Abstract
The current RNA-Seq method analyses fragments of mRNAs, from which it is occasionally difficult to reconstruct the entire transcript structure. Here, we performed and evaluated the recent procedure for full-length cDNA sequencing using the Nanopore sequencer MinION. We applied MinION RNA-Seq for various applications, which would not always be easy using the usual RNA-Seq by Illumina. First, we examined and found that even though the sequencing accuracy was still limited to 92.3%, practically useful RNA-Seq analysis is possible. Particularly, taking advantage of the long-read nature of MinION, we demonstrate the identification of splicing patterns and their combinations as a form of full-length cDNAs without losing precise information concerning their expression levels. Transcripts of fusion genes in cancer cells can also be identified and characterized. Furthermore, the full-length cDNA information can be used for phasing of the SNPs detected by WES on the transcripts, providing essential information to identify allele-specific transcriptional events. We constructed a catalogue of full-length cDNAs in seven major organs for two particular individuals and identified allele-specific transcription and splicing. Finally, we demonstrate that single-cell sequencing is also possible. RNA-Seq on the MinION platform should provide a novel approach that is complementary to the current RNA-Seq.
Keywords: nanopore sequencing, transcriptome, transcript isoform, allelic expression
1. Introduction
RNA-Seq has revolutionized transcriptome analysis. Without detailed information for each of the transcripts, that is, comprehensive information on the transcripts, the transcriptome information can be obtained by massively parallel sequencing of mRNA fragments.1 Indeed, there are numerous successful examples of the application of RNA-Seq for measuring gene expression levels and detecting splicing variants and mutations in transcripts.2,3 However, there is a large flaw in this method: it requires prior fragmentation of the mRNA molecules. When the entire transcript structure of a given transcript is studied, the fragmented sequence reads should be computationally assembled to form an ‘isoform’.4 However, precise isoform information is not always represented in the assembled transcripts. If repetitive sequences are included in its internal sequence or its expression level is low, providing an insufficient number of sequence tags, the obtained results are occasionally unreliable without further validation analyses. Moreover, it is essentially impossible to analyse the relationship between multiple sites of splicing, if any.
Recently, to overcome the limitations of the current short-read sequencing approach, several new long-read technologies have been developed, such as RSII and Sequel of Pacific Biosciences, MinION of Oxford Nanopore Technologies, and the linked-read method of 10X Genomics.5 In several aspects of genomic studies, long-read sequencing has appeared to have particular advantages. Namely, while it is occasionally difficult to align short reads uniquely to repetitive and duplicated regions, long reads can be effectively aligned even to these regions. Taking advantage of this feature, complementary use of long-read sequencing data analyses have now become authentic methods when de novo genome assembly or analysis of structural variations is contemplated. Since the long reads occasionally cover multiple SNPs in a single sequence, the mutual relationship of SNPs can also be obtained as ‘phasing information’.6
For the transcriptome study, PacBio RSII has been used for long-read sequencing of transcripts. In the ‘Iso-Seq’ method, the mRNAs are subjected to cDNA synthesis and amplification by the template-switching method.7 From the sequence data obtained from PacBio RSII, it is possible to determine the entire structure of the transcripts. However, in this protocol, extensive prior size fractionation is necessary, perhaps because of the bias of the sequencer toward reading shorter fragments.8 The procedure, including the sequencing reaction itself, is sometimes technically difficult and cannot be performed in a small laboratory. More importantly, current or provisioned sequencing throughput is inadequate to obtain sufficient numbers of sequences to extract the gene expression information.9
The MinION sequencer is a newly developed portable long-read sequencer. After several rounds of updates, its sequence accuracy has now exceeded 90%, and the expected number of reads per flow cell is over hundreds of thousands per flow cell.10 The read length has also been improved, having achieved a N50 read length of more than 100 kb.11 Pioneering work on the application of MinION to RNA-Seq using R7.3 MinION flow cell and single-cell RNA-Seq using R7.3 and 9.4 flow cells has been reported.12,13 However, in these study, the number of mapped reads was limited to only ∼100,000. Throughput of MinION is growing by improvement of squenecing chemistry and software, and further evaluation is needed.
In this study, we evaluated and optimized the RNA-Seq procedure using the MinION sequencer and applied the update procedure for transcriptome analyses for various purposes. First, we used seven lung cancer cell lines and MinION R9, R9.4, and R9.5 for developing the experimental and computational procedure. We employed LAST for mapping MinION reads to RefSeq transcripts or directly to the human genome. We demonstrate that reasonably accurate information on gene expression levels as well as unique information on the entire transcript structure, including the transcript products of fusion genes in cancers, can be obtained using this approach. We also successfully attempted phasing of SNP, which were identified by whole genome sequencing (WGS), using a series of RNA materials obtained from representative organs of particular male and female individuals.
2. Materials and methods
2.1. Total RNA and DNA
Total RNA purified from seven lung adenocarcinoma cell lines (PC-7, PC-9, H1975, H2228, VMRC-LCD, LC2/ad, and A549) were used. We purchased 14 total RNAs from seven organs, liver, kidney, skeletal muscle, pancreas, colon, heart, and lung, derived from two individuals (BioChain Institute). We also purchased two genomic DNAs derived from them. We conducted a quality check of the total RNA using the Agilent RNA Nano Kit.
2.2. FL-cDNA synthesis and amplification
Total RNA (50 ng) was used for FL-cDNA synthesis and amplification. We employed the SMART-Seq v4 Ultra Low Input RNA Kit (Takara Bio), except for the template switching oligo, poly-dT primer, and PCR primer. We used custom oligos with the same sequences as Smart-Seq2, instead of them.14 The synthesized first-strand cDNA was amplified by 16 cycles of PCR. FL-cDNA was quantified using the Agilent DNA 7500 Kit (Agilent Technologies).
2.3. FL-cDNA preparation from a single cell
LC2ad were dissociated to single cells by Accumax treatment (Innovative Cell Technologies). We employed the C1 single-cell auto prep system (Fluidigm) and SMART-Seq v4 Ultra Low Input RNA Kit with oligo DNA, identically to Smart-Seq2. We executed the C1 protocol ‘SMART-Seq v4 Rev B’. We acquired approximately 1 ng of FL-cDNA. As a control, we also conducted cDNA synthesis from bulk cells using a general thermal cycler with the same reaction conditions. Using SeqAmp DNA Polymerase (TAKARA Bio), we conducted a further amplification (1 cycle of 1 min at 96 °C, 5 cycles of 30 s at 95 °C, 65 °C for 30 s, 7 min at 68 °C, 1 cycle of 10 min at 72 °C). We purified the re-amplified cDNA using Agencourt AMPureXP (Beckman Coulter). Approximately 300 ng of FL-cDNA was obtained.
2.4. MinION sequencing of FL-cDNA
For 2D sequencing of FL-cDNA generated from total RNA, 1 μg of FL-cDNA was applied for library preparation of MinION with the protocol ‘2D genomic DNA by ligation’ and the Sequencing kit (SQK_MAP007), and the R9 flow cell (FLO-MAP104 or FLO-MIN105) or Sequencing kit (SQK-LSK208), and the R9.4 flow cell (FLO-MIN106). R9 flow cells were used for FL-cDNA-Seq of PC-7, PC-9, H1975, H2228, and VMRC-LCD. Both R9 and R9.4 flow cells were used for FL-cDNA-Seq of LC2ad. For 1D2 sequencing of FL-cDNA, 1 µg of FL-cDNA of A549 was applied for library preparation of MinION using the protocol ‘1D∘2 sequencing of genomic DNA’ with some modifications with the 1D2 sequencing kit (SQK-LSK308) and R9.5 flow cell (FLO-MIN107). For the purification steps after ligation of the 1D2 adapter and Barcode Adapter Mix, the same volume of AMPure XP beads was added to the sample, instead of adding 40% of its volume. For single cell samples, 300 ng of re-amplified FL-cDNA was applied using the protocol ‘Native barcoding genomic DNA’ with the Native Barcoding Kit (EXP-NBD002), Sequencing kit (SQK-LSK208), and R9.4 flow cell (FLO-MIN106). We conducted multiplex sequencing of 11 samples.
For direct RNA-Seq, we performed direct RNA-Seq of LC2/ad using the R9.5 flow cell, Direct RNA Sequencing Kit (SQK-RNA001), and 500 ng polyA-selected RNA of LC2/ad, following the manufacturer’s instructions.
2.5. Data analysis of MinION reads
We aligned the FL-cDNA-Seq reads of LC2/ad sequenced by R9.4 flow cells to all RefSeq transcripts of mRNA (NM) and ncRNA (NR) or the longest coding isoforms of each RefGene. BWA-MEM version 0.7.15 had the following parameters: ‘-x ont2d’. We also aligned the MinION reads to these references by lastal and last-split of LAST version 833 with the parameter tuned by last train. To calculate the coverage and identity, we used the MinION reads aligned to all RefSeq transcripts. Coverage was defined as the fraction of RefSeq transcript length covered by the single read. Identity was defined as the percentage of matched bases to the sum of matched bases, substitutions, insertions, and deletions. For gene expression estimation, we used all the MinION reads aligned to the longest coding isoforms of each RefGene. The number of reads mapped to each isoform was counted. We calculated the rpm (reads per million) value for each gene as the expression unit. MinION reads were also aligned to the hg38 analysis set by LAST with the parameter of lastal tuned by last-train and ‘-d90 -m50 -D10’ and last-split ‘-m1 -d2’.
For splice isoform detection, we used the alignments of all pass 2d reads of LC2/ad (R9.4) to the reference genome by LAST. To remove pseudogene mapping, low-quality reads and alignment results, we discarded the aligned reads with any of the following five conditions. (1) Reads did not have an intron, which was defined as a gap of more than 50 bp. (2) Strand score of LAST was zero. (3) Unmapped length of reads within splice junctions was more than 10 bp. (4) Reads overlapped with pseudogenes of Genocode v27. (5) Isoforms with the same exon–intron structures existed on the reverse strand. Junctions of the remaining reads compared with RefSeq transcripts allowed a gap within 20 bp. If the corresponding RefSeq transcripts did not exist and the isoform was covered by more than ten reads, we judged the read to be derived from a novel isoform.
For detection of fusion transcript, we mapped all pass 2d reads to the reference genome by Blat with the following parameter: ‘-noHead -mask=lower’.5,15We extracted reads with multiple alignments to multiple chromosomes, different strands of the same chromosome, or more than 1 Mb away from the same chromosome without an alternative alignment within 1 Mb of the same chromosome strand. If both ends of the reads were aligned with exons of RefSeq transcripts, we judged it as a fusion candidate.
For hetero SNP phasing of lung cancer cells, we used all pass 2d reads aligned to the genome by LAST and the hetero SNP data from our previous study.16 We selected reads with possible patterns of multiple SNP according to hetero SNPs and counted the number of reads with each SNP pattern. For each gene, we selected two major SNP patterns that did not mutually contradict one another, and were covered by more than five reads, respectively.
To detect differential allelic expression of two individuals, we used all 2d reads aligned to the genome by LAST and the hetero SNP data from this study (see also Supplementary Methods).17 First, we used all tissue-merged reads of each individual. We called the two major patterns of single or multiple SNPs from these reads as performed for the lung cancer cell line data. We counted the number of reads in each organ according to each phased pattern. To detect genes with significant differential allelic expression, we employed the binomial test.
3. Results and discussion
3.1. Optimization of the cDNA-Seq procedure
Using the SMART-Seq v4 Ultra Low Input RNA Kit from Takara Bio, which is based on the Smart-Seq2 method, we synthesized full-length cDNA (FL-cDNA) from 50 ng total RNA isolated from seven lung adenocarcinoma-derived cell lines: PC-7, PC-9, H1975, H2228, VMRC-LCD, LC2/ad, and A549 (Fig. 1A).14 In these cell lines, we have previously performed RNA-Seq and WGS.16 By cDNA amplification via 16 cycles of PCR, we obtained 5–9 μg of double-stranded FL-cDNA, starting from 50 nanograms of total RNA. Although we also conducted cDNA preparation by Smart-Seq2, the yield of FL-cDNA was lower than that of SMART-Seq v4 (data not shown). We used 1 μg of FL-cDNA for library preparation of the MinION sequencing. We conducted one or two runs of 2D sequencing for approximately each six cell lines except for A549 by R9 flow cells (Supplementary Table S1). We used four R9.4 flow cells for 2D sequencing of LC2/ad. Averages of all runs using the R9 and R9.4 flow cells were 161,325 and 302,059 reads of total reads and 42,761 and 180,938 reads of pass 2d reads, respectively. We used only pass 2d reads, which had a higher quality in the following analyses. As a result, we obtained 723,750 reads of pass 2d from four runs of LC2/ad by R9.4 flow cells in total. We also conducted a single run of 1D2 sequencing for A549 and obtained 994,111 1d reads and 931 pass 1dsq reads (Supplementary Table S2). Statistics for the FL-cDNA-Seq analysis of the other cell lines are summarized in Supplementary Table S1. For the following analyses, we mainly focused on the data obtained from LC2/ad but obtained essentially similar results from the other cell lines.
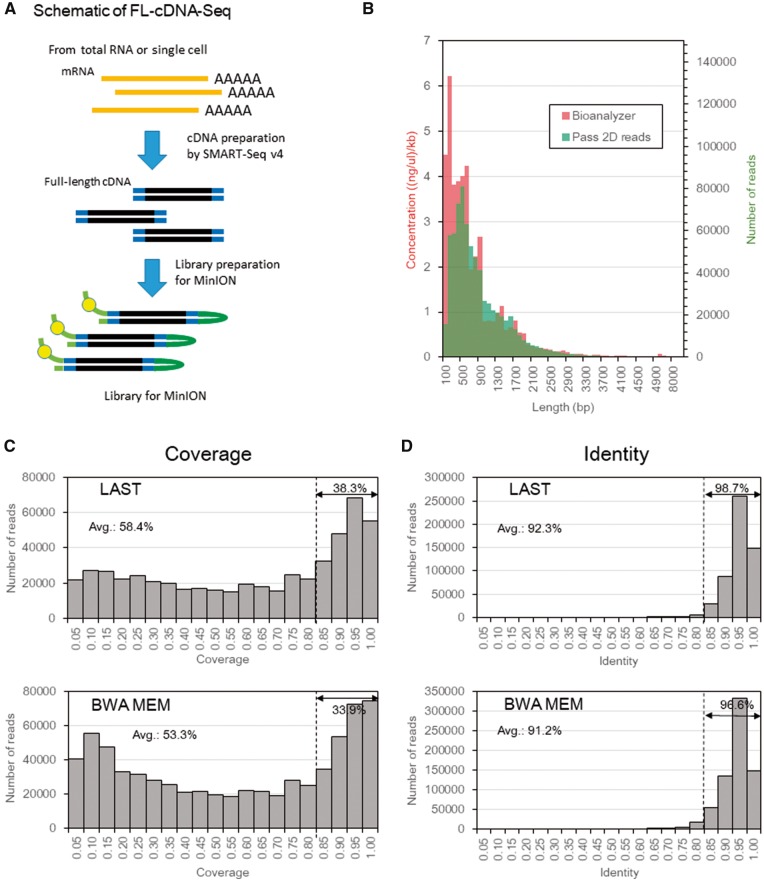
Figure 1.
MinION sequencing of full-length cDNA. (A) Schematic of the FL-cDNA-Seq method. (B) Distribution of the concentration bioanalyzer and frequency of pass 2d reads of LC2/ad (R9.4) in each range of lengths. (C, D) Distribution of the coverage (C) and identity (D) of pass 2D reads of LC2ad, aligned by LAST (top) and BWA MEM (bottom). The average coverage or identity and percentages of reads with a coverage or identity greater than 0.8 is shown on the graphs. The coverage was defined as the ratio of the length of the RefSeq transcript covered by a single MinION read.
First, we compared the results of LC2/ad between R9 and R9.4. The read lengths of R9 and R9.4 were distributed similarly to the average read lengths of 908 and 917 bp, respectively (Supplementary Table S1 and Fig. S1). The average read length of the other cell lines, which were sequenced by R9, was also approximately 1 kb long (Supplementary Table S1). We also compared the distributions of read length with the electropherogram of the Agilent Bioanalyzer (Fig. 1B). We found that the distributions mostly overlapped, except for short fragments. These results might indicate that base calling of short fragment is unfavorable for the base caller of MinION. These results collectively suggested that transcriptome information can be read on the MinION sequencer without imposing serious length bias.
3.2. The analytical pipeline
To associate the obtained sequences to the reference genes or transcripts, we constructed a computational pipeline. Currently, most alignment programs are optimized for short and accurate reads, assuming the process of Illumina reads. We examined and found that these programs, as they are, are not suited for mapping the longer and less-accurate MinION reads.
First, to select an appropriate mapping software for this purpose, the pass 2d reads of LC2ad sequenced by R9.4 were aligned to the RefSeq transcripts by LAST and BWA.18–20 For LAST, we employed the parameters tuned by LAST-TRAIN and aligned using last-al and -split. For BWA, we used the parameter optimized for pass 2d reads of MinION (-x ont2d option) and aligned them using BWA-MEM.20 The 532,956 reads (74%) and 690,528 reads (95%) were first aligned to the RefSeq transcripts using both LAST and BWA (Supplementary Table S3). We evaluated the obtained data regarding their coverages as the RefSeq coverage (the fraction of RefSeq transcript length covered by a single read). For LAST and BWA, the average coverage was 38.3% and 33.9% of the mapped reads, respectively, showing a RefSeq coverage of more than 0.80 (Fig. 1C). These results indicated that LAST could align the greater part of the read sequence to the RefSeq transcript, although the mapping rate of BWA was higher than LAST. Concomitantly, it is suggested that many of the transcripts were sequenced in their full-length forms. Based on these results, we decided to employ LAST as a default alignment software in the following analyses. When considering such alignment results, it is important to bear in mind that we can trivially obtain more and/or longer alignments, by using less-stringent alignment criteria.
3.3. The overall sequence accuracy
Based on the obtained sequence alignments, we evaluated the sequence accuracy (Fig. 1D). We found that the pass 2d reads obtained an overall accuracy of 92.3%. The mismatches consisted of 12.4%, 14.2%, and 73.4% base substitutions and insertions and deletions, respectively (Supplementary Fig. S2). We examined the base substitution pattern and found that (G to A) substitutions were the most frequent (29.4%), although the preference was not substantial. We further compared the accuracy and RefSeq coverage between the 2d of 2 D sequencing, 1d and pass 1dsq reads of recent 1D2 sequencing (Fig. 1D and Supplementary Fig. S3). Although the pass 1dsq reads (95.1%) were the most accurate compared with pass 2d reads (92.3%) and 1d reads (87.2%), there was no significant difference in the RefSeq coverage between them. However, the rate of pass 1dsq reads was quite low on this sequencing run (0.1%). In a typical run of FL-cDNA-Seq by 1D2 sequencing, the rate of pass 1dsq reads was 0.4-0.9% (data not shown).
3.4. Gene expression information
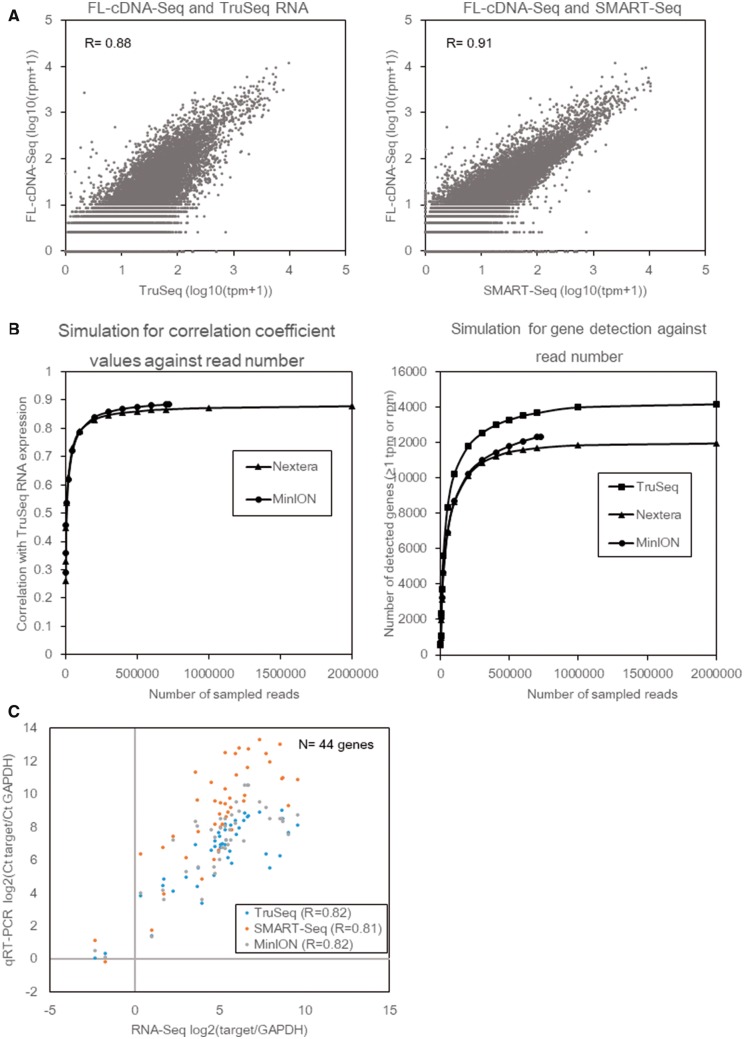
To evaluate the precise representation of the transcriptome in MinION reads, we compared the gene expression levels in LC2/ad between those measured by the MinION reads and by the conventional Illumina TruSeq RNA method, which had been read by HiSeq2500 in our previous study.16 To minimize the influence of multi-mapped reads and differences in alignment software, both reads were aligned to the longest RefSeq transcripts of each gene by LAST (Supplementary Table S3). We calculated the rpm (reads per million) and tpm (transcripts per million) as the gene expression unit for the MinION and Illumina data sets. A high correlation coefficient (R = 0.88) was observed between them (also see Supplementary Fig. S4, showing that similar results were also obtained for the BWA-processed data; R = 0.88). To further examine the influence of the mRNA fragmentation process, which is unique to TruSeq of Illumina, we used the same cDNA amplicons that were used for MinION sequencing and prepared an Illumina library by using the SMART-Seq v4 and the Nextera XT kits, referred to as SMART-Seq (Supplementary Table S4 and see also Supplementary Methods). The comparison revealed a higher correlation (R = 0.91), suggesting that an influence of the difference in template preparation procedure could partly explain the difference in the represented expression information (Fig. 2A). To further validate the influence regarding the generally smaller numbers of MinION reads (‘shallow sequencing depth’), we randomly sampled the reads of FL-cDNA-Seq and SMART-Seq and calculated the correlation efficiencies and number of detected genes as >1 rpm or tpm. On MinION, the correlation efficiency with TruSeq roughly reached a plateau at a sequencing depth of 40,000 reads, although the number of detected genes continued to increase thereafter (Fig. 2B). We also performed a similar comparison with SMART-Seq. SMART-Seq showed almost the same trend as FL-cDNA-Seq.
Figure 2.
Comparison of FL-cDNA-Seq and Illumina RNA-Seq. (A) The gene expression of FL-cDNA-Seq of LC2/ad (R9.4) was compared with that of TruSeq RNA (left) and SMART-Seq (right). Pearson correlation coefficients are shown on the graph. (B) Influence of sequencing depth on the estimation of gene expression level and gene detection. Reads for each method were randomly sampled in triplicate. The average of the Pearson correlation coefficients between TruSeq RNA and randomly sampled data for FL-cDNA-Seq and SMART-Seq is shown (left). The average number of genes with an expression level of more than 1 tpm or ppm is shown (right). (C) Comparison to qRT-qPCR. Forty-four genes detected by all methods were analyzed. The gene expression of these genes was normalized to GAPDH. Pearson correlation coefficients are shown on the graph. qRT-qPCR data of LC2/ad was obtained as in our previous study.16
Then, we validated the gene expression levels of 44 genes by RT-qPCR.16 We compared the obtained results with FL-cDNA-Seq and Illumina RNA-Seq methods. Although the number of FL-cDNA-Seq reads was generally smaller than that of Illumina reads, the FL-cDNA-Seq data showed a comparably high correlation efficiency with the RT-qPCR data [R = 0.82 (TruSeq), 0.81 (SMART-Seq), and 0.82 (FL-cDNA-Seq), respectively] (Fig. 2C). Taken together, we concluded that FL-cDNA-Seq is effective for the purposes of quantitative transcriptome analysis.
3.5. Comparison with direct RNA sequencing
We also conducted recent direct RNA-Seq of LC2/ad21 (Supplementary Table S5). We obtained 556,195 1d reads from a single run. The average length of the direct RNA-Seq reads was 926 bp, which was comparable to the average length of the above RNA-Seq. Similarly, the direct RNA-Seq reads were aligned to all RefSeq transcripts using LAST. The direct RNA-Seq reads showed comparable coverage (57.0%) of the reference transcripts to FL-cDNA-Seq (58.4%) (Supplementary Fig. S5A). Furthermore, the direct RNA-Seq showed a high correlation (R = 0.92 and 0.90) with TruSeq and the FL-cDNA-Seq, respectively (Supplementary Fig. S5B). In contrast, direct RNA-Seq showed lower fidelity (83.2%) than FL-cDNA-Seq (92.3%) (Fig. 1D and Supplementary Fig. S5A). Based on these results, we conclude that direct RNA-Seq is also useful to estimate expression levels of the whole transcriptome. An obvious advantage of direct RNA-Seq is that it is free from PCR bias and may be able to analyse mRNA for which reverse transcription is not efficient. However, there is a limitation to this method, which is that 500 ng of polyA RNA is required as starting material, which may limit its application to rare samples, such as the subtle amounts of RNA present in some clinical samples.
3.6. Applications of MinION transcriptome sequencing
In addition to the above applications, we found that FL-cDNA-Seq may have several applications that are not easily performed using Illumina RNA-Seq.
3.6.1. Detection of transcript variants and cancerous fusion genes
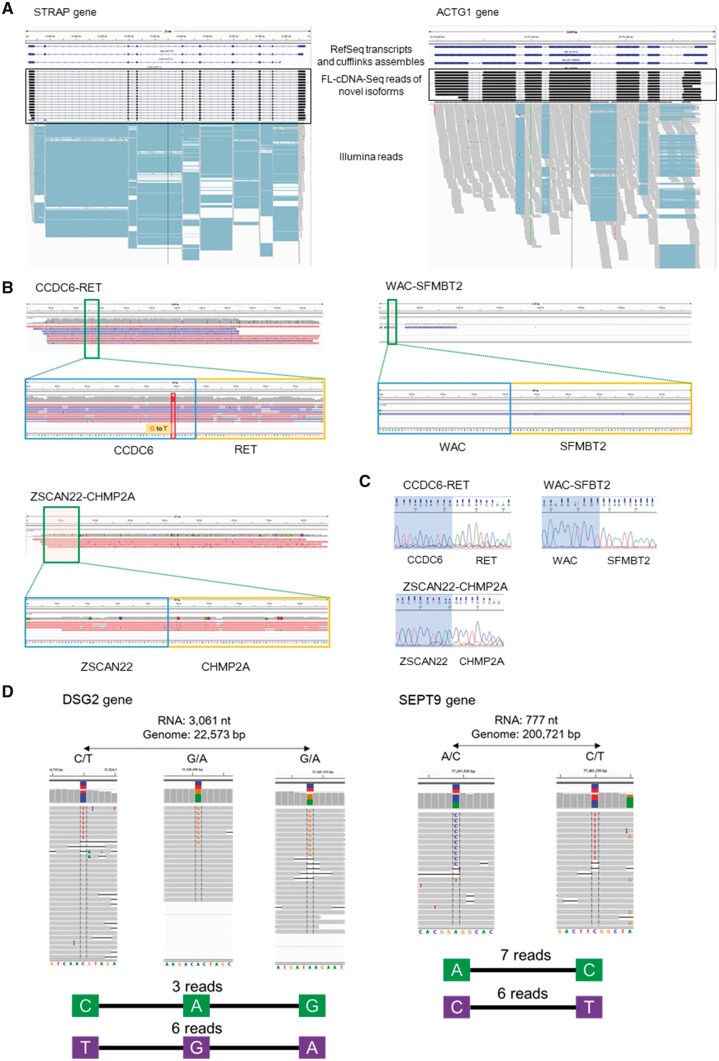
We examined whether the FL-cDNA-Seq reads could be used to detect transcript variants and mutations. For this purpose, the FL-cDNA-Seq reads of LC2/ad (R9.4) were aligned to the reference genome by LAST. We calculated the distance between the detected and RefSeq transcript TSS (transcription start site) and TTS (transcription termination site), and 82% and 63% of the TSS and TTS detected by MinION were located within ±500 bp from those of the RefSeq transcripts, respectively (Supplementary Figs S6 and S7). We compared the detected transcripts with transcript models of RefSeq. Although 6,018 of the detected transcripts matched the RefSeq models, 158 of them represented unannotated isoforms (Table 1 and Supplementary Table S6). We also compared the detected transcripts by FL-cDNA-Seq and RNA-Seq. Although only 33 (21%) of the novel isoforms detected by FL-cDNA-Seq were detected by cufflinks with guide of RefSeq annotation from the RNA-Seq data, all the splice junctions of 137 (87%) of them were covered by the RNA-Seq reads (Table 1). Examples are novel isoforms of STARP and ACTG1 genes that were difficult to detect by Illumina reads alone (Fig. 3A).
Table 1.
Application of MinION transcriptome sequencing: statistics for the isoform detected by FL-cDNA-Seq
| No. of mapped reads | No. of PF reads | % PF reads | Known isoform | Novel isoform | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| No. of reads | % reads | No. of isoforms | No. of reads | % reads | No. of isoforms | No. of cufflinks-supported isoforms | No. of Illumina-supported isoforms | |||
| 556,195 | 297,865 | 54% | 294,780 | 99% | 6,018 | 3,085 | 1% | 158 | 33 | 137 |
Figure 3.
Applications of MinION transcriptome sequencing. (A) Novel isoforms of the STRAP gene (left) and the ACTG1 gene (right). RefSeq transcripts and cufflink assembles using Illumina RNA-Seq are shown in the upper panel. FL-cDNA-Seq reads annotated as novel isoforms are shown in the middle panel. Illumina RNASeq reads are shown in the lower panel. (B) Three fusion genes detected by FL-cDNA. CCDC6-RET is known as a fusion gene of LC2/ad. Only the fusion chromosome of CCDC6-RET harbors a G to T SNP. WAC-SFMBT2 and ZSCAN22-CHMP2A were detected on LC2/ad and PC-9, respectively. (C) Sanger sequencing of the three fusion junctions. (D) Phased hetero SNP by FL-cDNA-Seq reads. We exemplified two phased genes, DSG2 (left) and SEPT9 (right), detected by the R9.4 reads of LC2ad. The distance between the phased SNP on the transcript and genome and hetero SNP patterns are shown at the top. The FL-cDNA-Seq reads are shown in the middle. The phased SNP pattern and number of reads covering all the SNPs are shown at the bottom.
We further attempted to detect fusion transcripts in cancer cells. We aligned the FL-cDNA-Seq reads of six cell lines to the reference genome using BLAT (Supplementary Table S7). The transcripts with two or more alignments that were mapped to different chromosomes, or those that were mapped to regions that were mutually separated by more than 1 Mb, were considered. A total 151 fusion transcript candidates were detected from the LC2/ad reads of R9.4 (Table 2 and Supplementary Table S8). The detected fusion transcripts included the previously reported cancer driver fusion transcript of the CCDC6-RET gene. As shown in Fig. 3B, seven reads directly represented the junction of the CCDC6-RET fusion transcript. Interestingly, the fusion point harbored a hetero germline SNP, which gave us a clue regarding the identities of the alleles in which the fusion event occurred. Considering all six cell lines together, we identified 194 fusion gene candidates (Table 2). For example, WAC-SFMBT2 and ZSCAN22-CHMP2A fusion transcripts were detected from LC2/ad and PC-9, respectively (Fig. 3B). These genes could be validated using Sanger sequencing (Fig. 3C and see also Supplementary Methods). These results indicate that MinION sequencing is a convenient method to detect fusion gene transcripts.
Table 2.
Application of MinION transcriptome sequencing: number of fusion gene candidates
| Cell line | Flow cell | No. of reads of fusion gene candidates | No. of fusion gene candidates |
|---|---|---|---|
| H1975 | R9 | 8 | 8 |
| PC-9 | R9 | 5 | 3 |
| PC-7 | R9 | 5 | 5 |
| H2228 | R9 | 7 | 7 |
| VMRC-LCD | R9 | 14 | 11 |
| LC2/ad | R9 | 10 | 10 |
| LC2/ad | R9.4 | 158 | 151 |
3.6.2. Haplotype phasing from MinION reads
In lung cancer, there are well-known driver mutations in the tyrosine kinase domain of the EGFR gene, which are targets for molecular target drugs such as gefitinib. There are also several secondary mutations known for resistance to drugs in the proximal region.22 For those mutations, it is important to obtain phase information (allele context of the mutations) since the clinical relevance will be distinct regardless of whether the mutations are located on the same allele. We expected long MinION reads to overcome the difficulties in phasing using Illumina reads. To avoid false detection due to the relatively high error rates of MinION, we employed the heterozygous SNV information for lung cancer cell lines previously detected from WGS using Illumina HiSeq.16 Considering all the cell lines together, multiple SNVs were phased on 291 genes (Table 3 and Supplementary Table S9). Interestingly, the mutual distance between those SNVs was occasionally greater than 100 kb because of the intron between them. This approach may enable rather efficient SNP phasing than that at the genomic level as far as the location in transcript regions (Fig. 3D).
Table 3.
Application of MinION transcriptome sequencing: phased gene number and heterozygous site distance
| Cell line | Flow cell | No. of phased gene | Average of phase distance | Average of phase distance without intron |
|---|---|---|---|---|
| H1975 | R9 | 17 | 5,791 | 737 |
| PC-9 | R9 | 5 | 5,256 | 570 |
| PC-7 | R9 | 2 | 1,546 | 725 |
| H2228 | R9 | 12 | 6,528 | 484 |
| VMRC-LCD | R9 | 2 | 2,246 | 102 |
| LC2/ad | R9 | 16 | 1,542 | 219 |
| LC2/ad | R9.4 | 237 | 8,872 | 569 |
3.6.3. FL-cDNA-Seq of multi-organs of two single individuals
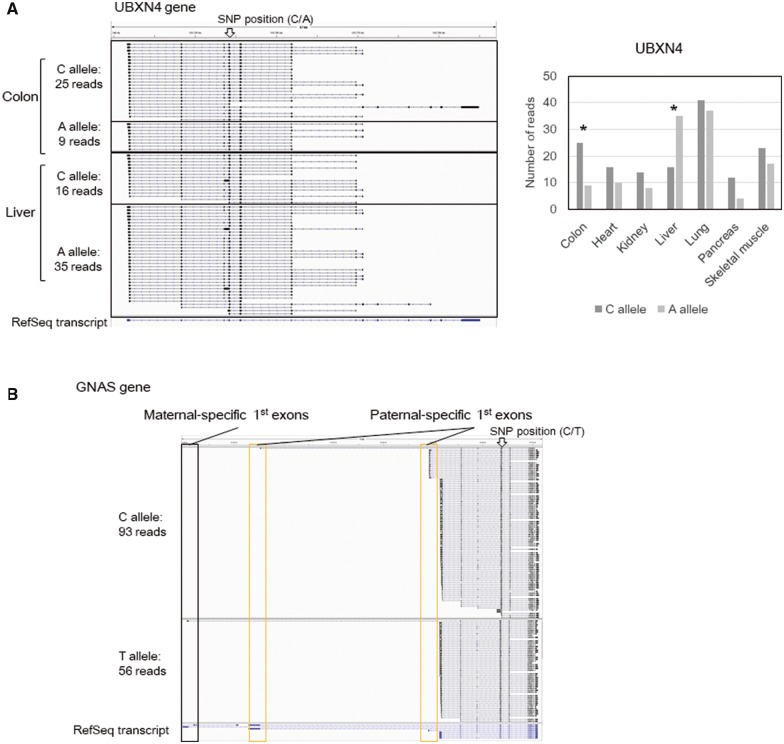
By using FL-cDNA-Seq, we attempted to detect allele-biased expression in various organs of given individuals. We obtained a total of 14 total RNA sets from seven organs of two healthy individuals, one male and one female. We also obtained their corresponding genomic DNAs. We performed FL-cDNA-Seq for these 14 RNA samples and aligned the reads to the human reference genome by LAST (Supplementary Tables S10 and S11). We also performed whole exome sequencing of two genomic DNA samples by Illumina (Supplementary Table S12 and see also Supplementary Methods). Using GATK best practices, 437,646 and 395,486 hetero SNPs were called from the genomic DNAs of the male and the female samples, respectively. Using the obtained hetero SNP data, we phased the FL-cDNA-Seq reads of all tissue-merged data. We phased 2,928 genes, and 306 genes showed significant allelic expression based on the binomial test (Table 4 and Supplementary Table S13). For instance, the UBXN4 gene was showed differential allelic expression in the female. Although the C allele showed higher expression than the A allele in most tissues of the female, only the liver showed the opposite allelic expression pattern (Fig. 4A). In the male, we also detected the GNAS gene, which has a known complex allelic imbalance expression pattern23,24 (Fig. 4B). This gene has one maternal chromosome-specific first exon, two paternal chromosome-specific first exons, and neutral first exons. Paternal first exons and the maternal first exon were only detected on the C allele and T allele, showing that the C and T alleles are on the paternal chromosome and maternal chromosome, respectively.
Table 4.
Number of genes with allelic imbalance expression
| Tissue | Male | Female | ||
|---|---|---|---|---|
| No. of phased gene | No. of genes with allelic expression (P < 0.01) | No. of phased gene | No. of genes with allelic expression (P < 0.01) | |
| Merged data | 1,219 | 105 | 1,707 | 201 |
| Colon | 625 | 64 | 1,394 | 161 |
| Heart | 836 | 78 | 1,227 | 161 |
| Kidney | 842 | 82 | 1,305 | 165 |
| Liver | 854 | 85 | 1,387 | 172 |
| Lung | 986 | 87 | 1,563 | 175 |
| Pancreas | 760 | 83 | 818 | 143 |
| Skeletal muscle | 792 | 63 | 1,215 | 157 |
Figure 4.
Detection of differential allelic expression using FL-cDNA-Seq reads. We exemplified two differentially expressed allelic genes: UBXN4 (A) in colon and liver of the female sample and GNAS (B) in merged data of the male sample. We showed the FL-cDNA-Seq reads separately by discrete SNP patterns. The positions of the SNPs are marked by arrows. (A) We also showed the expression patterns of UBXN4 in each tissues of the female sample. *P < 0.01.
3.6.4. Single cell FL-cDNA-Seq
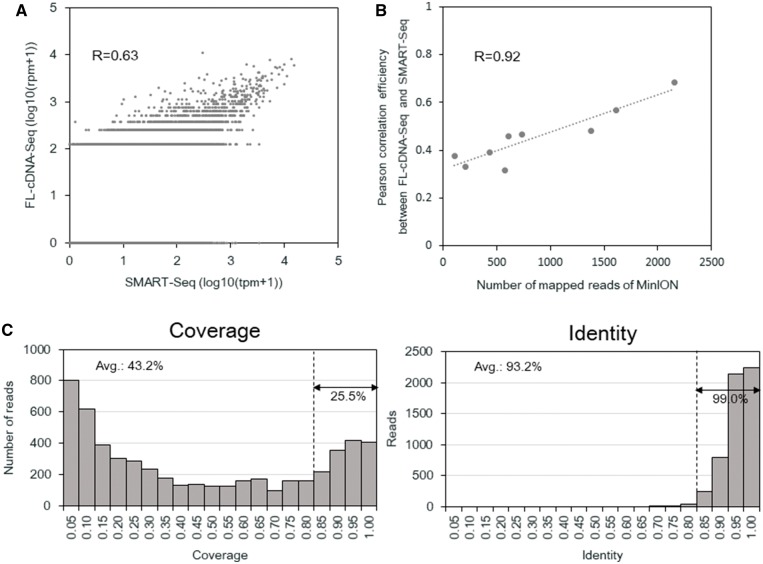
Although another group has already applied RNA-Seq using MinION to single-cell RNA-Seq, we also considered whether FL-cDNA-Seq is also applicable to this technique.13 Some existing methods for single-cell RNA-Seq also uses cDNA templates prepared by the same SMART-Seq v4.25,26 Full-length cDNA was synthesized and amplified using the Fluidigm C1 system. To obtain sufficient amounts of input template for FL-cDNA-Seq, five additional cycles of PCR were employed. Approximately 250–450 ng of cDNA templates were obtained from each of the single cells and subjected to FL-cDNA-Seq. The libraries obtained from one bulk and one doublet cell, as well as nine single cells, were barcoded and loaded onto a single R9.4 flow cell. A total of 24,817 reads (469–6,522 per samples) were obtained (Supplementary Table S14). For comparison, we also conducted Illumina sequencing using Nextera XT-processed cDNA obtained from the C1 platform (Supplementary Table S15). We compared the Illumina SMART-Seq and FL-cDNA-Seq of single cells. When we considered the correlation between SMART-Seq and FL-cDNA-Seq of the aggregated reads of nine single cells (‘virtual bulks’), the correlation was reasonably high (R = 0.63) (Fig. 5A). However, when calculated at the single cell level, the correlation was R = 0.32–0.69 (Supplementary Fig. S8). The low correlation efficiencies reflected the fact that the number of reads was small for the FL-cDNA-Seq reads (Fig. 5B). We also compared the single cell data and bulk data of FL-cDNA-Seq, prepared using the same protocol. Between the virtual bulk of single cells and bulk, the correlation was moderately high (R = 0.69) (Supplementary Fig. S9A). When calculated between the single cells and bulk, the correlation was R = 0.11–0.67 (Supplementary Fig. S9B). The overall accuracy and RefSeq coverage of FL-cDNA-Seq reads were 93.2% and 43.2% (Fig. 5C). Although the still limited throughput of the MinION sequencer is not suitable for gene expression analysis at the single cell level, we found that some full-length transcript variants were over-represented in particular individual cells. MinION sequencing can be complementarily used for this purpose.
Figure 5.
Single cell FL-cDNA-Seq using C1. (A) Comparison of the expression level of the virtual bulk of nine single cells of LC2/ad quantified by FL-cDNA-Seq and SMART-Seq. (B) Comparison of the expression level of bulk and virtual bulk of nine single cells of LC2/ad quantified by FL-cDNA-Seq. (C) Correlation of the number of mapped reads of FL-cDNA-Seq and the correlation coefficient between the same single cell data quantified by FL-cDNA-Seq using MinION. The Pearson correlation coefficient between them is shown in the graph. (A, B) The Pearson correlation coefficient was calculated using the genes detected for both data sets.
4. Conclusions
In this article, we evaluated current MinION sequencing and incorporated some modifications depending on the various applications. To our knowledge, this is the first article to survey the potential application of FL-cDNA-Seq, which may not be consistently easy with the current Illumina RNA-Seq. Indeed, our current knowledge regarding the transcriptome is almost limited to that obtained from Illumina or microarrays, which generally only represents information about fragmented transcripts. In contrast, FL-cDNA-Seq has enabled analysis of the entire transcript with respect to expression and base information at the isoform level. In this article, we demonstrated that these features first could enable one to take advantage of MinION long reads to gain novel insights in addition to the current transcriptome view. A no less important unique feature of the MinION sequencer is that it permits on-site sequencing without requiring any laboratory instruments for the sequencing. Because the flow cell as well as the data process software have been improved, now the sequencing yield has reached to more than 1 million reads. In our previous study, MinION sequencing of PCR amplicon from cDNA of clinical samples was performed.27 We could detect fusion genes and phase the hetero SNVs. It is assumed that FL-cDNA-Seq is also applicable for clinical samples. These advantages with the portable feature will further accelerated the compilation of transcriptome data. With the expansion of transcriptome information, we will be able to understand how the code of the genome and its mutations are linked to gene expression.
Supplementary Material
Acknowledgements
We sincerely thank Hiroyuki Wakaguri, Terumi Horiuchi, and Yuta Kuze for assistance with the data analysis, and Kazumi Abe, Kiyomi Imamura, Yuni Ishikawa, Mari Tsubaki, Sachie Shimazu, Megumi Kombu, Etsuko Kobayashi, and Yusuke Ogishi for their experimental support.
Accession numbers
The raw sequence reads generated in this study have been deposited at DDBJ under accession numbers RA006943D and DRA006942.
Funding
This work was supported by JSPS KAKENHI Grant Number 16H06279 and JST CREST Grant Number JPMJCR15G3.
Conflict of interest
None declared.
References
- 1. Wang Z., Gerstein M., Snyder M.. 2009, RNA-Seq: a revolutionary tool for transcriptomics, Nat. Rev. Genet., 10, 57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Pan Q., Shai O., Lee L. J., Frey B. J., Blencowe B. J.. 2008, Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing, Nat. Genet., 40, 1413–5. [DOI] [PubMed] [Google Scholar]
- 3. Piskol R., Ramaswami G., Li J. B.. 2013, Reliable identification of genomic variants from RNA-seq data, Am. J. Hum. Genet., 93, 641–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Steijger T., Abril J. F., Engström P. G., et al. 2013, Assessment of transcript reconstruction methods for RNA-seq, Nat. Methods, 10, 1177–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Goodwin S., McPherson J. D., McCombie W. R.. 2016, Coming of age: ten years of next-generation sequencing technologies, Nat. Rev. Genet., 17, 333–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Browning S., Browning B.. 2011, Haplotype phasing: existing methods and new developments, Nat. Rev. Genet., 12, 703–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Sharon D., Tilgner H., Grubert F., Snyder M.. 2013, A single-molecule long-read survey of the human transcriptome, Nat. Biotechnol., 31, 1009–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Rhoads A., Au K. F.. 2015, PacBio sequencing and its applications, Genomics. Proteomics Bioinformatics., 13, 278–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pacific Biosciences. Revolutionize Genomics with SMRT Sequencing. https://www.pacb.com/wp-content/uploads/SMRT-Sequencing-Brochure-Revolutionize-genomics-with-SMRT-Sequencing.pdf (accessed 21 June 2018).
- 10. Jain M., Olsen H. E., Paten B., et al. 2016, The Oxford Nanopore MinION: delivery of nanopore sequencing to the genomics community, Genome Biol., 17, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jain M., Koren S., Miga K. H., et al. 2018, Nanopore sequencing and assembly of a human genome with ultra-long reads, Nat. Biotechnol., 36, 338–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Oikonomopoulos S., Wang Y. C., Djambazian H., Badescu D., Ragoussis J.. 2016, Benchmarking of the Oxford Nanopore MinION sequencing for quantitative and qualitative assessment of cDNA populations, Sci. Rep., 6, 31602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Byrne A., Beaudin A. E., Olsen H. E., et al. 2017, Nanopore long-read RNAseq reveals widespread transcriptional variation among the surface receptors of individual B cells, Nat. Commun, 8, 16027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Picelli S., Björklund Å. K., Faridani O. R., Sagasser S., Winberg G., Sandberg R.. 2013, Smart-seq2 for sensitive full-length transcriptome profiling in single cells, Nat. Methods, 10, 1096–8. [DOI] [PubMed] [Google Scholar]
- 15. Kent W. J. 2002, BLAT—The BLAST-like alignment tool, Genome Res., 12, 656–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Suzuki A., Makinoshima H., Wakaguri H., et al. 2014, Aberrant transcriptional regulations in cancers: genome, transcriptome and epigenome analysis of lung adenocarcinoma cell lines, Nucleic Acids Res., 42, 13557–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Depristo M. A., Banks E., Poplin R., et al. 2011, A framework for variation discovery and genotyping using next-generation DNA sequencing data, Nat. Genet., 43, 491–501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hamada M., Ono Y., Asai K., Frith M. C.. 2017, Training alignment parameters for arbitrary sequencers with LAST-TRAIN, Bioinformatics, 33, 926–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Frith M. C., Kawaguchi R.. 2015, Split-alignment of genomes finds orthologies more accurately, Genome Biol., 16, 106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Li H., Durbin R.. 2009, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 25, 1754–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Garalde D. R., Snell E. A., Jachimowicz D., et al. 2018, Highly parallel direct RN A sequencing on an array of nanopores, Nat. Methods, 15, 201–6. [DOI] [PubMed] [Google Scholar]
- 22. Kobayashi S., Boggon T. J., Dayaram T., et al. 2005, EGFR mutation and resistance of non–small-cell lung cancer to Gefitinib, N. Engl. J. Med., 352, 786–92. [DOI] [PubMed] [Google Scholar]
- 23. Hayward B. E., Moran V., Strain L., Bonthron D. T.. 1998, Bidirectional imprinting of a single gene: gNAS1 encodes maternally, paternally, and biallelically derived proteins, Proc. Natl. Acad. Sci. U. S. A., 95, 15475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Turan S., Bastepe M.. 2013, The GNAS complex locus and human diseases associated with loss-of-function mutations or epimutations within this imprinted gene, Horm. Res. Paediatr., 80, 229–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Fish R. N., Bostick M., Lehman A., Farmer A.. 2016, Transcriptome analysis at the single-cell level using SMART technology In: Current Protocols in Molecular Biology. John Wiley & Sons, Inc: Hoboken, NJ, pp. 4.26.1–4.26.24. [DOI] [PubMed] [Google Scholar]
- 26. Yamamoto Y., Gotoh S., Korogi Y., et al. 2017, Long-term expansion of alveolar stem cells derived from human iPS cells in organoids, Nat. Methods, 14, 1097–106. [DOI] [PubMed] [Google Scholar]
- 27. Suzuki A., Suzuki M., Mizushima-Sugano J., et al. 2017, Sequencing and phasing cancer mutations in lung cancers using a long-read portable sequencer, DNA Res., 24, 585–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.