Abstract
In the early 1980s, while using purified glycosyltransferases to probe glycan structures on surfaces of living cells in the murine immune system, we discovered a novel form of serine/threonine protein glycosylation (O-linked β-GlcNAc; O-GlcNAc) that occurs on thousands of proteins within the nucleus, cytoplasm, and mitochondria. Prior to this discovery, it was dogma that protein glycosylation was restricted to the luminal compartments of the secretory pathway and on extracellular domains of membrane and secretory proteins. Work in the last 3 decades from several laboratories has shown that O-GlcNAc cycling serves as a nutrient sensor to regulate signaling, transcription, mitochondrial activity, and cytoskeletal functions. O-GlcNAc also has extensive cross-talk with phosphorylation, not only at the same or proximal sites on polypeptides, but also by regulating each other's enzymes that catalyze cycling of the modifications. O-GlcNAc is generally not elongated or modified. It cycles on and off polypeptides in a time scale similar to phosphorylation, and both the enzyme that adds O-GlcNAc, the O-GlcNAc transferase (OGT), and the enzyme that removes O-GlcNAc, O-GlcNAcase (OGA), are highly conserved from C. elegans to humans. Both O-GlcNAc cycling enzymes are essential in mammals and plants. Due to O-GlcNAc's fundamental roles as a nutrient and stress sensor, it plays an important role in the etiologies of chronic diseases of aging, including diabetes, cancer, and neurodegenerative disease. This review will present an overview of our current understanding of O-GlcNAc's regulation, functions, and roles in chronic diseases of aging.
Keywords: O-GlcNAcylation, O-linked N-acetylglucosamine (O-GlcNAc), O-linked N-acetylglucosamine (O-GlcNAc) transferase (OGT), phosphorylation, diabetes, cancer, Alzheimer's disease, neurodegeneration, O-GlcNAcase, kinases
Introduction
O-GlcNAc was discovered when bovine milk galactosyltransferase and UDP-[3H]galactose were used to probe the surfaces of living cells of the murine immune system for terminal GlcNAc moieties (1). Surprisingly, further analyses showed that nearly all of the incorporated [3H]galactose was added to single β-O-linked GlcNAc moieties attached to Ser(Thr) residues on polypeptides. Follow-up experiments showed that O-GlcNAc is highly enriched within the nucleus and cytoplasm (2), particularly on nuclear envelope and chromatin proteins. Thus, the labeling in initial experiments (1) detected O-GlcNAcylated proteins on the small percentage of lysed or damaged cells in the cultures, indicating that O-GlcNAc is quite abundant on nucleocytoplasmic proteins. Cytosolic localization of O-GlcNAc was further confirmed by identification of O-GlcNAc on cytoplasmic proteins in human erythrocytes (3). Subsequently, O-GlcNAc was found to be particularly enriched on the nuclear and cytosolic faces of the nuclear pore complex (4–6). Similar to its unusual localization in cells, O-GlcNAc was found to be on proteins associated with nucleic acids in viruses (7, 8) and also to be abundant on RNA polymerase II transcription factors (9, 10), some of which are well-known oncogenic factors (11, 12) or tumor suppressor proteins (13). The IIa (nonphosphorylated) form of RNA polymerase II is heavily O-GlcNAcylated on its C-terminal domain (CTD),2 in a region that reciprocally becomes heavily phosphorylated during the elongation phase of transcription (14). Early studies also suggested that O-GlcNAc is involved in the regulation of protein translation (15). Analyses of Drosophila polytene chromosomes showed that O-GlcNAc is particularly abundant on chromatin, especially at sites of active gene transcription (16). Pulse–chase studies found that O-GlcNAc cycles rapidly on the HSP27 family of heat shock proteins (17). O-GlcNAc was also shown to be highly dynamic on lymphocyte proteins, cycling rapidly in response to activation (18).
Using a synthetic peptide as a substrate, an assay for the O-GlcNAc transferase (OGT) was developed, and properties of the enzyme from rabbit reticulocytes were defined (19). Using the peptide substrate assay, combined with conventional and affinity chromatography, OGT was purified over 30,000-fold from rat liver and found to be a large multimeric enzyme with high affinity for its donor substrate, UDP-GlcNAc (20). An assay for O-GlcNAcase (OGA; the enzyme that removes O-GlcNAc) was also developed, and O-GlcNAcase was purified 22,000-fold from rat spleen (21) and shown to have enzymatic properties similar to those of previously described crude preparations of hexosaminidase C (22, 23). Subsequently, the OGT was cloned and sequenced from both rats and humans (24, 25) and was shown to be a unique enzyme with multiple tetratricopeptide repeats (protein-docking domains). OGT was shown to be an X-linked gene located near the centromere, and its sequence is very highly conserved from C. elegans to humans (26). Knockout of OGT showed that it is essential to the viability of murine embryonic stem cells and is required for embryonic development (27). Bovine brain OGA was purified, partially sequenced, and used to clone and sequence human OGA (28). OGA was shown to be identical to a previously cloned gene (MGEA5) associated with meningiomas and postulated to be a hyaluronidase (29). The O-GlcNAcase gene is also unique. It is located on human chromosome 10, is essential in mammals and plants, and is also very highly conserved from C. elegans to humans. Recent structural analyses of OGT have not only led to a much better understanding of its enzymatic mechanism, but also, these studies have helped us to understand how a single catalytic subunit can specifically target thousands of different substrates. These investigations have also led to the development of useful inhibitors (30–39) (for a recent review, see Ref. 40). Surprisingly, OGT was also recently shown to use a novel mechanism, involving UDP-GlcNAc at its active site, to proteolytically cleave the important transcription factor, host cell factor 1 (41, 42), into its active forms.
Whereas less is known about how OGA is targeted to it substrates, several recent studies have defined its detailed structure. These studies have also elucidated the molecular mechanisms of the enzyme, and they have led to the development of highly specific and potent OGA inhibitors that work in living cells (43–53).
Perhaps the greatest impediment to understanding the functions of O-GlcNAcylation is the enormous difficulty in detecting and mapping the sites of O-GlcNAc on proteins (54). Despite its abundance within the nucleus and cytoplasm, O-GlcNAc remained undetected until 1983 for many reasons. 1) Generally, the presence or absence of O-GlcNAc does not alter the electrophoretic migration of a polypeptide, even in two-dimensional electrophoresis; 2) O-GlcNAc is very labile at the source and in the gas phase in MS, making detection of O-GlcNAc peptides and site mapping very difficult (55, 56); 3) in mixtures, ion suppression of O-GlcNAc peptides by unmodified peptides in MS masks the presence of the O-GlcNAcylated species. Fortunately, pan-specific O-GlcNAc monoclonal antibodies have greatly improved methods for detection of O-GlcNAc (5, 57, 58), and specific enrichment methods have been developed to circumvent the ion suppression problem (55, 59). Perhaps the most important breakthrough in site mapping for O-GlcNAc on polypeptides has been the development of electron transfer dissociation fragmentation MS, which does not result in the cleavage of the very labile O-GlcNAc glycosidic linkage to serine or threonine (60, 61). Like other post-translational modifications, the functions of O-GlcNAc must be understood at the individual site level, making site mapping a key first step to elucidate its biological functions. Whereas inhibitors of OGT and OGA and genetic knockout experiments of OGA or OGT have allowed us to make great strides in understanding the functions of O-GlcNAc, the lack of methods to alter O-GlcNAcylation levels on a single protein or at a single site has greatly limited progress in this area. Site-directed mutagenesis of O-GlcNAc sites to alanine is useful, but effects are difficult to interpret if the same or proximal site is also subjected to phosphorylation or other modifications.
O-GlcNAc as a sensor of nutrients and stress
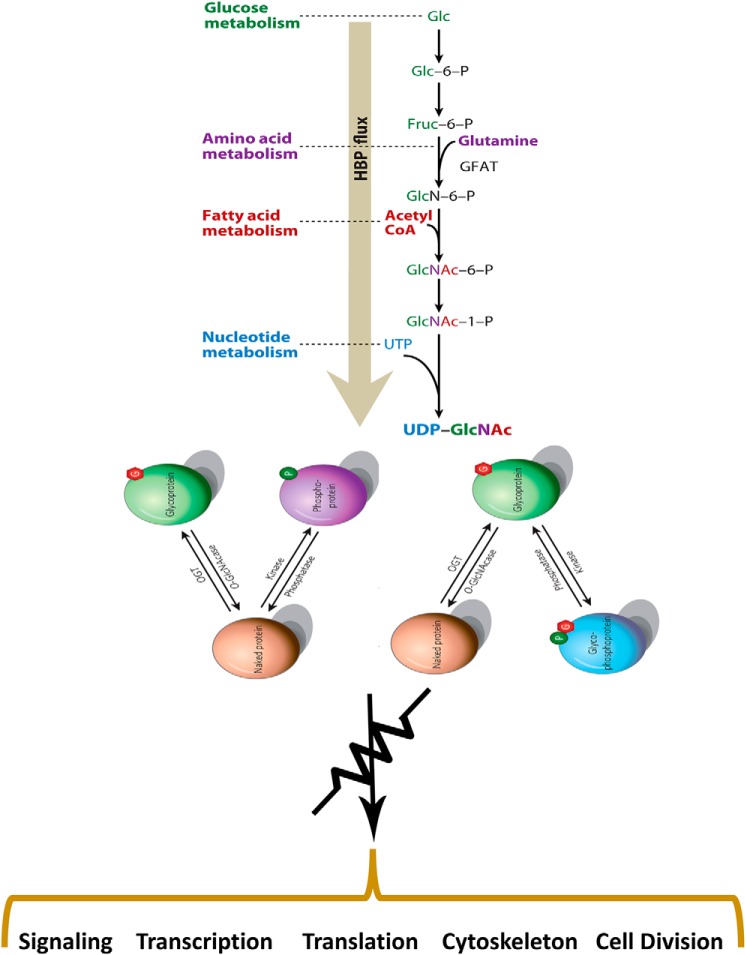
The concentration of UDP-GlcNAc (the donor substrate for OGT) in cells is highly responsive to nutrients and flux through the major metabolic pathways via their connectivity to the hexosamine biosynthetic pathway, including glucose metabolism, nitrogen metabolism, nucleotide metabolism, and fatty acid metabolism (Fig. 1) (62, 63). Both the activity of OGT and its substrate selectivity are highly responsive to UDP-GlcNAc concentrations across a large range (24, 64), indicating that O-GlcNAcylation at specific sites on polypeptides is highly responsive to the metabolic state of the cell. O-GlcNAcylation of nuclear pore proteins (65) and O-GlcNAcylation of many polypeptides within β-cells of the pancreas (66) are very responsive to extracellular glucose concentrations. Hyperglycemia qualitatively and quantitatively alters the O-GlcNAcylation or expression of many O-GlcNAc–modified proteins within the nucleus of rat aorta or smooth muscle cells (67). Hyperglycemia also inhibits vascular endothelial nitric-oxide synthase (eNOS) by O-GlcNAcylation, blocking its key regulatory AKT phosphorylation site (68). Elevated glucose and insulin both stimulate increased O-GlcNAcylation in L6 myotubes (a model of skeletal muscle) (69). Hyperglycemia-induced O-GlcNAcylation mediates plasminogen activator inhibitor-1 gene expression and Sp1 transcriptional activity in glomerular mesangial cells (70).
Figure 1.
The HBP links flux through major metabolic pathways, allowing O-GlcNAcylation to serve as a “rheostat” that modulates most cellular processes in response to nutrients. The biosynthesis of UDP-GlcNAc, the donor for the OGT, is directly coupled to flux through glucose, amino acid, fatty acid, and nucleotide metabolic pathways. OGT is highly sensitive to UDP-GlcNAc concentrations, both in terms of activity and selectivity. O-GlcNAcylation has extensive cross-talk with phosphorylation. Shown is the universal symbol for a rheostat, indicating that unlike phosphorylation, which is more analogous to a switch, O-GlcNAc serves in a more analog fashion as a rheostat to modulate processes in response to nutrients and stress. GFAT, glutamine:fructose-6-phosphate amidotransferase. Modified from Refs. 63 and 341. This research was originally published in Annual Review of Biochemistry. Hart, G. W., Slawson, C., Ramirez-Correa, G., and Lagerlof, O. Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 2011; 80:825–858 © Annual Reviews and Nature. Hart, G. W., Housley, M. P., and Slawson, C. Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 2007; 446:1017–1022 © Springer Nature.
O-GlcNAcylation of a large subset of proteins increases rapidly in response to almost any type of cellular stress, and this increased O-GlcNAcylation protects cells from cellular damage (71), including regulating DNA damage/repair (72). Paradoxically, several laboratories have shown that whereas short-term elevation of O-GlcNAcylation is cardioprotective (73–80), prolonged elevation of O-GlcNAcylation, as occurs in diabetes, contributes to cardiomyopathy and heart failure (81–86) (for a review, see Ref. 87).
Nutrient regulation of gene expression
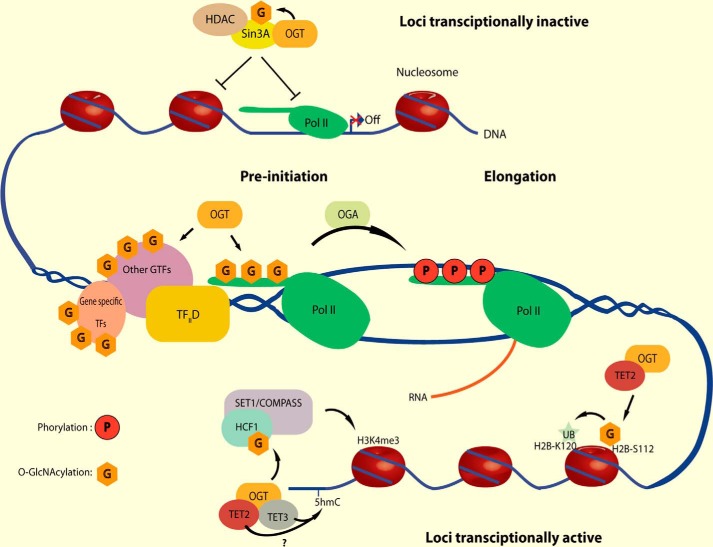
Cells must closely regulate gene expression in response to their metabolic state and the availability of building blocks and fuels. It is now clear that O-GlcNAcylation plays key roles in nutrient regulation of transcription, yet we know very little about the molecular mechanisms involved. Recent studies indicate that O-GlcNAcylation affects nearly every step of transcription (Fig. 2) (for reviews, see Refs. 88–96). Nearly all RNA polymerase II transcription factors are O-GlcNAcylated, often at multiple sites, and the functions of the modification depend not only on the specific transcription factor but also on the specific sites to which the sugar is attached (13, 96–143). OGT is an essential polycomb gene, which regulates expression of homeotic genes that control major morphogenetic events in development (144–147). Assembly of the preinitiation complex in the transcription cycle requires O-GlcNAcylation of the C-terminal domain of RNA polymerase II (95, 148, 149). OGA is a transcription elongation factor, which is required to remove O-GlcNAc from RNA polymerase II prior to the phosphorylation of the CTD (150). Phosphorylation of the CTD is required and precedes elongation (151). O-GlcNAc is part of the histone code (152–159). Whereas several O-GlcNAc sites on histones are in the tail regions, along with other epigenetic marks, some O-GlcNAc moieties are located at the histone:DNA interface. O-GlcNAc regulates both ubiquitination and methylation of histones (159–161). OGT and O-GlcNAc also regulate DNA methylation via their interactions and via regulation and modification of the TET proteins. Very recent studies have shown that the TATA-binding protein's (TBP's) cycling on and off of DNA is regulated by its O-GlcNAcylation.3 When bound to DNA, TBP is O-GlcNAcylated, and this modification reduces its interaction with the TFIID complex, specifically preventing binding to the BTAF1 subunit. Reducing the BTAF1:TBP interaction increases TBP's residence time at promoters, increasing overall promoter occupancy on several promoters and resulting in major changes in the expression of many metabolic enzymes.
Figure 2.
O-GlcNAcylation serves as a nutrient sensor to modulate nearly every step in transcription. Nearly every transcription factor is O-GlcNAcylated, often at multiple sites. OGT is a polycomb gene. Assembly of the preinitiation complex requires O-GlcNAcylation of RNA polymerase II. Elongation of mRNA requires removal of O-GlcNAc from RNA polymerase II. O-GlcNAc is part of the histone code. O-GlcNAc regulates ubiquitinylation and methylation of histones. O-GlcNAc regulates DNA methylation by the TET proteins. TATA-binding protein's residence time at promoters is regulated by O-GlcNAcylation. HDAC, histone deacetylase; Pol, polymerase; TF, transcription factor; H3K4me3, histone H3 Lys-4 trimethylation. Modified from Ref. 94. This research was originally published in Cell Metabolism. Hardivillé, S., and Hart, G. W. Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 2014; 20:208–213. © Cell Press.
Despite many descriptive studies of the roles of O-GlcNAc in transcription, there remain many questions about the functions of OGT, OGA, and O-GlcNAc in transcription. For example, 1) there are limited data suggesting that O-GlcNAcylation of transcription factors may affect their interactions with other components of the transcription machinery and alter their promoter specificity, but detailed studies of this possibility are lacking. This topic is of particular importance to molecular mechanisms underlying glucose toxicity, where abnormal gene expression occurs in many tissues exposed to prolonged hyperglycemia. 2) How O-GlcNAcylation regulates the basal machinery and the transcription cycle is not understood. 3) Are so-called “housekeeping transcription factors” O-GlcNAcylated differently in different cell types or in different metabolic states of the same cell type? These are but a few of the questions that need study. Clearly, how nutrients regulate transcription will be a fertile area for future research that will impact our understanding of disease etiologies.
We know even less about how dynamic O-GlcNAcylation regulates protein translation. However, data are emerging from several studies suggesting that nutrients also regulate protein synthesis via O-GlcNAcylation. Early studies suggested that O-GlcNAcylation of p67 protein plays a required role in p67's regulation of phosphorylation of the eIF-2 α subunit (15). Reticulocyte extracts, which are often used for the study of in vitro protein translation, are very efficient at O-GlcNAcylation of nascent polypeptides (162). It was proposed that OGT and O-GlcNAcylation protect proteins from aggregation during heat stress (163, 164). An unbiased RNA-mediated interference-based screen showed extensive O-GlcNAcylation of stress granules, which are ribonucleoprotein granules that regulate translation and mRNA decay, during cellular stress. O-GlcNAcylation of the translational machinery is required for aggregation of untranslated messenger ribonucleoproteins in the formation of stress granules (165). Gycomic analyses identified many O-GlcNAcylated translation factors and ribosome proteins (166). Over 20 core ribosome proteins are O-GlcNAcylated, and both OGT and OGA are tightly bound to purified ribosomal preparations. Even though most OGT is nuclear in dividing cells, the transferase is completely excluded from the nucleolus, the site of ribosome biogenesis (166). A mild overexpression of OGT causes some of the enzyme to “leak” into the nucleolus, resulting in profound disruption of nucleolar structure and an accumulation of 60S subunits and 80S monosomes. Upon inhibition of the proteasome, both OGT and OGA become very tightly bound to ribosomes. O-GlcNAcylation of many ribosome-associated proteins dramatically increases, and protein synthesis stops for a period of time. The significance of these observations needs further investigation. Recently, O-GlcNAcylation was found to be more extensive on nascent polypeptide chains, presumably protecting them from premature degradation by blocking co-translational ubiquitination, suggesting that O-GlcNAc might play a role in the cytosolic compartment that is similar to the role of N-glycans in the calnexin/calreticulin system for proteins in the secretory pathway (167).
Nutrient regulation of signaling
It is now clear that O-GlcNAcylation's interplay with phosphorylation plays a key role in modulating signaling pathways in response to nutrients and stress (168). One of the earliest studies suggesting interplay between protein phosphorylation and O-GlcNAcylation showed that the IIa form of RNA polymerase II is abundantly O-GlcNAcylated in its CTD, yet the heavily phosphorylated IIo form of RNA polymerase II CTD is completely lacking in O-GlcNAc moieties (14). In vitro assays using synthetic CTD repeats showed that O-GlcNAcylation and phosphorylation are mutually exclusive, with the presence of a single O-GlcNAc completely blocking the activity of CTD kinases and the presence of a single phosphate moiety completely blocking OGT's activity on CTD (169). O-GlcNAcylation at Ser-1177 of eNOS blocks its phosphorylation at this site, thus preventing its activation by AKT kinase (68). Short-term treatment of cells with the broad-spectrum phosphatase inhibitor, okadaic acid, or with phorbol esters, which activate protein kinase C, or treatment with adenosine monophosphate, which activates protein kinase A, all lead to global decreased O-GlcNAcylation (170). In contrast, treatment of cells with the nonspecific kinase inhibitor, staurosporine, increases global O-GlcNAcylation, supporting a “yin-yang” relationship between the two modifications on many proteins. One complexity in these types of studies is that if such treatments are performed at a high dose or for too long, they induce a stress response, which by itself elevates global O-GlcNAcylation.
Activation of protein kinase A or C in cerebellar neurons of post-natal mice results in reduced levels of O-GlcNAc, specifically in cytoskeletal and cytoskeleton-associated proteins, whereas inhibition of the same kinases results in increased levels of O-GlcNAc (171). Likewise, treatment of neuronal cells with the broad-spectrum phosphatase inhibitor, okadaic acid, which induces protein hyperphosphorylation, decreases the levels of O-GlcNAc in both nuclear and cytoplasmic proteins, but with a greater effect in the nuclear fraction (172). Other studies suggest that O-GlcNAc limits nucleoporin hyperphosphorylation during M-phase and hastens the resumption of regulated nuclear transport at the completion of cell division (173). Whereas there are numerous examples of phosphate and O-GlcNAc competing for the same hydroxyl moiety on a polypeptide (11, 12, 174–176), competition also occurs when they are located proximal to each other (100, 106, 177–185). The Stokes radius of an O-GlcNAc moiety is about 5 times that of a phosphate residue.
OGT occurs in a functional complex with protein phosphatases, suggesting that in some instances, the same protein complex both dephosphorylates and concomitantly O-GlcNAcylates polypeptides (186). Glycomic/proteomic analyses have shown that in terms of cross-talk between phosphorylation and O-GlcNAcylation at the site level, all possibilities exist (187, 188). In proteomic analyses of murine synaptosomes, 7% of mapped O-GlcNAc sites were modified reciprocally by phosphate at the same hydroxyl moiety (188). Just these two common post-translational modifications, which often occur at multiple sites on a polypeptide, greatly increase the molecular diversity of proteins. Even though both phosphorylation and O-GlcNAcylation are sub-stoichiometric at any single site on a polypeptide, it is likely that their competition does affect each other's cycling rates, which seems to be the most biologically relevant parameter.
Another aspect of cross-talk between protein phosphorylation and O-GlcNAcylation is the regulation of each other's cycling enzymes by the other modification. OGT and OGA are both regulated by phosphorylation. Calcium/calmodulin kinase IV (CaMKIV) activates OGT, and phosphorylation of OGT has an essential role in CaMKIV-dependent AP-1 activation upon depolarization of neuronal cells (189). As part of a feedback loop, OGT in turn O-GlcNAcylates CAMKIV in its ATP-binding pocket, inactivating the kinase (190). Insulin stimulates tyrosine phosphorylation of OGT by the insulin receptor and activates OGT's activity on many specific substrates (191). Other kinases also modify OGT and activate its catalytic activity, including Src, GSK3β, and CaMKII. OGA is phosphorylated on at least 10 different sites (Phosphosite Plus), but the functions of these modifications are unexplored.
Early studies identified at least 42 kinases that are substrates for OGT (192). Recent screens have shown that about 80% of all human kinases are substrates for OGT. To date, more than 100 kinases have been confirmed to be O-GlcNAcylated in living cells. Proteomic analyses found over 46 kinases modified by O-GlcNAc in murine synaptosomes alone (188).
Most importantly, every O-GlcNAcylated kinase studied to date, is regulated in some manner by the cycling sugar. O-GlcNAc at the ATP-binding pocket of CaMKIV completely inhibits the enzyme (190). The phosphorylated and O-GlcNAcylated forms of casein kinase II have different substrate selectivity (193). O-GlcNAc regulates the activity of the energy-sensing kinase, AMP-activated protein kinase, in skeletal muscle (194). Several members of the protein kinase C family are negatively regulated by O-GlcNAcylation (195). O-GlcNAcylation of phosphofructokinase II inhibits this key enzyme and increases flux into the pentose phosphate pathway, contributing to the “Warburg effect” in cancer cells (196). AKT (protein kinase B) is regulated by O-GlcNAcylation in hepatocytes (197). Activation of neurons increases O-GlcNAcylation of cyclin-dependent kinase 5, blocking its binding to p53 to suppress apoptosis (198). O-GlcNAcylation of p27 blocks cyclin/CDK–p27 binding, contributing to regulation of the cell cycle (199). O-GlcNAcylation of transforming growth factor-β–activated kinase (TAK1) regulates pro-inflammatory activation and M1 polarization of macrophages (200). O-GlcNAcylation of PKAcα and PKAcβ activates the enzymes in the brain and regulates their subcellular localizations (201). Loss of O-GlcNAc on these PKAs leads to impaired learning and memory associated with Alzheimer's disease. Elevated O-GlcNAcylation, as occurs in diabetes, causes CaMKII in the heart to become constitutively active and directly contributes to diabetes-associated cardiomyopathy and arrhythmias (202). Whereas only a handful of the over 400 O-GlcNAcylated kinases have been studied for the effects of O-GlcNAcylation on their functions or on their enzymatic activities, it is already evident that nutrients regulate kinase signaling in large part by modulating their O-GlcNAcylation, which affects kinase functions in many different ways.
Genetic studies also showed that O-GlcNAcylation regulates signaling in plants, especially the Gibbererellin growth hormone signaling pathway (203, 204). In Arabidopsis, O-GlcNAcylation of DELLA transcription factors, which are master growth repressors in plants, regulates and coordinates multiple signaling pathways during development (205). Plant OGT (Secret Agent) regulates flowering by activating histone methylation in Arabidopsis (206).
Nutrient regulation of cytokinesis and the cytoskeleton
Early studies showed that human Band 4.1, a protein that serves as a bridge joining the cytoskeleton to the inner surface of the plasma membrane in erythrocytes, is modified by O-GlcNAc (3). Cytokeratins 8 and 18 are O-GlcNAcylated at multiple sites, and pulse–chase analyses showed that the sugar is dynamically cycling on these intermediate filaments (207). Synapsin I, which anchors synaptic vesicles to the cytoskeleton at nerve terminals via a phosphorylation-regulated process, has at least seven O-GlcNAcylation sites clustered around its five phosphorylation sites. However, further analyses suggest that O-GlcNAc's roles in synapsin I's functions are more direct than simply controlling phosphorylation (178).
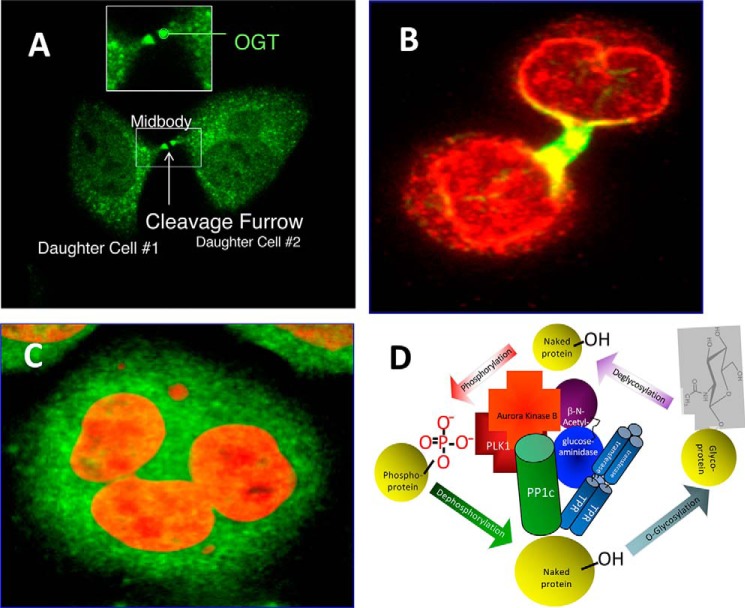
Several studies have shown that O-GlcNAcylation is involved in regulation of the cell cycle and cytokinesis (Fig. 3). Increased O-GlcNAcylation (induced pharmacologically or genetically) results in delayed G2/M progression, altered mitotic phosphorylation, and altered cyclin expression. Decreasing O-GlcNAcylation by overexpression of OGA induces a mitotic exit phenotype accompanied by a delay in mitotic phosphorylation, altered cyclin expression, and pronounced disruption in nuclear organization (208). Overexpression of OGT results in a polyploid phenotype with faulty cytokinesis, as is often seen in cancer cells (Fig. 3). Strikingly, at M-phase, OGT is highly concentrated at the mitotic spindle and mid-body, and a significant portion of OGT is transiently in a large molecular complex with cell cycle-regulated kinases, phosphatases, and OGA. Glycomic analyses identified 141 previously unknown O-GlcNAc sites on proteins that function in spindle assembly and cytokinesis. Many of these O-GlcNAcylation sites are either identical to known phosphorylation sites or are in close proximity to them. Increased O-GlcNAcylation also altered the phosphorylation of key proteins associated with the mitotic spindle and midbody. Overexpression of OGT increased the inhibitory phosphorylation of cyclin-dependent kinase 1 (CDK1) and reduced the phosphorylation of CDK1 target proteins. Increased phosphorylation of CDK1 resulted from increased activation of its upstream kinase, MYT1, and from a concomitant reduction in transcription of CDK1 phosphatase, CDC25C. OGT overexpression also caused a reduction in both mRNA expression and protein abundance of Polo-like kinase 1, which is upstream of both MYT1 and CDC25C. Pathway analyses of these data uncovered a cascade series of events that illustrate how nutrients regulate cell division via the complex interplay between O-GlcNAcylation and phosphorylation (Fig. 3) (209). Collectively, these studies indicate that O-GlcNAc cycling is a pivotal regulatory component of nutrient regulation of the cell cycle, controlling cell cycle progression by regulating mitotic phosphorylation, cyclin expression, and cell division (208).
Figure 3.
Nutrients regulate cytokinesis and the cell cycle by O-GlcNAcylation. A, OGT is highly concentrated at the mid-body during the late stages of cytokinesis. B, O-GlcNAcylated proteins are enriched at the midbody and at the nascent nuclear envelope during the late stages of cytokinesis. C, overexpression of OGT causes defective cytokinesis, resulting in polyploidy. D, during the late stages of cytokinesis, OGT, OGA, protein phosphatase I (PP1c), Polo-like kinase (PLK1), and Aurora kinase B (among other proteins) are in a transient molecular complex that modifies proteins involved in cell division.
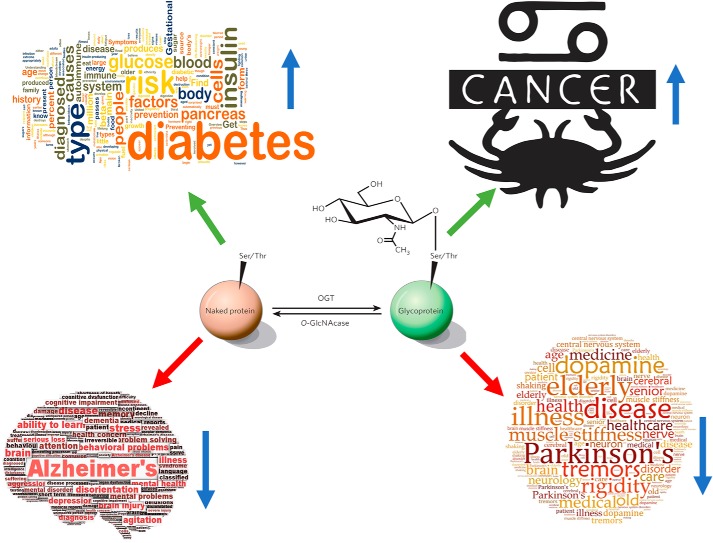
Abnormal O-GlcNAcylation underlies the etiologies of chronic diseases associated with aging (Fig. 4)
Figure 4.
O-GlcNAcylation is directly involved in etiologies of chronic diseases associated with aging. Prolonged elevation of O-GlcNAcylation contributes directly to glucose toxicity, insulin resistance, and β-cell dysfunctions in diabetes. Every cancer type studied to date has elevated O-GlcNAc cycling, and blocking O-GlcNAcylation prevents cancer progression. Decreased O-GlcNAcylation in the brain is associated with both Alzheimer's disease and Parkinson's disease.
Fundamental roles in diabetes
Data from several laboratories indicate that abnormal O-GlcNAcylation, such as occurs in hyperglycemia associated with diabetes, underlies molecular mechanisms of glucose toxicity, insulin resistance, mitochondrial dysfunction, and abnormal insulin synthesis and secretion by β-cells (for reviews, see Refs. 63, 94, and 210–217). Since the 1950s, there have been over 1400 papers linking the hexosamine biosynthetic pathway to the etiology of diabetes. Marshall et al. (218) showed that conversion of glucose to glucosamine by the hexosamine biosynthetic pathway (Fig. 1) is required for the desensitization of the insulin-responsive glucose transport system in adipocytes. Pre-exposure of isolated rat skeletal muscle to glucosamine induces insulin resistance of both glucose transport and glycogen synthesis (219). Increasing flux through the hexosamine biosynthetic pathway (HBP) in otherwise normal rats mimics the hallmarks of glucose toxicity, such as the inhibition of glucose-induced insulin secretion and reduced insulin stimulation of both glycolysis and glycogen synthesis (220). In a streptozotocin rat model of type I diabetes, prolonged hyperglycemia increased the flux through the hexosamine biosynthetic pathway, as determined by the UDP-hex/UDP-HexNAc ratio, by over 40% in skeletal muscle (221). Overexpression of glutamine:fructose-6-phosphate amidotransferase, the first and rate-limiting enzyme of the HBP, in skeletal muscle and adipose tissue of mice mimics the adverse regulatory and metabolic effects of hyperglycemia, specifically with respect to insulin resistance of glucose disposal (222). Even modest transgenic overexpression of OGT in muscle and fat of mice leads to insulin resistance and hyperleptinemia (223).
OGT has a phosphoinositide-binding domain. Upon insulin stimulation, phosphatidylinositol 3,4,5-trisphosphate recruits OGT from the nucleus to the plasma membrane, where OGT catalyzes increased O-GlcNAcylation of the insulin signaling pathway. This increased O-GlcNAcylation results in the altered phosphorylation on these signaling molecules and results in attenuated insulin signaling (224). In addition, hepatic overexpression of OGT reduces the expression of insulin-responsive genes and causes insulin resistance and dyslipidemia (224). Increased O-GlcNAcylation of glycogen synthase results in the retention of the enzyme in a glucose 6-phosphate–dependent state and contributes to reduced activation of the enzyme associated with insulin resistance (225).
Increased flux through the hexosamine biosynthetic pathway also appears to be involved in glucose toxicity and insulin resistance in humans with diabetes (226). Nucleotide polymorphisms in OGA are associated with diabetes in Mexican Americans (227). As might be expected from the role of the HBP as a central node of metabolism (Fig. 1), fat-induced insulin resistance is also associated with increased end products of the HBP, suggesting that elevated free fatty acids induce skeletal muscle insulin resistance by increasing the flux of fructose 6-phosphate into the hexosamine pathway (228). Palmitate also activates the HBP in human myotubes (229). Expression of the ob gene to make leptin, a potent adipokine released by adipocytes in response to increased energy storage, is controlled by end products of the HBP (88, 230, 231).
β-Cells of the pancreas have the highest relative amounts of OGT and O-GlcNAc of any tissue (232, 233). Prolonged elevation of O-GlcNAcylation contributes to β-cell death by apoptosis in diabetes (66). Elevated O-GlcNAc leads to deterioration of glucose-stimulated insulin secretion by the pancreas of diabetic Goto–Kakizaki rats (234). Key transcription factors that control expression of proinsulin are dynamically regulated by O-GlcNAcylation. Glucose regulates the nuclear transport of NeuroD1 via its O-GlcNAcylation. O-GlcNAc regulates the DNA binding by PDX-1 (102), and glucose controls the expression of the MAF-1 transcription factor via O-GlcNAcylation of unknown nuclear proteins (235). Collectively, these studies indicate a direct role for O-GlcNAcylation in regulating the production and secretion of insulin. It has been proposed that a chronic increase in O-GlcNAcylation may be a major factor leading to the deterioration of β-cell function (236).
Hyperglycemia qualitatively and quantitatively alters the O-GlcNAcylation or expression of many proteins within the nucleus. Abnormal O-GlcNAcylation of transcription factors, especially Sp1, appears to play a significant role in the abnormal expression of proteins observed in diabetes (90). Hyperglycemia increases O-GlcNAc on Sp1 and induces expression of plasminogen activator inhibitor-1 (237), which leads to the expression of genes that contribute to the pathogenesis of diabetic complications. In contrast, activation of PPARγ, a ligand-activated nuclear receptor that increases insulin sensitivity, reduces O-GlcNAcylation of Sp1 (238). Enhanced O-GlcNAcylation is associated with insulin resistance in GLUT1-overexpressing muscles (239). Hyperglycemia, via O-GlcNAcylation, impairs activation of the IR/IRS/PI3K/AKT pathway, resulting in deregulation of eNOS activity (240). Elevated O-GlcNAc also results in insulin resistance associated with defects in AKT activation in 3T3-L1 adipocytes (241).
Increased O-GlcNAcylation has also been implicated in diabetic complications of the eye. The elevated expression of O-GlcNAc–modified proteins and O-GlcNAc transferase plays a causative role in the corneal epithelial disorders of diabetic GK rats (242). There is growing evidence that increased O-GlcNAcylation contributes to the pathogenesis of diabetic retinopathy (243), including the early loss of retinal pericytes (244).
Abnormal O-GlcNAc modification of nucleocytoplasmic proteins appears to also be involved in glucose toxicity in vascular tissues (67, 245). O-GlcNAcylation of AKT kinase promotes vascular calcification (246). Prolonged elevation of O-GlcNAcylation impairs cardiac myocyte function and leads to the development of diabetic cardiomyopathy (247). O-GlcNAcylation of cardiac mitochondrial proteins appears to play a direct role in diabetic cardiomyopathy. Several proteins, which are components of the respiratory chain, including the subunit NDUFA9 of complex I, subunits core 1 and core 2 of complex III, and the mitochondrial DNA-encoded subunit I of complex IV (COX I), are O-GlcNAcylated. Hyperglycemia increases mitochondrial protein O-GlcNAcylation. Increased mitochondrial O-GlcNAcylation impairs activity of complex I, III, and IV in addition to lowering mitochondrial calcium and cellular ATP content. Mitochondrial function improves when O-GlcNAc is reduced by OGA expression, resulting in returning the activities of complex I, III, and IV, mitochondrial calcium, and cellular ATP content to control levels (248).
Glycomic analyses of purified rat heart mitochondria from normal and streptozocin-treated diabetic rats identified 88 O-GlcNAcylated proteins (84, 249, 250). OGT is strikingly increased in diabetic cardiac mitochondria, whereas mitochondrial OGA is concomitantly decreased. Most importantly, OGT is mislocalized in diabetic cardiac mitochondria. In normal cardiac mitochondria, OGT is mostly localized in complex IV of the respiratory chain. However, in diabetic cardiac mitochondria, much of the OGT is now found in the matrix and in complex III, resulting in altered O-GlcNAcylation on many proteins. Inhibition of OGT or OGA activity within neonatal rat cardiomyocytes significantly affects energy production, mitochondrial membrane potential, and mitochondrial oxygen consumption. Therefore, not only do cardiac mitochondria have robust O-GlcNAc cycling, but also dysregulation of O-GlcNAcylation likely plays a key role in mitochondrial dysfunction associated with diabetes (84).
Increased O-GlcNAcylation of an important kinase, CAMKII, that helps to regulate heart contractions contributes to arrhythmias associated with diabetes (202). Acute hyperglycemia increases O-GlcNAcylation of CaMKII at Ser-279, which activates CaMKII autonomously, even after Ca2+ concentration declines. O-GlcNAcylation of CaMKII is increased in the heart and brain of diabetic humans and rats. In cardiomyocytes, increased glucose concentration significantly enhances CaMKII-dependent activation of molecular processes that can contribute to cardiac mechanical dysfunction and arrhythmias. These effects are prevented by pharmacological inhibition of O-GlcNAc signaling or by genetic ablation of CaMKIIδ. In intact perfused hearts, arrhythmias are exacerbated by increased glucose concentration through O-GlcNAc– and CaMKII–dependent pathways. In diabetic animals, acute blockade of O-GlcNAcylation inhibited arrhythmogenesis. It was concluded that O-GlcNAcylation of CaMKII contributes critically to cardiac and neuronal pathophysiology in diabetes and other diseases (202).
O-GlcNAcylation also appears to play a role in diabetic nephropathy. Diabetic patients have significantly increased numbers of O-GlcNAc–positive cells in their glomeruli and significantly elevated staining in the tubuli (both in the nucleus and in the cytosol), suggesting that increased O-GlcNAcylation might contribute to the development of diabetic nephropathy (251). Increased O-GlcNAcylation of cytoskeletal proteins is closely associated with the morphological changes in the podocyte foot processes in the glomerulus and in microvilli of proximal tubules in the diabetic kidney (252). Hyperglycemia-induced elevation of O-GlcNAcylation also contributes to the progression of diabetic nephropathy via inhibition of AKT/eNOS phosphorylation and HSP72 induction (253).
Likewise, O-GlcNAcylation contributes to diabetic neuropathy. Hyper-O-GlcNAcylation of a neuronal protein, Milton, appears to contribute to diabetic neuropathy of the foot (254). To meet the high energy demands at the synapse, mitochondria need to be trafficked from the cell body to the nerve terminal. Milton serves as a bridge protein between the mitochondria and the motor protein, which carries the mitochondria along microtubules. When Milton is hyper-O-GlcNAcylated, as occurs in diabetes, mitochondrial motility is arrested. Because the longest axon in the body is from the spinal cord to the foot, neuropathy often manifests itself in the feet. It was proposed that OGT normally modulates mitochondrial dynamics in neurons based on nutrient availability, but when the modification becomes excessive, it abnormally arrests mitochondrial transport (254).
The relationship between Alzheimer's disease (AD) and diabetes is still unclear. However, based upon many common underlying mechanisms, some researchers refer to AD as “diabetes type 3” (255–258). Recent studies have shown that hyperglycemia in the diabetic patient appears to induce greater expression and activity of the mitochondrial isoform of OGT (mOGT). Changes in mOGT modify the structure and functionality of mitochondria in hippocampal cells, accelerate neuronal damage, and favor the early events in AD. It was proposed that mOGT activity could be a key point for AD development in patients with diabetes (259). An understanding of O-GlcNAc's roles in both diabetes and AD will clearly require many more detailed investigations.
Because O-GlcNAcylation is highly sensitive to glucose, it was postulated that monitoring O-GlcNAcylation of human erythrocyte proteins might serve as a biomarker for prediabetes prior to detectable changes in HbA1c. Indeed, O-GlcNAcylation of certain sites on human erythrocyte proteins (e.g. catalase) increases significantly in prediabetic patients prior to elevated HbA1c, reflecting the glycemic status of the individual. If validated on a larger clinical trial, O-GlcNAc site occupancy on erythrocyte proteins may eventually be useful as a diagnostic tool for the early detection of diabetes (260, 261).
O-GlcNAc and cancer
There is a rapidly growing literature suggesting that O-GlcNAcylation contributes to the properties and progression of cancer cells (Fig. 4) (for a review, see Refs. 262–266). Whereas it has generally been observed that O-GlcNAc cycling is universally elevated in cancer cells and, indeed, preventing increased O-GlcNAcylation can block cancer progression, it is still early days, and mechanistic details are often lacking.
Aberrant expression and activities of O-GlcNAc cycling enzymes, especially OGT, have been reported in all human cancers studied to date (267). Altered cellular metabolism is a major hallmark of cancer. Glucose uptake and glycolysis are accelerated in cancer cells (“Warburg Effect”), which gives cancer cells an advantage for intensive growth and proliferation. O-GlcNAc–dependent regulation of signaling pathways, transcription factors, enzymes, and epigenetic changes are all likely involved in metabolic reprograming of cancer (265, 266). Several researchers have proposed that inhibition of hyper-O-GlcNAcylation could be a potential novel therapeutic target for cancer treatment (264, 265). It has also been proposed that aberrant O-GlcNAcylated proteins might be novel biomarkers of cancer (268).
Recent studies suggest that O-GlcNAcylation plays a direct role in the Warburg effect (196). In response to hypoxia, O-GlcNAcylation at serine 529 of phosphofructokinase 1 (PFK1) is increased, inhibiting the enzyme and redirecting glucose flux through the pentose phosphate pathway. This metabolic switch confers a selective growth advantage on cancer cells. Blocking O-GlcNAcylation of PFK1 at serine 529 reduces cancer cell proliferation in vitro and impairs tumor formation in vivo. Other metabolic processes involving O-GlcNAc are also involved in cancer cell reprogramming. For example, O-GlcNAcylation also regulates glycolysis in cancer cells via hypoxia-inducible factor 1 (HIF-1α) and its transcriptional target GLUT1. Reducing O-GlcNAcylation in cancer cells results in endoplasmic reticulum stress and cancer cell apoptosis. Human breast cancers with high levels of HIF-1α contain elevated OGT and lower OGA levels, which correlate with poor prognosis (269). DNA methylation plays a direct role in aberrant silencing of tumor suppressor genes in cancer. Because OGT and O-GlcNAcylation regulate the TET proteins that hydroxymethylate DNA, it has been suggested that the cross-talk between OGT and TET proteins might play an important role in cancer cells (270).
Many oncogene and tumor suppressor gene products, such as c-Myc, SV40 large T antigen, Rb, and p53, are O-GlcNAcylated (271). In nongrowing cells, Thr-58, the major hot spot mutation site in Burkitt's lymphoma, is O-GlcNAcylated; however, in growing cells, this same site is phosphorylated. Mutation of Thr-58 to a nonhydroxy amino acid converts the transcription factor, c-Myc, into a potent oncoprotein (11, 12, 271). More recent studies have identified c-MYC as a key target of OGT's effects in prostate cancer cells (272). The tumor suppressor HIC1 (hypermethylated in cancer 1) is O-GlcNAcylated (273). O-GlcNAcylation regulates the nuclear localization and stabilization of β-catenin, a dual-function protein that is both a transcription factor and a regulator of E-cadherin trafficking, a protein that is key to the epithelial–mesenchymal transition in cancer (114).
Reduction of O-GlcNAcylation by RNAi knockdown of OGT in breast cancer cells inhibits tumor growth both in vitro and in vivo and concomitantly decreases cell cycle progression. Reducing O-GlcNAcylation in breast cancer cells decreases expression of the transcription factor, FoxM1, resulting in lower levels of multiple FoxM1-specific targets, including Skp2 and matrix metalloproteinase-2. Reducing O-GlcNAcylation decreased cancer cell invasion and growth (274). In human breast cancer, poorly differentiated tumors (grade II and III) have significantly higher OGT expression than grade I tumors. In contrast, OGA transcript levels are lower in grade II and III compared with grade I tumors. Lymph node metastasis is significantly associated with decreased OGA expression (275). In breast cancer cells, O-GlcNAcylation at Ser-108 of cofilin, a regulator of actin assembly, is required for its proper localization to invadopodia at the leading edge during cell invasion. Loss of O-GlcNAcylation of cofilin leads to destabilization of invadopodia and impairs cell invasion (276). Treatments that increase O-GlcNAcylation in MCF-7 breast cancer cells protect them from death induced by tamoxifen. In contrast, inhibition of OGT potentiates cell killing by tamoxifen (277).
O-GlcNAcylation is also universally elevated in prostate cancer. Reducing O-GlcNAcylation in prostate cancer cells decreases expression of matrix metalloproteinase (MMP)-2, MMP-9, and VEGF and inhibits invasion and angiogenesis. These effects are mediated by the degradation of the transcription factor, FoxM1, a known regulator of invasion and angiogenesis. Overexpression of a degradation-resistant FoxM1 mutant abrogated OGT RNAi-mediated effects on invasion, MMP levels, angiogenesis, and VEGF expression. In a mouse model of metastasis, reduction of OGT expression blocks bone metastasis (278). Strikingly, increasing O-GlcNAcylation by itself induces malignant transformation of nontumorigenic prostate cells. It was proposed that inhibiting the formation of the E-cadherin/catenin/cytoskeleton complex underlies O-GlcNAc–induced prostate cancer progression (279).
The roles of O-GlcNAcylation have now been examined in many different cancer types with similar findings. Elevation of OGA activity has been reported in thyroid cancers (280). Histochemical analyses showed that OGT and O-GlcNAcylation are significantly elevated in lung and colon cancer tissue, relative to adjacent normal tissue. O-GlcNAcylation also markedly enhanced the anchorage-independent growth of lung and colon cancer cells in vitro (281). Studies of colorectal cancer cells support the hypothesis that metabolic disorders underlying colorectal cancer occur by up-regulation of the hexosamine biosynthetic pathway that leads to abnormally high O-GlcNAcylation of β-catenin (282).
Colorectal cancer SW620 metastatic clones exhibit increased O-GlcNAcylation and decreased OGA expression compared with primary clone cells, SW480. Increasing global O-GlcNAcylation by RNAi knockdown of OGA results in phenotypic alterations that include acquisition of a fibroblast-like morphology, concomitant with epithelial metastatic progression and growth retardation. OGA silencing altered the expression of about 1300 genes, mostly involved in cell movement and growth, and specifically affected metabolic pathways of lipids and carbohydrates, suggesting that O-GlcNAcylation serves as a link between metabolic changes and cancer (283). O-GlcNAcylation is increased in primary colorectal cancer tissues on proteins including, cytokeratin 18, heterogeneous nuclear ribonucleoproteins A2/B1 (hnRNP A2/B1), hnRNP H, annexin A2, annexin A7, laminin-binding protein, α-tubulin, and protein DJ-1. It was proposed that aberrantly O-GlcNAc–modified proteins may provide novel biomarkers of cancer (284). It also has been proposed that the urinary content of OGA and OGT may be useful for bladder cancer diagnostics (285). Overexpression of OGT increases the aggressiveness of mass-forming cholangiocarcinomas (286).
O-GlcNAcylation, OGA, and OGT levels were examined in hepatocellular carcinoma (HCC) tissues of patients who underwent liver transplantation and compared with healthy liver tissues. Global O-GlcNAcylation levels were significantly elevated in HCC tissues more than that in healthy liver tissues. Importantly, low expression of OGA was an independent prognostic factor for predicting tumor recurrence of HCC following liver transplantation. In vitro assays demonstrated that O-GlcNAcylation plays important roles in migration, invasion, and viability of HCC cells, partly through regulating E-cadherin, MMP1, MMP2, and MMP3 expression (287).
Pancreatic cancer cells evade cell death, in part by up-regulating HSP70. A prodrug, Minnelide, down-regulates HSP70 to enhance killing of pancreatic cancer cells. Minnelide causes decreased O-GlcNAcylation of the transcription factor Sp1, preventing its nuclear localization and affecting its DNA binding. This in turn down-regulates prosurvival pathways in pancreatic cancer cells, allowing the drug to facilitate their killing (288). Hyper-O-GlcNAcylation in human pancreatic ductal adenocarcinoma (PDAC) results from elevation of OGT and reduction of OGA. Reducing hyper-O-GlcNAcylation has no effect on nontransformed pancreatic epithelial cell growth but inhibits PDAC cell proliferation, anchorage-independent growth, orthotopic tumor growth, and triggers apoptosis. Many of these effects appear to be mediated by O-GlcNAcylation of NF-κB (289).
Roles of O-GlcNAc in neurons and neurodegeneration
Except for the endocrine cells of the pancreas, O-GlcNAcylation, OGT, and OGA levels are the highest in the brain and in neurons. Functions ascribed to O-GlcNAcylation in the nervous system include modulation of circadian clocks (185, 290–293), synaptic functions and maturation (294–296), neuronal development (297–299), neuroprotection (300–302), learning and memory (303–307), neuronal apoptosis (245, 308–310), and appetite regulation (311). Mutations in OGT cause X-linked intellectual disability (312, 313). Given O-GlcNAc's abundance, cross-talk with phosphorylation, and involvement in many neuronal functions, it is not surprising that many researchers have reported a direct role for the cycling sugar in neurodegenerative diseases associated with aging (214, 314–317).
Although mostly indirect, the evidence that O-GlcNAcylation plays a major role in neurodegeneration is compelling: virtually every protein involved in AD or other forms of neurodegeneration is O-GlcNAcylated and phosphorylated, often reciprocally (168, 318). Glucose metabolism is impaired in AD neurons (319–321), which globally lowers O-GlcNAcylation. The Tau protein is normally O-GlcNAcylated, but O-GlcNAc is reduced and phosphorylation of Tau increases dramatically in AD (176). Amyloid precursor protein is O-GlcNAcylated, and the sugar regulates its trafficking and processing (322, 323). In addition, one of the subunits of the γ-secretase (nicastrin), which cleaves amyloid precursor protein to generate the toxic Aβ peptides, normally is O-GlcNAcylated. Many studies have documented the reciprocity between O-GlcNAcylation and phosphorylation of the Tau protein (179, 180, 324–327). Autopsies have shown reduced O-GlcNAcylation in patients with AD (180). The OGA gene maps to 10q24.1 coincident with the late AD locus in humans (328), and OGT maps to X13 coincident with the Parkinson dystonia locus (329). Overexpression of OGT in neurons increases O-GlcNAcylation at sites on Tau important to AD and concomitantly decreases protein phosphorylation at these same sites (180, 330). Cre-Lox brain targeted deletion of OGT causes hyperphosphorylation of Tau prior to neuronal death (27). Synaptic loss occurs in AD, and myriad synaptic proteins are modified by O-GlcNAc (188). Highly specific OGA inhibitors prevent hyperphosphorylation of Tau, decrease the production of toxic Aβ peptides, and improve memory in mice models of AD (48, 306, 331–333). It appears that one function of O-GlcNAcylation is to “cap” phosphorylation sites used at certain stages of development that are no longer required in the adult. We hypothesized (176) that as the brain ages, or due to vascular insufficiency, glucose utilization by certain neurons drops, leading to lower O-GlcNAcylation, which in turn uncovers phosphorylation sites recognized by ubiquitous kinases present in the brain. The exposure of these sites and/or the lack of O-GlcNAc itself causes the proteins to aggregate and form abnormal complexes and contributes to the abnormal processing of amyloid precursor protein.
Although much less studied, evidence is also emerging for O-GlcNAcylation's roles in Parkinson's disease (305, 317). As stated above, OGT maps to the Parkinson dystonia locus at Xq13 (329). Parkinson's disease results in degeneration of dopaminergic neurons in the substantia nigra, leading to a reduction of striatal dopamine levels. Tyrosine hydroxylase, which is the rate-limiting step in the biosynthesis of dopamine, is regulated by reciprocal O-GlcNAcylation/phosphorylation (334). α-Synuclein, a toxic aggregating protein involved in Parkinson's disease, is O-GlcNAcylated on at least eight sites, and even substoichiometric O-GlcNAcylation of α-synuclein affects its phosphorylation and blocks its degradation by calpain. O-GlcNAcylation also blocks the toxicity of α-synuclein added to cultured cells (335–338). Enzymatic O-GlcNAcylation by OGT also inhibits α-synuclein aggregation and promotes the formation of soluble SDS-resistant oligomers that stain negative for amyloid formation (339). However, O-GlcNAcylation's roles in Parkinson's disease may be much more complicated than preventing the formation of toxic protein aggregates. O-GlcNAcylation was found to be elevated in lysates from the post-mortem temporal cortex of Parkinson's disease patients compared with age-matched controls. Treatment of rat primary cortical neurons with a potent inhibitor of OGA significantly increased protein O-GlcNAcylation, activated MTOR signaling, decreased autophagic flux, and increased α-synuclein accumulation. Inhibition of MTOR by rapamycin decreased basal levels of protein O-GlcNAcylation, decreased AKT activation, and partially reversed the effect of the OGA inhibitor on α-synuclein monomer accumulation. It was postulated that excessive O-GlcNAcylation is detrimental to neurons by inhibiting autophagy and by increasing α-synuclein accumulation (340). Perhaps at moderate levels, O-GlcNAcylation is beneficial by preventing toxic protein aggregation, but when it is too high, the sugar modification alters other processes in a deleterious manner. Clearly, O-GlcNAcylation is a fertile area in which to explore new avenues for treatment of AD and other neurodegenerative diseases.
Conclusions and future directions
After more than 3 decades of research on O-GlcNAcylation, it is now clear that O-GlcNAcylation serves as a major nutrient sensor, acting like a rheostat, to modulate most cellular processes in response to the nutrient and stress status of the cell. Despite the efforts of many laboratories, this field is still in its infancy. The technological challenges of studying this protein modification remain a major hurdle, and there is an acute need for the development of new tools to study O-GlcNAc cycling in living cells. Tools to selectively modify O-GlcNAcylation on a single protein or at a specific site will be essential for elucidating molecular mechanisms underlying the sugar's physiological functions. Whereas nearly every protein involved in transcription is O-GlcNAcylated, and it is evident that nutrients regulate gene expression, in part, by the cycling of O-GlcNAc, we know very little about the molecular mechanistic roles that O-GlcNAcylation plays in the transcription machinery. O-GlcNAcylation is extraordinarily abundant in neurons and in the brain, and limited data suggest that the sugar plays major roles in neuronal functions and in neurodegeneration. However, so far, very little research with respect to the specific functions of O-GlcNAcylation in neuronal processes has been performed. The O-GlcNAcylation field is a major largely unexplored frontier that not only will have a huge effect on our understanding of fundamental biological processes, but also will impact future development of novel treatments for the chronic diseases of aging. The study of O-GlcNAcylation represents a wide open and exciting area for young researchers looking for a challenging yet important problem to tackle.
Acknowledgments
I thank the ASBMB for the great honor of receiving the 2018 Herbert Tabor award, named for a stellar scientist, a good friend, an outstanding citizen of our field, and someone I have admired for many years. I also recognize and thank the outstanding graduate students and postdoctoral fellows who have worked with me on O-GlcNAcylation for the past 3 decades. I have been blessed to have such creative, smart, and hard-working people in my laboratory.
This work was supported by National Institutes of Health Grants P01HL107153, R01GM116891, and R01DK61671 and the Georgia Research Alliance. The author declares that he has no conflicts of interest with the contents of this article. The content is solely the responsibility of the author and does not necessarily represent the official views of the National Institutes of Health.
S. Hardivillé, P. S. Banerjee, E. S. Selen Alpergin, G. Han, J. Ma, C. C. Talbot, Jr., P. Hu, M. J. Wolfgang, and G. W. Hart, submitted for publication.
- CTD
- C-terminal domain
- OGT
- O-GlcNAc transferase
- mOGT
- mitochondrial isoform of OGT
- OGA
- O-GlcNAcase
- eNOS
- endothelial nitric-oxide synthase
- CaMKII and CaMKIV
- calcium/calmodulin kinase II and IV, respectively
- TAK1
- transforming growth factor-β–activated kinase
- CDK
- cyclin-dependent kinase
- HBP
- hexosamine biosynthetic pathway
- AD
- Alzheimer's disease
- PFK1
- phosphofructokinase 1
- MMP
- matrix metalloproteinase
- VEGF
- vascular endothelial growth factor
- PDAC
- pancreatic ductal adenocarcinoma.
References
- 1. Torres C. R., and Hart G. W. (1984) Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes: evidence for O-linked GlcNAc. J. Biol. Chem. 259, 3308–3317 [PubMed] [Google Scholar]
- 2. Holt G. D., and Hart G. W. (1986) The subcellular distribution of terminal N-acetylglucosamine moieties: localization of a novel protein-saccharide linkage, O-linked GlcNAc. J. Biol. Chem. 261, 8049–8057 [PubMed] [Google Scholar]
- 3. Holt G. D., Haltiwanger R. S., Torres C. R., and Hart G. W. (1987) Erythrocytes contain cytoplasmic glycoproteins: O-linked GlcNAc on Band 4.1. J. Biol. Chem. 262, 14847–14850 [PubMed] [Google Scholar]
- 4. Hanover J. A., Cohen C. K., Willingham M. C., and Park M. K. (1987) O-Linked N-acetylglucosamine is attached to proteins of the nuclear pore: evidence for cytoplasmic and nucleoplasmic glycoproteins. J. Biol. Chem. 262, 9887–9894 [PubMed] [Google Scholar]
- 5. Holt G. D., Snow C. M., Senior A., Haltiwanger R. S., Gerace L., and Hart G. W. (1987) Nuclear pore complex glycoproteins contain cytoplasmically disposed O-linked N-acetylglucosamine. J. Cell Biol. 104, 1157–1164 10.1083/jcb.104.5.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Park M. K., D'Onofrio M., Willingham M. C., and Hanover J. A. (1987) A monoclonal antibody against a family of nuclear pore proteins (nucleoporins): O-linked N-acetylglucosamine is part of the immunodeterminant. Proc. Natl. Acad. Sci. U.S.A. 84, 6462–6466 10.1073/pnas.84.18.6462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Benko D. M., Haltiwanger R. S., Hart G. W., and Gibson W. (1988) Virion basic phosphoprotein from human cytomegalovirus contains O-linked N-acetylglucosamine. Proc. Natl. Acad. Sci. U.S.A. 85, 2573–2577 10.1073/pnas.85.8.2573 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Greis K. D., Gibson W., and Hart G. W. (1994) Site-specific glycosylation of the human cytomegalovirus tegument basic phosphoprotein (UL32) at serine 921 and serine 952. J. Virol. 68, 8339–8349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Jackson S. P., and Tjian R. (1988) O-Glycosylation of eukaryotic transcription factors: implications for mechanisms of transcriptional regulation. Cell 55, 125–133 10.1016/0092-8674(88)90015-3 [DOI] [PubMed] [Google Scholar]
- 10. Reason A. J., Morris H. R., Panico M., Marais R., Treisman R. H., Haltiwanger R. S., Hart G. W., Kelly W. G., and Dell A. (1992) Localization of O-GlcNAc modification on the serum response transcription factor. J. Biol. Chem. 267, 16911–16921 [PubMed] [Google Scholar]
- 11. Chou T. Y., Dang C. V., and Hart G. W. (1995) Glycosylation of the c-Myc transactivation domain. Proc. Natl. Acad. Sci. U.S.A. 92, 4417–4421 10.1073/pnas.92.10.4417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Chou T. Y., Hart G. W., and Dang C. V. (1995) c-Myc is glycosylated at threonine 58, a known phosphorylation site and a mutational hot spot in lymphomas. J. Biol. Chem. 270, 18961–18965 10.1074/jbc.270.32.18961 [DOI] [PubMed] [Google Scholar]
- 13. Wells L., Slawson C., and Hart G. W. (2011) The E2F-1 associated retinoblastoma-susceptibility gene product is modified by O-GlcNAc. Amino Acids 40, 877–883 10.1007/s00726-010-0709-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Kelly W. G., Dahmus M. E., and Hart G. W. (1993) RNA polymerase II is a glycoprotein: modification of the COOH-terminal domain by O-GlcNAc. J. Biol. Chem. 268, 10416–10424 [PubMed] [Google Scholar]
- 15. Datta B., Ray M. K., Chakrabarti D., Wylie D. E., and Gupta N. K. (1989) Glycosylation of eukaryotic peptide chain initiation factor 2 (eIF-2)-associated 67-kDa polypeptide (p67) and its possible role in the inhibition of eIF-2 kinase-catalyzed phosphorylation of the eIF-2 α-subunit. J. Biol. Chem. 264, 20620–20624 [PubMed] [Google Scholar]
- 16. Kelly W. G., and Hart G. W. (1989) Glycosylation of chromosomal proteins: localization of O-linked N-acetylglucosamine in Drosophila chromatin. Cell 57, 243–251 10.1016/0092-8674(89)90962-8 [DOI] [PubMed] [Google Scholar]
- 17. Roquemore E. P., Chevrier M. R., Cotter R. J., and Hart G. W. (1996) Dynamic O-GlcNAcylation of the small heat shock protein α B-crystallin. Biochemistry 35, 3578–3586 10.1021/bi951918j [DOI] [PubMed] [Google Scholar]
- 18. Kearse K. P., and Hart G. W. (1991) Lymphocyte activation induces rapid changes in nuclear and cytoplasmic glycoproteins. Proc. Natl. Acad. Sci. U.S.A. 88, 1701–1705 10.1073/pnas.88.5.1701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Haltiwanger R. S., Holt G. D., and Hart G. W. (1990) Enzymatic addition of O-GlcNAc to nuclear and cytoplasmic proteins. Identification of a uridine diphospho-N-acetylglucosamine:peptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 265, 2563–2568 [PubMed] [Google Scholar]
- 20. Haltiwanger R. S., Blomberg M. A., and Hart G. W. (1992) Glycosylation of nuclear and cytoplasmic proteins: purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide β-N-acetylglucosaminyltransferase. J. Biol. Chem. 267, 9005–9013 [PubMed] [Google Scholar]
- 21. Dong D. L., and Hart G. W. (1994) Purification and characterization of an O-GlcNAc selective N-acetyl-β-d-glucosaminidase from rat spleen cytosol. J. Biol. Chem. 269, 19321–19330 [PubMed] [Google Scholar]
- 22. Braidman I., Carroll M., Dance N., Robinson D., Poenaru L., Weber A., Dreyfus J. C., Overdijk B., and Hooghwinkel G. J. (1974) Characterisation of human N-acetyl-β-hexosaminidase C. FEBS Lett. 41, 181–184 10.1016/0014-5793(74)81206-8 [DOI] [PubMed] [Google Scholar]
- 23. Braidman I., Carroll M., Dance N., and Robinson D. (1974) Separation and properties of human brain hexosaminidase C. Biochem. J. 143, 295–301 10.1042/bj1430295 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kreppel L. K., Blomberg M. A., and Hart G. W. (1997) Dynamic glycosylation of nuclear and cytosolic proteins: cloning and characterization of a unique O-GlcNAc transferase with multiple tetratricopeptide repeats. J. Biol. Chem. 272, 9308–9315 10.1074/jbc.272.14.9308 [DOI] [PubMed] [Google Scholar]
- 25. Lubas W. A., Frank D. W., Krause M., and Hanover J. A. (1997) O-linked GlcNAc transferase is a conserved nucleocytoplasmic protein containing tetratricopeptide repeats. J. Biol. Chem. 272, 9316–9324 10.1074/jbc.272.14.9316 [DOI] [PubMed] [Google Scholar]
- 26. Shafi R., Iyer S. P., Ellies L. G., O'Donnell N., Marek K. W., Chui D., Hart G. W., and Marth J. D. (2000) The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc. Natl. Acad. Sci. U.S.A. 97, 5735–5739 10.1073/pnas.100471497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. O'Donnell N., Zachara N. E., Hart G. W., and Marth J. D. (2004) Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol. Cell. Biol. 24, 1680–1690 10.1128/MCB.24.4.1680-1690.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Gao Y., Wells L., Comer F. I., Parker G. J., and Hart G. W. (2001) Dynamic O-glycosylation of nuclear and cytosolic proteins: cloning and characterization of a neutral, cytosolic β-N-acetylglucosaminidase from human brain. J. Biol. Chem. 276, 9838–9845 10.1074/jbc.M010420200 [DOI] [PubMed] [Google Scholar]
- 29. Heckel D., Comtesse N., Brass N., Blin N., Zang K. D., and Meese E. (1998) Novel immunogenic antigen homologous to hyaluronidase in meningioma. Hum. Mol. Genet. 7, 1859–1872 10.1093/hmg/7.12.1859 [DOI] [PubMed] [Google Scholar]
- 30. Hurtado-Guerrero R., Dorfmueller H. C., and van Aalten D. M. (2008) Molecular mechanisms of O-GlcNAcylation. Curr. Opin. Struct. Biol. 18, 551–557 10.1016/j.sbi.2008.09.005 [DOI] [PubMed] [Google Scholar]
- 31. Martinez-Fleites C., Macauley M. S., He Y., Shen D. L., Vocadlo D. J., and Davies G. J. (2008) Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat. Struct. Mol. Biol. 15, 764–765 10.1038/nsmb.1443 [DOI] [PubMed] [Google Scholar]
- 32. Lazarus M. B., Nam Y., Jiang J., Sliz P., and Walker S. (2011) Structure of human O-GlcNAc transferase and its complex with a peptide substrate. Nature 469, 564–567 10.1038/nature09638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Vocadlo D. J. (2012) O-GlcNAc processing enzymes: catalytic mechanisms, substrate specificity, and enzyme regulation. Curr. Opin. Chem. Biol. 16, 488–497 10.1016/j.cbpa.2012.10.021 [DOI] [PubMed] [Google Scholar]
- 34. Ma X., Liu P., Yan H., Sun H., Liu X., Zhou F., Li L., Chen Y., Muthana M. M., Chen X., Wang P. G., and Zhang L. (2013) Substrate specificity provides insights into the sugar donor recognition mechanism of O-GlcNAc transferase (OGT). PLoS One 8, e63452 10.1371/journal.pone.0063452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pathak S., Alonso J., Schimpl M., Rafie K., Blair D. E., Borodkin V. S., Albarbarawi O., and van Aalten D. M. F. (2015) The active site of O-GlcNAc transferase imposes constraints on substrate sequence. Nat. Struct. Mol. Biol. 22, 744–750 10.1038/nsmb.3063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Trapannone R., Rafie K., and van Aalten D. M. (2016) O-GlcNAc transferase inhibitors: current tools and future challenges. Biochem. Soc. Trans. 44, 88–93 10.1042/BST20150189 [DOI] [PubMed] [Google Scholar]
- 37. Aquino-Gil M., Pierce A., Perez-Cervera Y., Zenteno E., and Lefebvre T. (2017) OGT: a short overview of an enzyme standing out from usual glycosyltransferases. Biochem. Soc. Trans. 45, 365–370 10.1042/BST20160404 [DOI] [PubMed] [Google Scholar]
- 38. Wang Y., Zhu J., and Zhang L. (2017) Discovery of cell-permeable O-GlcNAc transferase inhibitors via tethering in situ click chemistry. J. Med. Chem. 60, 263–272 10.1021/acs.jmedchem.6b01237 [DOI] [PubMed] [Google Scholar]
- 39. Ghirardello M., Perrone D., Chinaglia N., Sádaba D., Delso I., Tejero T., Marchesi E., Fogagnolo M., Rafie K., van Aalten D. M. F., and Merino P. (2018) UDP-GlcNAc analogs as inhibitors of O-GlcNAc transferase (OGT): spectroscopic, computational and biological studies. Chemistry 24, 7264–7272 10.1002/chem.201801083 [DOI] [PubMed] [Google Scholar]
- 40. Levine Z. G., and Walker S. (2016) The biochemistry of O-GlcNAc transferase: which functions make it essential in mammalian cells? Annu. Rev. Biochem. 85, 631–657 10.1146/annurev-biochem-060713-035344 [DOI] [PubMed] [Google Scholar]
- 41. Capotosti F., Guernier S., Lammers F., Waridel P., Cai Y., Jin J., Conaway J. W., Conaway R. C., and Herr W. (2011) O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell 144, 376–388 10.1016/j.cell.2010.12.030 [DOI] [PubMed] [Google Scholar]
- 42. Lazarus M. B., Jiang J., Kapuria V., Bhuiyan T., Janetzko J., Zandberg W. F., Vocadlo D. J., Herr W., and Walker S. (2013) HCF-1 is cleaved in the active site of O-GlcNAc transferase. Science 342, 1235–1239 10.1126/science.1243990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schultz J., and Pils B. (2002) Prediction of structure and functional residues for O-GlcNAcase, a divergent homologue of acetyltransferases. FEBS Lett. 529, 179–182 10.1016/S0014-5793(02)03322-7 [DOI] [PubMed] [Google Scholar]
- 44. Macauley M. S., Whitworth G. E., Debowski A. W., Chin D., and Vocadlo D. J. (2005) O-GlcNAcase uses substrate-assisted catalysis: kinetic analysis and development of highly selective mechanism-inspired inhibitors. J. Biol. Chem. 280, 25313–25322 10.1074/jbc.M413819200 [DOI] [PubMed] [Google Scholar]
- 45. Dennis R. J., Taylor E. J., Macauley M. S., Stubbs K. A., Turkenburg J. P., Hart S. J., Black G. N., Vocadlo D. J., and Davies G. J. (2006) Structure and mechanism of a bacterial β-glucosaminidase having O-GlcNAcase activity. Nat. Struct. Mol. Biol. 13, 365–371 10.1038/nsmb1079 [DOI] [PubMed] [Google Scholar]
- 46. Dorfmueller H. C., Borodkin V. S., Schimpl M., Shepherd S. M., Shpiro N. A., and van Aalten D. M. (2006) GlcNAcstatin: a picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J. Am. Chem. Soc. 128, 16484–16485 10.1021/ja066743n [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Clarke A. J., Hurtado-Guerrero R., Pathak S., Schüttelkopf A. W., Borodkin V., Shepherd S. M., Ibrahim A. F., and van Aalten D. M. (2008) Structural insights into mechanism and specificity of O-GlcNAc transferase. EMBO J. 27, 2780–2788 10.1038/emboj.2008.186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Yuzwa S. A., Macauley M. S., Heinonen J. E., Shan X., Dennis R. J., He Y., Whitworth G. E., Stubbs K. A., McEachern E. J., Davies G. J., and Vocadlo D. J. (2008) A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of Tau in vivo. Nat. Chem. Biol. 4, 483–490 10.1038/nchembio.96 [DOI] [PubMed] [Google Scholar]
- 49. Gloster T. M., and Vocadlo D. J. (2010) Mechanism, structure, and inhibition of O-GlcNAc processing enzymes. Curr. Signal Transduct. Ther. 5, 74–91 10.2174/157436210790226537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Alonso J., Schimpl M., and van Aalten D. M. (2014) O-GlcNAcase: promiscuous hexosaminidase or key regulator of O-GlcNAc signaling? J. Biol. Chem. 289, 34433–34439 10.1074/jbc.R114.609198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Li B., Li H., Hu C. W., and Jiang J. (2017) Structural insights into the substrate binding adaptability and specificity of human O-GlcNAcase. Nat. Commun. 8, 666 10.1038/s41467-017-00865-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Li B., Li H., Lu L., and Jiang J. (2017) Structures of human O-GlcNAcase and its complexes reveal a new substrate recognition mode. Nat. Struct. Mol. Biol. 24, 362–369 10.1038/nsmb.3390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roth C., Chan S., Offen W. A., Hemsworth G. R., Willems L. I., King D. T., Varghese V., Britton R., Vocadlo D. J., and Davies G. J. (2017) Structural and functional insight into human O-GlcNAcase. Nat. Chem. Biol. 13, 610–612 10.1038/nchembio.2358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Zachara N. E., Vosseller K., and Hart G. W. (2011) Detection and analysis of proteins modified by O-linked N-acetylglucosamine. Curr. Protoc. Protein Sci., Chapter 17, Unit 17.6 10.1002/0471142727.mb1706s95 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Wang Z., Udeshi N. D., O'Malley M., Shabanowitz J., Hunt D. F., and Hart G. W. (2010) Enrichment and site mapping of O-linked N-acetylglucosamine by a combination of chemical/enzymatic tagging, photochemical cleavage, and electron transfer dissociation mass spectrometry. Mol. Cell. Proteomics 9, 153–160 10.1074/mcp.M900268-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Ma J., and Hart G. W. (2017) Analysis of protein O-GlcNAcylation by mass spectrometry. Curr. Protoc. Protein Sci. 87, 24.10.1–24.10.16 10.1002/cpps.24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Snow C. M., Senior A., and Gerace L. (1987) Monoclonal antibodies identify a group of nuclear pore complex glycoproteins. J. Cell Biol. 104, 1143–1156 10.1083/jcb.104.5.1143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Comer F. I., Vosseller K., Wells L., Accavitti M. A., and Hart G. W. (2001) Characterization of a mouse monoclonal antibody specific for O-linked N-acetylglucosamine. Anal. Biochem. 293, 169–177 10.1006/abio.2001.5132 [DOI] [PubMed] [Google Scholar]
- 59. Alfaro J. F., Gong C. X., Monroe M. E., Aldrich J. T., Clauss T. R., Purvine S. O., Wang Z., Camp D. G. 2nd, Shabanowitz J., Stanley P., Hart G. W., Hunt D. F., Yang F., and Smith R. D. (2012) Tandem mass spectrometry identifies many mouse brain O-GlcNAcylated proteins including EGF domain-specific O-GlcNAc transferase targets. Proc. Natl. Acad. Sci. U.S.A. 109, 7280–7285 10.1073/pnas.1200425109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mikesh L. M., Ueberheide B., Chi A., Coon J. J., Syka J. E., Shabanowitz J., and Hunt D. F. (2006) The utility of ETD mass spectrometry in proteomic analysis. Biochim. Biophys. Acta 1764, 1811–1822 10.1016/j.bbapap.2006.10.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Myers S. A., Daou S., Affar E. B., Burlingame A. (2013) Electron transfer dissociation (ETD): the mass spectrometric breakthrough essential for O-GlcNAc protein site assignments—a study of the O-GlcNAcylated protein host cell factor C1. Proteomics 13, 982–991 10.1002/pmic.201200332 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Zachara N. E., and Hart G. W. (2004) O-GlcNAc a sensor of cellular state: the role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim. Biophys. Acta 1673, 13–28 10.1016/j.bbagen.2004.03.016 [DOI] [PubMed] [Google Scholar]
- 63. Hart G. W., Slawson C., Ramirez-Correa G., and Lagerlof O. (2011) Cross talk between O-GlcNAcylation and phosphorylation: roles in signaling, transcription, and chronic disease. Annu. Rev. Biochem. 80, 825–858 10.1146/annurev-biochem-060608-102511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kreppel L. K., and Hart G. W. (1999) Regulation of a cytosolic and nuclear O-GlcNAc transferase: role of the tetratricopeptide repeats. J. Biol. Chem. 274, 32015–32022 10.1074/jbc.274.45.32015 [DOI] [PubMed] [Google Scholar]
- 65. Han I., Oh E. S., and Kudlow J. E. (2000) Responsiveness of the state of O-linked N-acetylglucosamine modification of nuclear pore protein p62 to the extracellular glucose concentration. Biochem. J. 350, 109–114 10.1042/bj3500109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Liu K., Paterson A. J., Chin E., and Kudlow J. E. (2000) Glucose stimulates protein modification by O-linked GlcNAc in pancreatic beta cells: linkage of O-linked GlcNAc to beta cell death. Proc. Natl. Acad. Sci. U.S.A. 97, 2820–2825 10.1073/pnas.97.6.2820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Akimoto Y., Kreppel L. K., Hirano H., and Hart G. W. (2001) Hyperglycemia and the O-GlcNAc transferase in rat aortic smooth muscle cells: elevated expression and altered patterns of O-GlcNAcylation. Arch. Biochem. Biophys. 389, 166–175 10.1006/abbi.2001.2331 [DOI] [PubMed] [Google Scholar]
- 68. Du X. L., Edelstein D., Dimmeler S., Ju Q., Sui C., and Brownlee M. (2001) Hyperglycemia inhibits endothelial nitric oxide synthase activity by posttranslational modification at the Akt site. J. Clin. Invest. 108, 1341–1348 10.1172/JCI11235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Walgren J. L., Vincent T. S., Schey K. L., and Buse M. G. (2003) High glucose and insulin promote O-GlcNAc modification of proteins, including α-tubulin. Am. J. Physiol. Endocrinol. Metab. 284, E424–E434 10.1152/ajpendo.00382.2002 [DOI] [PubMed] [Google Scholar]
- 70. Goldberg H. J., Whiteside C. I., Hart G. W., and Fantus I. G. (2006) Posttranslational, reversible O-glycosylation is stimulated by high glucose and mediates plasminogen activator inhibitor-1 gene expression and Sp1 transcriptional activity in glomerular mesangial cells. Endocrinology 147, 222–231 10.1210/en.2005-0523 [DOI] [PubMed] [Google Scholar]
- 71. Zachara N. E., O'Donnell N., Cheung W. D., Mercer J. J., Marth J. D., and Hart G. W. (2004) Dynamic O-GlcNAc modification of nucleocytoplasmic proteins in response to stress: a survival response of mammalian cells. J. Biol. Chem. 279, 30133–30142 10.1074/jbc.M403773200 [DOI] [PubMed] [Google Scholar]
- 72. Zachara N. E., Molina H., Wong K. Y., Pandey A., and Hart G. W. (2011) The dynamic stress-induced “O-GlcNAc-ome” highlights functions for O-GlcNAc in regulating DNA damage/repair and other cellular pathways. Amino Acids 40, 793–808 10.1007/s00726-010-0695-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Liu J., Pang Y., Chang T., Bounelis P., Chatham J. C., and Marchase R. B. (2006) Increased hexosamine biosynthesis and protein O-GlcNAc levels associated with myocardial protection against calcium paradox and ischemia. J. Mol. Cell. Cardiol. 40, 303–312 10.1016/j.yjmcc.2005.11.003 [DOI] [PubMed] [Google Scholar]
- 74. Fülöp N., Marchase R. B., and Chatham J. C. (2007) Role of protein O-linked N-acetyl-glucosamine in mediating cell function and survival in the cardiovascular system. Cardiovasc. Res. 73, 288–297 10.1016/j.cardiores.2006.07.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Fülöp N., Zhang Z., Marchase R. B., and Chatham J. C. (2007) Glucosamine cardioprotection in perfused rat hearts associated with increased O-linked N-acetylglucosamine protein modification and altered p38 activation. Am. J. Physiol. Heart Circ. Physiol. 292, H2227–H2236 10.1152/ajpheart.01091.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Jones S. P., Zachara N. E., Ngoh G. A., Hill B. G., Teshima Y., Bhatnagar A., Hart G. W., and Marbán E. (2008) Cardioprotection by N-acetylglucosamine linkage to cellular proteins. Circulation 117, 1172–1182 10.1161/CIRCULATIONAHA.107.730515 [DOI] [PubMed] [Google Scholar]
- 77. Zou L., Yang S., Champattanachai V., Hu S., Chaudry I. H., Marchase R. B., and Chatham J. C. (2009) Glucosamine improves cardiac function following trauma-hemorrhage by increased protein O-GlcNAcylation and attenuation of NF-κB signaling. Am. J. Physiol. Heart Circ. Physiol. 296, H515–H523 10.1152/ajpheart.01025.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Jensen R. V., Zachara N. E., Nielsen P. H., Kimose H. H., Kristiansen S. B., and Bøtker H. E. (2013) Impact of O-GlcNAc on cardioprotection by remote ischaemic preconditioning in non-diabetic and diabetic patients. Cardiovasc. Res. 97, 369–378 10.1093/cvr/cvs337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Vibjerg Jensen R., Johnsen J., Buus Kristiansen S., Zachara N. E., and Bøtker H. E. (2013) Ischemic preconditioning increases myocardial O-GlcNAc glycosylation. Scand. Cardiovasc. J. 47, 168–174 10.3109/14017431.2012.756984 [DOI] [PubMed] [Google Scholar]
- 80. Marsh S. A., Collins H. E., and Chatham J. C. (2014) Protein O-GlcNAcylation and cardiovascular (patho)physiology. J. Biol. Chem. 289, 34449–34456 10.1074/jbc.R114.585984 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. McNulty P. H. (2007) Hexosamine biosynthetic pathway flux and cardiomyopathy in type 2 diabetes mellitus: focus on “Impact of type 2 diabetes and aging on cardiomyocyte function and O-linked N-acetylglucosamine levels in the heart”. Am. J. Physiol. Cell Physiol. 292, C1243–C1244 10.1152/ajpcell.00521.2006 [DOI] [PubMed] [Google Scholar]
- 82. Kim H. S., Park S. Y., Choi Y. R., Kang J. G., Joo H. J., Moon W. K., and Cho J. W. (2009) Excessive O-GlcNAcylation of proteins suppresses spontaneous cardiogenesis in ES cells. FEBS Lett. 583, 2474–2478 10.1016/j.febslet.2009.06.052 [DOI] [PubMed] [Google Scholar]
- 83. Aguilar H., Fricovsky E., Ihm S., Schimke M., Maya-Ramos L., Aroonsakool N., Ceballos G., Dillmann W., Villarreal F., and Ramirez-Sanchez I. (2014) Role for high-glucose-induced protein O-GlcNAcylation in stimulating cardiac fibroblast collagen synthesis. Am. J. Physiol. Cell Physiol. 306, C794–C804 10.1152/ajpcell.00251.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Banerjee P. S., Ma J., and Hart G. W. (2015) Diabetes-associated dysregulation of O-GlcNAcylation in rat cardiac mitochondria. Proc. Natl. Acad. Sci. U.S.A. 112, 6050–6055 10.1073/pnas.1424017112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Brahma M. K., Pepin M. E., and Wende A. R. (2017) My sweetheart is broken: role of glucose in diabetic cardiomyopathy. Diabetes Metab. J. 41, 1–9 10.4093/dmj.2017.41.1.1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Jia G., Hill M. A., and Sowers J. R. (2018) Diabetic cardiomyopathy: an update of mechanisms contributing to this clinical entity. Circ. Res. 122, 624–638 10.1161/CIRCRESAHA.117.311586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Zachara N. E. (2012) The roles of O-linked β-N-acetylglucosamine in cardiovascular physiology and disease. Am. J. Physiol. Heart Circ. Physiol. 302, H1905–H1918 10.1152/ajpheart.00445.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Wang J., Liu R., Hawkins M., Barzilai N., and Rossetti L. (1998) A nutrient-sensing pathway regulates leptin gene expression in muscle and fat. Nature 393, 684–688 10.1038/31474 [DOI] [PubMed] [Google Scholar]
- 89. Comer F. I., and Hart G. W. (1999) O-GlcNAc and the control of gene expression. Biochim. Biophys. Acta 1473, 161–171 10.1016/S0304-4165(99)00176-2 [DOI] [PubMed] [Google Scholar]
- 90. Issad T., and Kuo M. (2008) O-GlcNAc modification of transcription factors, glucose sensing and glucotoxicity. Trends Endocrinol. Metab. 19, 380–389 10.1016/j.tem.2008.09.001 [DOI] [PubMed] [Google Scholar]
- 91. Brimble S., Wollaston-Hayden E. E., Teo C. F., Morris A. C., and Wells L. (2010) The role of the O-GlcNAc modification in regulating eukaryotic gene expression. Curr. Signal Transduct. Ther. 5, 12–24 10.2174/157436210790226465 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Ho S. R., Wang K., Whisenhunt T. R., Huang P., Zhu X., Kudlow J. E., and Paterson A. J. (2010) O-GlcNAcylation enhances FOXO4 transcriptional regulation in response to stress. FEBS Lett. 584, 49–54 10.1016/j.febslet.2009.11.059 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Ozcan S., Andrali S. S., and Cantrell J. E. (2010) Modulation of transcription factor function by O-GlcNAc modification. Biochim. Biophys. Acta 1799, 353–364 10.1016/j.bbagrm.2010.02.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Hardivillé S., and Hart G. W. (2014) Nutrient regulation of signaling, transcription, and cell physiology by O-GlcNAcylation. Cell Metab. 20, 208–213 10.1016/j.cmet.2014.07.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Lewis B. A., and Hanover J. A. (2014) O-GlcNAc and the epigenetic regulation of gene expression. J. Biol. Chem. 289, 34440–34448 10.1074/jbc.R114.595439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Nagel A. K., and Ball L. E. (2015) Intracellular protein O-GlcNAc modification integrates nutrient status with transcriptional and metabolic regulation. Adv. Cancer Res. 126, 137–166 10.1016/bs.acr.2014.12.003 [DOI] [PubMed] [Google Scholar]
- 97. Liu S. H., Peng B. H., Ma J. T., Liu Y. C., and Ng S. Y. (1995) Serum response element associated transcription factors in mouse embryos: serum response factor, YY1, and PEA3 factor. Dev. Genet. 16, 229–240 10.1002/dvg.1020160303 [DOI] [PubMed] [Google Scholar]
- 98. Han I., and Kudlow J. E. (1997) Reduced O-glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol. Cell. Biol. 17, 2550–2558 10.1128/MCB.17.5.2550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Jiang M. S., and Hart G. W. (1997) A subpopulation of estrogen receptors are modified by O-linked N-acetylglucosamine. J. Biol. Chem. 272, 2421–2428 10.1074/jbc.272.4.2421 [DOI] [PubMed] [Google Scholar]
- 100. Juang Y. T., Solomou E. E., Rellahan B., and Tsokos G. C. (2002) Phosphorylation and O-linked glycosylation of Elf-1 leads to its translocation to the nucleus and binding to the promoter of the TCR ζ-chain. J. Immunol. 168, 2865–2871 10.4049/jimmunol.168.6.2865 [DOI] [PubMed] [Google Scholar]
- 101. Chalkley R. J., and Burlingame A. L. (2003) Identification of novel sites of O-N-acetylglucosamine modification of serum response factor using quadrupole time-of-flight mass spectrometry. Mol. Cell. Proteomics 2, 182–190 10.1074/mcp.M300027-MCP200 [DOI] [PubMed] [Google Scholar]
- 102. Gao Y., Miyazaki J., and Hart G. W. (2003) The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch. Biochem. Biophys. 415, 155–163 10.1016/S0003-9861(03)00234-0 [DOI] [PubMed] [Google Scholar]
- 103. Hiromura M., Choi C. H., Sabourin N. A., Jones H., Bachvarov D., and Usheva A. (2003) YY1 is regulated by O-linked N-acetylglucosaminylation (O-GlcNAcylation). J. Biol. Chem. 278, 14046–14052 10.1074/jbc.M300789200 [DOI] [PubMed] [Google Scholar]
- 104. Lamarre-Vincent N., and Hsieh-Wilson L. C. (2003) Dynamic glycosylation of the transcription factor CREB: a potential role in gene regulation. J. Am. Chem. Soc. 125, 6612–6613 10.1021/ja028200t [DOI] [PubMed] [Google Scholar]
- 105. Majumdar G., Harmon A., Candelaria R., Martinez-Hernandez A., Raghow R., and Solomon S. S. (2003) O-Glycosylation of Sp1 and transcriptional regulation of the calmodulin gene by insulin and glucagon. Am. J. Physiol. Endocrinol. Metab. 285, E584–E591 10.1152/ajpendo.00140.2003 [DOI] [PubMed] [Google Scholar]
- 106. Tsokos G. C., Nambiar M. P., and Juang Y. T. (2003) Activation of the Ets transcription factor Elf-1 requires phosphorylation and glycosylation: defective expression of activated Elf-1 is involved in the decreased TCR ζ chain gene expression in patients with systemic lupus erythematosus. Ann. N.Y. Acad. Sci. 987, 240–245 10.1111/j.1749-6632.2003.tb06054.x [DOI] [PubMed] [Google Scholar]
- 107. Gewinner C., Hart G., Zachara N., Cole R., Beisenherz-Huss C., and Groner B. (2004) The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J. Biol. Chem. 279, 3563–3572 10.1074/jbc.M306449200 [DOI] [PubMed] [Google Scholar]
- 108. Nanashima N., Asano J., Hayakari M., Nakamura T., Nakano H., Yamada T., Shimizu T., Akita M., Fan Y., and Tsuchida S. (2005) Nuclear localization of STAT5A modified with O-linked N-acetylglucosamine and early involution in the mammary gland of Hirosaki hairless rat. J. Biol. Chem. 280, 43010–43016 10.1074/jbc.M509481200 [DOI] [PubMed] [Google Scholar]
- 109. Ahmad I., Hoessli D. C., Walker-Nasir E., Rafik S. M., Shakoori A. R., and Nasir-ud-Din (2006) Oct-2 DNA binding transcription factor: functional consequences of phosphorylation and glycosylation. Nucleic Acids Res. 34, 175–184 10.1093/nar/gkj401 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Azuma Y., Miura K., Higai K., and Matsumoto K. (2007) Protein O-N-acetylglucosaminylation modulates promoter activities of cyclic AMP response element and activator protein 1 and enhances E-selectin expression on HuH-7 human hepatoma cells. Biol. Pharm. Bull. 30, 2284–2289 10.1248/bpb.30.2284 [DOI] [PubMed] [Google Scholar]
- 111. Noach N., Segev Y., Levi I., Segal S., and Priel E. (2007) Modification of topoisomerase I activity by glucose and by O-GlcNAcylation of the enzyme protein. Glycobiology 17, 1357–1364 10.1093/glycob/cwm105 [DOI] [PubMed] [Google Scholar]
- 112. Housley M. P., Rodgers J. T., Udeshi N. D., Kelly T. J., Shabanowitz J., Hunt D. F., Puigserver P., and Hart G. W. (2008) O-GlcNAc regulates FoxO activation in response to glucose. J. Biol. Chem. 283, 16283–16292 10.1074/jbc.M802240200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Kuo M., Zilberfarb V., Gangneux N., Christeff N., and Issad T. (2008) O-GlcNAc modification of FoxO1 increases its transcriptional activity: a role in the glucotoxicity phenomenon? Biochimie 90, 679–685 10.1016/j.biochi.2008.03.005 [DOI] [PubMed] [Google Scholar]
- 114. Sayat R., Leber B., Grubac V., Wiltshire L., and Persad S. (2008) O-GlcNAc-glycosylation of β-catenin regulates its nuclear localization and transcriptional activity. Exp. Cell Res. 314, 2774–2787 10.1016/j.yexcr.2008.05.017 [DOI] [PubMed] [Google Scholar]
- 115. Bachmaier R., Aryee D. N., Jug G., Kauer M., Kreppel M., Lee K. A., and Kovar H. (2009) O-GlcNAcylation is involved in the transcriptional activity of EWS-FLI1 in Ewing's sarcoma. Oncogene 28, 1280–1284 10.1038/onc.2008.484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Housley M. P., Udeshi N. D., Rodgers J. T., Shabanowitz J., Puigserver P., Hunt D. F., and Hart G. W. (2009) A PGC-1α-O-GlcNAc transferase complex regulates FoxO transcription factor activity in response to glucose. J. Biol. Chem. 284, 5148–5157 10.1074/jbc.M808890200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117. Jochmann R., Thurau M., Jung S., Hofmann C., Naschberger E., Kremmer E., Harrer T., Miller M., Schaft N., and Stürzl M. (2009) O-Linked N-acetylglucosaminylation of Sp1 inhibits the human immunodeficiency virus type 1 promoter. J. Virol. 83, 3704–3718 10.1128/JVI.01384-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Li X., Molina H., Huang H., Zhang Y. Y., Liu M., Qian S. W., Slawson C., Dias W. B., Pandey A., Hart G. W., Lane M. D., and Tang Q. Q. (2009) O-linked N-acetylglucosamine modification on CCAAT enhancer-binding protein β: role during adipocyte differentiation. J. Biol. Chem. 284, 19248–19254 10.1074/jbc.M109.005678 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Lim K., and Chang H. I. (2009) O-GlcNAc modification of Sp1 inhibits the functional interaction between Sp1 and Oct1. FEBS Lett. 583, 512–520 10.1016/j.febslet.2008.12.007 [DOI] [PubMed] [Google Scholar]
- 120. Anthonisen E. H., Berven L., Holm S., Nygård M., Nebb H. I., and Grönning-Wang L. M. (2010) Nuclear receptor liver X receptor is O-GlcNAc-modified in response to glucose. J. Biol. Chem. 285, 1607–1615 10.1074/jbc.M109.082685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121. Park S. Y., Kim H. S., Kim N. H., Ji S., Cha S. Y., Kang J. G., Ota I., Shimada K., Konishi N., Nam H. W., Hong S. W., Yang W. H., Roth J., Yook J. I., and Cho J. W. (2010) Snail1 is stabilized by O-GlcNAc modification in hyperglycaemic condition. EMBO J. 29, 3787–3796 10.1038/emboj.2010.254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122. Guinez C., Filhoulaud G., Rayah-Benhamed F., Marmier S., Dubuquoy C., Dentin R., Moldes M., Burnol A. F., Yang X., Lefebvre T., Girard J., and Postic C. (2011) O-GlcNAcylation increases ChREBP protein content and transcriptional activity in the liver. Diabetes 60, 1399–1413 10.2337/db10-0452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123. Xing D., Gong K., Feng W., Nozell S. E., Chen Y. F., Chatham J. C., and Oparil S. (2011) O-GlcNAc modification of NFκB p65 inhibits TNF-α-induced inflammatory mediator expression in rat aortic smooth muscle cells. PLoS One 6, e24021 10.1371/journal.pone.0024021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Allison D. F., Wamsley J. J., Kumar M., Li D., Gray L. G., Hart G. W., Jones D. R., and Mayo M. W. (2012) Modification of RelA by O-linked N-acetylglucosamine links glucose metabolism to NF-κB acetylation and transcription. Proc. Natl. Acad. Sci. U.S.A. 109, 16888–16893 10.1073/pnas.1208468109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125. Facundo H. T., Brainard R. E., Watson L. J., Ngoh G. A., Hamid T., Prabhu S. D., and Jones S. P. (2012) O-GlcNAc signaling is essential for NFAT-mediated transcriptional reprogramming during cardiomyocyte hypertrophy. Am. J. Physiol. Heart Circ. Physiol. 302, H2122–H2130 10.1152/ajpheart.00775.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126. Ido-Kitamura Y., Sasaki T., Kobayashi M., Kim H. J., Lee Y. S., Kikuchi O., Yokota-Hashimoto H., Iizuka K., Accili D., and Kitamura T. (2012) Hepatic FoxO1 integrates glucose utilization and lipid synthesis through regulation of Chrebp O-glycosylation. PLoS One 7, e47231 10.1371/journal.pone.0047231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127. Ji S., Park S. Y., Roth J., Kim H. S., and Cho J. W. (2012) O-GlcNAc modification of PPARγ reduces its transcriptional activity. Biochem. Biophys. Res. Commun. 417, 1158–1163 10.1016/j.bbrc.2011.12.086 [DOI] [PubMed] [Google Scholar]
- 128. Rexach J. E., Clark P. M., Mason D. E., Neve R. L., Peters E. C., and Hsieh-Wilson L. C. (2012) Dynamic O-GlcNAc modification regulates CREB-mediated gene expression and memory formation. Nat. Chem. Biol. 8, 253–261 10.1038/nchembio.770 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129. Hart G. W. (2013) Nutrient regulation of immunity: O-GlcNAcylation regulates stimulus-specific NF-κB-dependent transcription. Sci. Signal. 6, pe26 10.1126/scisignal.2004596 [DOI] [PubMed] [Google Scholar]
- 130. Kang J., Shen Z., Lim J. M., Handa H., Wells L., and Tantin D. (2013) Regulation of Oct1/Pou2f1 transcription activity by O-GlcNAcylation. FASEB J. 27, 2807–2817 10.1096/fj.12-220897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Ma Y. T., Luo H., Guan W. J., Zhang H., Chen C., Wang Z., and Li J. D. (2013) O-GlcNAcylation of BMAL1 regulates circadian rhythms in NIH3T3 fibroblasts. Biochem. Biophys. Res. Commun. 431, 382–387 10.1016/j.bbrc.2013.01.043 [DOI] [PubMed] [Google Scholar]
- 132. Ramakrishnan P., Clark P. M., Mason D. E., Peters E. C., Hsieh-Wilson L. C., and Baltimore D. (2013) Activation of the transcriptional function of the NF-κB protein c-Rel by O-GlcNAc glycosylation. Sci. Signal. 6, ra75 10.1126/scisignal.2004097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133. Chu C. S., Lo P. W., Yeh Y. H., Hsu P. H., Peng S. H., Teng Y. C., Kang M. L., Wong C. H., and Juan L. J. (2014) O-GlcNAcylation regulates EZH2 protein stability and function. Proc. Natl. Acad. Sci. U.S.A. 111, 1355–1360 10.1073/pnas.1323226111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134. Ha J. R., Hao L., Venkateswaran G., Huang Y. H., Garcia E., and Persad S. (2014) β-Catenin is O-GlcNAc glycosylated at serine 23: implications for β-catenin's subcellular localization and transactivator function. Exp. Cell Res. 321, 153–166 10.1016/j.yexcr.2013.11.021 [DOI] [PubMed] [Google Scholar]
- 135. Nagel A. K., and Ball L. E. (2014) O-GlcNAc modification of the runt-related transcription factor 2 (Runx2) links osteogenesis and nutrient metabolism in bone marrow mesenchymal stem cells. Mol. Cell. Proteomics 13, 3381–3395 10.1074/mcp.M114.040691 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136. Chen J., Liu X., Lü F., Liu X., Ru Y., Ren Y., Yao L., and Zhang Y. (2015) Transcription factor Nrf1 is negatively regulated by its O-GlcNAcylation status. FEBS Lett. 589, 2347–2358 10.1016/j.febslet.2015.07.030 [DOI] [PubMed] [Google Scholar]
- 137. Ha C., and Lim K. (2015) O-GlcNAc modification of Sp3 and Sp4 transcription factors negatively regulates their transcriptional activities. Biochem. Biophys. Res. Commun. 467, 341–347 10.1016/j.bbrc.2015.09.155 [DOI] [PubMed] [Google Scholar]
- 138. Kim S. J., Yoo W. S., Choi M., Chung I., Yoo J. M., and Choi W. S. (2016) Increased O-GlcNAcylation of NF-κB enhances retinal ganglion cell death in streptozotocin-induced diabetic retinopathy. Curr. Eye Res. 41, 249–257 10.3109/02713683.2015.1006372 [DOI] [PubMed] [Google Scholar]
- 139. Liu Q., Tao T., Liu F., Ni R., Lu C., and Shen A. (2016) Hyper-O-GlcNAcylation of YB-1 affects Ser102 phosphorylation and promotes cell proliferation in hepatocellular carcinoma. Exp. Cell Res. 349, 230–238 10.1016/j.yexcr.2016.10.011 [DOI] [PubMed] [Google Scholar]
- 140. Myers S. A., Peddada S., Chatterjee N., Friedrich T., Tomoda K., Krings G., Thomas S., Maynard J., Broeker M., Thomson M., Pollard K., Yamanaka S., Burlingame A. L., and Panning B. (2016) SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. eLife 5, e10647 10.7554/eLife.10647 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141. Constable S., Lim J. M., Vaidyanathan K., and Wells L. (2017) O-GlcNAc transferase regulates transcriptional activity of human Oct4. Glycobiology 27, 927–937 10.1093/glycob/cwx055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Li H., Liu X., Wang D., Su L., Zhao T., Li Z., Lin C., Zhang Y., Huang B., Lu J., and Li X. (2017) O-GlcNAcylation of SKN-1 modulates the lifespan and oxidative stress resistance in Caenorhabditis elegans. Sci. Rep. 7, 43601 10.1038/srep43601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Zhang Y., Qu Y., Niu T., Wang H., and Liu K. (2017) O-GlcNAc modification of Sp1 mediates hyperglycaemia-induced ICAM-1 up-regulation in endothelial cells. Biochem. Biophys. Res. Commun. 484, 79–84 10.1016/j.bbrc.2017.01.068 [DOI] [PubMed] [Google Scholar]
- 144. Gambetta M. C., Oktaba K., and Müller J. (2009) Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science 325, 93–96 10.1126/science.1169727 [DOI] [PubMed] [Google Scholar]
- 145. Sinclair D. A., Syrzycka M., Macauley M. S., Rastgardani T., Komljenovic I., Vocadlo D. J., Brock H. W., and Honda B. M. (2009) Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc. Natl. Acad. Sci. U.S.A. 106, 13427–13432 10.1073/pnas.0904638106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146. Hanover J. A., Krause M. W., and Love D. C. (2012) Bittersweet memories: linking metabolism to epigenetics through O-GlcNAcylation. Nat. Rev. Mol. Cell Biol. 13, 312–321 10.1038/nrm3334 [DOI] [PubMed] [Google Scholar]
- 147. Verrijzer C. P. (2014) Sugarcoating Polycomb repression. Dev. Cell 31, 521–522 10.1016/j.devcel.2014.11.027 [DOI] [PubMed] [Google Scholar]
- 148. Ranuncolo S. M., Ghosh S., Hanover J. A., Hart G. W., and Lewis B. A. (2012) Evidence of the involvement of O-GlcNAc-modified human RNA polymerase II CTD in transcription in vitro and in vivo. J. Biol. Chem. 287, 23549–23561 10.1074/jbc.M111.330910 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Lewis B. A., Burlingame A. L., and Myers S. A. (2016) Human RNA polymerase II promoter recruitment in vitro is regulated by O-linked N-acetylglucosaminyltransferase (OGT). J. Biol. Chem. 291, 14056–14061 10.1074/jbc.M115.684365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150. Resto M., Kim B. H., Fernandez A. G., Abraham B. J., Zhao K., and Lewis B. A. (2016) O-GlcNAcase is an RNA polymerase II elongation factor coupled to pausing factors SPT5 and TIF1β. J. Biol. Chem. 291, 22703–22713 10.1074/jbc.M116.751420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Svejstrup J. Q. (2012) Transcription: another mark in the tail. EMBO J. 31, 2753–2754 10.1038/emboj.2012.154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152. Sakabe K., Wang Z., and Hart G. W. (2010) β-N-Acetylglucosamine (O-GlcNAc) is part of the histone code. Proc. Natl. Acad. Sci. U.S.A. 107, 19915–19920 10.1073/pnas.1009023107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Zhang S., Roche K., Nasheuer H. P., and Lowndes N. F. (2011) Modification of histones by sugar β-N-acetylglucosamine (GlcNAc) occurs on multiple residues, including histone H3 serine 10, and is cell cycle-regulated. J. Biol. Chem. 286, 37483–37495 10.1074/jbc.M111.284885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Fong J. J., Nguyen B. L., Bridger R., Medrano E. E., Wells L., Pan S., and Sifers R. N. (2012) β-N-Acetylglucosamine (O-GlcNAc) is a novel regulator of mitosis-specific phosphorylations on histone H3. J. Biol. Chem. 287, 12195–12203 10.1074/jbc.M111.315804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155. Chen Q., Chen Y., Bian C., Fujiki R., and Yu X. (2013) TET2 promotes histone O-GlcNAcylation during gene transcription. Nature 493, 561–564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Xu Q., Yang C., Du Y., Chen Y., Liu H., Deng M., Zhang H., Zhang L., Liu T., Liu Q., Wang L., Lou Z., and Pei H. (2014) AMPK regulates histone H2B O-GlcNAcylation. Nucleic Acids Res. 42, 5594–5604 10.1093/nar/gku236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Delporte A., De Zaeytijd J., De Storme N., Azmi A., Geelen D., Smagghe G., Guisez Y., and Van Damme E. J. (2014) Cell cycle-dependent O-GlcNAc modification of tobacco histones and their interaction with the tobacco lectin. Plant Physiol. Biochem. 83, 151–158 10.1016/j.plaphy.2014.07.021 [DOI] [PubMed] [Google Scholar]
- 158. Dehennaut V., Leprince D., and Lefebvre T. (2014) O-GlcNAcylation, an epigenetic mark: focus on the histone code, TET family proteins, and polycomb group proteins. Front. Endocrinol. (Lausanne) 5, 155 10.3389/fendo.2014.00155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159. Wang P., Peng C., Liu X., Liu H., Chen Y., Zheng L., Han B., and Pei H. (2015) OGT mediated histone H2B S112 GlcNAcylation regulates DNA damage response. J. Genet. Genomics 42, 467–475 10.1016/j.jgg.2015.07.002 [DOI] [PubMed] [Google Scholar]
- 160. Leturcq M., Lefebvre T., Vercoutter-Edouart A. S. (2017) O-GlcNAcylation and chromatin remodeling in mammals: an up-to-date overview. Biochem. Soc. Trans. 45, 323–338 10.1042/BST20160388 [DOI] [PubMed] [Google Scholar]
- 161. Fujiki R., Hashiba W., Sekine H., Yokoyama A., Chikanishi T., Ito S., Imai Y., Kim J., He H. H., Igarashi K., Kanno J., Ohtake F., Kitagawa H., Roeder R. G., Brown M., and Kato S. (2011) GlcNAcylation of histone H2B facilitates its monoubiquitination. Nature 480, 557–560 10.1038/nature10656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Starr C. M., and Hanover J. A. (1990) Glycosylation of nuclear pore protein p62. Reticulocyte lysate catalyzes O-linked N-acetylglucosamine addition in vitro. The Journal of biological chemistry 265, 6868–6873 [PubMed] [Google Scholar]
- 163. Sohn K. C., Lee K. Y., Park J. E., and Do S. I. (2004) OGT functions as a catalytic chaperone under heat stress response: a unique defense role of OGT in hyperthermia. Biochem. Biophys. Res. Commun. 322, 1045–1051 10.1016/j.bbrc.2004.08.023 [DOI] [PubMed] [Google Scholar]
- 164. Lim K. H., and Chang H. I. (2006) O-linked N-acetylglucosamine suppresses thermal aggregation of Sp1. FEBS Lett. 580, 4645–4652 10.1016/j.febslet.2006.07.040 [DOI] [PubMed] [Google Scholar]
- 165. Ohn T., Kedersha N., Hickman T., Tisdale S., and Anderson P. (2008) A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat. Cell Biol. 10, 1224–1231 10.1038/ncb1783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Zeidan Q., Wang Z., De Maio A., and Hart G. W. (2010) O-GlcNAc cycling enzymes associate with the translational machinery and modify core ribosomal proteins. Mol. Biol. Cell 21, 1922–1936 10.1091/mbc.e09-11-0941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Zhu Y., Liu T. W., Cecioni S., Eskandari R., Zandberg W. F., and Vocadlo D. J. (2015) O-GlcNAc occurs cotranslationally to stabilize nascent polypeptide chains. Nat. Chem. Biol. 11, 319–325 10.1038/nchembio.1774 [DOI] [PubMed] [Google Scholar]
- 168. Zachara N. E., and Hart G. W. (2006) Cell signaling, the essential role of O-GlcNAc! Biochim. Biophys. Acta 1761, 599–617 10.1016/j.bbalip.2006.04.007 [DOI] [PubMed] [Google Scholar]
- 169. Comer F. I., and Hart G. W. (2001) Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry 40, 7845–7852 10.1021/bi0027480 [DOI] [PubMed] [Google Scholar]
- 170. Haltiwanger R. S., Busby S., Grove K., Li S., Mason D., Medina L., Moloney D., Philipsberg G., and Scartozzi R. (1997) O-Glycosylation of nuclear and cytoplasmic proteins: regulation analogous to phosphorylation? Biochem. Biophys. Res. Commun. 231, 237–242 10.1006/bbrc.1997.6110 [DOI] [PubMed] [Google Scholar]
- 171. Griffith L. S., and Schmitz B. (1999) O-Linked N-acetylglucosamine levels in cerebellar neurons respond reciprocally to pertubations of phosphorylation. Eur. J. Biochem. 262, 824–831 10.1046/j.1432-1327.1999.00439.x [DOI] [PubMed] [Google Scholar]
- 172. Lefebvre T., Alonso C., Mahboub S., Dupire M. J., Zanetta J. P., Caillet-Boudin M. L., and Michalski J. C. (1999) Effect of okadaic acid on O-linked N-acetylglucosamine levels in a neuroblastoma cell line. Biochim. Biophys. Acta 1472, 71–81 10.1016/S0304-4165(99)00105-1 [DOI] [PubMed] [Google Scholar]
- 173. Miller M. W., Caracciolo M. R., Berlin W. K., and Hanover J. A. (1999) Phosphorylation and glycosylation of nucleoporins. Arch. Biochem. Biophys. 367, 51–60 10.1006/abbi.1999.1237 [DOI] [PubMed] [Google Scholar]
- 174. Cheng X., Cole R. N., Zaia J., and Hart G. W. (2000) Alternative O-glycosylation/O-phosphorylation of the murine estrogen receptor β. Biochemistry 39, 11609–11620 10.1021/bi000755i [DOI] [PubMed] [Google Scholar]
- 175. Cheng X., and Hart G. W. (2001) Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor β: post-translational regulation of turnover and transactivation activity. J. Biol. Chem. 276, 10570–10575 10.1074/jbc.M010411200 [DOI] [PubMed] [Google Scholar]
- 176. Arnold C. S., Johnson G. V., Cole R. N., Dong D. L., Lee M., and Hart G. W. (1996) The microtubule-associated protein Tau is extensively modified with O-linked N-acetylglucosamine. J. Biol. Chem. 271, 28741–28744 10.1074/jbc.271.46.28741 [DOI] [PubMed] [Google Scholar]
- 177. Dong D. L., Xu Z. S., Hart G. W., and Cleveland D. W. (1996) Cytoplasmic O-GlcNAc modification of the head domain and the KSP repeat motif of the neurofilament protein neurofilament-H. J. Biol. Chem. 271, 20845–20852 10.1074/jbc.271.34.20845 [DOI] [PubMed] [Google Scholar]
- 178. Cole R. N., and Hart G. W. (1999) Glycosylation sites flank phosphorylation sites on synapsin I: O-linked N-acetylglucosamine residues are localized within domains mediating synapsin I interactions. J. Neurochem. 73, 418–428 [DOI] [PubMed] [Google Scholar]
- 179. Lefebvre T., Ferreira S., Dupont-Wallois L., Bussière T., Dupire M. J., Delacourte A., Michalski J. C., and Caillet-Boudin M. L. (2003) Evidence of a balance between phosphorylation and O-GlcNAc glycosylation of Tau proteins–a role in nuclear localization. Biochim. Biophys. Acta 1619, 167–176 10.1016/S0304-4165(02)00477-4 [DOI] [PubMed] [Google Scholar]
- 180. Liu F., Iqbal K., Grundke-Iqbal I., Hart G. W., and Gong C. X. (2004) O-GlcNAcylation regulates phosphorylation of Tau: a mechanism involved in Alzheimer's disease. Proc. Natl. Acad. Sci. U.S.A. 101, 10804–10809 10.1073/pnas.0400348101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Ahmad I., Hoessli D. C., Walker-Nasir E., Choudhary M. I., Rafik S. M., Shakoori A. R., and Nasir-ud-Din (2006) Phosphorylation and glycosylation interplay: protein modifications at hydroxy amino acids and prediction of signaling functions of the human β3 integrin family. J. Cell. Biochem. 99, 706–718 10.1002/jcb.20814 [DOI] [PubMed] [Google Scholar]
- 182. Chen Y. X., Du J. T., Zhou L. X., Liu X. H., Zhao Y. F., Nakanishi H., and Li Y. M. (2006) Alternative O-GlcNAcylation/O-phosphorylation of Ser16 induce different conformational disturbances to the N terminus of murine estrogen receptor beta. Chem. Biol. 13, 937–944 10.1016/j.chembiol.2006.06.017 [DOI] [PubMed] [Google Scholar]
- 183. Deng Y., Li B., Liu F., Iqbal K., Grundke-Iqbal I., Brandt R., and Gong C. X. (2008) Regulation between O-GlcNAcylation and phosphorylation of neurofilament-M and their dysregulation in Alzheimer disease. FASEB J. 22, 138–145 10.1096/fj.07-8309com [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Kang M. J., Kim C., Jeong H., Cho B. K., Ryou A. L., Hwang D., Mook-Jung I., and Yi E. C. (2013) Synapsin-1 and Tau reciprocal O-GlcNAcylation and phosphorylation sites in mouse brain synaptosomes. Exp. Mol. Med. 45, e29 10.1038/emm.2013.56 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185. Kaasik K., Kivimäe S., Allen J. J., Chalkley R. J., Huang Y., Baer K., Kissel H., Burlingame A. L., Shokat K. M., Ptáček L. J., and Fu Y. H. (2013) Glucose sensor O-GlcNAcylation coordinates with phosphorylation to regulate circadian clock. Cell Metab. 17, 291–302 10.1016/j.cmet.2012.12.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wells L., Kreppel L. K., Comer F. I., Wadzinski B. E., and Hart G. W. (2004) O-GlcNAc transferase is in a functional complex with protein phosphatase 1 catalytic subunits. J. Biol. Chem. 279, 38466–38470 10.1074/jbc.M406481200 [DOI] [PubMed] [Google Scholar]
- 187. Woo C. M., Lund P. J., Huang A. C., Davis M. M., Bertozzi C. R., and Pitteri S. J. (2018) Mapping and quantification of over 2000 O-linked glycopeptides in activated human T cells with isotope-targeted glycoproteomics (Isotag). Mol. Cell. Proteomics 17, 764–775 10.1074/mcp.RA117.000261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. Trinidad J. C., Barkan D. T., Gulledge B. F., Thalhammer A., Sali A., Schoepfer R., and Burlingame A. L. (2012) Global identification and characterization of both O-GlcNAcylation and phosphorylation at the murine synapse. Mol. Cell. Proteomics 11, 215–229 10.1074/mcp.O112.018366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189. Song M., Kim H. S., Park J. M., Kim S. H., Kim I. H., Ryu S. H., and Suh P. G. (2008) O-GlcNAc transferase is activated by CaMKIV-dependent phosphorylation under potassium chloride-induced depolarization in NG-108-15 cells. Cell. Signal. 20, 94–104 10.1016/j.cellsig.2007.09.002 [DOI] [PubMed] [Google Scholar]
- 190. Dias W. B., Cheung W. D., Wang Z., and Hart G. W. (2009) Regulation of calcium/calmodulin-dependent kinase IV by O-GlcNAc modification. J. Biol. Chem. 284, 21327–21337 10.1074/jbc.M109.007310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191. Whelan S. A., Lane M. D., and Hart G. W. (2008) Regulation of the O-linked β-N-acetylglucosamine transferase by insulin signaling. J. Biol. Chem. 283, 21411–21417 10.1074/jbc.M800677200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Dias W. B., Cheung W. D., and Hart G. W. (2012) O-GlcNAcylation of kinases. Biochem. Biophys. Res. Commun. 422, 224–228 10.1016/j.bbrc.2012.04.124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Tarrant M. K., Rho H. S., Xie Z., Jiang Y. L., Gross C., Culhane J. C., Yan G., Qian J., Ichikawa Y., Matsuoka T., Zachara N., Etzkorn F. A., Hart G. W., Jeong J. S., Blackshaw S., et al. (2012) Regulation of CK2 by phosphorylation and O-GlcNAcylation revealed by semisynthesis. Nat. Chem. Biol. 8, 262–269 10.1038/nchembio.771 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Bullen J. W., Balsbaugh J. L., Chanda D., Shabanowitz J., Hunt D. F., Neumann D., and Hart G. W. (2014) Cross-talk between two essential nutrient-sensitive enzymes: O-GlcNAc transferase (OGT) and AMP-activated protein kinase (AMPK). J. Biol. Chem. 289, 10592–10606 10.1074/jbc.M113.523068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195. Robles-Flores M., Meléndez L., García W., Mendoza-Hernández G., Lam T. T., Castañeda-Patlán C., and González-Aguilar H. (2008) Posttranslational modifications on protein kinase C isozymes: effects of epinephrine and phorbol esters. Biochim. Biophys. Acta 1783, 695–712 10.1016/j.bbamcr.2007.07.011 [DOI] [PubMed] [Google Scholar]
- 196. Yi W., Clark P. M., Mason D. E., Keenan M. C., Hill C., Goddard W. A. 3rd, Peters E. C., Driggers E. M., and Hsieh-Wilson L. C. (2012) Phosphofructokinase 1 glycosylation regulates cell growth and metabolism. Science 337, 975–980 10.1126/science.1222278 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Soesanto Y. A., Luo B., Jones D., Taylor R., Gabrielsen J. S., Parker G., and McClain D. A. (2008) Regulation of Akt signaling by O-GlcNAc in euglycemia. Am. J. Physiol. Endocrinol. Metab. 295, E974–E980 10.1152/ajpendo.90366.2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Chen R., Gong P., Tao T., Gao Y., Shen J., Yan Y., Duan C., Wang J., and Liu X. (2017) O-GlcNAc glycosylation of nNOS promotes neuronal apoptosis following glutamate excitotoxicity. Cell. Mol. Neurobiol. 37, 1465–1475 10.1007/s10571-017-0477-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 199. Qiu H., Liu F., Tao T., Zhang D., Liu X., Zhu G., Xu Z., Ni R., and Shen A. (2017) Modification of p27 with O-linked N-acetylglucosamine regulates cell proliferation in hepatocellular carcinoma. Mol. Carcinog. 56, 258–271 10.1002/mc.22490 [DOI] [PubMed] [Google Scholar]
- 200. Pathak S., Borodkin V. S., Albarbarawi O., Campbell D. G., Ibrahim A., and van Aalten D. M. (2012) O-GlcNAcylation of TAB1 modulates TAK1-mediated cytokine release. EMBO J. 31, 1394–1404 10.1038/emboj.2012.8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201. Xie S., Jin N., Gu J., Shi J., Sun J., Chu D., Zhang L., Dai C. L., Gu J. H., Gong C. X., Iqbal K., and Liu F. (2016) O-GlcNAcylation of protein kinase A catalytic subunits enhances its activity: a mechanism linked to learning and memory deficits in Alzheimer's disease. Aging Cell 15, 455–464 10.1111/acel.12449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202. Erickson J. R., Pereira L., Wang L., Han G., Ferguson A., Dao K., Copeland R. J., Despa F., Hart G. W., Ripplinger C. M., and Bers D. M. (2013) Diabetic hyperglycaemia activates CaMKII and arrhythmias by O-linked glycosylation. Nature 502, 372–376 10.1038/nature12537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203. Thornton T. M., Swain S. M., and Olszewski N. E. (1999) Gibberellin signal transduction presents … the SPY who O-GlcNAc'd me. Trends Plant Sci. 4, 424–428 10.1016/S1360-1385(99)01485-5 [DOI] [PubMed] [Google Scholar]
- 204. Olszewski N. E., West C. M., Sassi S. O., and Hartweck L. M. (2010) O-GlcNAc protein modification in plants: evolution and function. Biochim. Biophys. Acta 1800, 49–56 10.1016/j.bbagen.2009.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205. Zentella R., Hu J., Hsieh W. P., Matsumoto P. A., Dawdy A., Barnhill B., Oldenhof H., Hartweck L. M., Maitra S., Thomas S. G., Cockrell S., Boyce M., Shabanowitz J., Hunt D. F., Olszewski N. E., and Sun T. P. (2016) O-GlcNAcylation of master growth repressor DELLA by SECRET AGENT modulates multiple signaling pathways in Arabidopsis. Genes Dev. 30, 164–176 10.1101/gad.270587.115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 206. Xing L., Liu Y., Xu S., Xiao J., Wang B., Deng H., Lu Z., Xu Y., and Chong K. (2018) Arabidopsis O-GlcNAc transferase SEC activates histone methyltransferase ATX1 to regulate flowering. EMBO J. 37, e98115 10.15252/embj.201798115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207. Chou C. F., Smith A. J., and Omary M. B. (1992) Characterization and dynamics of O-linked glycosylation of human cytokeratin 8 and 18. J. Biol. Chem. 267, 3901–3906 [PubMed] [Google Scholar]
- 208. Slawson C., Zachara N. E., Vosseller K., Cheung W. D., Lane M. D., and Hart G. W. (2005) Perturbations in O-linked β-N-acetylglucosamine protein modification cause severe defects in mitotic progression and cytokinesis. J. Biol. Chem. 280, 32944–32956 10.1074/jbc.M503396200 [DOI] [PubMed] [Google Scholar]
- 209. Wang Z., Udeshi N. D., Slawson C., Compton P. D., Sakabe K., Cheung W. D., Shabanowitz J., Hunt D. F., and Hart G. W. (2010) Extensive crosstalk between O-GlcNAcylation and phosphorylation regulates cytokinesis. Sci. Signal. 3, ra2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 210. McClain D. A. (2002) Hexosamines as mediators of nutrient sensing and regulation in diabetes. J. Diabetes Complications 16, 72–80 10.1016/S1056-8727(01)00188-X [DOI] [PubMed] [Google Scholar]
- 211. Wells L., Vosseller K., and Hart G. W. (2003) A role for N-acetylglucosamine as a nutrient sensor and mediator of insulin resistance. Cell. Mol. Life Sci. 60, 222–228 10.1007/s000180300017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212. Akimoto Y., Hart G. W., Hirano H., and Kawakami H. (2005) O-GlcNAc modification of nucleocytoplasmic proteins and diabetes. Med. Mol. Morphol. 38, 84–91 10.1007/s00795-004-0264-1 [DOI] [PubMed] [Google Scholar]
- 213. Issad T., Masson E., and Pagesy P. (2010) O-GlcNAc modification, insulin signaling and diabetic complications. Diabetes Metab. 36, 423–435 10.1016/j.diabet.2010.09.001 [DOI] [PubMed] [Google Scholar]
- 214. Bond M. R., and Hanover J. A. (2013) O-GlcNAc cycling: a link between metabolism and chronic disease. Annu. Rev. Nutr. 33, 205–229 10.1146/annurev-nutr-071812-161240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215. Ma J., and Hart G. W. (2013) Protein O-GlcNAcylation in diabetes and diabetic complications. Expert Rev. Proteomics 10, 365–380 10.1586/14789450.2013.820536 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216. Banerjee P. S., Lagerlöf O., and Hart G. W. (2016) Roles of O-GlcNAc in chronic diseases of aging. Mol. Aspects Med. 51, 1–15 10.1016/j.mam.2016.05.005 [DOI] [PubMed] [Google Scholar]
- 217. Peterson S. B., and Hart G. W. (2016) New insights: a role for O-GlcNAcylation in diabetic complications. Crit. Rev. Biochem. Mol. Biol. 51, 150–161 10.3109/10409238.2015.1135102 [DOI] [PubMed] [Google Scholar]
- 218. Marshall S., Bacote V., and Traxinger R. R. (1991) Discovery of a metabolic pathway mediating glucose-induced desensitization of the glucose transport system: role of hexosamine biosynthesis in the induction of insulin resistance. J. Biol. Chem. 266, 4706–4712 [PubMed] [Google Scholar]
- 219. Robinson K. A., Sens D. A., and Buse M. G. (1993) Pre-exposure to glucosamine induces insulin resistance of glucose transport and glycogen synthesis in isolated rat skeletal muscles: study of mechanisms in muscle and in rat-1 fibroblasts overexpressing the human insulin receptor. Diabetes 42, 1333–1346 10.2337/diab.42.9.1333 [DOI] [PubMed] [Google Scholar]
- 220. Giaccari A., Morviducci L., Zorretta D., Sbraccia P., Leonetti F., Caiola S., Buongiorno A., Bonadonna R. C., and Tamburrano G. (1995) In vivo effects of glucosamine on insulin secretion and insulin sensitivity in the rat: possible relevance to the maladaptive responses to chronic hyperglycaemia. Diabetologia 38, 518–524 10.1007/BF00400719 [DOI] [PubMed] [Google Scholar]
- 221. Robinson K. A., Weinstein M. L., Lindenmayer G. E., and Buse M. G. (1995) Effects of diabetes and hyperglycemia on the hexosamine synthesis pathway in rat muscle and liver. Diabetes 44, 1438–1446 10.2337/diab.44.12.1438 [DOI] [PubMed] [Google Scholar]
- 222. Hebert L. F. Jr, Daniels M. C., Zhou J., Crook E. D., Turner R. L., Simmons S. T., Neidigh J. L., Zhu J. S., Baron A. D., and McClain D. A. (1996) Overexpression of glutamine:fructose-6-phosphate amidotransferase in transgenic mice leads to insulin resistance. J. Clin. Invest. 98, 930–936 10.1172/JCI118876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223. McClain D. A., Lubas W. A., Cooksey R. C., Hazel M., Parker G. J., Love D. C., and Hanover J. A. (2002) Altered glycan-dependent signaling induces insulin resistance and hyperleptinemia. Proc. Natl. Acad. Sci. U.S.A. 99, 10695–10699 10.1073/pnas.152346899 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224. Yang X., Ongusaha P. P., Miles P. D., Havstad J. C., Zhang F., So W. V., Kudlow J. E., Michell R. H., Olefsky J. M., Field S. J., and Evans R. M. (2008) Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature 451, 964–969 10.1038/nature06668 [DOI] [PubMed] [Google Scholar]
- 225. Parker G. J., Lund K. C., Taylor R. P., and McClain D. A. (2003) Insulin resistance of glycogen synthase mediated by O-linked N-acetylglucosamine. J. Biol. Chem. 278, 10022–10027 10.1074/jbc.M207787200 [DOI] [PubMed] [Google Scholar]
- 226. Yki-Järvinen H., Daniels M. C., Virkamäki A., Mäkimattila S., DeFronzo R. A., and McClain D. (1996) Increased glutamine:fructose-6-phosphate amidotransferase activity in skeletal muscle of patients with NIDDM. Diabetes 45, 302–307 10.2337/diab.45.3.302 [DOI] [PubMed] [Google Scholar]
- 227. Lehman D. M., Fu D. J., Freeman A. B., Hunt K. J., Leach R. J., Johnson-Pais T., Hamlington J., Dyer T. D., Arya R., Abboud H., Göring H. H., Duggirala R., Blangero J., Konrad R. J., and Stern M. P. (2005) A single nucleotide polymorphism in MGEA5 encoding O-GlcNAc-selective N-acetyl-β-d-glucosaminidase is associated with type 2 diabetes in Mexican Americans. Diabetes 54, 1214–1221 10.2337/diabetes.54.4.1214 [DOI] [PubMed] [Google Scholar]
- 228. Hawkins M., Barzilai N., Liu R., Hu M., Chen W., and Rossetti L. (1997) Role of the glucosamine pathway in fat-induced insulin resistance. J. Clin. Invest. 99, 2173–2182 10.1172/JCI119390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229. Weigert C., Klopfer K., Kausch C., Brodbeck K., Stumvoll M., Häring H. U., and Schleicher E. D. (2003) Palmitate-induced activation of the hexosamine pathway in human myotubes: increased expression of glutamine:fructose-6-phosphate aminotransferase. Diabetes 52, 650–656 10.2337/diabetes.52.3.650 [DOI] [PubMed] [Google Scholar]
- 230. Considine R. V., Cooksey R. C., Williams L. B., Fawcett R. L., Zhang P., Ambrosius W. T., Whitfield R. M., Jones R., Inman M., Huse J., and McClain D. A. (2000) Hexosamines regulate leptin production in human subcutaneous adipocytes. J. Clin. Endocrinol. Metab. 85, 3551–3556 10.1210/jcem.85.10.6916 [DOI] [PubMed] [Google Scholar]
- 231. McClain D. A., Alexander T., Cooksey R. C., and Considine R. V. (2000) Hexosamines stimulate leptin production in transgenic mice. Endocrinology 141, 1999–2002 10.1210/endo.141.6.7532 [DOI] [PubMed] [Google Scholar]
- 232. Hanover J. A., Lai Z., Lee G., Lubas W. A., and Sato S. M. (1999) Elevated O-linked N-acetylglucosamine metabolism in pancreatic beta-cells. Arch. Biochem. Biophys. 362, 38–45 10.1006/abbi.1998.1016 [DOI] [PubMed] [Google Scholar]
- 233. Akimoto Y., Kreppel L. K., Hirano H., and Hart G. W. (2000) Increased O-GlcNAc transferase in pancreas of rats with streptozotocin-induced diabetes. Diabetologia 43, 1239–1247 10.1007/s001250051519 [DOI] [PubMed] [Google Scholar]
- 234. Akimoto Y., Hart G. W., Wells L., Vosseller K., Yamamoto K., Munetomo E., Ohara-Imaizumi M., Nishiwaki C., Nagamatsu S., Hirano H., and Kawakami H. (2007) Elevation of the post-translational modification of proteins by O-linked N-acetylglucosamine leads to deterioration of the glucose-stimulated insulin secretion in the pancreas of diabetic Goto-Kakizaki rats. Glycobiology 17, 127–140 10.1093/glycob/cwl067 [DOI] [PubMed] [Google Scholar]
- 235. Vanderford N. L., Andrali S. S., and Ozcan S. (2007) Glucose induces MafA expression in pancreatic beta cell lines via the hexosamine biosynthetic pathway. J. Biol. Chem. 282, 1577–1584 10.1074/jbc.M605064200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236. Zraika S., Dunlop M., Proietto J., and Andrikopoulos S. (2002) The hexosamine biosynthesis pathway regulates insulin secretion via protein glycosylation in mouse islets. Arch. Biochem. Biophys. 405, 275–279 10.1016/S0003-9861(02)00397-1 [DOI] [PubMed] [Google Scholar]
- 237. Du X. L., Edelstein D., Rossetti L., Fantus I. G., Goldberg H., Ziyadeh F., Wu J., and Brownlee M. (2000) Hyperglycemia-induced mitochondrial superoxide overproduction activates the hexosamine pathway and induces plasminogen activator inhibitor-1 expression by increasing Sp1 glycosylation. Proc. Natl. Acad. Sci. U.S.A. 97, 12222–12226 10.1073/pnas.97.22.12222 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238. Chung S. S., Kim J. H., Park H. S., Choi H. H., Lee K. W., Cho Y. M., Lee H. K., and Park K. S. (2008) Activation of PPARγ negatively regulates O-GlcNAcylation of Sp1. Biochem. Biophys. Res. Commun. 372, 713–718 10.1016/j.bbrc.2008.05.096 [DOI] [PubMed] [Google Scholar]
- 239. Buse M. G., Robinson K. A., Marshall B. A., Hresko R. C., and Mueckler M. M. (2002) Enhanced O-GlcNAc protein modification is associated with insulin resistance in GLUT1-overexpressing muscles. Am. J. Physiol. Endocrinol. Metab. 283, E241–E250 10.1152/ajpendo.00060.2002 [DOI] [PubMed] [Google Scholar]
- 240. Federici M., Menghini R., Mauriello A., Hribal M. L., Ferrelli F., Lauro D., Sbraccia P., Spagnoli L. G., Sesti G., and Lauro R. (2002) Insulin-dependent activation of endothelial nitric oxide synthase is impaired by O-linked glycosylation modification of signaling proteins in human coronary endothelial cells. Circulation 106, 466–472 10.1161/01.CIR.0000023043.02648.51 [DOI] [PubMed] [Google Scholar]
- 241. Vosseller K., Wells L., Lane M. D., and Hart G. W. (2002) Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc. Natl. Acad. Sci. U.S.A. 99, 5313–5318 10.1073/pnas.072072399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242. Akimoto Y., Kawakami H., Yamamoto K., Munetomo E., Hida T., and Hirano H. (2003) Elevated expression of O-GlcNAc-modified proteins and O-GlcNAc transferase in corneas of diabetic Goto-Kakizaki rats. Invest. Ophthalmol. Vis. Sci. 44, 3802–3809 10.1167/iovs.03-0227 [DOI] [PubMed] [Google Scholar]
- 243. Semba R. D., Huang H., Lutty G. A., Van Eyk J. E., and Hart G. W. (2014) The role of O-GlcNAc signaling in the pathogenesis of diabetic retinopathy. Proteomics Clin. Appl. 8, 218–231 10.1002/prca.201300076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244. Gurel Z., and Sheibani N. (2018) O-Linked β-N-acetylglucosamine (O-GlcNAc) modification: a new pathway to decode pathogenesis of diabetic retinopathy. Clin. Sci. (Lond.) 132, 185–198 10.1042/CS20171454 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 245. Nakamura M., Barber A. J., Antonetti D. A., LaNoue K. F., Robinson K. A., Buse M. G., and Gardner T. W. (2001) Excessive hexosamines block the neuroprotective effect of insulin and induce apoptosis in retinal neurons. J. Biol. Chem. 276, 43748–43755 10.1074/jbc.M108594200 [DOI] [PubMed] [Google Scholar]
- 246. Heath J. M., Sun Y., Yuan K., Bradley W. E., Litovsky S., Dell'Italia L. J., Chatham J. C., Wu H., and Chen Y. (2014) Activation of AKT by O-linked N-acetylglucosamine induces vascular calcification in diabetes mellitus. Circ. Res. 114, 1094–1102 10.1161/CIRCRESAHA.114.302968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247. Clark R. J., McDonough P. M., Swanson E., Trost S. U., Suzuki M., Fukuda M., and Dillmann W. H. (2003) Diabetes and the accompanying hyperglycemia impairs cardiomyocyte calcium cycling through increased nuclear O-GlcNAcylation. J. Biol. Chem. 278, 44230–44237 10.1074/jbc.M303810200 [DOI] [PubMed] [Google Scholar]
- 248. Hu Y., Suarez J., Fricovsky E., Wang H., Scott B. T., Trauger S. A., Han W., Hu Y., Oyeleye M. O., and Dillmann W. H. (2009) Increased enzymatic O-GlcNAcylation of mitochondrial proteins impairs mitochondrial function in cardiac myocytes exposed to high glucose. J. Biol. Chem. 284, 547–555 10.1074/jbc.M808518200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 249. Ma J., Liu T., Wei A. C., Banerjee P., O'Rourke B., and Hart G. W. (2015) O-GlcNAcomic profiling identifies widespread O-linked β-N-acetylglucosamine modification (O-GlcNAcylation) in oxidative phosphorylation system regulating cardiac mitochondrial function. J. Biol. Chem. 290, 29141–29153 10.1074/jbc.M115.691741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250. Ma J., Banerjee P., Whelan S. A., Liu T., Wei A. C., Ramirez-Correa G., McComb M. E., Costello C. E., O'Rourke B., Murphy A., and Hart G. W. (2016) Comparative proteomics reveals dysregulated mitochondrial O-GlcNAcylation in diabetic hearts. J. Proteome Res. 15, 2254–2264 10.1021/acs.jproteome.6b00250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251. Degrell P., Cseh J., Mohás M., Molnár G. A., Pajor L., Chatham J. C., Fülöp N., and Wittmann I. (2009) Evidence of O-linked N-acetylglucosamine in diabetic nephropathy. Life Sci. 84, 389–393 10.1016/j.lfs.2009.01.007 [DOI] [PubMed] [Google Scholar]
- 252. Akimoto Y., Miura Y., Toda T., Wolfert M. A., Wells L., Boons G. J., Hart G. W., Endo T., and Kawakami H. (2011) Morphological changes in diabetic kidney are associated with increased O-GlcNAcylation of cytoskeletal proteins including α-actinin 4. Clin. Proteomics 8, 15 10.1186/1559-0275-8-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 253. Gellai R., Hodrea J., Lenart L., Hosszu A., Koszegi S., Balogh D., Ver A., Banki N. F., Fulop N., Molnar A., Wagner L., Vannay A., Szabo A. J., and Fekete A. (2016) Role of O-linked N-acetylglucosamine modification in diabetic nephropathy. Am. J. Physiol. Renal Physiol. 311, F1172–F1181 10.1152/ajprenal.00545.2015 [DOI] [PubMed] [Google Scholar]
- 254. Pekkurnaz G., Trinidad J. C., Wang X., Kong D., and Schwarz T. L. (2014) Glucose regulates mitochondrial motility via Milton modification by O-GlcNAc transferase. Cell 158, 54–68 10.1016/j.cell.2014.06.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255. Pilcher H. (2006) Alzheimer's disease could be “type 3 diabetes”. Lancet Neurol. 5, 388–389 10.1016/S1474-4422(06)70434-3 [DOI] [PubMed] [Google Scholar]
- 256. Kroner Z. (2009) The relationship between Alzheimer's disease and diabetes: type 3 diabetes? Altern. Med. Rev. 14, 373–379 [PubMed] [Google Scholar]
- 257. Kandimalla R., Thirumala V., and Reddy P. H. (2017) Is Alzheimer's disease a type 3 diabetes? A critical appraisal. Biochim. Biophys. Acta Mol. Basis Dis. 1863, 1078–1089 10.1016/j.bbadis.2016.08.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 258. Leszek J., Trypka E., Tarasov V. V., Ashraf G. M., and Aliev G. (2017) Type 3 diabetes mellitus: a novel implication of Alzheimers disease. Curr. Top. Med. Chem. 17, 1331–1335 10.2174/1568026617666170103163403 [DOI] [PubMed] [Google Scholar]
- 259. Lozano L., Lara-Lemus R., Zenteno E., and Alvarado-Vásquez N. (2014) The mitochondrial O-linked N-acetylglucosamine transferase (mOGT) in the diabetic patient could be the initial trigger to develop Alzheimer disease. Exp. Gerontol. 58, 198–202 10.1016/j.exger.2014.08.008 [DOI] [PubMed] [Google Scholar]
- 260. Wang Z., Park K., Comer F., Hsieh-Wilson L. C., Saudek C. D., and Hart G. W. (2009) Site-specific GlcNAcylation of human erythrocyte proteins: potential biomarker(s) for diabetes. Diabetes 58, 309–317 10.2337/db08-0994 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261. Park K., Saudek C. D., and Hart G. W. (2010) Increased expression of β-N-acetylglucosaminidase in erythrocytes from individuals with pre-diabetes and diabetes. Diabetes 59, 1845–1850 10.2337/db09-1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262. Lynch T. P., and Reginato M. J. (2011) O-GlcNAc transferase: a sweet new cancer target. Cell Cycle 10, 1712–1713 10.4161/cc.10.11.15561 [DOI] [PubMed] [Google Scholar]
- 263. Slawson C., and Hart G. W. (2011) O-GlcNAc signalling: implications for cancer cell biology. Nat. Rev. Cancer 11, 678–684 10.1038/nrc3114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264. Fardini Y., Dehennaut V., Lefebvre T., and Issad T. (2013) O-GlcNAcylation: a new cancer hallmark? Front. Endocrinol. 4, 99 10.3389/fendo.2013.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265. Ma Z., and Vosseller K. (2013) O-GlcNAc in cancer biology. Amino Acids 45, 719–733 10.1007/s00726-013-1543-8 [DOI] [PubMed] [Google Scholar]
- 266. Jóźwiak P., Forma E., Bryś M., and Krześlak A. (2014) O-GlcNAcylation and metabolic reprograming in cancer. Front. Endocrinol. (Lausanne) 5, 145 10.3389/fendo.2013.00099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 267. Forma E., Jóźwiak P., Bryś M., and Krześlak A. (2014) The potential role of O-GlcNAc modification in cancer epigenetics. Cell. Mol. Biol. Lett. 19, 438–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268. Chaiyawat P., Netsirisawan P., Svasti J., and Champattanachai V. (2014) Aberrant O-GlcNAcylated proteins: new perspectives in breast and colorectal cancer. Front. Endocrinol. (Lausanne) 5, 193 10.3389/fendo.2014.00193 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 269. Ferrer C. M., Lynch T. P., Sodi V. L., Falcone J. N., Schwab L. P., Peacock D. L., Vocadlo D. J., Seagroves T. N., and Reginato M. J. (2014) O-GlcNAcylation regulates cancer metabolism and survival stress signaling via regulation of the HIF-1 pathway. Mol. Cell 54, 820–831 10.1016/j.molcel.2014.04.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 270. Huang Y., and Rao A. (2014) Connections between TET proteins and aberrant DNA modification in cancer. Trends Genet. 30, 464–474 10.1016/j.tig.2014.07.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271. Chou T. Y., and Hart G. W. (2001) O-Linked N-acetylglucosamine and cancer: messages from the glycosylation of c-Myc. Adv. Exp. Med. Biol. 491, 413–418 10.1007/978-1-4615-1267-7_26 [DOI] [PubMed] [Google Scholar]
- 272. Itkonen H. M., Minner S., Guldvik I. J., Sandmann M. J., Tsourlakis M. C., Berge V., Svindland A., Schlomm T., and Mills I. G. (2013) O-GlcNAc transferase integrates metabolic pathways to regulate the stability of c-MYC in human prostate cancer cells. Cancer Res. 73, 5277–5287 10.1158/0008-5472.CAN-13-0549 [DOI] [PubMed] [Google Scholar]
- 273. Lefebvre T., Pinte S., Guérardel C., Deltour S., Martin-Soudant N., Slomianny M. C., Michalski J. C., and Leprince D. (2004) The tumor suppressor HIC1 (hypermethylated in cancer 1) is O-GlcNAc glycosylated. Eur. J. Biochem. 271, 3843–3854 10.1111/j.1432-1033.2004.04316.x [DOI] [PubMed] [Google Scholar]
- 274. Caldwell S. A., Jackson S. R., Shahriari K. S., Lynch T. P., Sethi G., Walker S., Vosseller K., and Reginato M. J. (2010) Nutrient sensor O-GlcNAc transferase regulates breast cancer tumorigenesis through targeting of the oncogenic transcription factor FoxM1. Oncogene 29, 2831–2842 10.1038/onc.2010.41 [DOI] [PubMed] [Google Scholar]
- 275. Krześlak A., Forma E., Bernaciak M., Romanowicz H., and Bryś M. (2012) Gene expression of O-GlcNAc cycling enzymes in human breast cancers. Clin. Exp. Med. 12, 61–65 10.1007/s10238-011-0138-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276. Huang X., Pan Q., Sun D., Chen W., Shen A., Huang M., Ding J., and Geng M. (2013) O-GlcNAcylation of cofilin promotes breast cancer cell invasion. J. Biol. Chem. 288, 36418–36425 10.1074/jbc.M113.495713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277. Kanwal S., Fardini Y., Pagesy P., N'Tumba-Byn T., Pierre-Eugène C., Masson E., Hampe C., and Issad T. (2013) O-GlcNAcylation-inducing treatments inhibit estrogen receptor α expression and confer resistance to 4-OH-tamoxifen in human breast cancer-derived MCF-7 cells. PLoS One 8, e69150 10.1371/journal.pone.0069150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278. Lynch T. P., Ferrer C. M., Jackson S. R., Shahriari K. S., Vosseller K., and Reginato M. J. (2012) Critical role of O-linked β-N-acetylglucosamine transferase in prostate cancer invasion, angiogenesis, and metastasis. J. Biol. Chem. 287, 11070–11081 10.1074/jbc.M111.302547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 279. Gu Y., Gao J., Han C., Zhang X., Liu H., Ma L., Sun X., and Yu W. (2014) O-GlcNAcylation is increased in prostate cancer tissues and enhances malignancy of prostate cancer cells. Mol. Med. Rep. 10, 897–904 10.3892/mmr.2014.2269 [DOI] [PubMed] [Google Scholar]
- 280. Krzeslak A., Pomorski L., and Lipinska A. (2010) Elevation of nucleocytoplasmic β-N-acetylglucosaminidase (O-GlcNAcase) activity in thyroid cancers. Int. J. Mol. Med. 25, 643–648 [DOI] [PubMed] [Google Scholar]
- 281. Mi W., Gu Y., Han C., Liu H., Fan Q., Zhang X., Cong Q., and Yu W. (2011) O-GlcNAcylation is a novel regulator of lung and colon cancer malignancy. Biochim. Biophys. Acta 1812, 514–519 10.1016/j.bbadis.2011.01.009 [DOI] [PubMed] [Google Scholar]
- 282. Olivier-Van Stichelen S., Guinez C., Mir A. M., Perez-Cervera Y., Liu C., Michalski J. C., and Lefebvre T. (2012) The hexosamine biosynthetic pathway and O-GlcNAcylation drive the expression of β-catenin and cell proliferation. Am. J. Physiol. Endocrinol. Metab. 302, E417–E424 10.1152/ajpendo.00390.2011 [DOI] [PubMed] [Google Scholar]
- 283. Yehezkel G., Cohen L., Kliger A., Manor E., and Khalaila I. (2012) O-Linked β-N-acetylglucosaminylation (O-GlcNAcylation) in primary and metastatic colorectal cancer clones and effect of N-acetyl-β-d-glucosaminidase silencing on cell phenotype and transcriptome. J. Biol. Chem. 287, 28755–28769 10.1074/jbc.M112.345546 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 284. Phueaouan T., Chaiyawat P., Netsirisawan P., Chokchaichamnankit D., Punyarit P., Srisomsap C., Svasti J., and Champattanachai V. (2013) Aberrant O-GlcNAc-modified proteins expressed in primary colorectal cancer. Oncol. Rep. 30, 2929–2936 10.3892/or.2013.2794 [DOI] [PubMed] [Google Scholar]
- 285. Rozanski W., Krzeslak A., Forma E., Brys M., Blewniewski M., Wozniak P., and Lipinski M. (2012) Prediction of bladder cancer based on urinary content of MGEA5 and OGT mRNA level. Clin. Lab. 58, 579–583 [PubMed] [Google Scholar]
- 286. Phoomak C., Silsirivanit A., Wongkham C., Sripa B., Puapairoj A., and Wongkham S. (2012) Overexpression of O-GlcNAc-transferase associates with aggressiveness of mass-forming cholangiocarcinoma. Asian Pac. J. Cancer Prev. 13, 101–105 [PubMed] [Google Scholar]
- 287. Zhu Q., Zhou L., Yang Z., Lai M., Xie H., Wu L., Xing C., Zhang F., and Zheng S. (2012) O-GlcNAcylation plays a role in tumor recurrence of hepatocellular carcinoma following liver transplantation. Med. Oncol. 29, 985–993 10.1007/s12032-011-9912-1 [DOI] [PubMed] [Google Scholar]
- 288. Banerjee S., Sangwan V., McGinn O., Chugh R., Dudeja V., Vickers S. M., and Saluja A. K. (2013) Triptolide-induced cell death in pancreatic cancer is mediated by O-GlcNAc modification of transcription factor Sp1. J. Biol. Chem. 288, 33927–33938 10.1074/jbc.M113.500983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 289. Ma Z., Vocadlo D. J., and Vosseller K. (2013) Hyper-O-GlcNAcylation is anti-apoptotic and maintains constitutive NF-κB activity in pancreatic cancer cells. J. Biol. Chem. 288, 15121–15130 10.1074/jbc.M113.470047 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290. Diernfellner A. C., and Brunner M. (2012) O-GlcNAcylation of a circadian clock protein: dPER taking its sweet time. Genes Dev. 26, 415–416 10.1101/gad.188524.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291. Kim E. Y., Jeong E. H., Park S., Jeong H. J., Edery I., and Cho J. W. (2012) A role for O-GlcNAcylation in setting circadian clock speed. Genes Dev. 26, 490–502 10.1101/gad.182378.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 292. Li M. D., Ruan H. B., Hughes M. E., Lee J. S., Singh J. P., Jones S. P., Nitabach M. N., and Yang X. (2013) O-GlcNAc signaling entrains the circadian clock by inhibiting BMAL1/CLOCK ubiquitination. Cell Metab. 17, 303–310 10.1016/j.cmet.2012.12.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 293. Fu M., and Yang X. (2017) The sweet tooth of the circadian clock. Biochem. Soc. Trans. 45, 871–884 10.1042/BST20160183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294. Skorobogatko Y., Landicho A., Chalkley R. J., Kossenkov A. V., Gallo G., and Vosseller K. (2014) O-linked β-N-acetylglucosamine (O-GlcNAc) site Thr-87 regulates synapsin I localization to synapses and size of the reserve pool of synaptic vesicles. J. Biol. Chem. 289, 3602–3612 10.1074/jbc.M113.512814 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295. Lagerlöf O., Hart G. W., and Huganir R. L. (2017) O-GlcNAc transferase regulates excitatory synapse maturity. Proc. Natl. Acad. Sci. U.S.A. 114, 1684–1689 10.1073/pnas.1621367114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296. Wang S., Yang F., Petyuk V. A., Shukla A. K., Monroe M. E., Gritsenko M. A., Rodland K. D., Smith R. D., Qian W. J., Gong C. X., and Liu T. (2017) Quantitative proteomics identifies altered O-GlcNAcylation of structural, synaptic and memory-associated proteins in Alzheimer's disease. J. Pathol. 243, 78–88 10.1002/path.4929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 297. Rex-Mathes M., Werner S., Strutas D., Griffith L. S., Viebahn C., Thelen K., and Schmitz B. (2001) O-GlcNAc expression in developing and ageing mouse brain. Biochimie 83, 583–590 10.1016/S0300-9084(01)01305-0 [DOI] [PubMed] [Google Scholar]
- 298. Farach A. M., and Galileo D. S. (2008) O-GlcNAc modification of radial glial vimentin filaments in the developing chick brain. Brain Cell Biol. 36, 191–202 10.1007/s11068-008-9036-5 [DOI] [PubMed] [Google Scholar]
- 299. Liu Y., Li X., Yu Y., Shi J., Liang Z., Run X., Li Y., Dai C. L., Grundke-Iqbal I., Iqbal K., Liu F., and Gong C. X. (2012) Developmental regulation of protein O-GlcNAcylation, O-GlcNAc transferase, and O-GlcNAcase in mammalian brain. PLoS One 7, e43724 10.1371/journal.pone.0043724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 300. Liu S., Sheng H., Yu Z., Paschen W., and Yang W. (2016) O-Linked β-N-acetylglucosamine modification of proteins is activated in post-ischemic brains of young but not aged mice: implications for impaired functional recovery from ischemic stress. J. Cereb. Blood Flow Metab. 36, 393–398 10.1177/0271678X15608393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301. He Y., Ma X., Li D., and Hao J. (2017) Thiamet G mediates neuroprotection in experimental stroke by modulating microglia/macrophage polarization and inhibiting NF-κB p65 signaling. J. Cereb. Blood Flow Metab. 37, 2938–2951 10.1177/0271678X16679671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 302. Jiang M., Yu S., Yu Z., Sheng H., Li Y., Liu S., Warner D. S., Paschen W., and Yang W. (2017) XBP1 (X-box-binding protein-1)-dependent O-GlcNAcylation is neuroprotective in ischemic stroke in young mice and its impairment in aged mice is rescued by thiamet-G. Stroke 48, 1646–1654 10.1161/STROKEAHA.117.016579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 303. Tallent M. K., Varghis N., Skorobogatko Y., Hernandez-Cuebas L., Whelan K., Vocadlo D. J., and Vosseller K. (2009) In vivo modulation of O-GlcNAc levels regulates hippocampal synaptic plasticity through interplay with phosphorylation. J. Biol. Chem. 284, 174–181 10.1074/jbc.M807431200 [DOI] [PubMed] [Google Scholar]
- 304. Din N., Ahmad I., Ul Haq I., Elahi S., Hoessli D. C., and Shakoori A. R. (2010) The function of GluR1 and GluR2 in cerebellar and hippocampal LTP and LTD is regulated by interplay of phosphorylation and O-GlcNAc modification. J. Cell. Biochem. 109, 585–597 [DOI] [PubMed] [Google Scholar]
- 305. Lagerlöf O., and Hart G. W. (2014) O-GlcNAcylation of neuronal proteins: roles in neuronal functions and in neurodegeneration. Adv. Neurobiol. 9, 343–366 10.1007/978-1-4939-1154-7_16 [DOI] [PubMed] [Google Scholar]
- 306. Yuzwa S. A., Shan X., Jones B. A., Zhao G., Woodward M. L., Li X., Zhu Y., McEachern E. J., Silverman M. A., Watson N. V., Gong C. X., and Vocadlo D. J. (2014) Pharmacological inhibition of O-GlcNAcase (OGA) prevents cognitive decline and amyloid plaque formation in bigenic Tau/APP mutant mice. Mol. Neurodegener. 9, 42 10.1186/1750-1326-9-42 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307. Jeon B. T., Heo R. W., Jeong E. A., Yi C. O., Lee J. Y., Kim K. E., Kim H., and Roh G. S. (2016) Effects of caloric restriction on O-GlcNAcylation, Ca2+ signaling, and learning impairment in the hippocampus of ob/ob mice. Neurobiol. Aging 44, 127–137 10.1016/j.neurobiolaging.2016.05.002 [DOI] [PubMed] [Google Scholar]
- 308. Shi J., Gu J. H., Dai C. L., Gu J., Jin X., Sun J., Iqbal K., Liu F., and Gong C. X. (2015) O-GlcNAcylation regulates ischemia-induced neuronal apoptosis through AKT signaling. Sci. Rep. 5, 14500 10.1038/srep14500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 309. Zhu L., Tao T., Zhang D., Liu X., Ke K., and Shen A. (2015) NOS1AP O-GlcNAc modification involved in neuron apoptosis induced by excitotoxicity. Int. J. Mol. Sci. 16, 16560–16575 10.3390/ijms160716560 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310. Ning X., Tao T., Shen J., Ji Y., Xie L., Wang H., Liu N., Xu X., Sun C., Zhang D., Shen A., and Ke K. (2017) The O-GlcNAc modification of CDK5 involved in neuronal apoptosis following in vitro intracerebral hemorrhage. Cell. Mol. Neurobiol. 37, 527–536 10.1007/s10571-016-0391-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 311. Lagerlöf O., Slocomb J. E., Hong I., Aponte Y., Blackshaw S., Hart G. W., and Huganir R. L. (2016) The nutrient sensor OGT in PVN neurons regulates feeding. Science 351, 1293–1296 10.1126/science.aad5494 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312. Vaidyanathan K., Niranjan T., Selvan N., Teo C. F., May M., Patel S., Weatherly B., Skinner C., Opitz J., Carey J., Viskochil D., Gecz J., Shaw M., Peng Y., Alexov E., et al. (2017) Identification and characterization of a missense mutation in the O-linked β-N-acetylglucosamine (O-GlcNAc) transferase gene that segregates with X-linked intellectual disability. J. Biol. Chem. 292, 8948–8963 10.1074/jbc.M116.771030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313. Willems A. P., Gundogdu M., Kempers M. J. E., Giltay J. C., Pfundt R., Elferink M., Loza B. F., Fuijkschot J., Ferenbach A. T., van Gassen K. L. I., van Aalten D. M. F., and Lefeber D. J. (2017) Mutations in N-acetylglucosamine (O-GlcNAc) transferase in patients with X-linked intellectual disability. J. Biol. Chem. 292, 12621–12631 10.1074/jbc.M117.790097 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314. Zhu Y., Shan X., Yuzwa S. A., and Vocadlo D. J. (2014) The emerging link between O-GlcNAc and Alzheimer disease. J. Biol. Chem. 289, 34472–34481 10.1074/jbc.R114.601351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 315. Gong C. X., Liu F., and Iqbal K. (2016) O-GlcNAcylation: a regulator of Tau pathology and neurodegeneration. Alzheimers Dement. 12, 1078–1089 10.1016/j.jalz.2016.02.011 [DOI] [PubMed] [Google Scholar]
- 316. Wani W. Y., Chatham J. C., Darley-Usmar V., McMahon L. L., and Zhang J. (2017) O-GlcNAcylation and neurodegeneration. Brain Res. Bull. 133, 80–87 10.1016/j.brainresbull.2016.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 317. Ma X., Li H., He Y., and Hao J. (2017) The emerging link between O-GlcNAcylation and neurological disorders. Cell. Mol. Life Sci. 74, 3667–3686 10.1007/s00018-017-2542-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318. Wells L., Vosseller K., and Hart G. W. (2001) Glycosylation of nucleocytoplasmic proteins: signal transduction and O-GlcNAc. Science 291, 2376–2378 10.1126/science.1058714 [DOI] [PubMed] [Google Scholar]
- 319. Schubert D. (2005) Glucose metabolism and Alzheimer's disease. Ageing Res. Rev. 4, 240–257 10.1016/j.arr.2005.02.003 [DOI] [PubMed] [Google Scholar]
- 320. Maher P. A., and Schubert D. R. (2009) Metabolic links between diabetes and Alzheimer's disease. Expert. Rev. Neurother. 9, 617–630 10.1586/ern.09.18 [DOI] [PubMed] [Google Scholar]
- 321. Liu F., Shi J., Tanimukai H., Gu J., Gu J., Grundke-Iqbal I., Iqbal K., and Gong C. X. (2009) Reduced O-GlcNAcylation links lower brain glucose metabolism and Tau pathology in Alzheimer's disease. Brain 132, 1820–1832 10.1093/brain/awp099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 322. Griffith L. S., Mathes M., and Schmitz B. (1995) β-Amyloid precursor protein is modified with O-linked N-acetylglucosamine. J. Neurosci. Res. 41, 270–278 10.1002/jnr.490410214 [DOI] [PubMed] [Google Scholar]
- 323. Chun Y. S., Kwon O. H., and Chung S. (2017) O-GlcNAcylation of amyloid-β precursor protein at threonine 576 residue regulates trafficking and processing. Biochem. Biophys. Res. Commun. 490, 486–491 10.1016/j.bbrc.2017.06.067 [DOI] [PubMed] [Google Scholar]
- 324. Li X., Lu F., Wang J. Z., and Gong C. X. (2006) Concurrent alterations of O-GlcNAcylation and phosphorylation of Tau in mouse brains during fasting. Eur. J. Neurosci. 23, 2078–2086 10.1111/j.1460-9568.2006.04735.x [DOI] [PubMed] [Google Scholar]
- 325. Smet-Nocca C., Broncel M., Wieruszeski J. M., Tokarski C., Hanoulle X., Leroy A., Landrieu I., Rolando C., Lippens G., and Hackenberger C. P. (2011) Identification of O-GlcNAc sites within peptides of the Tau protein and their impact on phosphorylation. Mol. Biosyst. 7, 1420–1429 10.1039/c0mb00337a [DOI] [PubMed] [Google Scholar]
- 326. Brister M. A., Pandey A. K., Bielska A. A., and Zondlo N. J. (2014) OGlcNAcylation and phosphorylation have opposing structural effects in Tau: phosphothreonine induces particular conformational order. J. Am. Chem. Soc. 136, 3803–3816 10.1021/ja407156m [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327. Gatta E., Lefebvre T., Gaetani S., dos Santos M., Marrocco J., Mir A. M., Cassano T., Maccari S., Nicoletti F., and Mairesse J. (2016) Evidence for an imbalance between Tau O-GlcNAcylation and phosphorylation in the hippocampus of a mouse model of Alzheimer's disease. Pharmacol. Res. 105, 186–197 10.1016/j.phrs.2016.01.006 [DOI] [PubMed] [Google Scholar]
- 328. Bertram L., Blacker D., Mullin K., Keeney D., Jones J., Basu S., Yhu S., McInnis M. G., Go R. C., Vekrellis K., Selkoe D. J., Saunders A. J., and Tanzi R. E. (2000) Evidence for genetic linkage of Alzheimer's disease to chromosome 10q. Science 290, 2302–2303 10.1126/science.290.5500.2302 [DOI] [PubMed] [Google Scholar]
- 329. Graeber M. B., Kupke K. G., and Müller U. (1992) Delineation of the dystonia-parkinsonism syndrome locus in Xq13. Proc. Natl. Acad. Sci. U.S.A. 89, 8245–8248 10.1073/pnas.89.17.8245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 330. Khidekel N., Ficarro S. B., Peters E. C., and Hsieh-Wilson L. C. (2004) Exploring the O-GlcNAc proteome: direct identification of O-GlcNAc-modified proteins from the brain. Proc. Natl. Acad. Sci. U.S.A. 101, 13132–13137 10.1073/pnas.0403471101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 331. Yuzwa S. A., Shan X., Macauley M. S., Clark T., Skorobogatko Y., Vosseller K., and Vocadlo D. J. (2012) Increasing O-GlcNAc slows neurodegeneration and stabilizes Tau against aggregation. Nat. Chem. Biol. 8, 393–399 10.1038/nchembio.797 [DOI] [PubMed] [Google Scholar]
- 332. Yuzwa S. A., and Vocadlo D. J. (2014) O-GlcNAc and neurodegeneration: biochemical mechanisms and potential roles in Alzheimer's disease and beyond. Chem. Soc. Rev. 43, 6839–6858 10.1039/C4CS00038B [DOI] [PubMed] [Google Scholar]
- 333. Hastings N. B., Wang X., Song L., Butts B. D., Grotz D., Hargreaves R., Fred Hess J., Hong K. K., Huang C. R., Hyde L., Laverty M., Lee J., Levitan D., Lu S. X., Maguire M., et al. (2017) Inhibition of O-GlcNAcase leads to elevation of O-GlcNAc Tau and reduction of tauopathy and cerebrospinal fluid Tau in rTg4510 mice. Mol. Neurodegener. 12, 39 10.1186/s13024-017-0181-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 334. Bork K., Kannicht C., Nöhring S., Reutter W., Weidemann W., Hart G. W., and Horstkorte R. (2008) N-Propanoylmannosamine interferes with O-GlcNAc modification of the tyrosine 3-monooxygenase and stimulates dopamine secretion. J. Neurosci. Res. 86, 647–652 10.1002/jnr.21526 [DOI] [PubMed] [Google Scholar]
- 335. Marotta N. P., Cherwien C. A., Abeywardana T., and Pratt M. R. (2012) O-GlcNAc modification prevents peptide-dependent acceleration of α-synuclein aggregation. Chembiochem 13, 2665–2670 10.1002/cbic.201200478 [DOI] [PubMed] [Google Scholar]
- 336. Marotta N. P., Lin Y. H., Lewis Y. E., Ambroso M. R., Zaro B. W., Roth M. T., Arnold D. B., Langen R., and Pratt M. R. (2015) O-GlcNAc modification blocks the aggregation and toxicity of the protein α-synuclein associated with Parkinson's disease. Nat. Chem. 7, 913–920 10.1038/nchem.2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 337. Lewis Y. E., Galesic A., Levine P. M., De Leon C. A., Lamiri N., Brennan C. K., and Pratt M. R. (2017) O-GlcNAcylation of α-synuclein at serine 87 reduces aggregation without affecting membrane binding. ACS Chem. Biol. 12, 1020–1027 10.1021/acschembio.7b00113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 338. Levine P. M., De Leon C. A., Galesic A., Balana A., Marotta N. P., Lewis Y. E., and Pratt M. R. (2017) O-GlcNAc modification inhibits the calpain-mediated cleavage of α-synuclein. Bioorg. Med. Chem. 25, 4977–4982 10.1016/j.bmc.2017.04.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 339. Zhang J., Lei H., Chen Y., Ma Y. T., Jiang F., Tan J., Zhang Y., and Li J. D. (2017) Enzymatic O-GlcNAcylation of α-synuclein reduces aggregation and increases SDS-resistant soluble oligomers. Neurosci. Lett. 655, 90–94 10.1016/j.neulet.2017.06.034 [DOI] [PubMed] [Google Scholar]
- 340. Wani W. Y., Ouyang X., Benavides G. A., Redmann M., Cofield S. S., Shacka J. J., Chatham J. C., Darley-Usmar V., and Zhang J. (2017) O-GlcNAc regulation of autophagy and α-synuclein homeostasis: implications for Parkinson's disease. Mol. Brain 10, 32 10.1186/s13041-017-0311-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341. Hart G. W., Housley M. P., and Slawson C. (2007) Cycling of O-linked β-N-acetylglucosamine on nucleocytoplasmic proteins. Nature 446, 1017–1022 10.1038/nature05815 [DOI] [PubMed] [Google Scholar]