Abstract
TMA20 (MCT-1), TMA22 (DENR) and TMA64 (eIF2D) are eukaryotic translation factors involved in ribosome recycling and re-initiation. They operate with P-site bound tRNA in post-termination or (re-)initiation translation complexes, thus participating in the removal of 40S ribosomal subunit from mRNA stop codons after termination and controlling translation re-initiation on mRNAs with upstream open reading frames (uORFs), as well as de novo initiation on some specific mRNAs. Here we report ribosomal profiling data of S.cerevisiae strains with individual deletions of TMA20, TMA64 or both TMA20 and TMA64 genes. We provide RNA-Seq and Ribo-Seq data from yeast strains grown in the rich YPD or minimal SD medium. We illustrate our data by plotting differential distribution of ribosomal-bound mRNA fragments throughout uORFs in 5′-untranslated region (5′ UTR) of GCN4 mRNA and on mRNA transcripts encoded in MAT locus in the mutant and wild-type strains, thus providing a basis for investigation of the role of these factors in the stress response, mating and sporulation. We also document a shift of transcription start site of the APC4 gene which occurs when the neighboring TMA64 gene is replaced by the standard G418-resistance cassette used for the creation of the Yeast Deletion Library. This shift results in dramatic deregulation of the APC4 gene expression, as revealed by our Ribo-Seq data, which can be probably used to explain strong genetic interactions of TMA64 with genes involved in the cell cycle and mitotic checkpoints. Raw RNA-Seq and Ribo-Seq data as well as all gene counts are available in NCBI Gene Expression Omnibus (GEO) repository under GEO accession GSE122039 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122039).
Specifications table
| Subject area | Biology |
| More specific subject area | Protein Synthesis, Translational Control, Ribosome, Bioinformatics, Transcriptomics, Translatomics |
| Type of data | Table and figures |
| How data was acquired | Ribosome profiling and RNA-Seq of wild-type or knockout yeast strains were performed. Sequences were obtained using Illumina HiSeq. 2000. |
| Data format | Raw and analyzed |
| Experimental factors | Saccharomyces cerevisiae BY4741 wild-type strain and BY4741-based strains with TMA20, TMA64 or both TMA20 and TMA64 knockouts were maintained in rich (YPD) or minimal (SD) media. |
| Experimental features | In the mid-log exponential phase, yeast cells were pretreated with cycloheximide and collected. cDNA libraries of ribosome-bound mRNA and total mRNA from wild-type and knockout strains were performed as described previously [1]. Sequenced reads were trimmed, read mapping and counting was performed. |
| Data source location | Moscow State University (Moscow, Russia) |
| Data accessibility | Analyzed data is presented in the article. Raw RNA-Seq and Ribo-Seq data as well as all gene counts are available in NCBI Gene Expression Omnibus (GEO) repository under GEO accession GSE122039 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122039). |
| Related research article | N/A |
Value of the data
-
•
The data provides a gene expression landscape of yeast strains lacking TMA20 and/or TMA64 proteins, which are orthologous to mammalian translation factors MCT-1 and eIF2D, thus expanding our knowledge about individual functional roles of these two translation factors in a living cell.
-
•
An abnormal translation of the MATa2 mRNA derived from the MAT locus of MATa yeast strain is detected, which can be used for explanation of sporulation defects previously detected in the TMA64 deletion strain.
-
•
Quantitative Ribo-Seq data provides essential information of translational changes in the knockout strains including altered uORFs translation in 5’ UTR of mRNA encoding important transcription regulator GCN4, thus providing a basis for investigating the role of these proteins in the stress response.
-
•
The RNA-Seq data highlights transcription abnormalities within the APC4 gene locus, caused by replacement of the adjacent TMA64 gene by the standard G418 or HYG resistance cassettes commonly used for generating gene deletions, which can be probably used to explain previously observed strong genetic interactions of TMA64 with genes involved in the cell cycle and mitotic checkpoints.
-
•
The deep sequenced Ribo-Seq and RNA-Seq are applicable for detailed bioinformatics analysis of translation events, such as prediction of alternative open reading frames.
1. Data
In this study we present ribosome profiling data generated from the wild-type BY4741 S.cerevisiae strain and strains lacking translation factors TMA20 (MCT-1), TMA64 (eIF2D) or both of them at the same time. Information on all performed experiments is shown in Table 1. Raw Ribo-Seq and RNA-Seq data are available online in the NCBI Gene Expression Omnibus repository (GEO accession: GSE122039, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122039). Supplementary Table S1 and Supplementary Figure S1 contain analyzed NGS data, as described below. Examples of differentially translated and transcribed genes in wild-type and knockout strains are presented in Fig. 1, Fig. 2, Fig. 3.
Table 1.
Summary of datasets obtained in the study.
| Sample | Sample name | Yeast strain name | Yeast strain genotype | Growth media | Sample type |
|---|---|---|---|---|---|
| 1 | wt1_ribo | wt | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 | YPD | Ribo-Seq |
| 2 | wt1_rna | YPD | RNA-Seq | ||
| 3 | wt2_ribo | YPD | Ribo-Seq | ||
| 4 | wt2_rna | YPD | RNA-Seq | ||
| 5 | wt_sd_ribo | SD | Ribo-Seq | ||
| 6 | wt_sd_rna | SD | RNA-Seq | ||
| 7 | tma20_ribo | Δtma20 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tma20Δ:KanMX4 | YPD | RiboSeq |
| 8 | tma20_rna | YPD | RNA-Seq | ||
| 9 | tma20_sd_ribo | SD | Ribo-Seq | ||
| 10 | tma20_sd_rna | SD | RNA-Seq | ||
| 11 | tma64_ribo | Δtma64 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tma64Δ:KanMX4 | YPD | Ribo-Seq |
| 12 | tma64_rna | YPD | RNA-Seq | ||
| 13 | tma64_sd_ribo | SD | Ribo-Seq | ||
| 14 | tma64_sd_rna | SD | RNA-Seq | ||
| 15 | tma20tma64_ribo | Δtma20Δtma64 | MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0 tma64Δ:HygMX4 tma20Δ:KanMX4 | YPD | Ribo-Seq |
| 16 | tma20tma64_rna | YPD | RNA-Seq | ||
| 17 | tma20tma64_sd_ribo | SD | Ribo-Seq | ||
| 18 | tma20tma64_sd_rna | SD | RNA-Seq |
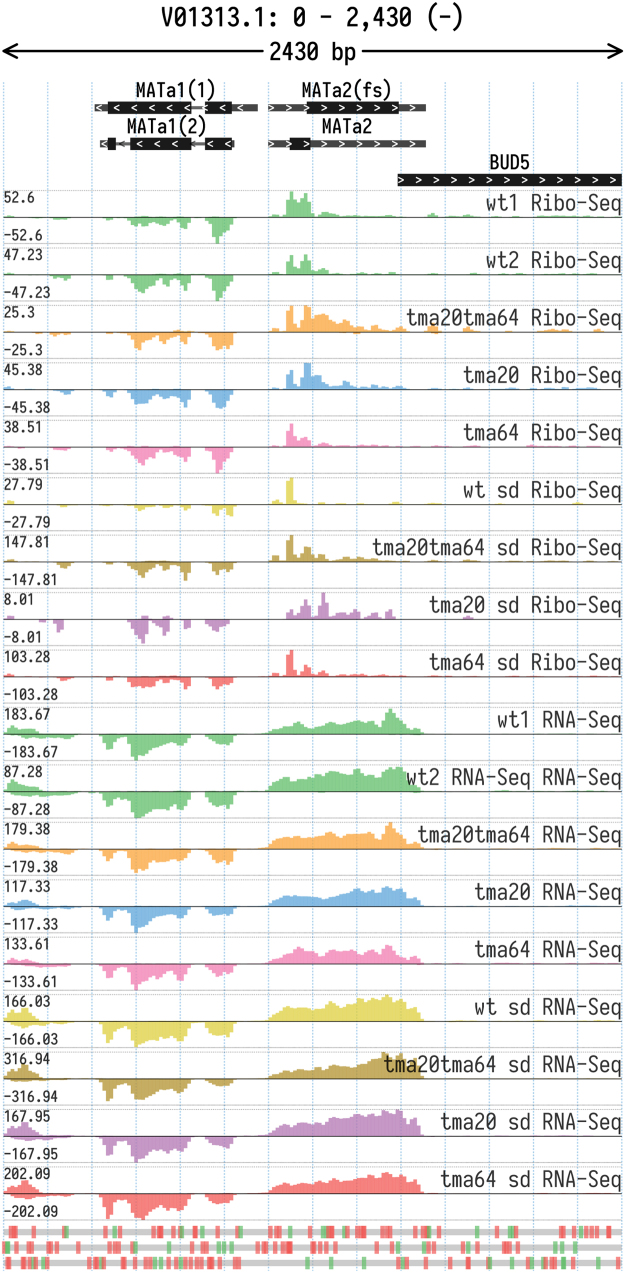
Fig. 1.
Ribo-Seq and RNA-Seq coverage of MATa locus in the studied yeast strains. According to Ribo-Seq signals, MATa2-2 ORF is probably translated in mutant strains. The Y axis tracks show total read coverage (positive and negative values correspond to the coverage of the direct and reverse complementary strands respectively).
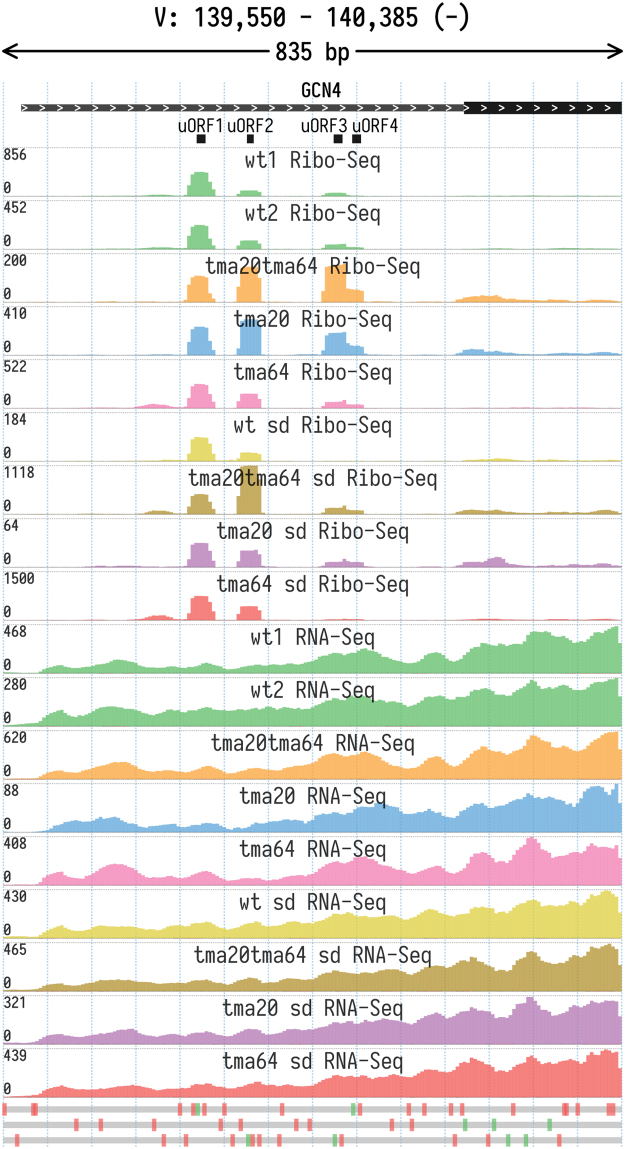
Fig. 2.
Ribo-Seq and RNA-Seq coverage of the 5′ UTR of the yeast GCN4 gene. Ribo-Seq profiles show ribosome occupancy at different uORFs that regulate translation of the main coding region. The Y axis tracks show total read coverage (positive and negative values correspond to the coverage of the forward and reverse complementary strands respectively).
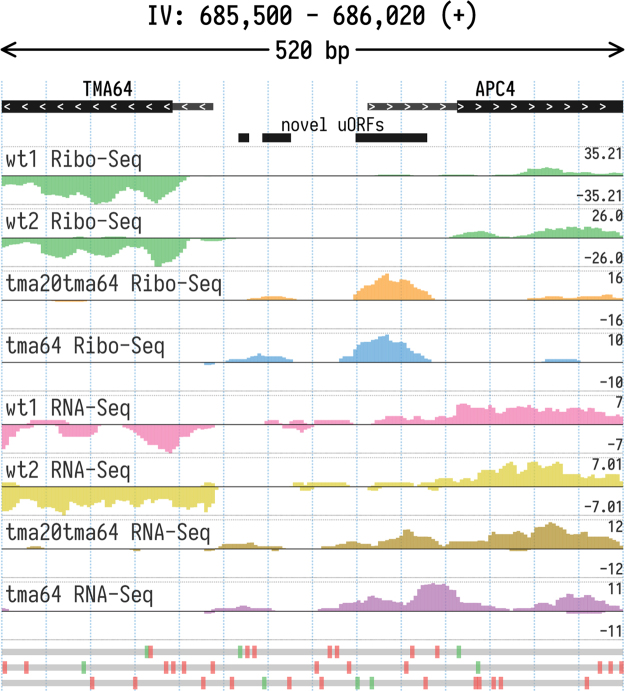
Fig. 3.
Ribo-Seq and RNA-Seq coverage of the region between the TMA64 and APC4 genes. Data suggests that strains with TMA64 knockout exhibit extended 5′ UTR of APC4 gene leading to translation of novel uORFs. The Y axis tracks show total read coverage (positive and negative values correspond to the coverage of the forward and reverse complementary strands respectively).
In contrast to a dataset previously obtained in a study by Young et al. [2], here we present ribosome profiling data not only for double knockout yeast strains, but also for strains with individual deletions of TMA20 and TMA64. This allows studying transcriptional and translational changes caused specifically by the absence of individual translation factors TMA20 (MCT-1) or TMA64 (eIF2D). Due to cycloheximide addition to yeast culture medium before harvesting the cells, distribution of mapped reads from Ribo-Seq sets may slightly vary from data described by Young et al. According to a previous study [3], use of the inhibitor in such a fashion is likely to cause blurring of local density effects, but also strengthens Ribo-Seq signals at translation initiation sites, facilitating analysis of ribosome distribution over uORFs.
2. Experimental design, materials, and methods
2.1. Cell maintenance and cDNA libraries preparation
RNA-Seq and Ribo-Seq сDNA libraries were prepared from total RNA samples or ribosome-bound RNA samples, respectively, for both the wild-type BY4741 yeast strain and three knockout strains (with individually deleted TMA20 or TMA64 genes, or with a double deletion of TMA20 and TMA64, hereafter referred as wt, Δtma20, Δtma64 and ΔΔtma20tma64, respectively). The libraries were sequenced, resulting in 9 RNA-Seq and 9 Ribo-Seq data sets. Table 1 summarizes information about all of the sequencing experiments.
The experimental procedure in general followed the ribosome profiling protocol described in [1]. Briefly, yeast cells were grown to an exponential phase in either rich YPD (1% yeast extract, 2% peptone, 2% glucose) or minimal SD (0,67% YNB w/o amino acids with ammonium sulfate, 2% glucose, complete amino acid supplementation) media. Cycloheximide was added to yeast media to a final concentration of 100 µg/ml and growth was continued for 3 more minutes; then cells were harvested by filtration, resuspended in polysome lysis buffer (20 mM Tris pH 8.0, 140 mM KCl, 1.5 mM MgCl2, 100 g/ml cycloheximide, 1% Triton), flash frozen in liquid nitrogen and homogenized by grinding. Then a portion of each cell lysate was used for total RNA isolation, while another part was treated with RNase I for polysome disassembly, applied to a sucrose gradient for fractionation, followed by isolation of a monosome fraction and extraction of ribosome-protected mRNA fragments for ribosome profiling. mRNA was isolated using Oligo(dT) beads and ribosome-bound RNA was isolated from sucrose fractions using acidic-phenol extraction. Further ribosome profiling and RNA-Seq library preparations were performed as described previously [1]. Two biological replicates indicated as WT1 and WT2 were performed for wild-type strain maintained in YPD.
2.2. Sequence data processing and analysis
Reads were trimmed with cutadapt v1.18 [4] and mapped to the Saccharomyces_cerevisiae.R64-1-1.90 (Ensembl) genome assembly using the respective genome annotation. The read mapping and counting were performed with STAR v2.5.3a [5]. The genomic signal plots (Fig. 1, Fig. 2, Fig. 3) were generated by the svist4get software [6] using bedGraph profiles constructed from bam alignments by samtools and bedtools [7], [8]. Transcription start site coordinates for MAT locus [9] and GCN4, TMA64 and APC4 genes [10] were used for 5’ UTR mapping in Fig. 2, Fig. 3.
3. Data analysis
3.1. Ribosome profiling of yeast strains lacking TMA20 and/or TMA64 genes
Translation factor TMA64 and homologs of its N- and C-terminal regions, TMA20 and TMA22 respectively (eIF2D, MCT-1, and DENR in mammals) are proteins involved in translation termination, re-initiation, and ribosome recycling. Initially, eIF2D and heterodimer MCT-1•DENR were assumed to provide a non-canonical translation initiation pathway as they facilitate GTP-independent delivery of Met-tRNAiMet and some elongator tRNAs to the 40S ribosomal P-site [11], [12]. In addition, in vitro and in vivo studies demonstrated that TMA64/eIF2D, TMA20/MCT-1, and TMA22/DENR are able to promote the post-terminational tRNA and mRNA release from the 40S ribosomal subunit both in yeast and mammals [2], [13]. The absence of these factors, together with the 40S recycling failure, led to deregulated translation re-initiation downstream of both short and full-size translated open reading frames in different organisms [2], [14], [15], [16].
The C-terminal regions of TMA64/eIF2D and TMA22/DENR contain the SUI1 domain, which is also present in the translation factor SUI1/eIF1. Structural data indicate that the SUI1 domains of all three factors have similar positions in the P-site of the 40S ribosomal subunit with a conserved β-loop protruding toward a codon-anticodon duplex formed by mRNA and a P-site tRNA [17], [18]. In accordance with biochemical data, this suggests that during recycling, TMA64/eIF2D and the heterodimer TMA20•TMA22 (MCT-1•DENR) may operate in a manner similar to SUI1/eIF1 in translation initiation, or control initiator tRNA access to re-initiating ribosomal complexes after uORF translation.
Raw and analyzed Ribo-Seq and RNA-Seq data sets for wild-type, individual Δtma20 and Δtma64, as well as double ΔΔtma20tma64 knockout yeast strains were obtained and uploaded into the NCBI Gene Expression Omnibus repository (GEO accession: GSE122039, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122039). The initial analysis revealed that they included deep RNA-Seq and Ribo-Seq data with more than 10 million uniquely mapped and counted reads within gene CDS. In general, in a half of the samples, ~50% of the total library length was composed of uniquely mapped reads. Across all samples, 60 to 90% of them were located within annotated CDS and included in the gene counts. Metagene start- and stop-centric profiles for Ribo-Seq data exhibited clear triplet periodicity (Supplementary Fig. S1). Supplementary Table S1 provides an overview of mapped reads as well as gene-level read counts for all experiments and descriptive statistics on the generated sequencing data.
3.2. Examples of data illustrating differential transcription and translation in wt and knockout strains
Two different gene cassettes, MATα and MATa, either of which can be present in the MAT locus of S.cerevisiae genome, define the mating type of yeast. Each mating-specific cassette encodes two transcripts directed from the opposite DNA strands by a shared bidirectional promoter: either MATa1 and MATa2 in a-type strains, or MATα1 and MATα2 in α-type strains (Fig. 1) [9]. All of the transcripts except MATa2 encode functional proteins – transcription factors that determine mating type or diploid phenotype (reviewed in [19], [20]). While MATa2 is considered to be non-functional [21], [22], it nevertheless contains two ORFs (Fig. 1), presumably originating from an original coding region with similarity to MATα2 via an internal frameshift [9], [23]. The second ORF could still encode a remarkably conserved amino acid sequence with a high similarity to a portion of MATα2 [9], [24] that represents its DNA-binding domain [25], [26]. The corresponding protein (MATa2-2) could compete with MATα2 for DNA binding or even have its own transcription factor activity [27]. However, its synthesis should be inhibited by presence of the first ORF in the MATa2 mRNA, which can be regarded as an uORF for the MATa2-2 coding region. Since TMA20 and TMA64 knockout strains have an upregulated translation re-initiation and/or readthrough activities [2], [16], [28], it was interesting to illustrate our ribosome profiling data with a footprint coverage of MATa locus present in BY4741 strain derivatives. As the sequenced S288C strain is MATα, Ribo-Seq and RNA-Seq reads were re-mapped to MATa locus sequence taken from GenBank (accession number V01313.1)[9]. Fig. 1 provides data on Ribo-Seq and RNA-Seq read coverage of MATa locus of the studied yeast strains. This data can be used to explain the role played by TMA20 and TMA64 translation factors in mating and sporulation programs [29], [30], [31], [32].
Another example involves GCN4, the global transcriptional regulator, which is activated during amino acid starvation. Expression of the GCN4 mRNA is controlled by a peculiar mechanism based on differential translation re-initiation on four short uORFs in its 5’ UTR (reviewed in [33]). Fig. 2 shows the 5’ proximal region of the GCN4 transcript, with differential ribosome footprint coverage of uORFs in different strains. Our data can be used for further investigation of TMA20 and TMA64 roles in uORF-mediated translational control of stress response.
APC4, the gene encoding a subunit of anaphase-promoting complex, is located in the same genetic locus as TMA64 and shares a 238-bp promoter region with it. The corresponding mRNAs are synthesized from opposite DNA strands. In the Δtma64 and ΔΔtma20tma64 strains the TMA64 coding sequence was replaced with G-418 or hygromycin resistance gene cassettes (KanMX or HygMX), respectively. Fig. 3 shows differential RNA-Seq coverage of the 238 bp region, flanked by segments of the APC4 and TMA64 coding regions or KanMX/HygMX cassettes, in different yeast strains. This data may likely account for the observed strong genetic interactions of TMA64 with genes involved in the cell cycle and mitotic checkpoints [34] and cell cycle abnormalities of TMA64 knockout strains [35].
Acknowledgments
We are grateful to Alan Hinnebusch (NIH) and Nicholas Guydosh (NIH) for providing the knockout yeast strains and helpful discussion. Ribosome profiling and library sequencing were supported by the Russian Science Foundation (grant RSF 18-14-00291 to S.E.D.). Adaptation of the visualization software was supported by the Russian Foundation for Basic Research (grant 18-34-20024 to I.V.K.). Bioinformatics pipeline was supported by the Program of Fundamental Research for State Academies for 2013–2020 years (No. 01201363825).
Footnotes
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2019.103701.
Supplementary data associated with this article can be found in the online version at doi:10.1016/j.dib.2019.103701.
Contributor Information
Desislava S. Makeeva, Email: desislava@belozersky.msu.ru.
Ivan V. Kulakovskiy, Email: ivan.kulakovskiy@gmail.com.
Sergey E. Dmitriev, Email: sergey.dmitriev@belozersky.msu.ru.
Transparency document. Supplementary material
Supplementary material
.
Appendix A. Supplementary material
Supplementary material
.
Supplementary material
.
References
- 1.Ingolia N.T., Ghaemmaghami S., Newman J.R., Weissman J.S. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–223. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Young D.J., Makeeva D.S., Zhang F., Anisimova A.S., Stolboushkina E.A., Ghobakhlou F., Shatsky I.N., Dmitriev S.E., Hinnebusch A.G., Guydosh N.R. Tma64/eIF2D, Tma20/MCT-1, and Tma22/DENR recycle post-termination 40S subunits in vivo. Mol. Cell. 2018;71:761–774. doi: 10.1016/j.molcel.2018.07.028. (e765) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gerashchenko M.V., Gladyshev V.N. Translation inhibitors cause abnormalities in ribosome profiling experiments. Nucleic Acids Res. 2014;42:e134. doi: 10.1093/nar/gku671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Martin M. Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.journal. 2011;2011(17):3. [Google Scholar]
- 5.Dobin A., Davis C.A., Schlesinger F., Drenkow J., Zaleski C., Jha S., Batut P., Chaisson M., Gingeras T.R. STAR: ultrafast universal RNA-seq aligner. Bioinformatics. 2013;29:15–21. doi: 10.1093/bioinformatics/bts635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.A.A. Egorov, svist4get: a simple visualization tool for genomic tracks from sequencing experiments, (2019): https://bitbucket.org/artegorov/svist4get/ [DOI] [PMC free article] [PubMed]
- 7.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Quinlan A.R., Hall I.M. BEDTools: a flexible suite of utilities for comparing genomic features. Bioinformatics. 2010;26:841–842. doi: 10.1093/bioinformatics/btq033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Astell C.R., Ahlstrom-Jonasson L., Smith M., Tatchell K., Nasmyth K.A., Hall B.D. The sequence of the DNAs coding for the mating-type loci of Saccharomyces cerevisiae. Cell. 1981;27:15–23. doi: 10.1016/0092-8674(81)90356-1. [DOI] [PubMed] [Google Scholar]
- 10.Miura F., Kawaguchi N., Sese J., Toyoda A., Hattori M., Morishita S., Ito T. A large-scale full-length cDNA analysis to explore the budding yeast transcriptome. Proc. Natl. Acad. Sci. USA. 2006;103:17846–17851. doi: 10.1073/pnas.0605645103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dmitriev S.E., Terenin I.M., Andreev D.E., Ivanov P.A., Dunaevsky J.E., Merrick W.C., Shatsky I.N. GTP-independent tRNA delivery to the ribosomal P-site by a novel eukaryotic translation factor. J Biol. Chem. 2010;285:26779–26787. doi: 10.1074/jbc.M110.119693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Skabkin M.A., Skabkina O.V., Dhote V., Komar A.A., Hellen C.U., Pestova T.V. Activities of Ligatin and MCT-1/DENR in eukaryotic translation initiation and ribosomal recycling. Genes Dev. 2010;24:1787–1801. doi: 10.1101/gad.1957510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Skabkin M.A., Skabkina O.V., Hellen C.U., Pestova T.V. Reinitiation and other unconventional post-termination events during eukaryotic translation. Mol. Cell. 2013;51:249–264. doi: 10.1016/j.molcel.2013.05.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Schleich S., Strassburger K., Janiesch P.C., Koledachkina T., Miller K.K., Haneke K., Cheng Y.S., Kuchler K., Stoecklin G., Duncan K.E., Teleman A.A. DENR-MCT-1 promotes translation re-initiation downstream of uORFs to control tissue growth. Nature. 2014;512:208–212. doi: 10.1038/nature13401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schleich S., Acevedo J.M., Clemm von Hohenberg K., Teleman A.A. Identification of transcripts with short stuORFs as targets for DENR*MCTS1-dependent translation in human cells. Sci. Rep. 2017;7:3722. doi: 10.1038/s41598-017-03949-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Torrance V., Lydall D. Overlapping open reading frames strongly reduce human and yeast STN1 gene expression and affect telomere function. PLoS Genet. 2018;14:e1007523. doi: 10.1371/journal.pgen.1007523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Weisser M., Schafer T., Leibundgut M., Bohringer D., Aylett C.H.S., Ban N. Structural and functional insights into human re-initiation complexes. Mol. Cell. 2017;67:447–456. doi: 10.1016/j.molcel.2017.06.032. (e447) [DOI] [PubMed] [Google Scholar]
- 18.Lomakin I.B., Stolboushkina E.A., Vaidya A.T., Zhao C., Garber M.B., Dmitriev S.E., Steitz T.A. Crystal structure of the human ribosome in complex with DENR-MCT-1. Cell Rep. 2017;20:521–528. doi: 10.1016/j.celrep.2017.06.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Haber J.E. Mating-type gene switching in Saccharomyces cerevisiae. Annu. Rev. Genet. 1998;32:561–599. doi: 10.1146/annurev.genet.32.1.561. [DOI] [PubMed] [Google Scholar]
- 20.Haber J.E. Mating-type genes and MAT switching in Saccharomyces cerevisiae. Genetics. 2012;191:33–64. doi: 10.1534/genetics.111.134577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatchell K., Nasmyth K.A., Hall B.D., Astell C., Smith M. in vitro mutation analysis of the mating-type locus in yeast. Cell. 1981;27:25–35. doi: 10.1016/0092-8674(81)90357-3. [DOI] [PubMed] [Google Scholar]
- 22.Dranginis A.M. Regulation of STA1 gene expression by MAT during the life cycle of Saccharomyces cerevisiae. Mol. Cell Biol. 1989;9:3992–3998. doi: 10.1128/mcb.9.9.3992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gordon J.L., Armisen D., Proux-Wera E., OhEigeartaigh S.S., Byrne K.P., Wolfe K.H. Evolutionary erosion of yeast sex chromosomes by mating-type switching accidents. Proc. Natl. Acad. Sci. USA. 2011;108:20024–20029. doi: 10.1073/pnas.1112808108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kellis M., Patterson N., Endrizzi M., Birren B., Lander E.S. Sequencing and comparison of yeast species to identify genes and regulatory elements. Nature. 2003;423:241–254. doi: 10.1038/nature01644. [DOI] [PubMed] [Google Scholar]
- 25.Li T., Jin Y., Vershon A.K., Wolberger C. Crystal structure of the MATa1/MATalpha2 homeodomain heterodimer in complex with DNA containing an A-tract. Nucleic Acids Res. 1998;26:5707–5718. doi: 10.1093/nar/26.24.5707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sauer R.T., Smith D.L., Johnson A.D. Flexibility of the yeast alpha 2 repressor enables it to occupy the ends of its operator, leaving the center free. Genes Dev. 1988;2:807–816. doi: 10.1101/gad.2.7.807. [DOI] [PubMed] [Google Scholar]
- 27.Hall M.N., Johnson A.D. Homeo domain of the yeast repressor alpha 2 is a sequence-specific DNA-binding domain but is not sufficient for repression. Science. 1987;237:1007–1012. doi: 10.1126/science.2887035. [DOI] [PubMed] [Google Scholar]
- 28.Fleischer T.C., Weaver C.M., McAfee K.J., Jennings J.L., Link A.J. Systematic identification and functional screens of uncharacterized proteins associated with eukaryotic ribosomal complexes. Genes Dev. 2006;20:1294–1307. doi: 10.1101/gad.1422006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Enyenihi A.H., Saunders W.S. Large-scale functional genomic analysis of sporulation and meiosis in Saccharomyces cerevisiae. Genetics. 2003;163:47–54. doi: 10.1093/genetics/163.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brar G.A., Yassour M., Friedman N., Regev A., Ingolia N.T., Weissman J.S. High-resolution view of the yeast meiotic program revealed by ribosome profiling. Science. 2012;335:552–557. doi: 10.1126/science.1215110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Primig M., Williams R.M., Winzeler E.A., Tevzadze G.G., Conway A.R., Hwang S.Y., Davis R.W., Esposito R.E. The core meiotic transcriptome in budding yeasts. Nat. Genet. 2000;26:415–423. doi: 10.1038/82539. [DOI] [PubMed] [Google Scholar]
- 32.Chu S., DeRisi J., Eisen M., Mulholland J., Botstein D., Brown P.O., Herskowitz I. The transcriptional program of sporulation in budding yeast. Science. 1998;282:699–705. doi: 10.1126/science.282.5389.699. [DOI] [PubMed] [Google Scholar]
- 33.Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005;59:407–450. doi: 10.1146/annurev.micro.59.031805.133833. [DOI] [PubMed] [Google Scholar]
- 34.Costanzo M., VanderSluis B., Koch E.N., Baryshnikova A., Pons C., Tan G., Wang W., Usaj M., Hanchard J., Lee S.D., Pelechano V., Styles E.B., Billmann M., van Leeuwen J., van Dyk N., Lin Z.Y., Kuzmin E., Nelson J., Piotrowski J.S., Srikumar T., Bahr S., Chen Y., Deshpande R., Kurat C.F., Li S.C., Li Z., Usaj M.M., Okada H., Pascoe N., San Luis B.J., Sharifpoor S., Shuteriqi E., Simpkins S.W., Snider J., Suresh H.G., Tan Y., Zhu H., Malod-Dognin N., Janjic V., Przulj N., Troyanskaya O.G., Stagljar I., Xia T., Ohya Y., Gingras A.C., Raught B., Boutros M., Steinmetz L.M., Moore C.L., Rosebrock A.P., Caudy A.A., Myers C.L., Andrews B., Boone C. A global genetic interaction network maps a wiring diagram of cellular function. Science. 2016;353 doi: 10.1126/science.aaf1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Watanabe M., Watanabe D., Nogami S., Morishita S., Ohya Y. Comprehensive and quantitative analysis of yeast deletion mutants defective in apical and isotropic bud growth. Curr. Genet. 2009;55:365–380. doi: 10.1007/s00294-009-0251-0. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material
Supplementary material
Supplementary material