Abstract
The most common pathogenic mutation in amyotrophic lateral sclerosis (ALS) and frontotemporal dementia (FTD) is an intronic GGGGCC (G4C2) repeat in the chromosome 9 open reading frame 72 (C9orf72) gene. Cellular toxicity due to RNA foci and dipeptide repeat (DPR) proteins produced by the sense and antisense repeat-containing transcripts is thought to underlie the pathogenesis of both diseases.
RNA sequencing (RNA-seq) data of C9orf72-ALS patients and controls were analyzed to better understand the sequence conservation of C9orf72 in patients. MicroRNAs were developed in conserved regions to silence C9orf72 (miC), and the feasibility of different silencing approaches was demonstrated in reporter overexpression systems. In addition, we demonstrated the feasibility of a bidirectional targeting approach by expressing two concatenated miC hairpins. The efficacy of miC was confirmed by the reduction of endogenously expressed C9orf72 mRNA, in both nucleus and cytoplasm, and an ∼50% reduction of nuclear RNA foci in (G4C2)44-expressing cells. Ultimately, two miC candidates were incorporated in adeno-associated virus vector serotype 5 (AAV5), and silencing of C9orf72 was demonstrated in HEK293T cells and induced pluripotent stem cell (iPSC)-derived neurons. These data support the feasibility of microRNA (miRNA)-based and AAV-delivered gene therapy that could alleviate the gain of toxicity seen in ALS and FTD patients.
Keywords: C9orf72, ALS, FTD, gene therapy, microRNA
Introduction
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease characterized by progressive degeneration of the upper and lower motor neurons, leading to muscle atrophy and paralysis. There is no disease-modifying therapy for ALS, and most patients die with respiratory failure within 3–5 years after the onset of symptoms.1, 2, 3 A significant number of ALS patients also develop frontotemporal dementia (FTD), a presenile dementia caused by progressive degeneration of the frontal and temporal lobes.3, 4 The most common genetic cause of familial and sporadic ALS and FTD is an expanded GGGGCC (G4C2) repeat in the first intron of the chromosome 9 open reading frame 72 (C9orf72) gene.2, 5 The G4C2 repeat in patients can be several hundred repeats long and is transcribed bidirectionally.1, 2 Generally, less than 30 repeats are considered non-pathogenic.1, 2, 6
The G4C2 repeat is found in approximately 40% of familial ALS cases and 9% of sporadic ALS.7, 8 The mechanisms underlying the involvement of C9orf72 in neurodegeneration have been a debate for several years, with loss of function, gain of toxicity, or a combination of both being implicated.9, 10 A reduction of C9orf72 transcripts is detected in a significant number of C9orf72-ALS (C9-ALS) patients, supporting loss of function.8, 11 However, complete elimination of C9orf72 in mice causes immune system-related pathology but no motor deficits. These conditions were rescued in mice hemizygous for C9orf72 that express 50% of C9orf72 mRNA.12, 13 In humans, a few loss-of-function mutations in C9orf72 were identified and all seemed to be non-pathogenic, thus C9orf72 reduction in humans is likely to be tolerable.14 Most evidence suggests that C9orf72-related pathogenicity is a result of gain of toxicity by the accumulation of sense and antisense RNA foci in the nucleus and deposition of dipeptide repeat (DPR) proteins in the cytoplasm.7, 15, 16, 17, 18, 19, 20 RNA foci can bind and sequester the function of RNA-binding proteins, leading to RNA-mediated toxicity.19, 21 DPR proteins are linked to a variety of toxic effects.9, 19, 21 Thus, lowering the accumulation of RNA foci and DPR proteins is an attractive therapeutic strategy in ALS and FTD.
Directly targeting the G4C2 repeat to exclusively silence the repeat-containing transcripts is preferable but challenging, due to the high GC content, bidirectional transcription, intronic position, and nuclear localization.22 The repeat-containing transcripts are also poorly characterized, and sequence variations downstream of the G4C2 repeat region suggest that the area close to the repeat may not be well conserved between patients.23
Fully complementary anti-GGGGCC or anti-CCCCGG single-stranded small intering RNAs (ss-siRNAs) reduced C9orf72 sense and antisense foci.24 Antisense oligonucleotides (ASOs) against C9orf72 intron 1 near the G4C2 repeat also resulted in the reduction of sense RNA foci and DPR proteins, while the overall C9orf72 levels were not affected.13, 17, 25 Interestingly, RNA foci and DPR proteins were also reduced by ASOs against exonic regions to target all C9orf72 transcripts. Thus, both mutant-specific or total C9orf72-silencing approaches can lower RNA foci and DPR proteins.
The administration of synthetic siRNAs and ASOs is promising but requires repeated administration. We and others have reported that siRNAs derived from short hairpin RNA (shRNA) or microRNA (miRNA) scaffolds delivered with adeno-associated virus (AAV) vectors have the advantage of providing long-lasting therapeutic effects in different disease models of the CNS.26, 27, 28, 29 High concentrations of shRNAs can bypass the nuclear RNase III Drosha processing and overload the cytoplasm with double-stranded RNA, leading to toxicity.30 miRNAs are safer, as their precursors are encoded in the genome, thus mimicking the natural processing pathway. Moreover, the miRNA-expression cassette can be driven by RNA polymerase II (Pol II) or Pol III promoters, allowing for tissue- or cell-specific expression.
We here report on the development of therapeutic miRNAs (miC) as a proof of concept to silence C9orf72 mRNA. We used a recently published RNA sequencing (RNA-seq) library from C9-ALS patients to investigate C9orf72 expression and sequence composition for the identification of potential miC target sites in intron 1 and exonic regions.31 miC constructs were designed to target the sense, antisense, or both strands of C9orf72. The natural cellular primary (pri)-miR-101-1 and pri-miR-451 scaffolds were selected to embed the miC sequences, based on our previous findings that both scaffolds produced guide strands that were highly active.29 pri-miRNAs are produced by Pol I or Pol II transcription, and they fold into hairpin-like scaffolds, which determine their further processing by the microprocessor complex.32 The tail of pri-miRNAs is cleaved by Drosha in the nucleus, generating a precursor miRNA (pre-miRNA) that is subsequently exported to the cytoplasm for further processing into a mature miRNA.33, 34, 35 The silencing efficacy of the miC-101 and miC-451 candidates was tested on luciferase (Luc) reporters, and it was confirmed with knockdown of the endogenously expressed C9orf72 mRNA. The processing pattern was determined by next-generation sequencing (NGS).
One major challenge for an effective RNAi approach is the nuclear localization of the repeat-containing transcripts, as the mature miC is produced in the cytoplasm. Therefore, the expression and silencing efficacy of the miC candidates in both nucleus and cytoplasm was investigated, and we studied the ability of these constructs to silence nuclear RNA foci formation caused by G4C2 repeats. Finally, two candidates were selected and incorporated into AAV5 vector to test their efficacy in vitro on HEK293T cells and in frontal brain-like neurons generated from induced pluripotent stem cells (iPSCs) from an FTD patient. The expression of total C9orf72 (all C9orf72 sense transcripts) was significantly reduced in both cell lines. Hence, our data provide evidence on the efficacy of artificial miRNAs against C9orf72 as a promising AAV-based gene therapy for ALS and FTD.
Results
Reduced C9orf72 Levels Detected by RNA-Seq in C9-ALS Patients
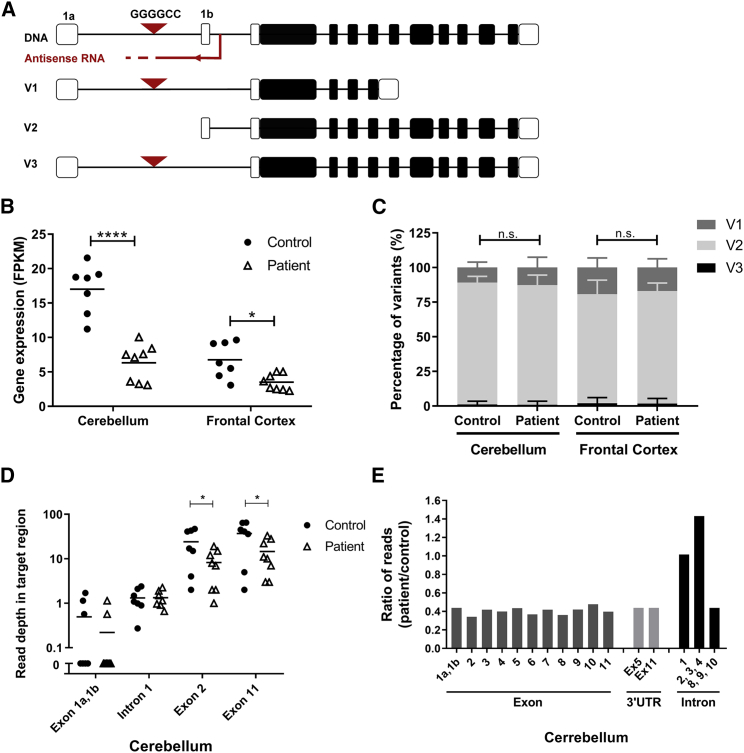
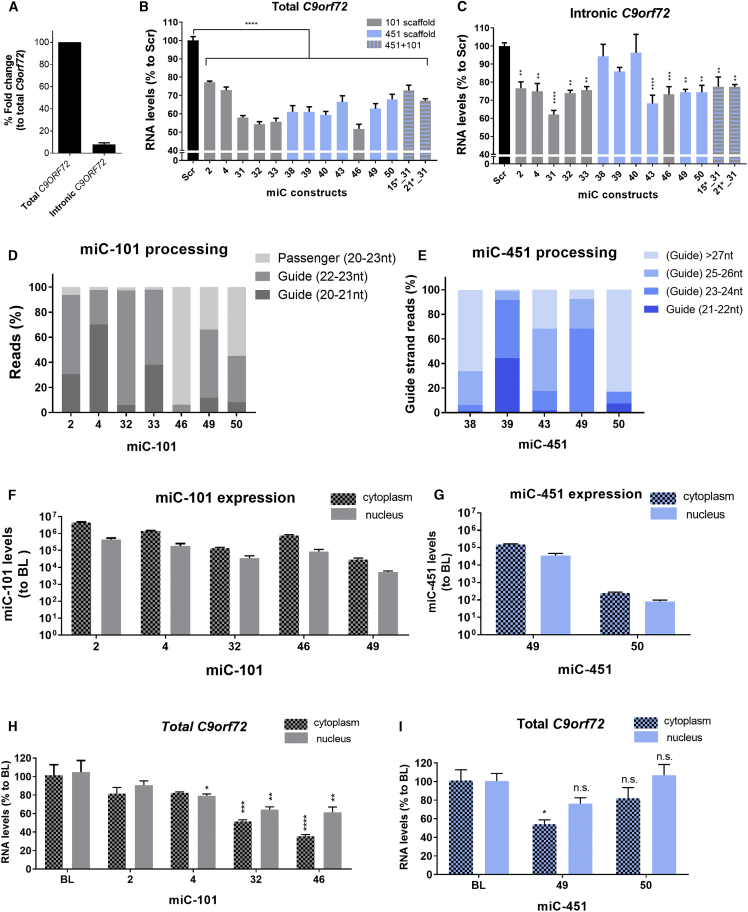
The human C9orf72 gene consists of 12 exons that can be transcribed in three different transcript variants (V1, V2, and V3) (Figure 1A). The G4C2 repeat in intron 1 is in the promoter region of V2 and the first intron of V1 and V3. We used a publicly available RNA-seq library to determine the expression of C9orf72 in cerebellar and cortical tissues of 8 C9-ALS patients and 7 healthy controls.31 Notably, we found that C9orf72 is more highly expressed (∼2-fold) in the cerebellum compared to cortex in both C9-ALS patients and controls (Figure 1B). It was also found that C9orf72 mRNA expression is consistently reduced in both cerebellum and cortex of C9-ALS patients. To determine if the reduction seen in C9-ALS patients is variant specific, we investigated the relative expressions of V1, V2, and V3 in patients and controls (Figure 1C). Expressions of V1, V2, and V3 were determined in percentages, relative to total (100%) C9orf72 expression. In both cerebellum and cortex, the proportions of the variants in patients and controls were similar, suggesting that the reduction of C9orf72 mRNA levels in C9-ALS patients is not variant specific.
Figure 1.
C9orf72 Expression and Target Regions
(A) Schematic of C9orf72 gene and transcript variants. The G4C2 repeat is in intron 1 of transcript variants V1 and V3 and in the promoter region of V2. (B) C9orf72 expression in C9-ALS patients and controls. C9orf72 gene expression was determined from RNA-seq libraries in cerebellum and frontal cortex from 8 C9-ALS patients and 7 control donors. The mean expression values are shown in fragments per kilobase of transcript per million mapped reads (FPKM). (C) Relative expression of predicted C9orf72 variants. The mRNA variants in C9-ALS and controls were predicted from the mapped and aligned RNA-seq data. Isoform V1 is predicted from read alignment from exon 1a to exon 5 (1,950 bp), isoform V2 from exon 1b to exon 11 (3,243 bp), and isoform V3 from exon 1a to exon 11 (3,338 bp). Expression of C9orf72 isoforms is presented in percentage of the total (100%) C9orf72 expression. t tests were performed between groups, and n.s. indicates no significant differences in the levels of all isoforms. (D) Estimation of the read depth per region in C9orf72. Read depth was estimated for exon 1a, intron 1, exon 1b, exon 2, and exon 11 in cerebellum from C9-ALS patient and controls. The total amount of reads per region was counted and corrected for the area size. Each dot or triangle represents a single sample. (E) Ratio of reads between C9-ALS patients and controls in cerebellum. The total amount of reads counted in different intronic and exonic regions from C9-ALS patients was divided by the total amount of reads from the same region of control donors. Data were evaluated using two-way ANOVA with Tukey’s multiple comparison: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Intronic Inclusions due to G4C2 Repeat Are Detected in C9-ALS Patients by RNA-Seq
C9orf72 intron 1 should be spliced out and degraded, but defective splicing may result in the accumulation of repeat-containing transcripts in the cell nucleus.36 These repeat-containing transcripts are poorly characterized, and there is little information on the conservation of this region in C9-ALS patients. To address this question, read alignments from C9-ALS patients and controls were compared to investigate the sequence conservation of intronic and exonic regions of C9orf72 transcripts (Figure 1D). The read depths in exon 1a, exon 1b, intron 1, exon 2, and exon 11 were estimated by correcting the total number of reads by the area size. We found a complete coverage of exon 2 to exon 11, though read depths in exon 1a, exon 1b, and intronic regions were very low in both C9-ALS and control groups. Exonic regions from exon 2 to exon 11 were less covered in patients, while the read depth for intron 1 in the patients was comparable with controls.
To estimate the relative coverage of the exons and introns, the ratio between the number of reads in C9-ALS patients and controls was determined (Figure 1E). All C9orf72 exonic regions were about 2-fold lower expressed in C9-ALS patients. Intron 1 had the same number of reads in patient samples compared to controls, while introns 2–4 were 1.4 times higher in C9-ALS patients than controls. Introns 5–7 were excluded, as these could potentially be 3′ UTR of the short C9orf72 variant, and coverage of introns 8–10 was not increased in C9-ALS patients. Similarly, in the frontal cortex samples of C9-ALS patients, the coverage ratio between intronic and exonic regions was increased (Figure S1). Thus, although exonic regions were expressed 2-fold lower in C9-ALS patients, this was not the case for introns 1–4, suggesting that intronic C9orf72 reads are relatively overexpressed in C9-ALS patients. Our data confirm previous findings that the higher amount of intronic sequences in the C9orf72 mRNA could be due to aberrant splicing.36, 37
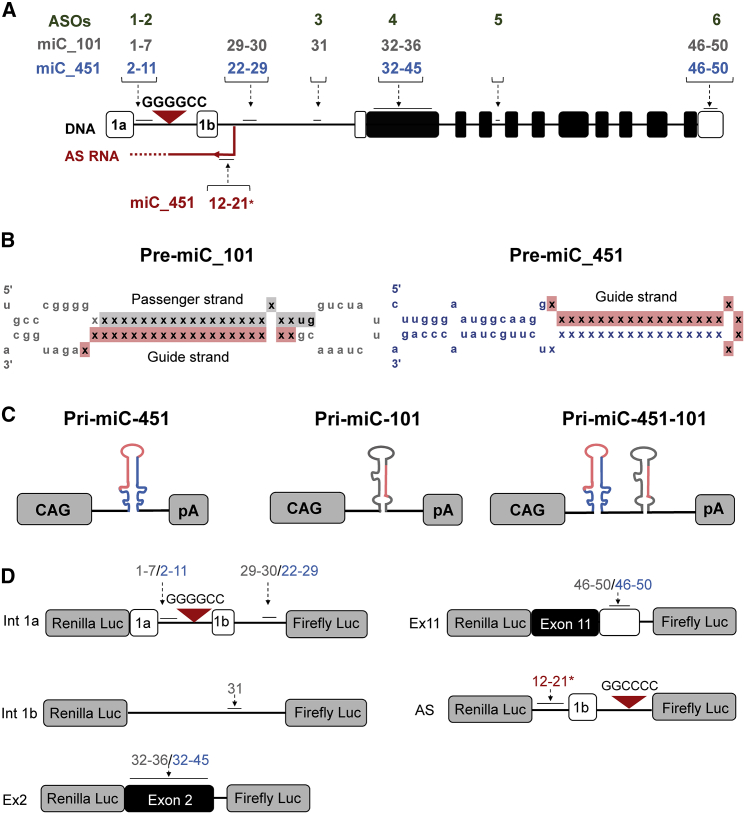
Design of miRNAs Targeting Conserved C9orf72 Regions
We selected the well-conserved exon 2 and exon 11 as target sites for a total silencing approach. Due to the poor coverage of intron 1, we selected regions that have been successfully targeted before by ASOs.13 For the antisense strand, a region that has been identified before by PCR was selected.21 We designed miC expression constructs miC1-miC11 and miC22-miC31 in intron 1 to target only the sense intronic transcripts (Figure 2A). miC32-miC50 was designed in exon 2 and exon 11 to target all sense C9orf72 transcripts. miC12*-miC22* was designed on the antisense strand to target only the antisense transcripts and is indicated with asterisks. The miC sequences were embedded in the pri-miR-101 and pri-miR-451 scaffold because of previous findings by us that both scaffolds produce high amount of active guide strands.29 The pri-miC-101 and pri-miC-451 structures were predicted to produce mature miC lengths of 21 and 22 nt, respectively (Figure 2B). The miC constructs were expressed by the synthetic cytomegalovirus (CMV) early enhancer and chicken β-actin (CAG) promoter (Figure 2C). This promoter is known to drive stable and high expression of a transgene and is highly active in the CNS.38
Figure 2.
Design of Artificial miC Expression Constructs Targeting Sense and Antisense C9orf72
(A) Schematic representation of the human C9orf72 gene consisting of 12 exons, including the alternatively spliced exons 1a and 1b and the intronic G4C2 repeat. The positions of the miC target sites are indicated with numbers. Numbers in green show positions of ASOs described by Lagier-Tourenne et al.17 In gray are the sense-targeting miC candidates embedded in the miR-101 scaffold, in blue are sense-targeting miC candidates embedded in the miR-451 scaffold, and in red are the antisense-targeting miC candidates embedded in the miR-451 scaffold. (B) Schematic of the miC-101 and miC-451 secondary structures. The scaffolds were selected from miRBase database (www.mirbase.org). The guide strand was replaced by the mature miC sequence, and the passenger strand was corrected in order to preserve pri-miC scaffolding. miR-101 can be processed into active guide strands (red) and in some cases passenger strands (gray). miR-451 produces only guide strands. (C) Schematic of the pri-miC-451 and pri-miC-101 constructs consisting of the CAG promoter, pri-miC sequence, and human growth hormone polyadenylation (hGH polyA) signal. (D) Schematic of the reporter genes. To represent the C9orf72 sense transcripts, sequences from C9orf72 intron 1 (int 1a and int 1b), exon 2 (ex2), and exon 11 (ex11) were cloned downstream of the RL gene. In addition, FL was co-expressed from the vector as an internal control. For the antisense reporter (AS), the intronic antisense sequence was cloned.
In Vitro Testing of miC-101 and miC-451 Constructs on Reporter Systems
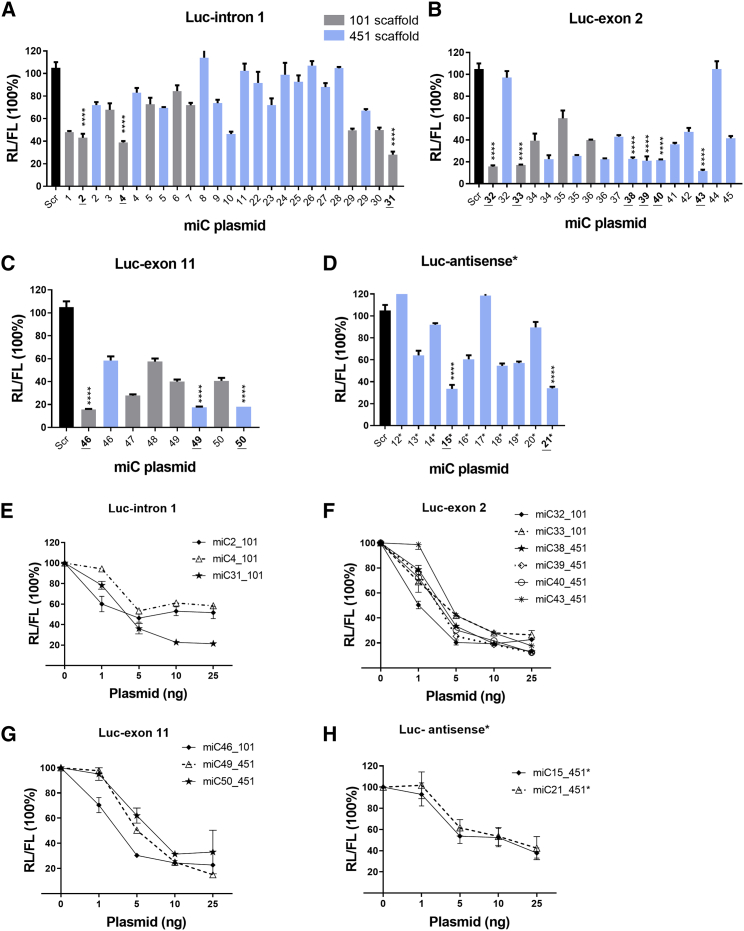
To test the efficacy of the miC candidates, we designed Luc reporters bearing complementary C9orf72 target regions fused to the renilla luciferase (RL) gene (Figure 2D). As targets, intron 1, exon 2, exon 11, and the antisense strand sequences were used. Independently from RL, firefly luciferase (FL) was expressed from the reporter vector to correct for transfection efficiency.
We first performed a prescreening for all the miC expression constructs by co-transfection with the corresponding Luc reporters in a 1:1 ratio. Of the miC variants designed to target the sense intronic transcripts, miC2_101 and miC4_101 showed a moderate knockdown (∼50%) and miC31_101 showed a strong knockdown (∼80%) (Figure 3A). These were selected for further optimization. Among candidates predicted to target total C9orf72, miC32_101, miC33_101, miC38_451, miC39_451, miC40_451, and miC43_451 targeting exon 2 showed a strong knockdown of >80% (Figure 3B). Similarly, miC46_101, miC49_451, and miC50_451 targeting exon 11 induced a strong knockdown (>80%) (Figure 3C). Dilution of the selected miC candidates demonstrated that the most effective candidates were miC31_101 against intron 1 (Figure 3E), miC32_101 against exon 2 (Figure 3F), and miC46_101 against exon 11 (Figure 3G).
Figure 3.
Silencing Efficacy of miC Candidates on Luc Reporters
(A) Knockdown by miC1 to miC11 and miC22 to miC31 targeting intron 1 either between exon 1a and the G4C2 repeat or between exon 1b and exon 2. HEK293T cells were co-transfected in a 1:1 ratio with Luc reporters and the different miC variants embedded in miR-101 (gray bars) or miR-451 (blue bars) scaffolds. RL and FL were measured 2 days post-transfection, and RL was normalized to FL expression. Scrambled miRNA (miScr) served as a negative control and was set at 100%. Candidates selected for further testing are underlined. (B and C) Knockdown of exon 2 reporter (B) by miC32 to miC45 and exon 11 reporter (C) by miC46 to miC50. (D) Knockdown of the antisense reporter by miC12_451* to miC21_451*. (E–H) Dose-dependent effects of the selected miC candidates on Luc reporters. HEK293T cells were co-transfected with 10 ng Luc reporters and 1, 5, 10, and 25 ng of the selected miC constructs for intron 1 (E), exon 2 (F), exon 11 (G), and the antisense transcript (H). RL and FL luciferases were measured as described above. Data were analyzed using a multiple-comparison one-way ANOVA to determine statistical significances for cells treated with scrambled and miC. The p values of miC candidates selected for further testing are listed in the graph by asterisks: ****p < 0.0001. Each bar represents the average and SD of 3 independent experiments.
For the antisense C9orf72 transcript, miC15_451* and miC21_451* were selected as the most effective candidates with a knockdown efficiency of ∼70% (Figure 3D). Both miC15_451* and miC21_451* showed an equal dose-dependent knockdown (Figure 3H).
Our data demonstrate that intron 1 remains a difficult target region, as all miC candidates in the highly structured repeat region between exons 1a and 1b failed to induce a strong knockdown. The most effective miC candidates were downstream of exon 1b or in exonic regions 2 and 11. Yet, if a moderate knockdown of the G4C2 repeat would be sufficient for a therapeutic effect, miC2_101 and miC4_101 could still be promising candidates to target the repeat-containing transcripts in C9-ALS patients.
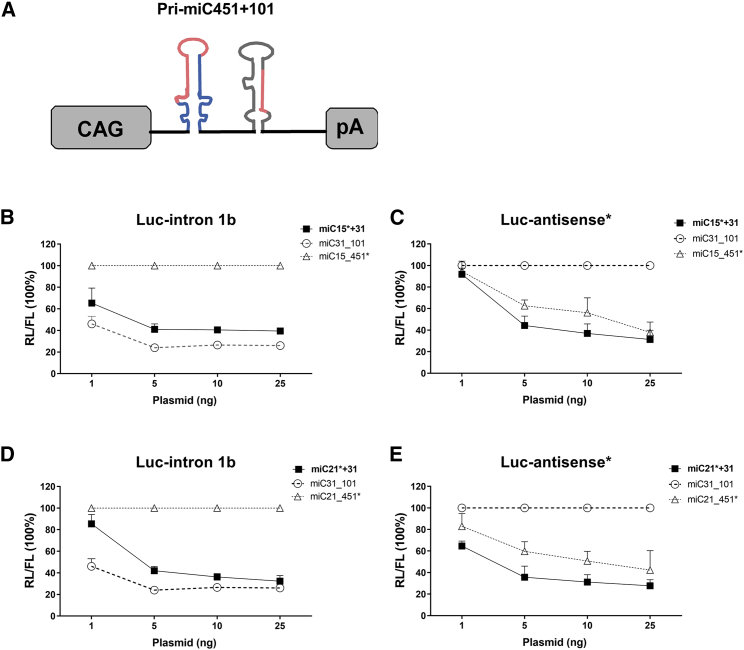
Bidirectional Targeting of C9orf72 Is Possible by the Introduction of a Second miC Concatenate
The region around the G4C2 repeat of C9orf72 is transcribed in both sense and antisense transcripts, and both strands have been linked to toxicity. Therefore, a therapy simultaneously targeting both strands could potentially add to therapeutic benefit. To investigate the feasibility for this approach using miRNAs, we made concatenated constructs expressing two hairpins predicted to target both transcripts under the control of the CAG-promoter (Figure 4A). The first hairpin from the concatenated hairpin miRNA construct was in a miR-451 scaffold and targets the antisense transcript. The second hairpin was in a miR-101 scaffold against the sense transcripts. The most effective candidates on Luc reporters for intron 1 sense and antisense were selected. The miC15*+31 construct was designed to express miC15_451* and miC31_101, and it was tested on the intron 1 and antisense reporters (Figures 4B and 4C). A silencing of up to 60% was observed on the intron 1 sense and on the antisense reporter. Similarly, miC21*+31 expressing miC21_451* and miC31_101 was made and tested, and up to a 60% knockdown was observed on both reporter constructs (Figures 4D and 4E). Both constructs showed a dose-dependent reduction of the intron 1 sense and antisense reporters containing Luc. Our data demonstrate that two different miCs can be properly processed and are active when expressed from a single promoter. Hence, a bidirectional miRNA-based approach to simultaneously target the sense and antisense transcripts of C9orf72 is feasible, using two miC variants expressed in a concatenated fashion.
Figure 4.
Efficacy of Double-Hairpin Constructs for Bidirectional Knockdown
(A) Schematic of the concatenated pri-miC-451 and pri-miC-101 construct consisting of the CAG promoter, two pri-miC sequences in miR-451 and miR-101 scaffolds, and human growth hormone polyadenylation (hGH polyA) signal. (B) Optimization of miC15*+31 on Luc-intron 1b reporter. HEK293T cells were co-transfected with 10 ng Luc-intron 1b reporter and 1, 5, 10, or 25 ng miC15*+31 construct designed to simultaneously target both sense and antisense C9orf72 transcripts. miC31_101 (targeting only sense) was used as a positive control and miC15_451* served as a negative control. RL and FL were measured as described in Figure 3. (C) Optimization of miC15*+31 on the antisense reporter. miC15*_451 was tested as a positive control for the antisense reporter and miC31_101 as a negative control. (D) Knockdown of Luc-intron 1b reporter by miC21*+31 with miC31_101 as a positive control and miC21_451* as a negative control. (E) Dose-dependent knockdown of the antisense reporter by miC21*+31. miC21_451* served as a positive control and miC31_101 as a negative control. Error bars represent the average and SD of 2 independent experiments.
Endogenous Knockdown of C9orf72 Expression in HEK293T Cells by miC Variants
We next investigated whether the selected miC candidates reduce the endogenous levels of C9orf72 in HEK293T cells. Cells were transfected with the selected miC candidates, and endogenous levels of C9orf72 mRNA were determined 2 days post-transfection by qRT-PCR. As HEK293T cells lack the G4C2 expansion linked to the C9orf72 pathology, we first determined the expression of total C9orf72 and intronic C9orf72 mRNA. We found good expression of total C9orf72, while the intronic C9orf72 was detectable but at very low levels (Figure 5A). For miC candidates in the miR-101 scaffold, total C9orf72 mRNA was reduced up to 50% by miC32_101, miC33_101, and miC46_101 (Figure 5B). The intronic C9orf72 transcripts were also decreased by ∼25% (Figure 5C). miC2_101 and miC4_101 targeting intron 1 were less effective in lowering total C9orf72, but the efficacy on intronic C9orf72 was comparable to miC candidates targeting total C9orf72. Thus, both candidates may be targeting the repeat-containing transcripts without significantly changing the total C9orf72 expression. miC31_101 targeting intron 1 showed the best efficacy for the intronic C9orf72 (40%); but, despite its intronic localization, the total C9orf72 RNA levels were also reduced by ∼40% (Figures 5B and 5C). Reduction of C9orf72 was also observed by the selected miC candidates in the miR-451 scaffold, but their efficacy was slightly lower. Altogether, we demonstrated the reduction of endogenous levels of C9orf72 in HEK293T cells, confirming that the miC candidates are functional in cells.
Figure 5.
Lowering of Endogenous C9orf72 mRNA
(A) Total and sense intronic C9orf72 expression in HEK293T cells. RNA was isolated, and qRT-PCR was performed for total and sense intronic C9orf72 transcripts. mRNA input levels were normalized to GAPDH and set relative to total C9orf72. (B and C) Endogenous knockdown of total C9orf72 (B) and sense intronic C9orf72 (C) by the selected miC candidates. qRT-PCR for total and sense C9orf72 was performed on RNA from HEK293T cells that were transfected with 250 ng of different miC plasmids. mRNA input levels were normalized to GAPDH mRNA. miScr served as a negative control and was set at 100%. (D) Processing of miC-101 candidates. HEK293T cells were transfected with 250 ng of the constructs. At 48 h post-transfection, RNA was isolated, and small RNA NGS was performed to determine the length and ratio of guide and passenger strands. (E) Processing of miC-451 candidates, performed as described in (D). (F) Cytoplasmic and nuclear expressions of miC candidates in the miR_101 scaffold. RNA was isolated from the cytoplasm and nucleus of HEK293T cells transfected with 250 ng miC plasmid. miC levels were measured by small RNA TaqMan, and mRNA input levels was normalized to u6 small nuclear RNA and set relative to untreated (BL) cells. (G) Expression of miC candidates in miR-451 in cytoplasm and nucleus, performed as described in (F). (H) Reduction of total C9orf72 by miC-101 in cytoplasm and nucleus. qRT-PCR for total C9orf72 was performed on cytoplasmic and nuclear fractions of transfected cells. mRNA input levels were corrected for GAPDH and BL was set at 100%. (I) Total C9orf72 reduction in the nucleus and cytoplasm by miC-451, performed as described in (H). Data were analyzed using a multiple-comparison one-way ANOVA to determine statistical significances for cells treated with scrambled or BL and miC. The p values are listed in the graph by asterisks: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Each graph represents the mean values with SD of 2 independent experiments.
Different Processing Pattern from the miR-101 and miR-451 Scaffolds
To assess the processing of the miC candidates, we analyzed the mature miC lengths and sequence composition of the guide and passenger strands by NGS for small transcriptome analysis (Figures 5D and 5E). NGS was performed on small RNAs isolated from HEK293T cells that were transfected with the selected miC constructs. For each sample, we obtained between 15 and 30 million small RNA reads that were subsequently adaptor trimmed and aligned against the corresponding reference sequence. All reads shorter than 10 nt, longer than 45 nt, or represented less than 10 times were excluded from the analysis.
miR-101 is processed into a miRNA duplex, first by Drosha cleavage and then by Dicer cleavage at the hairpin structure (Figure S2). The miRNA duplex is then separated, and the guide strand is usually incorporated into the RNA-induced silencing complex (RISC), while in most cases the passenger strand is degraded.39, 40, 41 The miC-101 candidates were processed into a 20- to 23-nt-long mature miRNA (Figure 5D). The most frequently found length of guide strands was 22 nt, which is 1 nt longer than the cleavage pattern predicted by miRBase (http://www.mirbase.org/) (Table S1). The length of the passenger strands ranged between 19 and 23 nt. In most cases, Drosha cleavage sites of the mature miC-101 at the 3′ end of the pre-miC-101 candidates were precise and consistent with the prediction from miRBase, except for miC33_101 and miC49_101. Following cleavage by Drosha, the hairpin of the pre-miC_101 was being cleaved by Dicer. Dicer cleavage in the hairpin generated more variability for almost all the miC variants. Processing of miC2_101, miC4_101, miC32_101, and miC33_101 yielded a high frequency of guide strands with very low percentages of the passenger strand. miC46_101 processing yielded more passenger strand, while miC49_101 and miC50_101 produced a relatively equal amount of guide and passenger strands.
The processing of the miC-451 candidates did not produce passenger strands but often generated longer guide strands than the predicted 22 nt obtained from miRbase (Figure 5E; Table S2). Drosha cleavage sites at the 5′ end of the mature miC-451 were precise, but the trimming of the 3′ ends of the mature miC-451 by poly(a)-specific ribonuclease (PARN) were different for most candidates, leading to a variety of mature lengths. miC39_451, miC43_451, and miC49_451 processing generated most often mature lengths between 21 and 26 nt long, and processing of miC38_451 and miC50_451 often resulted in mature lengths longer than 27 nt.
Overall, we demonstrated that expressing different C9orf72 target sequences from the miR-101 scaffold yields a differential processing of mature guide and passenger strands. Using the miC-451, no passenger strands were detected, but this scaffold often generated longer mature guide strands.
miC-101 and miC-451 Are Active in the Nucleus
pre-miRNAs are transported from the nucleus to the cytoplasm for further processing and incorporation into the RISC.39, 40, 41 However, because C9orf72-related ALS and FTD are characterized by accumulation of the G4C2-containing transcripts in the nucleus, mature miC should be active in the cell nucleus to achieve therapeutic effect. Based on the efficacy in vitro, we selected miC2_101 and miC4_101 as the most promising candidates to target only the intronic transcripts. Similarly, miC32_101, miC46_101, miC49_451, and miC50_451 were selected for a total silencing approach, based on their strong silencing efficacy in vitro. HEK293T cells were transfected with the different miC candidates; nuclear and cytoplasmic fractions were separated; and expressions of the mature miC2, miC4, miC32, miC46, miC49, and miC50 in nucleus and cytoplasm were evaluated.
We detected mature miC in both nucleus and cytoplasmic fractions for all miC candidates, but the expression levels in nucleus were consistently ∼5-fold lower compared to cytoplasm (Figures 5F and 5G). Next, we evaluated the silencing efficacy of the miC candidates in nucleus and cytoplasm by measuring the endogenous levels of total C9orf72 mRNA. miC32_101, miC46_101, and miC49_451 caused a reduction of total C9orf72 mRNA in both nucleus and cytoplasm (Figures 5H and 5I). However, the silencing efficacy in the nucleus was lower compared to cytoplasm, consistent with the lower miC levels detected in the nucleus. As expected, miC2_101 and miC4_101, which target the intronic C9orf72 transcripts, had limited to no effect on the total C9orf72 expression. Thus, our data suggest that the mature miC-101 and miC-451 can both shuttle from the cytoplasm to the cell nucleus and can actively induce knockdown. Reduction of C9orf72 was observed in both the nucleus and cytoplasm, suggesting that both scaffolds can be used for further development into gene therapy for ALS and FTD.
Reduction of Nuclear RNA Foci by miC Variants in (G4C2)44-Expressing Cells
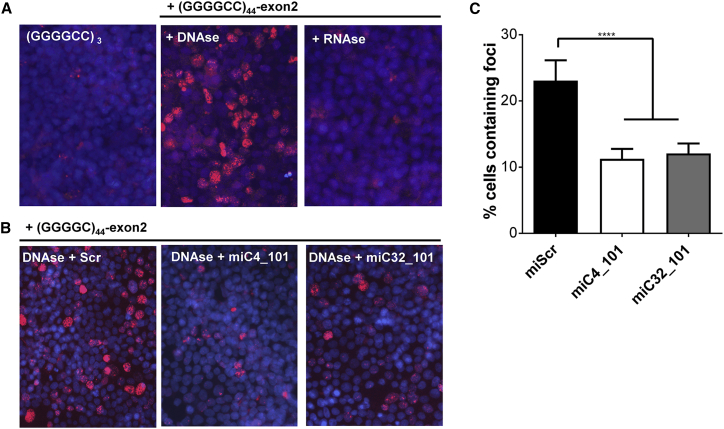
A hallmark of the RNA-mediated toxicity in ALS and/or FTD is the formation of toxic RNA foci by the repeat-containing transcripts. We generated a cell model that develops nuclear RNA foci using methods similar to those described previously.42, 43 We expressed constructs consisting of (G4C2)44 or (G4C2)3, including 150-nt 5′- and 50-nt 3′-flanking regions linked to C9orf72 exon 2 in HEK293T cells. Nuclear RNA foci were visualized by fluorescence in situ hybridization (FISH) using a TYE563-(C4G2)3 locked nucleic acid (LNA) probe. However, we were not able to detect DPR proteins in these cells, as the assay is technically challenging. Using a GFP construct, the transfection efficiency was determined to be ∼95%–100% (data not shown). We observed sense RNA foci at 2 days post-transfection in ∼40% of (G4C2)44 cells (Figure 6A), but antisense RNA foci were not detected (data not shown). RNA foci were primarily present in the nucleus. Control cells expressing a shorter (G4C2)3 repeat did not accumulate RNA foci. To evaluate whether the foci were RNA specific, transfected cells were treated with RNase or DNase (Figure 6A). Almost all observed foci were degraded by RNase, but not DNase, confirming that the observed foci were primarily composed of RNA.
Figure 6.
miC Variants Inhibit RNA Foci Formation in (G4C2)44-Expressing Cells
(A) RNA foci detected in HEK293T cells expressing (G4C2)44. Cells were transfected with 250 ng (G4C2)44 and (G4C2)3 plasmid and fixed 2 days post-transfection after treatment with DNase or RNase. RNA FISH was performed using a TYE563-(CCCCGG)3 LNA probe (red), and nuclei were stained with DAPI (blue). Nuclear foci were resistant to DNase but degraded by RNase indicating RNA foci. (B) Reduction of RNA foci by miC4_101 and miC32_101. Cells were co-transfected with 250 ng (G4C2)44 and 100 ng miC4_101, miC32_101, or miScr plasmid. Cells were fixed 2 days post-transfection, and RNA FISH was performed as described in (A). (C) Quantification of RNA foci in miC4_101- and miC32_101-transfected cells. A series of five images was made using 10× magnification to quantify the number of cells containing nuclear foci using ImageJ (mean ± SD, one-way ANOVA, multiple-comparison test, ***p < 0.001).
miC4_101 and miC32_101 were evaluated for efficacy on RNA foci formation by co-transfection. Both miC candidates significantly decreased the percentage of (G4C2)44 foci-positive cells by ∼50% after 24 h (Figures 6B and 6C). miScr served as the control and did not reduce the amount of foci in the cells. This confirms that our miC candidates were functional in reducing RNA foci in the cell nucleus.
Reduction of Endogenous C9orf72 in Cells by AAV5-miC
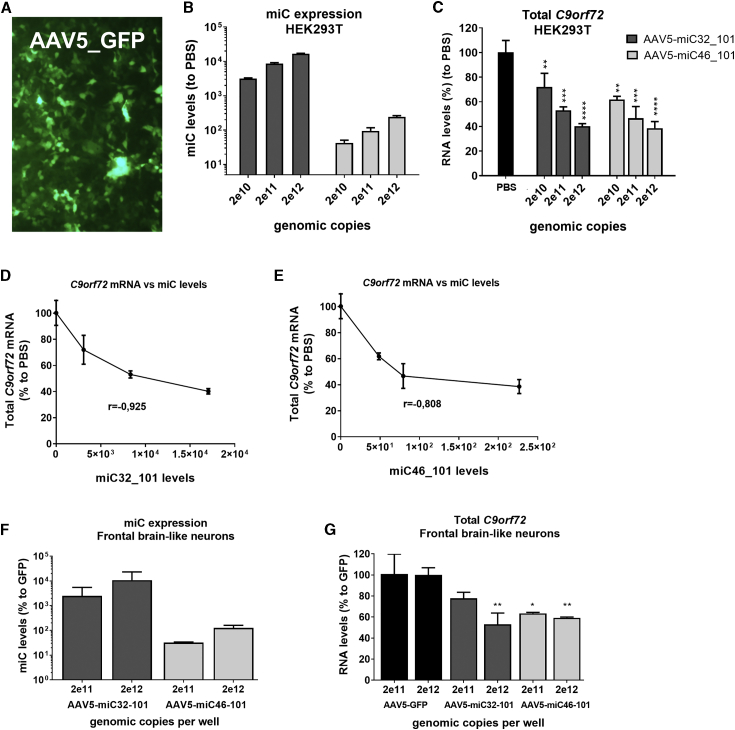
To further investigate the silencing of C9orf72 in the context of a gene therapy for ALS and FTD, we selected miC32_101 as the best candidate to target total C9orf72, based on the strong efficacy and low frequency of passenger strand formation. miC46_101 was selected as the most effective candidate based on silencing efficacy, but its high amount of passenger strand could make it less suitable to treat patients. Both candidates were incorporated into AAV5. Increasing doses of AAV5-miC32_101 and AAV5-miC46_101 were used to transduce HEK293T cells and iPSC-derived frontal brain-like neurons from an FTD patient. Expressions of the mature miC32 and miC46 were verified using TaqMan. Cells transduced with AAV5-GFP served as a control for the transduction efficiency. At 3 days post-transduction, AAV5-GFP transduced ∼80% of HEK293T cells (Figure 7A). The mature guide strand expressions of miC32 and miC46 were expressed in a dose-dependent manner and resulted in a dose-dependent reduction of total C9orf72 expression at a maximum of ∼40%–50% (Figures 7B and 7C). The levels of mature miC32_101 and miC46_101 produced in transduced cells correlated well with C9orf72 silencing (Figures 7D and 7E). Similarly, miC expression and up to 50% lowering of C9ORF72 was observed in transduced frontal brain-like neurons (Figures 7F and 7G). Hence, these data provide a strong rationale for further proof-of-concept studies in animal models of C9orf72-ALS, leading toward a miRNA-based gene therapy to treat ALS and FTD.
Figure 7.
Silencing of Endogenous C9orf72 by AAV5-miC
(A) Transduction efficiency by AAV5 in HEK293T cells. Cells were transduced with 1e12 genomic copies (gc) AAV5-GFP and visualized 3 days post-transduction. (B) Levels of mature miC32_101 and miC46_101 guide strands in transduced cells. Cells were transduced with 2e10, 2e11, and 2e12 gc AAV5-miC32_101 and AAV5-miC46_101. RNA was isolated 3 days post-transduction to determine expression of the mature miC32_101 and miC46_101 by TaqMan. MicroRNA input levels were normalized to U6 small nuclear RNA and set relative to PBS-treated cells. (C) Silencing of C9orf72 in transduced HEK293T cells, performed as described in (B). Total C9orf72 was determined by qRT-PCR. mRNA input was normalized to GAPDH and set relative to PBS-treated cells. (D) Correlation of miC32_101 expression levels and C9orf72 knockdown in cells upon transduction with AAV5-miC32_101. Pearson correlation (r) = −0,925. (E) Correlation of miC46_101 expression levels and C9orf72 knockdown in cells transduced with AAV5-miC46_101 (r = −0,808). (F) Levels of mature miC32_101 and miC46_101 guide strands in transduced iPSC-derived frontal brain-like neurons from an FTD patient. Cells were transduced with 2e11 and 2e12 gc AAV5-miC32_101 and AAV5-miC46_101. RNA was isolated 7 days post-transduction to determine expression of the mature miC32_101 and miC46_101 by TaqMan. MicroRNA input levels were normalized to U6 small nuclear RNA and set relative to AAV5-GFP-treated cells. (G) Reduction of C9orf72 in iPSC-derived frontal brain-like neurons from an FTD patient, performed as described in (F). Total C9orf72 levels were determined by qRT-PCR. mRNA input was normalized to GAPDH and set relative to cells treated with AAV5-GFP. Data were evaluated using two-way ANOVA, multiple-comparison test: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001. Each graph represents the mean values with SD of 2 independent experiments.
Our results indicate that miRNAs could be used as therapeutics to reduce the gain of toxicity caused by the G4C2 expanded repeat of C9orf72. We demonstrated the feasibility of different targeting approaches by miC to silence the sense, antisense, or both transcripts of C9orf72. In addition, the processing of miC in the miR-101 and miR-451 was demonstrated, and both scaffolds produced mature miC that was functional in the cell nucleus and cytoplasm. Silencing of C9orf72 by AAV5-miC was demonstrated in patient-derived frontal brain-like neurons, providing promising results for further development of a miRNA-based gene therapy for ALS and FTD.
Discussion
A G4C2 repeat expansion in intron 1 of C9orf72 is the most frequent cause of ALS and FTD, leading to the accumulation of sense and antisense RNA foci in the cell nucleus and DPR proteins in the cytoplasm. Here we report on the design and characterization of an AAV-delivered miRNA-based approach to target C9orf72 transcripts, with potentially a long-lived therapeutic effect following a single administration. We analyzed RNA-seq data from C9-ALS patients to investigate C9orf72 expression and sequence conservation to predict potential target regions for a miRNA-based targeting approach. We observed reduced levels of C9orf72 in cortex and cerebellum from C9-ALS patients. This finding is consistent with other studies where the significant reduction of C9orf72 was also reported in brain and spinal cord tissues and in iPSC-derived neurons from patients.2, 37, 44, 45, 46 Some studies reported that the V2 variant from C9orf72 was the most affected transcript variant.44, 47 We did not find evidence that the reduction seen in patients was variant specific. Reduced transcription of C9orf72 in ALS patients could be a result of epigenetic silencing due to CpG hypermethylation of the G4C2 repeat, and this raises the question whether haploinsufficiency also contributes to the pathology of the disease.47 However, a significant body of evidence also supports a G4C2-mediated toxicity.46, 47, 48, 49
To identify potential miC target sites, we investigated the sequence conservation of C9orf72. Whereas exonic regions from exon 2 to exon 11 were completely covered, intronic regions were poorly represented, and not much can be concluded about the conservation of intron 1-containing transcripts in patients. Despite the low coverage, read alignment for intronic regions was found in both patients and controls. Interestingly, the coverage of the introns 1–4 was relatively higher in C9-ALS patients than in controls, supporting findings that the impairment of full-length C9orf72 transcription in patients could be a result of the accumulation of aberrant or unspliced C9orf72 transcripts.36, 37 Directly targeting intron 1 is a challenging therapeutic strategy, due to the low expression and difficulty to detect intron 1-containing transcripts, as well as its high GC content. Moreover, GC-rich regions are common in the genome, and therapeutics targeting such regions should be carefully investigated for off-target effects. The coverage of the antisense transcript was very poor, and limited coverage was found only in the 5′ UTR of C9orf72. In contrast, exonic regions had complete coverage and, therefore, are much easier to target.
We designed miC candidates with intron 1-binding sites to target only the sense intronic transcripts and candidates binding within exon 2 and exon 11 to target all sense transcripts. miC candidates were also designed to specifically target the antisense transcripts of C9orf72.
In line with our difficulties to detect the repeat-containing transcripts with RNA-seq, the repeat region could also be less accessible for the mature miC due to its highly structured nature. Only two miC candidates targeting the G4C2 region showed a moderate knockdown of the intron 1 reporter. It is also possible that the high GC content of the mature miC itself interferes with strand loading and target recognition. miC candidates further downstream of the G4C2 repeat in intron 1 or in exon 2 and exon 11 showed a much stronger knockdown of the Luc reporters, and two miC candidates were also effective on the antisense reporter.
One challenge for our miC approach is the simultaneous targeting of sense and antisense transcripts. To test the feasibility of such an approach, we designed a construct that expresses two miC molecules in a concatenated fashion against both sense and antisense transcripts. Silencing of both the intron 1 reporter and the antisense reporter constructs was achieved, demonstrating the feasibility for a bidirectional silencing of C9orf72 by miC. Combining two or more miRNA hairpins in a single construct has been successfully performed previously, and one study showed increased silencing efficacy by miRNAs in a polycistron setting.50, 51 Thus, expressing different miC in a concatenated fashion could be promising to target both sense and antisense transcripts of C9orf72.
In this study, we used two differentially processed miRNA scaffolds, miR-101 and miR-451. The pre-miR-101 follows the canonical processing pathway, as the further downstream processing in the cytoplasm involved Dicer cleavage of the hairpin to generate 5′ arm strands and 3′ arm strands. The 5′ arm strand, which becomes the passenger strand, is usually degraded, though both strands could be incorporated into the RISC and loaded onto Argonaute (Ago), producing active guide and passenger strands that cleave the complimentary mRNA (Figure S2). The pre-miR-451 follows the non-canonical pathway as it escapes Dicer cleavage, producing only a 5′ arm that is subsequently cleaved by Ago and trimmed by PARN into 22- to 24-nt guide strands that are incorporated into the RISC (Figure S3). However, trimming of miR-451 can result in functional guide strands that are longer than the mature length.29, 52 Thus, both miR-101 and miR-451 are promising, but simultaneous guide and passenger strand expressions or mature miC much longer than the predicted length may increase the chance of hitting non-target genes due to miRNA-like effects. Hence, it is important to determine the processing of miC candidates in both scaffolds and assess target similarity with other genes to minimize off-target effects.
The processing of the miC candidates was determined by NGS analysis. We confirmed a differential processing of the various miC-101 candidates, resulting in different ratios of guide and passenger strands. miC2_101, miC4_101, miC32_101, and miC33_101 produced low amounts of passenger strand and are predicted to have few off-target effects. miC46_101, miC49_101, and miC50_101 may have an increased risk for off-target effects due to the high amount of passenger strands. The finding that the miC-101 candidates were differently processed, even in the same miRNA scaffold, supports previous observations that not only the pri-miRNA scaffold but also the miRNA-sequence are critical for choosing which sequences of the duplex enter the RISC.29, 53, 54, 55, 56 This strand selection has been extensively studied, and it seems to be also dependent on the difference in stability of the miRNA duplex at the 5′ ends of each strand.29, 54, 55, 56, 57, 58, 59 miC-451 candidates did not show any passenger strand activity. However, miC38_451 and miC50_451 produced mature guide strands longer than 27 nt, and they could also have increased risk for off-target effects. The different miC-processing pattern suggests that selection of C9orf72-silencing candidates should be based on a balanced assessment of silencing efficacy and NGS processing data.
The efficacy of the best miC candidates was further confirmed by their ability to reduce endogenously expressed C9orf72 mRNA. We found up to ∼50% reduction of total C9orf72 mRNA and ∼25% reduction of intronic transcripts by miC candidates targeting exon 2 or exon 11. The silencing efficacy of miC-451 was slightly lower compared to miC-101, consistent with the lower levels of mature miC-451 detected by NGS. Two miC candidates targeting the repeat region had limited effect on the total C9orf72 mRNA expression, but the reduction of the intronic transcripts was comparable with candidates targeting the total C9orf72 mRNA. Thus, a total silencing approach and a mutant-specific approach can both result in lowering G4C2 repeat-containing C9orf72 transcripts.
Another challenge for therapeutic miC is the nuclear localization of the toxic transcripts. Thus, recognition of the transcripts by the miC candidates within the nucleus is required, whereas the mature miC is produced in the cytoplasm. Nuclear RNA foci have been found in different neuronal cells of patients, including spinal motor neurons, cerebellar Purkinje cells, cerebellar granule cells, hippocampal neurons, and pyramidal cells in the motor cortex.17 We report that the mature miC-101 and miC-451 candidates are detected in both nucleus and cytoplasm, and they lower C9orf72 mRNA in both cellular structures. There is increasing evidence that miRNA-RISC components, such as Ago and Dicer, are present in the cell nucleus and retain their catalytic activity in the nucleus.60, 61 The current assumption is that miRNA-AGO complexes that are formed in the cytoplasm can travel back into the nucleus.61 Indeed, we detected a reduction of nuclear RNA foci after treatment with miC candidates targeting intronic C9orf72 or total C9orf72 transcripts.
A critical factor determining the success of a gene therapy is the selected delivery system to the target cells or tissues. It is likely that a widespread transduction of neurons in the whole brain and spinal cord, including glia cells, will be required to achieve disease-modifying therapeutic effect in ALS and/or FTD patients. AAV vectors exhibit important advantages, including high transgene expression, long-term stability, and positive safety profile due to the non-pathogenic nature of the wild-type form.62 We demonstrated high expression of miC levels and up to 50% reduction of total C9orf72 by AAV5-miC candidates in iPSC-derived frontal brain-like neurons from an FTD patient. It remains questionable how much reduction is required to achieve a rescue effect on the disease phenotype. However, previous findings using ASOs in an ALS mouse model without a clear clinical phenotype showed between 40%–60% reduction of C9orf72, which appears to be sufficient to reduce RNA foci formation by ∼50% and DPR proteins by ∼80%.13
The delivery of RNAi-based AAV treatment strategies in ALS have predominantly been explored in preclinical studies to treat SOD1-ALS and have demonstrated to be promising.63, 64, 65, 66 AAV9 resulted in the effective transduction of spinal cord and motor neurons, but sufficient widespread distribution in the adult CNS upon systemic injection remains challenging.66, 67, 68 In children with spinal muscular atrophy type 1, a single intravenous infusion of AAV9 for gene replacement therapy resulted in widespread transduction of neurons within the spinal cord and significant clinical improvement.69 However, for an RNAi-based approach in adult ALS and FTD patients, the blood-brain barrier (BBB) could still be a major obstacle for sufficient transduction of the CNS after systemic delivery. We have previously demonstrated strong transduction of the striatum and cortex and lowering of mutant huntingtin protein, in a Huntington’s disease minipig model, after direct injection of AAV5 into the striatum.70 Furthermore, intrastriatal injections of AAV5 have been shown to distribute via axons through anterograde and/or retrograde transport and resulted in the transduction of different types of neurons, astrocytes, microglia, and oligodendrocytes.70, 71 Thus, a local delivery to the brain parenchyma is promising in patients with FTD for treatment of the symptoms associated with cortical degeneration. Parenchymal delivery of AAV also resulted in the transduction of motor neurons along the corticospinal tract in non-human primates.72 However, it needs to be assessed whether the transduction of motor neurons is sufficient to achieve therapeutic concentrations of AAV. An alternative approach could be intrathecal administration, but further studies in large animals are required to predict the efficacy in the brain and spinal cord. Ultimately, intrathecal administration combined with parenchymal administration to the brain may be required to obtain sufficient transduction of the affected areas in ALS and ALS and/or FTD patients.
In summary, following an RNA sequence analysis of C9orf72 in ALS patients and controls, we rationally designed miC candidates using the miR-101 and miR-451 scaffolds to target total, intronic, and antisense C9orf72 transcripts. We demonstrated that specifically targeting the G4C2 repeat by miC is possible but challenging. The feasibility of a bidirectional approach was also demonstrated by expressing two miC hairpins targeting the sense and antisense C9orf72 in a concatenated fashion. The efficacy of the miC candidates was evaluated based on their ability to lower C9orf72 mRNA, and the processing was determined by NGS. In addition, we also provided evidence that miC can be active in the nucleus by the reduction of nuclear C9orf72 mRNA and RNA foci.
Materials and Methods
RNA-Seq and C9orf72 Target Sequences
An RNA-seq library published by Prudencio et al.31 was downloaded. The data were analyzed by BaseClear B.V. In brief, we downloaded RNA-seq samples from the Sequence Read Archive (SRA) from NCBI GEO: GSE67196 (https://www.ncbi.nlm.nih.gov/geo/).31 These samples were individually mapped to the human reference genome (GRCh38.p10) with TopHat version 2.1.1.73 The mapped reads from TopHat are fragments per kilobase of transcript per million mapped reads (FPKM) values. Differential gene and isoform expressions were estimated on their relative abundance by Cufflinks 2.2.1.74 With the CummeRbund 2.16 R package, we visualized gene and isoform expressions of C9orf72.75 The alignment (.bam) and junctions files (.bed) generated by TopHat were used by the Integrative Genomics Viewer (IGV) 2.3.94.76
DNA Constructs
To generate the miC vectors, we searched for sequences in intron 1, exon 2, and exon 11 of C9orf72 that were mostly conserved among human and non-human primates and mouse. The miC sequences were incorporated into the cellular pri-miRNA miR-101-1 or miR-451 scaffold of the human. 200-nt 5′- and 3′-flanking regions were included with EcoRV and BamHI restriction sites, and the mfold program (http://unafold.rna.albany.edu/?q=mfold) was used to determine if the miC candidates were folded correctly into their secondary structures. The complete sequences were ordered from GeneArt gene synthesis (Invitrogen). These constructs were subsequently cloned into an expression vector containing the CMV immediate-early enhancer fused to chicken β-actin (CAG) promoter (Inovio, Plymouth Meeting, PA) using the EcoRV and BamHI sites. For generation of the Luc reporters, sequences from C9orf72 intron 1 (sense and antisense), exon 2, or exon 11 were synthesized at GeneArt gene synthesis and cloned in the 3′ UTR of the RL gene of the psiCHECK-2 vector (Promega, Madison, WI). The FL gene was also expressed in this vector and served as an internal control (Figure 2D).
Culture and Transfections of HEK293T Cells
HEK293T cells were maintained in DMEM (Invitrogen) containing 10% fetal calf serum (Greiner, Kremsmünster, Austria), 100 U/mL penicillin, and 100 U/mL streptomycin (Thermo Fisher Scientific, Waltham, MA), at 37°C and 5% CO2. For all assays, cells were seeded in 24-well plates at a density of 0.1*106 cells per well in DMEM. Transfections in HEK293T cells were performed 1 day post-plating with Lipofectamine 2000 reagent (Invitrogen), according to the manufacturer’s instructions.
Culture of iPSC Neurons
FTD (ND42765) iPSCs derived from fibroblasts were ordered from Coriell Biorepository (https://www.coriell.org/), and they were cultured on Matrigel (Corning)-coated 6-well plates in mTeSR1 (STEMCELL Technologies). For embryoid body-based neural induction, iPSCs were seeded on AggreWell800 plates and cultured in STEMdiff Neural Induction Medium (STEMCELL Technologies) for 5 days with daily medium changes. Embryoid bodies were harvested and plated on 6-well plates coated with poly-D-lysine (Sigma-Aldrich) and laminin (Sigma-Aldrich) in STEMdiff Neural Induction Medium for 7 days with daily medium changes. Rosettes were selected with rosette selection medium and plated on poly-D-lysine- and laminin-coated 6-well plates in STEMdiff Neural Induction Medium for 24 h. For differentiation into frontal brain-like neurons, STEMdiff Neural Induction Medium was replaced with STEMdiff Neuron Differentiation Medium (STEMCELL Technologies), and neuroprogenitor cells were differentiated for 5 days. The neuroprogenitor cells were then plated on poly-D-lysine- and laminin-coated plates in STEMdiff Neuron Maturation Medium (STEMCELL Technologies) for 1 week. The mature frontal brain-like neurons were stored in liquid nitrogen in Neuroprogenitor Freezing Medium (STEMCELL Technologies).
Luciferase Assays
HEK293T cells co-transfected with the miC expression constructs and Luc reporters were assayed at 48 h post-transfection in 100 μL 1× passive lysis buffer (Promega) by gentle rocking for 15 min at room temperature. The cell lysates were centrifuged for 5 min at 4,000 rpm to get rid of cell debris, and 10 μL supernatant was used to measure FL and RL activities with the Dual-Luciferase Reporter Assay System (Promega). Relative luciferase activity was calculated as the ratio between RL and FL activities.
RNA Isolation
For all RNA isolation, cells were lysed in 200 μL Tryzol, and RNA isolation was performed using the Direct-zol kit (R2061, Zymo Research), according to the manufacturer’s protocol.
qRT-PCR and miRNA TaqMan Assay
To determine C9orf72 mRNA knockdown, HEK293T cells were transfected with different concentrations of the miC variants. Total RNA was isolated from the cells at 2 days post-transfection. Genomic DNA contamination was removed by DNase treatment using recombinant shrimp DNase (Thermo Fisher Scientific). First-strand complementary DNA was reverse transcribed using random hexamer primers with the Dynamo kit (Finnzymes, Espoo, Finland). Real-time PCR amplification was performed with primers targeting total C9orf72 (forward 5′-CGGAAAGGAAGAATATGGATGC-3′, reverse 5′-CCATTACAGGAATCACTTCTCCA-3′, and probe 5′-AGCATTGGAATAATACTCTGACCCTGATCTTC-3′) or sense intronic C9orf72 (forward 5′-ACGCCTGCACAATTTCAGCCCAA-3′, reverse 5′-CAAGTCTGTGTCATCTCGGAGCTG-3′, and probe 5′-TGAGGGCAGCAATGCAAGTCGGTGTG-3′).77 The assays were performed on ABI 7000 or ABI 7500 (Applied Biosystems, Foster City, CA, USA). The mRNA expression levels were normalized to human GAPDH (forward 5′-GAAGGTGAAGGTCGGAGTC-3′, reverse 5′-GAAGATGGTGATGGGATTTC-3′, and probe 5′-CAAGCTTCCCGTTCTCAGCC-3′) as an internal control, and the level of gene expression was calculated relative to cells transfected with a scrambled plasmid.
To determine the expression of miC, a Custom TaqMan Small RNA Assay Design Tool (Thermo Fisher Scientific) was used to design miC2 (assay ID CTEPR3R), miC4 (assay ID CTFVKNN), miC32 (assay ID CSGJPRB), miC46 (assay ID CSHSNXJ), miC49 (assay ID CTGZE9K), and miC50 (assay ID CTH49UH). All RT reactions and TaqMan for small RNAs were performed according to the manufacturer’s protocols.
RNA Isolation from Nucleus and Cytoplasm Separation
Nucleus and cytoplasm were isolated 2 days post-transfections. Cells were washed with cold 1× PBS, and 100 μL cold Nuclei EZ Lysis Buffer was added. Cells were incubated on a shaker on ice for 5 min, and nucleus was collected by centrifugation at 500 × g for 5 min. The supernatant contained cytoplasmic components and was stored. The pellets were washed twice in 300 μL cold Nuclei EZ Lysis Buffer, and the pellets containing nuclei components were stored. 200 μL Tryzol was added to both cytoplasmic and nuclei lysates, and RNA was isolated as described in the previous section.
NGS and Data Analysis
Small RNA-seq libraries for the Illumina sequencing platform were generated using high-quality total RNA as input and the NEXTflex Small RNA-seq kit (Bioo Scientific, Austin, TX) as described before.29 Briefly, the small RNA species were subjected to ligation with 3′ and 5′ RNA adapters, first-strand reverse transcription, and PCR amplification. Sample-specific barcodes were introduced in the PCR step. The PCR products were separated on Tris/Borate/EDTA (TBE)-PAGE electrophoresis, and the expected band around 30 bp was recovered for each sample. The resulting sequencing libraries were quantified on a BioAnalyzer (Agilent, Santa Clara, CA). The libraries were multiplexed, clustered, and sequenced on an Illumina HiSeq 2000 (TruSeq version [v.]3 chemistry) with a single-read 36-cycle sequencing protocol and indexing. The sequencing run was analyzed with the Illumina CASAVA pipeline (v.1.8.2), with demultiplexing based on sample-specific barcodes. The raw sequencing data produced were processed, removing the sequence reads that were of too low quality (only “passing filter” reads were selected). In total, between 15 and 35 million reads per sample were generated. NGS small RNA raw datasets were analyzed using the CLC Genomics Workbench 8 (QIAGEN). The obtained reads were adaptor trimmed, which decreased the average read size from ∼50 to ∼25 bp. All reads containing ambiguity N symbols, shorter than 10 nt, longer than 45 nt, and represented less than 10 times were discarded. Next, the obtained unique small RNA reads were aligned to the reference sequences of the pre-miC constructs with a maximum of 3-nt mismatches allowed. The percentages of reads based on the total number of reads matching the reference sequence were calculated (Table S1).
Cloning of (G4C2)44-Exon2 and (G4C2)3-Exon2 Expression Vectors
To generate (G4C2)44 and (G4C2)3 expression vectors, we designed a sequence consisting of the intronic C9orf72 repeat region. The sequence contained either 44 or 3 repeats of the G4C2 hexanucleotide, and 150 nt of 5′- and 50 nt of 3′-flanking regions were added. A restriction site ApaI was included at the 3′ end to serve as a cloning site. These sequences were synthesized and ordered from GeneArt gene synthesis (Invitrogen). To link the (G4C2)44 and (G4C2)3 vectors to exon 2, the C9orf72 exon 2 sequence was ordered at GeneArt gene synthesis and cloned in the (G4C2)44 and (G4C2)3 vectors using the ApaI site.
RNA Foci FISH
RNA FISH was performed as described previously with some adjustments.42 In brief, HEK293T cells were grown on the poly-D-lysine-coated 8-well Nunc Lab-Tek Chamber Slide System at a density of 80,000 cells/well. The cells were transfected on day 2 with 200 ng (G4C2)44 or (G4C2)3 plasmid. On day 4, cells were fixed in 4% paraformaldehyde for 20 min, permeabilized in ice-cold methanol for 10 min, and washed 3 times with diethylpyrocarbonate (DEPC)-treated PBS (DEPC-PBS). Optionally, cells were treated for 30 min with 5 mg/mL RNase A (QIAGEN) or 100 U RNase-free DNase (Invitrogen). The cells were incubated for 1 h in hybridization buffer (50% formamide, 10% dextran sulfate, 0.1 mg/mL yeast tRNA,2×saline-sodium citrate (SSC), and 50 mM sodium phosphate) at 37°C and hybridized overnight with 40 nm TYE563-(C4G2)3 LNA probe in hybridization buffer at 37°C. Cells were then washed once with 40% formamide and 1×SSC for 30 min at 37°C and 3 times with DEPC-PBS at room temperature for 5 min. The slides were mounted with ProLong Gold Antifade Mountant with DAPI (Invitrogen) and visualized using a LEICA DM2500 fluorescence microscope.
To determine the effect of miC4 and miC32 on foci formation, HEK293T cells were co-transfected with 200 ng (G4C2)44 and 400 ng miC4, miC32, or miScr plasmid. At 2 days post-transfection, cells were fixed and subjected to FISH as described above. To quantify foci-bearing cells, ten fields were randomly selected under 10× magnification. For each field, the number of foci-positive nuclei and the total number of nuclei were counted using ImageJ software. These counts were used to determine the average percentage of foci-positive cells for each condition.
AAV5 Vector Production and Transductions
AAV5 encoding miC32_101 and miC46_101 was produced by a baculovirus-based AAV production system, as described previously.29 Briefly, the miC cassettes were obtained by digestion with restriction enzymes HindIII and PvuI and cloned in a uniQure transfer plasmid named pVD789 in order to generate an entry plasmid. The presence of the two inverted terminal repeats (ITRs) was confirmed by restriction digestion with SmaI. The ITR-CAG-miC cassettes were inserted into a recombinant baculovirus vector by homologous recombination in Spodoptera frugiperda Sf9 cells, and clones were selected and screened by plague purification and PCR. The recombinant baculovirus containing the ITR-CAG-miC cassettes was further amplified until passage 6 (P6) in Sf+ cells and screened for the best production and stability by PCR and qRT-PCR. To generate AAV5, Sf+ cells were triple infected with three different recombinant baculoviruses expressing the ITRs-CAG-miC, the replicon enzyme, and the capsid protein. The cells were lysed 72 h after the triple infection, and the crude lysate was treated with 50 U/mL Benzonase (Merck, Darmstadt, Germany) for 1 h at 37°C. AAV5 was purified on an AVB Sepharose column (GE Healthcare, Little Chalfont, UK) using an AKTA purification system (GE Healthcare). The final titer concentration was determined by qRT-PCR with primers amplifying a 95-bp fragment from the CAG promoter region.
For transductions with AAV, HEK293T cells were plated in 24-well plates at 0.1 × 106 cells/well in DMEM and transduced with AAV after 24 h. Mature frontal brain-like neurons were plated at 0.3 × 106 cells/well and transduced with AAV after 1 week. RNA isolation and TaqMan for miC32_101, miC46_101, and total C9orf72 qRT-PCR were performed as described previously.
Statistical Analysis
Data were analyzed using the one-way ANOVA or Student’s t test to determine statistical significances between control and treated cells. A two-way ANOVA was used to determine statistical significances among multiple treated groups. The p values were either listed or represented by the following number of asterisks: *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Author Contributions
Conceptualization, P.K. and R.M.; Investigation, R.M., J.M.L., T.v.d.Z., J.M., and J.S.; Supervision, P.K.; Formal Analysis, R.M., J.M.L., and I.K.; Visualization and Writing – Initial Draft, R.M.; Project Administration and Writing – Review and Editing, P.K., S.J.v.D., M.M.E., and H.P.; Funding Acquisition, P.K.
Acknowledgments
The authors would like to thank Olivier ter Brake and Eileen Sawyer for reviewing the manuscript.
Footnotes
Supplemental Information includes three figures and two tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2019.01.010.
Supplemental Information
References
- 1.Renton A.E., Majounie E., Waite A., Simón-Sánchez J., Rollinson S., Gibbs J.R., Schymick J.C., Laaksovirta H., van Swieten J.C., Myllykangas L., ITALSGEN Consortium A hexanucleotide repeat expansion in C9ORF72 is the cause of chromosome 9p21-linked ALS-FTD. Neuron. 2011;72:257–268. doi: 10.1016/j.neuron.2011.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.DeJesus-Hernandez M., Mackenzie I.R., Boeve B.F., Boxer A.L., Baker M., Rutherford N.J., Nicholson A.M., Finch N.A., Flynn H., Adamson J. Expanded GGGGCC hexanucleotide repeat in noncoding region of C9ORF72 causes chromosome 9p-linked FTD and ALS. Neuron. 2011;72:245–256. doi: 10.1016/j.neuron.2011.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mancuso R., Navarro X. Amyotrophic lateral sclerosis: Current perspectives from basic research to the clinic. Prog. Neurobiol. 2015;133:1–26. doi: 10.1016/j.pneurobio.2015.07.004. [DOI] [PubMed] [Google Scholar]
- 4.Nolan M., Talbot K., Ansorge O. Pathogenesis of FUS-associated ALS and FTD: insights from rodent models. Acta Neuropathol. Commun. 2016;4:99. doi: 10.1186/s40478-016-0358-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Heutink P., Jansen I.E., Lynes E.M. C9orf72; abnormal RNA expression is the key. Exp. Neurol. 2014;262(Pt B):102–110. doi: 10.1016/j.expneurol.2014.05.020. [DOI] [PubMed] [Google Scholar]
- 6.Gijselinck I., Van Mossevelde S., van der Zee J., Sieben A., Engelborghs S., De Bleecker J., Ivanoiu A., Deryck O., Edbauer D., Zhang M. The C9orf72 repeat size correlates with onset age of disease, DNA methylation and transcriptional downregulation of the promoter. Mol. Psychiatry. 2016;21:1112–1124. doi: 10.1038/mp.2015.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van Blitterswijk M., DeJesus-Hernandez M., Rademakers R. How do C9ORF72 repeat expansions cause amyotrophic lateral sclerosis and frontotemporal dementia: can we learn from other noncoding repeat expansion disorders? Curr. Opin. Neurol. 2012;25:689–700. doi: 10.1097/WCO.0b013e32835a3efb. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mis M.S.C., Brajkovic S., Tafuri F., Bresolin N., Comi G.P., Corti S. Development of Therapeutics for C9ORF72 ALS/FTD-Related Disorders. Mol. Neurobiol. 2017;54:4466–4476. doi: 10.1007/s12035-016-9993-0. [DOI] [PubMed] [Google Scholar]
- 9.Gendron T.F., Belzil V.V., Zhang Y.J., Petrucelli L. Mechanisms of toxicity in C9FTLD/ALS. Acta Neuropathol. 2014;127:359–376. doi: 10.1007/s00401-013-1237-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ling S.C., Polymenidou M., Cleveland D.W. Converging mechanisms in ALS and FTD: disrupted RNA and protein homeostasis. Neuron. 2013;79:416–438. doi: 10.1016/j.neuron.2013.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ciura S., Lattante S., Le Ber I., Latouche M., Tostivint H., Brice A., Kabashi E. Loss of function of C9orf72 causes motor deficits in a zebrafish model of amyotrophic lateral sclerosis. Ann. Neurol. 2013;74:180–187. doi: 10.1002/ana.23946. [DOI] [PubMed] [Google Scholar]
- 12.Koppers M., Blokhuis A.M., Westeneng H.J., Terpstra M.L., Zundel C.A.C., Vieira de Sá R., Schellevis R.D., Waite A.J., Blake D.J., Veldink J.H. C9orf72 ablation in mice does not cause motor neuron degeneration or motor deficits. Ann. Neurol. 2015;78:426–438. doi: 10.1002/ana.24453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jiang J., Zhu Q., Gendron T.F., Saberi S., McAlonis-Downes M., Seelman A., Stauffer J.E., Jafar-Nejad P., Drenner K., Schulte D. Gain of Toxicity from ALS/FTD-Linked Repeat Expansions in C9ORF72 Is Alleviated by Antisense Oligonucleotides Targeting GGGGCC-Containing RNAs. Neuron. 2016;90:535–550. doi: 10.1016/j.neuron.2016.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Harms M.B., Cady J., Zaidman C., Cooper P., Bali T., Allred P., Cruchaga C., Baughn M., Libby R.T., Pestronk A. Lack of C9ORF72 coding mutations supports a gain of function for repeat expansions in amyotrophic lateral sclerosis. Neurobiol. Aging. 2013;34:2234.e13–2234.e19. doi: 10.1016/j.neurobiolaging.2013.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.O’Rourke J.G., Bogdanik L., Muhammad A.K.M.G., Gendron T.F., Kim K.J., Austin A., Cady J., Liu E.Y., Zarrow J., Grant S. C9orf72 BAC Transgenic Mice Display Typical Pathologic Features of ALS/FTD. Neuron. 2015;88:892–901. doi: 10.1016/j.neuron.2015.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Almeida S., Gascon E., Tran H., Chou H.J., Gendron T.F., Degroot S., Tapper A.R., Sellier C., Charlet-Berguerand N., Karydas A. Modeling key pathological features of frontotemporal dementia with C9ORF72 repeat expansion in iPSC-derived human neurons. Acta Neuropathol. 2013;126:385–399. doi: 10.1007/s00401-013-1149-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lagier-Tourenne C., Baughn M., Rigo F., Sun S., Liu P., Li H.-R., Jiang J., Watt A.T., Chun S., Katz M. Targeted degradation of sense and antisense C9orf72 RNA foci as therapy for ALS and frontotemporal degeneration. Proc. Natl. Acad. Sci. USA. 2013;110:E4530–E4539. doi: 10.1073/pnas.1318835110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gendron T.F., Bieniek K.F., Zhang Y.J., Jansen-West K., Ash P.E.A., Caulfield T., Daughrity L., Dunmore J.H., Castanedes-Casey M., Chew J. Antisense transcripts of the expanded C9ORF72 hexanucleotide repeat form nuclear RNA foci and undergo repeat-associated non-ATG translation in c9FTD/ALS. Acta Neuropathol. 2013;126:829–844. doi: 10.1007/s00401-013-1192-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Peters O.M., Cabrera G.T., Tran H., Gendron T.F., McKeon J.E., Metterville J., Weiss A., Wightman N., Salameh J., Kim J. Human C9ORF72 Hexanucleotide Expansion Reproduces RNA Foci and Dipeptide Repeat Proteins but Not Neurodegeneration in BAC Transgenic Mice. Neuron. 2015;88:902–909. doi: 10.1016/j.neuron.2015.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ash P.E.A., Bieniek K.F., Gendron T.F., Caulfield T., Lin W.L., Dejesus-Hernandez M., van Blitterswijk M.M., Jansen-West K., Paul J.W., 3rd, Rademakers R. Unconventional translation of C9ORF72 GGGGCC expansion generates insoluble polypeptides specific to c9FTD/ALS. Neuron. 2013;77:639–646. doi: 10.1016/j.neuron.2013.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zu T., Liu Y., Bañez-Coronel M., Reid T., Pletnikova O., Lewis J., Miller T.M., Harms M.B., Falchook A.E., Subramony S.H. RAN proteins and RNA foci from antisense transcripts in C9ORF72 ALS and frontotemporal dementia. Proc. Natl. Acad. Sci. USA. 2013;110:E4968–E4977. doi: 10.1073/pnas.1315438110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hu J., Liu J., Li L., Gagnon K.T., Corey D.R. Engineering Duplex RNAs for Challenging Targets: Recognition of GGGGCC/CCCCGG Repeats at the ALS/FTD C9orf72 Locus. Chem. Biol. 2015;22:1505–1511. doi: 10.1016/j.chembiol.2015.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nordin A., Akimoto C., Wuolikainen A., Alstermark H., Forsberg K., Baumann P., Pinto S., de Carvalho M., Hübers A., Nordin F. Sequence variations in C9orf72 downstream of the hexanucleotide repeat region and its effect on repeat-primed PCR interpretation: a large multinational screening study. Amyotroph. Lateral Scler. Frontotemporal Degener. 2017;18:256–264. doi: 10.1080/21678421.2016.1262423. [DOI] [PubMed] [Google Scholar]
- 24.Hu J., Rigo F., Prakash T.P., Corey D.R. Recognition of c9orf72 Mutant RNA by Single-Stranded Silencing RNAs. Nucleic Acid Ther. 2017;27:87–94. doi: 10.1089/nat.2016.0655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gendron T.F., Chew J., Stankowski J.N., Hayes L.R., Zhang Y.J., Prudencio M., Carlomagno Y., Daughrity L.M., Jansen-West K., Perkerson E.A. Poly(GP) proteins are a useful pharmacodynamic marker for C9ORF72-associated amyotrophic lateral sclerosis. Sci. Transl. Med. 2017;9:eaai7866. doi: 10.1126/scitranslmed.aai7866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Drouet V., Perrin V., Hassig R., Dufour N., Auregan G., Alves S., Bonvento G., Brouillet E., Luthi-Carter R., Hantraye P., Déglon N. Sustained effects of nonallele-specific Huntingtin silencing. Ann. Neurol. 2009;65:276–285. doi: 10.1002/ana.21569. [DOI] [PubMed] [Google Scholar]
- 27.Stanek L.M., Sardi S.P., Mastis B., Richards A.R., Treleaven C.M., Taksir T., Misra K., Cheng S.H., Shihabuddin L.S. Silencing mutant huntingtin by adeno-associated virus-mediated RNA interference ameliorates disease manifestations in the YAC128 mouse model of Huntington’s disease. Hum. Gene Ther. 2014;25:461–474. doi: 10.1089/hum.2013.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Boudreau R.L., Rodríguez-Lebrón E., Davidson B.L. RNAi medicine for the brain: progresses and challenges. Hum. Mol. Genet. 2011;20(R1):R21–R27. doi: 10.1093/hmg/ddr137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Miniarikova J., Zanella I., Huseinovic A., van der Zon T., Hanemaaijer E., Martier R., Koornneef A., Southwell A.L., Hayden M.R., van Deventer S.J. Design, Characterization, and Lead Selection of Therapeutic miRNAs Targeting Huntingtin for Development of Gene Therapy for Huntington’s Disease. Mol. Ther. Nucleic Acids. 2016;5:e297. doi: 10.1038/mtna.2016.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Snøve O., Jr., Rossi J.J. Toxicity in mice expressing short hairpin RNAs gives new insight into RNAi. Genome Biol. 2006;7:231. doi: 10.1186/gb-2006-7-8-231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prudencio M., Belzil V.V., Batra R., Ross C.A., Gendron T.F., Pregent L.J., Murray M.E., Overstreet K.K., Piazza-Johnston A.E., Desaro P. Distinct brain transcriptome profiles in C9orf72-associated and sporadic ALS. Nat. Neurosci. 2015;18:1175–1182. doi: 10.1038/nn.4065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee Y., Kim M., Han J., Yeom K.-H., Lee S., Baek S.H., Kim V.N. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 34.Han J., Lee Y., Yeom K.H., Kim Y.K., Jin H., Kim V.N. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Auyeung V.C., Ulitsky I., McGeary S.E., Bartel D.P. Beyond secondary structure: primary-sequence determinants license pri-miRNA hairpins for processing. Cell. 2013;152:844–858. doi: 10.1016/j.cell.2013.01.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Niblock M., Smith B.N., Lee Y.-B., Sardone V., Topp S., Troakes C., Al-Sarraj S., Leblond C.S., Dion P.A., Rouleau G.A. Retention of hexanucleotide repeat-containing intron in C9orf72 mRNA: implications for the pathogenesis of ALS/FTD. Acta Neuropathol. Commun. 2016;4:18. doi: 10.1186/s40478-016-0289-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Haeusler A.R., Donnelly C.J., Periz G., Simko E.A.J., Shaw P.G., Kim M.S., Maragakis N.J., Troncoso J.C., Pandey A., Sattler R. C9orf72 nucleotide repeat structures initiate molecular cascades of disease. Nature. 2014;507:195–200. doi: 10.1038/nature13124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Klein R.L., Hamby M.E., Gong Y., Hirko A.C., Wang S., Hughes J.A., King M.A., Meyer E.M. Dose and promoter effects of adeno-associated viral vector for green fluorescent protein expression in the rat brain. Exp. Neurol. 2002;176:66–74. doi: 10.1006/exnr.2002.7942. [DOI] [PubMed] [Google Scholar]
- 39.Grimm D., Streetz K.L., Jopling C.L., Storm T.A., Pandey K., Davis C.R., Marion P., Salazar F., Kay M.A. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 40.McBride J.L., Boudreau R.L., Harper S.Q., Staber P.D., Monteys A.M., Martins I., Gilmore B.L., Burstein H., Peluso R.W., Polisky B. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl. Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cifuentes D., Xue H., Taylor D.W., Patnode H., Mishima Y., Cheloufi S., Ma E., Mane S., Hannon G.J., Lawson N.D. A novel miRNA processing pathway independent of Dicer requires Argonaute2 catalytic activity. Science. 2010;328:1694–1698. doi: 10.1126/science.1190809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Su Z., Zhang Y., Gendron T.F., Bauer P.O., Chew J., Yang W.Y., Fostvedt E., Jansen-West K., Belzil V.V., Desaro P. Discovery of a biomarker and lead small molecules to target r(GGGGCC)-associated defects in c9FTD/ALS. Neuron. 2014;83:1043–1050. doi: 10.1016/j.neuron.2014.07.041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stepto A., Gallo J.M., Shaw C.E., Hirth F. Modelling C9ORF72 hexanucleotide repeat expansion in amyotrophic lateral sclerosis and frontotemporal dementia. Acta. Neuropathol. 2014;127:377–389. doi: 10.1007/s00401-013-1235-1. [DOI] [PubMed] [Google Scholar]
- 44.van Blitterswijk M., Gendron T.F., Baker M.C., DeJesus-Hernandez M., Finch N.A., Brown P.H., Daughrity L.M., Murray M.E., Heckman M.G., Jiang J. Novel clinical associations with specific C9ORF72 transcripts in patients with repeat expansions in C9ORF72. Acta. Neuropathol. 2015;130:863–876. doi: 10.1007/s00401-015-1480-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Donnelly C.J., Zhang P.W., Pham J.T., Haeusler A.R., Mistry N.A., Vidensky S., Daley E.L., Poth E.M., Hoover B., Fines D.M. RNA toxicity from the ALS/FTD C9ORF72 expansion is mitigated by antisense intervention. Neuron. 2013;80:415–428. doi: 10.1016/j.neuron.2013.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Shi Y., Lin S., Staats K.A., Li Y., Chang W.-H., Hung S.-T., Hendricks E., Linares G.R., Wang Y., Son E.Y. Haploinsufficiency leads to neurodegeneration in C9ORF72 ALS/FTD human induced motor neurons. Nat. Med. 2018;24:313–325. doi: 10.1038/nm.4490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu E.Y., Russ J., Wu K., Neal D., Suh E., McNally A.G., Irwin D.J., Van Deerlin V.M., Lee E.B. C9orf72 hypermethylation protects against repeat expansion-associated pathology in ALS/FTD. Acta Neuropathol. 2014;128:525–541. doi: 10.1007/s00401-014-1286-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fratta P., Poulter M., Lashley T., Rohrer J.D., Polke J.M., Beck J., Ryan N., Hensman D., Mizielinska S., Waite A.J. Homozygosity for the C9orf72 GGGGCC repeat expansion in frontotemporal dementia. Acta Neuropathol. 2013;126:401–409. doi: 10.1007/s00401-013-1147-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu Y., Pattamatta A., Zu T., Reid T., Bardhi O., Borchelt D.R., Yachnis A.T., Ranum L.P. C9orf72 BAC Mouse Model with Motor Deficits and Neurodegenerative Features of ALS/FTD. Neuron. 2016;90:521–534. doi: 10.1016/j.neuron.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 50.Liu Y.P., Haasnoot J., ter Brake O., Berkhout B., Konstantinova P. Inhibition of HIV-1 by multiple siRNAs expressed from a single microRNA polycistron. Nucleic Acids Res. 2008;36:2811–2824. doi: 10.1093/nar/gkn109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sun D., Melegari M., Sridhar S., Rogler C.E., Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 52.Yoda M., Cifuentes D., Izumi N., Sakaguchi Y., Suzuki T., Giraldez A.J., Tomari Y. Poly(A)-specific ribonuclease mediates 3′-end trimming of Argonaute2-cleaved precursor microRNAs. Cell Rep. 2013;5:715–726. doi: 10.1016/j.celrep.2013.09.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Schwarz D.S., Hutvágner G., Du T., Xu Z., Aronin N., Zamore P.D. Asymmetry in the assembly of the RNAi enzyme complex. Cell. 2003;115:199–208. doi: 10.1016/s0092-8674(03)00759-1. [DOI] [PubMed] [Google Scholar]
- 54.Vitsios D.M., Davis M.P., van Dongen S., Enright A.J. Large-scale analysis of microRNA expression, epi-transcriptomic features and biogenesis. Nucleic Acids Res. 2017;45:1079–1090. doi: 10.1093/nar/gkw1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Maczuga P., Verheij J., van der Loos C., van Logtenstein R., Hooijer G., Martier R., Borel F., Lubelski J., Koornneef A., Blits B. Therapeutic expression of hairpins targeting apolipoprotein B100 induces phenotypic and transcriptome changes in murine liver. Gene Ther. 2014;21:60–70. doi: 10.1038/gt.2013.58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Maczuga P., Koornneef A., Borel F., Petry H., van Deventer S., Ritsema T., Konstantinova P. Optimization and comparison of knockdown efficacy between polymerase II expressed shRNA and artificial miRNA targeting luciferase and Apolipoprotein B100. BMC Biotechnol. 2012;12:42. doi: 10.1186/1472-6750-12-42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Slezak-Prochazka I., Durmus S., Kroesen B.-J., van den Berg A. MicroRNAs, macrocontrol: regulation of miRNA processing. RNA. 2010;16:1087–1095. doi: 10.1261/rna.1804410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Guo L., Lu Z. The fate of miRNA* strand through evolutionary analysis: implication for degradation as merely carrier strand or potential regulatory molecule? PLoS ONE. 2010;5:e11387. doi: 10.1371/journal.pone.0011387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diederichs S., Haber D.A. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 60.Gagnon K.T., Li L., Chu Y., Janowski B.A., Corey D.R. RNAi factors are present and active in human cell nuclei. Cell Rep. 2014;6:211–221. doi: 10.1016/j.celrep.2013.12.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Catalanotto C., Cogoni C., Zardo G. MicroRNA in control of gene expression: An overview of nuclear functions. Int. J. Mol. Sci. 2016;17:E1712. doi: 10.3390/ijms17101712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Saraiva J., Nobre R.J., Pereira de Almeida L. Gene therapy for the CNS using AAVs: The impact of systemic delivery by AAV9. J. Control. Release. 2016;241:94–109. doi: 10.1016/j.jconrel.2016.09.011. [DOI] [PubMed] [Google Scholar]
- 63.Borel F., Gernoux G., Cardozo B., Metterville J.P.T., Toro Cabrera G.C., Song L., Su Q., Gao G.P., Elmallah M.K., Brown R.H., Jr., Mueller C. Therapeutic rAAVrh10 Mediated SOD1 Silencing in Adult SOD1(G93A) Mice and Nonhuman Primates. Hum. Gene Ther. 2016;27:19–31. doi: 10.1089/hum.2015.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Biferi M., Cohen-Tannoudji M., Cappelletto A., Giroux B., Roda M., Astord S., Marais T., Ferry A., Voit T., Barkats M. A new AAV10-mediated gene therapy for SOD1-linked ALS. Neuromuscul. Disord. 2017;27(Suppl 2):S246. doi: 10.1016/j.ymthe.2017.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Dirren E., Aebischer J., Rochat C., Towne C., Schneider B.L., Aebischer P. SOD1 silencing in motoneurons or glia rescues neuromuscular function in ALS mice. Ann. Clin. Transl. Neurol. 2015;2:167–184. doi: 10.1002/acn3.162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Foust K.D., Salazar D.L., Likhite S., Ferraiuolo L., Ditsworth D., Ilieva H., Meyer K., Schmelzer L., Braun L., Cleveland D.W., Kaspar B.K. Therapeutic AAV9-mediated suppression of mutant SOD1 slows disease progression and extends survival in models of inherited ALS. Mol. Ther. 2013;21:2148–2159. doi: 10.1038/mt.2013.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Stoica L., Todeasa S.H., Cabrera G.T., Salameh J.S., ElMallah M.K., Mueller C., Brown R.H., Jr., Sena-Esteves M. Adeno-associated virus-delivered artificial microRNA extends survival and delays paralysis in an amyotrophic lateral sclerosis mouse model. Ann. Neurol. 2016;79:687–700. doi: 10.1002/ana.24618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wang W., Wen D., Duan W., Yin J., Cui C., Wang Y., Li Z., Liu Y., Li C. Systemic administration of scAAV9-IGF1 extends survival in SOD1G93A ALS mice via inhibiting p38 MAPK and the JNK-mediated apoptosis pathway. Brain Res. Bull. 2018;139:203–210. doi: 10.1016/j.brainresbull.2018.02.015. [DOI] [PubMed] [Google Scholar]
- 69.Mendell J.R., Al-Zaidy S., Shell R., Arnold W.D., Rodino-Klapac L.R., Prior T.W., Lowes L., Alfano L., Berry K., Church K. Single-Dose Gene-Replacement Therapy for Spinal Muscular Atrophy. N. Engl. J. Med. 2017;377:1713–1722. doi: 10.1056/NEJMoa1706198. [DOI] [PubMed] [Google Scholar]
- 70.Evers M.M., Miniarikova J., Juhas S., Vallès A., Bohuslavova B., Juhasova J., Skalnikova H.K., Vodicka P., Valekova I., Brouwers C. AAV5-miHTT Gene Therapy Demonstrates Broad Distribution and Strong Human Mutant Huntingtin Lowering in a Huntington’s Disease Minipig Model. Mol. Ther. 2018;26:2163–2177. doi: 10.1016/j.ymthe.2018.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Aschauer D.F., Kreuz S., Rumpel S. Analysis of transduction efficiency, tropism and axonal transport of AAV serotypes 1, 2, 5, 6, 8 and 9 in the mouse brain. PLoS ONE. 2013;8:e76310. doi: 10.1371/journal.pone.0076310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Samaranch L., Blits B., San Sebastian W., Hadaczek P., Bringas J., Sudhakar V., Macayan M., Pivirotto P.J., Petry H., Bankiewicz K.S. MR-guided parenchymal delivery of adeno-associated viral vector serotype 5 in non-human primate brain. Gene Ther. 2017;24:253–261. doi: 10.1038/gt.2017.14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Trapnell C., Pachter L., Salzberg S.L. TopHat: discovering splice junctions with RNA-Seq. Bioinformatics. 2009;25:1105–1111. doi: 10.1093/bioinformatics/btp120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Goff L.A., Trapnell C., Kelley D. CummeRbund: visualization and exploration of Cufflinks high-throughput sequencing data. 2012. http://compbio.mit.edu/cummeRbund/
- 76.Robinson J.T., Thorvaldsdóttir H., Winckler W., Guttman M., Lander E.S., Getz G., Mesirov J.P. Integrative genomics viewer. Nat. Biotechnol. 2011;29:24–26. doi: 10.1038/nbt.1754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Liu J., Hu J., Ludlow A.T., Pham J.T., Shay J.W., Rothstein J.D., Corey D.R. c9orf72 Disease-Related Foci Are Each Composed of One Mutant Expanded Repeat RNA. Cell Chem. Biol. 2017;24:141–148. doi: 10.1016/j.chembiol.2016.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.