Abstract
Summary
Recombinase polymerase amplification (RPA), an isothermal nucleic acid amplification method, is enhancing our ability to detect a diverse array of pathogens, thereby assisting the diagnosis of infectious diseases and the detection of microorganisms in food and water. However, new bioinformatics tools are needed to automate and improve the design of the primers and probes sets to be used in RPA, particularly to account for the high genetic diversity of circulating pathogens and cross detection of genetically similar organisms. PrimedRPA is a python-based package that automates the creation and filtering of RPA primers and probe sets. It aligns several sequences to identify conserved targets, and filters regions that cross react with possible background organisms.
Availability and implementation
PrimedRPA was implemented in Python 3 and supported on Linux and MacOS and is freely available from http://pathogenseq.lshtm.ac.uk/PrimedRPA.html.
Supplementary information
Supplementary data are available at Bioinformatics online.
1 Introduction
The last decade has seen a prodigious increase in the development and adaptation of novel and existing isothermal amplification technologies for molecular diagnostics. Recombinase Polymerase Amplification (RPA) enables both sensitive and rapid isothermal DNA amplification (Piepenburg et al., 2006). RPA is establishing itself as a robust alternative to PCR, and becoming a molecular tool of choice for the rapid, specific, and cost-effective identification of pathogens. Its minimal sample preparation requirements, low operation temperature (25–42°C), and commercial availability of freeze-dried reagents, mean this method has been applied in field laboratory settings and on-board automated sample-to-answer microfluidic devices. Further, this technique can be performed directly in non-processed samples, such as whole blood (Magro et al., 2017).
There is no automated software for designing primer-probe sets for RPA. Identifying candidate regions for assay development can be difficult as regions need to be conserved, with little homology to potential background DNA. Also, the sequence for primers and a probe to bind should create as small an amplicon as possible. Any DNA in the reaction that is not the target can be considered as background. Existing primer design software such as Primer3 (Untergasser et al., 2012) and RExPrimer (Piriyapongsa et al., 2009) cannot be used to automate TwistAmp® exo probe design, as they are typically longer than what these programs allow and specific requirements need to be met, including the positioning of two thymidine residues in the probe to which the fluorophore and quencher are attached. To overcome these issues, we developed Primer design for RPA (PrimedRPA), which automates the RPA primer and exo probe design process. In addition, as RPA is permissive to the presence of SNPs, the software can input and align several target sequences to account for the high genetic diversity of circulating pathogens. The software can also input several background sequences to avoid the design of primer/probes that can cross react with genetically similar organisms. Here we test the software against several pathogens and validate some of the resulting primers in the laboratory.
2 Materials and methods
The PrimedRPA package, developed in python, creates and filters RPA primer-probe sets specific for target DNA sequence(s). An overview of the package is presented in Figure 1. The user defines the input sequence(s), the parameters for filtering, and the sequence files for the background binding check, through altering the PrimedRPA_Parameters.txt file. The filtering parameters include primer, probe and amplicon lengths, GC content, ability to form a secondary structure and heterodimerise, and the tolerance of binding to background DNA (Fig. 1, Red and Brown). The user can input a single target sequence or multiple sequences. When the user inputs a sequence file (‘fasta format’) containing multiple sequences, an initial alignment is produced and conserved regions are extracted as target DNA. If a single sequence is inputted it will be taken as the target DNA (Fig. 1, Green). Candidate RPA primer-probe sets are then generated. These preliminary sets undergo filtering based on user-defined parameters. If a background binding check is required a filtered set is presented in ascending order of a score that reflects the primer-probe sets ability to bind to background DNA, where smaller scores reflect a lower probability of binding. The probes are exported as raw sequences allowing the user to choose where to insert the fluorophore, dSpacer and quencher. The script guarantees the presence of two thymidine residues in the middle region of the probe for the fluorophore and quencher to be attached to.
Fig. 1.
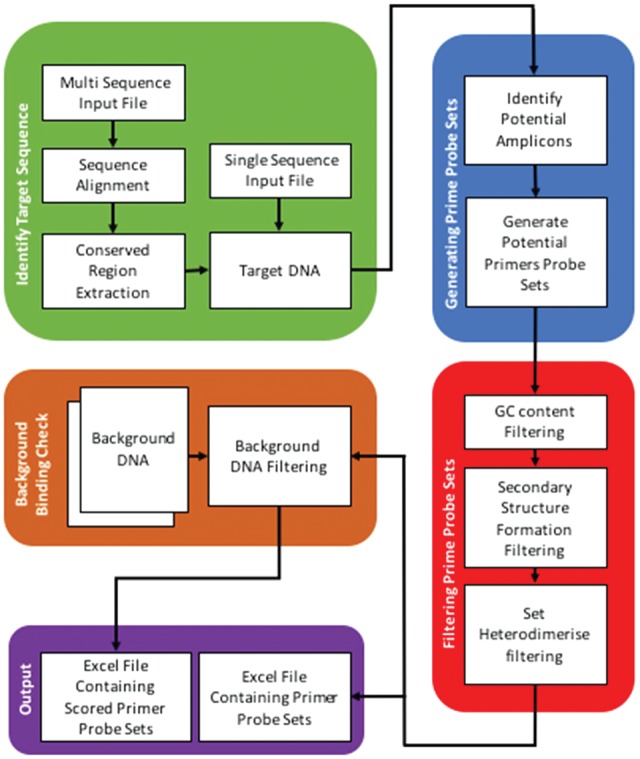
The analytical pipeline of PrimedRPA
3 Results
To assess the performance of PrimedRPA, we attempted to identify primers-probe sets in pathogens that had previously been published. For Streptococcus pneumoniae we used the lepA gene as a target (3170 bp) and for the Bovine ephemeral fever virus (BEFV) a terminal region in the genome (460 bp). Within 6 s, we identified 71 and 138 primers-probes sets for S. pneumoniae and BEF, respectively, including some overlapping with previously published RPA sets (Hou et al., 2017; Clancy et al., 2015) (Supplementary Tables S1 and S2 for parameters and output examples). We also tested the software to identify primers-probes that could amplify Zika virus from any geographical region. By using 105 Zika sequences sourced globally, PrimedRPA identified 140 potential primer-probes sets that would bind to Zika independently of the genetic diversity. To demonstrate the utility in a setting where there is high inter-species similarities, the mitochondrial (mt) sequence (6 kb) of the Plasmodium vivax malaria parasite was processed with a background check using 495 mt sequences from the five other human infecting plasmodium species. Several potential sets were generated and we validated one set of primers in the laboratory (Supplementary Table S1 and File S1) that passed the background binding check. Sanger capillary sequencing confirmed that primers were specific for P. vivax, even in samples with mixed P. vivax and P falciparum DNA (Supplementary File S1).
4 Discussion
Automating the primer and exo probe design process for RPA will assist with implementing this technique and provide a stepping stone for its broader application in diagnostic tests. We have developed an in silico assay design tool, which provides multiple possible primers and probes that can be screened and optimized in vitro with the RPA technology. TwistAmp® exo fluorescent probes can be converted into lateral flow probes, and therefore the PrimedRPA package could be used to design such applications. Further, the software can be extended as nucleic acid amplification detection kits continue to evolve and their applications in biomedical settings increase.
Supplementary Material
Acknowledgements
The MRC eMedlab was used for computational work.
Funding
This work was supported by BBSRC LiDO PHD studentship (M.H. and M.R.), MRC LID PhD studentship (A.I.) and Bloomsbury Research Fund PhD studentship (D.W.). T.G.C. and S.C. are funded by MRC UK grants (MR/K000551/1, MR/M01360X/1, MR/N010469/1).
Conflict of Interest: MSF is an employee of TwistDx, the developer and manufacturer of RPA technology.
References
- Clancy E. et al. (2015) Development of a rapid recombinase polymerase amplification assay for the detection of Streptococcus pneumoniae in whole blood. BMC Infect. Dis., 15, 481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou P. et al. (2017) Development of a recombinase polymerase amplification combined with lateral-flow dipstick assay for detection of bovine ephemeral fever virus. Mol. Cell. Probes, 38, 31–37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magro L. et al. (2017) Paper-based RNA detection and multiplexed analysis for Ebola virus diagnostics. Sci. Rep., 7, 1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piepenburg O. et al. (2006) DNA detection using recombination proteins. PLoS Biol, 4, e204.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piriyapongsa J. et al. (2009) RExPrimer: an integrated primer designing tool increases PCR effectiveness by avoiding 3’ SNP-in-primer and mis-priming from structural variation. BMC Genomics, 10, S4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Untergasser A. et al. (2012) Primer3—new capabilities and interfaces. Nucleic Acids Res., 40, e115.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


