Abstract
Myocyte enhancer factor 2 (MEF2) transcription factors are key regulators of the development and adult phenotype of diverse tissues, including skeletal and cardiac muscles. Controlled by multiple post-translational modifications, MEF2D is an effector for the Ca2+/calmodulin-dependent protein phosphatase calcineurin (CaN, PP2B, and PPP3). CaN-catalyzed dephosphorylation promotes the desumoylation and acetylation of MEF2D, increasing its transcriptional activity. Both MEF2D and CaN bind the scaffold protein muscle A-kinase–anchoring protein β (mAKAPβ), which is localized to the nuclear envelope, such that C2C12 skeletal myoblast differentiation and neonatal rat ventricular myocyte hypertrophy are inhibited by mAKAPβ signalosome targeting. Using immunoprecipitation and DNA-binding assays, we now show that the formation of mAKAPβ signalosomes is required for MEF2D dephosphorylation, desumoylation, and acetylation in C2C12 cells. Reduced MEF2D phosphorylation was coupled to a switch from type IIa histone deacetylase to p300 histone acetylase binding that correlated with increased MEF2D-dependent gene expression and ventricular myocyte hypertrophy. Together, these results highlight the importance of mAKAPβ signalosomes for regulating MEF2D activity in striated muscle, affirming mAKAPβ as a nodal regulator in the myocyte intracellular signaling network.
Keywords: phosphatase, skeletal muscle, signal transduction, scaffold protein, phosphorylation, calcineurin, histone deacetylase, mAKAPβ, MEF2D, HDAC5, transcription factor, gene regulation, myoblast, A-kinase–anchoring protein, signalosome
Introduction
The MADS (minichromosome maintenance gene-1, agamous, deficiens, and serum response factor) box transcription factor MEF2 family is an important regulator of both tissue specification in development and the response to disease (1–3). For example, triple gene deletion of MEF2A, -C, and -D impaired myoblast differentiation and skeletal muscle regeneration in mice (4). In addition, MEF2D was required in vivo for the induction of pathological cardiac remodeling by pressure overload and chronic catecholamine infusion (5). As a transcription factor family that directly binds DNA, MEF2 serves to nucleate chromatin complexes at promoters and enhancers that activate or repress gene expression through the recruitment of histone acetylases (e.g. p300) and class IIa histone deactylases (HDAC4, -5, -7, and -9),3 respectively (6).
The activity of MEF2 transcription factors is extensively regulated by post-translational modifications, such that they serve as nodal integrators of intracellular signaling by Ca2+, mitogen-activated protein kinase, cyclic nucleotide, and phospholipid-dependent pathways (1–3). One key regulator of MEF2 activity is the Ca2+/calmodulin-dependent protein phosphatase calcineurin (CaN), which can potentiate MEF2-dependent transcriptional activity through residue-specific dephosphorylation. An elegant model has been proposed for the activation of MEF2A in which CaN dephosphorylation of MEF2A Ser-408 promotes the desumoylation and acetylation of Lys-403 (7, 8). A similar model has been proposed for MEF2D residues Ser-444 and Lys-439 (9). Despite extensive information regarding the regulation of MEF2-dependent gene expression by chromatin complexes and their modification by signaling enzymes, how cross-talk between the multiple relevant upstream signaling pathways is coordinated remains obscure. Given the prominent role of MEF2 transcription factors in disease, a better understanding of MEF2 regulatory mechanisms is not only of basic scientific interest, but also direct translational importance.
We have published that MEF2A and MEF2D (which can strongly form heterodimers), CaN, and class IIa HDACs all associate with the scaffold protein muscle A-kinase–anchoring protein (mAKAP) (10–13). mAKAP is expressed as two alternatively spliced isoforms that are highly conserved among vertebrate species, mAKAPα (250 kDa) in neurons and mAKAPβ (230 kDa) in striated myocytes (Fig. 1A) (14, 15). Localized to the nuclear envelope, mAKAPβ function has been extensively studied and plays key roles in the regulation of gene expression controlling pathological cardiac remodeling and skeletal muscle regeneration (15–17). Following early studies regarding the coordination of cAMP and mitogen-activated protein kinase signaling by mAKAPβ signalosomes (18, 19), mAKAPβ was shown to bind also protein kinase D and its activators phospholipase Cϵ and protein kinase Cϵ, conferring mAKAPβ-dependent regulation of type II HDAC nuclear–cytosolic translocation by both phosphoinositide and protein kinase A signaling (12, 20, 21). As currently defined, the mAKAP domain that binds HDACs overlaps that for MEF2A/D (mAKAP amino acid residues 301–500) (11, 12), whereas CaN binds a distinct mAKAP domain (aa 1286–1346) (13). Notably, MEF2–CaN complexes were only detected in cardiac myocytes in the presence of mAKAPβ expression, demonstrating the importance of the bridging scaffold for signalosome formation (10). Unlike the binding of inactive CaN to other scaffold proteins (22), CaN is recruited to mAKAPβ in hypertrophic agonist-stimulated myocytes in an active conformation (13). Accordingly, we have shown that MEF2 transcriptional activity is inhibited in cells by anchoring disruptor peptide–based blockade of either mAKAPβ–MEF2 or mAKAPβ–CaN binding (10, 11). Taken together, the literature supports a model in which mAKAPβ signalosomes coordinate the regulation of MEF2 and class IIa HDACs by upstream signals, thereby modulating MEF2-dependent gene expression in striated myocytes.
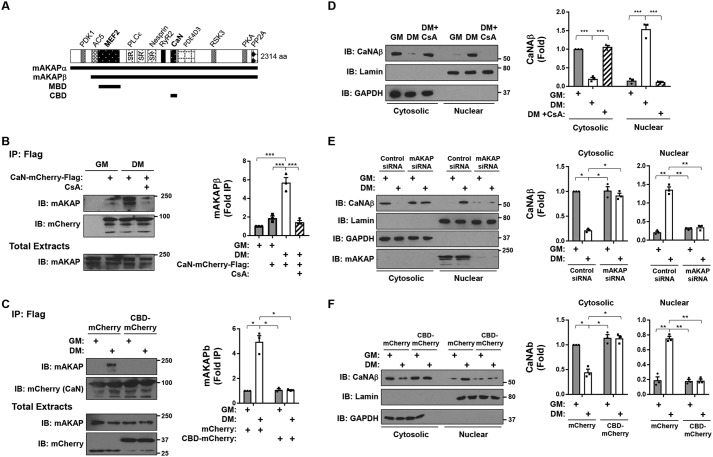
Figure 1.
Recruitment of active calcineurin to mAKAPβ signalosomes in differentiating C2C12 skeletal myoblasts. A, mAKAPβ in striated myocyte is identical to aa residues 245–2314 of the neuronal mAKAPα (14). The three spectrin repeats (SR) required for nuclear envelope targeting are indicated (40). Binding sites are shown for those mAKAP binding partners for which there is evidence of direct binding: 3-phosphoinositide-dependent kinase-1 (PDK1) (14); adenylyl cyclase 5 (AC5) (41); MEF2 (11); phospholipase Cϵ (PLCϵ) (21); nesprin-1α (23); ryanodine receptor (RyR) (42); CaN (13); phosphodiesterase 4D3 (PDE4D3) (38); p90 ribosomal S6 kinase 3 (RSK3) (43); protein kinase A (PKA) (40); protein phosphatase 2A (PP2A) (44). HDAC5 binding to mAKAPβ has been mapped to the MEF2D site (12). MEF2D (aa 301–500) and CaN (aa 1285–1345) binding domain peptides are designated MBD and CBD, respectively. B, C2C12 cells transfected with an expression plasmid for mCherry- and FLAG-tagged CaN were cultured in GM or DM in the absence or presence of cyclosporin A (500 nm) for 3 h before immunoprecipitation using FLAG antibodies and detection of associated mAKAPβ. p (one-way ANOVA) < 0.0001. C, same as in B except using cells co-expressing CBD-mCherry or mCherry control. Note that CaN-mCherry-FLAG and the smaller CBD-mCherry (37 kDa) and mCherry (29 kDa) were readily separated by SDS-PAGE. p (two-way ANOVA for both factors and interaction) < 0.02. D, C2C12 cells cultured as in B were fractionated into cytosolic and nuclear fractions. GAPDH and lamin A antibodies were used to show the efficiency of fractionation. Detection of endogenous CaNAβ in each fraction was determined by Western blotting. p (two-way ANOVA for culture conditions and interaction) < 0.01. E, C2C12 cells were transfected with mAKAP or control siRNA before fractionation and analysis as in D. F, C2C12 cells were transfected with mCherry or CBD-mCherry expression plasmids before fractionation and analysis as in D. p (two-way ANOVA for both factors and interaction) < 0.02 for both cytosolic and nuclear fractions for E and F. n = 3 independent experiments for all panels. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Error bars, S.E.
Whereas a role for mAKAPβ in regulating MEF2-target genes has been established for both skeletal myocyte differentiation and pathological cardiac remodeling (11, 15), it remains unclear how mAKAPβ-bound CaN affects MEF2 activity. As for nuclear factor of activated T-cells (NFATc) transcription factors, CaN dephosphorylation can increase MEF2D mobility in SDS-PAGE (8, 9). mAKAPβ gene deletion blocked the increase in MEF2D mobility in SDS-PAGE detected using heart extracts obtained from mice subjected to long-term pressure overload (15), a condition when MEF2D would be expected to be active and promote adverse remodeling (5). We now reveal that mAKAPβ scaffolding is required in myocytes for CaN-dependent MEF2D Ser-444 dephosphorylation, resulting in a switch between sumoylation and acetylation and between HDAC5 and p300 association, ultimately effecting MEF2D-dependent gene transcription. This study provides mechanistic insight into how MEF2D is regulated by mAKAPβ signalosomes, affecting the composition of MEF2D chromatin complexes.
Results
Recruitment of calcineurin to mAKAβ signalosomes during myoblast differentiation
CaN is a constitutive heterodimer of catalytic (CaNA) and regulatory (CaNB) subunits (22). We have previously published that unlike CaN scaffolds containing a consensus PXIXIT CaN binding motif that can constitutively bind an allosteric site on the CaNA subunit, CaNA binding to mAKAPβ is increased in the presence of Ca2+/calmodulin and induced in cardiac myocytes by adrenergic stimulation (13). We now show that CaN-mAKAPβ binding is similarly regulated during myoblast differentiation. C2C12 mouse skeletal myoblasts were transfected with an expression plasmid for FLAG- and mCherry-tagged CaNAβ and cultured in growth (GM) or differentiation (DM) medium. FLAG tag antibody–mediated immunoprecipitation of mAKAPβ was significantly increased using cells cultured in DM (Fig. 1B; note that the duration of culture in DM was insufficient to increase mAKAPβ expression (11)). Consistent with the binding of activated CaNAβ to mAKAPβ, mAKAPβ–CaNAβ co-immunoprecipitation was inhibited by the CaN inhibitor cyclosporin A. CaNAβ binding to mAKAPβ in C2C12 cells was apparently to the same mAKAPβ site described previously (13), as expression of a CaN anchoring disruptor based upon the previously defined mAKAP CaN-binding domain (CBD; mAKAP aa 1285–1345), which does not inhibit CaN activity but only CaN-mAKAP binding (13), competed CaN-mAKAPβ co-immunoprecipitation (Fig. 1C). mAKAPβ is localized to the nuclear envelope by binding to the transmembrane protein nesprin-1α (23). Consistent with the recruitment of CaN to mAKAPβ in differentiating myoblasts, CaNAβ was enriched in a nuclear fraction (containing the nuclear envelope) only in C2C12 cells in DM (Fig. 1D). Notably, CaN enrichment in the nuclear fraction was inhibited by cyclosporin A, depletion of mAKAPβ using siRNA, and expression of the CaN anchoring disruptor peptide (Fig. 1, D–F). Together, these results imply that active CaN is recruited to perinuclear mAKAPβ signalosomes by DM stimulation.
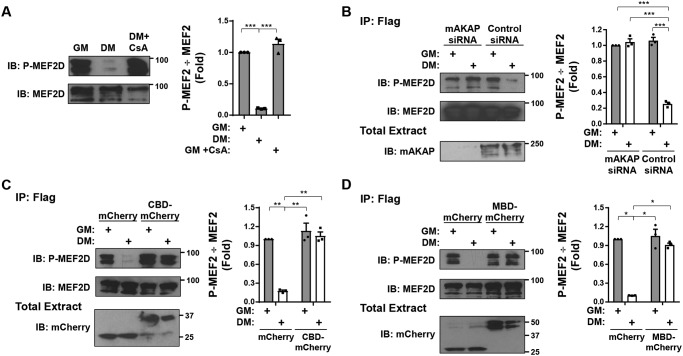
mAKAPβ is required for calcineurin-catalyzed MEF2D Ser-444 dephosphorylation
Ser-444 is the primary CaN-dephosphorylated MEF2D residue for transcriptional activation (8). We have previously reported that MEF2D transcriptional activity in C2C12 cells is induced by DM, such that the activation is inhibited by expression of the mAKAP-derived CaN anchoring disruptor peptide (10). Consistent with that report, we determined that phosphorylation of MEF2D detected with a phospho-Ser-444–specific antibody was decreased in cells cultured in DM via a CaN-dependent mechanism (Fig. 2A). In addition, as mAKAPβ was required for CaN and MEF2D association in myocytes (10), Ser-444 dephosphorylation was inhibited by siRNA-mediated suppression of mAKAPβ expression in cells (Fig. 2B). Just as CaN-mAKAPβ binding can be inhibited using CBD, MEF2 binding to mAKAPβ can be competed by expression of the mAKAP MEF2-binding domain (MBD; mAKAP 301–500; Fig. 1A) (11). The importance of mAKAPβ signalosome formation for Ser-444 dephosphorylation was thereby corroborated using the CaN and MEF2D anchoring disruptor peptides to prevent DM-induced dephosphorylation (Fig. 2, C and D), demonstrating the importance of active CaN at mAKAPβ signalosomes for this post-translational modification.
Figure 2.
Regulation of MEF2D Ser-444 phosphorylation. A, C2C12 cells transfected with an expression plasmid for GFP- and FLAG-tagged MEF2D were cultured in GM or DM in the absence or presence of cyclosporin A (500 nm) for 3 h before immunoprecipitation using FLAG antibodies and Western blotting using MEF2D and phospho-Ser-444 (P-MEF2D) antibodies. p (one-way ANOVA) = 0.0002. B–D, same as in A except using cells co-transfected with mAKAP or control siRNA (B), mCherry or CBD-mCherry expression plasmids (C), or mCherry or MBD-mCherry expression plasmids. n = 3 independent experiments for all panels. p (two-way ANOVA for both factors and interaction) < 0.03 for B–D. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Error bars, S.E.
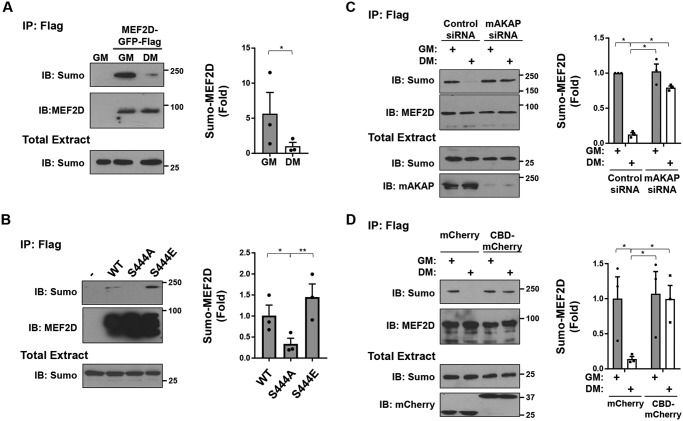
CaN-dependent MEF2D dephosphorylation induces a switch between sumoylation and acetylation
In neurons and heterologous cells, MEF2D Ser-444 dephosphorylation promotes the desumoylation and acetylation of Lys-439 (7–9). We found that culturing of C2C12 cells in DM also promoted the desumoylation of MEF2D (Fig. 3A). Likewise, a Ser-444 alanine MEF2D phosphoablative mutant (S444A) was decreased in sumoylation, whereas a Ser-444 glutamic acid phosphomimetic mutant (S444E) was increased in sumoylation when expressed in heterologous cells (Fig. 3B). Importantly, mAKAP depletion using siRNA inhibited the desumoylation induced by DM in C2C12 cells (Fig. 3C). Furthermore, inhibition of CaN–mAKAPβ binding by the CaN anchoring disruptor peptide also inhibited DM-induced desumoylation in C2C12 cells (Fig. 3D), demonstrating the importance of complex formation for this posttranslational modification event.
Figure 3.
Regulation of MEF2D sumoylation. A, C2C12 cells co-transfected with expression plasmids for GFP- and FLAG-tagged MEF2D and EYFP-tagged SUMO1 were cultured in GM or DM for 3 h before immunoprecipitation using FLAG antibodies and Western blotting using MEF2D and SUMO1 antibodies. Note that sumoylated MEF2D was not detectable in total extracts (not shown). To prevent desumoylation, 25 mm N-ethylmaleimide was added to the lysis buffer. B, HEK293 cells were co-transfected with GFP- and FLAG-tagged MEF2D mutants and EYFP-tagged SUMO1 before immunoprecipitation using FLAG antibodies and Western blotting as in A. p (one-way ANOVA) = 0.004. C, same as A except C2C12 cells transfected with mAKAP or control siRNA. p (two-way ANOVA for both factors and interaction) < 0.03. D, same as A except using C2C12 cells co-expressing mCherry or CBD-mCherry. p (two-way ANOVA for mCherry proteins expressed and interaction) < 0.04. n = 3 independent experiments for all panels. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Error bars, S.E.
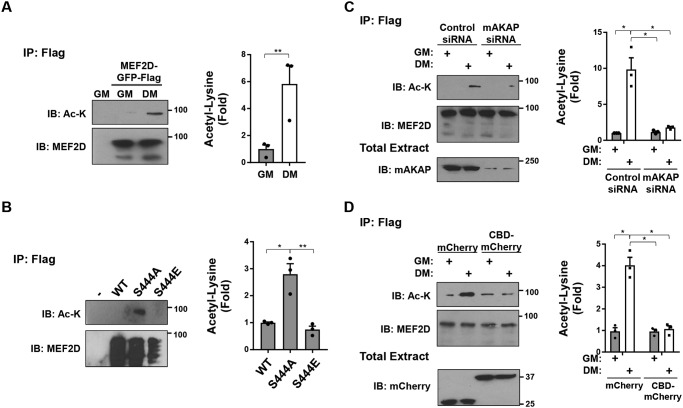
As expected, MEF2D desumoylation was concurrent with MEF2D lysine acetylation in C2C12 cells (Fig. 4A). Likewise, the MEF2D S444A, but not the S444E, mutant was increased in acetylation in HEK293 cells (Fig. 4B). In addition, both expression of mAKAPβ and CaN anchoring to mAKAPβ in C2C12 cells was required for the DM-induced MEF2D acetylation (Fig. 4, C and D). Together, these data show that the previously described sumoylation–acetylation switch occurs in C2C12 cells and, moreover, via a mAKAPβ-anchored CaN-dependent mechanism.
Figure 4.
Regulation of MEF2D acetylation. A, C2C12 cells transfected with expression plasmids for GFP- and FLAG-tagged MEF2D were cultured in GM or DM containing 5 μm trichostatin A for 3 h before immunoprecipitation using FLAG antibodies and Western blotting using MEF2D and acetylated lysine (Ac-K) antibodies. B, HEK293 cells were transfected with GFP- and FLAG-tagged MEF2D mutants before immunoprecipitation using FLAG antibodies and Western blotting as in A. p (one-way ANOVA) = 0.008. C, same as A except using C2C12 cells transfected with mAKAP or control siRNA. D, same as A except using C2C12 cells co-expressing mCherry or CBD-mCherry. n = 3 independent experiments for all panels. p (two-way ANOVA for both factors and interaction) < 0.04 for C and D. *, p ≤ 0.05; **, p ≤ 0.01. Error bars, S.E.
Desumoylation–acetylation drives a switch in MEF2D transcriptional co-activator/co-repressor binding
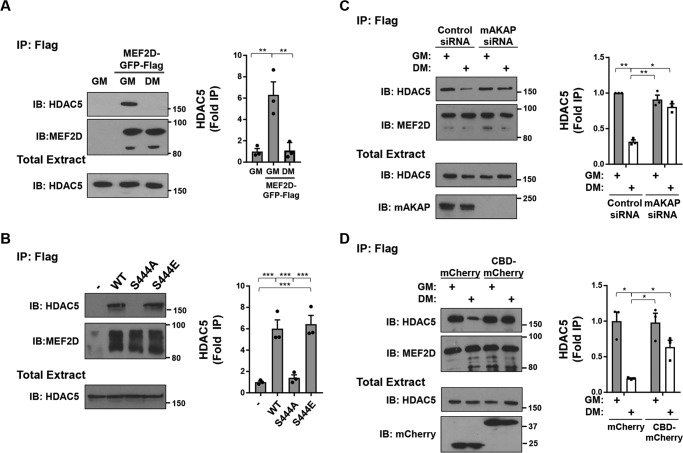
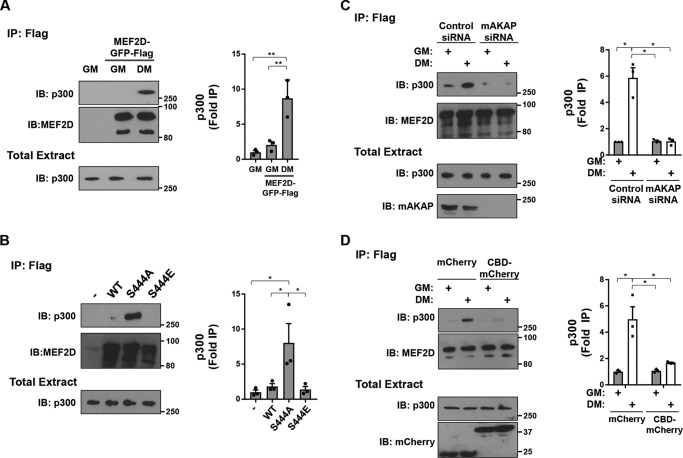
As MEF2D dephosphorylation, desumoylation, and acetylation correlated with increased MEF2D transcriptional activity in C2C12 cells in DM (11), we considered that the altered post-translational modifications might impact the binding of transcriptional co-regulators. To test this hypothesis, we co-immunoprecipitated HDAC5 co-repressor and p300 co-activator with MEF2D in C2C12 extracts. GFP-tagged HDAC5 was readily co-immunoprecipitated with FLAG- and GFP-tagged MEF2D from cells in growth, but not differentiation, medium (Fig. 5A). Likewise, MEF2D WT and S444E, but not S444A, mutant was associated with HDAC5 in HEK293 cells (Fig. 5B). Further, the loss of HDAC5 binding in C2C12 cells cultured in DM depended upon mAKAPβ expression and mAKAPβ-anchored CaN, consistent with the aforementioned regulation of Ser-444 dephosphorylation (Fig. 5, C and D). Conversely, culture in DM induced the association of p300 with MEF2D (Fig. 6A). Likewise, MEF2D S444A, but not WT and S444E, bound p300 in HEK293 cells (Fig. 6B). This association was dependent on the expression of mAKAPβ (Fig. 6C). Moreover, the recruitment of p300 depended upon CaN binding to the mAKAPβ scaffold, as shown by expression of the anchoring disruptor peptide in C2C12 cells (Fig. 6D).
Figure 5.
Regulation of MEF2D-HDAC5 binding. A, C2C12 cells co-transfected with expression plasmids for GFP- and FLAG-tagged MEF2D and GFP-tagged HDAC5 were cultured in GM or DM for 3 h before immunoprecipitation using FLAG antibodies and Western blotting using MEF2D and HDAC5 antibodies. p (one-way ANOVA) = 0.003. B, HEK293 cells were transfected with GFP- and FLAG-tagged MEF2D mutants and GFP-HDAC5 before immunoprecipitation using FLAG antibodies and Western blotting as in A. p (one-way ANOVA) < 0.0001. C, same as A except using C2C12 cells transfected with mAKAP or control siRNA. p (two-way ANOVA for both factors and interaction) < 0.04. D, same as A except using C2C12 cells co-expressing mCherry or CBD-mCherry. p (two-way ANOVA for growth media and interaction) < 0.04. n = 3 independent experiments for all panels. *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Error bars, S.E.
Figure 6.
Regulation of MEF2D–p300 binding. A, C2C12 cells co-transfected with expression plasmids for GFP- and FLAG-tagged MEF2D and p300 were cultured in GM or DM for 3 h before immunoprecipitation using FLAG antibodies and Western blotting using MEF2D and p300 antibodies. p (one-way ANOVA) = 0.003. B, HEK293 cells were transfected with GFP- and FLAG-tagged MEF2D mutants and GFP-HDAC5 before immunoprecipitation using FLAG antibodies and Western blotting as in A. p (one-way ANOVA) = 0.02. C, same as A except C2C12 cells transfected with mAKAP or control siRNA. p (two-way ANOVA for both factors and interaction) < 0.04. D, same as A except using C2C12 cells co-expressing mCherry or CBD-mCherry. p (two-way ANOVA for growth media and interaction) < 0.05. n = 3 independent experiments for all panels. *, p ≤ 0.05; **, p ≤ 0.01. Error bars, S.E.
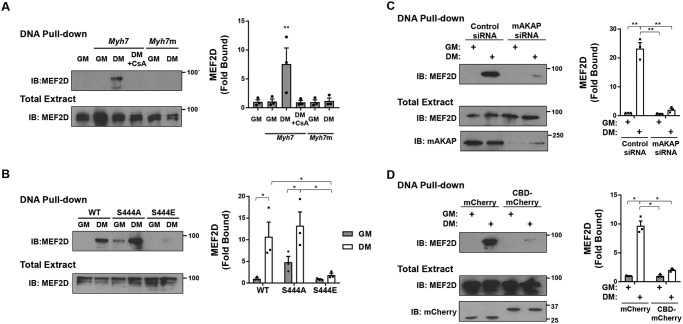
Whereas MEF2D can directly bind DNA, where it may recruit either co-repressors or co-activators to individual promoters and enhancers, MEF2D can also be recruited via other transcription factors to participate in higher-order chromatin complexes (24). One example is the recruitment in the presence of activated CaN of MEF2D and p300 to the MyoD cis-active element at −462 to −457 bp of the Myh7 (β-myosin heavy chain) promoter (24). Using a biotinylated oligonucleotide encompassing that site in a pulldown assay, MEF2D-binding activity for the MyoD cis-active element was found to be enhanced in C2C12 cells in DM (Fig. 7A). Consistent with the above results, recruitment of MEF2D was inhibited by the CaN inhibitor cyclosporin A, by S444E mutation, by mAKAPβ depletion using siRNA, and by expression of the CaN anchoring disruptor peptide (Fig. 7, B–D). Taken together, these results show that dephosphorylation of MEF2D Ser-444 promoting desumoylation and acetylation of the transcription factor results in a switch between MEF2D binding to transcriptional co-repressors and co-activators, all requiring association with the mAKAPβ scaffold in differentiating myoblasts.
Figure 7.
Calcineurin-dependent regulation of MEF2D–p300 DNA complexes. A, C2C12 cells expressing GFP- and FLAG-tagged MEF2D were cultured in GM or DM in the absence or presence of cyclosporin A (500 nm) for 3 h before pulldown assay using a biotinylated oligonucleotide based upon a Myh7 MyoD cis-active element (24) and Western blotting using MEF2D antibodies. Myh7m is a mutant oligonucleotide control. p (one-way ANOVA) = 0.004. B, same as in A except using HEK293 cells co-expressing mutant MEF2D proteins. p (two-way ANOVA for MEF2D mutants and interaction) < 0.04. C, same as A except C2C12 cells transfected with mAKAP or control siRNA. D, same as A except using C2C12 cells co-expressing mCherry or CBD-mCherry. p (two-way ANOVA for both factors and interaction) < 0.02 for C and D. n = 3 independent experiments for all panels. *, p ≤ 0.05; **, p ≤ 0.01. Error bars, S.E.
Functional consequences of CaN-dependent MEF2D dephosphorylation
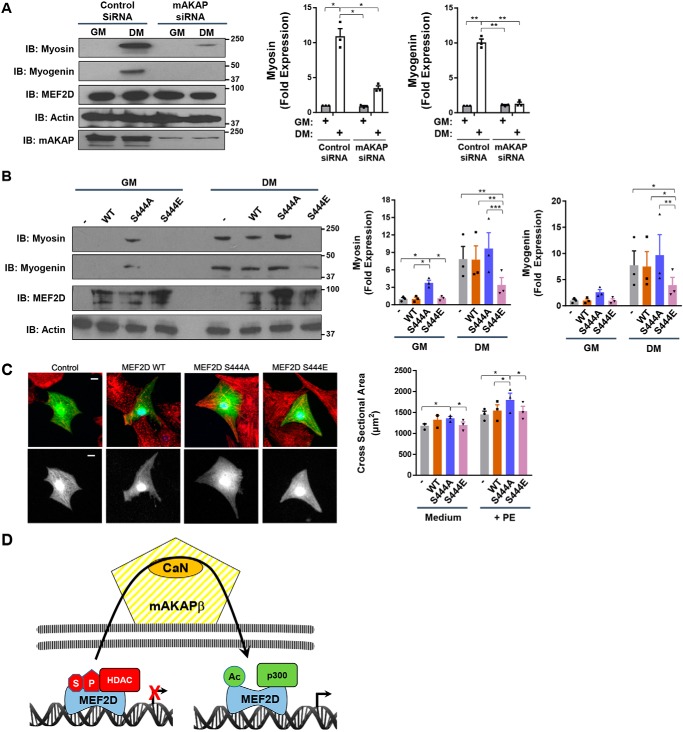
The biochemical studies described above revealed how altered MEF2D post-translational modifications result in altered transcriptional co-regulator binding. To show the functional consequence of these post-translational modifications in C2C12 cells, the expression of myocyte markers known to be directly regulated by MEF2D and induced during myoblast differentiation was assayed (1, 24). Consistent with the recruitment of p300 co-activator to MEF2D in DM via a mAKAPβ-dependent mechanism, mAKAPβ siRNA attenuated the DM-induced expression of myosin heavy chain and myogenin (Fig. 8A). In support of these findings, expression of MEF2D S444A, mimicking MEF2D activation by DM, induced the expression of myosin heavy chain in C2C12 cells cultured in GM (Fig. 8B). Conversely, expression of MEF2D S444E mutant significantly repressed the expression in C2C12 cells in DM of myosin heavy chain and myogenin (Fig. 8C).
Figure 8.
Regulation of myocyte phenotype by MEF2D Ser-444 phosphorylation. A, C2C12 cells transfected with mAKAP or control siRNA were cultured for 24 h in GM and DM before analysis by Western blotting. p (two-way ANOVA for both factors and interaction) < 0.03 for both genes. B, same as in A except with cells expressing GFP- and FLAG-tagged MEF2D WT and mutant proteins. p (two-way ANOVA for MEF2D mutants and interaction) < 0.01 for myosin; p (two-way ANOVA for MEF2D mutants) = 0.02 for myogenin. C, neonatal rat ventricular myocytes were transfected with plasmids expressing FLAG-tagged MEF2D mutants and marker GFP and stained with α-actinin (red) and FLAG (blue) antibodies. The GFP channel is shown also in grayscale below. Bar, 10 μm. Myocyte cross-section areas measured using GFP images are shown. n = 3 independent experiments for A–C. p (two-way ANOVA for both factors) < 0.05. D, model for regulation of MEF2D chromatin complexes by perinuclear mAKAPβ signalosomes. We propose that MEF2D dynamically associates with chromatin, such that transient association with mAKAPβ facilitates its regulation by CaN. HDAC nuclear export is also promoted by mAKAPβ-dependent signaling (12).
To determine whether CaN-dependent MEF2D dephosphorylation was also important for cardiac myocyte hypertrophy, the MEF2D mutants were expressed in cultured rat neonatal ventricular myocytes (RNVMs). Expression of MEF2D S444A resulted in increased RNVM cross-section area regardless of whether the cells were cultured in the presence of the hypertrophic α-adrenergic agonist phenylephrine (PE) or in minimal medium (Fig. 8C). Whereas S444E mutation had no effect in presence of PE, unstimulated RNVMs expressing the S444E mutant tended to be smaller than those overexpressing WT MEF2D (p = 0.11). Together, these results show that mAKAPβ-facilitated, CaN-catalyzed alterations in MEF2D post-translational modification can have significant effects upon the phenotype of striated myocytes.
Discussion
This study provides two major insights into the regulation of MEF2D transcription factor (Fig. 8D): 1) association of MEF2D and CaN with the scaffold protein mAKAPβ is required for dephosphorylation of Ser-444 and its consequences in myocytes, and 2) Ser-444 dephosphorylation and subsequent MEF2D desumoylation and acetylation result in a switch between binding of HDAC5 co-repressor and p300 co-activator that correlates with an increase in MEF2D-dependent gene transcription.
We previously published that MEF2 activity in myocytes depends upon mAKAPβ-associated CaN (10). Unlike at other scaffolds where CaN is bound in an inactive conformation ready for local action (22), we found that catalytically active CaN is recruited to mAKAPβ upon myocyte stimulation, whether in cardiac myocytes with norepinephrine (13) or as shown now in C2C12 cells with DM (Fig. 1). Binding of CaN to mAKAPβ was required for NFATc3 activation in cardiac myocytes (13) and MEF2D dephosphorylation at Ser-444 (Fig. 2). It has been reported that CaN translocates into the nucleus with NFAT transcription factors when activated (25). Remarkably, disruption of mAKAPβ signalosomes, using mAKAP siRNA or the CBD anchoring disruptor, prevented CaN accumulation in nuclear fractions (Fig. 1). Our results imply that mAKAPβ is important not just for CaN association with the perinuclear scaffold, but also general CaN action in the nucleus.
We show that association of the phosphatase and transcription factor with the scaffold are required for MEF2D Ser-444 dephosphorylation and the resulting critical changes in MEF2D post-translational modification that affect transcriptional regulation. In proliferating skeletal myoblasts (Fig. 1) and unstimulated cardiac myocytes (13) in which CaN is not highly bound to mAKAPβ, MEF2D-HDAC repressor complexes apparently predominate (Figs. 5 and 6). Besides MEF2D regulation by post-translational modifications that affect HDAC binding, class IIa HDACs are regulated by nuclear–cytoplasmic translocation (6). Following cellular stimulation, protein kinase D and Ca2+/calmodulin-dependent protein kinases catalyze HDAC phosphorylation, 14-3-3 binding to HDACs, and HDAC nuclear export (26). Notably, mAKAPβ also serves as a scaffold for both class IIa HDACs and protein kinase D and is required for HDAC nuclear export in cardiac myocytes (12). In this regard, mAKAPβ signalosomes not only inhibit MEF2 binding to HDACs via Ser-444 CaN-catalyzed dephosphorylation, but also promote HDAC export from the nucleus, permitting the formation of active MEF2D transcriptional complexes on nuclear chromatin. Whereas mAKAPβ-dependent signaling explains how MEF2–HDAC complexes can be dissociated, the protein kinase(s) responsible for phosphorylating MEF2D Ser-444 in myocytes and maintaining HDAC binding is unknown. One candidate is cyclin-dependent protein 5 (Cdk5), which phosphorylates Ser-444 in neurons and heterologous cells (9, 27) and is inhibited by nestin during skeletal myoblast differentiation (28). It is worth noting that whereas the class IIa HDACs HDAC4 and HDAC5 can both bind MEF2D, neither is a MEF2D deacetylase (29). Instead, class I HDAC3 (but not HDAC1, -2, and -8) and class 3 NAD+-dependent deacetylase SIRT1 (but not SIRT2) deacetylate MEF2D (29, 30), consistent with the known distinct functions of different classes of HDACs (6). It is currently unknown where MEF2D is deacetylated (and Ser-444 is phosphorylated) and whether class I or 3 HDACs associate with mAKAPβ signalosomes.
Dephosphorylation of MEF2D Ser-444 permits MEF2D Lys-439 desumoylation by SUMO protease and acetylation of that residue (31). We show herein that CaN-dephosphorylated MEF2D preferentially binds p300 (Fig. 6). p300 lysine acetyltransferase is a prototypical histone acetylase recruited by many transcription factors to chromatin, where p300 preferentially associates with promoters and enhancers in active transcription units (32). p300 serves both to acetylate histones and to form protein–protein interactions with basal transcription factors, resulting in RNA polymerase II recruitment. p300 can also acetylate MEF2D and is presumably responsible for MEF2D Lys-439 acetylation (29). Thus, in contrast to the lack of class IIa HDAC MEF2D deacetylase activity, the binding of p300 should promote the maintenance of MEF2D co-activator complexes through continued MEF2D acetylation. In addition, because p300 can serve as a large scaffold that associates multiple transcription factors, MEF2D–p300 binding can result in the formation of larger transcriptional complexes. For example, MEF2D–p300 binding induces the association of MEF2D with MyoD and NFATc1 at the Myh7 promoter (24), consistent with our observation that dephosphorylated MEF2D stably associates with the Myh7 MyoD site in a pulldown assay (Fig. 7). mAKAPβ-dependent MEF2D post-translational modification is likely to be a general mechanism for MEF2D complex formation as the class IV POU domain protein Emb, MEF2D, and p300 form a complex in the mouse Actc1 (cardiac actin) distal enhancer (33). Further, because mAKAPβ regulates other transcription factors besides MEF2 (16), including the nuclear import of NFATc family members by mAKAPβ-bound CaN (13, 15), mAKAPβ signalosomes may contribute more broadly to the formation of MEF2D co-activator complexes.
This study reveals the regulation of MEF2D by a cascade of altered post-translational modifications initiated by CaN-catalyzed dephosphorylation at mAKAPβ signalosomes. Whereas one might assume that enhancing MEF2D-activated gene expression would be favorable in skeletal muscle, where MEF2D and mAKAPβ promote maturation of myocytes (17), inhibition of mAKAPβ–MEF2D signaling may be advantageous in the heart, where it drives pathological remodeling (5, 15). Inhibition of CaN is problematic as a heart therapy due to its requirement for myocyte survival following an ischemic event and because current CaN inhibitors are immunosuppressant (34). Targeting of mAKAPβ signalosomes using tissue-specific delivery of anchoring disruptor peptides, including those based upon the CaN- and MEF2D-binding peptides, may provide an approach to achieve inhibition of cardiac remodeling and prevent the development of heart failure that bypasses the use of current CaN inhibitors that have significant side effects.
Experimental procedures
Reagents
Antibodies included mouse anti-MEF2D (Santa Cruz Biotechnology, Inc., sc-271153), rabbit anti-phospho-Ser-444 MEF2D (Thermo Fisher Scientific, PA5–38293), mouse anti-HDAC5 (Santa Cruz Biotechnology, sc-133225), rabbit anti-p300 (Santa Cruz Biotechnology, sc-48343), rabbit anti-mAKAP (23), mouse anti-FLAG tag (Sigma, A220), mouse anti-CaNAβ (Santa Cruz Biotechnology, sc-365612), mouse anti-lamin (Santa Cruz Biotechnology, sc-7293), mouse anti-GAPDH (Santa Cruz Biotechnology, sc-20358), mouse anti-Sumo1 (Santa Cruz Biotechnology, sc-5308), rabbit anti-acetylated lysine (Cell Signaling, 9441), rabbit anti-mCherry (BioVision Inc., 5993100), mouse anti-GFP (Santa Cruz Biotechnology, sc-9996), rabbit anti-GFP (Invitrogen, A11122), mouse anti-α-actinin (Sigma A7811), rabbit anti-actin (Cell Signaling, 8456), and mouse anti-myogenin (Santa Cruz Biotechnology, sc-12732). The MF-20 antibody developed by Donald A. Fischman, M.D. was obtained from the Developmental Studies Hybridoma Bank developed under the auspices of the NICHD, National Institutes of Health, and maintained by the Department of Biology, University of Iowa (Iowa City, IA). ON-TARGETplus siRNA oligonucleotides were mAKAP (5′-GGAGGAAAUAGCAAGGUUAUU-3′, NM_198111.2 bp 7809–7827) and Nontargeting siRNA 1 from Dharmacon. The expression plasmid for FLAG and mCherry-tagged CaN (Aβ isoform) was constructed by Genewiz by inserting a CaN cDNA with a C-terminal FLAG tag into the EcoRI and BamHI sites of pmCherry-C1. Expression vectors (pcDNA3.1) for FLAG-tagged human MEF2D WT, S444A, and S444E were generously provided by Dr. Xiang-Jiao Yang (31). An expression plasmid for FLAG- and GFP-tagged MEF2D (pEGFPN1-FLAG-MEF2D) was constructed by inserting a PCR-amplified cDNA for mouse MEF2D α1β isoform (GenBankTM number S68893) with a C-terminal FLAG tag into the HindIII and AgeI sites of pEGFPN1 (Clontech). S444A and S444E mutations were generated by site-directed mutagenesis. The expression plasmid for EYFP-tagged SUMO was a gift from Mary Dasso (Addgene plasmid 13380 (35)). The expression plasmid for GFP-tagged HDAC5 was a gift from Reuben Shaw (Addgene plasmid 32211 (36)). The expression plasmid pcDNA3.1-p300 was a gift from Warner Greene (Addgene plasmid 23252 (37)). Expression vectors for the CaN (mAKAP aa 1285–1345) and MEF (mAKAP aa 301–500) anchoring disruptor peptides were as described previously (10, 12).
C2C12 cell culture
C2C12 mouse skeletal myoblasts were passaged at low density in GM (Dulbecco's modified Eagle's medium (DMEM; Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin (Invitrogen)). Cells were monitored for spontaneous differentiation due to overgrowth. To induce differentiation, cells at ∼70% confluence were washed with PBS and cultured in DM (DMEM supplemented with 2% horse serum and 1% penicillin/streptomycin). Two days prior to transfection, cells were plated at 40% confluence in GM. Cells were transfected using Viromer Red transfection reagent for plasmids (Origene TT100302) or Viromer Blue for siRNA (Origene TT100300) following the manufacturer's instructions. Briefly, DNA was diluted to 11 ng/μl in 300 μl of RED Buffer while 2.4 μl of Viromer RED was incubated in 57.6 μl of RED Buffer. After mixing, the solution was incubated for 15 min at room temperature before the addition to one 35-mm dish. The next day, cells were washed extensively and incubated in DM for 3–24 h.
Culture of primary neonatal rat ventricular myocytes
Myocytes were prepared as described previously (38). Briefly, cardiac ventricles free of atria and connective tissue were isolated from 1–3-day-old rat pups euthanized by decapitation. Myocytes were dissociated by several cycles of trypsin treatment and serum neutralization. After dissociation, the cells were collected by centrifugation, passed through a 70-μm mesh cell strainer to remove clumps, and preplated in culture dishes to remove fibroblasts. After 1 h, the medium containing the unattached myocytes was removed, and the cells were collected by centrifugation and plated again in 6-well dishes at a density of 500,000 myocytes/plate. Plating medium was DMEM with 17% Medium 199, 1% penicillin/streptomycin (Gibco-BRL), 10% horse serum, and 5% fetal bovine serum. The following day, the plates were washed and then incubated with plating medium containing neither horse serum nor fetal bovine serum (maintenance medium). Myocytes in maintenance medium supplemented with 4% horse serum were transfected with plasmids using Transfast (Promega) as recommended by the manufacturer. Myocytes were cultured for 2 days before analysis in the absence or presence of agonist, before immunocytochemistry and fluorescent imaging were performed as described previously (10). At least 25 individual cells were measured for each condition for each biological replicate. The Stanford University Administrative Panel on Laboratory Animal Care approved all animal procedures used in this work.
HEK293 cell culture
HEK293 cells were cultured in GM. Transfection of HEK293 cells was performed using MegaTran transfection reagent (Origene TT200002). Briefly, 1 μg of DNA was incubated in 200 μl of Opti-MEM and 3 μl of MegaTran for 10 min before the addition to HEK293 cells previously plated on 60-mm dishes containing 2 ml of Opti-MEM without antibiotics. Cells were used for immunoprecipitation experiments the following day.
Subcellular fractionation
Nuclear and cytosolic fractions were prepared using the NE-PER Nuclear and Cytoplasmic Extraction Kit from Thermo Fisher Scientific. Purity of the fractions was determined by Western blotting for lamin A/C and GAPDH.
Immunoprecipitation assays
Cells were washed twice with PBS and then lysed with 1 ml of HSE buffer (20 mm HEPES, pH 7.4, 150 mm NaCl, 5 mm EDTA, 1% Triton X-100, 10% glycerol) supplemented with protease inhibitors (4-(2-aminoethyl)-benzenesulfonyl fluoride, benzamidine, leupeptin/pepstatin). For experiments to detect sumoylation of MEF2D, 25 mm N-ethylmaleimide was added to the lysis buffer. For detection of MEF2D acetylation, cells were incubated with 5 μm trichostatin A HDAC inhibitor for 3 h before lysis, and 5 μm trichostatin A was also added to the lysis buffer. Following centrifugation at 13,200 rpm at 4 °C, soluble cell lysates were incubated with 15 μl of Protein G beads and 2 μg of antibody overnight at 4 °C. Beads were pelleted and washed three times with HSE buffer and boiled in 25 μl of 2× SDS loading buffer. Samples were separated on either 7.5 or 10% SDS-polyacrylamide gels and transferred to nitrocellulose membranes. Blots were blocked in 5% milk for 1 h, followed by incubation in primary antibody overnight at 4 °C. Following washes, secondary antibodies (horseradish peroxidase–conjugated rabbit or mouse IgG, Santa Cruz Biotechnology) were incubated with the membranes (1:10,000 for MEF2D and 1:5000 for all others) overnight. Signals were visualized with an enhanced chemiluminescence reagent (Pierce) and exposed to X-ray film.
Avidin biotin-conjugated DNA-binding (ABCD) assay
Detection of MEF2D–DNA complexes was by the ABCD pulldown assay described by Glass et al. (39). Double-stranded, oligonucleotides biotinylated at the 3′ end of the sense strand were obtained for −472 to −423 bp of the rabbit Myh7 promoter (−472 to −423) (24). Oligonucleotides containing random sequence were used as a control. Oligonucleotide probes (0.1 μm) were incubated with extracts derived from transfected C2C12 cells and 20 μl of Neutravadin agarose (Thermo Fisher Scientific, 29200). After an overnight incubation at 4 °C while rocking, the beads were washed extensively in lysis buffer, and bound proteins were eluted for SDS-PAGE. Bound MEF2D was detected by Western blotting.
Statistics
Matched one- or two-way ANOVA was performed as appropriate, followed by Tukey or Dunnett post hoc testing using GraphPad Prism version 7. For single comparisons, two-tailed t-tests were performed. All data are presented as mean ± S.E.: *, p ≤ 0.05; **, p ≤ 0.01; ***, p ≤ 0.001. Note that for multiple comparisons, not all post hoc tests are indicated in the figures, such that a lack of symbols does not imply p > 0.05.
Author contributions
J. L., S. A. P., and K. L. D.-K. investigation; J. L., S. A. P., and K. L. D.-K. methodology; J. L., S. A. P., and K. L. D.-K. writing-review and editing; H. T. resources; M. S. K. and K. L. D.-K. conceptualization; M. S. K. and K. L. D.-K. funding acquisition; M. S. K. writing-original draft; M. S. K. and K. L. D.-K. project administration; K. L. D.-K. formal analysis.
This work was supported, in whole or in part, by State of Connecticut Department of Public Health Grant 2014-0133 (to K. D. K.) and National Institutes of Health Grants HL126825 (to K. D. K. and M. S. K.) and HL126950, EY026766, and EY026877 (to M. S. K). K. L. D.-K., J. L., and M. S. K. are co-inventors of patented intellectual property concerning the targeting of mAKAP for the treatment of heart failure. M. S. K. owns equity in Anchored RSK3 Inhibitors, LLC, and Cardiac RSK3 Inhibitors, LLC, companies interested in developing mAKAPβ-based therapies. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
- HDAC
- histone deacetylase
- CaN
- calcineurin
- mAKAP
- muscle A-kinase–anchoring protein
- aa
- amino acids
- NFATc
- nuclear factor of activated T-cells
- GM
- growth medium
- DM
- differentiation medium
- RNVM
- rat neonatal ventricular myocyte
- PE
- phenylephrine
- SUMO
- small ubiquitin-like modifier
- DMEM
- Dulbecco's modified Eagle's medium
- GAPDH
- glyceraldehyde-3-phosphate dehydrogenase
- ABCD
- avidin biotin-conjugated DNA-binding
- ANOVA
- analysis of variance
- CBD
- CaN-binding domain
- MBD
- MEF2-binding domain
- IP
- immunoprecipitation
- IB
- immunoblotting
- EYFP
- enhanced yellow fluorescent protein.
References
- 1. Potthoff M. J., and Olson E. N. (2007) MEF2: a central regulator of diverse developmental programs. Development 134, 4131–4140 10.1242/dev.008367 [DOI] [PubMed] [Google Scholar]
- 2. Taylor M. V., and Hughes S. M. (2017) Mef2 and the skeletal muscle differentiation program. Semin. Cell Dev. Biol. 72, 33–44 10.1016/j.semcdb.2017.11.020 [DOI] [PubMed] [Google Scholar]
- 3. Dietrich J. B. (2013) The MEF2 family and the brain: from molecules to memory. Cell Tissue Res. 352, 179–190 10.1007/s00441-013-1565-2 [DOI] [PubMed] [Google Scholar]
- 4. Liu N., Nelson B. R., Bezprozvannaya S., Shelton J. M., Richardson J. A., Bassel-Duby R., and Olson E. N. (2014) Requirement of MEF2A, C, and D for skeletal muscle regeneration. Proc. Natl. Acad. Sci. U.S.A. 111, 4109–4114 10.1073/pnas.1401732111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kim Y., Phan D., van Rooij E., Wang D. Z., McAnally J., Qi X., Richardson J. A., Hill J. A., Bassel-Duby R., and Olson E. N. (2008) The MEF2D transcription factor mediates stress-dependent cardiac remodeling in mice. J. Clin. Invest. 118, 124–132 10.1172/JCI33255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Xie M., and Hill J. A. (2013) HDAC-dependent ventricular remodeling. Trends Cardiovasc. Med. 23, 229–235 10.1016/j.tcm.2012.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Shalizi A., Gaudillière B., Yuan Z., Stegmüller J., Shirogane T., Ge Q., Tan Y., Schulman B., Harper J. W., and Bonni A. (2006) A calcium-regulated MEF2 sumoylation switch controls postsynaptic differentiation. Science 311, 1012–1017 10.1126/science.1122513 [DOI] [PubMed] [Google Scholar]
- 8. Flavell S. W., Cowan C. W., Kim T. K., Greer P. L., Lin Y., Paradis S., Griffith E. C., Hu L. S., Chen C., and Greenberg M. E. (2006) Activity-dependent regulation of MEF2 transcription factors suppresses excitatory synapse number. Science 311, 1008–1012 10.1126/science.1122511 [DOI] [PubMed] [Google Scholar]
- 9. Grégoire S., Tremblay A. M., Xiao L., Yang Q., Ma K., Nie J., Mao Z., Wu Z., Giguère V., and Yang X. J. (2006) Control of MEF2 transcriptional activity by coordinated phosphorylation and sumoylation. J. Biol. Chem. 281, 4423–4433 10.1074/jbc.M509471200 [DOI] [PubMed] [Google Scholar]
- 10. Li J., Vargas M. A., Kapiloff M. S., and Dodge-Kafka K. L. (2013) Regulation of MEF2 transcriptional activity by calcineurin/mAKAP complexes. Exp. Cell Res. 319, 447–454 10.1016/j.yexcr.2012.12.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Vargas M. A., Tirnauer J. S., Glidden N., Kapiloff M. S., and Dodge-Kafka K. L. (2012) Myocyte enhancer factor 2 (MEF2) tethering to muscle selective A-kinase anchoring protein (mAKAP) is necessary for myogenic differentiation. Cell. Signal. 24, 1496–1503 10.1016/j.cellsig.2012.03.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dodge-Kafka K. L., Gildart M., Li J., Thakur H., and Kapiloff M. S. (2018) Bidirectional regulation of HDAC5 by mAKAPβ signalosomes in cardiac myocytes. J. Mol. Cell Cardiol. 118, 13–25 10.1016/j.yjmcc.2018.03.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Li J., Negro A., Lopez J., Bauman A. L., Henson E., Dodge-Kafka K., and Kapiloff M. S. (2010) The mAKAPβ scaffold regulates cardiac myocyte hypertrophy via recruitment of activated calcineurin. J. Mol. Cell Cardiol. 48, 387–394 10.1016/j.yjmcc.2009.10.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Michel J. J., Townley I. K., Dodge-Kafka K. L., Zhang F., Kapiloff M. S., and Scott J. D. (2005) Spatial restriction of PDK1 activation cascades by anchoring to mAKAPα. Mol. Cell 20, 661–672 10.1016/j.molcel.2005.10.013 [DOI] [PubMed] [Google Scholar]
- 15. Kritzer M. D., Li J., Passariello C. L., Gayanilo M., Thakur H., Dayan J., Dodge-Kafka K., and Kapiloff M. S. (2014) The scaffold protein muscle A-kinase anchoring protein β orchestrates cardiac myocyte hypertrophic signaling required for the development of heart failure. Circ. Heart Fail. 7, 663–672 10.1161/CIRCHEARTFAILURE.114.001266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Passariello C. L., Li J., Dodge-Kafka K., and Kapiloff M. S. (2015) mAKAP-a master scaffold for cardiac remodeling. J. Cardiovasc. Pharmacol. 65, 218–225 10.1097/FJC.0000000000000206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Lee S. W., Won J. Y., Yang J., Lee J., Kim S. Y., Lee E. J., and Kim H. S. (2015) AKAP6 inhibition impairs myoblast differentiation and muscle regeneration: positive loop between AKAP6 and myogenin. Sci. Rep. 5, 16523 10.1038/srep16523 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Pare G. C., Bauman A. L., McHenry M., Michel J. J., Dodge-Kafka K. L., and Kapiloff M. S. (2005) The mAKAP complex participates in the induction of cardiac myocyte hypertrophy by adrenergic receptor signaling. J. Cell Sci. 118, 5637–5646 10.1242/jcs.02675 [DOI] [PubMed] [Google Scholar]
- 19. Dodge-Kafka K. L., Soughayer J., Pare G. C., Carlisle Michel J. J., Langeberg L. K., Kapiloff M. S., and Scott J. D. (2005) The protein kinase A anchoring protein mAKAP coordinates two integrated cAMP effector pathways. Nature 437, 574–578 10.1038/nature03966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Zhang L., Malik S., Pang J., Wang H., Park K. M., Yule D. I., Blaxall B. C., and Smrcka A. V. (2013) Phospholipase Cϵ hydrolyzes perinuclear phosphatidylinositol 4-phosphate to regulate cardiac hypertrophy. Cell 153, 216–227 10.1016/j.cell.2013.02.047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zhang L., Malik S., Kelley G. G., Kapiloff M. S., and Smrcka A. V. (2011) Phospholipase C ϵ scaffolds to muscle-specific A kinase anchoring protein (mAKAPβ) and integrates multiple hypertrophic stimuli in cardiac myocytes. J. Biol. Chem. 286, 23012–23021 10.1074/jbc.M111.231993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Parra V., and Rothermel B. A. (2017) Calcineurin signaling in the heart: the importance of time and place. J. Mol. Cell Cardiol. 103, 121–136 10.1016/j.yjmcc.2016.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Pare G. C., Easlick J. L., Mislow J. M., McNally E. M., and Kapiloff M. S. (2005) Nesprin-1α contributes to the targeting of mAKAP to the cardiac myocyte nuclear envelope. Exp. Cell Res. 303, 388–399 10.1016/j.yexcr.2004.10.009 [DOI] [PubMed] [Google Scholar]
- 24. Meissner J. D., Umeda P. K., Chang K. C., Gros G., and Scheibe R. J. (2007) Activation of the β myosin heavy chain promoter by MEF-2D, MyoD, p300, and the calcineurin/NFATc1 pathway. J. Cell Physiol. 211, 138–148 10.1002/jcp.20916 [DOI] [PubMed] [Google Scholar]
- 25. Shibasaki F., Price E. R., Milan D., and McKeon F. (1996) Role of kinases and the phosphatase calcineurin in the nuclear shuttling of transcription factor NF-AT4. Nature 382, 370–373 10.1038/382370a0 [DOI] [PubMed] [Google Scholar]
- 26. Dewenter M., von der Lieth A., Katus H. A., and Backs J. (2017) Calcium signaling and transcriptional regulation in cardiomyocytes. Circ. Res. 121, 1000–1020 10.1161/CIRCRESAHA.117.310355 [DOI] [PubMed] [Google Scholar]
- 27. Gong X., Tang X., Wiedmann M., Wang X., Peng J., Zheng D., Blair L. A., Marshall J., and Mao Z. (2003) Cdk5-mediated inhibition of the protective effects of transcription factor MEF2 in neurotoxicity-induced apoptosis. Neuron 38, 33–46 10.1016/S0896-6273(03)00191-0 [DOI] [PubMed] [Google Scholar]
- 28. Lindqvist J., Torvaldson E., Gullmets J., Karvonen H., Nagy A., Taimen P., and Eriksson J. E. (2017) Nestin contributes to skeletal muscle homeostasis and regeneration. J. Cell Sci. 130, 2833–2842 10.1242/jcs.202226 [DOI] [PubMed] [Google Scholar]
- 29. Grégoire S., Xiao L., Nie J., Zhang X., Xu M., Li J., Wong J., Seto E., and Yang X. J. (2007) Histone deacetylase 3 interacts with and deacetylates myocyte enhancer factor 2. Mol. Cell Biol. 27, 1280–1295 10.1128/MCB.00882-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Zhao X., Sternsdorf T., Bolger T. A., Evans R. M., and Yao T. P. (2005) Regulation of MEF2 by histone deacetylase 4- and SIRT1 deacetylase-mediated lysine modifications. Mol. Cell Biol. 25, 8456–8464 10.1128/MCB.25.19.8456-8464.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Grégoire S., and Yang X. J. (2005) Association with class IIa histone deacetylases upregulates the sumoylation of MEF2 transcription factors. Mol. Cell Biol. 25, 2273–2287 10.1128/MCB.25.6.2273-2287.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Holmqvist P. H., and Mannervik M. (2013) Genomic occupancy of the transcriptional co-activators p300 and CBP. Transcription 4, 18–23 10.4161/trns.22601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Molinari S., Relaix F., Lemonnier M., Kirschbaum B., Schäfer B., and Buckingham M. (2004) A novel complex regulates cardiac actin gene expression through interaction of Emb, a class VI POU domain protein, MEF2D, and the histone transacetylase p300. Mol. Cell Biol. 24, 2944–2957 10.1128/MCB.24.7.2944-2957.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Bueno O. F., Lips D. J., Kaiser R. A., Wilkins B. J., Dai Y. S., Glascock B. J., Klevitsky R., Hewett T. E., Kimball T. R., Aronow B. J., Doevendans P. A., and Molkentin J. D. (2004) Calcineurin Aβ gene targeting predisposes the myocardium to acute ischemia-induced apoptosis and dysfunction. Circ. Res. 94, 91–99 10.1161/01.RES.0000107197.99679.77 [DOI] [PubMed] [Google Scholar]
- 35. Ayaydin F., and Dasso M. (2004) Distinct in vivo dynamics of vertebrate SUMO paralogues. Mol. Biol. Cell 15, 5208–5218 10.1091/mbc.e04-07-0589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Mihaylova M. M., Vasquez D. S., Ravnskjaer K., Denechaud P. D., Yu R. T., Alvarez J. G., Downes M., Evans R. M., Montminy M., and Shaw R. J. (2011) Class IIa histone deacetylases are hormone-activated regulators of FOXO and mammalian glucose homeostasis. Cell 145, 607–621 10.1016/j.cell.2011.03.043 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Chen L. F., Mu Y., and Greene W. C. (2002) Acetylation of RelA at discrete sites regulates distinct nuclear functions of NF-κB. EMBO J. 21, 6539–6548 10.1093/emboj/cdf660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Dodge K. L., Khouangsathiene S., Kapiloff M. S., Mouton R., Hill E. V., Houslay M. D., Langeberg L. K., and Scott J. D. (2001) mAKAP assembles a protein kinase A/PDE4 phosphodiesterase cAMP signaling module. EMBO J. 20, 1921–1930 10.1093/emboj/20.8.1921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Glass C. K., Holloway J. M., Devary O. V., and Rosenfeld M. G. (1988) The thyroid hormone receptor binds with opposite transcriptional effects to a common sequence motif in thyroid hormone and estrogen response elements. Cell 54, 313–323 10.1016/0092-8674(88)90194-8 [DOI] [PubMed] [Google Scholar]
- 40. Kapiloff M. S., Schillace R. V., Westphal A. M., and Scott J. D. (1999) mAKAP: an A-kinase anchoring protein targeted to the nuclear membrane of differentiated myocytes. J. Cell Sci. 112, 2725–2736 [DOI] [PubMed] [Google Scholar]
- 41. Kapiloff M. S., Piggott L. A., Sadana R., Li J., Heredia L. A., Henson E., Efendiev R., and Dessauer C. W. (2009) An adenylyl cyclase-mAKAPβ signaling complex regulates cAMP levels in cardiac myocytes. J. Biol. Chem. 284, 23540–23546 10.1074/jbc.M109.030072 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Marx S. O., Reiken S., Hisamatsu Y., Gaburjakova M., Gaburjakova J., Yang Y. M., Rosemblit N., and Marks A. R. (2001) Phosphorylation-dependent regulation of ryanodine receptors: a novel role for leucine/isoleucine zippers. J. Cell Biol. 153, 699–708 10.1083/jcb.153.4.699 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Passariello C. L., Gayanilo M., Kritzer M. D., Thakur H., Cozacov Z., Rusconi F., Wieczorek D., Sanders M., Li J., and Kapiloff M. S. (2013) p90 ribosomal S6 kinase 3 contributes to cardiac insufficiency in alpha-tropomyosin Glu180Gly transgenic mice. Am. J. Physiol. Heart Circ. Physiol. 305, H1010–H1019 10.1152/ajpheart.00237.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dodge-Kafka K. L., Bauman A., Mayer N., Henson E., Heredia L., Ahn J., McAvoy T., Nairn A. C., and Kapiloff M. S. (2010) cAMP-stimulated protein phosphatase 2A activity associated with muscle A kinase-anchoring protein (mAKAP) signaling complexes inhibits the phosphorylation and activity of the cAMP-specific phosphodiesterase PDE4D3. J. Biol. Chem. 285, 11078–11086 10.1074/jbc.M109.034868 [DOI] [PMC free article] [PubMed] [Google Scholar]