Abstract
Mammalian sperm undergo a series of biochemical and physiological changes collectively known as capacitation in order to acquire the ability to fertilize. Although the increase in phosphorylation associated with mouse sperm capacitation is well established, the identity of the proteins involved in this signaling cascade remains largely unknown. Tandem mass spectrometry (MS/MS) has been used to identify the exact sites of phosphorylation and to compare the relative extent of phosphorylation at these sites. In the present work, we find that a novel site of phosphorylation on a peptide derived from the radial spoke protein Rsph6a is more phosphorylated in capacitated mouse sperm. The Rsph6a gene has six exons, five of which are conserved during evolution in flagellated cells. The exon containing the capacitation-induced phosphorylation site was found exclusively in eutherian mammals. Transcript analyses revealed at least two different testis-specific splicing variants for Rsph6a.Rsph6a mRNA expression was restricted to spermatocytes. Using antibodies generated against the Rsph6a N-terminal domain, western blotting and immunofluorescence analyses indicated that the protein remains in mature sperm and localizes to the sperm flagellum. Consistent with its role in the axoneme, solubility analyses revealed that Rsph6 is attached to cytoskeletal structures. Based on previous studies in Chlamydomonas reinhardtii, we predict that Rsph6 participates in the interaction between the central pair of microtubules and the surrounding pairs. The findings that Rsph6a is more phosphorylated during capacitation and is predicted to function in axonemal localization make Rsph6a a candidate protein mediating signaling processes in the sperm flagellum.
Keywords: sperm, capacitation, axoneme, radial spoke, phosphorylation
A sequence corresponding to the N-terminal domain of the radial spoke protein Rsph6 was found phosphorylated in capacitated sperm. Rsph6 expression is a testis-specific gene. Rsph6 protein remains present in mature sperm, and is localized to the sperm flagellum.
Introduction
During epididymal maturation, mammalian sperm acquire the ability to move progressively [1]. However, to fertilize metaphase-II eggs, sperm require an additional maturation step in the female reproductive tract known as capacitation. At the cell biological level, capacitation is characterized by changes in the sperm motility pattern known as hyperactivation and by the preparation for an exocytotic reaction of the acrosomal vesicle [2]. Molecularly, capacitation is associated with a fast HCO3−-dependent activation of the atypical adenylyl cyclase Adcy10 (aka sAC) and a consequent increase in cAMP levels and Prkac (aka PKA) activation. Although it is well established that PKA starts a phosphorylation cascade leading to capacitation, the identity of downstream effectors is not well understood. Capacitation-associated changes in the phosphorylation status of proteins have been studied by western blot analyses using anti-phospho antibodies [3–5]. While this approach is relatively straightforward, it is not sufficient to identify de novo phosphorylation sites and can only be used when a particular phosphorylation site is already known [6].
Post-translational modifications such as phosphorylation can also be determined and quantified using proteomic approaches [6–10]. Previously, our group used immobilized metal affinity chromatography (IMAC) for phosphopeptide enrichment combined with MS/MS to identify 63 phosphorylation sites in capacitated human sperm [7]. However, this study did not reveal which of these sites undergo phosphorylation during capacitation. To identify sequences that show a change in their phosphorylation status, we used a differential isotopic labeling strategy in a subsequent manuscript to compare phosphopeptides derived from both capacitated and noncapacitated mouse sperm proteins [8]. As part of that study, the relative quantification of 53 phosphorylation sites was reported. In the present work a similar approach was used, which led to the identification of an additional peptide sequence showing increased phosphorylation in capacitated sperm. This peptide corresponded to a PKA-consensus sequence (RRXS/T) in the N-terminal region of the mouse radial spoke head protein 6a (Rsph6a). Although homologs of this protein could be found throughout evolution, including in the flagellated algae Chlamydomonas reinhardtii, the N-terminal domain containing the above-mentioned phosphosite was exclusively present in eutherian mammals.
Radial spokes are T-shaped structures formed by multiple protein subunits, and are found in the axoneme of eukaryotic cilia and flagella [11]. Radial spoke proteins play a role connecting the central pair of microtubules with the outer ones and are essential for motility and for the characteristic bending pattern of any given axoneme [11]. As part of this work we investigated the evolution of Rsph6a, its gene expression pattern, and its localization in mature mouse sperm. We confirmed that Rsph6a mRNA is expressed exclusively in testicular germ cells during meiosis and showed that this gene protein product, Rsph6a, remains in mature sperm associated with the cytoskeletal flagellar structures. Finally, 2D PAGE western analyses revealed that in capacitated sperm a portion of this protein displays more acidic isoelectric points (IP) and that these changes in IP were blocked in the presence of cAMP pathway inhibitors.
Materials and Methods
Ethics statement
Animal care and use of experimental animals was conducted in accordance with specific guidelines and standards dictated by the Office of Laboratory Animal Welfare (OLAW) and approved by the Institutional Animal Use and Care Committee (IACUC), University of Massachusetts-Amherst (Protocol #2016–0026).
Materials
Materials and reagents were purchased from various sources. Luria broth, DL-dithiothreitol (DTT), iodoacetamide, urea, thiourea, Trizma base, sodium chloride (NaCl), magnesium chloride (MgCl2), potassium chloride (KCl), sodium orthovanadate (NaVO4), β-glycerophosphate (βGP), β-mercaptoethanol (βME), 3-[(3-cholamidopropyl) dimethylammonio] -1-propanesulfonate hydrate (CHAPS), sodium bicarbonate (NaHCO3) and bovine serum albumin (BSA, fatty acid free, cat. # A0281) were purchased from Sigma-Aldrich (St. Louis, MO). HEPES (cat. # BP310) and glycerol were obtained from Fisher Scientific. Kanamycin sulfate (cat. # 420311) was purchased from Millipore (Burlington, MA). [Isopropyl-β-D-thiogalactopyranoside] (IPTG) was obtained from Research Product International (IL). Ready-strip IPG strips (cat. # 163–200), Bio-lyte ® (3/10, 40%, pH range: 3.5–9.4) and sodium dodecyl sulfate (SDS) were purchased from Bio Rad (BioRad Laboratories, Hercules, CA). H-89 (cat. #130964–39) was purchased from Cayman Chemical (Ann Arbor, MI). LRE1 (6-chloro-N4-cyclopropyl-N4-(2-thienylmethyl)-2,4-pyrimidinediamine), a selective allosteric inhibitor of soluble adenylyl cyclase (sAC), was purchased from Nanosyn (Santa Clara, CA). Protease inhibitor cocktail tablets (cat. # 11697498001) were purchased from Roche (Indianapolis, IN). Polyclonal His-Tag rabbit antibody (cat. #2365S) was purchased from Cell Signaling (Danvers, MA). Two Rsph6a monoclonal antibodies E2 and E3 were custom-made by Abmart (Shanghai, China). Horseradish peroxidase-conjugated anti-mouse IgG was purchased from Jackson ImmunoResearch laboratories (West Grove, PA). Peroxidase-conjugated anti-rabbit IgG and ECL Prime Western blotting detection reagents were purchased from GE Healthcare (Pittsburgh, PA). Alexa Fluor 488-conjugated secondary anti-mouse antibody was purchased from Invitrogen, ThermoFisher Scientific (Waltham, MA).
Animals
CD1 retired breeder male mice were used (Charles River Laboratories, Wilmington, MA). For Rsph4a and Rsph6a expression analyses during mouse development, tissues from 1-, 2-, 3-, 3.5-, 4-, and 8-week-old mice, and from 7-month-old mice were used.
Mouse sperm preparation
Mouse cauda epididymal sperm were collected from CD1 retired breeder male mice by the “swim out” method in TYH/Hepes medium as described previously [12]. The TYH/Hepes medium contained the following compounds (concentrations in parenthesis are given in mM): NaCl (100), KCl (4.7), KH2PO4 (1.2), MgSO4 (1.2), glucose (5.5), pyruvic acid (0.8), CaCl2 (1.7), HEPES (20). Mouse sperm capacitation was achieved by incubation in TYH/Hepes medium supplemented with 15 mM NaHCO3 and 5 mg/mL BSA for 1 h at 37°C; TYH/Hepes medium with no additives served as noncapacitating medium. After washing in PBS, sperm pellets were either processed immediately or saved at –80°C.
Differential isotopic labeling and global quantification of phosphopeptides from sperm total protein digests
Procedures used for the identification of phosphopeptides and the quantification of the relative extent of phosphorylation on sperm proteins have been thoroughly described previously [8]. Briefly, extracts were prepared from sperm incubated in conditions that either support or do not support capacitation (see below) and subsequent trypsin proteolytic digestion was conducted as described [8]. Fisher esterification was carried out in either noncap and cap samples using methyl alcohol labeled with either hydrogen (d0) or deuterium (d3). At the end of the procedure, four distinct samples were obtained: noncap (d0), noncap (d3), cap (d0), and cap (d3). Differentially labeled aliquots of the dissimilar, esterified samples (i.e. noncap (d0 esters) and cap (d3 esters); or noncap (d3 esters) and cap (d0 esters)) were combined and subjected to IMAC. The IMAC precolumn was connected to an analytical reverse-phase HPLC column containing an integrated microelectrospray (ESI) emitter tip. Peptides were then gradient-eluted directly into an LTQ-FT linear ion trap-Fourier transform mass spectrometer as described [8]. Phosphopeptide sequences were identified using the SEQUEST algorithm within Proteome Discoverer (ThermoFisher Scientific) and manually verified. Quantification of the relative extent of phosphorylation was done by calculating the ratio of the capacitated peptide peak (d0 esters) area to the corresponding noncapacitated peptide (d3 esters) peak area. This procedure was repeated with the alternatively labeled extracts, cap (d3 esters) and noncap (d0 esters). Final values were calculated by taking the square root of the ratio of the two individual ratios, which served to compensate for potential systematic errors inherent in comparing the deuterated and the nondeuterated peptides.
Sequence alignment and phylogenetic analysis
Using mouse Rspha6a gene as query, we searched for homologs in the ENSEMBL database (release 90) within chordates. Only sequences with complete coding sequences (CDS) were kept for further analysis, and aligned using CLUSTAL Omega at EMBL-EBI [13, 14]. Sequences that did not start with an ATG codon or that had multiple ambiguities were discarded. Protein and nucleotide alignments were visually inspected using Wasabi web browser [15]. Protein conservation was analyzed using PlotCon (http://www.bioinformatics.nl/cgi-bin/emboss/plotcon), using the EBLOSUM62 aminoacidic similarity matrix, with a window size of 100 residues to account for conservation in neighboring sites. Phylogenetic analyses were performed with MrBayes 3.2.6 [16, 17] through the CIPRES Science Gateway [18] under the following parameters: using a GTR + InvGamma model with exponential and Dirichlet priors, performing two runs for 107 generations, sampling every 103 generations, and discarding the first 25% generations considered as the burn-in phase. The resulting consensus tree was inspected and formatted using the Interactive Tree Of Life (https://itol.embl.de/).
RNA extraction and reverse transcriptase polymerase chain reaction
Mouse tissue from various aged male mice was dissected and homogenized. Total RNA was extracted using High Pure RNA Isolation Kit (Roche), and quality and total RNA concentration was checked with a Nanodrop spectrophotometer (BioDrop, Cambridge, UK), as previously described [19]. Synthesis of cDNA from RNA samples was performed by iScriptTM cDNA Synthesis Kit (BIO-RAD, Hercules, CA) in a 20 μL reaction volume; the starting amount of total RNA was 500 ng. The reverse transcriptase polymerase chain reaction (RT-PCR) reaction was carried out with Ruby-Taq master mix (Affimetrix, Santa Clara, CA) using β-Actin [(forward primer (F)- AGAGGGAAATCGTGCGTGAC; reverse primer (R)- CAATAGTGATGACCTGGCCGT), Rsph4a (F-AGCCTACCTGCTGAGTACCA; R-TAAGTTGCTTGAGGGCGAGG), and Rsph6a specific primers (F1-TCAGCCTGTATGAGCACCTG; R1-TCCTCCTCCCCTAGCTCTTC, F3-GAGAAGGTGGTGGACAGCGT; R3-GCCGAACTGGTAGAAGCCC, F4-CAGCACGGACCCTACCTGAG; R4-TCTTCCATTTCCTGTTCGCC, and F5-AAGAGAAGGCAGACGAGGCA; R5-GGAAGGGCTATGGGAAGGAG). All PCR reactions consisted of initial denaturation of 95°C for 2 min, followed by 35 cycles of 30 s denaturation at 95°C; annealing for 30 s at 57°C for F1/R1, 59°C for F3/R3, 56°C for F4/R4, and 58°C for F5/R5; and extension for 30 s at 72°C. The final extension of the reactions was carried out at 72°C for 2 min. Total cDNA used to perform RT-PCR reaction was 3 ng. Finally, PCR products were run on 2% quick dissolve agarose gel (BioExpress, Kaysville, UT) for 30 min at 100 V, stained with ethidium bromide, and visualized with a G: Box imaging system (Syngene, Frederick, MD).
Analysis of mouse Rsph6a expression by quantitative PCR
Quantitative PCR analysis on N-terminus region of Rsph6a gene was performed using the PerfeCta® SYBR Green® SuperMix, Low RoxTM cocktail (Quanta Biosciences, Gaithersburg, MD) and F4/R4 Rsph6a N-terminus oligonucleotide primers as described above. Reaction mixtures were assembled in 20-μL volume (3 ng cDNA and 500 nM primers) and incubated in a Stratagene MX300p qPCR system (Agilent Technologies, Santa Clara, CA). Conditions used for thermal detection consisted of initial denaturation at 95°C for 3 min, followed by 40 cycles of 30 s denaturation at 95°C, annealing at 56°C for 45 s, and extension at 72°C for 1 min. Dissociation curves were run simultaneously to assess the occurrence of unspecific binding of SYBR green to primer dimers, or any contaminating DNA, and observed no additional peaks other than the desired single products. Conditions used to evaluate dissociation curves were 95°C for 1 min, 55°C for 30 s, 95°C for 30 s. Quantitative PCR analysis was performed on three biological replicates. For each biological replicate, reactions were run in triplicate. For normalization, analysis of a housekeeping gene (β-actin) was conducted in parallel with all tissues, and CT values for each reaction were collected and averaged as described before [20]. Then, CT values from mouse tissues were deduced from their corresponding β-Actin CT values and calibrated with CT values obtained from data obtained in 4-week-old mouse testis, and the 2 -ΔΔCT method was applied to report mRNA expression. Error bars indicate ± standard error of the means (±SEM).
-ΔΔCT method was applied to report mRNA expression. Error bars indicate ± standard error of the means (±SEM).
Molecular cloning and recombinant protein production
Two of the predicted full-length Rsph6a transcripts named I1 and I2 were amplified using Phusion® High-Fidelity DNA polymerase (New England Biolabs, Ipswich, MA) from 100 ng cDNA synthesized from 8-week-old mouse testis tissues. Primers used to amplify both full-length cDNAs were F8- GGAATTCCATATGGGGGAACCACCGCCCAAT and R8-CCGCTCGAGGTCCTCCAGGTCTTCATCCTC (underlined sequences indicate the NdeI and XhoI restriction sites included in primers). The PCR products were then separated on 1.5% agarose gels and fragments (1369 and 2144 bp) were excised and purified using PrepEase® Gel extraction Kit (USB Corporation, Cleveland, OH), and directly cloned into a pET-30a (+) expression vector containing a kanamycin-resistance gene and a C-terminal His-Tag sequence, as previously described [21]. For this purpose, common restriction sites containing PCR products and pET vector were digested, purified, and subsequently ligated. Digestion was carried out with 1 μg plasmid DNA (PCR product or pET vector) with 10 units NdeI and XhoI in CutSmartTM Buffer (New England BioLabs) at 37°C. Ligation reactions were performed at 14°C overnight using Express Link TM T4 DNA Ligase (5 units) (ThermoFisher Scientific). For the production of the His-tagged proteins, we followed a modified protocol [22]. A pET vector carrying Rsph6a inserts was transformed into T7 expression BL21 (DE3) Escherichia coli strain (NEB). After confirming insert sizes by colony PCR and sequencing (Genewiz, South Plainfield, NJ), BL21 cells were grown overnight at 37°C in a shaking incubator (250 rpm) in LB broth containing 100 μg/mL kanamycin. After cultures had reached the desired optical density (OD) of 0.61, protein expression was induced with 1 mM IPTG for 4 h at 37°C with constant shaking. Cells were then harvested at 14,000 rpm for 15 min, and cell pellets saved at –80°C for later analysis by western blotting.
Probe design and in situ hybridization
RNA probes (antisense and sense) were designed for Rsph6a testis-specific N-terminus analysis by in situ hybridization; Tssk6 amplimers were used as control. Complementary DNA from mouse testis (100 ng) was amplified with F (antisense)-TCTTCCATTTCCTGTTCGCC, R (antisense)-TAATACGACTCACTATAGGGTCTTCCATTTCCTGTTCGCC and F(sense)- TAATACGACTCACTATAGGGCAGCACGGACCCTACCTGAG, R(sense)- TCTTCCATTTCCTGTTCGCC for Rsph6a and F (antisense)- AAACTCCTGAGCGAACTTGG and R (antisense)- TAATACGACTCACTATAGGGACCGAAATCCGTGATCTTGA for Tssk6. In vitro transcription and analysis by in situ hybridization were performed as previously described [19, 23] with minor modifications. After probe hybridization, tissue sections were washed, treated with blocking solution, and incubated with anti-Digoxigenin-AP (Fab fragments antibody, Roche). After color development by incubation with BM Purple AP substrate (Roche), slides were prepared for mounting in Cytoseal-60 (ThermoFisher Scientific).
Custom-made Rsph6a antibodies
Two mouse monoclonal antibodies against Rsph6a were generated by Abmart (Shanghai, China). Briefly, Rsph6a protein sequence was subjected to a proprietary algorithm analysis (SEALTM method) to identify two 12-residue fragments (E2: PSQTRRASQGSE; E3: QRGSRSSQGSQD) with the highest “epitope score” in the N-terminus region of Rsph6a protein. These antigenic sequences were then used to immunize Balb/c mice to generate hybridoma cells after fusion of splenocytes with a selected myeloma cell line. After subcloning (2–4 rounds) of these hybridoma cells, positive subclones selected by ELISA screening were injected into mouse peritoneal cavity for production of ascites fluid. Ascites were then purified for each antibody, and mAb E2 and mAb E3 (corresponding to E2 and E3, respectively) were selected and further characterized for their ability to recognize recombinant Rsph6a isoforms by western blotting.
Rsph6a protein extraction and solubility analysis
To analyze Rsph6a solubility, aliquots of approximately 3 million sperm were suspended in noncapacitating media. After 15 min, sperm were centrifuged at 10,000 × g for 5 min. Sperm pellets were then resuspended in a buffer containing: Tris/HCl 40 mM pH 7.5, complete protease inhibitor cocktail (Roche), and 1 % Triton X-100. This suspension was left on ice for 30 min and then centrifuged again at the same speed. Supernatants were kept for western blot analysis and pellets were resuspended in nonreducing SDS sample buffer (without added β-mercaptoethanol or DTT), boiled for 5 min, and centrifuged. The resulting supernatant was kept for western blot analysis. In an independent experiment, sperm pellets were directly suspended in nonreducing sample buffer, boiled for 5 min, and then centrifuged. Supernatants were kept for western blot analysis and the sperm pellet boiled once more in nonreducing sample buffer. After centrifugation at 10,000 × g, supernatants were saved for western blot analysis and the resulting pellet was suspended in reducing sample buffer containing 50 mM DTT and boiled once more for 5 min. After centrifugation, the supernatant was saved for western blotting analysis.
SDS-PAGE and western blot analysis
For western blot analysis, sperm proteins extracted with a reducing agent were boiled for 5 min and subjected to SDS-PAGE while the protein samples that were extracted without a reducing agent were further denatured by boiling in the presence of 50 mM DTT. Separated proteins were then transferred to PVDF membranes (Millipore), blocked with 5% dry fat-free milk powder for 1 h, and incubated overnight at 4°C with mAbs E2 or E3 (1:300–500 dilution from a 10 μg/ml stock concentration), followed by HRP-conjugated secondary anti-mouse antibody (1:10,000 dilution from a 0.8 μg/ml stock concentration). Protein detection was performed by chemiluminescence with ECL Prime (GE Healthcare). When necessary, membrane stripping was done at 55°C in β-mercaptoethanol-containing solution, followed by several washes with PBS before probing for phospho-tyrosine proteins by anti-pY (1:10,000) (clone 4G10, Millipore) and anti-tubulin (1:2000) from the University of Iowa Hybridoma Bank (http://dshb.biology.uiowa.edu/). To confirm specificity and binding affinity of antibodies, peptide competition assays were used [20]. Briefly, mAbs E2 and E3 were incubated in the presence of the corresponding antigenic peptides E2/P6 and E3/P1 (200-fold molar excess) for 2 h at room temperature with gentle rocking in TBS prior to use for immunoblotting or immunofluorescence.
Immunolocalization of Rsph6a in mouse sperm
Indirect immunofluorescence analysis was performed as previously described [20]. Briefly, sperm samples were collected in TYH/Hepes buffer and washed gently. Cells were then pipetted onto polylysine-coated coverslips, air-dried, and permeabilized with 0.5% Triton X-100 in PBS. Following washings and blocking for nonspecific binding, incubation with E2 or E3 antibodies (1:50 dilution from a 10 μg/ml stock concentration) in 1% BSA was done overnight at 4°C, followed by incubation with Alexa-488-conjugated goat anti-mouse secondary antibody (1:200 in 3% BSA) (ThermoFisher Scientific (Invitrogen)). Coverslips were thoroughly washed and mounted onto slides using Vectashield-H1000 mounting solution (Vector Laboratories, Burlingame, CA). Immunofluorescence images were acquired with a Nikon Eclipse TE300 fluorescence microscope (Melville, NY).
Two-dimensional gel electrophoresis (2D-PAGE) and 2D western blotting
Sperm pellets (equivalent to 4–6 × 106) were lysed in nonreducing sample buffer, boiled, centrifuged, and supernatants were removed, as described above, to eliminate significant amount of proteins. Subsequently, the remaining pellet was resuspended in rehydration buffer containing 8M urea, 2M thiourea, 4% CHAPS, 20 mM DTT, 0.5% (w/v) Bio-Lyte® 3/10 ampholytes, and 0.001% bromophenol blue for 30 min in a slow rocking shaker at room temperature as previously described [24] with slight modifications. Briefly, after complete solubilization of sperm pellets, 7-cm IPG strips (pI 3–10, BioRad) were passively rehydrated for 16 h at RT. Isoelectric focusing (IEF) of IPG strips was then performed in a PROTEAN IEF Cell (Bio-Rad) in six steps as follows: 50 V for 15 min (linear), 100 V for 15 min (linear), 150 V for 15 min (linear), 250 V for 15 min (linear), 4000 V for 2 h (linear); the final focusing step was maintained for 24,000 Vh. After completion of IEF, equilibrated IPG strips were briefly rinsed in SDS running buffer, and subjected to the second dimension, SDS-PAGE, as described above. Once completed, gels were transferred to PVDF membranes and western blotting conducted with anti-Rsph6a antibodies as described above.
In silico Rsph6a phosphorylation analysis
We selected the human and mouse Rsph6a protein sequences as references, and performed phosphorylation site predictions using three online tools: NetPhos 3.1 Server, pkaPS, and the GPS Web Server. The NetPhos 3.1 Server [25, 26] predicts serine, threonine, or tyrosine phosphorylation sites in eukaryotic proteins using ensembles of neural networks. Both generic and kinase-specific predictions can be performed with this tool. For our analysis, only scores above 0.9 were recorded. The pkaPS predicts the existence of PKA phosphorylation sites using a simplified kinase binding model [27].The GPS 3.0 [28] predicts protein phosphorylation sites and their cognate kinases, and is able to classify protein kinases into a hierarchical structure with four levels (including group, family, subfamily, and single protein kinase). The Netphos 3.1 Server was used to predict generic and kinase-specific phosphorylation sites, while pkaPS and GPS Web Server were applied to find PKA-specific phosphosites.
Statistical analysis
Statistical analysis was performed using the Infostat 2011 software (www.infostat.com.ar). In order to accomplish the assumptions of parametric tests, quantitative PCR data in percentages were converted to ratios and then subjected to arcsine square root transformation. One-way ANOVA was then applied followed by the Tukey test, and differences considered significant when P-values were given as indicated in the figure legends.
Results
Relative quantification of phosphopeptides by differential isotopic labeling
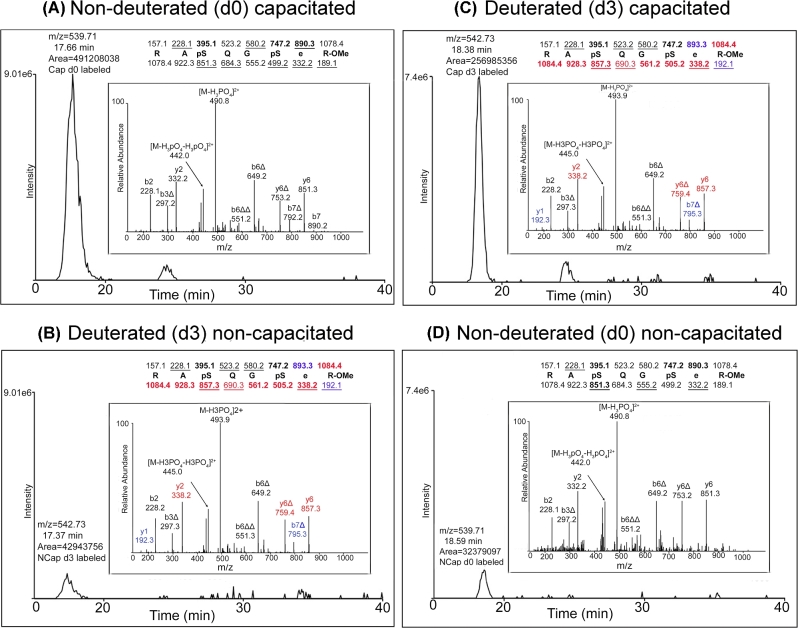
To increase our understanding of the extent of protein phosphorylation in mouse sperm incubated in conditions that either support or do not support capacitation, we used the approach of Fisher esterification with differential isotopic labeling as described previously [8]. An additional phosphopeptide (R.RApSQGpSeR.A) which had not been observed in our previous analyses was found in this study. The relative extent of phosphorylation on the serine residues within this peptide was calculated based on three independent biological replicates of capacitated and noncapacitated sperm. In each replicate, samples were analyzed in two different experiments as previously described [8]. First, capacitated sperm extracts were labeled with nondeuterated Fisher esterification reagent (Figure 1A, d0 cap) and noncapacitated sperm extracts with deuterated reagent (Figure 1B, d3 noncap). The second analysis was carried out using the reverse isotopic labeling scheme, in this case capacitated sperm extracts were labeled with deuterated reagent (Figure 1C, d3 cap) and noncapacitated sperm extracts with nondeuterated reagent (Figure 1D, d0 noncap). The square root of the product of the ratios: 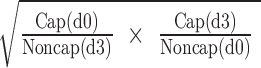 was then calculated for each of the three independent replicates. This analysis indicated an average increase in phosphorylation of 9.53-fold ± 0.57 at these sites in capacitated mouse Rsph6a relative to a noncapacitated extract. As control, in each of the replicates, the change in phosphorylation at the tyrosine residue in the peptide K.YLpYAMR.L from hexokinase type I was also calculated. The average square root of the product in this case was 0.95-fold ± 0.07 which is consistent with previous findings indicating that the extent of tyrosine phosphorylation in hexokinase type I does not change during capacitation [8, 29].
was then calculated for each of the three independent replicates. This analysis indicated an average increase in phosphorylation of 9.53-fold ± 0.57 at these sites in capacitated mouse Rsph6a relative to a noncapacitated extract. As control, in each of the replicates, the change in phosphorylation at the tyrosine residue in the peptide K.YLpYAMR.L from hexokinase type I was also calculated. The average square root of the product in this case was 0.95-fold ± 0.07 which is consistent with previous findings indicating that the extent of tyrosine phosphorylation in hexokinase type I does not change during capacitation [8, 29].
Figure 1.
Enrichment of Rsph6a phosphopeptide in capacitated mouse sperm. Single ion chromatograms for the differentially labeled phosphopeptide RApSQGpSeR from (A) the d0 capacitated/d3 noncapacitated analysis and (B) the d3 capacitated/d0 noncapacitated analysis. Predicted monoisotopic masses for the ions of type b and y are shown above and below the sequences, respectively. Ions observed in the associated MS/MS spectrum are underlined and those that lose phosphoric acid are presented in bold type. The label “Δ” denotes loss of phosphoric acid from the corresponding ion of type b or y. “R-OMe” represents the Fisher esterified C-terminal arginine, while “e” represents the Fisher esterified glutamic acid. Masses for b and y ions, which are shifted by +3 amu in the deuterated spectra, are presented in blue, while those ions shifted by +6 amu are presented in red. The Cap/NCap ratio for each analysis (11.44 in experiment A and 7.94 in experiment B) was used to calculate an overall odds ratio of 9.53, in order to remove potential errors associated with the comparison of the deuterated and nondeuterated peptide species [8]. Spectra shown are representative of at least three separate biological replicates, run under two conditions (d0 and d3) as explained above.
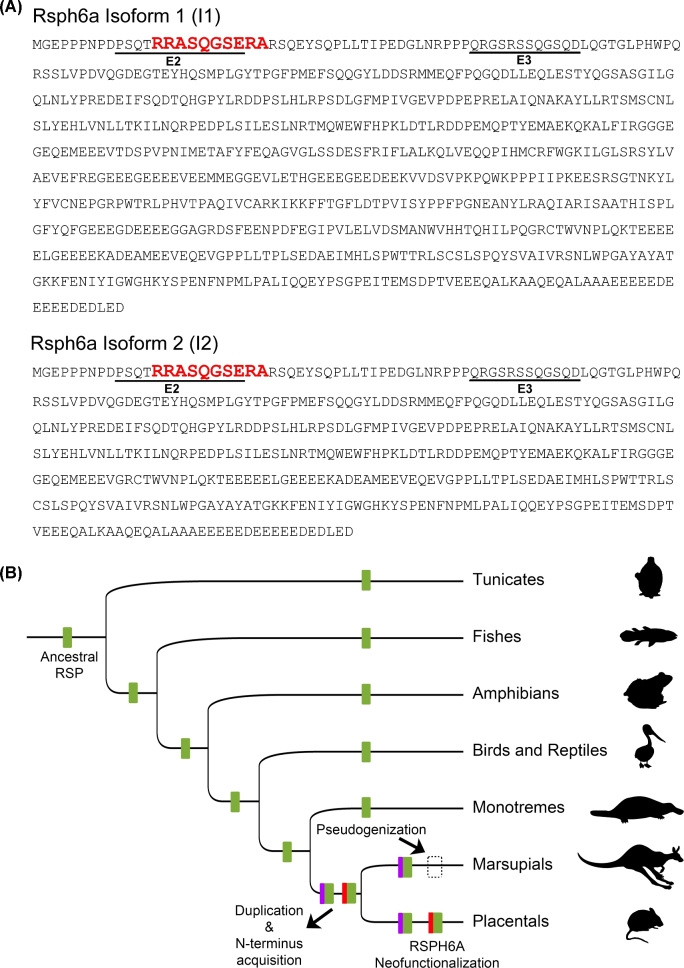
The identified phosphopeptide sequence R.RApSQGpSeR.A was then used to search for candidate proteins in the NCBI database, using protein–protein BLAST. Two isoforms of the mouse Rsph6a (aka RSHL1) demonstrated 100% identity with the query sequence (Figure 2A, see the phosphopeptide sequence in red, boldface, and with larger font). Both isoforms corresponded to the Rsph6a gene sequence (Gene ID: 83434). The mouse Rsph6a gene homologs could then be traced back to the metazoan-protozoan divergence (Ensembl Genomes gene tree), and it was found that the Rsph6 gene had gone through several duplications along lineages (Figure 2B). The Bayesian phylogenetic analysis (Supplemental Figure S1A) yielded two Rsph eutherian subtrees, which reflect the occurrence of a significant duplication event. The presence of two of these sequences in the wallaby and in the Tasmanian devil (Wasabi alignment and tree) indicates that the duplication occurred in the Therian (placentals and marsupials) common ancestor. In these two latter cases, however, the corresponding coding sequence does not start with an ATG, indicating a loss of function and pseudogenization in marsupials (and thus were excluded from the phylogenetic analysis). As a result of this event, Eutherian species have two paralogs: Rsph6a and Rsph4a. Both copies have a longer N-terminus compared to non-Therian species (ca. 200 additional residues in both proteins) (Supplemental Figure S1B). In addition, it is worth noting that, despite both paralogs being highly conserved, the Rsph6a N-terminus has little homology with that of Rsph4a in placental mammals (Supplemental Figure S2A and B). The rest of the protein is highly conserved among Chordata.
Figure 2.
(A) Rsph6a isoform 1 and isoform 2 amino acid sequences. Antibodies epitopes are underlined in each isoform (E2 or E3). The sequence corresponding to the phosphopeptide detected by MS/MS is shown in red, boldface, and larger font. (B) Chordates phylogeny showing the origin of mammalian RSPH6A and RSPH4A. The chordates common ancestor had one RSPH gene (indicated as ancestral RSP), which was duplicated along the Therian (marsupials and placental mammals) branch. Each copy acquired an extra N-terminus. Within placentals, one copy retained the canonical function (RSPH4A) while the other is tissue-specific (RSPH6A) as a result of a neofunctionalization process. In marsupials, RSPH6A is no longer a functional gene as a consequence of a pseudogenization process.
Rsph6a is expressed exclusively in testicular germ cells
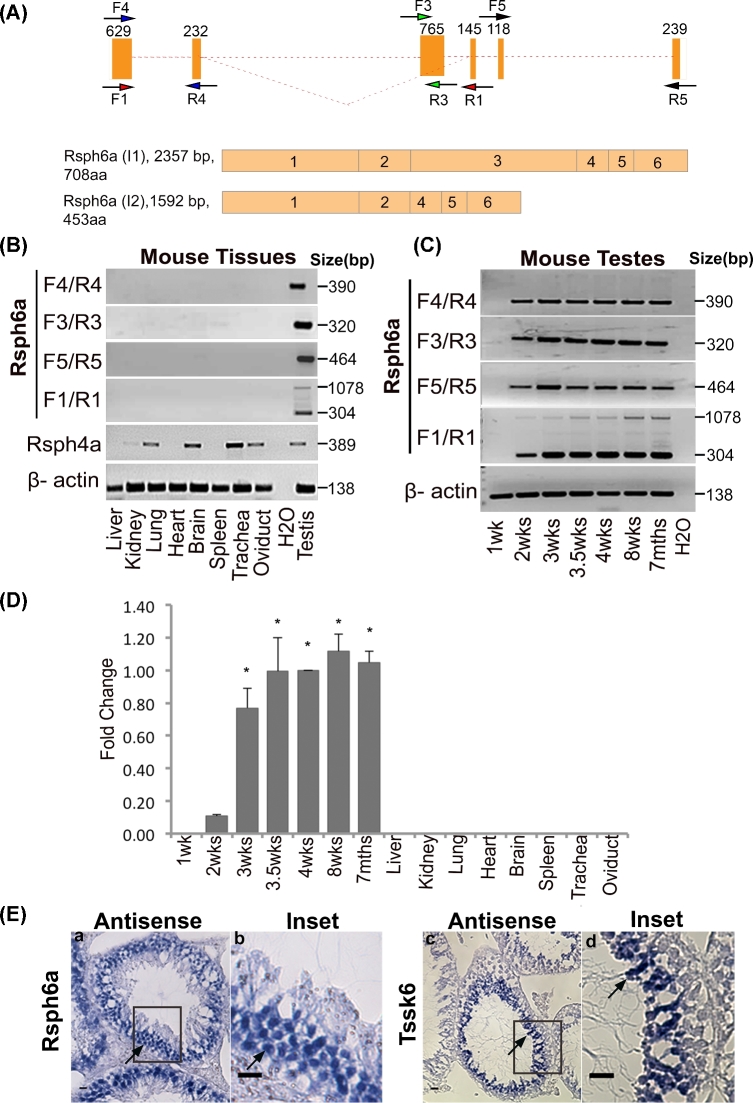
Rsph6 is found in two splicing variants in the NCBI database. The first one, Rsph6a isoform I1, consists of six exons, while in the alternative form, the third 765-bp exon is skipped to produce a second isoform, Rsph6a I2 (Figure 3A). Forward and reverse oligonucleotides, encoding for sequences in the six exons (see Figure 3A), were used to evaluate Rsph6a mRNA expression by RT-PCR. Confirming previous reports [30, 31], all Rsph6a amplicons were found exclusively in the testis (Figure 3B). On the other hand, the somatic homolog Rsph4a was expressed in brain, kidney, and in cilia-containing tissues such as lung, trachea, and oviduct (Figure 3B). When testicular mRNA was obtained from mice of different ages, Rsph6a transcripts were found in mice that were 2 weeks old or older (Figure 3C) suggesting that Rsph6a expression starts during meiosis [32]. RT-PCR amplification using F1-R1 primers revealed at least two bands with expected molecular sizes (considering the primer annealing site) representing I1 and I2 transcripts. These results were further validated by quantitative RT-PCR analysis using one of the primer sets (F4/R4) (Figure 3D). In situ hybridization analysis indicated that Rshp6a mRNA expression is restricted to spermatocytes (Figure 3E). As positive control for the in situ hybridization experiments, we confirmed exclusive postmeiotic expression of testis-specific serine kinase 6 (Tssk6) mRNA in elongating spermatids (Figure 3E) as previously described [20, 33]. In both cases, negative controls were conducted using the respective sense probes (Supplemental Figure S3A and B).
Figure 3.
Rsph6a is expressed in spermatocytes. (A) Schematic diagrams showing two mouse Rsph6a isoforms (I1 and I2). Isoform I2 is formed by splicing of a 765-bp exon in isoform I1. Numbers inside exons indicate exon order (bottom). Same color arrows indicate exon-specific forward and reverse primers designed along the length of the +Rsph6a gene (top). Numbers above each exon represent exon size. (B) Rsph6a gene expression analysis of several mouse tissues by RT-PCR using exon-specific primers as shown in (A). Primers used for Rsph4a are listed in the Methods section. (C) Rsph6a gene expression analysis of mouse testes at various developmental ages by RT-PCR using exon-specific primers as shown in (A). For both RT-PCR reactions, β-actin was used as a loading control. (D) Quantitative PCR analysis on mouse testes using F4/R4 primers. Shown are pooled results from three biological replicates. For each biological replicate, three reactions were run per assay, and the same experiment was repeated on separate dates at least two times. For normalization, β-actin was run in parallel along with other tissues and the values obtained for 4.0-week-old mouse testis were used as the “calibrator”; the 2 -ΔΔCT method (fold change) was applied to report mRNA expression. Error bars represent the standard error of the means (±). Statistical differences in expression are indicated by asterisks (P < 0.05) when compared to Rsph6a expression found in 1- to 2-week-old mouse testes. (E) In situ hybridization showing expression of Rsph6a (left panel a and b) and Tssk6 mRNA (right panel c and d) in mouse testis sections. Black rectangular boxes represent enlarged portion (insets); black arrows show Rsph6a-positive secondary spermatocytes, and Tssk6-positive postmeiotic spermatids, respectively. Scale bars, 50 μm.
-ΔΔCT method (fold change) was applied to report mRNA expression. Error bars represent the standard error of the means (±). Statistical differences in expression are indicated by asterisks (P < 0.05) when compared to Rsph6a expression found in 1- to 2-week-old mouse testes. (E) In situ hybridization showing expression of Rsph6a (left panel a and b) and Tssk6 mRNA (right panel c and d) in mouse testis sections. Black rectangular boxes represent enlarged portion (insets); black arrows show Rsph6a-positive secondary spermatocytes, and Tssk6-positive postmeiotic spermatids, respectively. Scale bars, 50 μm.
Rsph6a protein remains in mature sperm and localizes to the flagellum
To investigate the fate of Rsph6a after spermatogenesis, monoclonal antibodies were custom-made against two different N-terminal peptide sequences (mAb E3 raised against QRGSRSSQGSQD and mAb E2 raised against PSQTRRASQGSE) (see underlined sequences in Figure 2A). To validate these antibodies, the two Rsph6a splicing variants (isoforms I1 and I2) were expressed in E. coli as His-tagged recombinant proteins. In control experiments, both protein isoforms of an apparent molecular weight (MW) of 81 KDa (isoform I1) and 60 KDa (isoform I2) were recognized by the anti-His antibody (Supplemental Figure S4). Both Rsph6a isoforms were also recognized by mAb E3 (Figure 4A, left panel) and by mAb E2 (Supplemental Figure S5A, left panel) and, in both cases, the signal was competed out by incubation with the respective antigenic peptide (Figure 4A and Supplemental Figure S5A, right panels).
Figure 4.
Validation of a custom-made anti-Rsph6a antibody (E3) in recombinant Rsph6a isoforms and in mouse sperm. (A) Representative western blot showing detection of recombinant Rsph6a isoforms (I1 and I2) when incubated with E3 antibody alone (A, left panel) or in the in the presence of 200-molar excess of blocking peptide (P1) (A, right panel). (B) Protein lysates extracted from mouse sperm treated under capacitating (C) or noncapacitating (N) conditions were subjected to western analysis and detection of Rsph6a was done with mAb E3 antibody. In B (left panel), three distinct proteins are shown upon detection with mAb E3. Antibody competition with the E3 antigenic peptide (P1) is shown in B (right panel). PVDF membranes were stripped and sequentially probed with anti-tubulin antibody as loading control. (C) Immunofluorescence detection of Rsph6a in mouse sperm when probed with mAb E3 alone or in the presence of blocking peptide: (a, e) DIC images; (b) immunofluorescence with E3 alone or (f) when incubated with blocking peptide (E3-P1); (c) merged a-b; (d) inset showing magnified image from (c). Scale bars, 20 μm. Western blotting and Immunofluorescence images shown are representative of at least three independent experiments.
When used to evaluate the presence of Rsph6 in noncapacitated and capacitated sperm samples, the E3 antibody recognized three proteins of apparent MW of approximately 110, 60, and 37 KDa, respectively (Figure 4B, left panel). All these proteins were no longer detected after blocking the antibody with the respective antigenic peptide (Figure 4B, right panel). The approximately 110 and the 60 KDa proteins were also recognized by E2; however, this antibody recognized an additional protein of approximately 75 KDa and did not recognize the approximately 37 KDa band (Supplemental Figure S5B, left panel). Both antibodies were made against different epitopes and it is possible that differences in the western blots are due to nonspecific cross-reaction. Similar to results with mAb E3, these bands were fully competed out by co-incubating mAb E2 with the corresponding antigenic peptide (Supplemental Figure S5B, right panel). Consistent with a role in the axoneme, Rsph6a was found to be insoluble in 1% Triton X-100. Moreover, a significant fraction of Rsph6a, including the 60-KDa band, remained insoluble even after the sperm were boiled in Laemmli sample buffer without the addition of reducing agents (Supplemental Figure S6). Finally, in immunofluorescence assays, both antibodies stained the whole sperm flagellum and the signal was eliminated by pre-incubation with their respective antigenic peptide (Figure 4C and Supplemental Figure S5C).
Rsph6a phosphorylation during sperm capacitation is dependent on PKA
Our MS/MS analysis indicated that the N-terminal domain of Rsph6a undergoes increased phosphorylation during capacitation. Therefore, we performed phosphorylation site predictions using three online tools: the NetPhos 3.1 Server, the pkaPS, and the GPS Web Server. Supplementary Table S1 shows all phosphosites predicted by Netphos 3.1, and PKA-specific phosphorylation sites predicted by pkaPS and GPS, both for mouse and human Rsph6a. Five of these sites were predicted by all three methods and showed the highest phosphorylation scores in all of them. Although there were more candidate sites predicted by one or two methods, the aforementioned phosphosites are predicted to be PKA phosphorylation sites. The first site on the N-terminus is a serine located in the 17th residue of the mouse Rsph6a and coincides with the serine at site 21 in the human protein. This site matches the phosphorylation site found using mass spectrometry (Figure 2A, see sequence in boldface, red, and larger font) and is conserved in the unique eutherian mammalian species Rsph6a N-terminus sequence. Together, these data suggest that this sequence has an important role in Rsph6a post-translational modification. The second site found corresponds to the serine at the 29th position of the human RSPH6A, and although it is a conserved serine, it is predicted to be phosphorylated by PKA in human but not in mouse. The third site found using bioinformatics in mouse corresponds to the serine at the 52nd residue of Rsph6a. This site is not ubiquitous among mammals, but instead is restricted to rodents. Similarly, although the serine at the 62nd position of the human RSPH6A is conserved among most placental mammals, it is predicted to form part of a PKA cognate in human but not in mouse. The last site is common to both human and mouse (residues 519 and 510, respectively), and it corresponds to a serine conserved among 36 out of 38 Rsph6a protein sequences analyzed.
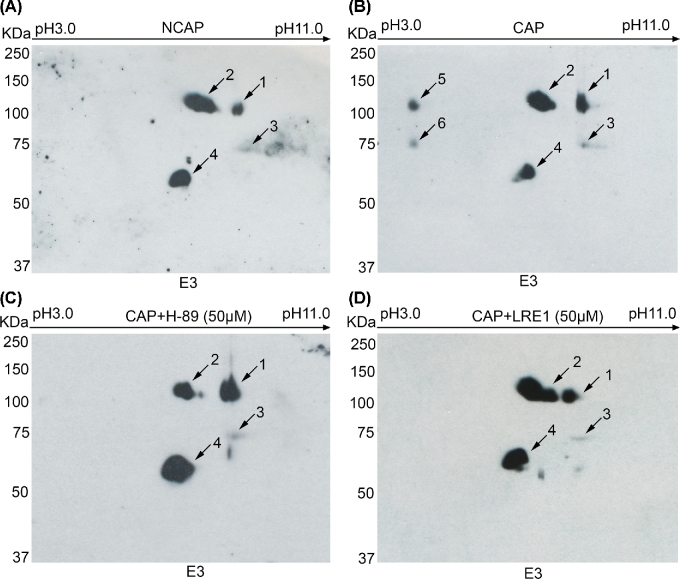
Because phosphorylated proteins have more acidic IPs [34], we used 2D gels to analyze Rsph6a behavior in mouse sperm incubated in media that either supported or did not support capacitation. The low solubility of proteins under nonreducing conditions was used to eliminate a significant amount of protein background, as described in the Methods section. The remaining (cell pellet) fraction was extracted with 2D urea buffer containing DTT. In these conditions four spots were detected in noncapacitated sperm lysates (Figure 5A), in the MW range corresponding to the approximately 110 KDa and the approximately 60–75 KDa Rsph6a isoforms. When the same experiments were conducted using sperm incubated in conditions that support capacitation (addition of BSA 5 mg/ml and HCO3− 15 mM), the approximately 110 and the 75 KDa spots showed a partial protein shift to a lower, more acidic IP (Figure 5B). The lower IP is consistent with phosphorylation of these protein isoforms in capacitated sperm. Considering phosphosite predictions, 2D analyses were also conducted in sperm capacitated in the presence of H-89, a known PKA inhibitor, and of LRE1, a recently described inhibitor of the atypical adenylyl cyclase Adcy10 (aka sAC) [35] (Figure 5C and D, respectively). Both inhibitors blocked the observed changes in Rsph6a IP.
Figure 5.
Identification of phosphorylated Rsph6a by two-dimensional gel electrophoresis. (A) 2D-PAGE of mouse sperm lysates under noncapacitating (NCAP) or (B) capacitating (CAP) conditions probed with mAb E3 antibody. (C) 2D-PAGE analysis of mouse sperm lysates incubated under capacitating conditions in the presence of 50 μM H-89 inhibitor (CAP + H-89) or (D) 50 μM of LRE1 inhibitor (CAP + LRE1) probed with mAb E3 antibody. Numbers indicate the Rsph6a spots at different isoelectric points, and changes in phosphorylated Rsph6a are observed upon each treatment. Images are representative results of at least three independent experiments.
Discussion
Capacitation is a lengthy process associated with biochemical changes occurring in different sperm compartments. At the molecular level, cAMP-dependent phosphorylation triggers an increase in flagellar beating and initiates additional phosphorylation cascades leading to hyperactivation and preparation for the acrosome reaction [36, 37]. However, the identity and role of phosphorylated proteins has not been fully established. Toward this goal, we previously described a differential isotopic labeling strategy coupled with IMAC-based phosphopeptide enrichment and tandem mass spectrometry [8]. This approach allowed us to estimate the relative extent of phosphorylation on 42 different phosphopeptides from capacitated mouse sperm. In the present work, an additional phosphopeptide was identified using the same methodology. This phosphopeptide (R.RApSQGpSeR.A) corresponded to the radial spoke head protein Rsph6a.
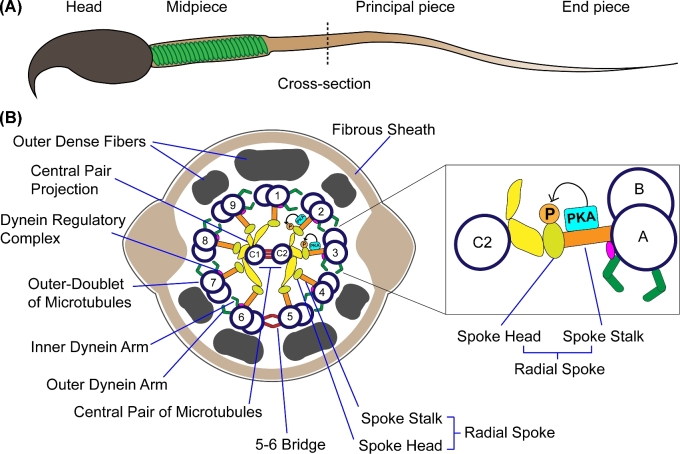
Radial spoke proteins form a multi-unit protein structure present in eukaryotic cilia and flagella [38]. Among these structures, the sperm flagellum is typically divided into three sections: the mid-piece, the principal piece, and the end piece (Figure 6A). These compartments have a central axoneme, which is a 9 + 2 structure consisting of a central pair of microtubules surrounded by nine peripheral microtubule doublets (Figure 6B) [39]. Throughout the mid- and principal pieces, the axoneme is surrounded by nine outer dense fibers (ODF). However, while the mid-piece ODF are wrapped in a helix of mitochondria, the principal piece ODF is covered by a fibrous sheath (shown in Figure 6B) [39]. In addition to the central pair and outer doublets, the axonemal structure has dynein motors and radial spoke proteins [38]. Dyneins are minus-ended microtubule motors responsible for translating the energy released by ATP hydrolysis into movement by active sliding of the microtubules. Radial spokes connect the central microtubule pair with the outer doublets throughout the axoneme and play an essential role in converting the dynein-dependent microtubule sliding into flagellar bending. The radial spoke structure is composed by at least 23 proteins, which are classified into two subfamilies, following their localization in the general spoke structure. Each spoke is T-shaped with an enlarged “head” projecting toward the axonemal central pair, and a thinner “stalk” which binds to the A microtubule of the outer doublets (Figure 6B). Studies using the flagellated algae C. reinhardtii provided evidence indicating that radial spoke proteins act as signal transducers that control protein phosphorylation and motility [40]. In addition, radial spoke defects have been shown in human patients presenting primary cilia dyskinesia, a pathology that causes respiratory problems and infertility due to immotile cilia and flagella [30, 41, 42].
Figure 6.
Model. (A) Schematic representation of the mouse sperm. The head and flagellar regions mid-piece, principal piece, and end piece are shown. A dotted line indicates the cross-section schematized in B. (B) Schematic representation of the principal piece cross-section. Fibrous sheath, outer dense fibers, and axonemal components are indicated. Numbers 1 to 9 indicate the microtubule doublet pair. C1 and C2 indicate the central pair of microtubules. Protein kinase A (PKA) is shown in the radial spoke stalk, and potential sites of phosphorylation of Rsph6a by PKA during sperm capacitation are indicated by a (P).
Although radial spoke morphology has many similarities across species, the proteins that form this structure have diverged during evolution [11]. In the case of Rsph6a, phylogenetic analysis revealed that this protein originated by duplication in a Therian common ancestor. This event gave rise to two paralogs, Rsph6a and Rsph4a, which produce two reciprocally monophyletic subtrees in the phylogeny (Supplemental Figure S1A, Wasabi tree and alignment). The outcome of this process was the acquisition of a novel first exon leading to the enlargement of the N-terminus of both proteins, approximately 200 residues longer than non-Therian species. This novel N-terminus shows poor levels of identity between both paralogs (Supplemental Figure S2).
After duplication, a new gene copy might acquire either a new function (neofunctionalization), a complementary partitioning of the original function between two duplicates (subfunctionalization), or pseudogenization (nonfunctionalization) by mutational decay [43, 44]. Rsph4a retained the canonical function, reflected by the fact that it is ubiquitously expressed in multiple tissues, and also because it is the homolog retained by marsupials. In contrast, Rsph6a is only present in placental mammals, its expression is restricted to the testes, and it has a novel Eutherian-specific function. The Rsph6a N-terminus may have modified the protein's original function, but its differential expression may have evolved due to modifications in transcriptional regulation. In marsupials, which have a homologous sequence but in which the ORF was lost, Rsph6a underwent mutations resulting in a nonfunctional pseudogene. Being a radial spoke protein, Rsph6a would be expected to be incorporated into the axoneme in spermatids during spermiogenesis. However, our RT-PCR and in situ hybridization data indicate that Rsph6a mRNA is expressed in spermatocytes. A similar expression pattern has also been observed for murine Tmem146, a gene that encodes for CATSPERδ, a protein subunit proposed to be required for proper assembly and/or transport of the CATSPER ion channel complex in mature sperm. One of the alternative splice variants of mouse CATSPERδ, encoded by Tmem146-s mRNA, only appeared at postnatal day 17 of testis development and its expression was restricted to spermatocytes [45]. More research will be necessary to understand when exactly Rsph6a protein is translated and how it is incorporated into the sperm flagellum.
The interaction between radial spoke proteins and the central microtubule pair is essential for modulation of ciliary and flagellar motility by cyclic nucleotides, phosphorylation, calcium, and calmodulin [11]. Coincidently, mouse sperm capacitation is regulated by molecular pathways that involve cAMP [46], phosphorylation events [29, 36, 47], and calcium [48]. We found Rsph6a to be phosphorylated more extensively in the N-terminal sequence R.RApSQGpSeR.A during capacitation. This sequence presents the consensus PKA substrate motif RRXS/TY [49] suggesting that Rsph6A is a PKA substrate. This hypothesis was supported by two-dimensional electrophoresis results indicating that the shift in Rsph6a IP observed in capacitated sperm was prevented by incubation with the PKA inhibitor H-89 and with LRE1, a recently described Adcy10 inhibitor [35]. In addition, bioinformatics analyses predict this Rsph6a N-terminal site to be a PKA consensus sequence. This analysis also predicts three additional phosphorylation sites for this protein, all of them containing consensus sequences for PKA phosphorylation. Altogether, these results suggest that Rsph6a is phosphorylated by PKA during sperm capacitation.
Another second messenger intrinsically connected to the regulation of flagellar and ciliary movement is Ca2+. Studies in Tetrahymena have shown that the Rsph6a and Rsph4a paralogs, p62 and p66, bind to Ca2+/Calmodulin columns [50]. Sequence comparisons indicate that at least one of the Ca2+/calmodulin-predicted binding sites for Tetrahymena has been conserved in the same position in both Rsph4a and Rsph6a (Supplemental Figure S2A, gray-boxed sequence). Intracellular Ca2+ concentrations modulate waveforms originated by axonemal bending in many flagellated eukaryotic systems including Paramecium [51], Chlamydomonas [52, 53], and sea urchin sperm [54]. In mammalian sperm, Ca2+ is involved in the regulation of hyperactivated movement [55]. Altogether, there is compelling evidence that Ca2+ ions play essential roles in the regulation of flagellar motility; however, the high number of Ca2+-binding proteins in eukaryotic flagella and cilia makes it difficult to assign particular roles to any given protein. Therefore, the most probable scenario is that multiple Ca2+ sensors, among them radial spoke proteins, are required for motility control [11].
Functional characterization of each of the radial spoke proteins will require loss-of-function studies such as the generation of knockout models or analysis of naturally occurring genetic mutations. In this regard, human ciliopathies have provided important insight into the role of axonemal proteins. In particular, Rsph1 [31], Rsph9, and Rsph4a [56] have been linked to primary ciliar dyskinesia, a disease characterized by defects in motile cilia. Although no other radial spoke proteins have been knocked out yet, specific axonemal defects have been mapped by eliminating other axonemal proteins (for review, see [38]). Many of these transgenic models have unique phenotypes in motile cilia and have been instrumental in elucidating radial spoke and central pair functions. However, in most of these knockout models, elimination of a particular protein affects the overall organization of the axoneme, making it difficult to determine the specific role of any given protein. In addition, these approaches are generally silent regarding signaling events involved in motility regulation. The use of newly discovered gene editing technologies such as Crispr/Cas9 [57] will allow the generation of point mutation, knock-in transgenic models that target specific amino acids in axonemal proteins. Identification of those sites that undergo post-translational modification in these proteins will be essential to determine which residues are key targets for further experimentation and mutation to elucidate the signaling pathways controlling capacitation and normal sperm function.
Supplementary data
Supplemental Figure S1. (A) Bayesian phylogenetic tree of a total of 91 RSPH coding sequences, representing all major chordates groups. Branches derived from nodes with posterior probability values under 0.7 are shown in light gray, while black branches depict nodes with posterior probabilities equal or over 0.7. The RSPH sequence of Ciona was set as outgroup. In Eutherian mammals RSPH4a and a produce two reciprocally monophyletic subtrees, reflecting a duplication event that can be traced back to the Therian ancestor (see http://wasabiapp.org:8000/?id=i0mphn). Marsupial RSPH6a sequences were excluded in this analysis due to coding sequence degeneration. The interactive version of the tree has been uploaded to the iTOL server (https://itol.embl.de/tree/2001689155162511505223012). (B) Phylogenetic tree indicative of the relationship between radial spoke proteins. The black boxes are representative of a duplication event; white boxes are representative of the loss of one copy. Representation of Rsph4a and Rsph6a exons is separated by space. White blocks are representative of 5'UTR and 3'UTR regions. Blocks in either red, blue, green, magenta, ochre, or cyan correspond to homologous codifying regions of exons 1, 2, 3, 4, 5, and 6 of the mouse Rsph4a protein. The yellow block represents the region of exon 1 from Rsph6a, conserved in all placental mammals. Gray blocks represent regions without homology using the ORF of mouse Rsph4. The asterisk (*) indicates partial homology of these exons with their respective mouse ones. Long exons are indicated with a double-forward slash symbol (//).
Supplemental Figure S2. (A) Analysis of the amino acid sequence homology between full-length mouse Rsph4a and Rsph6a. Red residues indicate conserved amino acids, which are signalized with an asterisk (*), while blue ones indicate amino acid similarity, and are signalized with a dot (.). Gray shaded region indicates Ca2+/calmodulin-predicted binding site for Rsph6a and Rsph4a at the same position (Supplemental Figure S2A, gray-boxed sequences). (B) Conservation plot showing the average similarity score at individual amino acid positions from multiple sequence alignments of Rsph4a and Rsph6a from major Vertebrates groups. The plots were generated with EMBOSS PlotCon using the EBLOSUM62 comparison matrix and a window size of 100.
Supplemental Figure S3. In situ analysis of Rsph6a and Tssk6 mRNA. (A)Rsph6a mRNA signal obtained in mouse testis sections when incubated with the corresponding sense (control) RNA probes. (B)Tssk6 mRNA in mouse testis sections when incubated with the corresponding sense (control) RNA probes. Scale bars, 50 μm.
Supplemental Figure S4. Production of Rsph6a isoforms as His-tagged recombinant proteins. BL21 E. coli cells containing Rsph6a inserts were induced with 1 mM IPTG at 37°C for 4 h. Cell pellet harvested from 1 mL of bacteria culture was directly lysed in 50 μL 5X sample buffer and loaded onto 8% SDS-PAGE. A representative set of western blots of the experiments repeated at least three times and probed with Anti-His antibody is shown. Arrows indicate expected protein sizes for recombinant I1 (left panel) and I2 (right panel).
Supplemental Figure S5. Validation of anti-Rsph6a antibody (E2). Western blot showing detection of Rsph6a in recombinants (A) and in mouse sperm extracts (B) when probed with E2 antibody alone or in the presence of blocking peptide P6. (C) Immunofluorescence showing localization of Rsph6a in mouse when probed with E2 antibody. (a, e) DIC images, (b) observed immunofluorescence with E2 alone or (f) when incubated with blocking peptide (E2-P6), (c) merged, (d) inset showing magnified image from (c). Scale bars, 20 μm.
Supplemental Figure S6. Rsph6a protein extraction and solubility analysis. For TritonX-100 extraction, sperm pellets were lysed in lysis buffer containing 1% TritonX-100, centrifuged, and separated into soluble (S) and insoluble (P) fractions. Separated fractions were then processed for western blotting as described in the Methods section. For sequential sample buffer extraction, sperm proteins were extracted in nonreducing (NR) condition; boiled, centrifuged, and soluble (S) and insoluble (P) fractions were separated. Proteins from insoluble fractions (P) were then further extracted with nonreducing (NR) SDS sample buffer (without added β-mercaptoethanol or DTT) or reducing (R) SDS sample buffer (with added 50 mM DTT). Proteins extracted in these conditions were then subjected to 8% SDS-PAGE for western blot analysis as described above in the Methods section. A representative western blot image of three independent experiments probed with E3 antibody is shown. For solubility controls, PVDF membranes were stripped and sequentially probed with anti-phosphotyrosine antibody (pY) for Hexokinase, and with anti-tubulin antibody, respectively.
Supplemental Table S1. In silico prediction of Rshp6a phosphorylation sites by PKA. Total phosphosites predicted by Netphos 3.1, and PKA-specific phosphorylation sites predicted with pkaPS and GPS, in human RSPH6A (left) or mouse Rsph6a (right). Gray boxes indicate positions detected by the three methods.
Acknowledgments
The authors would like to thank Dr Dominique Alfandari and Dr Genevieve Abbruzzese for their help with expression and purification of recombinant His-Rsph6a proteins, and to Dr Kimberly Tremblay and Dr Pablo Ortiz-Pineda for their advice on in situ hybridization technique. We also want to acknowledge the contribution of Quinn Murphy during the initial phase of this study.
Footnotes
Grant Support: This study was supported by a grant R01-HD-038082 (to P.E.V.) from the National Institutes of Health (NIH), USA.
Authors contribution
Contribution of each author towards completion of this article is as follows: AMS: experimental design and supervision, research compliance, writing and revision of manuscript; BP: experimental design, performed most of the experiments, writing and revision of manuscript; DC: phylogenetic analysis, in silico analysis of protein phosphorylation, writing and revision of manuscript; DT: performed some of in situ hybridization analysis, and revision of manuscript; JM: experimental design; revision of manuscript; JP: identification and analysis of mouse sperm phosphopeptides, revision of manuscript; MDP: identification and analysis of mouse sperm phosphopeptides, writing and revision of manuscript; MGG: experimental design and assistance with immunofluorescence and in situ hybridization imaging, writing and revision of manuscript; PEV: experimental design and supervision, writing and revision of manuscript, provided research funding.
References
- 1. Gervasi MG, Visconti PE. Molecular changes and signaling events occurring in spermatozoa during epididymal maturation. Andrology 2017; 5:204–218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gervasi MG, Visconti PE. Chang's meaning of capacitation: A molecular perspective. Mol Reprod Dev 2016; 83:860–874. [DOI] [PubMed] [Google Scholar]
- 3. Jha KN, Salicioni AM, Arcelay E, Chertihin O, Kumari S, Herr JC, Visconti PE. Evidence for the involvement of proline-directed serine/threonine phosphorylation in sperm capacitation. Mol Hum Reprod 2006; 12:781–789. [DOI] [PubMed] [Google Scholar]
- 4. Krapf D, Arcelay E, Wertheimer EV, Sanjay A, Pilder SH, Salicioni AM, Visconti PE. Inhibition of Ser/Thr phosphatases induces capacitation-associated signaling in the presence of Src kinase inhibitors. J Biol Chem 2010; 285:7977–7985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Stival C, La Spina FA, Baro Graf C, Arcelay E, Arranz SE, Ferreira JJ, Le Grand S, Dzikunu VA, Santi CM, Visconti PE, Buffone MG, Krapf D. Src kinase is the connecting player between Protein Kinase A (PKA) activation and hyperpolarization through SLO3 potassium channel regulation in mouse sperm. J Biol Chem 2015; 290:18855–18864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Porambo JR, Salicioni AM, Visconti PE, Platt MD. Sperm phosphoproteomics: historical perspectives and current methodologies. Expert Rev Proteomics 2012; 9:533–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ficarro S, Chertihin O, Westbrook VA, White F, Jayes F, Kalab P, Marto JA, Shabanowitz J, Herr JC, Hunt DF, Visconti PE. Phosphoproteome analysis of capacitated human sperm. J Biol Chem 2003; 278:11579–11589. [DOI] [PubMed] [Google Scholar]
- 8. Platt MD, Salicioni AM, Hunt DF, Visconti PE. Use of differential isotopic labeling and mass spectrometry to analyze capacitation-associated changes in the phosphorylation status of mouse sperm proteins. J Proteome Res 2009; 8:1431–1440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Baker MA. Proteomics of post-translational modifications of mammalian spermatozoa. Cell Tissue Res 2016; 363:279–287. [DOI] [PubMed] [Google Scholar]
- 10. Chung JJ, Shim SH, Everley RA, Gygi SP, Zhuang X, Clapham DE. Structurally distinct Ca2+ signaling domains of sperm flagella orchestrate tyrosine phosphorylation and motility. Cell 2014; 157:808–822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhu X, Liu Y, Yang P. Radial Spokes-A snapshot of the motility regulation, assembly, and evolution of cilia and flagella. Cold Spring Harb Perspect Biol 2017; 9:a028126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Alvau A, Battistone MA, Gervasi MG, Navarrete FA, Xu XR, Sanchez-Cardenas C, De la Vega-Beltran L, Da Ros VG, Greer PA, Darszon A, Krapf D, Salicioni AM et al. The tyrosine kinase FER is responsible for the capacitation-associated increase in tyrosine phosphorylation in murine sperm. Development 2016; 143:2325–2333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res 2010; 38:W695–W699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Sievers F, Wilm A, Dineen D, Gibson TJ, Karplus K, Li W, Lopez R, McWilliam H, Remmert M, Soding J, Thompson JD, Higgins DG. Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol Syst Biol 2014; 7:539–539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Veidenberg A, Medlar A, Loytynoja A. Wasabi: An integrated platform for evolutionary sequence analysis and data visualization. Mol Biol Evol 2016; 33:1126–1130. [DOI] [PubMed] [Google Scholar]
- 16. Huelsenbeck JP, Ronquist F. MRBAYES: Bayesian inference of phylogenetic trees. Bioinformatics 2001; 17:754–755. [DOI] [PubMed] [Google Scholar]
- 17. Ronquist F, Huelsenbeck JP. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics 2003; 19:1572–1574. [DOI] [PubMed] [Google Scholar]
- 18. Miller MA, Schwartz T, Pickett BE, He S, Klem EB, Scheuermann RH, Passarotti M, Kaufman S, O’Leary MA. A RESTful API for access to phylogenetic tools via the CIPRES science gateway. Evol Bioinform Online 2015; 11:43–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wallingford MC, Filkins R, Adams D, Walentuk M, Salicioni AM, Visconti PE, Mager J. Identification of a novel isoform of the leukemia-associated MLLT1 (ENL/LTG19) protein. Gene Expr Patterns 2015; 17:11–15. [DOI] [PubMed] [Google Scholar]
- 20. Li Y, Sosnik J, Brassard L, Reese M, Spiridonov NA, Bates TC, Johnson GR, Anguita J, Visconti PE, Salicioni AM. Expression and localization of five members of the testis-specific serine kinase (Tssk) family in mouse and human sperm and testis. Mol Hum Reprod 2011; 17:42–56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Abbruzzese G, Becker SF, Kashef J, Alfandari D. ADAM13 cleavage of cadherin-11 promotes CNC migration independently of the homophilic binding site. Dev Biol 2016; 415:383–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Arnau J, Lauritzen C, Pedersen J. Cloning strategy, production and purification of proteins with exopeptidase-cleavable His-tags. Nat Protoc 2006; 1:2326–2333. [DOI] [PubMed] [Google Scholar]
- 23. Mendelsohn C, Batourina E, Fung S, Gilbert T, Dodd J. Stromal cells mediate retinoid-dependent functions essential for renal development. Development 1999; 126:1139–1148. [DOI] [PubMed] [Google Scholar]
- 24. Arcelay E, Salicioni AM, Wertheimer E, Visconti PE. Identification of proteins undergoing tyrosine phosphorylation during mouse sperm capacitation. Int J Dev Biol 2008; 52:463–472. [DOI] [PubMed] [Google Scholar]
- 25. Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J Mol Biol 1999; 294:1351–1362. [DOI] [PubMed] [Google Scholar]
- 26. Blom N, Sicheritz-Ponten T, Gupta R, Gammeltoft S, Brunak S. Prediction of post-translational glycosylation and phosphorylation of proteins from the amino acid sequence. Proteomics 2004; 4:1633–1649. [DOI] [PubMed] [Google Scholar]
- 27. Neuberger G, Schneider G, Eisenhaber F. pkaPS: prediction of protein kinase A phosphorylation sites with the simplified kinase-substrate binding model. Biol Direct 2007; 2:1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xue Y, Ren J, Gao X, Jin C, Wen L, Yao X. GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol Cell Proteomics 2008; 7:1598–1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Visconti PE, Bailey JL, Moore GD, Pan D, Olds-Clarke P, Kopf GS. Capacitation of mouse spermatozoa. I. Correlation between the capacitation state and protein tyrosine phosphorylation. Development 1995; 121:1129–1137. [DOI] [PubMed] [Google Scholar]
- 30. Eriksson M, Ansved T, Anvret M, Carey N. A mammalian radial spokehead-like gene, RSHL1, at the myotonic dystrophy-1 locus. Biochem Biophys Res Commun 2001; 281:835–841. [DOI] [PubMed] [Google Scholar]
- 31. Kott E, Legendre M, Copin B, Papon JF, Dastot-Le Moal F, Montantin G, Duquesnoy P, Piterboth W, Amram D, Bassinet L, Beucher J, Beydon N et al. Loss-of-function mutations in RSPH1 cause primary ciliary dyskinesia with central-complex and radial-spoke defects. Am J Hum Genet 2013; 93:561–570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Griswold MD. Spermatogenesis: The commitment to meiosis. Physiol Rev 2016; 96:1–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Jha KN, Tripurani SK, Johnson GR. TSSK6 is required for gammaH2AX formation and the histone-to-protamine transition during spermiogenesis. J Cell Sci 2017; 130:1835–1844. [DOI] [PubMed] [Google Scholar]
- 34. Garrison JC, Wagner JD. Glucagon and the Ca2+-linked hormones angiotensin II, norepinephrine, and vasopressin stimulate the phosphorylation of distinct substrates in intact hepatocytes. J Biol Chem 1982; 257:13135–13143. [PubMed] [Google Scholar]
- 35. Ramos-Espiritu L, Kleinboelting S, Navarrete FA, Alvau A, Visconti PE, Valsecchi F, Starkov A, Manfredi G, Buck H, Adura C, Zippin JH, van den Heuvel J et al. Discovery of LRE1 as a specific and allosteric inhibitor of soluble adenylyl cyclase. Nat Chem Biol 2016; 12:838–844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Visconti PE, Johnson LR, Oyaski M, Fornes M, Moss SB, Gerton GL, Kopf GS. Regulation, localization, and anchoring of protein kinase A subunits during mouse sperm capacitation. Dev Biol 1997; 192:351–363. [DOI] [PubMed] [Google Scholar]
- 37. Nolan MA, Babcock DF, Wennemuth G, Brown W, Burton KA, McKnight GS. Sperm-specific protein kinase A catalytic subunit Calpha2 orchestrates cAMP signaling for male fertility. Proc Natl Acad Sci USA 2004; 101:13483–13488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Inaba K. Sperm flagella: comparative and phylogenetic perspectives of protein components. Mol Hum Reprod 2011; 17:524–538. [DOI] [PubMed] [Google Scholar]
- 39. Lindemann CB, Lesich KA. Functional anatomy of the mammalian sperm flagellum. Cytoskeleton 2016; 73:652–669. [DOI] [PubMed] [Google Scholar]
- 40. Yang P, Yang C, Sale WS. Flagellar radial spoke protein 2 is a calmodulin binding protein required for motility in Chlamydomonas reinhardtii. Eukaryotic Cell 2004; 3:72–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sturgess JM, Chao J, Wong J, Aspin N, Turner JA. Cilia with defective radial spokes: a cause of human respiratory disease. N Engl J Med 1979; 300:53–56. [DOI] [PubMed] [Google Scholar]
- 42. Knowles MR, Daniels LA, Davis SD, Zariwala MA, Leigh MW. Primary ciliary dyskinesia. Recent advances in diagnostics, genetics, and characterization of clinical disease. Am J Respir Crit Care Med 2013; 188:913–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Innan H, Kondrashov F. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet 2010; 11:97–108. [DOI] [PubMed] [Google Scholar]
- 44. Innan H. Population genetic models of duplicated genes. Genetica 2009; 137:19–37. [DOI] [PubMed] [Google Scholar]
- 45. Chung JJ, Navarro B, Krapivinsky G, Krapivinsky L, Clapham DE. A novel gene required for male fertility and functional CATSPER channel formation in spermatozoa. Nat Commun 2011; 2:153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Wertheimer E, Krapf D, de la Vega-Beltran JL, Sanchez-Cardenas C, Navarrete F, Haddad D, Escoffier J, Salicioni AM, Levin LR, Buck J, Mager J, Darszon A et al. Compartmentalization of distinct cAMP signaling pathways in mammalian sperm. J Biol Chem 2013; 288:35307–35320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Visconti PE, Moore GD, Bailey JL, Leclerc P, Connors SA, Pan D, Olds-Clarke P, Kopf GS. Capacitation of mouse spermatozoa. II. Protein tyrosine phosphorylation and capacitation are regulated by a cAMP-dependent pathway. Development 1995; 121:1139–1150. [DOI] [PubMed] [Google Scholar]
- 48. Navarrete FA, Garcia-Vazquez FA, Alvau A, Escoffier J, Krapf D, Sanchez-Cardenas C, Salicioni AM, Darszon A, Visconti PE. Biphasic role of calcium in mouse sperm capacitation signaling pathways. J Cell Physiol 2015; 230:1758–1769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pearson RB, Kemp BE. Protein kinase phosphorylation site sequences and consensus specificity motifs: tabulations. Methods Enzymol 1991; 200:62–81. [DOI] [PubMed] [Google Scholar]
- 50. Ueno H, Iwataki Y, Numata O. Homologues of radial spoke head proteins interact with Ca2+/calmodulin in Tetrahymena cilia. J Biochem 2006; 140:525–533. [DOI] [PubMed] [Google Scholar]
- 51. Nakaoka Y, Tanaka H, Oosawa F. Ca2+-dependent regulation of beat frequency of cilia in Paramecium. J Cell Sci 1984; 65:223–231. [DOI] [PubMed] [Google Scholar]
- 52. Kamiya R, Witman GB. Submicromolar levels of calcium control the balance of beating between the two flagella in demembranated models of Chlamydomonas. J Cell Biol 1984; 98:97–107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Bessen M, Fay RB, Witman GB. Calcium control of waveform in isolated flagellar axonemes of Chlamydomonas. J Cell Biol 1980; 86:446–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Wood CD, Nishigaki T, Furuta T, Baba SA, Darszon A. Real-time analysis of the role of Ca(2+) in flagellar movement and motility in single sea urchin sperm. J Cell Biol 2005; 169:725–731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Chang H, Suarez SS. Two distinct Ca(2+) signaling pathways modulate sperm flagellar beating patterns in mice. Biol Reprod 2011; 85:296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Castleman VH, Romio L, Chodhari R, Hirst RA, de Castro SC, Parker KA, Ybot-Gonzalez P, Emes RD, Wilson SW, Wallis C, Johnson CA, Herrera RJ et al. Mutations in radial spoke head protein genes RSPH9 and RSPH4A cause primary ciliary dyskinesia with central-microtubular-pair abnormalities. Am J Hum Genet 2009; 84:197–209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 2012; 337:816–821. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.