Abstract
The Zika virus presents a major public health concern due to severe fetal neurological disorders associated with infections in pregnant women. In addition to vaccine development, the discovery of selective antiviral drugs is essential to combat future epidemic Zika virus outbreaks. The Zika virus NS2B-NS3 protease, which performs replication-critical cleavages of the viral polyprotein, is a promising drug target. We report the first macrocyclic peptide-based inhibitors of the NS2B-NS3 protease, discovered de novo through in vitro display screening of a genetically reprogrammed library including noncanonical residues. Six compounds were selected, resynthesized, and isolated, all of which displayed affinities in the low nanomolar concentration range. Five compounds showed significant protease inhibition. Two of these were validated as hits with submicromolar inhibition constants and selectivity toward Zika over the related proteases from dengue and West Nile viruses. The compounds were characterized as noncompetitive inhibitors, suggesting allosteric inhibition.
Keywords: Flavivirus, macrocyclic peptides, protease inhibitors, noncanonical amino acids, display screening
In recent years, the Zika virus has emerged from a neglected member of the flavivirus genus to a health-threatening pathogen.1 Although most infections are asymptomatic, neurological complications such as the Guillain–Barré syndrome have been reported in a small proportion of patients. The link between severe disorders in fetuses (microcephaly) and Zika virus infections in pregnant woman prompted the WHO to declare Zika virus a Public Health Emergency of International Concern in 2016.1,2 Since then, the virus has circulated in almost all Caribbean and Latin American countries, continental USA (e.g., Florida and Texas), Africa, Southeast Asia, and several Pacific islands with infections reported in 84 countries and territories around the globe. In contrast to other prominent examples of flaviviruses, such as dengue or West Nile, the Zika virus can be transmitted not only by Aedes aegypti and Aedes albopictus mosquitos, but also through sexual contact.1,2 About 30 potential Zika virus vaccines are currently being evaluated, out of which only four have entered phase 1 clinical trials.2 Particularly concerning for Zika (and dengue) virus vaccination campaigns are potential cross-reactions between Zika and dengue virus antibodies, where the resulting antibody-dependent enhancements can lead to increased viremia and severity of the disease, as observed previously for consecutive infections with different dengue virus serotypes.2 Therefore, alternative specific antiviral therapeutic options are needed for the treatment of symptomatic patients and infected pregnant women.
Like other flaviviruses, Zika virus comprises a single-stranded positive sense RNA genome that encodes a viral polyprotein, which is post-translationally processed by host-cell proteases and the viral NS2B-NS3 protease into three structural (C, prM/M, E) and seven nonstructural proteins (NS1, NS2A, NS2B, NS3, NS4A, NS4B, NS5). The NS2B-NS3 protease of flaviviruses is considered a promising antiviral drug target, and several lead compounds have already been discovered for the corresponding dengue virus proteases.3 NS2B-NS3 is a serine protease, which consists of the N-terminal domain of NS3 and a short cofactor from the hydrophilic core sequence of NS2B. For in vitro screening campaigns, three different Zika virus NS2B-NS3 protease (ZIKVpro) constructs have been proposed and crystallized. First, a construct with a covalent linker peptide between NS2B and NS3 (gZiPro) was adopted based on previous successful dengue and West Nile virus protease constructs.4 Two additional unlinked versions have been described, which are based on either NS2B/NS3 coexpression (bZiPro)5 or an autocleavage site in the linker peptide between NS2B and NS3 (eZiPro).6 The C-terminal tetrapeptide of NS2B in eZiPro was shown to interfere with access of substrate to the active site.6
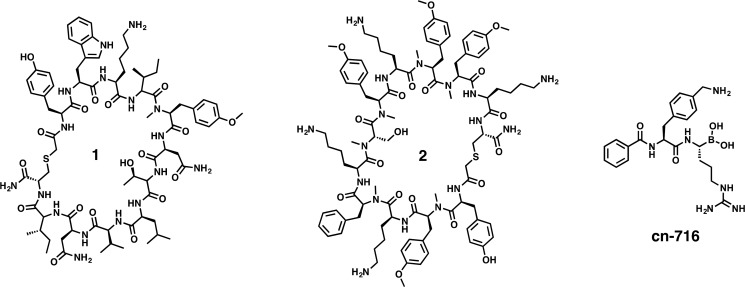
Few ZIKVpro inhibitors have been described so far.7 The most effective ones are substrate-derived peptide analogs that bind covalently to the catalytically active serine residue (e.g., cn-716, Figure 2).4,8,9 However, due to conserved features in substrate recognition among serine proteases, these compounds display only poor selectivity between flaviviral and host proteases.8 Therefore, alternative non-active-site inhibitors that do not mimic the substrate or transition state may exhibit decreased off-target effects. Recently, natural products as well as compounds derived from previous West Nile virus screening campaigns have been reported to act as micromolar allosteric inhibitors of ZIKVpro.10,11 In view of the limited coverage of chemical space offered by natural products and compound libraries, as well as common bias for promiscuous binders, we set out to identify completely new structural scaffolds de novo by capitalizing on recent advances in mRNA display techniques.
Figure 2.
Chemical structures of synthesized hit compounds 1 and 2 that were identified as nanomolar noncompetitive inhibitors of the Zika virus NS2B-NS3 protease. Compound cn-716 is a previously published4 covalent active-site inhibitor of the Zika virus NS2B-NS3 protease, which has been used in this study for comparison.
Small (<2 kDa) macrocyclic peptides are appealing starting points for such drug discovery. A key strength of macrocyclic peptides is that high-affinity ligands can be isolated for nearly any target rapidly using display screening approaches (phage display, mRNA display, etc.).12 Moreover, display screening can be combined with genetic code reprogramming techniques, allowing the screening of libraries incorporating structural characteristics such as backbone N-methylation and/or d-stereochemistry, which have been shown to improve biostability and membrane permeability, and therefore making the resulting hit compounds better starting points for drug discovery efforts than canonical peptides.12−14 In the present study, we have used such a screening approach (Random nonstandard Peptide Integrated Discovery, RaPID; Figure S1) to identify potent macrocyclic peptide ligands of ZIKVpro. These peptide ligands were specific noncompetitive inhibitors for ZIKVpro, and several were found to display submicromolar inhibition constants (Ki). This study highlights the ability of such screening approaches to rapidly identify potent inhibitors of newly discovered targets such as those from emerging viral diseases.
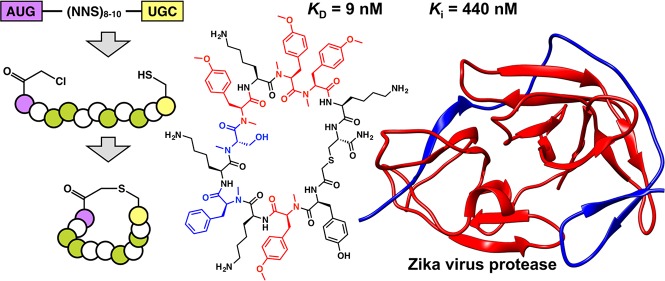
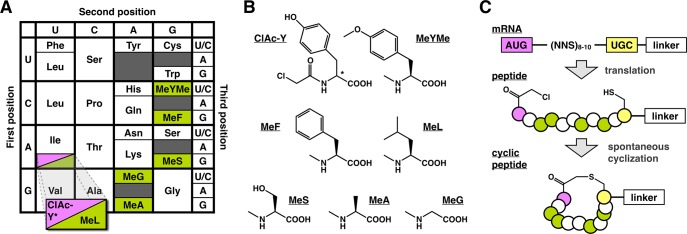
In order to identify novel inhibitors of ZIKVpro, we designed two macrocyclic peptide libraries for affinity screening by RaPID display. The libraries were constructed using a genetically reprogrammed in vitro translation approach and were designed to include five N-methyl residues (MeG, MeL, MeA, MeS, MeF) and the unnatural amino acid N-methyl-4-O-methyl-tyrosine (MeYMe) (Figure 1A,B). Each library was initiated with either l- or d-N-chloroacetyl tyrosine, in order to induce spontaneous macrocyclization following translation (Figure 1C).15 The genetic code was designed such that the noncanonical amino acids replaced Met (allowing for reprogramming of translation initiation) and all of the canonical residues except Lys, which have charged side chains at neutral pH (i.e., Asp, Glu, and Arg). Additionally, the random region of the peptide library was limited to a maximum of 10 codons, making the overall library both less charged and smaller than any previously reported comparable cyclic peptide libraries.12,13,16−21 The total theoretical diversity of both libraries combined exceeded 5 × 1013 macrocyclic peptides, making the total number of compounds screened several orders of magnitude greater than is typical using conventional high-throughput methods.
Figure 1.
Design of macrocyclic peptide libraries for screening. The peptide libraries for screening were synthesized by genetically reprogrammed translation of a new mRNA library that was more focused on hydrophobic residues than previous libraries. The codon table for the genetic code used is shown in panel A with Met replaced with N-chloroacetyl-tyrosine (ClAc-Y) for the initiation AUG codon or N-methyl-leucine (MeL) for all downstream AUG codons. (B) Structures of the noncanonical amino acids used. (C) Peptide libraries were synthesized by translation of a semirandom mRNA template comprising an AUG start codon, 8–10 NNS codons (N = A, G, C or U; S = G or C), a UGC (Cys) codon, and a linker sequence for covalent linkage of each peptide and mRNA. Translation of this library under the genetic code shown leads to formation of a semirandomized peptide library that cyclizes spontaneously to produce a macrocyclic peptide library. *Two libraries were synthesized, one initiated with ClAc-l-Y and one initiated with ClAc-d-Y.
Iterative affinity screening of these libraries against the linked ZIKVpro construct (gZiPro) immobilized on magnetic beads led to the identification of six families of macrocyclic peptide ligands (alignments of the 100 most frequent sequences from each library are shown in Figure S2). The most abundant member of each of these was synthesized by solid phase methodology omitting the C-terminal linker sequence (i.e., all residues C-terminal of the cyclizing cysteine). All six of these displayed high affinity for ZIKVpro with dissociation constants (KD) in the range of 5–168 nM as determined by surface plasmon resonance experiments (Table 1, Table S1 and Figure S4). All of these compounds contain at least one unnatural modification in the peptide backbone and five out of six include the unnatural amino acid N-methyl-4-O-methyl-tyrosine (MeYMe), suggesting that these features are crucial for high affinity.
Table 1. Affinity and Inhibitory Activity of Macrocyclic Peptides against Flaviviral Proteases.
| ZIKV (gZiPro)b |
IC50/μM |
|||||
|---|---|---|---|---|---|---|
| Cpd. | sequencea | Ki/μMc (IC50/μM) | KD/μMd | ZIKV (bZiPro)e | DENVprof | WNVprog |
| 1 | -S-Ac-YWKIMeYMeNTLVNIC-NH2 | 0.80 ± 0.08 | 0.02 | 1.5 ± 0.1 | >10 | >10 |
| (1.32 ± 0.03) | ||||||
| 2 | -S–Ac-YMeYMeKMeFKMeSMeYMeKMeYMeMeYMeKC-NH2 | 0.44 ± 0.03 | 0.009 | 0.25 ± 0.01 | 1.7 ± 0.1 | 3.9 ± 0.4 |
| (0.62 ± 0.04) | ||||||
| 3 | -S-Ac-YTNFYLYPYMeYMeFC-NH2 | 2.0 ± 0.2 | 0.008 | 2.2 ± 0.1 | >20 | >20 |
| (3.3 ± 0.2) | ||||||
| 4 | -S–Ac-YMeGIAKYNMeYMeMeYMeIPC-NH2 | 20.4 ± 2.3 | 0.009 | 4.6 ± 0.3 | ≥20 | ≥20 |
| (16.5 ± 0.9) | ||||||
| 5 | -S-Ac-YTLPFHNMeGTFFC-NH2 | >100 | 0.005 | ≥50 | >20 | >20 |
| 6 | -S-Ac-d-YAIIMeYMeYNKYMeLNC-NH2 | 3.5 ± 0.4 | 0.168 | 3.2 ± 0.4 | >20 | >20 |
| (7.1 ± 0.2) | ||||||
The N-terminal thioether-acyl moiety (−S-Ac−) forms a macrocycle through the side chain of the underlined cysteine residue. MeYMe, N-methyl-4-O-methyl-tyrosine; MeF, N-methyl-phenylalanine; MeS, N-methyl-serine; MeG, N-methyl-glycine; MeL, N-methyl-leucine; d-Y, d-tyrosine.
Zika virus NS2B-NS3 protease (linked construct) C80S/C143S (1 nM). Substrate: Bz-Nle-Lys-Lys-Arg-AMC.
Inhibition constants (Ki) were calculated from measurements at four different substrate concentrations using a noncompetitive inhibition model (nonlinear least-squares fits). Half maximal inhibitory concentrations (IC50) for a substrate concentration of 15 μM are reported in parentheses.
Dissociation constants (KD) were determined by surface plasmon resonance using immobilized Zika virus NS2B-NS3 protease (gZiPro).
Zika virus NS2B-NS3 protease (unlinked construct; 0.5 nM). Substrate: Bz-Nle-Lys-Lys-Arg-AMC. Half maximal inhibitory concentrations (IC50) were calculated for a substrate concentration of 15 μM.
Dengue virus serotype 2 NS2B-NS3 protease (100 nM). Substrate: Abz-Nle-Lys-Arg-Arg-Ser-3-(NO2)Tyr. Half maximal inhibitory concentrations (IC50) were calculated for a substrate concentration of 50 μM.
West Nile virus NS2B-NS3 protease (150 nM). Substrate: Abz-Gly-Leu-Lys-Arg-Gly-Gly-3-(NO2)Tyr. Half maximal inhibitory concentrations (IC50) were calculated for a substrate concentration of 50 μM.
The selected compounds were further analyzed with respect to their actual potential to inhibit the catalytic activity of the linked ZIKVpro construct (gZiPro) in a biochemical assay. All compounds except 5 showed significant inhibition of ZIKVpro substrate processing below a 100 μM cutoff (Table 1, Figure S5). Notably, 5 is the only analyzed peptide macrocycle that lacks the unnatural O-methyl-tyrosine side chain. Despite having no influence on catalytic activity, compound 5 binds to ZIKVpro with extraordinary affinity (KD = 5 nM), indicating a tight binding event that is likely to be distant from the substrate binding site without any impact on the structural integrity of the catalytic center. Therefore, compound 5 linked to any reporter molecules (e.g., via a C-terminal amide bond) might have applications in selective probing of ZIKVpro without perturbing its activity. This exemplifies a major advantage of the presented affinity screening over conventional high-throughput screening methods for protease inhibitors, which usually focus on inhibition only.
To exclude any construct-specific artifacts, we further assessed the activity of the unlinked ZIKVpro construct (bZiPro) in the presence of compounds 1–6 using the same biochemical assay. The IC50 values observed for gZiPro and bZiPro were mostly similar (Table 1, Figure S6), indicating that the linked (gZiPro) and unlinked (bZiPro) constructs are equally suitable templates for drug discovery campaigns. Compounds 1 and 2 displayed the strongest inhibition of ZIKVpro with inhibition constants firmly in the nanomolar range (Table 1). These two macrocyclic peptides are structurally unrelated (Figure 2) and may therefore serve as independent starting points for further optimizations. In contrast to the previously reported ZIKVpro active-site inhibitor cn-716, which showed strong inhibition of thrombin (IC50 = 0.5 μM) and trypsin (IC50 = 0.05 μM),8 neither compound 1 nor 2 displayed thrombin or trypsin inhibition at the highest assayed concentrations of 25 and 50 μM, respectively (data not shown), highlighting their specificity toward ZIKVpro.
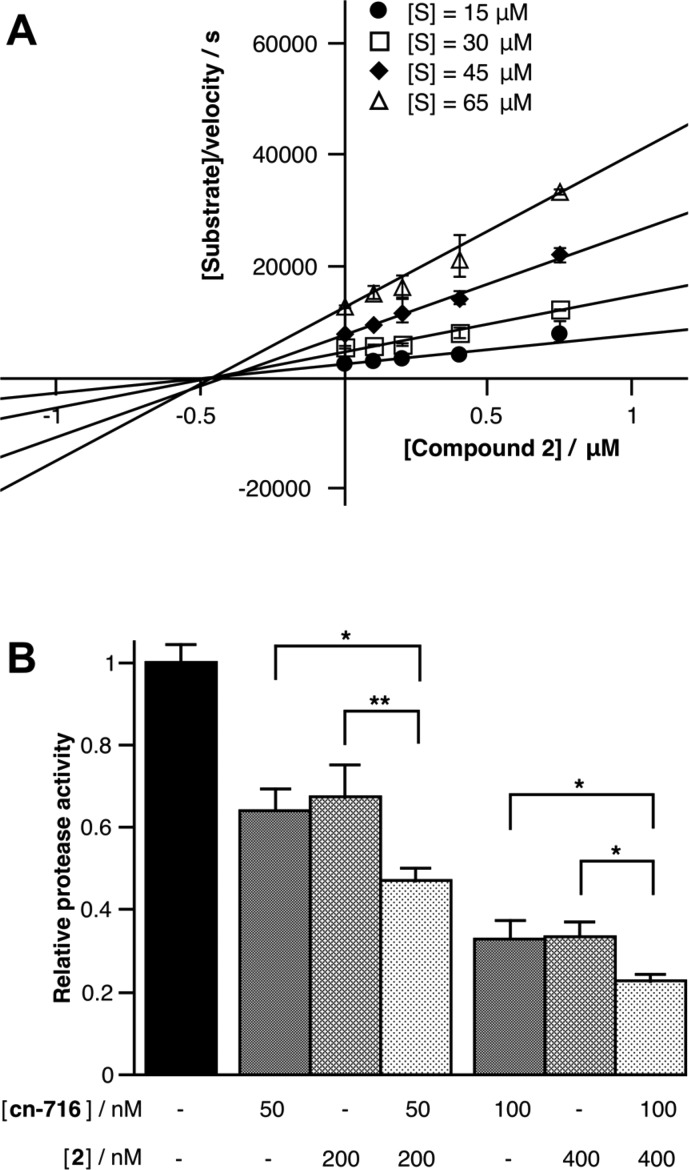
Mechanistic studies with gZiPro at various substrate and inhibitor concentrations revealed a noncompetitive inhibition mode for compounds 1–4 and 6, as evidenced both by nonlinear least-squares data fits (Table S3) and Cornish–Bowden plots. As an example, Figure 3A shows the Cornish–Bowden plot for the most active peptide 2, where the abscissa intersection point clearly indicates noncompetitive binding.22 As expected for the noncompetitive binding model, the degree of inhibition was independent of the substrate concentration, even at multiples of the Km value. Due to the size of substrate and macrocyclic inhibitor, both cannot occupy the active site simultaneously, suggesting that compounds 1–4 and 6 may act as allosteric antagonists of ZIKVpro activity. Allosteric inhibitors have been reported previously for dengue virus NS2B-NS3 proteases at different sites of NS3,23,24 which may also act by perturbing the essential interactions between NS2B and NS3.25 The Ki values were all larger than the KD values determined by SPR (Table 1), suggesting induced conformational changes in the protease that affect catalysis to different degrees (in the most extreme case, compound 5 binds with high affinity but does not affect the catalytic rate at all). This observation is consistent with the hypothesis that the peptides might be allosteric inhibitors.
Figure 3.
(A) Cornish–Bowden plot for compound 2 and Zika virus NS2B-NS3 protease (gZiPro). The intersection point (−Ki) of the four independent linear fits with the abscissa indicates a noncompetitive inhibition mechanism with a Ki value of 0.44 μM.22 The same value was obtained from nonlinear least-squares fits using a noncompetitive binding model (Graphpad Prism 7.0; Table S3). (B) Inhibition of Zika virus NS2B-NS3 protease (bZiPro) activity by the noncompetitive inhibitor 2 and the covalently binding active-site inhibitor cn-716. Addition of compound 2 in the presence of cn-716 results in synergistically decreased protease activity, suggesting that 2 binds at an allosteric site.
Although, in principle, resistance may easily evolve against allosteric inhibitors of viral proteases, such inhibitors can be valuable therapeutics in combination with substrate-derived active-site inhibitors. Established reverse transcriptase inhibitors for the treatment of chronic HIV infections are a prominent example.26 As drug-like active-site inhibitors of flaviviral proteases suffer from low affinity, their combination with allosteric antagonists is a promising route toward antiflaviviral therapeutics with adequate pharmacokinetic properties. To support the potential allosteric binding mode indicated by the noncompetitive inhibition data, we performed the activity assay with bZiPro in the presence and absence of compound 2 and the covalent active-site inhibitor cn-716 (Figure 3B). As expected, the simultaneous presence of inhibitor 2 and cn-716 inhibited the ZIKVpro activity more strongly than the individual compounds at identical concentrations, indicating synergistic inhibition of ZIKVpro (Figure 3B). As cn-716 is known to occupy the active site by binding covalently to the active-site serine residue 135,4 this is strong evidence for compound 2 binding to a site distant from the substrate-binding site. Unfortunately, attempts to verify the allosteric binding mode by NMR spectroscopy or X-ray crystallography have failed to date because of the low solubility of the inhibitors.
To further assess the specificity of the discovered inhibitors, we tested their performance with dengue and West Nile virus proteases (Table 1). Only compound 2 showed significant inhibition of both proteases in the low micromolar range with selectivity indices of 4 and 9 in favor of ZIKVpro over the dengue and West Nile virus proteases, respectively. Compounds 1 and 3 displayed selectivity indices greater than 10 in favor of ZIKVpro. Only compound 4 showed similarly weak inhibition constants around 20 μM for all three viral proteases. The selectivity of our new inhibitors toward ZIKVpro is in stark contrast to the broad cross-reactivity of previously described flaviviral protease inhibitors, which are predominantly active-site inhibitors.7 As the active center is highly conserved, selective targeting has been difficult in the past. Consequently, several compounds that were previously discovered as inhibitors of dengue and West Nile virus proteases have also been reported to inhibit ZIKVpro or even human serine proteases.7 The completely new structural motifs of the present study thus not only allow an increase in inhibition by noncompetitive interactions but also selective probing of ZIKVpro.
The most active compound 2 (Ki = 0.44 μM, KD = 0.009 μM; gZiPro) exhibits multiple noncanonical structural features. Half of all backbone nitrogen atoms of peptide 2 are methylated, and it contains four unnatural O-methyl-tyrosine side chains, representing a chemical complexity that would require several optimization cycles underpinned by successful structure–activity relationships if starting from a canonical peptide library. None of the selected screening hits showed evidence for competitive inhibition at the active site of the protease. This bias toward the enrichment of sequences that may target exosites rather than the active site of ZIKVpro suggests that active-site interactions of macrocycles were of significantly lower affinity during the screening (preventing enrichment of those sequences). This in turn may suggest low druggability of the active site of ZIKVpro (and closely related flaviviruses) by classical competitive inhibitors. This conclusion is supported by a previous study that aimed to identify high-affinity active-site inhibitors in a library of substrate analogues, where interactions with key residues in the active site could be established, but only an additional covalent bond with the catalytic serine residue yielded affinities in the nanomolar range (as in cn-716, Figure 2).4,8 Alternatively, the absence of competitive inhibitors in the present work may be related to the deliberate exclusion of arginine (to avoid pharmacokinetic liabilities). All flaviviral proteases share the recognition of basic residues (arginine and lysine) in the nonprime substrate site with a particular preference for arginine in P1, and most peptide-based inhibitors address this recognition motif.3,8,9,27−30 By excluding arginine from our screening library, we not only avoided an amino acid with a potentially negative impact on drug-likeness but apparently also favored the selection of noncompetitive inhibitors. Therefore, the observed bias may simply be attributed to the experimental setup.
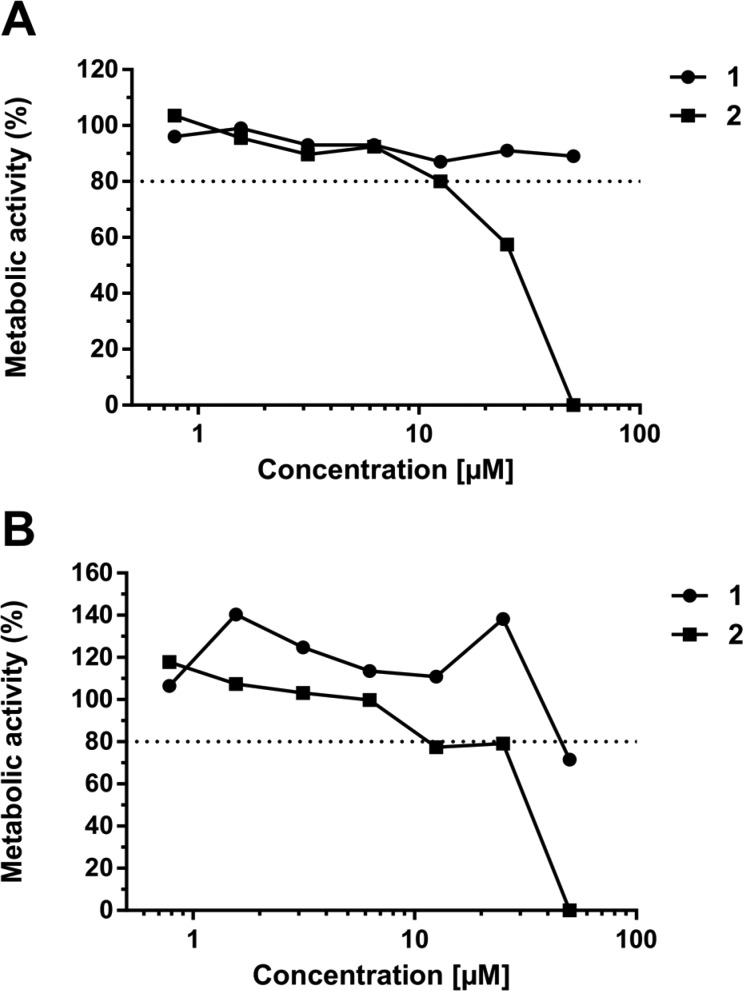
The two most active compounds from this screening campaign, 1 and 2, displayed no cytotoxic effects at concentrations up to 12.5 μM after 24 and 48 h of incubation with Huh-7 cells (Figure 4). At higher concentrations, cytotoxic effects became apparent only for compound 2. In summary, both macrocyclic peptides, 1 and 2, combine nanomolar dissociation and inhibition constants with unusual (noncompetitive) inhibition mechanisms, excellent selectivity against human proteases, and low cytotoxicity. Our study demonstrates that display screenings offer an excellent way to rapidly generate selective probes and tool compounds that can modulate the enzymatic activity of the target protein from fully active (5) to highly inhibited (2). Future attempts will focus on further derivatizations of the identified and validated hits 1 and 2 to allow deeper insights into structure–activity relationships and facilitate in-cell studies and the generation of structural data by X-ray crystallography and NMR spectroscopy. These studies will capitalize on chemical opportunities provided by the exocyclic C-termini of these compounds that can readily be tagged with reporter molecules or moieties that modulate solubility or cellular uptake without altering the macrocyclic peptide structure.
Figure 4.
Effect of compounds 1 and 2 on the metabolic activity of Huh-7 cells (% reduction compared to nontreated). The dotted lines indicate an 80% cell-viability cutoff. Neither compound shows significant cytotoxic effects at concentrations up to 12.5 μM: (A) 24 h and (B) 48 h incubation.
In conclusion, we present the first macrocyclic peptide inhibitors of ZIKVpro identified through a RaPID screening technique that is orthogonal to conventional high-throughput approaches. With a single selection screen and no further optimizations, we identified unnatural peptides of remarkable affinity, inhibitory activity, and target selectivity. Most importantly, our results indicate that noncompetitive inhibitors can be made that are highly active and may present excellent options for probes, tools, and potential drug candidates in comparison to active-site inhibitors that have been unsuccessfully pursued for a very long time. Future work will assess whether a combination of competitive and noncompetitive inhibitors indeed offers a superior approach for the development of antiviral agents against Zika, dengue, and related viral infectious diseases.
Acknowledgments
C.N. thanks the Alexander von Humboldt Foundation for a Feodor Lynen Research Fellowship. Financial support by the Australian Research Council, including a Laureate Fellowship for G.O. is gratefully acknowledged. This work was also partially supported by CREST for Molecular Technologies, JST, and JSPS KAKENHI (16H06444 and 26220204) to H.S. C.K. acknowledges support by the Deutsche Forschungsgemeinschaft (KL-1356/3-2). We thank Mrs. Natascha Stefan for technical support.
Biographies
Christoph Nitsche studied chemistry and business administration. He obtained his Ph.D. from Heidelberg University on the discovery of dengue virus protease inhibitors in 2014, for which he was awarded the Ph.D. prize for medicinal chemistry by the German Chemical Society (2015). He has since worked as a Feodor Lynen Fellow at the Australian National University (2015–2018) and as a Rising Star Fellow at the Free University of Berlin (2018–2019). In 2019, he received a Discovery Early Career Research Award (DECRA) from the Australian Research Council to start his research group on new technologies for drug discovery and bioconjugation.
Hiroaki Suga is a Professor of the Department of Chemistry, Graduate School of Science in the University of Tokyo. He received Ph.D. at MIT (1994) followed by post-doctoral fellow in Mass General Hospital. He was Assistant and tenured Associate Professor in the SUNY at Buffalo (1997–2003) and Professor in the Research Center for Advanced Science and Technology in the University of Tokyo (2003–2010). Since 2010, he has remained in his current position. He is the recipient of Akabori Memorial Award 2014, Max-Bergmann Medal 2016, and Nagoya Medal Silver 2017. He is also a founder of PeptiDream Inc., Tokyo.
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00535.
Experimental details for Zika, dengue, and West Nile virus protease constructs and preparation, synthesis and screening of macrocyclic peptide library, solid phase peptide synthesis, surface plasmon resonance, enzymatic protease assays (Zika, dengue, West Nile, thrombin, trypsin), cellular assays. Figure S1 (RaPID screening process), Figure S2 (alignment of peptide sequences), Figure S3 (MS data), Figure S4 (SPR binding curves), Figures S5 and S6 (dose–response curves), Figure S7 (Huh-7 cell viability data), Table S1 (SPR kinetic parameters), Table S2 (DNA oligonucleotide sequences), Table S3 (nonlinear least-squares fitting parameters) (PDF)
Author Contributions
# These authors contributed equally. All authors have given approval to the final version of the manuscript.
The authors declare no competing financial interest.
Supplementary Material
References
- Baud D.; Gubler D. J.; Schaub B.; Lanteri M. C.; Musso D. An update on Zika virus infection. Lancet 2017, 390, 2099–2109. 10.1016/S0140-6736(17)31450-2. [DOI] [PubMed] [Google Scholar]
- Poland G. A.; Kennedy R. B.; Ovsyannikova I. G.; Palacios R.; Ho P. L.; Kalil J. Development of vaccines against Zika virus. Lancet Infect. Dis. 2018, 18, e211–e219. 10.1016/S1473-3099(18)30063-X. [DOI] [PubMed] [Google Scholar]
- Nitsche C.; Holloway S.; Schirmeister T.; Klein C. D. Biochemistry and medicinal chemistry of the dengue virus protease. Chem. Rev. 2014, 114, 11348–11381. 10.1021/cr500233q. [DOI] [PubMed] [Google Scholar]
- Lei J.; Hansen G.; Nitsche C.; Klein C. D.; Zhang L.; Hilgenfeld R. Crystal structure of Zika virus NS2B-NS3 protease in complex with a boronate inhibitor. Science 2016, 353, 503–505. 10.1126/science.aag2419. [DOI] [PubMed] [Google Scholar]
- Zhang Z.; Li Y.; Loh Y. R.; Phoo W. W.; Hung A. W.; Kang C.; Luo D. Crystal structure of unlinked NS2B-NS3 protease from Zika virus. Science 2016, 354, 1597–1600. 10.1126/science.aai9309. [DOI] [PubMed] [Google Scholar]
- Phoo W. W.; Li Y.; Zhang Z.; Lee M. Y.; Loh Y. R.; Tan Y. B.; Ng E. Y.; Lescar J.; Kang C.; Luo D. Structure of the NS2B-NS3 protease from Zika virus after self-cleavage. Nat. Commun. 2016, 7, 13410. 10.1038/ncomms13410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche C.Strategies towards protease inhibitors for emerging flaviviruses. In Dengue and Zika: Control and Antiviral Treatment Strategies; Hilgenfeld R., Vasudevan S. G., Eds.; Springer: Singapore, 2018; pp 175–186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitsche C.; Zhang L.; Weigel L. F.; Schilz J.; Graf D.; Bartenschlager R.; Hilgenfeld R.; Klein C. D. Peptide–boronic acid inhibitors of flaviviral proteases: medicinal chemistry and structural biology. J. Med. Chem. 2017, 60, 511–516. 10.1021/acs.jmedchem.6b01021. [DOI] [PubMed] [Google Scholar]
- Li Y.; Zhang Z.; Phoo W. W.; Loh Y. R.; Wang W.; Liu S.; Chen M. W.; Hung A. W.; Keller T. H.; Luo D.; Kang C. Structural dynamics of Zika virus NS2B-NS3 protease binding to dipeptide inhibitors. Structure 2017, 25, 1242–1250. 10.1016/j.str.2017.06.006. [DOI] [PubMed] [Google Scholar]
- Roy A.; Lim L.; Srivastava S.; Lu Y.; Song J. Solution conformations of Zika NS2B-NS3pro and its inhibition by natural products from edible plants. PLoS One 2017, 12, e0180632 10.1371/journal.pone.0180632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiryaev S. A.; Farhy C.; Pinto A.; Huang C. T.; Simonetti N.; Elong Ngono A.; Dewing A.; Shresta S.; Pinkerton A. B.; Cieplak P.; Strongin A. Y.; Terskikh A. V. Characterization of the Zika virus two-component NS2B-NS3 protease and structure-assisted identification of allosteric small-molecule antagonists. Antiviral Res. 2017, 143, 218–229. 10.1016/j.antiviral.2017.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Passioura T.; Katoh T.; Goto Y.; Suga H. Selection-based discovery of druglike macrocyclic peptides. Annu. Rev. Biochem. 2014, 83, 727–752. 10.1146/annurev-biochem-060713-035456. [DOI] [PubMed] [Google Scholar]
- Passioura T.; Watashi K.; Fukano K.; Shimura S.; Saso W.; Morishita R.; Ogasawara Y.; Tanaka Y.; Mizokami M.; Sureau C.; Suga H.; Wakita T. De novo macrocyclic peptide inhibitors of hepatitis B virus cellular entry. Cell Chem. Biol. 2018, 25, 906–915. 10.1016/j.chembiol.2018.04.011. [DOI] [PubMed] [Google Scholar]
- Yamagishi Y.; Shoji I.; Miyagawa S.; Kawakami T.; Katoh T.; Goto Y.; Suga H. Natural product-like macrocyclic N-methyl-peptide inhibitors against a ubiquitin ligase uncovered from a ribosome-expressed de novo library. Chem. Biol. 2011, 18, 1562–1570. 10.1016/j.chembiol.2011.09.013. [DOI] [PubMed] [Google Scholar]
- Goto Y.; Ohta A.; Sako Y.; Yamagishi Y.; Murakami H.; Suga H. Reprogramming the translation initiation for the synthesis of physiologically stable cyclic peptides. ACS Chem. Biol. 2008, 3, 120–129. 10.1021/cb700233t. [DOI] [PubMed] [Google Scholar]
- Song X.; Lu L. Y.; Passioura T.; Suga H. Macrocyclic peptide inhibitors for the protein-protein interaction of Zaire Ebola virus protein 24 and karyopherin alpha 5. Org. Biomol. Chem. 2017, 15, 5155–5160. 10.1039/C7OB00012J. [DOI] [PubMed] [Google Scholar]
- Jongkees S. A. K.; Caner S.; Tysoe C.; Brayer G. D.; Withers S. G.; Suga H. Rapid discovery of potent and selective glycosidase-inhibiting de novo peptides. Cell Chem. Biol. 2017, 24, 381–390. 10.1016/j.chembiol.2017.02.001. [DOI] [PubMed] [Google Scholar]
- Matsunaga Y.; Bashiruddin N. K.; Kitago Y.; Takagi J.; Suga H. Allosteric inhibition of a semaphorin 4D receptor plexin B1 by a high-affinity macrocyclic peptide. Cell Chem. Biol. 2016, 23, 1341–1350. 10.1016/j.chembiol.2016.09.015. [DOI] [PubMed] [Google Scholar]
- Hacker D. E.; Hoinka J.; Iqbal E. S.; Przytycka T. M.; Hartman M. C. Highly constrained bicyclic scaffolds for the discovery of protease-stable peptides via mRNA display. ACS Chem. Biol. 2017, 12, 795–804. 10.1021/acschembio.6b01006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kale S. S.; Villequey C.; Kong X. D.; Zorzi A.; Deyle K.; Heinis C. Cyclization of peptides with two chemical bridges affords large scaffold diversities. Nat. Chem. 2018, 10, 715–723. 10.1038/s41557-018-0042-7. [DOI] [PubMed] [Google Scholar]
- Rentero Rebollo I.; McCallin S.; Bertoldo D.; Entenza J. M.; Moreillon P.; Heinis C. Development of potent and selective S. aureus sortase A inhibitors based on peptide macrocycles. ACS Med. Chem. Lett. 2016, 7, 606–611. 10.1021/acsmedchemlett.6b00045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornish-Bowden A. A simple graphical method for determining the inhibition constants of mixed, uncompetitive and non-competitive inhibitors. Biochem. J. 1974, 137, 143–144. 10.1042/bj1370143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yildiz M.; Ghosh S.; Bell J. A.; Sherman W.; Hardy J. A. Allosteric inhibition of the NS2B-NS3 protease from dengue virus. ACS Chem. Biol. 2013, 8, 2744–2752. 10.1021/cb400612h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Bock S.; Snitko M.; Berger T.; Weidner T.; Holloway S.; Kanitz M.; Diederich W. E.; Steuber H.; Walter C.; Hofmann D.; Weissbrich B.; Spannaus R.; Acosta E. G.; Bartenschlager R.; Engels B.; Schirmeister T.; Bodem J. Novel dengue virus NS2B/NS3 protease inhibitors. Antimicrob. Agents Chemother. 2015, 59, 1100–1109. 10.1128/AAC.03543-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brecher M.; Li Z.; Liu B.; Zhang J.; Koetzner C. A.; Alifarag A.; Jones S. A.; Lin Q.; Kramer L. D.; Li H. A conformational switch high-throughput screening assay and allosteric inhibition of the flavivirus NS2B-NS3 protease. PLoS Pathog. 2017, 13, e1006411 10.1371/journal.ppat.1006411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Clercq E. The design of drugs for HIV and HCV. Nat. Rev. Drug Discovery 2007, 6, 1001–1018. 10.1038/nrd2424. [DOI] [PubMed] [Google Scholar]
- Phoo W. W.; Zhang Z.; Wirawan M.; Chew E. J. C.; Chew A. B. L.; Kouretova J.; Steinmetzer T.; Luo D. Structures of Zika virus NS2B-NS3 protease in complex with peptidomimetic inhibitors. Antiviral Res. 2018, 160, 17–24. 10.1016/j.antiviral.2018.10.006. [DOI] [PubMed] [Google Scholar]
- Nitsche C.; Behnam M. A.; Steuer C.; Klein C. D. Retro peptide-hybrids as selective inhibitors of the dengue virus NS2B-NS3 protease. Antiviral Res. 2012, 94, 72–79. 10.1016/j.antiviral.2012.02.008. [DOI] [PubMed] [Google Scholar]
- Nitsche C.; Schreier V. N.; Behnam M. A.; Kumar A.; Bartenschlager R.; Klein C. D. Thiazolidinone-peptide hybrids as dengue virus protease inhibitors with antiviral activity in cell culture. J. Med. Chem. 2013, 56, 8389–8403. 10.1021/jm400828u. [DOI] [PubMed] [Google Scholar]
- Weigel L. F.; Nitsche C.; Graf D.; Bartenschlager R.; Klein C. D. Phenylalanine and phenylglycine analogues as arginine mimetics in dengue protease inhibitors. J. Med. Chem. 2015, 58, 7719–7733. 10.1021/acs.jmedchem.5b00612. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.