Abstract
Despite the essential role of the active metabolite of vitamin A, all-trans retinoic acid (atRA) in spermatogenesis, the enzymes, and cellular populations responsible for its synthesis in the postnatal testis remain largely unknown. The aldehyde dehydrogenase 1A (ALDH1A) family of enzymes residing within Sertoli cells is responsible for the synthesis of atRA, driving the first round of spermatogenesis. Those studies also revealed that the atRA required to drive subsequent rounds of spermatogenesis is possibly derived from the ALDH1A enzymes residing within the meiotic and post-meiotic germ cells. Three ALDH1A isozymes (ALDH1A1, ALDH1A2, and ALDH1A3) are present in the testis. Although, ALDH1A1 is expressed in adult Sertoli cells and is suggested to contribute to the atRA required for the pre-meiotic transitions, ALDH1A2 is proposed to be the essential isomer involved in testicular atRA biosynthesis. In this report, we first examine the requirement for ALDH1A2 via the generation and analysis of a conditional Aldh1a2 germ cell knockout and a tamoxifen-induced Aldh1a2 knockout model. We then utilized the pan-ALDH1A inhibitor (WIN 18446) to test the collective contribution of the ALDH1A enzymes to atRA biosynthesis following the first round of spermatogenesis. Collectively, our data provide the first in vivo evidence demonstrating that animals severely deficient in ALDH1A2 postnatally proceed normally through spermatogenesis. Our studies with a pan-ALDH1A inhibitor (WIN 18446) also suggest that an alternative source of atRA biosynthesis independent of the ALDH1A enzymes becomes available to maintain atRA levels for several spermatogenic cycles following an initial atRA injection.
Keywords: spermatogenesis; Aldh1A; Aldh1a2; WIN 18,446; testis; spermatogonia; retinoic acid
Elimination of ALDH1A enzymatic activity following a single pulse of retinoic acid does not immediately ablate spermatogenesis due to the presence of an additional source of atRetinal oxidation.
Introduction
Spermatogenesis is the process whereby spermatogonia proliferate and differentiate to generate the male reproductive gametes, spermatozoa [1–4]. The transition of A undifferentiated spermatogonia into A1 differentiating spermatogonia (A to A1 transition, or spermatogonial differentiation) is a critical regulatory step during spermatogenesis [1–4]. The A to A1 transition occurs at periodic intervals of 8.6 days along the length of testis tubules, resulting in the appearance of 12 reoccurring sets of cellular associations called stages [5–7]. The stepwise appearance of each of these stages ensures the asynchronous nature of the spermatogenetic wave and maintains the continuous release of sperm [5, 6]. Extensive evidence exists demonstrating that all-trans retinoic acid (atRA) is absolutely required for the initiation and maintenance of the spermatogenic wave [8]. While the necessity of atRA for spermatogenesis has been widely validated, the cellular source and the enzymatic families responsible for its synthesis in the postnatal testis are not completely understood.
A role for atRA, the active derivative of vitamin A, in spermatogenesis was initially identified in rodents fed a vitamin A-deficient (VAD) diet [9]. Histological analysis of VAD testes revealed an arrest at the A to A1 transition that was readily reversed by an exogenous atRA injection. Surprisingly, spermatogenesis was restored in a synchronous manner, resulting in the pulsatile release of sperm [10–12]. Although a thorough understanding of the retinoid metabolizing enzymes are still being shaped in the mammalian testis, the currently accepted model for the conversion of vitamin A to atRA involves a two-step enzymatic pathway. The first step involves the oxidation of all-trans retinol (atROL) to all-trans retinaldehyde (atRAL) by the cytosolic alcohol dehydrogenases or the microsomal retinol dehydrogenases [13–19]. This step is generally considered to be reversible and rate limiting. In contrast, the second reaction is irreversible and involves the conversion of atRAL to atRA by the aldehyde dehydrogenase 1A (ALDH1A) family [20–22]. The ALDH1A family of enzymes includes three isozymes that have roles in atRA synthesis: ALDH1A1 [23], ALDH1A2 [24], and ALDH1A3 [13, 25–27]. Localization studies have shown that the ALDH1A isoforms exhibit cell-specific expression patterns within the testis, suggestive of distinct biological functions [28–30]. Specifically, Aldh1a1 transcripts were detected in Leydig cells, and transcripts for Aldh1a2 were detected in spermatogonia, spermatocytes, and spermatids [31–36]. Transcripts for Aldh1a1 and Aldh1a2 were also detected in prepubertal and adult Sertoli cells [31, 32]. ALDH1A3 was detected at very low levels in Leydig cells, spermatocytes, and spermatids within the 30 dpp testis [35, 37, 38]. Studies with a pan-ALDH1A inhibitor, WIN 18446 (WIN), revealed that the ALDH1A enzymes are collectively responsible for more than 95% of atRA biosynthesis in the wild-type mouse testis, with ALDH1A2 estimated to contribute to over 61% [23, 39–43]. Inhibition of the ALDH1A enzymes in neonatal mice via treatment with WIN followed by an atRA injection results in testes that are enriched for germ cells synchronously proceeding through development [44]. The strong inhibitory effects of WIN on the ALDH1A enzymes makes this chemical treatment a powerful tool to address questions regarding the contributions of the ALDH1A enzymes to retinoid-dependent processes such as spermatogenesis.
Accumulating evidence suggests that the sources of the atRA signal are differentially regulated between the first round and subsequent rounds of spermatogenesis. Specifically, it has been shown that the atRA required to drive the first round of spermatogenesis is derived from the ALDH1A enzymes residing within Sertoli cells [31]. The enzymatic and cellular sources of atRA biosynthesis driving the subsequent rounds of spermatogonial differentiation remain to be identified; however, it is widely hypothesized that the ALDH1A enzymes, specifically ALDH1A2, residing within the meiotic and postmeiotic germ cells are involved [31, 32, 38, 45]. The in vivo role that Aldh1a2 plays within germ cells has yet to be determined, as global Aldh1a2-null mice die early in embryonic development (E9.5–10.5) [39, 40, 46]. To overcome the embryonic lethality associated with the null model, we utilized traditional Cre-lox technology to generate animals in which Aldh1a2 was excised postnatally beginning in the differentiating germ cells (via the Stra8-Cre) and globally (via the tamoxifen-inducible Cre). To our knowledge, these genetic models are the first in vivo demonstration that severe deficiency of ALDH1A2 within the postnatal testis does not negatively affect spermatogenic progression. To address the collective contribution of the three major ALDH1A isozymes to testicular atRA biosynthesis, we also utilized the pan-ALDH1A inhibitor, WIN 18446. Surprisingly, potent inhibition of the ALDH1A enzymes (via WIN 18446 treatment) was not sufficient to immediately block subsequent rounds of spermatogonial differentiation, nor did it result in significantly reduced atRA levels. Additional in vivo studies simultaneously using WIN 18446 and hydralazine (HYD), a potent inhibitor of the aldehyde oxidases (AOXs) (another family of enzymes capable of catalyzing the atRAL to atRA oxidation reaction), suggested that the AOX family of enzymes may play a previously unexplored role in testicular atRA biosynthesis. Collectively, our studies suggest that multiple enzyme families may be capable of synthesizing atRA in the postnatal testis driving the second and subsequent rounds of spermatogenesis.
Materials and methods
Animal care and ethics statement
All animal care and procedures were conducted according to protocols and guidelines approved by the Washington State University Committee on the Use and Care of Animals. Studies were performed with male mice that were housed in a humidity- and temperature-controlled room with access to water and food ad libitum. Mice were euthanized by CO2 asphyxiation followed by cervical dislocation.
Mouse lines, mouse breeding, and PCR genotype analysis
The Aldh1a2fl/fl transgenic line was generated by the Mouse Biology Program (MBP) at the University of California, Davis. The targeting construct and the replacement vector strategy used to generate the Aldh1a2fl/fl transgenic line is provided in Supplemental Figure S1 and details regarding the generation of the line can also be found in Supplemental Methods (Supplemental Data are available online at www.biolreprod.org). Aldh1a2fl/fl females were mated with Aldh1a2fl/+, Stra8-Cre [47] male mice to generate Aldh1a2fl/fl, Stra8-Cre negative (control) and Aldh1a2fl/fl, Stra8-Cre positive (cKO) animals. The Stra8-Cre (Stock number #008208) line was purchased from the Jackson Laboratory (Bar Harbor, ME). To generate the tamoxifen-inducible global knockout of Aldh1a2, we utilized the B6.129-Gt(Rosa)26Sortm1(cre/ERT2)Tyj (Stock number #008463, hereafter referred to as CreERT2) transgenic line, purchased from the Jackson Laboratory (Bar Harbor). Administration of tamoxifen (via intraperitoneal injection) induces the CreERT2 fusion protein to translocate to the nucleus and excise floxed DNA fragments. For simplicity, mice whose floxed allele was excised following tamoxifen injection will hereafter be denoted by Aldh1a2Δ. Adult Aldh1a2fl/fl female mice were crossed with Aldh1a2+/+, CreERT2 homozygous males to generate F1 animals heterozygous for both genes. These heterozygous animals were mated together to generate F2 animals. F2 animals that were heterozygous for Aldh1a2 (Aldh1a2fl/+) and homozygous for the CreERT2 were mated to generate F3 experimental animals. This breeding scheme generated animals with the following genotypes within the F3 generation: Aldh1a2+/+, CreERT2; Aldh1a2fl/+, CreERT2; and Aldh1a2fl/fl, CreERT2. To determine the genotypes of experimental mice, genomic DNA samples were extracted from a small piece of tail or ear tissue, which served as templates for PCR reactions. Allele-specific PCR reactions were performed using primer sets described in Supplemental Table S1. PCR detection of the excised allele was performed on all experimental mice before (obtained from tail clip) and after (obtained from ear/tail chip) tamoxifen injection. Prior to the tamoxifen injection, the excised band was never detected in any genomic samples regardless of the animal's genotype. No excision bands were detected in any of the Aldh1a2+/+, CreERT2 animals injected with tamoxifen. However, a prominent excision band was detected in all Aldh1a2Δ/+, CreERT2 and Aldh1a2Δ/Δ, CreERT2 animals.
Tamoxifen preparation and administration
Tamoxifen (Tm, Sigma T5648) was dissolved in 10% ethanol and 90% sesame oil at a concentration of 20 mg/ml, which was subsequently wrapped in foil to protect the solution from light. Aldh1a2+/+, CreERT2; Aldh1a2fl/+, CreERT2; and Aldh1a2fl/fl, CreERT2 animals aged 8 or 21 dpp were administered 80 mg/kg tamoxifen via an intraperitoneal injection for either three or five consecutive days, respectively [48]. For all injection strategies, animals were left to recover for 60 additional days prior to analysis.
Fertility evaluation
Adult Aldh1a2fl/fl, Stra8-Cre control and cKO males (n = 4 per genotype) were housed with 129/B6 wild-type adult females of known fertility at a male-to-female sex ratio of 1:2. Males were observed for normal mounting behavior and female mice were checked each morning for the presence of a copulatory plug. The sex of the offspring, the total number of offspring, and the number of litters produced over a 4-month fertility trial were recorded.
RNA isolation and quantitative real-time polymerase chain reaction
Testis samples for quantitative real-time polymerase chain reaction (qRT-PCR) analysis were snap-frozen on dry ice upon removal and stored at –80°C. Tissues were homogenized and total RNA was extracted using TRIzol reagent (15596018, Ambio Life Technologies) according to the manufacturer's directions. RNA quality and quantity was determined using a NanoDrop 1000 Spectrophotometer (Thermo Scientific). Total RNA (250–500 ng) was reverse-transcribed using the iScript cDNA synthesis kit (1708891, Bio-Rad). Quantitative RT-PCR was performed using Fast SYBR Green PCR Mastermix (4385612, Applied Biosystems) on a 7500 Fast PCR System (Applied Biosystems). Primer sequences for the genes analyzed are presented in Supplemental Table S2. The expression of these genes was determined from three to six mice, each of which was measured in triplicate and averaged. The relative mRNA levels in each sample were calculated using the comparative CT method (ΔΔCT) [49]. The values were normalized using the expression of the ribosomal protein S2 (Rps2).
WIN, hydralazine treatments, and atRA injections
We utilized a previously published WIN7D + RA synchrony protocol with minor modifications [44, 45]. Briefly, 2 dpp 129/B6 male mice were pipette fed 100 mg/kg/bw (body weight) WIN (gift from Dr John Amory) daily for 7 days, given an atRA injection on day 9 and maintained on one of the following treatment regimens during the injection recovery: (1) 1% gum tragacanth (WIN7D + RA), (2) 150 mg/kg/bw WIN (WIN7D + RA + WIN), (3) 25 mg/kg/bw of HYD (Sigma-Aldrich, H1753) (WIN7D + RA + HYD), or (4) 150 mg/kg/bw WIN and 25 mg/kg/bw HYD (WIN7D + RA + WIN/HYD) (Supplemental Figure S2). Depending on the experiment, the lengths of the WIN maintenance treatments were 8, 16, 24, 32, or 40 days. A group of animals were fed 100 mg/kg/bw WIN from 2–8 dpp, injected with vehicle control (dimethylsulfoxide, DMSO) injection on day 9, and then maintained on 150 mg/kg/bw WIN during injection recovery (WIN7D + DMSO + WIN; Supplemental Figure S2). An additional group of animals were pipette fed 1% gum tragacanth for the duration of the treatment regimen (Supplemental Figure S2). Mice given atRA injections were subcutaneously injected with 200 μg atRA (Sigma-Aldrich, R2625) suspended in 10 μl DMSO (Fisher Scientific, BP231-1). The WIN compound was suspended in 1% gum tragacanth and HYD was suspended in ddH2O. Irrespective of the type of injection that the animals received (atRA or DMSO), WIN or HYD-maintained animals also received WIN or HYD on the day of the injection.
Immunohistochemistry
Immunohistochemistry (IHC) was performed as previously described [50] with the exception that the primary antibodies used were aldehyde dehydrogenase family 1 member A2 (ALDH1A2; 1:100 dilution; 13951-1-AP; Proteintech Group), stimulated by retinoic acid 8 (STRA8; antibody produced in-house) [51], zinc finger and BTB domain containing 16 (ZBTB16; 1:500 dilution; sc-22839; Santa Cruz Biotechnology), germ cell nuclear antigen (GCNA; gift from Dr. George Enders), and SRY (sex dertermining region Y) - box 9 (SOX9; 1:500 dilution; AB5535; EMD Millipore). Control sections were incubated in blocking solution with the primary antibody omitted.
Measurement of testicular atRA levels
For testicular atRA measurement of the Aldh1a2, Stra8-Cre and Aldh1a2, CreERT2 animals (n = 3−4 per treatment), testis tissue (30–50 mg) was homogenized in 5X tissue-weight saline. Tissue homogenates (120 μL) were transferred into 1.7 ml Eppendorf tubes followed by the addition of 5 μL of 2 μM at-RA-d5 as an internal standard. Acetonitrile (240 μL) was added to tissue homogenates to precipitate proteins. After 5–10 s of vortexing, samples were left on ice for 10 min and then centrifuged at 18 000 × g at 4°C for 30 min. Supernatant was transferred to glass vials for liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis as described in published methods [36, 52]
For testicular atRA measurement of WIN treated mice, each data point (n = 3–4 per treatment group) was obtained from pooled testes of 8–15 treated mice (188–226 mg per sample). The extraction protocol and LC-MS/MS method used to measure WIN-treated animals were performed as described in references [36, 51–53].
Cell quantification
Serial sections for cell quantifications were separated by a minimum of 20 μm. Immunostaining of testis sections for ZBTB16, STRA8, GCNA, or SOX9 was performed as described above. The different cell types were identified based on (1) ZBTB16-, STRA8-, GCNA-, or SOX9-positive immunostaining; (2) the shape and size of the nuclei; and (3) the location of the cells within the seminiferous tubules. Quantification was performed to count (1) the number of ZBTB16-, STRA8-, or SOX9-immunopositive spermatogonia, or (2) the number of tubules displaying normal versus absent/missing layers of GCNA-positive germ cells within 200 tubules per animal. All quantification was performed using a minimum of three animals per treatment group for each experiment.
Statistical analysis
Statistical significance between two experimental groups was evaluated using the Student t-test (Prism; Version 7.0c; Graphpad), while statistical significance between more than two groups was determined using a one-way ANOVA (Tukey's HSD post hoc test; Prism; Version 7.0c; Graphpad). A P-value of 0.05 or less was considered statistically significant, and all data are presented as mean ± standard error of the mean (SEM).
Results
Germ cell-specific deletion of Aldh1a2 does not result in abnormal histological phenotypes
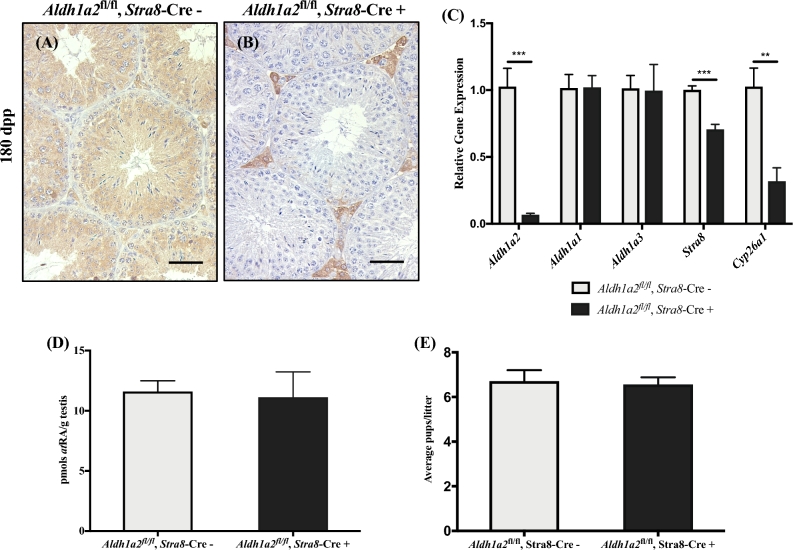
To explore the role of Aldh1a2 specifically in postnatal germ cells, we generated an animal with a germ cell-specific deletion of Aldh1a2 using the Stra8-Cre. The Stra8-Cre was chosen because it is expressed around 3 dpp in a select population of Aal spermatogonia and in all A1 differentiating spermatogonia [47]. Expression of the Cre protein should eliminate Aldh1a2 within the germ cell populations of interest (namely the preleptotene spermatocytes, pachytene spermatocytes, and spermatid populations). Aldh1a2 control and cKO animals were obtained at the expected Mendelian frequencies, and cKO mice were morphologically indistinguishable from their control littermates. To verify that we had generated a genuine germ cell-specific Aldh1a2 cKO, we used a previously characterized antibody against ALDH1A2 to localize the protein on testis sections of control and cKO mice at 60 and 180 dpp by IHC [37]. Consistent with published results, we observed abundant ALDH1A2-positive spermatogonia, spermatocytes, spermatids, and Leydig cells in all control testis sections (Figure 1A and Supplemental Figure S3A and B; n = 4). The ALDH1A2 protein was undetectable within the seminiferous epithelium of cKO mice; however, we detected ALDH1A2-positive cells within the interstitial space (Figure 1B and Supplemental Figure S3C and D; n = 4). Quantitative RT-PCR analysis also demonstrated that Aldh1a2 expression was significantly reduced in cKO animals at 60 and 180 dpp compared to controls (Figure 1C and Supplemental Figure S3E; n = 3–6). To determine whether Aldh1a1 and Aldh1a3 are upregulated in response to reduced levels of Aldh1a2, we performed qRT-PCR in 60 and 180 dpp testes from cKO mice and found no statistically significant changes in the expression of Aldh1a1 or Aldh1a3 between the cKO and control testes at either age point (Figure 1C and Supplemental Figure S3E; n = 3–6).
Figure 1.
Elimination of Aldh1a2 using the Stra8-Cre. Control (Aldh1a2fl/fl, Stra8-Cre–) and cKO (Aldh1a2fl/fl, Stra8-Cre+) animals analyzed at 180 dpp. Representative control (A) and cKO (B) cross-section stained for ALDH1A2. Immunopositive cells are indicated by brown precipitate. (C) qRT-PCR analysis of Aldh1a2, Aldh1a1, Aldh1a3, Stra8, and Cyp26a1. (D) Graphical representation of atRA measurements. (E) Graphical representation of the average number of pups per litter. Scale bars = 100 μM. n = 3–4. * P < 0.05, ** P < 0.01, and *** P < 0.001.
Elimination of Aldh1a2 via Stra8-Cre does not significantly reduce atRA levels
To get an indirect measure of the atRA levels in the Aldh1a2 cKO testes, we performed qRT-PCR for the expression of atRA target genes Stra8 (stimulated by retinoic acid 8) and Cyp26a1 (cytochrome P450 family 26 subfamily A member 1) [54]. A significant reduction in the expression of Stra8 was observed in the 180 dpp testis (Figure 1C; n = 4). We also detected a significant reduction in the expression of Cyp26a1 in the 60 and 180 dpp testes (Figure 1C and Supplemental Figure S3E; n = 4). As Cyp26a1 has been used as an indirect indicator of tissue retinoid levels [55], these gene expression differences prompted us to quantify the atRA levels within these testes via LC-MS/MS. No significant reduction in the levels of atRA was measured in the 60 or 180 dpp cKO animals compared to the age-matched controls (Figure 1D and Supplemental Figure S3F; n = 3–4).
Elimination of Aldh1a2 within germ cells does not alter the reproductive capacity of cKO mice
To determine whether elimination of Aldh1a2 within the germ cells resulted in any adverse effects on the reproductive capacity of cKO mice, Aldh1a2 control (n = 4) and cKO (n = 4) male mice (approximately 60 days old) were paired with two wild-type female mice each of known fertility for a 4-month fertility study. All males, regardless of genotype, displayed normal breeding behavior (i.e. normal mounting behavior and copulatory plug production). Female mice (n = 8) mated with Aldh1a2, Stra8-Cre control males produced an average of 5.25 litters, with an average of 6.7 ± 0.4 pups per litter (Supplemental Table S3). Female mice (n = 8) mated with Aldh1a2, Stra8-Cre cKO males produced an average of 5 litters, with an average of 6.6 ± 0.3 pups per litter (Supplemental Table S3). The average number of pups sired per litter was not significantly different between the control and cKO males (Figure 1E).
Tamoxifen-inducible postnatal global deletion of Aldh1a2 results in normal testicular histology and no significant reduction in atRA levels
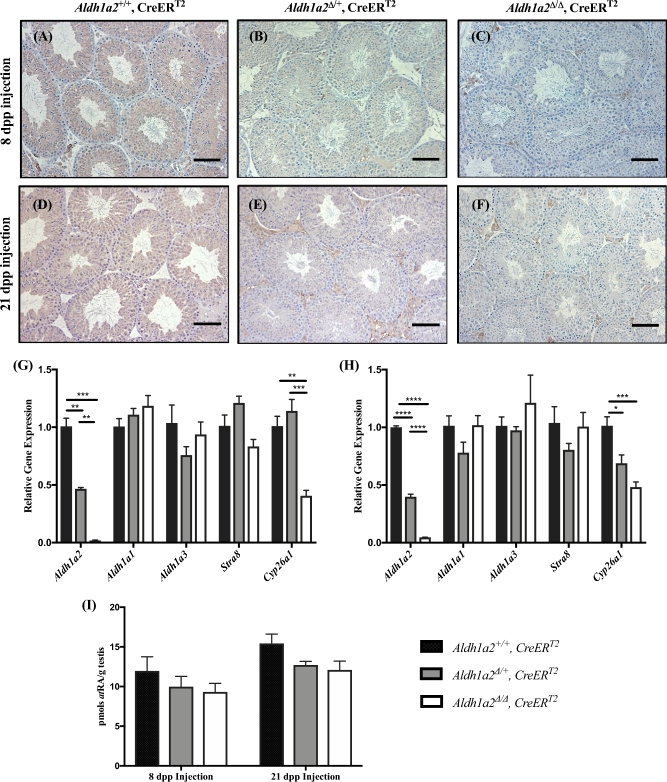
Our IHC analysis of Aldh1a2 cKO testes demonstrated ALDH1A2 expression within the interstitial space (Figure 1B and Supplemental Figure S3C and D; n = 4). As ALDH1A2 has been localized to Leydig cells and macrophages, we wanted to rule out the possibility that the activity of the ALDH1A2 enzymes within the interstitial space was sufficient to drive the qualitatively normal spermatogenesis observed in the Aldh1a2 cKO [37, 56]. To accomplish this, we utilized the CreERT2 transgenic line to generate animals in which global excision of the Aldh1a2 allele can be induced by the administration of tamoxifen. No significant differences in the body weights of the tamoxifen-treated animals were observed on the day of euthanasia, regardless of their genotype or the age at which tamoxifen was administered (Supplemental Figure S4; n = 5–8). We used IHC to compare ALDH1A2 protein expression in testis cross sections from Aldh1a2+/+, CreERT2; Aldh1a2Δ/+, CreERT2; and Aldh1a2Δ/Δ, CreERT2 animals injected with tamoxifen at 8 and 21 dpp (Figure 2A–F; n = 3–7). ALDH1A2 protein expression was robustly detected in the interstitial space, spermatocytes, and spermatids of the Aldh1a2+/+, CreERT2, and Aldh1a2Δ/+, CreERT2 animals (Figure 2A, B, D and E; n = 3–7). In contrast, ALDH1A2 protein expression was not detected in the seminiferous epithelium of Aldh1a2Δ/Δ, CreERT2 animals; however, we were able to detect faint ALDH1A2 staining within the interstitial space (Figure 2C and F; n = 3–7). We performed qRT-PCR to determine the degree of Aldh1a2 reduction in the tamoxifen-treated testes. As anticipated, Aldh1a2 transcript levels were decreased by approximately 50% in all Aldh1a2Δ/+, CreERT2 animals and 96–99% in all Aldh1a2Δ/Δ, CreERT2 animals (Figure 2G and H; n = 4–5). No significant changes in the expression levels of Aldh1a1 and Aldh1a3 were measured by qRT-PCR (Figure 2G and H; n = 4–5). The expression of Stra8 was not significantly different between the three genotypes in age-matched animals (Figure 2G and H; n = 4–5). However, expression of Cyp26a1 was significantly reduced in the Aldh1a2Δ/Δ, CreERT2 animals compared to Aldh1a2+/+, CreERT2, and Aldh1a2Δ/+ animals (Figure 2G and H; n = 4–5). No significant reduction in atRA levels was observed in the testes of mice injected with tamoxifen at 8 or 21 dpp as measured by LC-MS/MS (Figure 2I; n = 3–7).
Figure 2.
Elimination of Aldh1a2 using the inducible CreERT2. Analysis of Aldh1a2+/+, CreERT2; Aldh1a2▵/+, CreERT2; and Aldh1a2▵/▵, CreERT2 animals injected with tamoxifen. Representative immunohistochemistry from animals injected at 8 (A, B, and C; 68 dpp at euthanasia) and 21 dpp (D, E and F; 81 dpp at euthanasia). Cross-sections are stained for ALDH1A2 protein and immunopositive cells are indicated by brown precipitate. qRT-PCR analysis was performed to determine the relative expression of Aldh1a2, Aldh1a1, Aldh1a3, Stra8, and Cyp26a1 in animals injected with tamoxifen at 8 (G) and 21 dpp (H). (I) Graphical representation of atRA measurements. Scale bars = 100 μm. n = 3–7. *P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001.
Maintenance on WIN for one spermatogenic cycle following the WIN7D + RA synchrony protocol significantly increases the number of ZBTB16-positive spermatogonia but does not result in reduced atRA levels
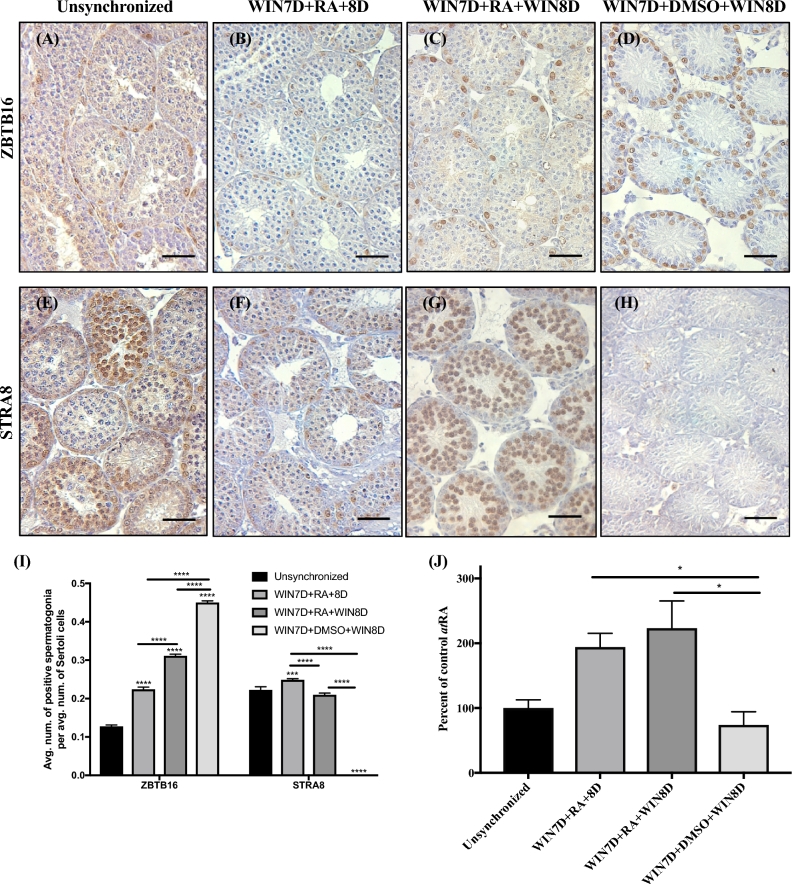
To determine the contribution of all the ALDH1A enzymes to atRA biosynthesis, we utilized a WIN7D + RA synchrony protocol previously established in our laboratory [44]. This protocol involves the maintenance of 2 dpp mice on WIN 18446 for 7 days followed by an injection of atRA. WIN is a potent inhibitor of the ALDH enzymes, and treatment of neonatal mice with WIN for 7 consecutive days reduces atRA levels and results in testes that are enriched for undifferentiated spermatogonia [44]. An exogenous injection of atRA induces the arrested undifferentiated spermatogonia to simultaneously differentiate [44]. If the WIN treatment was continued for 1 cycle or 8 days following the injection of atRA (WIN7D + RA + WIN8D), we could address the contribution of the ALDH1A enzymes following the initial atRA pulse synthesized by the Sertoli cells during normal spermatogenesis. Additionally, this protocol generates testes that are synchronously undergoing spermatogenesis, eliminating the complexity associated with the heterogeneous nature of spermatogenesis in wild-type rodents. Our experimental design included four treatment groups: (1) unsynchronized animals, (2) animals given WIN7D + RA + 8D resulting in synchronized testes, (3) animals given WIN7D + RA + WIN8D (8 additional days of WIN 18446 treatment), and (4) animals given WIN7D + DMSO + WIN8D (continuous WIN 18446 treatment and no atRA) (Supplemental Figure S5A). The 8-day WIN maintenance protocol did not significantly alter the body weights of animals (Supplemental Figure S5B; n = 28–46). However, on the day of euthanasia, the testis weights of unsynchronized, WIN7D + RA + 8D, and WIN7D + RA + WIN8D treated animals were significantly greater than those of the WIN7D + DMSO + WIN8D treated animals (Supplemental Figure S5C; n = 8–46). We hypothesized that if the ALDH1A family of enzymes is the major enzymatic family involved in atRA synthesis during subsequent rounds of spermatogenesis, then WIN7D + RA + WIN8D treated animals would display reduced spermatogonial differentiation compared to the unsynchronized and WIN7D + RA + 8D treated animals. To assess this, we first immunostained and quantified testis sections for the ZBTB16 protein that marks undifferentiated spermatogonia and SOX9 protein which marks Sertoli cells (Figure 3A–D; n = 3) [57, 58]. There was an average of 0.127 ± 0.003 ZBTB16-positive spermatogonia per average number of Sertoli cells in unsynchronized animals (Figure 3I; n = 3). WIN7D + RA + 8D, WIN7D + RA + WIN8D, and WIN7D + DMSO + WIN8D treated animals displayed a 1.8-, 2.4-, and 3.5-fold increase in the average number of ZBTB16-positive spermatogonia per average number of Sertoli cells, respectively, relative to the unsynchronized animals (Figure 3I; n = 3). We also immunostained testis sections for the STRA8 protein that marks differentiating spermatogonia and quantified the proportion of STRA8-positive spermatogonia per average number of Sertoli cells (Figure 3E–H and I; n = 3) [59]. STRA8 was expressed in a heterogeneous manner in testis sections obtained from the unsynchronized animals, whereas synchronous expression of STRA8 was detected in WIN7D + RA + 8D and WIN7D + RA + WIN8D treated animals (Figure 3E–G; n = 3). No detectable STRA8-positive differentiating spermatogonia were detected in the WIN7D + DMSO + WIN8D treated animals (Figure 3H; n = 3). The average number of STRA8-positive differentiating spermatogonia in the unsynchronized, WIN7D + RA + 8D, and WIN7D + RA + WIN8D treated groups were significantly increased compared to the WIN7D + DMSO + WIN8D treated animals (Figure 3I; n = 3). To determine whether the STRA8 counts were reflective of the atRA levels within treated testes, we measured atRA levels by LC-MS/MS. The amounts of atRA measured in the WIN7D + RA + 8D and WIN7D + RA + WIN8D treated groups were significantly greater than those measured for the WIN7D + DMSO + WIN8D treated animals (Figure 3J; n = 3–4).
Figure 3.
Maintenance on WIN7D + RA + WIN8D increases the number of ZBTB16-positive spermatogonia but does not decrease the number of STRA8-positive spermatogonia or atRA levels. Immunostaining of testis sections from unsynchronized (A and E), WIN7D + RA + 8D (B and F), WIN7D + RA + WIN8D (C and G), and WIN7D + DMSO + WIN8D treated mice (D and H). Testis sections were immunostained for ZBTB16 (A–D) and STRA8 (E–H) protein. For each treatment group, immmunopositive cells are denoted by brown precipitate. (I) Graphical representation of ZBTB16 and STRA8 counts. (J) Graphical representation of atRA measurements reported as percent of control. Asterisks represent statistical difference of a treatment group compared to control. Black lines represent statistical significance between treatment groups. Animals were 17 dpp at euthanasia. Scale bar = 100 μM. n = 3–4. *P < 0.05, ***P < 0.001, ****P < 0.0001.
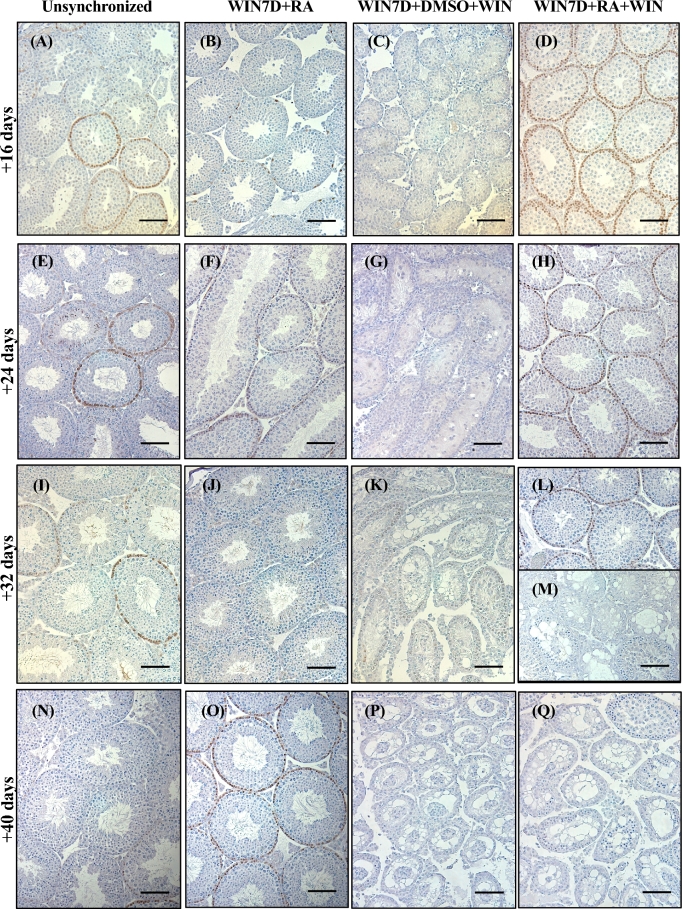
Maintenance on WIN for several spermatogenic cycles following the WIN7D + RA synchrony protocol eventually results in degenerative seminiferous tubules
Since 1 cycle or 8 days of additional WIN treatment was unable to arrest spermatogonial differentiation following the atRA injection, we hypothesized that several additional cycles of treatment were required. To test this, we maintained several groups of animals on WIN for 16, 24, 32, or 40 additional days following the atRA or DMSO injections (Supplemental Figure S6). For comparison purposes, we also maintained unsynchronized and WIN7D + RA treated animals for 16, 24, 32, and 40 day recovery periods (Supplemental Figure S6). Histological analyses were performed to examine STRA8 expression (Figure 4; n = 3–4). Untreated, unsynchronized animals displayed a heterogeneous pattern of STRA8 expression within spermatogonia and preleptotene/leptotene spermatocytes as expected (Figure 4A, E, I, and N; n = 3–4). STRA8 expression was detected in spermatogonia and preleptotene spermatocytes in WIN7D + RA + 16D, WIN7D + RA + 24D, and WIN7D + RA + 40D treated animals but was rare in the WIN7D + RA + 32D treated animals (Figure 4B, F, J, and O; n = 3–4). A high degree of phenotypic heterogeneity was observed in the WIN7D + RA + WIN32D group, as some males displayed testis tubules with normal histology while others had a mixture of morphologically normal tubules adjacent to those with apoptotic cells and missing advanced germ cells (Figure 4 L and M; n = 4). In all WIN7D + RA + WIN40D animals, severe testis degeneration was observed (Figure 4Q; n = 4). No STRA8-positive spermatogonia were detected in any of the animals given a DMSO injection and maintained on WIN for 16, 24, 32, or 40 days (Figure 4C, G, K, and P; n = 3–4).
Figure 4.
Maintenance on WIN for several spermatogenic cycles eventually results in degenerative seminiferous tubules. Immunostaining of testis sections from unsynchronized (A, E, I, and N), WIN7D + RA (B, F, J, and O), WIN7D + DMSO + WIN (C, G, K, and P), and WIN7D + RA + WIN (D, H, L, M, and Q) treated mice for 16, 24, 32, and 40 additional maintenance days. Testis sections were immunostained for STRA8. Immunopositive cells are indicated by brown precipitate. Scale bar = 100 μM. n = 3–4.
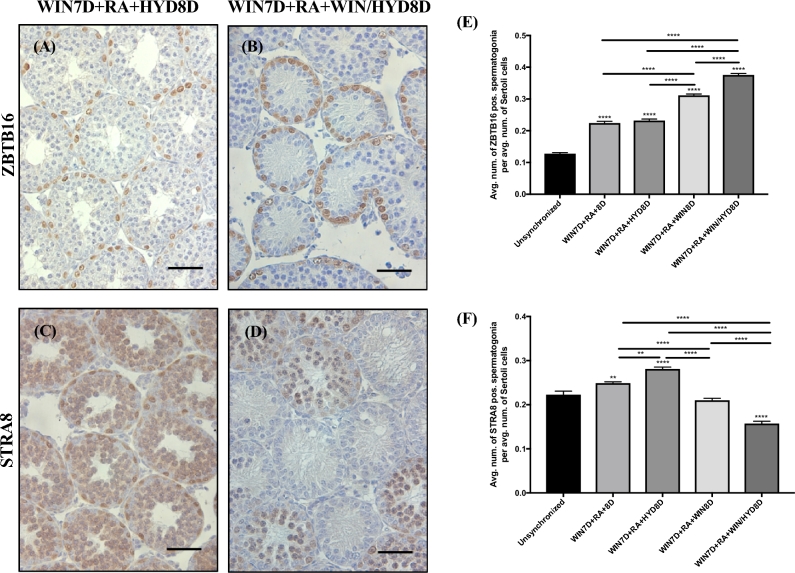
Maintenance on WIN and HYD for one spermatogenic cycle following the WIN7D + RA synchrony protocol increases the number of ZBTB16-positive spermatogonia
Although WIN is a potent pan-ALDH1A inhibitor, approximately 32–40 days of WIN maintenance was required for severe seminiferous tubule degeneration when administered after an atRA pulse. Based on these findings, we hypothesized that an alternative source of atRA synthesis, independent of the ALDH1A enzymes, becomes available following the atRA injection. Of the enzyme families capable of catalyzing the atRAL to atRA reaction, transcripts for members of the AOX family were identified in microarray sequencing experiments previously performed in our laboratory [60, 61]. To test their potential contribution to atRA biosynthesis, we maintained animals on a potent inhibitor of the AOX family of enzymes, HYD (Supplemental Figure S7) [62, 63]. Compared to unsynchronized animals, significant differences in the number of STRA8-positive spermatogonia per average number of Sertoli cells were detected in animals maintained on the WIN7D + RA + 8D, WIN7D + RA + HYD8D, and WIN7D + RA + WIN/HYD8D treatment schemes (Figure 5F; n = 3). No significant differences in the number of STRA8-positive spermatogonia per average number of Sertoli cells were detected in the WIN7D + RA + WIN8D treatment group (Figure 5F; n = 3). A significant increase in the number of ZBTB16-positive spermatogonia was detected in all treatment groups compared to the unsynchronized animals (Figure 5E; n = 3).
Figure 5.
WIN7D + RA + HYD8D animals do not show any spermatogenic defects, although WIN7D + RA + WIN/HYD8D animals show an increased number of ZBTB16-positive spermatogonia. Immunostaining of testis sections for ZBTB16 and STRA8 for WIN7D + RA + HYD8D (A and B) and WIN7D + RA + WIN/HYD8D treated mice (C and D). Graphical representation of the average number of ZBTB16- (E) and STRA8-positive (F) spermatogonia per tubule within each treatment group. Asterisks represent statistical difference of a treatment group compared to control. Black lines indicate statistical significance between treatment groups. Animals were 17 dpp at euthanasia. Scale bar = 100 μM. n = 3. *P < 0.05, **P < 0.01, ****P < 0.0001.
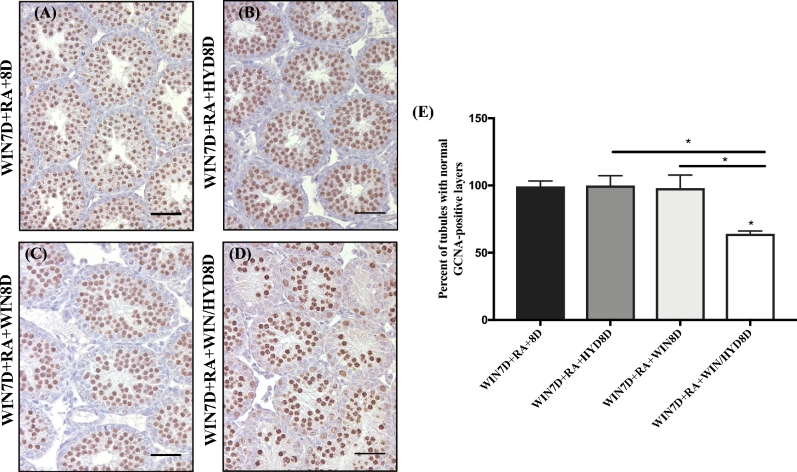
Concurrent maintenance on WIN/HYD for one spermatogenic cycle following the WIN7D + RA synchrony protocol increases the number of testis tubules missing germ cells expressing germ cell nuclear antigen
Our histological observations of the WIN7D + RA + WIN/HYD8D treated animals prompted us to quantify the number of testis tubules missing germ cells. Testis sections from WIN7D + RA + 8D, WIN7D + RA + WIN8D, WIN7D + RA + HYD8D, and WIN7D + RA + WIN/HYD8D treated animals were immunostained for the germ cell-specific marker GCNA (Figure 6A–D) [57]. Quantification revealed that 2% of the testis tubules within the WIN7D + RA + 8D, WIN7D + RA + HYD8D, and WIN7D + RA + WIN8D treated groups were missing cells or layers of GCNA-positive cells (Figure 6E; n = 3–4). In contrast, 36% of WIN7D + RA + WIN/HYD8D treated animals had testis tubules missing cells or layers of GCNA-positive cells (Figure 6E; n = 3–4).
Figure 6.
WIN7D + RA + WIN/HYD8D animals have an increased number of tubules with abnormal or missing layers of GCNA-positive germ cells. Representative testis sections from WIN7D + RA + 8D (A), WIN7D + RA + HYD8D (B), WIN7D + RA + WIN8D (C), and WIN7D + RA + WIN/HYD8D (D) treated animals immunostained for GCNA. Immunopositive cells are indicated by brown precipitate. (E) Graphical representation of the number of tubules from WIN7D + RA + 8D, WIN7D + RA + HYD8D, WIN7D + RA + WIN8D, and WIN7D + RA + WIN/HYD8D treated testes containing normal GCNA-positive germ cell layers. Asterisks represent statistical difference of a treatment group compared to control. Black lines represent statistical significance between treatment groups. Animals were 17 dpp at euthanasia. n = 3–4. *P < 0.05.
Discussion
It is well established that atRA is absolutely required for spermatogonial differentiation [9, 11]; however, many gaps still remain in our understanding of the enzymatic and cellular sources responsible for generating atRA. Using a genetic model, Raverdeau and colleagues demonstrated that atRA in the first round of spermatogenesis is synthesized by the ALDH1A enzymes within Sertoli cells [31]. Importantly, if the mice with all three ALDH1A enzymes deleted in Sertoli cells were given a single injection of atRA, spermatogenesis resumed and was maintained suggesting that the atRA injection resulted in an alternate source of continuous atRA synthesis [31]. However, the cellular and enzymatic source(s) of atRA driving these subsequent rounds of spermatogonial differentiation is a key research question that remains to be fully addressed.
ALDH1A2 is a cytoplasmic atRA-generating enzyme enriched in meiotic and postmeiotic germ cells within the murine testis [24, 64, 65]. Previous publications have provided evidence to suggest that this enzyme may be the primary source of atRA synthesis driving subsequent rounds of spermatogenesis. However, our present findings do not support a role for ALDH1A2 as an essential enzyme to testicular atRA biosynthesis, as severe deficiency of Aldh1a2 within the postnatal testis in either the germ cell cKO or the Aldh1a2Δ/Δ, CreERT2 animals analyzed in this study did not result in significantly reduced atRA levels. Interestingly, Cyp26a1 has been suggested to be a reliable indicator of endogenous atRA levels, yet the significant reduction in Cyp26a1 expression in both the germ cell cKO and the Aldh1a2Δ/Δ, CreERT2 animals was not reflective of endogenous levels of total testicular atRA. The biological function of the CYP26 enzymes is to metabolize atRA into inactive metabolites [20, 66–68]. Therefore, the reduction in Cyp26a1 expression may actually be suggestive of a feedback mechanism that is lowering degradation activities in order to sustain higher levels atRA within the Aldh1a2 cKO and Aldh1a2Δ/Δ, CreERT2 testes. It is also possible that the reduction in Cyp26a1 may allow for circulating extratesticular atRA to enter the testis, although this possibility seems unlikely as it has recently been reported that CYP26B1, not CYP26A1, is the major isomer involved in the regulation of atRA levels within the seminiferous epithelium [55]. Therefore, future investigations using radiolabeled atRA within these Aldh1a2 cKO testes are required to further investigate this possibility.
It has been widely hypothesized that the atRA required for spermatogenesis is made inside the epithelium [35]. In support of this hypothesis, Kurlandsky and colleagues found that less than 1% of circulating atRA enters the seminiferous epithelium [69], which is not enough to elicit the responses that we observed. Therefore, we hypothesized that the levels of atRA detected within the Aldh1a2 germ cell cKO and Aldh1a2Δ/Δ, CreERT2 testes could be due to compensation by the other ALDH1A enzymes (Aldh1a1 and Aldh1a3). We tested this hypothesis pharmacologically by inhibiting the activities of the ALDH1A enzymes using the pan-ALDH1A inhibitor WIN 18446. In the absence of an atRA pulse or injection, the ZBTB16-positive spermatogonia accumulated but no differentiated STRA8-positive spermatogonia appeared. If given a single injection of atRA, our analysis revealed that the ZBTB16-positive population was sensitive to continued maintenance on WIN; however, we observed no significant reductions in the number of STRA8-positive spermatogonia or atRA levels. Further, we found that approximately 32–40 days of continuous WIN treatment was required to induce a severe degenerative phenotype if a single atRA pulse was given at 9 dpp. It is presently unclear why treatment with WIN requires such a prolonged period of time before cessation of spermatogenesis occurs. However, our data demonstrate that once the testis receives an atRA pulse, produced in situ by Sertoli cells or by injection, potent inhibition of the ALDH1A enzymes is insufficient to immediately inhibit subsequent rounds of spermatogonial differentiation.
Although the ALDH1A enzymes are the only family of enzymes currently implicated in the oxidation of atRAL to atRA in the mammalian testis, multiple enzyme families are able to perform this same metabolic step in other retinoid-dependent organs, such as the liver [43, 70–72]. The AOX enzymes are of particular interest as transcripts for two of the AOX isomers were significantly upregulated in testis microarray studies previously performed in our laboratory. Aox4 was upregulated in germ cells compared to Sertoli cells during a synchronized first round of spermatogenesis [60]. Aox3 was significantly upregulated in another microarray study aimed at identifying transcripts differentially expressed in spermatogenic stages in the adult testis, with maximal expression observed in stages VI–VIII [61]. Aox3 mRNA has also been detected in germ cells via in situ hybridization [73]. However, the Km values for the AOX enzymes is a magnitude higher than those reported for the ALDH1A enzymes, and because of this the AOX enzymes may only play a physiologically relevant role in testicular atRA biosynthesis when the ALDH1A enzymes are inactive [74–76]. Accordingly, the Aox4-null mouse is fully fertile, supporting a secondary role for these enzymes in testicular atRA biosynthesis [72]. Presently, very little is known about the role of AOX3 within the postnatal testis, as no genetic knockout models have been produced to date. Future studies of the Aox3-null mouse and ALDH1A/AOX compound animals will further elucidate the role that these enzymes play in testicular atRA biosynthesis.
We found the WIN7D + DMSO + WIN animals particularly interesting, as histological analysis of these animals demonstrated that the dosage of WIN utilized in our studies was sufficient to block spermatogonial differentiation for prolonged periods of time. Surprisingly, atRA measurements revealed residual levels of atRA remaining in the WIN7D + DMSO + WIN8D treated testes. This was consistent with other studies, in which residual amounts of atRA were detected even though histological analysis confirmed that WIN treatment resulted in the complete loss of advanced germ cells and cessation of spermatogenesis [37, 77, 78]. These findings lead us to hypothesize that the A to A1 transition may require a certain threshold of atRA that can only be met when the ALDH1A enzymes are fully functional.
The requirement for atRA in spermatogonial differentiation, meiotic initiation, blood–testis barrier formation, spermiogenesis, and spermiation has been well studied. However, investigations of how and where atRA is produced to drive these processes have been complicated by gaps in our knowledge of the enzymes responsible for atRA biosynthesis. Although the expression of ALDH1A1 residing within adult Sertoli cells may contribute to the biosynthesis of atRA driving the premeiotic transitions, several groups have provided evidence to suggest that ALDH1A2 residing within the meiotic and postmeiotic germ cells is the major isomer involved [31, 32, 38, 45]. In this report, we have examined for the first time the in vivo contribution of ALDH1A2 to postnatal testicular atRA levels using two complementary genetic approaches. Our results demonstrate that severe deficiency of Aldh1a2 did not result in any adverse effects on male fertility or health. This report also provides evidence suggesting that the AOX family of enzymes may play a physiologically relevant role in testicular atRA biosynthesis, specifically following an injection of atRA when the ALDH1A enzymes are inhibited.
Supplementary data
Supplemental Figure S1. Generation of conditional allele to knock out the Aldh1a2 gene. The conditional allele for Aldh1a2 was generated via a sequence replacement strategy. The diagram shows the wild-type Aldh1a2 allele (top row), targeting construct (second row), homologous recombinant (third row), targeted conditional Aldh1a2 allele without the neo cassette (fourth row), and the Cre-mediated deletion of the Aldh1a2 gene (fifth row). The targeting construct contained (1) loxP sites that flanked exon 4, (2) a 2.5 kb 5′ short arm of homology, (3) a 5.6 kb 3′ long arm of homology, (4) a Diphtheria Toxin A (DTA) cassette, and (6) a Neomycin (Neo) cassette flanked by FRT sites for selective deletion. The Neo element allowed for positive selection in ES cells, while the DTA element allowed for negative selection in ES cells. After homologous recombination of the conditional knockout construct, the Aldh1a2 gene will have normal expression until Cre-mediated deletion of exon 4. Deletion of exon 4 creates a frameshift mutation and a premature stop, which renders the Aldh1a2 gene inactive.
Supplemental Figure S2. Schematic of the WIN maintenance regimens. Control animals were fed 1% gum tragacanth for the entirety of the treatment regimen. Two dpp male mice were given 100 mg/kg WIN daily for seven consecutive days, given an atRA injection at 9 dpp and then maintained on 1% gum tragacanth (WIN7D + RA), 150 mg/kg WIN (WIN7D + RA + WIN), 25 mg/kg HYD (WIN7D + RA + HYD) or 150 mg/kg WIN and 25 mg/kg HYD (WIN7D + RA + WIN/HYD). Another group of animals were also fed 100 mg/kg WIN from 2–8 dpp, followed by a DMSO injection at 9 dpp and maintained on 150 mg/kg WIN (WIN7D + DMSO + WIN). Depending on the experiment, maintenance animals were maintained on their treatment schemes for 8, 16, 24, 32, or 40 days.
Supplemental Figure S3. Elimination of Aldh1a2 using the Stra8-Cre. Control (Aldh1a2fl/fl, Stra8-Cre–) and cKO (Aldh1a2fl/fl, Stra8-Cre+) animals analyzed at 60 dpp. Representative cross-sections for control (A, B) and cKO (C, D) animals, stained for ALDH1A2. Immunopositive cells are indicated by brown precipitate. (E) qRT-PCR analysis of Aldh1a2, Aldh1a1, Aldh1a3, Stra8, and Cyp26a1. (F) Graphical representation of atRA measurements. Scale bars = 100 μM. for n = 3–6. *P < 0.05, and **P < 0.01.
Supplemental Figure S4. Body weight in tamoxifen injected Aldh1a2, CreERT2 animals. Body weight of Aldh1a2+/+, CreERT2; Aldh1a2+/Ä, CreERT2; and Aldh1a2Ä/Ä, CreERT2 animals injected with tamoxifen at 8 and 21 dpp. Body weights were measured in grams (g) on the day of euthanasia, n = 5–8.
Supplemental Figure S5. WIN maintenance regimen, changes in body weight, and average testis weight. (A) Schematic of the WIN maintenance regimen across one spermatogenic cycle. (B) Average body weight measured in grams (g) over the 8-day maintenance regimen (n = 28–46). (C) Average testis weight measured in grams (g) on the day of euthanasia (n = 8–46). Asterisks represent statistical difference of a treatment group compared to control. Black lines represent statistical significance between treatment groups. ****P < 0.0001.
Supplemental Figure S6. Schematic of the WIN maintenance regimen across several spermatogenic cycles. Unsynchronized animals were fed 1% gum tragacanth for 16, 24, 32, or 40 days. Two dpp male mice were given 100 mg/kg WIN daily for seven consecutive days and then placed in one of the following three treatment groups: (1) atRA injection at 9 dpp and maintained on 1% gum tragacanth for 16, 24, 32, or 40 days (WIN7D + RA); (2) atRA injection at 9 dpp and maintained on 150 mg/kg WIN for 16, 24, 32, or 40 days (WIN7D + RA + WIN); or (3) DMSO injection at 9 dpp and maintained on 150 mg/kg WIN for 16, 24, 32, or 40 days (WIN7D + DMSO + WIN).
Supplemental Figure S7. Schematic of the treatment regimen for animals maintained solely on HYD or concurrently on WIN and HYD following the WIN + RA synchrony protocol. Two dpp male mice were given 100 mg/kg WIN daily for seven consecutive days, injected with atRA at 9 dpp and maintained either on 25 mg/kg HYD (WIN7D + RA + HYD8D) or maintained concurrently on 150 mg/kg WIN and 25 mg/kg HYD for 8 additional days (WIN7D + RA + WIN/HYD8D).
Supplemental Table S1. PCR primers for genotyping.
Supplemental Table S2. Primers for quantitative RT-PCR.
Supplemental Table S3. Male fertility data for Aldhla2fl/fl, Stra8-Cre mutant mice.
Acknowledgments
We are most grateful to Dr Joseph L. Napoli for the donation of the Aldh1a2fl/fl transgenic line as well as the technical support provided by the Mouse Biology Program (MBP) at the University of California, Davis. We would like to thank Dr John Amory for the kind gift of the WIN 18,446 compound. We would also like to acknowledge Rachel Gewiss, Debra Mitchell, Zane Rivers, and Quinton Beedle for their technical assistance, helpful comments, and critical reading of the manuscript.
Notes
Conference presentation: Presented in part at the 50th Annual Meeting of the Society for the Study of Reproduction, 13–16 July 2017, Washington D.C., USA.
Footnotes
Grant support: This research was supported by U.S. National Institutes of Health grants RO1 10808 to MDG and U54 HD 42454 to MDG.
References
- 1. Yang QE, Oatley JM. Spermatogonial stem cell functions in physiological and pathological conditions. Curr Top Dev Biol 2014; 107:235–267. [DOI] [PubMed] [Google Scholar]
- 2. Busada JT, Geyer CB. The role of retinoic acid (RA) in spermatogonial differentiation. Biol Reprod 2016; 94:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Drumond AL, Meistrich ML, Chiarini-Garcia H. Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction 2011; 142:145–155. [DOI] [PubMed] [Google Scholar]
- 4. Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction 2015; 149:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Oakberg EF. A description of spermiogenesis in the mouse and its use in analysis of the cycle of the seminiferous epithelium and germ cell renewal. Am J Anat 1956; 99:391–413. [DOI] [PubMed] [Google Scholar]
- 6. Russell L, Ettlin R, Sinha HA, Clegg E. Histological and Histopathological Evaluation of the Testis Clearwater. Florida: Cache River Press; 1990. [Google Scholar]
- 7. Clermont Y, Trott M. Duration of the cycle of the seminiferous epithelium in the mouse and hamster determined by means of 3H-thymidine and radioautography. Fertil Steril 1969; 20:805–817. [DOI] [PubMed] [Google Scholar]
- 8. Hogarth C, Griswold M. Driving asynchronous spermatogenenesis: is retinoic acid the answer. Anim Reprod 2012; 9:742–750. [Google Scholar]
- 9. Wolbach SB, Howe PR. Tissue changes following deprivation of fat-soluble a vitamin. J Exp Med 1925; 42:753–777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. van Pelt AM, de Rooij DG. Retinoic acid is able to reinitiate spermatogenesis in vitamin A-deficient rats and high replicate doses support the full development of spermatogenic cells. Endocrinology 1991; 128:697–704. [DOI] [PubMed] [Google Scholar]
- 11. Griswold MD, Bishop PD, Kim KH, Ping R, Siiteri JE, Morales C. Function of vitamin A in normal and synchronized seminiferous tubules. Ann NY Acad Sci 1989; 564:154–172. [DOI] [PubMed] [Google Scholar]
- 12. van Pelt AM, de Rooij DG. Synchronization of the seminiferous epithelium after vitamin A replacement in vitamin A-deficient mice. Biol Reprod 1990; 43:363–367. [DOI] [PubMed] [Google Scholar]
- 13. Duester G. Families of retinoid dehydrogenases regulating vitamin A function. Eur J Biochem 2000; 267:4315–4324. [DOI] [PubMed] [Google Scholar]
- 14. Pares X, Farres J, Kedishvili N, Duester G. Medium- and short-chain dehydrogenase/reductase gene and protein families. Cell Mol Life Sci 2008; 65:3936–3949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Boerman MH, Napoli JL. Effects of sulfhydryl reagents, retinoids, and solubilization on the activity of microsomal retinol dehydrogenase. Arch Biochem Biophys 1995; 321:434–441. [DOI] [PubMed] [Google Scholar]
- 16. Mertz JR, Shang E, Piantedosi R, Wei S, Wolgemuth DJ, Blaner WS. Identification and characterization of a stereospecific human enzyme that catalyzes 9-cis-retinol oxidation. J Biol Chem 1997; 272:11744–11749. [DOI] [PubMed] [Google Scholar]
- 17. Romert A, Tuvendal P, Simon A, Dencker L, Eriksson U. The identification of a 9-cis retinol dehydrogenase in the mouse embryo reveals a pathway for synthesis of 9-cis retinoic acid. Proc Natl Acad Sci USA 1998; 95:4404–4409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lapshina EA, Belyaeva OV, Chumakova OV, Kedishvili NY. Differential recognition of the free versus bound retinol by human microsomal retinol/sterol dehydrogenases: characterization of the holo-CRBP dehydrogenase activity of RoDH-4. Biochemistry 2003; 42:776–784. [DOI] [PubMed] [Google Scholar]
- 19. Lee MO, Manthey CL, Sladek NE. Identification of mouse liver aldehyde dehydrogenases that catalyze the oxidation of retinaldehyde to retinoic acid. Biochem Pharmacol 1991; 42:1279–1285. [DOI] [PubMed] [Google Scholar]
- 20. Duester G. Retinoic acid synthesis and signaling during early organogenesis. Cell 2008; 134:921–931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Theodosiou M, Laudet V, Schubert M. From carrot to clinic: an overview of the retinoic acid signaling pathway. Cell Mol Life Sci 2010; 67:1423–1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Niederreither K, McCaffery P, Drager UC, Chambon P, Dolle P. Restricted expression and retinoic acid-induced downregulation of the retinaldehyde dehydrogenase type 2 (RALDH-2) gene during mouse development. Mech Dev 1997; 62:67–78. [DOI] [PubMed] [Google Scholar]
- 23. Fan X, Molotkov A, Manabe S, Donmoyer CM, Deltour L, Foglio MH, Cuenca AE, Blaner WS, Lipton SA, Duester G. Targeted disruption of Aldh1a1 (Raldh1) provides evidence for a complex mechanism of retinoic acid synthesis in the developing retina. Mol Cell Biol 2003; 23:4637–4648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Wang X, Penzes P, Napoli JL. Cloning of a cDNA encoding an aldehyde dehydrogenase and its expression in Escherichia coli. J Biol Chem 1996; 271:16288–16293. [DOI] [PubMed] [Google Scholar]
- 25. Sima A, Parisotto M, Mader S, Bhat PV. Kinetic characterization of recombinant mouse retinal dehydrogenase types 3 and 4 for retinal substrates. Biochim Biophys Acta 2009; 1790:1660–1664. [DOI] [PubMed] [Google Scholar]
- 26. Grun F, Hirose Y, Kawauchi S, Ogura T, Umesono K. Aldehyde dehydrogenase 6, a cytosolic retinaldehyde dehydrogenase prominently expressed in sensory neuroepithelia during development. J Biol Chem 2000; 275:41210–41218. [DOI] [PubMed] [Google Scholar]
- 27. Vasiliou V, Pappa A, Estey T. Role of human aldehyde dehydrogenases in endobiotic and xenobiotic metabolism. Drug Metab Rev 2004; 36:279–299. [DOI] [PubMed] [Google Scholar]
- 28. Moretti A, Li J, Donini S, Sobol RW, Rizzi M, Garavaglia S. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid. Sci Rep 2016; 6:35710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Moretti A, Li J, Donini S, Sobol WR, Garavaglia S. Crystal structure of human aldehyde dehydrogenase 1A3 complexed with NAD+ and retinoic acid. Sci Rep 2016; 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Hsu LC, Chang WC, Yoshida A. Mouse type-2 retinaldehyde dehydrogenase (RALDH2): genomic organization, tissue-dependent expression, chromosome assignment and comparison to other types. Biochim Biophys Acta 2000; 1492:289–293. [DOI] [PubMed] [Google Scholar]
- 31. Raverdeau M, Gely-Pernot A, Feret B, Dennefeld C, Benoit G, Davidson I, Chambon P, Mark M, Ghyselinck NB. Retinoic acid induces Sertoli cell paracrine signals for spermatogonia differentiation but cell autonomously drives spermatocyte meiosis. Proc Natl Acad Sci USA 2012; 109:16582–16587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Endo T, Freinkman E, de Rooij DG, Page DC. Periodic production of retinoic acid by meiotic and somatic cells coordinates four transitions in mouse spermatogenesis. Proc Natl Acad Sci USA 2017; 114:E10132–E10141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Wu JW, Wang RY, Guo QS, Xu C. Expression of the retinoic acid-metabolizing enzymes RALDH2 and CYP26b1 during mouse postnatal testis development. Asian J Androl 2008; 10:569–576. [DOI] [PubMed] [Google Scholar]
- 34. Zhai Y, Sperkova Z, Napoli JL. Cellular expression of retinal dehydrogenase types 1 and 2: effects of vitamin A status on testis mRNA. J Cell Physiol 2001; 186:220–232. [DOI] [PubMed] [Google Scholar]
- 35. Vernet N, Dennefeld C, Rochette-Egly C, Oulad-Abdelghani M, Chambon P, Ghyselinck NB, Mark M. Retinoic acid metabolism and signaling pathways in the adult and developing mouse testis. Endocrinology 2006; 147:96–110. [DOI] [PubMed] [Google Scholar]
- 36. Arnold SL, Kent T, Hogarth CA, Schlatt S, Prasad B, Haenisch M, Walsh T, Muller CH, Griswold MD, Amory JK, Isoherranen N. Importance of ALDH1A enzymes in determining human testicular retinoic acid concentrations. J Lipid Res 2015; 56:342–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kent T, Arnold SL, Fasnacht R, Rowsey R, Mitchell D, Hogarth CA, Isoherranen N, Griswold MD. ALDH enzyme expression is independent of the spermatogenic cycle, and their inhibition causes misregulation of murine spermatogenic processes. Biol Reprod 2016; 94:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Sugimoto R, Nabeshima Y, Yoshida S. Retinoic acid metabolism links the periodical differentiation of germ cells with the cycle of Sertoli cells in mouse seminiferous epithelium. Mech Dev 2012; 128:610–624. [DOI] [PubMed] [Google Scholar]
- 39. Niederreither K, Subbarayan V, Dolle P, Chambon P. Embryonic retinoic acid synthesis is essential for early mouse post-implantation development. Nat Genet 1999; 21:444–448. [DOI] [PubMed] [Google Scholar]
- 40. Mic FA, Haselbeck RJ, Cuenca AE, Duester G. Novel retinoic acid generating activities in the neural tube and heart identified by conditional rescue of Raldh2 null mutant mice. Development 2002; 129:2271–2282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Dupe V, Matt N, Garnier JM, Chambon P, Mark M, Ghyselinck NB. A newborn lethal defect due to inactivation of retinaldehyde dehydrogenase type 3 is prevented by maternal retinoic acid treatment. Proc Natl Acad Sci USA 2003; 100:14036–14041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Molotkova N, Molotkov A, Duester G. Role of retinoic acid during forebrain development begins late when Raldh3 generates retinoic acid in the ventral subventricular zone. Dev Biol 2007; 303:601–610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Arnold SL, Kent T, Hogarth CA, Griswold MD, Amory JK, Isoherranen N. Pharmacological inhibition of ALDH1A in mice decreases all-trans retinoic acid concentrations in a tissue specific manner. Biochem Pharmacol 2015; 95:177–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Hogarth CA, Evanoff R, Mitchell D, Kent T, Small C, Amory JK, Griswold MD. Turning a spermatogenic wave into a tsunami: synchronizing murine spermatogenesis using WIN 18,4461. Biol Reprod 2013; 88:40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Kasimanickam VR. Expression of retinoic acid-metabolizing enzymes, ALDH1A1, ALDH1A2, ALDH1A3, CYP26A1, CYP26B1 and CYP26C1 in canine testis during post-natal development. Reprod Dom Anim 2016; 51:901–909. [DOI] [PubMed] [Google Scholar]
- 46. Niederreither K, Vermot J, Le Roux I, Schuhbaur B, Chambon P, Dolle P. The regional pattern of retinoic acid synthesis by RALDH2 is essential for the development of posterior pharyngeal arches and the enteric nervous system. Development 2003; 130:2525–2534. [DOI] [PubMed] [Google Scholar]
- 47. Sadate-Ngatchou PI, Payne CJ, Dearth AT, Braun RE. Cre recombinase activity specific to postnatal, premeiotic male germ cells in transgenic mice. Genesis 2008; 46:738–742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Feil S, Valtcheva N, Feil R. Inducible Cre mice. Methods Mol Biol 2009; 530:343–363. [DOI] [PubMed] [Google Scholar]
- 49. Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods 2001; 25:402–408. [DOI] [PubMed] [Google Scholar]
- 50. Hogarth C, Griswold M. Immunohistochemical approaches for the study of spermatogenesis. Methods Mol Biol 2013; 927:309–320. [DOI] [PubMed] [Google Scholar]
- 51. Hogarth CA, Arnold S, Kent T, Mitchell D, Isoherranen N, Griswold MD. Processive pulses of retinoic acid propel asynchronous and continuous murine sperm production. Biol Reprod 2015; 92:37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Arnold SL, Amory JK, Walsh TJ, Isoherranen N. A sensitive and specific method for measurement of multiple retinoids in human serum with UHPLC-MS/MS. J Lipid Res 2012; 53:587–598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Stevison F, Hogarth C, Tripathy S, Kent T, Isoherranen N. Inhibition of the all-trans retinoic acid (at RA) hydroxylases CYP26A1 and CYP26B1 results in dynamic, tissue-specific changes in endogenous at RA signaling. Drug Metab Dispos 2017; 45:846–854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Hogarth CA, Evanoff R, Snyder E, Kent T, Mitchell D, Small C, Amory JK, Griswold MD. Suppression of Stra8 expression in the mouse gonad by WIN 18,4461. Biol Reprod 2011; 84:957–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ross AC. Retinoid production and catabolism: role of diet in regulating retinol esterification and retinoic acid oxidation. J Nutr 2003; 133:291S–296S. [DOI] [PubMed] [Google Scholar]
- 56. DeFalco T, Potter SJ, Williams AV, Waller B, Kan MJ, Capel B. Macrophages contribute to the spermatogonial niche in the adult testis. Cell Rep 2015; 12:1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36:647–652. [DOI] [PubMed] [Google Scholar]
- 58. Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004; 36:653–659. [DOI] [PubMed] [Google Scholar]
- 59. Zhou Q, Li Y, Nie R, Friel P, Mitchell D, Evanoff RM, Pouchnik D, Banasik B, McCarrey JR, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) and maturation of murine gonocytes and spermatogonia induced by retinoic acid in vitro. Biol Reprod 2008; 78:537–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Evans E, Hogarth C, Mitchell D, Griswold M. Riding the spermatogenic wave: profiling gene expression within neonatal germ and sertoli cells during a synchronized initial wave of spermatogenesis in mice. Biol Reprod 2014; 90:108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jauregui EJ, Mitchell D, Garza SM, Topping T, Hogarth CA, Griswold MD. Leydig cell genes change their expression and association with polysomes in a stage-specific manner in the adult mouse testis. Biol Reprod 2018; 98:722–738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Strelevitz TJ, Orozco CC, Obach RS. Hydralazine as a selective probe inactivator of aldehyde oxidase in human hepatocytes: estimation of the contribution of aldehyde oxidase to metabolic clearance. Drug Metab Dispos 2012; 40:1441–1448. [DOI] [PubMed] [Google Scholar]
- 63. Johnson C, Stubley-Beedham C, Stell JG. Hydralazine: a potent inhibitor of aldehyde oxidase activity in vitro and in vivo. Biochem Pharmacol 1985; 34:4251–4256. [DOI] [PubMed] [Google Scholar]
- 64. Nishimura M, Naito S. Tissue-specific mRNA expression profiles of human phase I metabolizing enzymes except for cytochrome P450 and phase II metabolizing enzymes. Drug Metab Pharmacokinet 2006; 21:357–374. [DOI] [PubMed] [Google Scholar]
- 65. Alnouti Y, Klaassen CD. Tissue distribution, ontogeny, and regulation of aldehyde dehydrogenase (Aldh) enzymes mRNA by prototypical microsomal enzyme inducers in mice. Toxicol Sci 2008; 101:51–64. [DOI] [PubMed] [Google Scholar]
- 66. Thatcher JE, Isoherranen N. The role of CYP26 enzymes in retinoic acid clearance. Expert Opin Drug Metab Toxicol 2009; 5:875–886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Kedishvili NY. Enzymology of retinoic acid biosynthesis and degradation. J Lipid Res 2013; 54:1744–1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Fiorella PD, Napoli JL. Microsomal retinoic acid metabolism. Effects of cellular retinoic acid-binding protein (type I) and C18-hydroxylation as an initial step. J Biol Chem 1994; 269:10538–10544. [PubMed] [Google Scholar]
- 69. Kurlandsky SB, Gamble MV, Ramakrishnan R, Blaner WS. Plasma delivery of retinoic acid to tissues in the rat. J Biol Chem 1995; 270:17850–17857. [DOI] [PubMed] [Google Scholar]
- 70. Vila R, Kurosaki M, Barzago MM, Kolek M, Bastone A, Colombo L, Salmona M, Terao M, Garattini E. Regulation and biochemistry of mouse molybdo-flavoenzymes. The DBA/2 mouse is selectively deficient in the expression of aldehyde oxidase homologues 1 and 2 and represents a unique source for the purification and characterization of aldehyde oxidase. J Biol Chem 2004; 279:8668–8683. [DOI] [PubMed] [Google Scholar]
- 71. Huang DY, Furukawa A, Ichikawa Y. Molecular cloning of retinal oxidase/aldehyde oxidase cDNAs from rabbit and mouse livers and functional expression of recombinant mouse retinal oxidase cDNA in Escherichia coli. Arch Biochem Biophys 1999; 364:264–272. [DOI] [PubMed] [Google Scholar]
- 72. Terao M, Kurosaki M, Marini M, Vanoni MA, Saltini G, Bonetto V, Bastone A, Federico C, Saccone S, Fanelli R, Salmona M, Garattini E. Purification of the aldehyde oxidase homolog 1 (AOH1) protein and cloning of the AOH1 and aldehyde oxidase homolog 2 (AOH2) genes. Identification of a novel molybdo-flavoprotein gene cluster on mouse chromosome 1. J Biol Chem 2001; 276:46347–46363. [DOI] [PubMed] [Google Scholar]
- 73. Terao M, Kurosaki M, Saltini G, Demontis S, Marini M, Salmona M, Garattini E. Cloning of the cDNAs coding for two novel molybdo-flavoproteins showing high similarity with aldehyde oxidase and xanthine oxidoreductase. J Biol Chem 2000; 275:30690–30700. [DOI] [PubMed] [Google Scholar]
- 74. Garattini E, Fratelli M, Terao M. The mammalian aldehyde oxidase gene family. Hum Genomics 2009; 4:119–130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Chen Y, Koppaka V, Thompson DC, Vasiliou V. Focus on molecules: ALDH1A1: from lens and corneal crystallin to stem cell marker. Exp Eye Res 2012; 102:105–106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Yoshida A, Hsu LC, Dave V. Retinal oxidation activity and biological role of human cytosolic aldehyde dehydrogenase. Enzyme 1992; 46:239–244. [DOI] [PubMed] [Google Scholar]
- 77. Paik J, Haenisch M, Muller CH, Goldstein AS, Arnold S, Isoherranen N, Brabb T, Treuting PM, Amory JK. Inhibition of retinoic acid biosynthesis by the bisdichloroacetyldiamine WIN 18,446 markedly suppresses spermatogenesis and alters retinoid metabolism in mice. J Biol Chem 2014; 289:15104–15117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Amory JK, Muller CH, Shimshoni JA, Isoherranen N, Paik J, Moreb JS, Amory DW Sr, Evanoff R, Goldstein AS, Griswold MD. Suppression of spermatogenesis by bisdichloroacetyldiamines is mediated by inhibition of testicular retinoic acid biosynthesis. J Androl 2011; 32:111–119. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.