Abstract
Summary
Synthetic lethality is a state when simultaneous loss of two genes is lethal to a cancer cell, while the loss of the individual genes is not. We developed an R package DiscoverSL to predict and visualize synthetic lethality in cancers using multi-omic cancer data. Mutation, copy number alteration and gene expression data from The Cancer Genome Atlas project were combined to develop a multi-parametric Random Forest classifier. The effects of selectively targeting the predicted synthetic lethal genes is tested in silico using shRNA and drug screening data from cancer cell line databases. The clinical outcome in patients with mutation in primary gene and over/under-expression in the synthetic lethal gene is evaluated using Kaplan–Meier analysis. The method helps to identify new therapeutic approaches by exploiting the concept of synthetic lethality.
Availability and implementation
DiscoverSL package with user manual and sample workflow is available for download from github url: https://github.com/shaoli86/DiscoverSL/releases/tag/V1.0 under GNU GPL-3.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
In synthetic lethality, the mutant tumor cells are dependent upon their synthetic lethal (SL) interactors for survival. So, in tumors where the driver or oncogenes cannot be targeted, SL interactors can potentially serve as a drug target (Ashworth, 2008). Some recently published algorithms use cancer genomic data for prediction of SL interactions (Jerby-Arnon et al., 2014; Sinha et al., 2017; Srihari et al., 2015), still the problem of identifying clinically relevant SL interactors persists. Cancers are driven by mutations and, there is a need for an integrative resource that identifies targetable and clinically relevant SL interactions based on cancer gene mutations, along with potential drugs for targeting them. Here we present an integrative R package ‘DiscoverSL’ that predicts cancer-specific SL interactions of any given susceptibility gene using a machine learning approach. The clinical relevance of the predictions was assessed in silico using cell line and patient data using the functional modules provided, to estimate the relative sensitivity to shRNA silencing, copy number changes, drugs and Kaplan–Meier (KM) analysis. The package also includes additional plot modules for intuitive visualization. Together, DiscoverSL R package offers an integrative approach to discover mutation-specific SL interactions in cancers.
2 The algorithm and modules
The potential SL interactions were predicted using a Random Forest (RF) model trained on positive and negative SL genes collected from published screens and applied to cancer data (see Supplementary Methods). As shown in Figure 1, the training model includes, reported SL interactions and select cancer types. Four features were estimated using the following modules as shown in Step1: DiffExp (differential expression of Gene2 +/− mutation in Gene1), ExpCorrelation (correlation of expression), Mutex (mutual exclusivity) and SharedPathway (association with common pathways) derived from mutation, gene expression, copy number alteration (CNA) profiles from The Cancer Genome Atlas (TCGA) patient samples and associated gene-pathway information using the MsigDB (Subramanian et al., 2005). The predictive power is tested using cross-validation and an independent test set from SynLethDB (Guo et al., 2015) (Supplementary Fig. S1 and Supplementary Table S1) (Step 2). To reinforce the importance of patient-specific SL interactions in making therapeutic decisions, in-silico validation was provided as shown in Step3. These include, (i) conditional essentiality calculated from shRNA screens using module DiffRNAi (visualization: plotRNAi), (ii) relative targetability calculated from TCGA CNA data using module TTestMutAmp (visualization: plotAmplificationDiff), (iii) drug sensitivity from cell line data using module DrugSensitivity (visualization: plotSensitivitybyDrug) and (iv) KM curve to show the effect of change in expression of SL interactor gene in patient samples carrying mutation in the primary gene which is available as plotSurvivalCurveSL. Detailed description of the dataset and calculation of all functions are available in Supplementary Material.
Fig. 1.
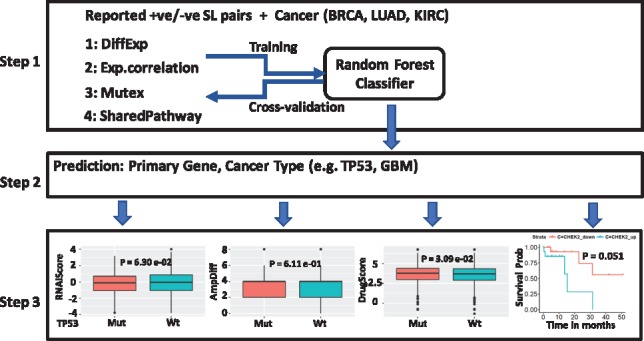
The workflow of the DiscoverSL showing the trained RF model on combined multiple data types (Step1), applied to new data for prediction (Step 2) and validation (Step 3)
3 Case study
The utility of DiscoverSL is demonstrated with a case study on the SL interactions for the known tumor suppressor gene TP53 in Glioblastoma Multiforme (GBM). The model predicted 313 potential SL interactors for TP53 in GBM, filtered for conditional essentiality or drug sensitivity (P-value <0.1). The predicted SL genes included previously reported SL interactors Checkpoint Kinase 1 and 2 (CHEK1/CHEK2) (Origanti et al., 2013) (Supplementary Fig. S2). KM analysis of disease-free survival for CHEK1 and CHEK2 under-expression versus over-expression in the presence of TP53 mutation was shown in GBM (P = 0.06 and P = 0.05) (Supplementary Fig. S2e–f). Drug Sensitivity for targeting CHEK1/CHEK2 could not be tested in TP53 mutated cells due to unavailability of data. However, the predicted SL interactors of TP53 included cancer-related genes, targeted by drugs Ponatinib (FGFR2, FGFR3, FGFR4, FLT3, RET), Pazopanib (FGFR3, FLT1, FLT4), Axitinib (FLT1, FLT4) and Olaparib (PARP1) that show relative sensitivity in presence of TP53 mutation in GDSC cancer cell lines (Yang et al., 2013) (Supplementary Fig. S3a–d). Pathway enrichment analysis showed that the predicted SL interactors of TP53 in GBM were associated with PI3k-Akt signaling, Rap1 signaling, RAF/MAP kinase cascade and Ras signaling pathways (Supplementary Table S2). A recent publication reported that inhibition of PI3K/Akt pathway selectively radiosensitized p53 mutant GBM cell lines compared to cells with wild-type p53 (Palanichamy et al., 2018). These observations show that DiscoverSL can be used to identify SL candidates with potential clinical relevance.
4 Conclusion
Investigating cancer-specific SL interactions offers immense potential for targeted drug treatment for patients based on their mutation profiles. DiscoverSL offers an integrative computational pipeline for prediction and in-silico validation of SL interactions derived from patient-specific mutations in cancer. It can be an excellent resource for discovering clinically relevant and targetable synthetic lethal interactions in cancer.
Funding
This work was funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute.
Conflict of Interest: none declared.
Supplementary Material
References
- Ashworth A. (2008) A synthetic lethal therapeutic approach: poly(ADP) ribose polymerase inhibitors for the treatment of cancers deficient in DNA double-strand break repair. J. Clin. Oncol., 26, 3785–3790. [DOI] [PubMed] [Google Scholar]
- Guo J. et al. (2015) SynLethDB: synthetic lethality database toward discovery of selective and sensitive anticancer drug targets. Nucleic Acids Res., gkv1108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jerby-Arnon L. et al. (2014) Predicting cancer-specific vulnerability via data-driven detection of synthetic lethality. Cell, 158, 1199–1209. [DOI] [PubMed] [Google Scholar]
- Origanti S. et al. (2013) Synthetic lethality of Chk1 inhibition combined with p53 and/or p21 loss during a DNA damage response in normal and tumor cells. Oncogene, 32, 577–588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palanichamy K. et al. (2018) Lack of Constitutively Active DNA Repair Sensitizes Glioblastomas to Akt Inhibition and Induces Synthetic Lethality with Radiation Treatment in a p53-Dependent Manner. Mol. Cancer Ther., 17, 336–346. [DOI] [PubMed] [Google Scholar]
- Sinha S. et al. (2017) Systematic discovery of mutation-specific synthetic lethals by mining pan-cancer human primary tumor data. Nat. Commun., 8, 15580.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srihari S. et al. (2015) Inferring synthetic lethal interactions from mutual exclusivity of genetic events in cancer. Biol. Direct., 10, 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subramanian A. et al. (2005) Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. USA, 102, 15545–15550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W. et al. (2013) Genomics of Drug Sensitivity in Cancer (GDSC): a resource for therapeutic biomarker discovery in cancer cells. Nucleic Acids Res., 41, D955–D961. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


