Abstract
The self-renewal, proliferation, and differentiation of the spermatogonial populations must be finely coordinated in the mammalian testis, as dysregulation of these processes can lead to subfertility, infertility, or the formation of tumors. There are wide gaps in our understanding of how these spermatogonial populations are formed and maintained, and our laboratory has focused on identifying the molecular and cellular pathways that direct their development. Others and we have shown, using a combination of pharmacologic inhibitors and genetic models, that activation of mTOR complex 1 (mTORC1) is important for spermatogonial differentiation in vivo. Here, we extend those studies to directly test the germ cell-autonomous requirement for mTORC1 in spermatogonial differentiation. We created germ cell conditional knockout mice for “regulatory associated protein of MTOR, complex 1” (Rptor), which encodes an essential component of mTORC1. While germ cell KO mice were viable and healthy, they had smaller testes than littermate controls, and no sperm were present in their cauda epididymides. We found that an initial cohort of Rptor KO spermatogonia proliferated, differentiated, and entered meiosis (which they were unable to complete). However, no self-renewing spermatogonia were formed, and thus the entire germline was lost by adulthood, resulting in Sertoli cell-only testes. These results reveal the cell autonomous requirement for RPTOR in the formation or maintenance of the foundational self-renewing spermatogonial stem cell pool in the mouse testis and underscore complex roles for mTORC1 and its constituent proteins in male germ cell development.
Keywords: spermatogenesis, testis, fertility, Mtor, mTORC1, Rptor
SSC maintenance requires RPTOR
Introduction
Steady-state spermatogenesis in adult mammals depends on the capacity of spermatogonial stem cells (SSCs) to both maintain their population through self-renewing divisions and continuously produce transit-amplifying progenitor spermatogonia, which initiate differentiation in response to retinoic acid (RA). Throughout the male reproductive lifespan, ratios of the three subtypes of spermatogonia (stem, progenitor, and differentiating) are maintained in a consistent balance. Perturbations in this balance can lead to testicular cancer or infertility, the latter of which is often manifest in humans as nonobstructive azoospermia (NOA). Currently, NOA is diagnosed in approximately 8–12% of male infertility cases (reviewed in [1]), although little is known about the underlying causes. What is clear is that gamete production is lost if SSCs either fail to self-renew (resulting in loss of the germline and Sertoli cell-only [SCO] syndrome) or to proliferate and differentiate (resulting in maturation arrest).
In mice, the foundational pool of SSCs forms in the neonatal testis at the initiation of spermatogenesis as quiescent precursor prospermatogonia transition to spermatogonia [2–4]. Functional evidence for this comes from homologous transplantation experiments demonstrating that spermatogonia from mice as young as postnatal day (P)3 can colonize older germ cell-depleted recipient testes to produce donor-derived sperm [5]. Undifferentiated progenitor and differentiating spermatogonial populations arise concurrently with the SSC population. The initial cohort of progenitor and differentiating spermatogonia give rise to the first round, or wave, of spermatogenesis by entering meiosis as preleptotene spermatocytes on P8, completing meiosis as haploid spermatids on P21, and becoming the first functional sperm by approximately P35–50 (reviewed in [6, 7]). This first wave of spermatogenesis is not unique to mice; indeed, all mammals must initiate spermatogenesis from a precursor prospermatogonial pool, although there are significant differences in the temporal dynamics between species [7].
One primary goal of our laboratory is to define the molecular pathways that direct spermatogonial differentiation in response to RA. We previously reported that gene products KIT, SOHLH1, and SOHLH2, each of which are required for differentiation, were upregulated in vivo by RA at the level of polyribosome occupancy, resulting in an increase in protein levels without a change in mRNA abundance [8–10]. This RA-stimulated translation required activation of the “mechanistic, or mammalian target of rapamycin” (MTOR), which is a serine/threonine protein kinase capable of integrating a variety of intracellular and extracellular stimuli to regulate a diverse set of cellular processes such as growth, proliferation, differentiation, autophagy, metabolism, protein synthesis, and cytoskeletal regulation [11–16]. MTOR acts through two distinct protein complexes (mTORC1 and mTORC2); one major role of mTORC1 is stimulation of cap-dependent protein synthesis by phosphorylating substrates including “eukaryotic translation initiation factor 4E binding protein 1” (EIF4EBP1) and “ribosomal protein S6 kinase polypeptides 1 and 2” (RPS6KB1/2) [13, 16].
Several recent studies from our laboratory and others have highlighted mTORC1 as a critical mediator of spermatogonial development. In two recent studies, germ cell conditional knockouts (KO) were created for Tsc1 and Tsc2 [17, 18], which are known upstream negative regulators of mTORC1 and mTORC2 [19–23]. Deletion of Tsc1 and Tsc2 in spermatogenic cells resulted in MTOR hyperactivation, increased spermatogonial differentiation, and partial depletion of the germline [17, 18]. Our laboratory reported that global inhibition of mTORC1 by rapamycin blocked spermatogonial differentiation, preleptotene spermatocyte formation, and the RA-induced translation of KIT, SOHLH1, and SOHLH2 in neonatal mice [24]. Further, our laboratory recently generated male germ cell Mtor KO mice [25], and found that testes of all ages contained only singly isolated undifferentiated spermatogonia, revealing a critical role for MTOR in spermatogonial differentiation and fertility. Additionally, we observed that a small population of undifferentiated spermatogonia survived even in aged Mtor KO mice. This reveals that MTOR is dispensable for the genesis and survival of SSCs, but is required for the proliferation of undifferentiated progenitor spermatogonia [25]. The similar spermatogenic phenotype of Mtor KO and rapamycin-treated mice implies that mTORC1, rather than mTORC2, is the major regulator of spermatogonial proliferation and differentiation.
Here, we further test the role of mTORC1 in mouse male germ cell development by examining mice with a germ cell deletion of “regulatory associated protein of MTOR, complex 1” (Rptor), which encodes an essential scaffolding component of mTORC1 [26, 27]. To our surprise, we found that the reproductive phenotype of Rptor KO mice was distinct from those of either rapamycin-treated or Mtor KO mice [25, 28]. A robust population of undifferentiated and differentiating spermatogonia formed during the first wave of spermatogenesis in neonatal testes of Rptor KO mice; these cells entered, but were unable to successfully complete meiosis, leading to infertility due to an absence of epididymal spermatozoa. However, the spermatogonia population was quickly exhausted in the juvenile testis, revealing that RPTOR is dispensable for spermatogonial proliferation and differentiation. This is the first example, to the best of our knowledge, of a protein that is absolutely required for formation or maintenance of the foundational SSC pool in the mouse testis, and clearly supports previous reports suggesting that the first wave of spermatogenesis is an SSC-independent event.
Materials and Methods
Generation and care of experimental animals
All animal procedures were carried out in adherence with the guidelines of the National Research Council Guide for the Care and Use of Laboratory Animals and using protocols approved by the Animal Care and Use Committee of East Carolina University (AUP #A194). Rptor male germ cell KO mice were created by crossing female mice homozygous for a floxed Rptor allele (#013188, The Jackson Laboratory) with young (<P60) male mice carrying one floxed allele as well as the Ddx4-Cre transgene (#006954, The Jackson Laboratory). The resulting WT and KO pups were on a mixed background (C57Bl/6/129S4/SvJae/FVB). Transgenic mice harboring floxed Rptor alleles and/or Cre recombinase were identified by PCR-based genotyping (Primers: Rptor Forward 5′-CTCAGTAGTGGTATGTGCTCAG, Rptor Reverse- 5′-GGGTACAGTATGTCAGCACAG, Cre Forward 5′-CTAAACATGCTTCATCGTCGGTCC, and Cre Reverse 5′-GGATTAACATTCTCCCACCGTCAG). In all experiments, age-matched littermates were used for comparison with PCR-verified germ cell KO animals. Littermates heterozygous for the floxed Rptor allele with or without the Ddx4-Cre allele and Cre-negative mice homozygous for the floxed Rptor allele were considered WT and analyzed together. The following numbers of mice were analyzed at each of these ages: P8 = 5 WT and 2 KO, P18 = 4 WT and 2 KO, P33 = 1 WT and 1 KO, P > 60 = 4 WT and 4 KO. Amplified products were resolved on 3% agarose gels.
Cell quantitation from frozen testis sections
Immunostaining was performed to mark the entire germ cell population (DDX4 = cytoplasmic pan germ cell marker in the neonatal testis [29, 30]; TRA98 = nuclear pan germ cell marker at all developmental stages [30, 31]). All Sertoli cells were labeled using anti-GATA4 [32]. Specific antibody information is provided in Table 1. Testes from two Rptor germ cell KO mice at each age were used for quantitation. Quantitation of germ cells expressing various fate markers was carried out as previously described [9, 25]. Labeled cells were deemed positive or negative for a specific marker using the threshold tool in Image J (U.S. National Institutes of Health) with the program's default algorithm. Thresholds used were as follows: DDX4 = 100–255, KIT = 40–255, GFRA1 = 90–255, STRA8 = 70–255, SYCP3 = 39–255, CDH1 = 47–255, p-AKT 30–255 TRA98 = 25–255. The numbers of cells positive for each marker were divided by the total numbers of germ cells labeled by DDX4 or TRA98, and the percentage was obtained by multiplying by 100. To quantitate the proportion of the germ cell population labeled by markers of spermatogonial fate, cells were counted across two to three discrete microscope fields per animal represented a systematically selected area containing at least 1000 germ cells per mouse and protein marker of interest. The total numbers of DDX4 + germ cells and GATA4+ Sertoli cells were reported per testis cord or tubule cross section. DDX4+ and GATA4+ cells were counted from > 20 cross sections across distinct microscopic fields. Apoptotic germ cells were identified by staining for cleaved PARP1 and a pan germ cell marker (DDX4 or TRA98) and quantitated across two discrete microscope fields (at ×200) per animal. Images for quantitation were acquired with a Fluoview FV1000 laser scanning confocal microscope (Olympus America).
Table 1.
Antibodies.
| Protein | Vendor (Catalog Number) | Dilution |
|---|---|---|
| STRA8 | Abcam (ab49602) | 1:3000 |
| c-PARP1 | Cell Signaling Technology (94 885) | 1:100 |
| DDX4 | R&D Systems (AF2030), Abcam (ab13840) | 1:800, 1:1000 |
| GATA4 | Santa Cruz (sc-1237) | 1:100 |
| GFRA1 | R&D Systems (AF560) | 1:800 |
| KIT | Cell Signaling Technology (#3074) | 1:1000 |
| CRE | Cell Signaling Technology (#15 036) | 1:100 |
| p-RPS6 | Cell Signaling Technology (#5364P) | 1:800 |
| TRA98 | Abcam (ab82527) | 1:1000 |
| H1T | Gift from Mary Ann Handel [33] | 1:500 |
| MKI67 | Abcam (ab15580) | 1:400 |
| SYCP3 | Proteintech (#23024–1-AP) | 1:250 |
| FOXO1 | Cell Signaling Technology (#2880) | 1:100 |
| p-AKT | Cell Signaling Technology (#4060) | 1:200 |
| CDH1 | Cell Signaling Technology (#3195) | 1:200 |
Cauda epididymal sperm counts
Cauda epididymides were dissected from adult animals immediately following euthanasia. Each epididymis was placed in 1 ml of room temperature 1X PBS, minced into small pieces, and repeatedly mixed by pipetting. Samples were diluted 1:10 with distilled water to immobilize sperm, and duplicate counts were obtained per epididymis using a hemocytometer.
Histological analysis and indirect immunofluorescence
Histologic analyses were carried out on sections of paraffin-embedded Bouin-fixed testes. Nuclear and cytoplasmic structures were visualized following hematoxylin and eosin staining. Images were captured using an Axio Observer A1 microscope (Carl Zeiss Microscopy) equipped with an XL16C digital camera and Exponent version 1.3 software (Dage-MTI). To prepare frozen testis sections, testes were immersion-fixed in fresh 4% paraformaldehyde, washed in 1X PBS, incubated overnight in 30% sucrose at 4°C, and frozen in O.C.T. Five micrometer sections were incubated in blocking reagent (1X PBS containing 3% BSA (Fisher Scientific) + 0.1% Triton X-100) for 30 min at room temperature. Primary antibodies were diluted with blocking reagent and incubated on tissue sections for 1 h at room temperature. The primary antibodies used are listed in Table 1. Primary antibody was omitted from one section in each procedural replicate to serve as a negative control. Following three stringency washes in 1X PBS + 0.1% Triton X-100, sections were incubated in blocking reagent containing secondary antibody (Alexa Fluor-405, -488, -555, or -559 at a 1:1000 dilution, Invitrogen) and/or phalloidin-635 (1:500, Invitrogen) for 1 h at room temperature. Stringency washes were performed as described above. Coverslips were mounted using Vectastain containing DAPI (Vector Laboratories), except in cases when acid treatment was performed or the Alexa Fluor-405 secondary antibody was utilized. Images of stained tissues were obtained using a Fluoview FV1000 laser scanning confocal microscope (Olympus America).
Statistics
Statistical differences between experimental groups were evaluated using the Student t-test, and the level of significance was set at P < 0.05. Error bars indicate one standard deviation. Mendelian birth ratios were evaluated by Chi-square analysis.
Results
Germ cell development is disrupted in Rptor KO testes
Global deletion of Rptor caused embryonic lethality shortly after implantation [34]. To assess its role in spermatogenesis and formation of spermatogonia, we created male germline KO mice by crossing female mice homozygous for a floxed Rptor allele with male mice heterozygous for a floxed Rptor allele and the Ddx4-Cre transgene (Figure 1A [35]). Obtaining sufficient numbers of Rptor germ cell KO mice was challenging; from 16 breeding pairs the numbers of KO mice were consistently underrepresented within litters, resulting in a non-Mendelian birth ratio (Chi-square, P < 0.005). Only 13 male Rptorfl/fl; Ddx4-Cre (hereafter designated KO) mice were born out of 285 pups. Breeding pairs that successfully generated germ cell KO pups only produced an average of 4.6 pups per litter. This reduced litter size, combined with an absence of observed stillborn pups, led us to suspect that Cre expression outside of the germline was causing the embryonic lethality characteristic of Rptor conventional (whole-body) KO mice [34].
Figure 1.
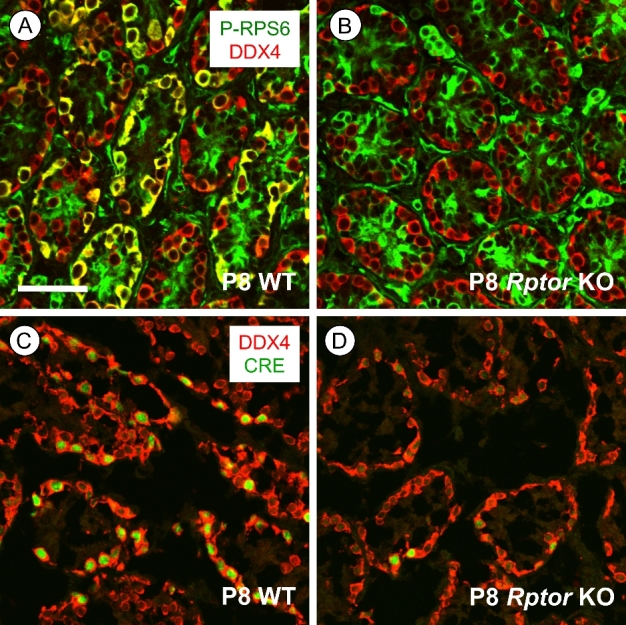
mTORC1 activity is lost specifically in germ cells of Rptor KO mice. (A, B) Immunostaining for p-RPS6 (green) is present in P8 WT DDX4+ spermatogonia (in red), but not in Rptor KO spermatogonia; areas of co-labeling are yellow. (C, D) Representative P8 WT and Rptorfl/+; Ddx4-Cre testis sections stained for Cre recombinase (green) along with DDX4 (red). Scale bar (in B) = 50 μm.
We verified the loss of RPTOR protein in KO testes by co-immunostaining at P8 for phospho(p)-RPS6, a well-characterized downstream substrate of mTORC1 and indicator of its activity [13, 14, 16]. In contrast to WT, where it is detectable in both differentiating and undifferentiated spermatogonia (data not shown), p-RPS6 was undetectable in male germ cells from Rptor KO mice (Figure 1B). This absence of p-RPS6 detectability indicated that Ddx4-Cre mediated Rptor deletion occurred specifically in germ cells with high efficiency, as we and others have reported previously for other floxed genes [25, 35, 36]. Immunofluorescent staining for Cre recombinase also verified its expression was detectable exclusively in germ cells in P8 Rptor heterozygous (Rptorfl/+; Ddx4-Cre) and germ cell KO mice (Figure 1C and D). Together, absence of CRE protein in testicular somatic cells as well as consistent absence of p-RPS6 in Rptor KO germ cells indicated that Ddx4-Cre mediated Rptor deletion was occurring solely in male germ cells.
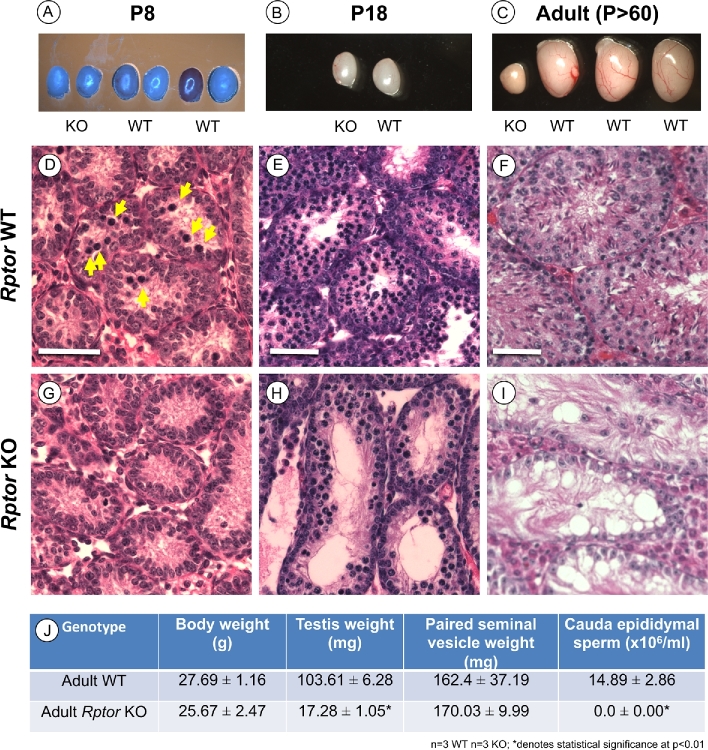
Rptor male germ cell KO mice appeared healthy and were indistinguishable from their WT littermates. At P8 and P18, there were no significant differences in testis sizes or weights (Figure 2A, B, and J). However, this changed as the mice aged; adult KO testes were dramatically smaller than those of WT littermates (Figure 2C and J, P < 0.01). We next examined the histology of P8, P18, and P > 60 adult testes. As expected, P8 WT mouse testes contained abundant spermatogonia as well as germ cells with condensed nuclei characteristic of the first preleptotene spermatocytes (Figure 2D). P18 and adult WT animals also showed normal histology; spermatocytes were abundant as the most advanced germ cell type at P18, and all germ cell types were present in adult testes (Figure 2E and F). In contrast, P8 Rptor KO testes appeared to lack preleptotene spermatocytes (Figure 2G). Surprisingly, numerous spermatocytes were present by P18, although total germ cell numbers were noticeably decreased (Figure 2H). Adult KO testes had an apparent SCO phenotype, with no obvious remaining germ cells (Figure 2I). Together, these observations revealed that absence of RPTOR in the germline delayed the appearance of spermatocytes at P8, and this was followed by a complete loss of the germline by early adulthood. Adult Rptor KO animals were clearly infertile, as there were no detectable cauda epididymal sperm (Figure 2J).
Figure 2.
Male germ cell deletion of Rptor causes a progressive decline in spermatogenesis leading to infertility. (A–C) Representative images of unfixed whole testes from Rptor KO and WT littermates aged P8, P18, and adult P > 60 were acquired immediately post dissection. Note: there was a lighting issue on the day P8 testes were dissected (see A); actual colors were the same as in B nd C. (D–I) Hematoxylin and eosin-stained representative sections from Rptor WT (D–F) and KO (G–I) testes from P8, P18, and P > 60 animals. Yellow arrows in “D” point to condensed nuclei of preleptotene spermatocytes. (J) Average body weight, testis weight, seminal vesicle weight, and cauda epididymal sperm counts from adult (P > 60) Rptor WT and KO animals. Mean values are shown with standard deviation, and asterisks indicate statistical significance at P < 0.01. Scale bars = 50 μm.
Fewer undifferentiated spermatogonia reside in P8 Rptor KO testes
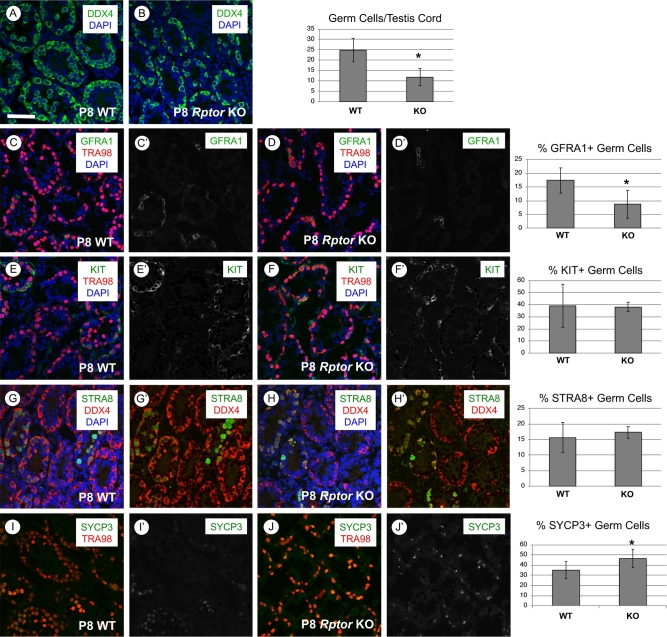
We next sought to investigate the ontogeny of the above-described defects during the first wave of spermatogenesis. Immunostaining for DDX4, a pan germ cell marker in the neonatal testis [29], revealed a statistically significant twofold reduction in the number of germ cells per testis cord in P8 Rptor KO mice (Figure 3A and B, P = 0.047). This reduction in germ cell number was observed without a significant increase in the number of apoptotic c-PARP1 + spermatogonia (Supplementary Figure S1A and B). Immunostaining for MKI67, a marker of proliferating cells, revealed that loss of RPTOR did not change the proportion of germ cells actively participating in the cell cycle (Supplementary Figure S1C and D). In addition, numbers of GATA4+ Sertoli cells per testis cord were not statistically different between Rptor KO and WT testes (Supplementary Figure S1E and F).
Figure 3.
P8 Rptor KO testes have reduced germ cell numbers but normal percentages of undifferentiated and differentiating spermatogonia. In each row, representative images of immunostaining for specific proteins and accompanying quantitation are shown for P8 WT and KO testes. (A, B) The entire germ cell population is DDX4+ (green), and nuclei are stained with DAPI (blue). (C, D) Undifferentiated stem and progenitor spermatogonia are GFRA1+ (green), all germ cells DDX4+ (red), and nuclei are stained with DAPI (blue). (E, F) Differentiating spermatogonia are KIT+ (green), all germ cells are TRA98+ (red), and nuclei are stained with DAPI (blue). (G, H) Germ cells responding to retinoic acid are STRA8+ (green), all germ cells are DDX4+ (red), and nuclei are stained with DAPI (blue). (I, J) Germ cells staining positive for the meiotic marker SYCP3 (green), all germ cells are TRA98+ (red). Error bars indicate one standard deviation, and asterisks denote statistical significance at P < 0.05. Scale bar (in A) = 50 μm.
We next assessed the ratios of undifferentiated and differentiating spermatogonial subpopulations by immunostaining for markers of spermatogonial fate [37–50]. There was a significant reduction in the numbers of GFRA1+ and CDH1+ undifferentiated spermatogonia in Rptor KO testes (Figure 3C and D, Supplementary Figure S1G and H). Although the GFRA1+ germ cell population was reduced in Rptor KO testes, the proliferation marker MKI67 was detectable in an increased proportion of these cells (74% in KO vs 53% in WT, P = 0.0001). This reveals that the reduction in GFRA1+ cells was not due to increased quiescence among this specific spermatogonial population (Supplementary Figure S4A and B). In addition, spermatogonia in developing Rptor KO testes had increased levels of cytoplasmic FOXO1 (Supplementary Figure S2A–E), which correlates with the differentiating state [36, 51]. FOXO1 localization was shown to be regulated by AKT kinase activity downstream of mTORC1 phosphorylation [34, 52]. Consistent with this pathway model, we found that the percentage of germ cells staining positive for AKT phosphorylation at the mTORC2-specific residue (S473) was increased from 16 to 28% (P = 0.001) in Rptor KO germ cells (Supplementary Figure S3A and B), suggesting an increase in mTORC2 and thus AKT activity in the absence of RPTOR.
In the developing testis at P8, there were similar numbers of KIT+ differentiating spermatogonia in WT and Rptor KO mice (Figure 3E and F). We found that “stimulated by retinoic acid gene 8” (STRA8), a marker of the RA response in differentiating spermatogonia in the developing testis and preleptotene spermatocytes [9, 53, 54], was also detectable in similar proportions of germ cells in P8 Rptor WT and KO testes (Figure 3G and H). These results together suggest that RA responsiveness was unaffected in Rptor KO spermatogonia. SYCP3, which is expressed in differentiating spermatogonia in the developing testis as well as in early meiotic cells, was detectable in a higher percentage of germ cells in P8 KO than WT testes (46.6% vs 35.1%, P = 0.0429; Figure 2I and J). Taken together, we conclude that the reduction in the germ cell population in P8 Rptor KO testes was due to a combination of reduced numbers of undifferentiated spermatogonia along with the aforementioned absence of preleptotene spermatocytes (Figure 2G). However, the presence of normal numbers of STRA8+, KIT+, and SYCP3+ spermatogonia implies that although preleptotene spermatocytes do not appear abundant in P8 KO testes, germ cells lacking RPTOR are differentiating and progressing toward meiosis in the developing testis.
First-wave Rptor KO germ cells initiate, but fail to complete meiosis
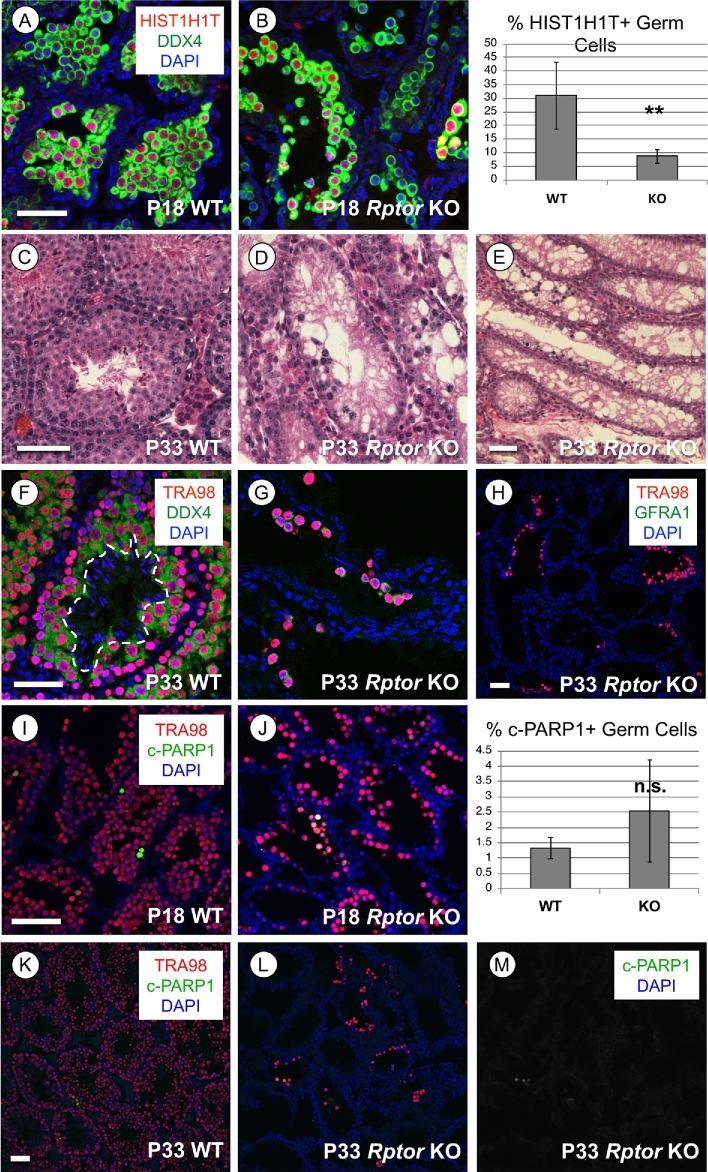
We next analyzed testes from WT and Rptor KO mice at P18, P33, and P > 60 to assess the progression of spermatogenesis defects due to the absence of RPTOR. Histological analyses suggested that P18 Rptor KO seminiferous epithelia appeared to contain few spermatogonia, but numerous spermatocytes adjacent to the nascent lumina of the seminiferous tubules (Figure 2H). We verified that these were indeed HIST1H1T+ (also termed H1T; [33]) spermatocytes, although there were significantly fewer in Rptor KO vs WT testes (8% vs 31%, P = 0.0014; Figure 4A and B). At P33, Rptor KO testes contained sparse small clusters of spermatocytes, but no round or elongating spermatids were ever observed (Figure 4C–G). No GFRA1+ undifferentiated spermatogonia were detectable among the P33 KO germ cell population (Figure 4H). Clusters of c-PARP1+ apoptotic cells were detectable throughout P18 and P33 Rptor KO testes, although their numbers were not statistically different between P18 WT and KO (Figure 4I–M). Together, these results reveal that Rptor KO germ cells initiated but failed to complete meiosis during the first wave of spermatogenesis, and the loss of spermatogonia caused germline extinction by adulthood.
Figure 4.
Spermatocytes in Rptor KO testes fail to complete meiosis. In each row, representative images of immunofluorescence for specific proteins or hematoxylin & eosin staining are shown in WT and KO testis sections from mice of indicated ages. (A, B) Spermatocytes are identified by positive signal for the spermatocyte-specific histone H1ST1H1T (red), all germ cells are DDX4+ (green), and nuclei are stained with DAPI (blue). (C–E) Hematoxylin & eosin staining of paraffin-embedded tissue sections from P33 mice. (F, G) Germ cell nuclei are TRA98+ (red) and cytoplasm of germ cells is DDX4+ (green). Dashed line surrounds the condensed nuclei of elongating spermatids (in E). (H) Broad field image demonstrating that GFRA1+ undifferentiated spermatogonia (green) are absent among the remaining KO germline. Germ cell nuclei are TRA98+ (red), and all nuclei are stained with DAPI (blue). (I–M) Cells undergoing apoptosis are cleaved(c)-PARP1 + (green), and germ cell nuclei are stained TRA98 + (red) at P18 (I–J) and P33 (K–M). Asterisks indicate statistical significance P < 0.01. Scale bars (in A and C) = 50 μm.
The entire spermatogonial population is lost during the first wave of spermatogenesis
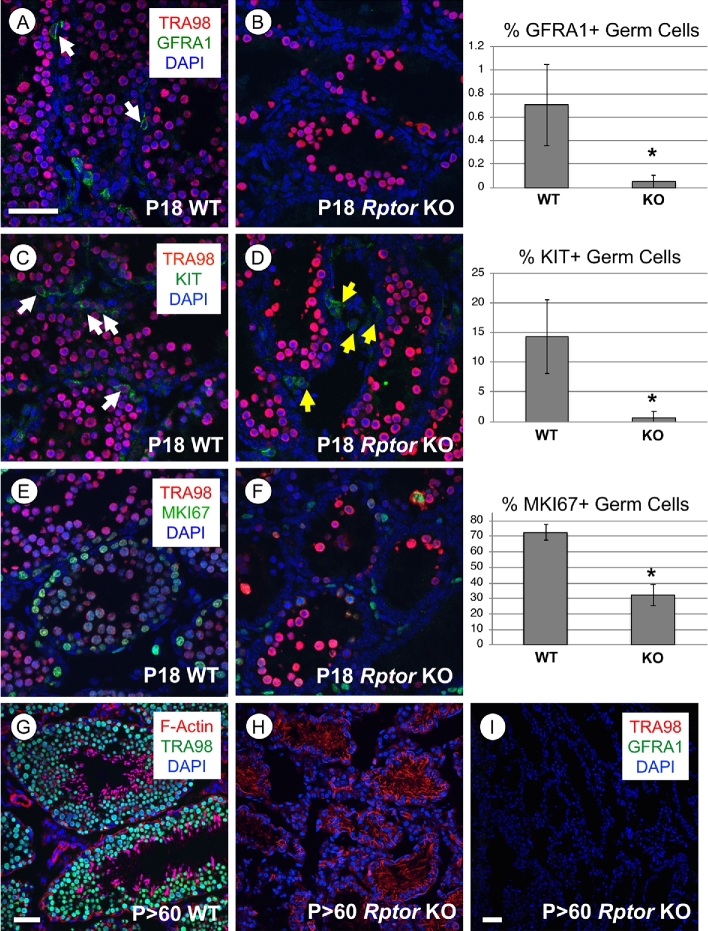
We found that numbers of GFRA1+ undifferentiated spermatogonia in P18 Rptor KO testes were dramatically reduced in comparison to their WT littermates (P = 0.007, Figure 5A and B). The percentage of KIT+ differentiating spermatogonia was also significantly reduced in Rptor KO testes (14% in WT vs. 0.5% in the KO population, Figure 5C and D, P = 0.0006). Taken together, these results revealed few spermatogonia remained in P18 Rptor KO as compared to WT testes. Additionally, the proportion of germ cells containing detectable MKI67 was reduced in KO testes, supporting the observation of decreased numbers of proliferating spermatogonia (Figure 5E and F). By P > 60, Rptor KO testes exhibited an apparent SCO phenotype, with numerous vacuoles present between Sertoli cell cytoplasmic projections (Figure 2I). We verified this SCO phenotype by immunostaining for the pan germ cell marker TRA98 and GFRA1, a marker of undifferentiated spermatogonia and SSCs (Figure 5G–I).
Figure 5.
Spermatogonia are lost during the first wave of spermatogenesis in Rptor KO testes, and the germline is lost by adulthood. In the top three rows, representative immunostaining images are shown for specified proteins, and the accompanying quantitation is shown. White arrows indicate germ cells labeled with the indicated protein marker. (A, B) Undifferentiated progenitor spermatogonia are GFRA1+ (green), all germ cells are TRA98+ (red), and nuclei are stained with DAPI (blue). (C, D) Differentiating spermatogonia are KIT+ (green), all germ cells are TRA98+ (red), and nuclei are stained with DAPI (blue). Yellow arrows (in D) indicate TRA98-/KIT+ interstitial cells. (E, F) Proliferative cells are MKI67+ (green), all germ cells are TRA98+ (red), and nuclei are stained with DAPI (blue). (G, H) Nuclei of all germ cells are labeled by TRA98 (green) and F-Actin is labeled by phalloidin (red). (I) Broad field image of an adult Rptor KO testis section devoid of TRA98+ germ cells (red) and GFRA1+ undifferentiated spermatogonia (green). All nuclei are labeled with DAPI (blue). Error bars indicate one standard deviation. Asterisks denote statistical significance at P < 0.05. Scale bar (in A, G–I) = 50 μm.
Discussion
Here, we generated male germline-specific Rptor KO mice and reported a germ cell autonomous requirement for RPTOR in male fertility. The loss of RPTOR caused distinct defects: (1) failure to complete meiosis and form haploid round spermatids and (2) failure to maintain a functional SSC pool during the first wave of spermatogenesis. Because of the meiotic defect, Rptor KO males failed to produce any gametes during the first wave of spermatogenesis, and because of the SSC defect the entire germline was lost by early adulthood.
The reproductive defects in Rptor KO mice were initially observed during the first wave of spermatogenesis. Although spermatogonia proliferated, differentiated, and entered meiosis as primary spermatocytes, they were unable to complete meiosis and form haploid spermatids. These observations support those of another group, whose manuscript was published while we were completing this study. They utilized Neurog3-Cre (expressed in a subset of undifferentiated spermatogonia by P7 [55, 56]) to conditionally delete Rptor in the germline. They reported that Rptorfl/−; Neurog3-Cre mice were infertile due to failure to progress through pachynema [57]. Therefore, we shifted our focus to assess the role of RPTOR in spermatogonial development. We utilized Ddx4-Cre, which efficiently targets floxed alleles in prospermatogonia of the fetal testis as early as E15 [35]. This early deletion provides adequate time for the targeted deletion to occur and for mRNAs and their encoded proteins to disappear before the formation of postnatal spermatogonia, thus allowing for a clear assessment of the resulting reproductive phenotype.
The observation that Rptor KO spermatogonia differentiated and entered meiosis was surprising, given previous reports characterizing mTORC1 as a key mediator of spermatogonial proliferation and differentiation [8, 15, 16, 22]. We previously reported that RA-induced translation of the differentiation-required proteins KIT, SOHLH1, and SOHLH2 required activation of the PI3K/AKT/mTORC1 pathway [6–8]. mTORC1 is known to stimulate the cap-dependent protein synthesis of 5′-TOP mRNAs through phosphorylation of its downstream targets EIF4EBP1 and RPS6KB1/2 [11, 12]. However, a recent study demonstrated that the insulin-stimulated synthesis of 5′TOP mRNAs required the MTOR kinase but was only moderately affected by Rptor or Rictor KO in cultured mouse embryonic fibroblasts [55]. This suggests that enhanced translation of suppressed mRNAs in spermatogonia does not require RPTOR; perhaps functional redundancy exists that allows sufficient KIT protein to be produced via RPTOR-independent mTORC1, or another uncharacterized parallel pathway.
We discovered that the foundational SSC pool was not maintained in testes of developing Rptor germ cell KO mice. This result was unexpected, as our previously published studies revealed that the undifferentiated spermatogonial population containing SSCs was maintained following global inhibition of mTORC1 with rapamycin or germline deletion of Mtor [8, 22]. Additionally, other groups reported that increased mTORC1 activity in germ cells drove excessive spermatogonial differentiation at the expense of the undifferentiated spermatogonial population, but did not exhaust the SSC population [15, 16]. We envision two potential explanations for this observation. In the first scenario, the loss of RPTOR would be predicted to create a detrimental imbalance between mTORC1 and mTORC2 in SSCs. Since multiple proteins are shared between mTORC1 and mTORC2, more mTORC2 complexes may assemble in the absence of RPTOR, resulting in increased mTORC2 activity. Additionally, some mTORC1 phosphorylation targets (GRB10 and S6K1) operate in negative feedback loops to inhibit PI3K/AKT pathway activation by insulin signaling, and thus inhibit mTORC2 activation [58–61]. One of the best-characterized roles for mTORC2 is phosphorylation, and subsequent full activation, of AKT [52]. AKT has been demonstrated to be a critical mediator of spermatogonial development downstream of both self-renewal signals such as GDNF (first shown in [69] and later confirmed in [70]) and the differentiation signal provided by RA [8, 28]. AKT phosphorylation by mTORC2 at S473 is necessary for its ability to phosphorylate the transcription factors FOXO1/3, resulting in their exclusion from the nucleus and preventing their transcriptional gene regulation [34, 62, 63]. Consistent with mTORC2 overactivation, we found that AKT phosphorylated at S473 was detectable in an increased percentage of Rptor KO germ cells, and FOXO1 was predominantly cytoplasmic in KO testes. Analyses of germ cell-specific KO lines demonstrated that FOXO1 mediates multiple aspects of germ cell development, including spermatogonial differentiation as well as SSC establishment and self-renewal [36]. The same study also reported that germ cell deletion of PTEN, an upstream negative regulator of the PI3K/AKT pathway, resulted in AKT hyperactivity and FOXO1 cytoplasmic localization in germ cells [36]. The phenotype of the PTEN KO mice resembled Rptor KO in that meiosis was incomplete, spermatogonia were lost during the first wave of spermatogenesis, and the germ line was severely depleted by P28, indicating loss of SSC function. Based on our results, it appears that absence of RPTOR skews the balance of mTOR signaling toward mTORC2 hyperactivity. The resultant dysregulation of AKT signaling may underlie the loss of SSC function observed in germ cell Rptor KO testes. In the second scenario, RPTOR may have an mTORC1-independent role critical for the function of the SSC pool. There is precedence for this from a recent study using mouse hepatocytes. In that study, abundant mTORC1-free RPTOR was discovered, and the ratio of free to mTORC1-bound RPTOR decreased with age and obesity. In addition, increasing the amount of free RPTOR reduced liver triglycerides and body weights in aged or obese mice [56]. These observations demonstrate that mTORC1-free RPTOR regulated cell behavior in both normal and pathologic conditions, indicating that RPTOR may not function exclusively as a scaffolding protein for mTORC1.
The results presented here support the concept that the first wave of mammalian spermatogenesis does not rely on SSCs, but rather that the initial populations of progenitor and differentiating spermatogonia arise directly from precursor prospermatogonia and subsequent waves are dependent on SSCs. There are three types of existing evidence in the literature using mice as a model organism. First, the characteristic morphological features of undifferentiated and differentiating spermatogonia such as nuclear size and the appearance of chromatin and nucleoli have been used to determine that spermatogonia resembling both subtypes appeared, nearly simultaneously as early as P3–4 [3, 64–66]. Second, an indirect lineage-tracing approach was employed using a tamoxifen-inducible “neurogenin 3” (Neurog3, also termed Ngn3) gene promoter sequence to direct Cre activation of a ß-galactosidase reporter gene [67]. Since Neurog3 mRNAs were present in undifferentiated spermatogonia (but not precursor prospermatogonia or later differentiating spermatogonia), the resultant ß-gal- gametes observed in young adult mice supported the concept that Neurog3- prospermatogonia became Neurog3- differentiating spermatogonia without transitioning through a Neurog3+ undifferentiated (e.g. SSC or progenitor) state [67]. Third, several germ cell KO mouse models displayed relatively normal first waves of spermatogenesis, followed by progressive loss of the germline during the aging process that was partial (Rb1/Rb [68, 69], Zbtb16/Plzf [70, 71], Taf4b [72]) or complete (Etv5/Erm [73, 74]). These KO models reveal distinct roles and differential requirements for these proteins in SSC function. Specifically, ETV5 is required for SSC survival, while proteins such as RB1, ZBTB16, and TAF4B are involved in, but not absolutely required for, SSC maintenance and/or self-renewal.
In conclusion, our data demonstrate the germ cell autonomous requirement for RPTOR, during the first wave of spermatogenesis, for completion of meiosis as well as for the formation of a functional, self-renewing population of SSCs. Our results here provide additional evidence supporting the concept that the first wave of spermatogenesis is SSC-independent.
Supplementary data
Supplementary Figure S1. Germ cell Rptor KO did not affect germ cell quiescence, apoptosis, or the number of Sertoli cells in P8 testes. In each row, representative images of immunofluorescence for specific proteins and accompanying quantitation are shown for P8 WT and KO testes. (A, B) Cells undergoing apoptosis are labeled with cleaved PARP1 (green), all germ cells are labeled with DDX4 (blue). (C, D) Cells which are not quiescent are labeled with the proliferation marker MKI67 (green), all germ cells are labeled with DDX4 (red), and nuclei are stained with DAPI (blue). (E, F) Sertoli cell nuclei are labeled by GATA4 (red) and nuclei are stained with DAPI. (G, H) Undifferentiated spermatogonia are labeled with CDH1 (green) and nuclei are stained with DAPI (blue). Error bars indicate one standard deviation. Asterisks denote statistical significance at P < 0.05. Scale bar = 50 μm.
Supplementary Figure S2. FOXO1 localization is predominantly cytoplasmic in Rptor KO germ cells. (A, B) Representative immunofluorescent images of P8 WT and Rptor KO testis sections stained for FOXO1 (green) and the pan germ cell marker TRA98 (red). Nuclei are labeled with DAPI (blue). Arrows indicate example cells containing nuclear FOXO1 staining. (C, D) The same images from “A” and “B” with the FOXO1 signal isolated. (E) Corresponding quantitation of the percent of germ cells with nuclear or cytoplasmic-localized FOXO1 staining in WT and KO animals. Asterisks denote statistical significance between WT and KO groups by pairwise comparison at P < 0.01. Scale bar = 50 μm.
Supplementary Figure S3. AKT phosphorylation by mTORC2 is detectable in an increased proportion of Rptor KO germ cells. (A, B) Representative immunofluorescent images of P8 WT and Rptor KO testis sections stained for p-AKT (S473) (green) isolated in (C, D), the pan germ cell marker DDX4 (red), and corresponding quantitation of the percent of germ cells labeled. Arrows indicate example germ cells containing detectable p-AKT. Asterisks denote statistical significance at P < 0.01. Scale bar = 50 μm.
Supplementary Figure S4. The proliferation marker MKI67 is detectable in an increased percentage of GFRA1+ undifferentiated spermatogonia in Rptor KO testes. (A, B) Representative immunofluorescent images of P8 WT and Rptor KO testis sections stained for GFRA1+ undifferentiated spermatogonia (green) and the marker of cells in active phases of the cell cycle MKI67 (red). Corresponding quantitation of the percent of GFRA1+ cells with detectable MKI67 is shown. Arrows indicate example GFRA1+ MKI67- cells. Asterisks denote statistical significance at P < 0.01. Scale bar = 50 μm.
Acknowledgements
The authors thank Joani Zary-Oswald (ECU) for technical assistance.
Notes
Edited by Dr. Sarah Kimmins, PhD, McGill University
Footnotes
Grant support. This work was supported by grants from the NIH/NICHD (HD072552 and HD090083 to C.B.G.).
References
- 1. Gassei K, Orwig KE. Experimental methods to preserve male fertility and treat male factor infertility. Fertil Steril 2016; 105:256–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. de Rooij DG, Grootegoed JA. Spermatogonial stem cells. Curr Opin Cell Biol 1998; 10:694–701. [DOI] [PubMed] [Google Scholar]
- 3. Drumond AL, Meistrich ML, Chiarini-Garcia H. Spermatogonial morphology and kinetics during testis development in mice: a high-resolution light microscopy approach. Reproduction 2011; 142:145–155. [DOI] [PubMed] [Google Scholar]
- 4. Nagano R, Tabata S, Nakanishi Y, Ohsako S, Kurohmaru M, Hayashi Y. Reproliferation and relocation of mouse male germ cells (gonocytes) during prespermatogenesis. Anat Rec 2000; 258:210–220. [DOI] [PubMed] [Google Scholar]
- 5. McLean DJ, Friel PJ, Johnston DS, Griswold MD. Characterization of spermatogonial stem cell maturation and differentiation in neonatal mice. Biol Reprod 2003; 69:2085–2091. [DOI] [PubMed] [Google Scholar]
- 6. Busada JT, Geyer CB. The role of retinoic acid (RA) in spermatogonial differentiation. Biol Reprod 2016; 94:10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Geyer CB. Setting the stage: the first round of spermatogenesis. In: Oatley JM, Griswold MD (eds.) The Biology of Mammalian Spermatogonia. New York: Springer; 2017: 39–63. [Google Scholar]
- 8. Busada JT, Chappell VA, Niedenberger BA, Kaye EP, Keiper BD, Hogarth CA, Geyer CB. Retinoic acid regulates Kit translation during spermatogonial differentiation in the mouse. Dev Biol 2015; 397:140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Busada JT, Kaye EP, Renegar RH, Geyer CB. Retinoic acid induces multiple hallmarks of the prospermatogonia-to-spermatogonia transition in the neonatal mouse. Biol Reprod 2014; 90:64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chappell VA, Busada JT, Keiper BD, Geyer CB. Translational activation of developmental messenger rnas during neonatal mouse testis development. Biol Reprod 2013; 89:61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Eyre TA, Collins GP, Goldstone AH, Cwynarski K. Time now to TORC the TORC? New developments in mTOR pathway inhibition in lymphoid malignancies. Br J Haematol 2014; 166:336–351. [DOI] [PubMed] [Google Scholar]
- 12. Hay N, Sonenberg N. Upstream and downstream of mTOR. Genes Dev 2004; 18:1926–1945. [DOI] [PubMed] [Google Scholar]
- 13. Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell 2012; 149:274–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ma XM, Blenis J. Molecular mechanisms of mTOR-mediated translational control. Nat Rev Mol Cell Biol 2009; 10:307–318. [DOI] [PubMed] [Google Scholar]
- 15. Shimobayashi M, Hall MN. Making new contacts: the mTOR network in metabolism and signalling crosstalk. Nat Rev Mol Cell Biol 2014; 15:155–162. [DOI] [PubMed] [Google Scholar]
- 16. Thoreen CC, Chantranupong L, Keys HR, Wang T, Gray NS, Sabatini DM. A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 2012; 485:109–113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Hobbs RM, La HM, Makela JA, Kobayashi T, Noda T, Pandolfi PP. Distinct germline progenitor subsets defined through Tsc2-mTORC1 signaling. EMBO Rep 2015; 16:467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang C, Wang Z, Xiong Z, Dai H, Zou Z, Jia C, Bai X, Chen Z. mTORC1 activation promotes spermatogonial differentiation and causes subfertility in mice. Biol Reprod 2016; 95:97–97. [DOI] [PubMed] [Google Scholar]
- 19. Huang J, Manning BD. The TSC1–TSC2 complex: a molecular switchboard controlling cell growth. Biochem J 2008; 412:179–190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Huang J, Manning BD. A complex interplay between Akt, TSC2 and the two mTOR complexes: Figure 1. Biochm Soc Trans 2009; 37:217–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Mieulet V, Lamb RF. Tuberous sclerosis complex: linking cancer to metabolism. Trends Mol. Med. 2010; 16:329–335. [DOI] [PubMed] [Google Scholar]
- 22. Orlova KA, Crino PB. The tuberous sclerosis complex. Ann N Y Acad Sci 2010; 1184:87–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tomasoni R, Mondino A. The tuberous sclerosis complex: balancing proliferation and survival. Biochm Soc Trans 2011; 39:466–471. [DOI] [PubMed] [Google Scholar]
- 24. Busada JT, Niedenberger BA, Velte EK, Keiper BD, Geyer CB. Mammalian target of rapamycin complex 1 (mTORC1) Is required for mouse spermatogonial differentiation in vivo. Dev Biol 2015; 407:90–102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Serra ND, Velte EK, Niedenberger BA, Kirsanov O, Geyer CB. Cell-autonomous requirement for mammalian target of rapamycin (Mtor) in spermatogonial proliferation and differentiation in the mouse. Biol Reprod 2017; 96:816–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hara K, Maruki Y, Long X, Yoshino K, Oshiro N, Hidayat S, Tokunaga C, Avruch J, Yonezawa K. Raptor, a binding partner of target of rapamycin (TOR), mediates TOR action. Cell 2002; 110:177–189. [DOI] [PubMed] [Google Scholar]
- 27. Kim DH, Sarbassov DD, Ali SM, King JE, Latek RR, Erdjument-Bromage H, Tempst P, Sabatini DM. mTOR interacts with raptor to form a nutrient-sensitive complex that signals to the cell growth machinery. Cell 2002; 110:163–175. [DOI] [PubMed] [Google Scholar]
- 28. Busada JT, Niedenberger BA, Velte EK, Keiper BD, Geyer CB. Mammalian target of rapamycin complex 1 (mTORC1) Is required for mouse spermatogonial differentiation in vivo. Dev Biol 2015; 407:90–102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Niedenberger BA, Busada JT, Geyer CB. Marker expression reveals heterogeneity of spermatogonia in the neonatal mouse testis. Reproduction 2015; 149:329–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tanaka SS, Toyooka Y, Akasu R, Katoh-Fukui Y, Nakahara Y, Suzuki R, Yokoyama M, Noce T. The mouse homolog of Drosophila Vasa is required for the development of male germ cells. Genes Dev 2000; 14:841–853. [PMC free article] [PubMed] [Google Scholar]
- 31. Inoue N, Omohara Y, Yokota S. Expression of a testis-specific nuclear protein, tra98, in mouse testis during spermatogenesis. a quantitative and qualitative immunoelectron microscopy (iem) analysis. A quantitative and qualitative immunoelectron microscopy (TEM) analysis. OJCB 2011; 1:11–20. [Google Scholar]
- 32. Viger RS, Mertineit C, Trasler JM, Nemer M. Transcription factor GATA-4 is expressed in a sexually dimorphic pattern during mouse gonadal development and is a potent activator of the Mullerian inhibiting substance promoter. Development 1998; 125:2665–2675. [DOI] [PubMed] [Google Scholar]
- 33. Inselman A, Eaker S, Handel MA. Temporal expression of cell cycle-related proteins during spermatogenesis: establishing a timeline for onset of the meiotic divisions. Cytogenet Genome Res 2003; 103:277–284. [DOI] [PubMed] [Google Scholar]
- 34. Guertin DA, Stevens DM, Thoreen CC, Burds AA, Kalaany NY, Moffat J, Brown M, Fitzgerald KJ, Sabatini DM. Ablation in mice of the mTORC components raptor, rictor, or mLST8 reveals that mTORC2 is required for signaling to Akt-FOXO and PKCalpha, but not S6K1. Dev Cell 2006; 11:859–871. [DOI] [PubMed] [Google Scholar]
- 35. Gallardo T, Shirley L, John GB, Castrillon DH. Generation of a germ cell-specific mouse transgenic Cre line,Vasa-Cre. Genesis 2007; 45:413–417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Goertz MJ, Wu Z, Gallardo TD, Hamra FK, Castrillon DH. Foxo1 is required in mouse spermatogonial stem cells for their maintenance and the initiation of spermatogenesis. J Clin Invest 2011; 121:3456–3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Besmer P, Manova K, Duttlinger R, Huang EJ, Packer A, Gyssler C, Bachvarova RF. The kit-ligand (steel factor) and its receptor c-kit/W: pleiotropic roles in gametogenesis and melanogenesis. Dev Suppl 1993:125–137. [PubMed] [Google Scholar]
- 38. Grasso M, Fuso A, Dovere L, de Rooij DG, Stefanini M, Boitani C, Vicini E. Distribution of GFRA1-expressing spermatogonia in adult mouse testis. Reproduction 2012; 143:325–332. [DOI] [PubMed] [Google Scholar]
- 39. Hofmann MC, Braydich-Stolle L, Dym M. Isolation of male germ-line stem cells; influence of GDNF. Dev Biol 2005; 279:114–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Kissel H, Timokhina I, Hardy MP, Rothschild G, Tajima Y, Soares V, Angeles M, Whitlow SR, Manova K, Besmer P. Point mutation in kit receptor tyrosine kinase reveals essential roles for kit signaling in spermatogenesis and oogenesis without affecting other kit responses. EMBO J 2000; 19:1312–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kubota H, Avarbock MR, Brinster RL. Growth factors essential for self-renewal and expansion of mouse spermatogonial stem cells. Proc Natl Acad Sci USA 2004; 101:16489–16494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Meng X, Lindahl M, Hyvonen ME, Parvinen M, de Rooij DG, Hess MW, Raatikainen-Ahokas A, Sainio K, Rauvala H, Lakso M, Pichel JG, Westphal H et al. . Regulation of cell fate decision of undifferentiated spermatogonia by GDNF. Science 2000; 287:1489–1493. [DOI] [PubMed] [Google Scholar]
- 43. Mithraprabhu S, Loveland KL. Control of KIT signalling in male germ cells: what can we learn from other systems? Reproduction 2009; 138:743–757. [DOI] [PubMed] [Google Scholar]
- 44. Oatley JM, Avarbock MR, Telaranta AI, Fearon DT, Brinster RL. Identifying genes important for spermatogonial stem cell self-renewal and survival. Proc Natl Acad Sci USA 2006; 103:9524–9529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Oatley JM, Brinster RL. Spermatogonial stem cells. Methods Enzymol 2006; 419:259–282. [DOI] [PubMed] [Google Scholar]
- 46. Packer AI, Besmer P, Bachvarova RF. Kit ligand mediates survival of type A spermatogonia and dividing spermatocytes in postnatal mouse testes. Mol Reprod Dev 1995; 42:303–310. [DOI] [PubMed] [Google Scholar]
- 47. Prabhu SM, Meistrich ML, McLaughlin EA, Roman SD, Warne S, Mendis S, Itman C, Loveland KL. Expression of c-Kit receptor mRNA and protein in the developing, adult and irradiated rodent testis. Reproduction 2006; 131:489–499. [DOI] [PubMed] [Google Scholar]
- 48. Schrans-Stassen BH, van de Kant HJ, de Rooij DG, van Pelt AM. Differential expression of c- kit in mouse undifferentiated and differentiating type A spermatogonia. Endocrinology 1999; 140:5894–5900. [DOI] [PubMed] [Google Scholar]
- 49. Tokuda M, Kadokawa Y, Kurahashi H, Marunouchi T. CDH1 is a specific marker for undifferentiated spermatogonia in mouse testes. Biol Reprod 2007; 76:130–141. [DOI] [PubMed] [Google Scholar]
- 50. Yoshinaga K, Nishikawa S, Ogawa M, Hayashi S, Kunisada T, Fujimoto T. Role of c-kit in mouse spermatogenesis: identification of spermatogonia as a specific site of c-kit expression and function. Development 1991; 113:689–699. [DOI] [PubMed] [Google Scholar]
- 51. Ngo D, Cheng Q, O’Connor AE, DeBoer KD, Lo CY, Beaulieu E, De Seram M, Hobbs RM, O’Bryan MK, Morand EF. Glucocorticoid-induced leucine zipper (GILZ) regulates testicular FOXO1 activity and spermatogonial stem cell (SSC) function. PLoS One 2013; 8:e59149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Sarbassov DD, Guertin DA, Ali SM, Sabatini DM. Phosphorylation and regulation of Akt/PKB by the rictor-mTOR complex. Science 2005; 307:1098–1101. [DOI] [PubMed] [Google Scholar]
- 53. Oulad-Abdelghani M, Bouillet P, Decimo D, Gansmuller A, Heyberger S, Dolle P, Bronner S, Lutz Y, Chambon P. Characterization of a premeiotic germ cell-specific cytoplasmic protein encoded by Stra8, a novel retinoic acid-responsive gene. J Cell Biol 1996; 135:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zhou Q, Nie R, Li Y, Friel P, Mitchell D, Hess RA, Small C, Griswold MD. Expression of stimulated by retinoic acid gene 8 (Stra8) in spermatogenic cells induced by retinoic acid: an in vivo study in vitamin a-sufficient postnatal murine testes. Biol Reprod 2008; 79:35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Schonhoff SE, Giel-Moloney M, Leiter AB. Neurogenin 3-expressing progenitor cells in the gastrointestinal tract differentiate into both endocrine and non-endocrine cell types. Dev Biol 2004; 270:443–454. [DOI] [PubMed] [Google Scholar]
- 56. Zheng K, Wang PJ. Blockade of pachytene piRNA biogenesis reveals a novel requirement for maintaining post-meiotic germline genome integrity. PLoS Genet 2012; 8:e1003038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Xiong M, Zhu Z, Tian S, Zhu R, Bai S, Fu K, Davis JG, Sun Z, Baur JA, Zheng K, Ye L. Conditional ablation of Raptor in the male germline causes infertility due to meiotic arrest and impaired inactivation of sex chromosomes. FASEB J. 2017; 31:3934–3949. [DOI] [PubMed] [Google Scholar]
- 58. Hsu PP, Kang SA, Rameseder J, Zhang Y, Ottina KA, Lim D, Peterson TR, Choi Y, Gray NS, Yaffe MB, Marto JA, Sabatini DM. The mTOR-regulated phosphoproteome reveals a mechanism of mTORC1-mediated inhibition of growth factor signaling. Science 2011; 332:1317–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Yu Y, Yoon SO, Poulogiannis G, Yang Q, Ma XM, Villen J, Kubica N, Hoffman GR, Cantley LC, Gygi SP, Blenis J. Phosphoproteomic analysis identifies Grb10 as an mTORC1 substrate that negatively regulates insulin signaling. Science 2011; 332:1322–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Shah OJ, Wang Z, Hunter T. Inappropriate activation of the TSC/Rheb/mTOR/S6K cassette induces IRS1/2 depletion, insulin resistance, and cell survival deficiencies. Curr Biol 2004; 14:1650–1656. [DOI] [PubMed] [Google Scholar]
- 61. Harrington LS, Findlay GM, Gray A, Tolkacheva T, Wigfield S, Rebholz H, Barnett J, Leslie NR, Cheng S, Shepherd PR, Gout I, Downes CP et al. . The TSC1-2 tumor suppressor controls insulin–PI3K signaling via regulation of IRS proteins. J Cell Biol 2004; 166:213–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brunet A, Bonni A, Zigmond MJ, Lin MZ, Juo P, Hu LS, Anderson MJ, Arden KC, Blenis J, Greenberg ME. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999; 96:857–868. [DOI] [PubMed] [Google Scholar]
- 63. Jacinto E, Facchinetti V, Liu D, Soto N, Wei S, Jung SY, Huang Q, Qin J, Su B. SIN1/MIP1 maintains rictor-mTOR complex integrity and regulates Akt phosphorylation and substrate specificity. Cell 2006; 127:125–137. [DOI] [PubMed] [Google Scholar]
- 64. Kluin PM, de Rooij DG. A comparison between the morphology and cell kinetics of gonocytes and adult type undifferentiated spermatogonia in the mouse. Int J Androl 1981; 4:475–493. [DOI] [PubMed] [Google Scholar]
- 65. Kluin PM, Kramer MF, de Rooij DG. Spermatogenesis in the immature mouse proceeds faster than in the adult. Int J Androl 1982; 5:282–294. [DOI] [PubMed] [Google Scholar]
- 66. Kluin PM, Kramer MF, de Rooij DG. Proliferation of spermatogonia and Sertoli cells in maturing mice. Anat Embryol 1984; 169:73–78. [DOI] [PubMed] [Google Scholar]
- 67. Yoshida S, Sukeno M, Nakagawa T, Ohbo K, Nagamatsu G, Suda T, Nabeshima Y. The first round of mouse spermatogenesis is a distinctive program that lacks the self-renewing spermatogonia stage. Development 2006; 133:1495–1505. [DOI] [PubMed] [Google Scholar]
- 68. Hu YC, de Rooij DG, Page DC. Tumor suppressor gene Rb is required for self-renewal of spermatogonial stem cells in mice. Proc Natl Acad Sci USA 2013; 110:12685–12690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Yang QE, Gwost I, Oatley MJ, Oatley JM. Retinoblastoma protein (RB1) controls fate determination in stem cells and progenitors of the mouse male germline. Biol Reprod 2013; 89:113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Buaas FW, Kirsh AL, Sharma M, McLean DJ, Morris JL, Griswold MD, de Rooij DG, Braun RE. Plzf is required in adult male germ cells for stem cell self-renewal. Nat Genet 2004; 36:647–652. [DOI] [PubMed] [Google Scholar]
- 71. Costoya JA, Hobbs RM, Barna M, Cattoretti G, Manova K, Sukhwani M, Orwig KE, Wolgemuth DJ, Pandolfi PP. Essential role of Plzf in maintenance of spermatogonial stem cells. Nat Genet 2004; 36:653–659. [DOI] [PubMed] [Google Scholar]
- 72. Falender AE, Freiman RN, Geles KG, Lo KC, Hwang K, Lamb DJ, Morris PL, Tjian R, Richards JS. Maintenance of spermatogenesis requires TAF4b, a gonad-specific subunit of TFIID. Genes Dev 2005; 19:794–803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Chen C, Ouyang W, Grigura V, Zhou Q, Carnes K, Lim H, Zhao GQ, Arber S, Kurpios N, Murphy TL, Cheng AM, Hassell JA et al. . ERM is required for transcriptional control of the spermatogonial stem cell niche. Nature 2005; 436:1030–1034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Tyagi G, Carnes K, Morrow C, Kostereva NV, Ekman GC, Meling DD, Hostetler C, Griswold M, Murphy KM, Hess RA, Hofmann MC, Cooke PS. Loss of Etv5 decreases proliferation and RET levels in neonatal mouse testicular germ cells and causes an abnormal first wave of spermatogenesis. Biol Reprod 2009; 81:258–266. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.