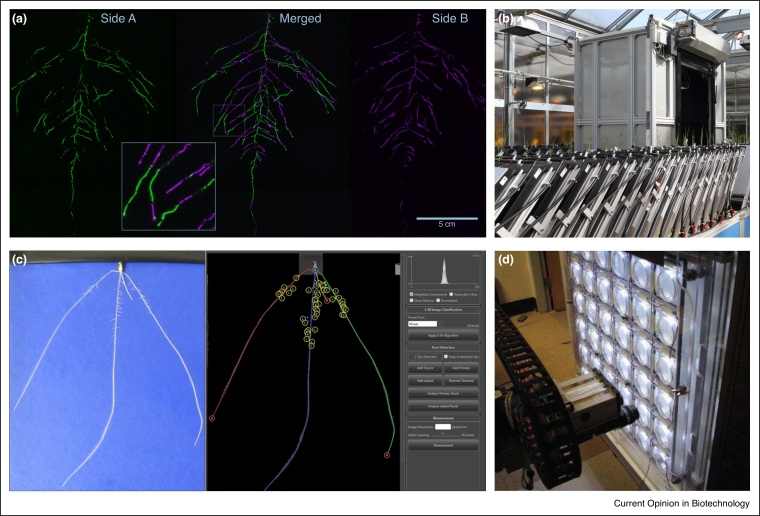
2D imaging of plant roots. (a) GLO-Roots [5••]. Arabidopsis plant expressing a luminescent reporter imaged on each side of the rhizotron (coloured green and magenta respectively) at 21 days after sowing (DAS). (b) GROWSCREEN-Rhizo [12]. A high-throughput automated root phenotyping platform using soil-filled rhizotrons. (c) Pouch system [9] for cereal seedlings (left panel). RootNav [51] analysis software (right panel). (d) Phytomorph [13] A high-throughput robotic imaging platform for Arabidopsis growing on agar plates.
Box 1. Glossary of technologies applied to plant root phenotyping.
Electrical resistance tomography (ERT) — imaging of sub-surface soil structure from maps of electrical resistivity measured via buried probes.
Electromagnetic inductance (EMI) — mapping of spatial soil electrical conductivity using sensors held above the soil surface.
Ground penetrating radar (GPR) — mapping of sub-surface structure by measuring reflection, refraction, and scattering of pulses of high-frequency radio waves. Receiving antennae may be positioned in contact with the soil or above the soil surface.
Magnetic resonance imaging (MRI) — imaging technique based on absorption and re-emission of electromagnetic radiation from nuclei in a magnetic field.
Positron emission tomography (PET) — imaging technique based on detection of gamma radiation from tracer molecules.
Rhizotron — a growth chamber with transparent or removable observation windows through which roots can be imaged.
X-ray computed tomography (X-ray CT) — imaging technique based on attenuation of X-rays to create cross-sections for reconstruction into a 3D model.
Alt-text: Box 1
Technologies for root phenotyping under controlled conditions
The opaque nature of soil makes phenotyping root systems in situ challenging compared to analysing above-ground plant organs. Non-destructive techniques under controlled conditions have traditionally relied on rhizotrons (enclosures with transparent or removable observation windows), growth pouches, or transparent artificial growth media (Figure 1). Images are usually two-dimensional (2D) and, if soil is used, often fail to capture the complete root system architecture as many roots will be occluded by soil particles. The GLO-Root system designed for Arabidopsis [5••] mitigates these effects by using luminescence-based reporters for visualisation of architecture and gene expression patterns, and by combining images from both sides of the rhizotron (Figure 1). Soil-free techniques such as hydroponics, aeroponics, gel plates, and growth pouches provide greater contrast between root and substrate allowing accurate extraction of root system architecture, although the root systems of plants grown in artificial media can vary considerably from those grown in soil [6]. Pouch systems using plants grown vertically on germination paper have been successfully used in seedling screens for many species including, bean [7], maize [8], wheat [9], oilseed rape [10], and pearl millet [11]. Despite their limitations, 2D soil and artificial media systems are widely used due to their suitability for incorporation into high-throughput root phenotyping platforms such as GrowScreen-Rhizo [12], Phytomorph [13], GrowScreen-PaGe [10], RADIX [14] and RhizoTubes [15].
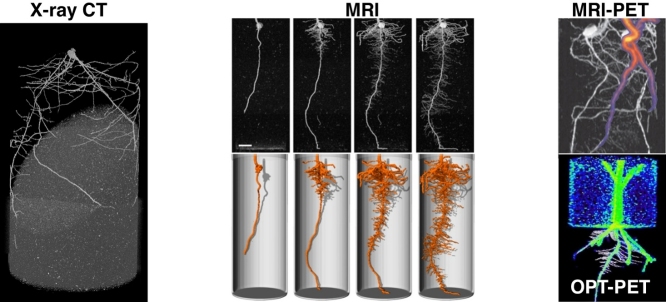
Plant root systems are three-dimensional (3D) structures with many features that are difficult to quantify in 2D [3•] such as the arrangement of seminal roots at the root crown of cereals (that are often asymmetrically distributed), and the angle and number of roots and root whorls in maize crowns. Dynamic growth responses such as gravitropism and circumnutation are also more readily studied in 3D [2]. Three-dimensional representations of root systems can be produced from multiple-viewpoint imaging of plants grown in optically transparent media [2,3•,4] or hydroponically using a support system [16]. One such system was successfully used to uncover the underlying genetic basis for several 3D root architectural traits in rice not revealed by 2D phenotyping [3•]. Non-destructive, 3D phenotyping of roots in soil is currently achievable using three tomographic techniques originally developed for medical applications (Figure 2): X-ray computed tomography (X-ray CT), magnetic resonance imaging (MRI) and positron emission tomography (PET).
Figure 2.
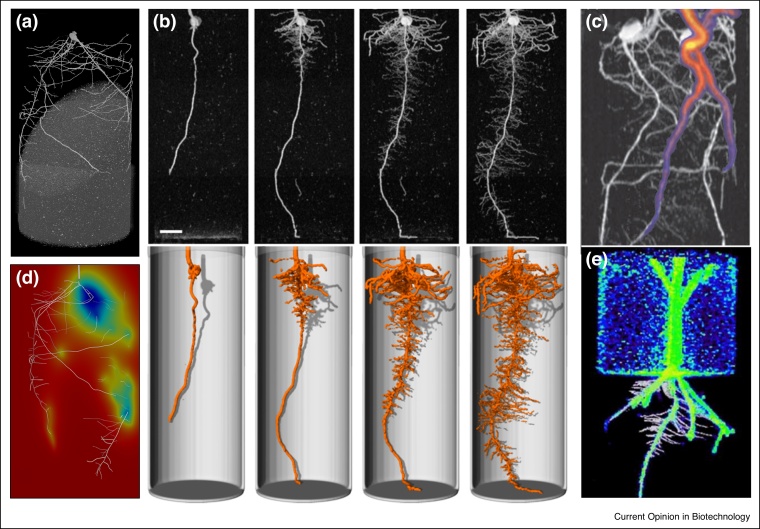
3D tomographic imaging of plant roots. (a) X-ray CT micrograph of a wheat seedling 12 DAS. (b) MRI imaging of a maize root system at 6, 9, 12, and 15 DAS [23••]. Upper panel, MRI data (2D maximum intensity projection). Lower panel, 3D surface render. Scale bar: 20 mm. (c) Maize roots imaged using MRI-PET [24]. Two plants are growing in the same pot. The greyscale image is MRI, the colour is 11C PET data following application to a leaf of one plant. (d) OpenSimRoot [65] simulation using output from (a) to model rhizosphere N depletion. (e) Maize root imaged at 9 DAS using optical projection tomography (OPT) and PET [4]. The black and white image is OPT, the colour is 11C PET data.
X-ray CT allows the visualisation of 3D volumes based on differential X-ray attenuation. Although first demonstrated in plant roots over 30 years ago [17], only recently has advances in scan time, resolution, reconstruction times, and image segmentation software made X-ray CT a viable technology for root phenotyping in soil [18]. X-ray CT has been used to examine the cultivar-specific response of rice root systems to growth medium texture [19]; patterning of lateral roots in Arabidopsis, maize, and rice [20]; inter-specific interactions between aspen and spruce [21]; and to quantify roots of prairie dropseed to parameterise computational fluid dynamics simulations [22]. MRI employs radio-frequency waves and strong magnetic fields to stimulate atoms (usually of hydrogen in water) and produce a 3D spatial map [23••]. MRI has been employed to image the root systems of soil-grown maize, bean, sugar beet, and barley [23••,24,25]. PET scanning visualizes the distribution of short half-life radioactive tracers, such as carbon isotopes used in plant metabolic processes [26]. Despite a high sensitivity for tracers, PET is currently limited to a relatively coarse resolution of ∼1.4 mm [24]. To overcome this limitation, PET is often combined with other tomographic techniques including MRI [24], X-ray CT [26], and optical projection tomography [4].
Both X-ray CT and MRI can be used for monitoring root system growth over time, with no adverse effects of repeated scanning on plant development [23••,27], although care should be taken in X-ray CT experiments to monitor any impact of repeated scans on root development and soil biota [27]. MRI and X-ray CT can be seen as complimentary technologies, with their own advantages and limitations [28]. However, MRI is more dependent than X-ray CT on the choice of substrate with initial experiments requiring specific soils or the removal of ferromagnetic particles [29]. A recent study of eight substrates (6 natural soils and 2 artificial mixtures) reported five suitable for root segmentation via MRI in barley plants (including the two artificial mixtures). Of the three natural soils deemed unsuitable to MRI, two were able to be used to resolve thicker roots [29]. Both MRI and X-ray CT imaging are confounded by high soil moisture, although both techniques are suitable for soils held at field capacity or lower [29,30]. In their medical applications, CT scanners and MRI machines are usually arranged for horizontal sample loading. The availability of non-medical X-ray CT scanners with vertical sample loading has, combined with the lower equipment cost, led to a wider adoption of this technology for plant phenotyping compared to MRI.
Root phenotyping in the field
Field-based phenotyping has seen significant advances in recent years with sensor technologies to quantify canopy traits (including LIDAR, multi- and hyperspectral imaging, thermography, and RGB imaging) deployed on drones, tractor mounts and gantry systems [31, 32, 33, 34, 35]. Phenotyping for root traits in the field has seen comparatively less advancement, largely due to the difficulties associated with imaging below-ground. Classic methods such as the soil core-break are widely used, with this protocol being recently improved by employing UV illumination and fluorescence spectroscopy to enhance soil-root image contrast and allowing automated image capture and processing of core-break faces [36].
Shovelomics [37], or root crown phenotyping, is one of the most widely adopted high-throughput field methods. The protocol, originally designed for maize, has been adapted for other species including legumes [38] and wheat [39]. Shovelomics generates a number of key root architecture parameters including crown root number and angle [37]. Automatic image analysis software such as DIRT [40] and REST [41] have further increased throughput; however, the rate-limiting step is still the manual excavation of the crown root system. Automation of this labour-intensive process is being addressed by researchers at Pennsylvania State University in the DEEPER project, part of the ARPA-E funded ROOTS program that includes inter alia development of field-deployable X-ray CT and MRI platforms [42].
Development of non-invasive geophysical techniques to study the root and soil profile has also advanced in recent years. This includes Electrical Resistance Tomography (ERT) that measures soil water profiles. Most commonly used to analyse large diameter root profiles (e.g. trees [43]), ERT has seen limited adoption to date in the areas of crop phenotyping [44]. Although ERT is non-destructive and significantly higher throughput than traditional coring methods, it is limited by the number of probe arrays that can be placed in the field and is still considered low throughput compared to other geophysical approaches such as electromagnetic inductance (EMI). EMI is significantly higher throughput than ERT as it does not require probes or direct contact with the soil [45] and was recently implemented to quantify root activity in wheat [46••]. For a detailed comparison of ERT, EMI and penetrometer methods for measuring differences in soil water profiles at the plot scale between genotypes, see [46••].
Ground penetrating radar (GPR), another geophysical technique of similar throughput to EMI, uses high frequency radio waves to detect objects or boundaries between materials in the ground based on their permittivity. GPR, like EMI, has been used to detect and quantify tree roots, but does not currently have the resolution to detect roots less than 2 mm in diameter (reviewed in [47]). Despite this, GPR has recently been used to detect bulk root biomass in wheat and sugar cane (although with limited ability to detect differences between genotypes [48]) and shows potential as a future phenotyping tool, perhaps in combination with other geophysical sensors.
Advances in root image analysis
As high-throughput image capture of root systems has become mainstream and generates ever larger datasets, there is a requirement for fast and accurate software solutions to reliably derive traits. Until fairly recently, software has focused on 2D imaging paradigms, resulting in a large number of tools, exhibiting a mixture of manual (e.g. DART [49]), semi-automatic (e.g. SmartRoot [50], RootNav [51]), and fully automated (e.g. EZ-Rhizo [52], GiA Roots [53], DIRT [40]) approaches (Figure 3). Software in this area has traditionally relied heavily on the assumption that root images are consistent across an experiment, and that the root system exhibits high contrast against the background. Where this is the case, image thresholding to identify root material as reliably lighter or darker than the background has proven effective [51,53]. Thresholding alone can be susceptible to image noise, where image pixels are incorrectly assigned as either root material, or background. It has been common practice to perform corrective filtering of segmented images prior to analysis of the RSA. Morphological operations such as erosion and dilation can be used to correct small errors, and skeletonisation may be used to simplify the root structure to make topological analysis and trait measurement more straightforward. [52,53] are examples of tools that take this approach.
Figure 3.
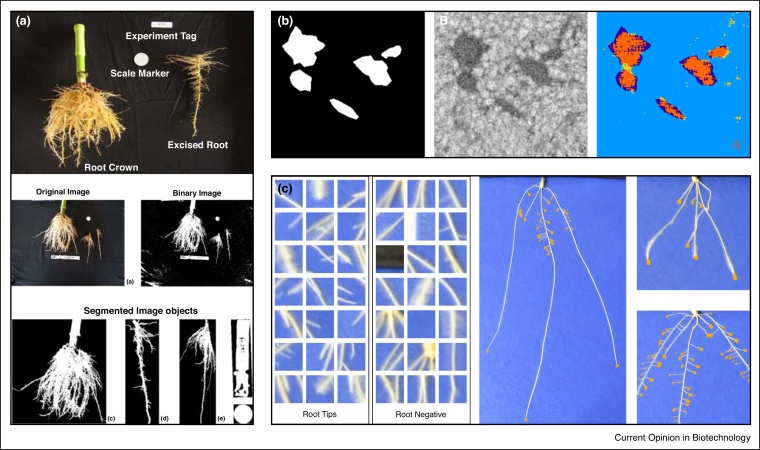
Automated root image analysis software. (a) DIRT [40] measures traits based on the ‘shovelomics’ approach [37]. Root systems are washed, and imaged from above in front of a dark background. Root systems are separated from background via thresholding, and RSA traits derived from each segmented object. (b) Root-soil segmentation in X-Ray CT [61]. Root and soil pixels are identified via a Support Vector Machine classifier trained on deep-learned features. Images show the ground-truth, original image, and SVM classifier output. (c) End-to-end deep learning for root tip identification [59•]. A deep network trained on thousands of instances of root tips and negative samples can be passed over an entire image to obtain likely root tip locations.
Following the identification of root material within an image, RSA traits such as width are easily derived. Some tools, such as EZRhizo [52] and WinRhizo [54] utilise pixel-distance transforms to approximate the width of each root, deriving a frequency distribution of root size within an image. These tools operate automatically but become less reliable where root systems exhibit complex topology, including bunched roots and crossovers. Some tools have attempted to “track” the root system, maintaining a link between the bases and apices of roots to form coherent geometries rather than isolated pixels. RootTrace [55], for example, uses a particle filtering framework to track Arabidopsis roots from base to apex on agar plates. SmartRoot [50] uses a semi-automatic approach to follow root material once first identified by a user. RootNav [51] is also semi-automated, seeking the shortest path on the image between user-identified seed and root tip locations.
3D root analysis software is less developed, with fewer tools available. Multi-view reconstruction from RGB images has been applied to roots grown in transparent media [2,3•]. For MRI data, [56] used Frangi filters to highlight tubular structures in the 3D image, before applying a shortest path search to obtain root topology. The root structure is then cleaned using a tree-pruning approach. In X-ray CT images, RooTrak [57] utilises a level set tracking approach to follow roots within a soil column and has been extended to handle multiple competing root systems [58].
Deep machine learning is becoming a standard technique for many computer vision problems as large annotated datasets become available. Within plant science, the majority of deep learning has been applied to plant shoots. For 2D root images, tip locations have been identified using a deep network-based classifier, scanned over an image to produce a location map [59•]. After training, deep learning algorithms delivered impressive improvements in tip detection (>99%) compared other approaches (<60%) [59•]. Deep learning thus has the potential to reduce the reliance on user-input to increase both the throughput and quality of phenotypic data [60]. For 3D images, deep learning has been applied to the root-soil segmentation problem, where deep learned features are used to drive a Support Vector Machine classifying root/soil pixels [61].
Future perspectives
This review has focused on recent advances in phenotyping plant root architecture, particularly in the field of non-destructive 3D imaging of root systems. The next challenge is to incorporate these techniques into phenotyping platforms with throughputs comparable to 2D systems to allow large-scale quantitative genetic studies. Selecting crops based on their root anatomical traits also represents a promising approach. For example, maize lines with increased root airspaces (root cortical aerenchyma; RCA) grown under drought stress in smallholder plots in Malawi had 78–143% greater yield than crops with less RCA [62•]. Similarly, root xylem diameter has been linked with conservation of water resources to aid grain filling in wheat [63•]. To date, screening for root anatomical traits is very time consuming. However, the recent development of high-throughput 2D and 3D anatomical phenotyping approaches employing microtoming [64] and laser ablation tomography [62•], respectively, makes it possible to profile the thousands of root samples required for breeding programmes.
Advances in data acquisition, handling and processing are becoming increasingly important to plant phenotyping [65]. Integrating innovative approaches such as deep learning into root phenotyping pipelines will require researchers to actively engage with computer scientists. Similar challenges and opportunities face researchers wanting to employ in silico models to probe how multiple root architectural and anatomical traits interact to impact plant performance. The development of open-source software such as OpenSimRoot and RootBox [66•,67,68] promises to greatly aid modelling efforts by root researchers and provide invaluable insight into the highly non-linear interactions between root phenotypes. Such multi-disciplinary approaches will underpin efforts to develop crops with improved root systems and help address the urgent need for future crops better adapted to the challenge of climate change.
Conflicts of interest statement
None declared.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
•• of outstanding interest
Acknowledgements
This work was supported by the Biotechnology and Biological Sciences Research Council [grant number BB/M001806/1, BB/L026848/1, BB/P026834/1] (MJB, DMW, and MPP) and Designing Future Wheat Institute Strategic Programme (JAA); the Leverhulme Trust [grant number RPG-2016-409] (MJB and DMW); the European Research Council FUTUREROOTS Advanced Investigator grant [grant number 294729] to JAA, DMW, and MJB; and the University of Nottingham Future Food Beacon of Excellence.
Contributor Information
Malcolm J Bennett, Email: malcolm.bennett@nottingham.ac.uk.
Darren M Wells, Email: darren.wells@nottingham.ac.uk.
References
- 1.Lynch J.P. Roots of the second green revolution. Aust J Bot. 2007;55:493–512. [Google Scholar]
- 2.Clark R.T., MacCurdy R.B., Jung J.K., Shaff J.E., McCouch S.R., Aneshansley D.J., Kochian L.V. Three-dimensional root phenotyping with a novel imaging and software platform. Plant Physiol. 2011;156:455–465. doi: 10.1104/pp.110.169102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3•.Topp C.N., Iyer-Pascuzzi A.S., Anderson J.T., Lee C.-R., Zurek P.R., Symonova O., Zheng Y., Bucksch A., Mileyko Y., Galkovskyi T. 3D phenotyping and quantitative trait locus mapping identify core regions of the rice genome controlling root architecture. Proc Natl Acad Sci U S A. 2013;110:E1695–E1704. doi: 10.1073/pnas.1304354110. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors developed a 3D phenotyping platform based on transparent media and used it to identify QTL for root architecture in rice.
- 4.Wang Q., Komarov S., Mathews A.J., Li K., Topp C., O’Sullivan J.A., Y-C Tai. 2014. Combined 3D PET and optical projection tomography techniques for plant root phenotyping. ArXiv150100242 Phys Q-Bio. [Google Scholar]
- 5••.Rellán-Álvarez R., Lobet G., Lindner H., Pradier P.-L., Sebastian J., Yee M.-C., Geng Y., Trontin C., LaRue T., Schrager-Lavelle A. GLO-Roots: an imaging platform enabling multidimensional characterization of soil-grown root systems. eLife. 2015;4:e07597. doi: 10.7554/eLife.07597. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors developed an innovative luciferase reporter based approach to image transgenic roots in rhizotrons. The GLO-ROOTS approach provides novel opportunities to study the impact of soil properties on root gene expression.
- 6.Kuijken R.C., van Eeuwijk F.A., Marcelis L.F., Bouwmeester H.J. Root phenotyping: from component trait in the lab to breeding. J Exp Bot. 2015;66:5389–5401. doi: 10.1093/jxb/erv239. [DOI] [PubMed] [Google Scholar]
- 7.Bonser A.M., Lynch J., Snapp S. Effect of phosphorus deficiency on growth angle of basal roots in Phaseolus vulgaris. New Phytol. 1996;132:281–288. doi: 10.1111/j.1469-8137.1996.tb01847.x. [DOI] [PubMed] [Google Scholar]
- 8.Hund A., Trachsel S., Stamp P. Growth of axile and lateral roots of maize: i development of a phenotying platform. Plant Soil. 2009;325:335–349. [Google Scholar]
- 9.Atkinson J.A., Wingen L.U., Griffiths M., Pound M.P., Gaju O., Foulkes M.J., Gouis J.L., Griffiths S., Bennett M.J., King J. Phenotyping pipeline reveals major seedling root growth QTL in hexaploid wheat. J Exp Bot. 2015;66:2283–2292. doi: 10.1093/jxb/erv006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gioia T., Galinski A., Lenz H., Müller C., Lentz J., Heinz K., Briese C., Putz A., Fiorani F., Watt M. GrowScreen-PaGe, a non-invasive, high-throughput phenotyping system based on germination paper to quantify crop phenotypic diversity and plasticity of root traits under varying nutrient supply. Funct Plant Biol. 2017;44:76–93. doi: 10.1071/FP16128. [DOI] [PubMed] [Google Scholar]
- 11.Passot S., Gnacko F., Moukouanga D., Lucas M., Guyomarc’h S., Ortega B.M., Atkinson J.A., Belko M.N., Bennett M.J., Gantet P. Characterization of pearl millet root architecture and anatomy reveals three types of lateral roots. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.00829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nagel K.A., Putz A., Gilmer F., Heinz K., Fischbach A., Pfeifer J., Faget M., Blossfeld S., Ernst M., Dimaki C. GROWSCREEN-Rhizo is a novel phenotyping robot enabling simultaneous measurements of root and shoot growth for plants grown in soil-filled rhizotrons. Funct Plant Biol. 2012;39:891–904. doi: 10.1071/FP12023. [DOI] [PubMed] [Google Scholar]
- 13.Subramanian R., Spalding E.P., Ferrier N.J. A high throughput robot system for machine vision based plant phenotype studies. Mach Vis Appl. 2013;24:619–636. [Google Scholar]
- 14.Le Marié C., Kirchgessner N., Flütsch P., Pfeifer J., Walter A., Hund A. RADIX: rhizoslide platform allowing high throughput digital image analysis of root system expansion. Plant Methods. 2016;12:40. doi: 10.1186/s13007-016-0140-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jeudy C., Adrian M., Baussard C., Bernard C., Bernaud E., Bourion V., Busset H., Cabrera-Bosquet L., Cointault F., Han S. RhizoTubes as a new tool for high throughput imaging of plant root development and architecture: test, comparison with pot grown plants and validation. Plant Methods. 2016;12 doi: 10.1186/s13007-016-0131-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Piñeros M.A., Larson B.G., Shaff J.E., Schneider D.J., Falcão A.X., Yuan L., Clark R.T., Craft E.J., Davis T.W., Pradier P.‐L. Evolving technologies for growing, imaging and analyzing 3D root system architecture of crop plants. J Integr Plant Biol. 2015;58:230–241. doi: 10.1111/jipb.12456. [DOI] [PubMed] [Google Scholar]
- 17.Hainsworth J.M., Aylmore L.A.G. The use of computer assisted tomography to determine spatial distribution of soil water content. Soil Res. 1983;21:435–443. [Google Scholar]
- 18.Mooney S.J., Pridmore T.P., Helliwell J., Bennett M.J. Developing X-ray computed tomography to non-invasively image 3-D root systems architecture in soil. Plant Soil. 2012;352:1–22. [Google Scholar]
- 19.Rogers E.D., Monaenkova D., Mijar M., Nori A., Goldman D.I., Benfey P.N. X-ray computed tomography reveals the response of root system architecture to soil texture. Plant Physiol. 2016;171:2028–2040. doi: 10.1104/pp.16.00397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bao Y., Aggarwal P., Robbins N.E., Sturrock C.J., Thompson M.C., Tan H.Q., Tham C., Duan L., Rodriguez P.L., Vernoux T. Plant roots use a patterning mechanism to position lateral root branches toward available water. Proc Natl Acad Sci U S A. 2014 doi: 10.1073/pnas.1400966111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Paya A.M., Silverberg J.L., Padgett J., Bauerle T.L. X-ray computed tomography uncovers root–root interactions: quantifying spatial relationships between interacting root systems in three dimensions. Front Plant Sci. 2015;6 doi: 10.3389/fpls.2015.00274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yang X., Varga T., Liu C., Scheibe T.D. What can we learn from in-soil imaging of a live plant: X-ray computed tomography and 3D numerical simulation of root-soil system. Rhizosphere. 2017;3:259–262. [Google Scholar]
- 23••.van Dusschoten D., Metzner R., Kochs J., Postma J.A., Pflugfelder D., Bühler J., Schurr U., Jahnke S. Quantitative 3D analysis of plant roots growing in soil using magnetic resonance imaging. Plant Physiol. 2016;170:1176–1188. doi: 10.1104/pp.15.01388. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors present a detailed study of the development and evaluation of an MRI system capable of automated imaging of roots of maize and barley in soil
- 24.Jahnke S., Menzel M.I., van Dusschoten D., Roeb G.W., Bühler J., Minwuyelet S., Blümler P., Temperton V.M., Hombach T., Streun M. Combined MRI-PET dissects dynamic changes in plant structures and functions. Plant J Cell Mol Biol. 2009;59:634–644. doi: 10.1111/j.1365-313X.2009.03888.x. [DOI] [PubMed] [Google Scholar]
- 25.Metzner R., van Dusschoten D., Bühler J., Schurr U., Jahnke S. Belowground plant development measured with magnetic resonance imaging (MRI): exploiting the potential for non-invasive trait quantification using sugar beet as a proxy. Front Plant Sci. 2014;5 doi: 10.3389/fpls.2014.00469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garbout A., Munkholm L.J., Hansen S.B., Petersen B.M., Munk O.L., Pajor R. The use of PET/CT scanning technique for 3D visualization and quantification of real-time soil/plant interactions. Plant Soil. 2012;352:113–127. [Google Scholar]
- 27.Zappala S., Helliwell J.R., Tracy S.R., Mairhofer S., Sturrock C.J., Pridmore T., Bennett M., Mooney S.J. Effects of X-ray dose on rhizosphere studies using X-ray computed tomography. PLOS ONE. 2013;8 doi: 10.1371/journal.pone.0067250. e67250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Metzner R., Eggert A., van Dusschoten D., Pflugfelder D., Gerth S., Schurr U., Uhlmann N., Jahnke S. Direct comparison of MRI and X-ray CT technologies for 3D imaging of root systems in soil: potential and challenges for root trait quantification. Plant Methods. 2015;11:17. doi: 10.1186/s13007-015-0060-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pflugfelder D., Metzner R., van Dusschoten D., Reichel R., Jahnke S., Koller R. Non-invasive imaging of plant roots in different soils using magnetic resonance imaging (MRI) Plant Methods. 2017;13:102. doi: 10.1186/s13007-017-0252-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zappala S., Mairhofer S., Tracy S., Sturrock C.J., Bennett M., Pridmore T., Mooney S.J. Quantifying the effect of soil moisture content on segmenting root system architecture in X-ray computed tomography images. Plant Soil. 2013;370:35–45. [Google Scholar]
- 31.Deery D.M., Rebetzke G.J., Jimenez-Berni J.A., James R.A., Condon A.G., Bovill W.D., Hutchinson P., Scarrow J., Davy R., Furbank R.T. Methodology for high-throughput field phenotyping of canopy temperature using airborne thermography. Front Plant Sci. 2016;7 doi: 10.3389/fpls.2016.01808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barker J., Zhang N., Sharon J., Steeves R., Wang X., Wei Y., Poland J. Development of a field-based high-throughput mobile phenotyping platform. Comput Electron Agric. 2016;122:74–85. [Google Scholar]
- 33.Bai G., Ge Y., Hussain W., Baenziger P.S., Graef G. A multi-sensor system for high throughput field phenotyping in soybean and wheat breeding. Comput Electron Agric. 2016;128:181–192. [Google Scholar]
- 34.Holman F.H., Riche A.B., Michalski A., Castle M., Wooster M.J., Hawkesford M.J. High throughput field phenotyping of wheat plant height and growth rate in field plot trials using uav based remote sensing. Remote Sens. 2016;8:1031. [Google Scholar]
- 35.Virlet N., Sabermanesh K., Sadeghi-Tehran P., Hawkesford M.J. Field Scanalyzer: An automated robotic field phenotyping platform for detailed crop monitoring. Funct Plant Biol. 2017;44:143–153. doi: 10.1071/FP16163. [DOI] [PubMed] [Google Scholar]
- 36.Wasson A., Bischof L., Zwart A., Watt M. A portable fluorescence spectroscopy imaging system for automated root phenotyping in soil cores in the field. J Exp Bot. 2016;67:1033–1043. doi: 10.1093/jxb/erv570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Trachsel S., Kaeppler S.M., Brown K.M., Lynch J.P. Shovelomics: high throughput phenotyping of maize (Zea mays L.) root architecture in the field. Plant Soil. 2011;341:75–87. [Google Scholar]
- 38.Burridge J., Jochua C.N., Bucksch A., Lynch J.P. Legume shovelomics: high-throughput phenotyping of common bean (Phaseolus vulgaris L.) and cowpea (Vigna unguiculata subsp, unguiculata) root architecture in the field. Field Crops Res. 2016;192:21–32. [Google Scholar]
- 39.York L.M., Slack S., Bennett M.J., Foulkes M.J. Wheat shovelomics I: a field phenotyping approach for characterising the structure and function of root systems in tillering species. bioRxiv. 2018 [Google Scholar]
- 40.Bucksch A., Burridge J., York L.M., Das A., Nord E., Weitz J.S., Lynch J.P. Image-based high-throughput field phenotyping of crop roots. Plant Physiol. 2014 doi: 10.1104/pp.114.243519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colombi T., Kirchgessner N., Marié C.A.L., York L.M., Lynch J.P., Hund A. Next generation shovelomics: set up a tent and REST. Plant Soil. 2015;388:1–20. [Google Scholar]
- 42.ARPA-E. ROOTS — Rhizosphere Observations Optimizing Terrestrial Sequestration; https://arpa-e.energy.gov/?q=arpa-e-programs/roots.
- 43.Amato M., Basso B., Celano G., Bitella G., Morelli G., Rossi R. In situ detection of tree root distribution and biomass by multi-electrode resistivity imaging. Tree Physiol. 2008;28:1441–1448. doi: 10.1093/treephys/28.10.1441. [DOI] [PubMed] [Google Scholar]
- 44.Srayeddin I., Doussan C. Estimation of the spatial variability of root water uptake of maize and sorghum at the field scale by electrical resistivity tomography. Plant Soil. 2009;319:185–207. [Google Scholar]
- 45.Shanahan P.W., Binley A., Whalley W.R., Watts C.W. The use of electromagnetic induction to monitor changes in soil moisture profiles beneath different wheat genotypes. Soil Sci Soc Am J. 2015;79:459–466. [Google Scholar]
- 46••.Whalley W.R., Binley A., Watts C.W., Shanahan P., Dodd I.C., Ober E.S., Ashton R.W., Webster C.P., White R.P., Hawkesford M.J. Methods to estimate changes in soil water for phenotyping root activity in the field. Plant Soil. 2017;415:407–422. doi: 10.1007/s11104-016-3161-1. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors present the first use of EMI to quantify genotypic differences in root activity, as well as a detailed comparison between ERT, EMI and penetrometer measurements.
- 47.Liu X., Dong X., Leskovar D.I. Ground penetrating radar for underground sensing in agriculture: a review. Int Agrophys. 2016;30:533–543. [Google Scholar]
- 48.Liu X., Dong X., Xue Q., Leskovar D.I., Jifon J., Butnor J.R., Marek T. Ground penetrating radar (GPR) detects fine roots of agricultural crops in the field. Plant Soil. 2018;423:517–531. [Google Scholar]
- 49.Bot J.L., Serra V., Fabre J., Draye X., Adamowicz S., Pagès L. DART: a software to analyse root system architecture and development from captured images. Plant Soil. 2010;326:261–273. [Google Scholar]
- 50.Lobet G., Pagès L., Draye X. A novel image-analysis toolbox enabling quantitative analysis of root system architecture. Plant Physiol. 2011;157:29–39. doi: 10.1104/pp.111.179895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pound M.P., French A.P., Atkinson J.A., Wells D.M., Bennett M.J., Pridmore T. RootNav: navigating images of complex root architectures. Plant Physiol. 2013;162:1802–1814. doi: 10.1104/pp.113.221531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Armengaud P. EZ-Rhizo software. Plant Signal Behav. 2009;4:139–141. doi: 10.4161/psb.4.2.7763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Galkovskyi T., Mileyko Y., Bucksch A., Moore B., Symonova O., Price C.A., Topp C.N., Iyer-Pascuzzi A.S., Zurek P.R., Fang S. GiA roots: software for the high throughput analysis of plant root system architecture. BMC Plant Biol. 2012;12:116. doi: 10.1186/1471-2229-12-116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Arsenault J.-L., Poulcur S., Messier C., Guay R. WinRHlZO™, a root-measuring system with a unique overlap correction method. Hort Science. 1995;30:906. [Google Scholar]
- 55.Naeem A., French A.P., Wells D.M., Pridmore T.P. High-throughput feature counting and measurement of roots. Bioinformatics. 2011;27:1337–1338. doi: 10.1093/bioinformatics/btr126. [DOI] [PubMed] [Google Scholar]
- 56.Schulz H., Postma J.A., van Dusschoten D., Scharr H., Behnke S. Computer Vision, Imaging and Computer Graphics. Theory and Application. Springer; Berlin, Heidelberg: 2013. Plant root system analysis from MRI images; pp. 411–425. [Google Scholar]
- 57.Mairhofer S., Zappala S., Tracy S.R., Sturrock C., Bennett M., Mooney S.J., Pridmore T. RooTrak: automated recovery of three-dimensional plant root architecture in soil from X-ray microcomputed tomography images using visual tracking. Plant Physiol. 2012;158:561–569. doi: 10.1104/pp.111.186221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mairhofer S., Sturrock C.J., Bennett M.J., Mooney S.J., Pridmore T.P. Extracting multiple interacting root systems using X-ray microcomputed tomography. Plant J. 2015;84:1034–1043. doi: 10.1111/tpj.13047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59•.Pound M.P., Atkinson J.A., Townsend A.J., Wilson M.H., Griffiths M., Jackson A.S., Bulat A., Tzimiropoulos G., Wells D.M., Murchie E.H. Deep machine learning provides state-of-the-art performance in image-based plant phenotyping. GigaScience. 2017;6:1–10. doi: 10.1093/gigascience/gix083. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors report the use of Deep Learning approaches to automate root and canopy phenotyping image analysis tasks.
- 60.Atkinson J.A., Lobet G., Noll M., Meyer P.E., Griffiths M., Wells D.M. Combining semi-automated image analysis techniques with machine learning algorithms to accelerate large-scale genetic studies. GigaScience. 2017;6:1–7. doi: 10.1093/gigascience/gix084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Douarre C., Schielein R., Frindel C., Gerth S., Rousseau D. Deep learning based root-soil segmentation from X-ray tomography. bioRxiv. 2016 [Google Scholar]
- 62•.Chimungu J.G., Maliro M.F.A., Nalivata P.C., Kanyama-Phiri G., Brown K.M., Lynch J.P. Utility of root cortical aerenchyma under water limited conditions in tropical maize (Zea mays L.) Field Crops Res. 2015;171:86–98. [Google Scholar]; The authors report how an anatomical scale root (cortical aerenchyma) trait has a major impact to improve yield stability under drought conditions.
- 63•.Richards R.A., Passioura J.B. A breeding program to reduce the diameter of the major xylem vessel in the seminal roots of wheat and its effect on grain yield in rain-fed environments. Aust J Agric Res. 1989;40:943–950. [Google Scholar]; The authors report how an anatomical scale root (xylem diameter) trait has a major impact to improve yield stability under drought conditions.
- 64.Atkinson J.A., Wells D.M. An updated protocol for high throughput plant tissue sectioning. Front Plant Sci. 2017;8 doi: 10.3389/fpls.2017.01721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Tardieu F., Cabrera-Bosquet L., Pridmore T., Bennett M. Plant phenomics, from sensors to knowledge. Curr Biol. 2017;27:R770–R783. doi: 10.1016/j.cub.2017.05.055. [DOI] [PubMed] [Google Scholar]
- 66•.Postma J.A., Kuppe C., Owen M.R., Mellor N., Griffiths M., Bennett M.J., Lynch J.P., Watt M. OpenSimRoot: widening the scope and application of root architectural models. New Phytol. 2017;215:1274–1286. doi: 10.1111/nph.14641. [DOI] [PMC free article] [PubMed] [Google Scholar]; The authors describe how they have enabled the SimRoot root architecture model to become an open-source platform better able to be used by life science researchers.
- 67.Leitner D., Klepsch S., Bodner G., Schnepf A. A dynamic root system growth model based on L-Systems. Plant Soil. 2010;332:177–192. [Google Scholar]
- 68.Schnepf A., Leitner D., Landl M., Lobet G., Mai T.H., Morandage S., Sheng C., Zörner M., Vanderborght J., Vereecken H. CRootBox: a structural–functional modelling framework for root systems. Ann Bot. 2018;121:1033–1053. doi: 10.1093/aob/mcx221. [DOI] [PMC free article] [PubMed] [Google Scholar]