Abstract
DNA methylation is known as the prima donna epigenetic mark for its critical role in regulating local gene transcription. Changes in the landscape of DNA methylation across the genome occur during cellular transition, such as differentiation and altered neuronal plasticity, and become dysregulated in disease states such as cancer. The TET family of enzymes is known to be responsible for catalyzing the reverse process that is DNA demethylation by recognizing 5-methylcytosine and oxidizing the methyl group via an Fe(II)/alpha-ketoglutarate-dependent mechanism. Here, we describe the design, synthesis, and evaluation of novel cytosine-based TET enzyme inhibitors, a class of small molecule probes previously underdeveloped but broadly desired in the field of epigenetics. We identify a promising cytosine-based lead compound, Bobcat339, that has mid-μM inhibitor activity against TET1 and TET2, but does not inhibit the DNA methyltransferase, DNMT3a. In silico modeling of the TET enzyme active site is used to rationalize the activity of Bobcat339 and other cytosine-based inhibitors. These new molecular tools will be useful to the field of epigenetics and serve as a starting point for new therapeutics that target DNA methylation and gene transcription.
Keywords: TET inhibitors, TET enzymes, DNA methylation, cytosine methylation, epigenetics, neuroepigenetics
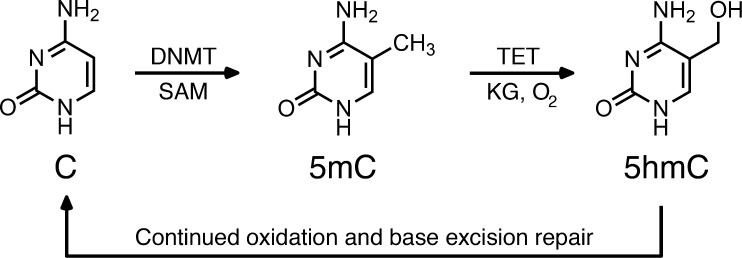
DNA methylation of cytosine is a long-lived and self-perpetuating epigenetic mark necessary for maintaining cell-type transcriptional programs.1 Methylation of cytosine at cytidine-guanosine (CpG) sites in promoters is an inhibitory epigenetic mechanism capable of completely silencing a gene in perpetuity. In addition, patterns of DNA methylation contribute to the creation of precise epigenetic landscapes associated with active gene transcription.2 Such durability is owed to a rigorous system wherein de novo DNA methyltransferases (DNMTs) catalyze the methyl transfer from S-adenosyl methionine to cytosines on previously unmethylated DNA strands, while maintenance DNMTs methylate the cytosine on the complementary strand of the CpG site, producing a dimethyl epigenetic mark.3−6
DNA methylation patterns tend to be maintained in differentiated cells; however, the methylation of DNA exists as a dynamic process, reversible by the Ten-11 translocation methylcytosine dioxygenase (TET) family of dioxygenases, coded by three separate genes (Tet1, Tet2, and Tet3). These isoenzymes recognize and oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC), 5-formylcytosine (5fC), and 5-carboxylcytosine (5caC) via an Fe(II)/alpha-ketoglutarate-dependent mechanism.7−10 These oxidized cytosine derivatives, themselves persistent epigenetic marks,7,8,11−13 can then function as intermediates subject to deamination and glycosylase-dependent excision and repair, leading to the reversal back to unmodified cytosine.14 The result is a cyclical epigenetic mechanism that is enduring, yet dynamically regulated (Scheme 1).
Scheme 1. Active DNA Methylation and Demethylation.
DNA methylation is dynamically regulated by methyl writing enzymes (DNMTs) and methyl erasing enzymes (TETs), allowing the mechanism to govern local gene expression, a process important for cell function and identity.
Disruption of DNA methylation patterns is known to be a hallmark of cancer with Tet2 being one of the most frequently mutated genes in hematopoietic malignancies.15 Likewise, mutations in Tet1, Tet2, and Tet3, eliciting reduced expression, impaired enzymatic activity, and concomitant decrease in levels of 5hmC, appear to impart a diverse range of mutational landscapes in a wide variety of different cancer types including liver, lung, gastric, prostate, and breast cancer as well as melanoma and glioblastoma.15−20 However, the precise impact of altered TET activity on the progression and maintenance of these cancers is largely unknown, contributing to the need for further molecular tools for intensifying investigative efforts surrounding premalignant and malignant transformation of TET-mutated cells.15
With growing consensus, it is hypothesized that epigenetic mechanisms, namely, DNA methylation, can help explain the long-lasting changes in gene expression associated with long-term memory in the central nervous system. Active DNA methylation has been found to occur at memory-associated genes within memory circuits21,22 and is required for long-term memory formation.6,23−25 Consequently, studies using DNMT inhibitors have shown DNMT activity to be crucial for memory formation and stabilization.6,21,27−29 Moreover, support for active DNA methylation as a positive regulator is in accordance with evidence situating TET-mediated active DNA demethylation in a negative regulatory role within memory systems. Viral overexpression of Tet1 has been shown to generate deficits in long-term contextual fear memory,29 and Tet1 knockout mice exhibit impaired memory extinction and enhanced long-term memory.30,31 With implications for other cognitive disorders such as Alzheimer’s disease, understood to be associated with altered 5mC patterns at a number of disease-specific genes,32,33 as well as for mechanisms of drug addiction,23,34 a deeper interrogation into TET-mediated demethylation of DNA has become increasingly imperative.
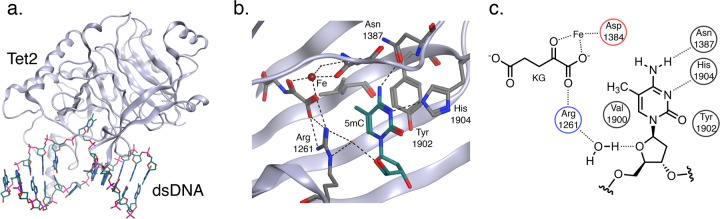
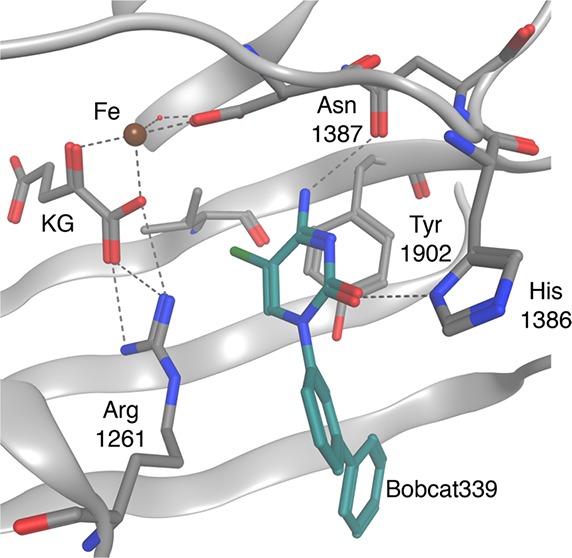
Unfortunately, there is currently no class of selective TET inhibitors to pharmacologically probe these biochemical processes sufficiently. Thus, the design and development of potent inhibitors of the TET enzymes were undertaken, starting with an assessment of the solved crystal structure of human TET2 (Figure 1a).26 The structure, which shows the TET2 enzyme bound to methylated double-stranded DNA (dsDNA), reveals how the enzyme isolates and recognizes 5mC by orientating its methyl substituent proximal to the oxidative Fe center. Several critical hydrogen bonds are formed for enzymatic recognition of 5mC. Asn1387 accepts a hydrogen bond from N7, and His 1904 donates a hydrogen bond to N3 of the 5mC ring (Figure 1b,c). Mutating either residue leads to the loss of enzymatic function and reduced binding affinities to methylated DNA.26 Therefore, it was considered desirable to maintain these contacts in the de novo design of competitive inhibitors based on cytosine. The deoxyribose also makes positive contacts with the active site in the form of a water-mediated hydrogen bond between the hydrofuran oxygen and Arg1261, a critical residue that also binds alpha-ketoglutarate. The methyl substituent on cytosine increases binding affinity to TET2;26 however, installing a methyl group into the design would likely produce a competitive substrate rather than an inhibitor. Thus, a search was undertaken to identify a suitable bioisostere for the methyl at the 5-position.
Figure 1.
Crystal structure of TET2-DNA complex.26 (a) TET2 binds dsDNA, breaks the double helix, and inserts 5mC into its active site. (b) View of the TET2 active site binding 5mC by forming hydrogen bonds with Asn1387, His1904, and Arg1261, all of which are critical residues for TET2 catalytic activity and methylated DNA binding. The oxidative iron center is shown in proximity to the methyl group on 5mC. (c) A 2D rendering of the 5mC-bound TET2 active site and critical residue interactions; blue = basic residue, red = acidic residue, and black = neutral residue.
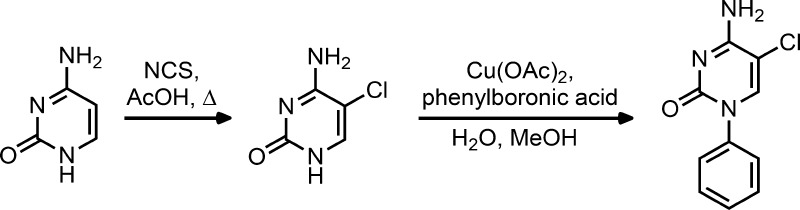
Initial candidates included halides, particularly chlorine, which is able to approximate the size of a methyl group. Additionally, a CF3 group was contemplated for its ability to mimic the tetrahedral shape of the methyl as well as remain protected against oxidation via fluorination. Finally, to aid in the ease of analog synthesis and refrain from designing compounds that may be incorporated into DNA, such as the cytosine-based DNMT inhibitors azacitidine and decitabine, the replacement of the deoxyribose for a phenyl group was considered as a starting point. Therefore, a two-step synthesis was undertaken (Scheme 2): First, the 5-position of cytosine was halogenated (or alkylated) by taking advantage of the preference for cytosine to undergo electrophilic aromatic substitution at this position. For example, 5-chlorosytosine was synthesized using N-chlorosuccinimide (NCS) in acetic acid at reflux.35 Then, the N1 position of the cytosine derivative was coupled using copper-mediated Ullman conditions36 to phenylboronic acid.
Scheme 2. Cytosine-Based TET Enzyme Inhibitor Synthesis.
The 5-position of cytosine was chlorinated using NCS in acetic acid and then coupled to phenylboronic acid using copper-mediated Ullman conditions. All other N1 and 5-substituted cytosine derivatives were synthesized using similar conditions.
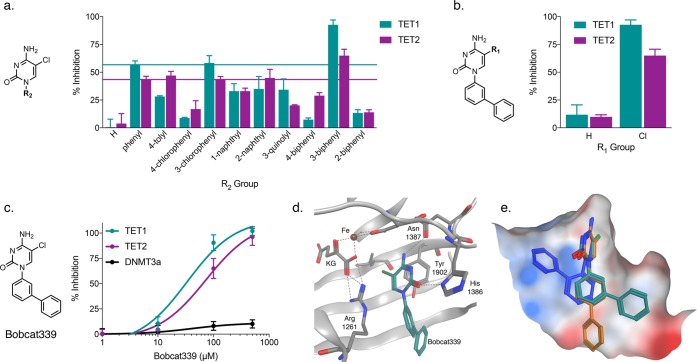
Next, these simple 5mC derivatives were tested for inhibition of recombinant human TET1 or TET2 enzyme-mediated oxidation of methylated dsDNA in an ELISA with an inhibitor concentration of 100 μM (Figure 2). As expected, hydrogen substitution at the R1 position was insufficient to elicit significant inhibition. Three bioisosteres of a methyl group, chlorine, bromine, and trifluoromethyl, were then tested as substituents. Our original hypothesis was that the trifluoromethyl group, with its similar size and tetrahedral geometry, would be the most effective at mimicking the binding conformation of the endogenous methyl group, while being protected from oxidation by fluorine substitution. However, this derivative failed to perform with any greater potency than no substitution at all. The 5-chloro substitution (Bobcat212) proved the most effective with 57% and 43% inhibition of TET1 and TET2, respectively.
Figure 2.
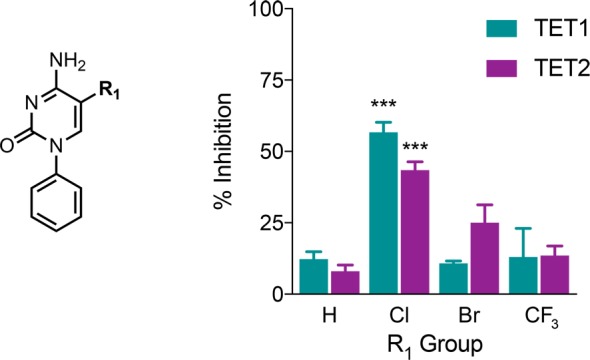
Examining possible methyl bioisosteres at the R1 position. Chloro, bromo, and trifluoromethyl-substituted derivatives were tested for inhibition of TET1- or TET2-mediated oxidation of methylated dsDNA. Each compound was tested at 100 μM in an ELISA. All data presented are N = 3, error bars indicate ± SEM. Two-way ANOVA, *P < 0.05, **P < 0.01, ***P < 0.001.
Our attention then turned to the optimization of the aryl substituent R2 position on the N1 of the 5-chlorocytosine. Using the same synthetic strategy as the phenyl series, 5-chlorocytosine was coupled to a variety of arylboronic acids under Ullman conditions. These R2 derivatives, which included tolyl, chlorophenyl, naphthyl, quinolyl, and biphenyl substitution, were then tested for inhibitory activity against TET1 and TET2 (Figure 3a). Most performed with similar or worse potency to the original phenyl derivative; however, results demonstrate R2 substitution to be vital for activity, as most aryl derivatives displayed significantly stronger inhibition than unsubstituted 5-chlorocytosine. Notably, compound Bobcat339, substituted at the R2 position with 3-biphenyl, showed significantly enhanced activity at TET1 (P = 0.002, two-way ANOVA) with comparable activity to the original phenyl derivative at TET2. This observed activity was also 5-chlorocytosine-dependent (Figure 3b), and removal of the chlorine substituent significantly reduced activity at both TET1 (P < 0.0001) and TET2 (P = 0.0003).
Figure 3.
Inhibitor optimization. (a) Several aryl groups were tested at the R2 position for inhibition of TET1 and TET2 at 100 μM. 3-Biphenyl substitution significantly increased TET1 inhibition (P = 0.002) over simple phenyl substitution, while 2-biphenyl (P = 0.0001) and 4-biphenyl (P < 0.0001) substitution significantly reduced TET1 inhibition as compared to a phenyl substitution. (b) 5-Chloro substitution at the R1 position is necessary to maintain the activity of 3-biphenyl substitution at the R2 position for both TET1 (P < 0.0001) and TET2 (P = 0.0003). (c) Bobcat339 inhibits TET1 (IC50 = 33 μM) and TET2 (IC50 = 73 μM), but not DNMT3a. (d) Bobcat339 docked into a homology model of TET1. (e) Predicted binding conformations of Bobcat339 (teal), its 2-biphenyl isomer (blue), and its 4-biphenyl isomer (gold). Molecular surface; white = hydrophobic, blue = electropositive, and red = electronegative. All data presented are N = 3, error bars indicate ± SEM.
Since any impediment to active DNA methylation may counteract the potential therapeutic effects of inhibiting the TET enzymes, Bobcat339 was evaluated for its desired role as a selective inhibitor of TET1 and TET2 using an inhibitory assay for DNMT3a. Bobcat339 elicited a dose–response relationship for both TET1 (IC50 = 33 μM) and TET2 (IC50 = 73 μM), while failing to show substantial inhibition of DNMT3a at a concentration of 500 μM (Figure 3c). These results underscore the potential of these compounds to inhibit the removal, but not the placement of methyl marks on the genome.
The activity of Bobcat339 was not shared by its constitutional isomers, the 2-biphenyl and 4-biphenyl derivatives, suggesting that the three-dimensional structure of the 3-biphenyl isomer is preferentially accommodated by the TET active site. To interrogate this hypothesis computationally, we docked these cytosine derivatives into the active site of the TET2 crystal structure and a homology model of TET1 using the Molecular Operating Environment (MOE) software package. To generate the TET1 homology model, its primary protein sequence was aligned to that of the residues present in the published crystal structure of TET2 (Supporting Information). Based on the binding orientation of 5mC, our cytosine derivatives were placed into the active site of the TET1 homology model. Then, allowing all bonds to rotate, a series of potential binding conformations was created. These conformations were next docked into both the TET1 and TET2 models, allowed to relax to a local energy minimum using the Amber10:EHT force field, and scored using the London ΔG algorithm. The proposed binding mode of Bobcat339 for both isoenzymes situates the 5-chlorocytosine headgroup directly into the active site in a fashion that mimics the arrangement of 5mC, forming two base-pairing-like hydrogen bonds with the enzyme and placing the chlorine into the small pocket that typically accommodates the methyl substrate (Figure 3d). Furthermore, the 3-biphenyl group is oriented at an angle to make hydrophobic contacts with the side wall of the binding pocket, which is disrupted when the biphenyl substitution pattern is altered. There is a high degree of homology between the active sites of both isoenzymes, and the calculated binding affinities of Bobcat339 are similar for TET1 (calc. ΔGBinding = −10.23 kcal/mol) and TET2 (calc. ΔGBinding = −10.08 kcal/mol). The 4-biphenyl derivative fails to take advantage of these interactions, while the 2-biphenyl derivative is forced into a different binding orientation entirely due to what would be steric clashing with Tyr 1902 (Figure 3e), leading to a decreased association with the active site for both the 4-biphenyl (calc. ΔΔGBinding = +0.99 kcal/mol) and the 2-biphenyl (calc. ΔΔGBinding = +0.52 kcal/mol) derivatives compared to Bobcat339 when docked into the TET1 model. Similar weaker binding affinities were also observed for the 4-biphenyl (calc. ΔΔGBinding = +0.37 kcal/mol) and the 2-biphenyl (calc. ΔΔGBinding = +0.72 kcal/mol) derivatives when docked into the TET2 model. These calculated differences in binding affinity align with the observed differences in enzyme inhibition elicited by these isomers.
Lastly, to determine whether these TET inhibitors could act as molecular probes for reducing 5hmC levels in live cells, Bobcat 339 and Bobcat212 were dosed onto HT-22 cells, an immortalized mouse hippocampal neuronal cell line. HT-22 cells were treated with 10 μM Bobcat339 or Bobcat212 in 1% DMSO for 24 h and lysed. DNA was extracted, and 5hmC levels were detected using a 5hmC-specific antibody, allowing for percent 5hmC of all cytosine species to be determined by colorimetric detection (Figure 4). Cells treated with Bobcat339 had significantly reduced global 5hmC levels as compared to the vehicle control, demonstrating its ability to reduce DNA 5hmC abundance by inhibiting TET enzyme function in living cells and providing support for its utility as a viable pharmacological probe.
Figure 4.
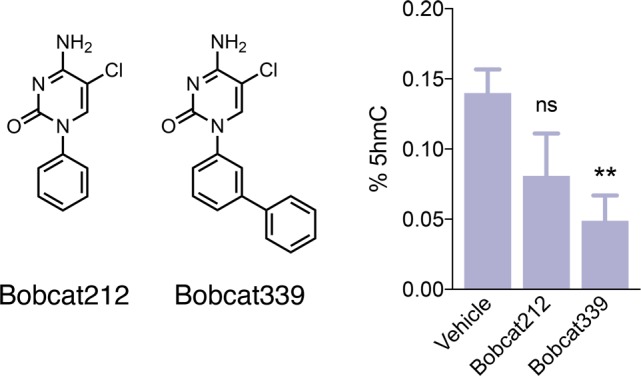
Bobcat339 reduces DNA 5hmC levels in hippocampal neurons. Lead compounds Bobcat212 and Bobcat339 were dosed at 10 μM in 1% DMSO onto HT-22 mouse hippocampal neurons for 24 h. Cells were then lysed, DNA extracted, and TET enzyme inhibition determined by 5hmC-specfic antibody binding. Data presented are N = 6 biological replicates per group, error bars indicate ± SEM. One-way ANOVA, ns = not significant, *P < 0.05, **P < 0.01.
Here, we have identified and evaluated a new class of cytosine-based TET enzyme inhibitors. The most potent in this series, Bobcat339, generated mid-μM IC50s for TET1 and TET2 without inhibiting DNMT3a, a cytosine-recognizing DNA methyltransferase, and reduced 5hmC abundance in the DNA of cultured neurons. The TET enzymes are critical epigenetic modifiers that regulate gene imprinting to consequently govern cell differentiation and development. Namely, TET enzymes have been implicated in the epigenetic dysregulation driving several cancer states as well as in the modulation of neuronal plasticity and memory. As there does not currently exist a selective pharmacological tool for probing TET enzyme function, Bobcat339 and the other 5-chlorocytosine derivatives described here are important contributions toward such a reagent.
Acknowledgments
This work is dedicated to the memory of Kindal Kivisto.
Glossary
ABBREVIATIONS
- 5caC
5-carboxylcytosine
- 5fC
5-formylcytosine
- 5hmC
5-hydroxymethylcytosine
- 5mC
5-methylcytosine
- CpG
cytidine-guanosine
- DNMT
DNA methyltransferase
- dsDNA
double-stranded DNA
- MOE
Molecular Operating Environment
- TET
Ten-11 translocation methylcytosine dioxygenase
Supporting Information Available
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsmedchemlett.8b00474.
Experimental procedures and data for all new compounds synthesized, in vitro inhibition assays, cell culture, molecular modeling, and docking studies (PDF)
Author Contributions
G.N.L.C. and A.J.K. wrote the manuscript. Docking studies were performed by G.N.L.C. Chemical synthesis was performed by K.L.W., H.S., J.A.A., E.I.J., and N.J.K. Chemical characterization was performed by N.J.K. In vitro biological analysis was performed by K.L.W. and H.S. Cell culture was performed by M.J.B. and M.K. B.G.M. provided research support for the project. All authors have given approval to the final version of the manuscript.
Research reported in this publication was supported by an Institutional Development Award (IDeA) from the National Institute of General Medical Sciences of the National Institutes of Health under grant number P20GM103423, the Pitt–Hopkins Research Foundation, the Orphan Disease Center’s 2018 MDBR Pilot Grant Program, the Sherman Fairchild Foundation, and Bates College.
The authors declare no competing financial interest.
Supplementary Material
References
- Kennedy A. J.; Sweatt J. D. Drugging the methylome: DNA methylation and memory. Crit. Rev. Biochem. Mol. Biol. 2016, 51 (3), 185–194. 10.3109/10409238.2016.1150958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J. J.; Kennedy A. J.; Sweatt J. D. DNA methylation and its implications and accessibility for neuropsychiatric therapeutics. Annu. Rev. Pharmacol. Toxicol. 2015, 55, 591–611. 10.1146/annurev-pharmtox-010814-124527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okano M.; Xie S.; Li E. Cloning and characterization of a family of novel mammalian DNA (cytosine-5) methyltransferases. Nat. Genet. 1998, 19 (3), 219–220. 10.1038/890. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation de novo. Science 1999, 286 (5448), 2287–2288. 10.1126/science.286.5448.2287. [DOI] [PubMed] [Google Scholar]
- Bird A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16 (1), 6–21. 10.1101/gad.947102. [DOI] [PubMed] [Google Scholar]
- Feng J.; Zhou Y.; Campbell S. L.; Le T.; Li E.; Sweatt J. D.; Silva A. J.; Fan G. Dnmt1 and Dnmt3a maintain DNA methylation and regulate synaptic function in adult forebrain neurons. Nat. Neurosci. 2010, 13 (4), 423–430. 10.1038/nn.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tahiliani M.; Koh K. P.; Shen Y.; Pastor W. A.; Bandukwala H.; Brudno Y.; Agarwal S.; Iyer L. M.; Liu D. R.; Aravind L.; Rao A. Conversion of 5-Methylcytosine to 5-Hydroxymethylcytosine in Mammalian DNA by MLL Partner TET1. Science 2009, 324 (5929), 930–935. 10.1126/science.1170116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito S.; Shen L.; Dai Q.; Wu S. C.; Collins L. B.; Swenberg J. A.; He C.; Zhang Y. Tet proteins can convert 5-methylcytosine to 5-formylcytosine and 5-carboxylcytosine. Science 2011, 333 (6047), 1300–1303. 10.1126/science.1210597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pastor W. A.; Aravind L.; Rao A. TETonic shift: biological roles of TET proteins in DNA demethylation and transcription. Nat. Rev. Mol. Cell Biol. 2013, 14 (6), 341–356. 10.1038/nrm3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Y.-F.; Li B.-Z.; Li Z.; Liu P.; Wang Y.; Tang Q.; Ding J.; Jia Y.; Chen Z.; Li L.; Sun Y.; Li X.; Dai Q.; Song C.-X.; Zhang K.; He C.; Xu G.-L. Tet-mediated formation of 5-carboxylcytosine and its excision by TDG in mammalian DNA. Science 2011, 333 (6047), 1303–1307. 10.1126/science.1210944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Globisch D.; Münzel M.; Müller M.; Michalakis S.; Wagner M.; Koch S.; Brückl T.; Biel M.; Carell T. Tissue distribution of 5-hydroxymethylcytosine and search for active demethylation intermediates. PLoS One 2010, 5 (12), e15367 10.1371/journal.pone.0015367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szwagierczak A.; Bultmann S.; Schmidt C. S.; Spada F.; Leonhardt H. Sensitive enzymatic quantification of 5-hydroxymethylcytosine in genomic DNA. Nucleic Acids Res. 2010, 38 (19), e181–e181. 10.1093/nar/gkq684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.; Zhang Y. Reversing DNA methylation: mechanisms, genomics, and biological functions. Cell 2014, 156 (1–2), 45–68. 10.1016/j.cell.2013.12.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J. U.; Su Y.; Zhong C.; Ming G.-l.; Song H. Hydroxylation of 5-methylcytosine by TET1 promotes active DNA demethylation in the adult brain. Cell 2011, 145 (3), 423–434. 10.1016/j.cell.2011.03.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rasmussen K. D.; Helin K. Role of TET enzymes in DNA methylation, development, and cancer. Genes Dev. 2016, 30 (7), 733–750. 10.1101/gad.276568.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kudo Y.; Tateishi K.; Yamamoto K.; Yamamoto S.; Asaoka Y.; Ijichi H.; Nagae G.; Yoshida H.; Aburatani H.; Koike K. Loss of 5-hydroxymethylcytosine is accompanied with malignant cellular transformation. Cancer science 2012, 103 (4), 670–676. 10.1111/j.1349-7006.2012.02213.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lian C. G.; Xu Y.; Ceol C.; Wu F.; Larson A.; Dresser K.; Xu W.; Tan L.; Hu Y.; Zhan Q.; Lee C.-W.; Hu D.; Lian B. Q.; Kleffel S.; Yang Y.; Neiswender J.; Khorasani A. J.; Fang R.; Lezcano C.; Duncan L. M.; Scolyer R. A.; Thompson J. F.; Kakavand H.; Houvras Y.; Zon L. I.; Mihm M. C.; Kaiser U. B.; Schatton T.; Woda B. A.; Murphy G. F.; Shi Y. G. Loss of 5-hydroxymethylcytosine is an epigenetic hallmark of melanoma. Cell 2012, 150 (6), 1135–1146. 10.1016/j.cell.2012.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcan S.; Rohle D.; Goenka A.; Walsh L. A.; Fang F.; Yilmaz E.; Campos C.; Fabius A. W. M.; Lu C.; Ward P. S.; Thompson C. B.; Kaufman A.; Guryanova O.; Levine R.; Heguy A.; Viale A.; Morris L. G. T.; Huse J. T.; Mellinghoff I. K.; Chan T. A. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature 2012, 483 (7390), 479–483. 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu C.; Liu L.; Chen X.; Shen J.; Shan J.; Xu Y.; Yang Z.; Wu L.; Xia F.; Bie P.; Cui Y.; Bian X.-w.; Qian C. Decrease of 5-hydroxymethylcytosine is associated with progression of hepatocellular carcinoma through downregulation of TET1. PLoS One 2013, 8 (5), e62828 10.1371/journal.pone.0062828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang H.; Liu Y.; Bai F.; Zhang J.-Y.; Ma S.-H.; Liu J.; Xu Z.-D.; Zhu H.-G.; Ling Z.-Q.; Ye D.; Guan K.-L.; Xiong Y. Tumor development is associated with decrease of TET gene expression and 5-methylcytosine hydroxylation. Oncogene 2013, 32 (5), 663–669. 10.1038/onc.2012.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lubin F. D.; Roth T. L.; Sweatt J. D. Epigenetic regulation of BDNF gene transcription in the consolidation of fear memory. J. Neurosci. 2008, 28 (42), 10576–10586. 10.1523/JNEUROSCI.1786-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duke C. G.; Kennedy A. J.; Gavin C. F.; Day J. J.; Sweatt J. D. Experience-dependent epigenomic reorganization in the hippocampus. Learning & memory (Cold Spring Harbor, N.Y.) 2017, 24 (7), 278–288. 10.1101/lm.045112.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J. J.; Childs D.; Guzman-Karlsson M. C.; Kibe M.; Moulden J.; Song E.; Tahir A.; Sweatt J. D. DNA methylation regulates associative reward learning. Nat. Neurosci. 2013, 16 (10), 1445–1452. 10.1038/nn.3504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miller C. A.; Gavin C. F.; White J. A.; Parrish R. R.; Honasoge A.; Yancey C. R.; Rivera I. M.; Rubio M. D.; Rumbaugh G.; Sweatt J. D. Cortical DNA methylation maintains remote memory. Nat. Neurosci. 2010, 13 (6), 664–666. 10.1038/nn.2560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monsey M. S.; Ota K. T.; Akingbade I. F.; Hong E. S.; Schafe G. E. Epigenetic alterations are critical for fear memory consolidation and synaptic plasticity in the lateral amygdala. PLoS One 2011, 6 (5), e19958 10.1371/journal.pone.0019958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu L.; Li Z.; Cheng J.; Rao Q.; Gong W.; Liu M.; Shi Y. G.; Zhu J.; Wang P.; Xu Y. Crystal structure of TET2-DNA complex: insight into TET-mediated 5mC oxidation. Cell 2013, 155 (7), 1545–1555. 10.1016/j.cell.2013.11.020. [DOI] [PubMed] [Google Scholar]
- Miller C. A.; Sweatt J. D. Covalent modification of DNA regulates memory formation. Neuron 2007, 53 (6), 857–869. 10.1016/j.neuron.2007.02.022. [DOI] [PubMed] [Google Scholar]
- Levenson J. M.; Roth T. L.; Lubin F. D.; Miller C. A.; Huang I.-C.; Desai P.; Malone L. M.; Sweatt J. D. Evidence that DNA (cytosine-5) methyltransferase regulates synaptic plasticity in the hippocampus. J. Biol. Chem. 2006, 281 (23), 15763–15773. 10.1074/jbc.M511767200. [DOI] [PubMed] [Google Scholar]
- Miller C. A.; Campbell S. L.; Sweatt J. D. DNA methylation and histone acetylation work in concert to regulate memory formation and synaptic plasticity. Neurobiol. Learn. Mem. 2008, 89 (4), 599–603. 10.1016/j.nlm.2007.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaas G. A.; Zhong C.; Eason D. E.; Ross D. L.; Vachhani R. V.; Ming G.-l.; King J. R.; Song H.; Sweatt J. D. TET1 controls CNS 5-methylcytosine hydroxylation, active DNA demethylation, gene transcription, and memory formation. Neuron 2013, 79 (6), 1086–1093. 10.1016/j.neuron.2013.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D.; Aggarwal M.; Kaas G. A.; Lewis J.; Wang J.; Ross D. L.; Zhong C.; Kennedy A.; Song H.; Sweatt J. D. Tet1 Oxidase Regulates Neuronal Gene Transcription, Active DNA Hydroxy-methylation, Object Location Memory, and Threat Recognition Memory. Neuroepigenetics 2015, 4, 12–27. 10.1016/j.nepig.2015.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohgi H.; Utsugisawa K.; Nagane Y.; Yoshimura M.; Genda Y.; Ukitsu M. Reduction with age in methylcytosine in the promoter region −224 approximately −101 of the amyloid precursor protein gene in autopsy human cortex. Brain research. Mol. Brain Res. 1999, 70 (2), 288–292. 10.1016/S0169-328X(99)00163-1. [DOI] [PubMed] [Google Scholar]
- Lardenoije R.; Iatrou A.; Kenis G.; Kompotis K.; Steinbusch H. W. M.; Mastroeni D.; Coleman P.; Lemere C. A.; Hof P. R.; van den Hove D. L. A.; Rutten B. P. F. The epigenetics of aging and neurodegeneration. Prog. Neurobiol. 2015, 131, 21–64. 10.1016/j.pneurobio.2015.05.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng J.; Shao N.; Szulwach K. E.; Vialou V.; Huynh J.; Zhong C.; Le T.; Ferguson D.; Cahill M. E.; Li Y.; Koo J. W.; Ribeiro E.; Labonte B.; Laitman B. M.; Estey D.; Stockman V.; Kennedy P.; Couroussé T.; Mensah I.; Turecki G.; Faull K. F.; Ming G.-l.; Song H.; Fan G.; Casaccia P.; Shen L.; Jin P.; Nestler E. J. Role of Tet1 and 5-hydroxymethylcytosine in cocaine action. Nat. Neurosci. 2015, 18 (4), 536–544. 10.1038/nn.3976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar V.; Yap J.; Muroyama A.; Malhotra S. V. Highly Efficient Method for C-5 Halogenation of Pyrimidine-Based Nucleosides in Ionic Liquids. Synthesis 2009, 2009 (23), 3957–3962. 10.1055/s-0029-1217042. [DOI] [Google Scholar]
- Yue Y.; Zheng Z. G.; Wu B.; Xia C. Q.; Yu X. Q. Copper-Catalyzed Cross-Coupling Reactions of Nucleobases with Arylboronic Acids: An Efficient Access to N-Arylnucleobases. Eur. J. Org. Chem. 2005, 2005 (24), 5154–5157. 10.1002/ejoc.200500589. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.