Abstract
Hanseanispora species, including H. guilliermondii, are long known to be abundant in wine grape-musts and to play a critical role in vinification by modulating, among other aspects, the wine sensory profile. Despite this, the genetics and physiology of Hanseniaspora species remains poorly understood. The first genomic sequence of a H. guilliermondii strain (UTAD222) and the discussion of its potential significance are presented in this work. Metabolic reconstruction revealed that H. guilliermondii is not equipped with a functional gluconeogenesis or glyoxylate cycle, nor does it harbours key enzymes for glycerol or galactose catabolism or for biosynthesis of biotin and thiamine. Also, no fructose-specific transporter could also be predicted from the analysis of H. guilliermondii genome leaving open the mechanisms underlying the fructophilic character of this yeast. Comparative analysis involving H. guilliermondii, H. uvarum, H. opuntiae and S. cerevisiae revealed 14 H. guilliermondii-specific genes (including five viral proteins and one β-glucosidase). Furthermore, 870 proteins were only found within the Hanseniaspora proteomes including several β-glucosidases and decarboxylases required for catabolism of biogenic amines. The release of H. guilliermondii genomic sequence and the comparative genomics/proteomics analyses performed, is expected to accelerate research focused on Hanseniaspora species and to broaden their application in the wine industry and in other bio-industries in which they could be explored as cell factories.
Keywords: Hanseniaspora guilliermondii, non-Saccharomyces wine yeasts, genome sequencing and annotation
1. Introduction
Wine fermentation is a complex biochemical process in which yeasts are the pivotal players. Although Saccharomyces cerevisiae is the leading microorganism in conducting alcoholic fermentation of the grape must, it is being increasingly acknowledged the critical role of the remaining microbiota in shaping the properties of the produced wines giving rise to the concept of ‘microbial terroir’.1–3 In particular, the activity of many other yeast species belonging to various non-Saccharomyces genera has been found to greatly influence the course of the fermentation and also the sensory characteristics of the final wine.4,5 Apiculate yeasts of the Hanseniaspora genus are among the non-Saccharomyces species more abundant in grape-musts, particularly in the early stages of the fermentation.3,6–10 Although H. uvarum is usually the more abundant species,6,9,11 several studies also report a high abundance of H. guilliermondii.8,9,12–15 As observed with other Hanseniaspora species, the presence of H. guilliermondii has a positive effect in the aroma profile of wines16–19 as well as of other fermented beverages.20,21 In general, the positive contribution of non-Saccharomyces species (and of H. guilliermondii in particular) to the aroma profile of the wine is believed to result from their ability to produce esters, higher alcohols or glycerol, but also through the synthesis of enzymes (such as β-glucosidases) that release flavor and aroma compounds (usually terpenols) present in the grapes as flavourless glycoconjugate precursors.4,5Hanseniaspora species have also been used to improve the content in glycerol or mannoproteins22,23 or to reduce acidity or alcohol level.24–26
When co-inoculated with S. cerevisae in grape-must H. guilliermondii predominates in an initial stage, however, as the fermentation proceeds its abundance decreases prominently.8,27–29 The low tolerance to ethanol has been suggested to account for the reduced fitness of H. guilliermondii in the later stages of the fermentation (in analogy to what was proposed for other non-Saccharomyces yeasts); however, the relatively high tolerance observed in some strains30 along with the demonstration that death of H. guilliermondii cells occurred in the presence of S. cerevisiae regardless of the amount of ethanol present in the fermentation broth,29 puts in question this simple model by which S. cerevisae and H. guilliermondii interact.31 Indeed, it was recently demonstrated that H. guilliermondii death in the presence of S. cerevisiae results from this latter species producing antimicrobial peptides that induce loss of cell viability in the H. guilliermondii cells upon cell–cell contacts.32
Although it is clear that the presence of H. guilliermondii affects the performance of alcoholic fermentation, the exact molecular mechanisms underlying its interaction with S. cerevisiae cells are still elusive questions. In a first attempt to address this issue, Barbosa et al. have studied the effect exerted by H. guilliermondii on the genomic expression of S. cerevisiae during a mixed-culture fermentation of a natural grape-must.27 The expression of about 300 S. cerevisiae genes was altered by the presence of H. guilliermondii, in comparison with the corresponding transcript levels registered in single-cultures.27Saccharomyces cerevisiae genes involved in the biosynthesis of vitamins were enriched among those found to be more up-regulated in the mixed S. cerevisiae–H. guilliermondii fermentation, while genes related with the uptake and biosynthesis of amino acids were enriched among those more expressed in the single-culture.27 Notably, the differences in the aroma profiles of the wines obtained in mixed-culture fermentations, appeared to be tightly correlated with the changes observed in the transcript profile of S. cerevisae genes related with formation of the aroma compounds observed in mixed-culture fermentations and attributable to the presence of H. guilliermondii.27
The first annotated genomic sequence of a H. guilliermondii strain (the UTAD222 strain, also focused in this work) was recently released33 accompanying the releases of genomic sequences for H. uvarum (strains AWRI3580 and DSM2768)34,35 and H. opuntiae (strain AWRI3578).34 However, in those short studies, the information coming from the release of these genomic sequences was not explored, this being an essential aspect to better understand the biology and physiology of the Hanseniaspora species, specially in the context of wine fermentation. In this work, a thorough analysis of the H. guilliermondii genomic sequence was performed, involving not only extensive functional analysis of the H. guilliermondii genome but also a comparative analysis with H. uvarum, H. opuntiae and also S. cerevisiae. Exploring those comparative genomic results a chromosomal map for the H. guilliermondii UTAD222 strain is proposed, this being the first time that this information is given for a strain of this species.
2. Materials and methods
2.1. Strains
In this work, 33 indigenous strains from the UTAD culture collection having a presumptive identification of belonging to the Hanseniaspora genus were used, these being listed in Supplementary Table S1. The strains were recovered from 10 different wineries of the Douro Demarcated Region in Portugal and were isolated from grapes, grape-must or wine. Pre-selection of these autochthons was based on the ability to grow on l-lysine agar selective medium followed by microscopic examination of the size, morphology and reproduction mode of the cells. Although two non-Saccharomyces genera associated with winemaking environment are characterized by bipolar budding, Hanseniaspora and Saccharomycodes, the first can be easily distinguished from the second by the smaller size of the cells. Seven reference strains were also used: H. guilliermondii CECT11027T, H. occidentalis CECT11341T, H. osmophila CECTT 11206, H. uvarum CECT1444T and H. vineae CECT1471T, from the Spanish Type Culture Collection (CECT); and H. guilliermondii CBS465T and H. opuntiae CBS8733T obtained from CBS-KNAW fungal collection (Supplementary Table S1).
2.2. Genotypic characterization of a cohort of indigenous Hanseniaspora strains
Thirty-two autochthonous Hanseniaspora strains along with the above-mentioned reference strains were subjected to (GTG)5-PCR fingerprinting for species identification. To extract the DNA from the isolates, two loopfulls of freshly grown cultures were resuspended in 500 µl of lysis buffer (50 mM Tris–HCl, 250 mM. NaCl, 50 mM EDTA, 0.3% SDS, pH 8.0). After adding one volume of phenol (pH 8.0), cells were disrupted by bead-beating for 30 s at speed 4.0 using a Fastprep cell disrupter (Bio101, Inc), followed by standard phenol: chloroform extraction. Total DNA was precipitated from the aqueous phase by the addition of 2 vol of ice-cold ethanol and of 1/10 volume of 3 M Na-acetate for 60 min at −70°C. The precipitated DNA was recovered by centrifugation at 15,000 g for 10 min at 4°C; the pellet was washed with 70% ethanol and resuspended in water. All PCR reactions were performed in 25 µl reaction volumes containing ∼10 ng of genomic DNA, 1× PCR buffer, 0.4 mM of each of the four dNTPs, 1 µM primer (GTG)5 5′-GTGGTGGTGGTGGTG-3′ and 1 U of Taq DNA polymerase (Invitrogen). The amplification conditions used were: initial template denaturation at 94°C for 5 min, 40 cycles of denaturation for 1 min at 94°C, annealing at 31°C for 1 min and extension at 72°C for 2 min. A final elongation at 72°C for 7 min was performed in the end. PCR products were separated on a 1.2% TBE agarose gel and were visualized after staining with ethidium bromide (0.5 μg/ml) and the DNA banding patterns analysed using the BioNumerics software (Applied Maths). Similarities among isolates were estimated using the Pearson coefficient and clustering was based on the UPGMA method. The identification of the autochthonous isolates was achieved by comparing their genotype profiles with those of the reference strains. To further support the results obtained, representatives of each cluster identified in the PCR fingerprinting were selected for 26S rDNA D1/D2 sequencing using primers NL-1 (5′-GCA TAT CAA TAA GCG GAG GAA AAG-3′) and NL-4 (5′-GGT CCG TGT TTC AAG ACG G-3′). The obtained 26S rDNA D1/D2 sequences were deposited at NCBI under accession numbers (MG832576 to MG832583, MG877743 and MG87744). The sequences obtained were compared with other sequences reported in the GenBank using the BLAST algorithm and a phylogenetic tree was obtained using a neighbor-joining algorithm. Species identification was ascribed to an isolate if its 26S rDNA D1/D2 sequence differed by no more than 2–3 base substitutions (i.e. ≥ 99.0% sequence identity) to that of a taxonomically accepted species type strain. Differentiation of H. opuntiae and H. guilliermondii was achieved by 5.8S-ITS-RFLP analysis, in which ITS1/ITS4 amplification products were digested with restriction enzyme DraI, and separated on 2% agarose gel on 1× TBE buffer as previously described.36
2.3. Phenotypic screening of the cohort of indigenous Hanseniaspora strains
The autochthonous Hanseniaspora strains were phenotypically profiled for relevant oenological traits, including their ability to excrete β-glucosidases, proteases and pectinases, potential to produce H2S and resistance to ethanol, sulphur dioxide (SO2), cerulenine and 5,5,5-trifluoro-dl-leucine (TFL). All assays were performed using 3 μl of a cell suspension (at an OD600nm of 0.1) prepared from an overnight pre-culture in YPD growth medium (containing, per liter, 20 g of glucose, 10 g yeast extract and 20 g bactopeptone). β-glycosidase activity was tested as previously described.37 Briefly, yeast cells were grown in a medium containing 0.5% of arbutine as the sole carbon source, 0.67% yeast nitrogen base and 2% agar (adjusted at pH 5.0 prior sterilization) and supplemented with 2 ml (per 100 ml of medium) ferric ammonium citrate solution (1% w/v). The plates were incubated at 30°C and β-glycosidase activity was observed after 2, 4, 6 and 8 h by the appearance of a dark brown color in the colonies. A non-inoculated plate of arbutin agar was used as a negative control. Proteolytic activity was determined assessing growth on skim milk agar plates, after incubation for 3–8 days at 30°C38 while the potential ability to produce H2S was evaluated by growing yeast cells in Biggy agar.37 The strains were classified in different categories according with the appearance of a brown-black coloration of the colonies, which is directly correlated with sulfite-reductase activity. Resistance to SO2 and ethanol were evaluated on YPD agar medium (buffered at pH 3.5), supplemented with increasing concentrations of SO2 (0.25, 0.5, 0.75, 1.0 and 1.5 mM) or ethanol (5.0, 6.0, 7.5, 9.0 and 10% v/v). Susceptibility phenotypes were registered after incubation at 30°C during 3 days, after visual inspection of colonies growth and comparison with growth observed in non-supplemented YPD agar. Resistance to TFL and cerulenin was assessed by determining growth on agar plates containing 0.67% of yeast nitrogen base (Difco), 2% of agar supplemented with glucose (2% w/v) in combination with TFL (1 mM) or cerulenin (25 or 100 µM). The plates were maintained at 30°C for 5 days and colonies that developed on the plates containing the inhibitors were considered resistant to TFL or cerulenin.39 Based on all the phenotypic data gathered, cluster analysis was carried out using SAS JMP 11.0 (JMP, 2011, Cary, NC) and a phenogram of yeast strains was obtained with Euclidean distance as an association measure and complete linkage as the agglomeration method.
2.4. Genome sequencing, assembly and comparison of H. guilliermondii UTAD222 genome with those available for other Hanseniaspora species
The genome of the UTAD222 strain was sequenced at StabVida (Portugal) using a whole-genome shotgun approach that explored paired-end Illumina MiSeq platform, as briefly described before.33 In specific, the genomic DNA of H. guilliermondii UTAD222 was extracted and used to construct a DNA library having inserts in the range of 250–350 bp. Library amplification was performed on the cluster generation station of the GAIIx and using the Illumina cluster generation kit. To obtain the paired-end reads, primers were designed to hybridized with Illumina specific adaptors resulting in reading of each end as a separate run. The sequencing reaction was run for 100 cycles (tagging, imaging and cleavage of one terminal base at a time), and four images of each tile on the chip were taken in different wavelengths for exciting each base-specific fluorophore. The obtained reads were afterwards assembled into contigs using the de novo assembler available on the CLC Genomics Workbench (version 9; Quiagen) set at default parameters. The obtained 208 contigs were organized into seven chromosomes, based on the identification of seven chromosomal bands in PFGE analysis of H. guilliermondii UTAD222 genomic DNA and using as a reference the proposed organization for the genome of H. uvarum DSM2768.35 To perform this organization, an iterative process was used: at first, whole-genome alignments of the UTAD222 contigs and the contigs available described for H. uvarum AWRI3580 and H. opuntiae AWRI357834 were performed using Mauve Contig Mover.40 These whole-genome alignments were performed using the default parameters (match seed weight 15, min LBC weight 200, scoring matrix HOXD). Whenever possible the whole-genome alignments were also performed using the contigs available for the H. uvarum DSM2768 strain.35 To reinforce the organization proposed by the whole-genome alignments, it was investigated colinearity between the genomes of H. guilliermondii, H. uvarum AWRI3580 and H. opuntiae AWRI3578. For this, pairwise alignments between the proteins predicted for the three species were performed and the best hits selected. These best hit pairs were then mapped in the contig organization proposed by the whole-genome alignments giving rise to the figures shown in Supplementary Fig. S3. As shown, in the vast majority of the cases it was possible to confirm colinearity with the best gene orthologues between the three species being located contiguously. To get further confirmation that the proposed organization for UTAD222 contigs was correct a number of junctions (chosen in all chromosomes) were experimentally validated by PCR. While some these PCR reactions were randomly chosen, others aimed at solving divergences identified in the whole-genome alignments. This is, for example, the case of contigs 4 and 5 which were mapped to chromosomes G and F, respectively, in H. guilliermondii, but that in H. uvarum and H. opuntiae are located in other genomic regions (as highlighted in Supplementary Fig. S3). All contig junctions experimentally verified are highlighted in Supplementary Fig. S3.
2.5. Annotation of H. guilliermondii UTAD222 genome and subsequent comparison with the proteome described for other Hanseniaspora species and for S. cerevisiae
Primary structural annotation of the H. guilliermondii UTAD222 genome was achieved by applying three de novo prediction programs: (i) Fgenesh with different matrices (trained on Aspergillus nidulans, Neurospora crassa and a mixed matrix based on different species);41 (ii) GeneMark-ES42 and (iii) Augustus.43 The different gene structures and evidences were displayed in GBrowse allowing manual validation and, whenever needed, correction of the predicted coding sequences. In specific, gene models differently predicted by the algorithms were manually curated based on the structure obtained for homologues found in related annotated genomes including H. uvarum, H. opuntiae and S. cerevisiae. The final call set comprises 4,070 protein coding genes. tRNA-encoding genes were predicted using tRNAscan-SE.44 The protein coding genes were analysed and functionally annotated using the Pedant system.45 The genome and annotation were submitted to the European Nucleotide Archive, ENA at http://www.ebi.ac.uk/ena/data/view/FQNF01000001–FQNF01000208. Metabolic reconstruction of the H. guilliermondii UTAD222 metabolic network was performed using KEGG Koala reconstruction tool,46 choosing Fungi as the taxonomy group of the genome and allowing the KEEG database to be searched for both eukaryotic and prokaryotic sequences. The comparison of the proteomes of H. guilliermondii UTAD222 with the proteomes of H. uvarum, H. opuntiae or S. cerevisae EC1118 was based on BLASTP using the sets of proteins available for each one of these species. Hanseniaspora guilliermondii UTAD222 proteins were consider similar to those present in other Hanseniaspora spp (or in S. cerevisiae EC1118) when the associated alignment had an associated e-value below e-20 and a minimum of identity of 30%. Pairwise alignments between H. uvarum/H. guilliermondii (or H. opuntiae/H. guilliermondii) having an identity between 30 and 50% were considered similar, while those having an associated identity >50% were considered highly similar. Under these conditions, we could identify 3,958 highly similar and 65 similar H. guilliermondii/H. opuntiae protein pairs; 3,790 highly similar and 169 similar H. guilliermondii/H. uvarum protein pairs; and 1,906 highly similar and 771 similar H. guilliermondii/S. cerevisiae protein pairs.
3. Results and discussion
3.1. Genotypic screening of a cohort of autochthonous Hanseniaspora strains
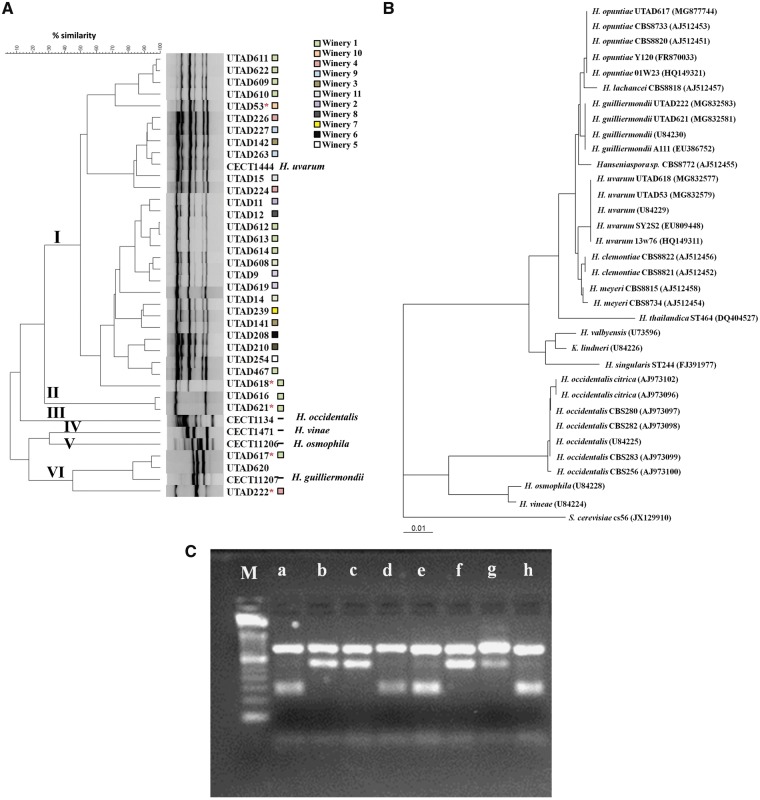
Prior studies have addressed the genetic diversity of Hanseniaspora strains in French and Italian wine producing regions, for example,8,10,15 however, no equivalent information was available for Portuguese wine producing areas. Recent reports describing the microbiomes of Portuguese wine regions have unveiled a high frequency for members of the Hanseniaspora genus,47,48 but the data gathered did not allowed discrimination at the strain level. To get insights into this, we have examined the distribution of species in a cohort of 30 strains presumptively identified as belonging to the Hanseaniaspora genus, and that also included the UTAD222 strain focused in this work. These presumed Hanseniaspora strains were isolated from grapes, grape-must or wine obtained in different wineries across the Demarcated Douro region (details in Supplementary Fig. S1) and that also included the herein focused UTAD222 strain. To characterize the genetic diversity of the strains, PCR fingerprinting was used. As controls, five reference strains from the Spanish Culture Collection representative of Hanseniaspora species already described to be present in grape-musts were included in this screening: H. guilliermondii CECT11027T, H. occidentalis CECT11341T, H. osmophila CECT11206T, H. uvarum CECT1444T and H. vineae CECT1471T. The results obtained clearly showed that the strains under examination could be divided in six groups (at a cut-off value of 50% similarity, Fig. 1A). The method used had a good discriminating power at the species level since the five reference strains were clustered separately (Fig. 1A). To obtain further information concerning the identification of the strains, the conserved ITS region of a randomly selected set of strains representative of each one of the groups (UTAD53, UTAD222, UTAD617, UTAD618 e UTAD621) was obtained and compared with sequences attributed to Hanseniaspora strains deposited at NCBI generating the phylogenetic tree that is shown in Fig. 1B. Strains UTAD53 and UTAD618 were identified as belonging to the H. uvarum species, while strain UTAD222 was identified as H. guilliermondii (Fig. 1B), this being in agreement with the results of the PCR fingerprinting (Fig. 1A). Strikingly, sequencing of the D1/D2 region of strain UTAD617 that clustered with the H. guilliermondii reference strain in the PCR fingerprinting, identified this strain as belonging to the H. opuntiae species (Fig. 1B). Previous reports showed a close genetic relatedness between H. guilliermondii and H. opuntiae,36,49 which could account for a difficulty in discriminating these two species with the PCR fingerprinting used. Thus, we have compared the profile of bands obtained upon digestion with DraI of the ITS region of strains UTAD616, UTAD617, UTAD620, UTAD621, UTAD222 since this method allows a clear distinction between H. opuntiae (three bands with expected sizes of 420, 300 and 30 bp) and H. guilliermondii (four bands with expected sizes of 420, 150, 130 and 30 bp).49 The reference strains H. guilliermondii CECT11027T, H. opuntiae CBS8733T and H. guilliermondii CBS465T were also used in this assay as controls (Fig. 1C). The pattern of bands obtained for strain UTAD222 is similar to the one of CBS465T, confirming the identification of the isolate as H. guilliermondii (Fig. 1C). UTAD616 and UTAD621 isolates, whose species identification could not be achieved in the PCR fingerprinting, were also identified as H. guilliermondii. UTAD617 and UTAD620 were identified as H. opuntiae (Fig. 1C), as well as the reference strain CECT11027T, which is in accordance with the close similarity of these three strains observed by PCR fingerprinting (Fig. 1A). Surprisingly, the reference strain CECT11027T was identified as H. opuntiae. The genetic analysis performed rendered clear the higher predominance of H. uvarum strains in our small cohort of strains, this predominance being in line with what is reported in Iberian wine producing regions.6,12,48 It was also clear the absence of a correlation between the genotype affiliation and the geographical origin of the Hanseniaspora isolates recovered, indicating that grape-musts from a particular grape variety or winery do not appear to be preferentially colonized by a specific strain (Fig. 1A). Consistently, the absence of a specific genetic signature in a given winery or in grape varieties was also reported in other studies focusing the genetic diversity of H. uvarum and H. guilliermondii isolates.10,50
Figure 1.
Genetic characterization, based on PCR-fingerprinting, of the 33 UTAD Hanseniaspora strains recovered from the Douro demarcated region and of the selected reference strains H. guilliermondii CECT11027T, H. occidentalis CECT11341T, H. osmophila CECT 11206T, H. uvarum CECT1444T and H. vineae CECT1471T; (A) Dendrogram obtained by hierarchical analysis of PCR (GTG)5 patterns using Pearson’s correlation coefficient and UPGMA clustering method; (B) Neighbour-joining phylogenetic tree showing the relationships of selected Hanseniaspora strains (marked with an asterisk in A), inferred from the sequences of the D1/D2 domain of the LSU RNA gene. Bootstrap percentages >50% from 1,000 bootstrap replicates are shown. The outgroup species was Saccharomyces cerevisiae. Bar, 1% sequence divergence; (C) Restriction patterns obtained upon restriction with DraI of the ITS region of Hanseniaspora isolates or references strain: M, molecular marker; (a) isolate UTAD616, (b) isolate UTAD617, (c) isolate UTAD620, (d) isolate UTAD621, (e) isolate UTAD222, (f) reference strain H. guilliermondii CECT11027, (g) type strain H. opuntiae CBS8733T and (h) type strain H. guilliermondii CBS465T. The squares designate the different wineries from where the isolates were retrieved.
3.2. Phenotypic screening of the autochthonous Hanseniaspora strains for interesting oenological properties
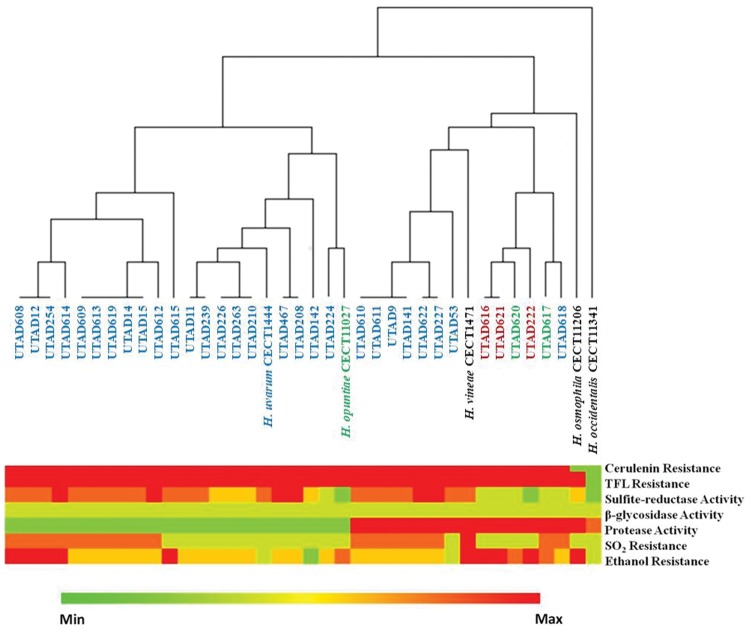
The set of strains previously used in the genotypic screening was subjected to a phenotypic characterization focused on relevant oenological properties such as tolerance to sulphur dioxide and ethanol, ability to grow in presence of 5,5,5-trifluoro-dl-leucine (TFL) or cerulenin and activity of sulphite reductase, proteases or β-glucosidases (Fig. 2). The results obtained showed a significant variation in the phenotypic profile of the H. uvarum isolates tested (e.g. strains showed a marked difference in tolerance to ethanol or to SO2), while the three H. guilliermondii isolates (UTAD616, UTAD621, UTAD222) exhibited, in general, a more similar phenotypic profile (Fig. 2). The H. opuntiae isolates exhibited similar phenotypic traits to those exhibited by the H. guilliermondii isolates, with the exception of the reference strain CECT11027T that differed by having lower sulphite reductase activity and higher resistance to ethanol (Fig. 2). Another aspect that emerged from this phenotypic analysis was the considerably low protease activity and low tolerance to ethanol of most H. uvarum isolates, in comparison with the isolates gathered from the other Hanseniaspora species and, in particular, with the H. guilliermondii isolates (Fig. 2). This observation is in line with previous studies reporting a higher resilience of H. guilliermondii to ethanol stress, specially when compared with other non-Saccharomyces species.30 The interesting oenological properties herein observed for the H. guilliermondii UTAD222 strain, namely the low activity of sulphite reductase, the high protease activity and the high tolerance to ethanol, reinforce the interest in deepening the study of this strain specially in what concerns to the analysis of its genome sequence.
Figure 2.
Phenomic characterization of the Hanseniaspora indigenous strains concerning relevant oenological properties. Besides the isolates, the type strains H. opuntiae CECT11027T (formerly designated as H. guilliermondii but herein demonstrated to be H. opuntiae), H. occidentalis CECT11341T, H. osmophila CECT 11206T, H. uvarum CECT1444T and H. vineae CECT1471T. After conducting the phenotypic tests (in triplicate), the results were used to build the heat map shown in the figure and used for the subsequent clustering analysis shown. Strains classification was based on the results of the PCR fingerprinting and, in some cases, also based on results of the D1/D2 sequencing and subsequent analysis of the DraI restriction profile (to distinguish between the H. opuntiae and H. guilliermondii strains). Hanseniaspora uvarum strains are identified in light blue, H. opuntiae strains in green and H. guilliermondii in red.
3.2.1. Genome structure of H. guilliermondii UTAD222 and comparative analysis with the genomes of other Hanseniaspora species
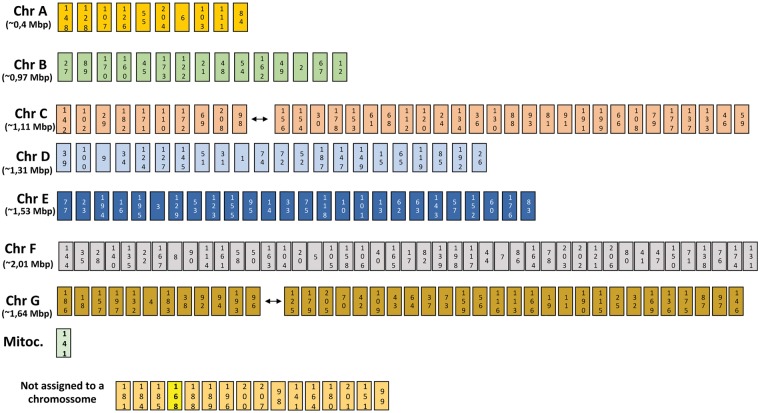
To obtain the genome sequence of the H. guilliermondii UTAD222 strain, two independent rounds of paired-end MiSeq Illumina-sequencing were used.33 The assembly of the ca. 54 million reads obtained resulted in 208 contigs, yielding a total of 9,037,850 assembled bases (Table 1). This size predicted for the genome of the H. guilliermondii UTAD222 strain is in line with the genome size described for H. uvarum (ranging between 8.8. and 9.7 Mb34,35) and H. opuntiae (8.8 Mb).34 The predicted GC content of H. guilliermondii UTAD222 is ∼31%, also in line with the percentages reported for other Hanseniaspora species (34.7% for H. opuntiae and 31.6% for H. uvarum) and for S. cerevisiae strains (38%). Prior karyotyping of H. guilliermondii wine strains from different geographical origins suggested that that this species is equipped with seven chromosomes, the same number that was attributed to most H. uvarum strains.35,51,52 In line with these results, karyotyping of the UTAD222 strain also revealed seven chromosomal bands (Supplementary Fig. S2). Using this information and also taking advantage of the recently proposed chromosomal map for H. uvarum DSM2768 strain35 and the close similarity between H. guilliermondii, H. opuntiae and H. uvarum species, we have built an organized chromosomal map of the 208 contigs obtained for the UTAD222 strain (Fig. 3 and further information provided in Supplementary Fig. S3). With this approach 192 out of the 208 contigs obtained for the H. guilliermondii UTAD222 strain were assigned to the 7 H. guilliermondii chromosomes (Fig. 3 and Supplementary Fig. S3). Among the non-assigned contigs was contig 141 (with an approximate size of 12 kb) which is predicted to be the mitochondrial chromosome, based on the high similarity with the mitochondrial sequence described for H. uvarum.53 Comparison with the genomes H. uvarum AWRI3580 and H. opuntiae AWRI3578 proteins evidenced a significant colinearity since in the large majority of the cases orthologues between these species were found to be located contiguously. Despite the large genomic similarity, two relevant genomic alterations were observed in the genome of H. guilliermondii UTAD222 corresponding to presumable translocations in chromosomes C, G and F, as detailed in Supplementary Fig. S3. Another aspect that was also rendered clear by our comparative analysis was a higher divergence of the H. uvarum DSM2768 strain, comparing to H. opuntiae AWRI3578, H. guilliermondii UTAD222 and even to H. uvarum AWRI3580 (Supplementary Fig. S3). With the data available, it is not possible to conclude if this divergence corresponds to a different genomic architectures of this strain or if it result from misleading attribution of contigs in the DSM2768 chromosomes since this assignment was very much based on adjusting the size of the chromosomes to the size of the contigs, thereby differing from the herein presented comparative genome analysis.
Table 1.
General features of the H. guilliermondii UTAD222 genome obtained after the sequencing and the subsequent manually curated annotation
| H. guilliermondii genomic features | |
|---|---|
| Genome assembly statistics | |
| Total number of reads | 53,913,308 |
| Nr of contigs | 208 |
| Coverage | x819 |
| N50 (bp) | 91.417 |
| Maximum contig length (bp) | 247,000 |
| Minimum contig length (bp) | 1.023 |
| Average contig length (bp) | 43.451 |
| Assembly size (bp) | 9,037,850 |
| Average GC content (%) | 30.9 |
| Annotation | |
| Total nr of CDS | 4,070 |
| tRNAs | 79 |
| rRNAs | 3 |
| Genes with 1 exon | 3,998 |
| Genes with 2 exons | 69 |
| Genes with more than 2 exons | 3 |
Figure 3.
Proposed chromosomal map for the H. guilliermondii UTAD222, based on data gathered from comparative genomic analysis herein gathered with those reported for H. uvarum (strains AWRI3580 and DSM7230)34,35 and H. opuntiae AWRI3578.34 Further details on how this map was obtained are provided in Supplementary Fig. S3 and in Materials and methods. In yellow, the contig including information predicted to harbour viral DNA is highlighted, while in green, the mitochondrial chromosome is highlighted.
3.2.2. Annotation of the H. guilliermondii UTAD222 genomic sequence
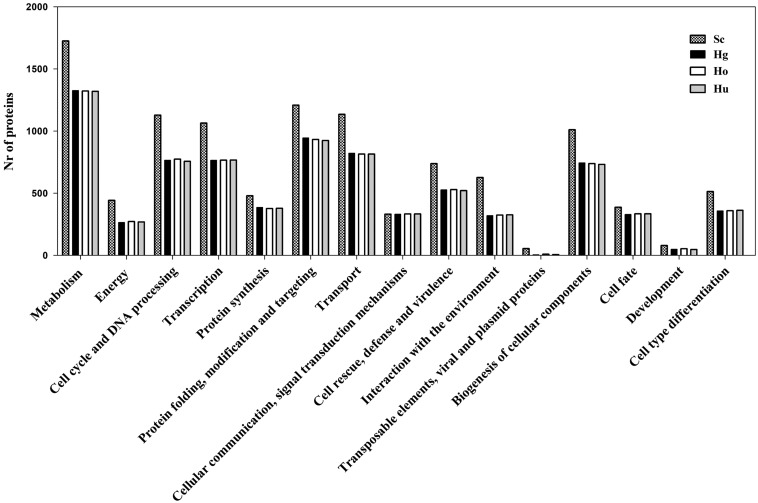
Automatic annotation of the genome sequence of H. guilliermondii UTAD222 was firstly performed using ab initio gene detection, afterwards manually curated to refine the results obtained by the informatics analysis. The predicted ORFeome of H. guilliermondii UTAD222 is estimated in 4,070 protein-encoding genes and 79 tRNAs (Table 1). The incidence of introns is ∼1.8%, in line with the low percentages described for other yeast species.54 In specific, 98% (corresponding to 3,998 genes) of the genes were predicted to be intron-free, 69 genes predicted to have two exons and three genes predicted to have three exons. The 4,070 genes that compose the predicted UTAD222 ORFeome were clustered according to their physiological function using MIPS-based Functional Catalogue. As depicted in Fig. 4, the number of H. guilliermondii genes (as well as of H. opuntiae and H. uvarum) included in each functional category was, on an average 70% the number of genes considered for S. cerevisiae, used herein as reference considering the extensive information available on gene functional analysis for this yeast species. This observation along with the fact that many of the H. guilliermondii proteins are similar to more than one S. cerevisiae protein (as detailed in Supplementary Table S5), strongly indicates that H. guilliermondii is a pre-genome duplication species, a trait that has been proposed for this species55 and recently demonstrated for H. uvarum.35
Figure 4.
Functional clustering of the proteins predicted to be encoded by H. guilliermondii UTAD222. Using the annotated and manually validated H. guilliermondii gene models (Hg), functional clustering was performed using MIPS functional catalogue. For the sake of comparison, a similar analysis was also performed for H. uvarum AWRI3578 (Hu), H. opuntiae AWRI3580 (Ho) and S. cerevisiae S288c (Sc) proteomes. For this, the different sets of proteins predicted to be encoded by the genomes of these three species were imported into Pedant database and then clustered, based on physiological function, using MIPS functional catalogue.
3.2.3. Reconstruction of H. guilliermondii UTAD222 metabolic network
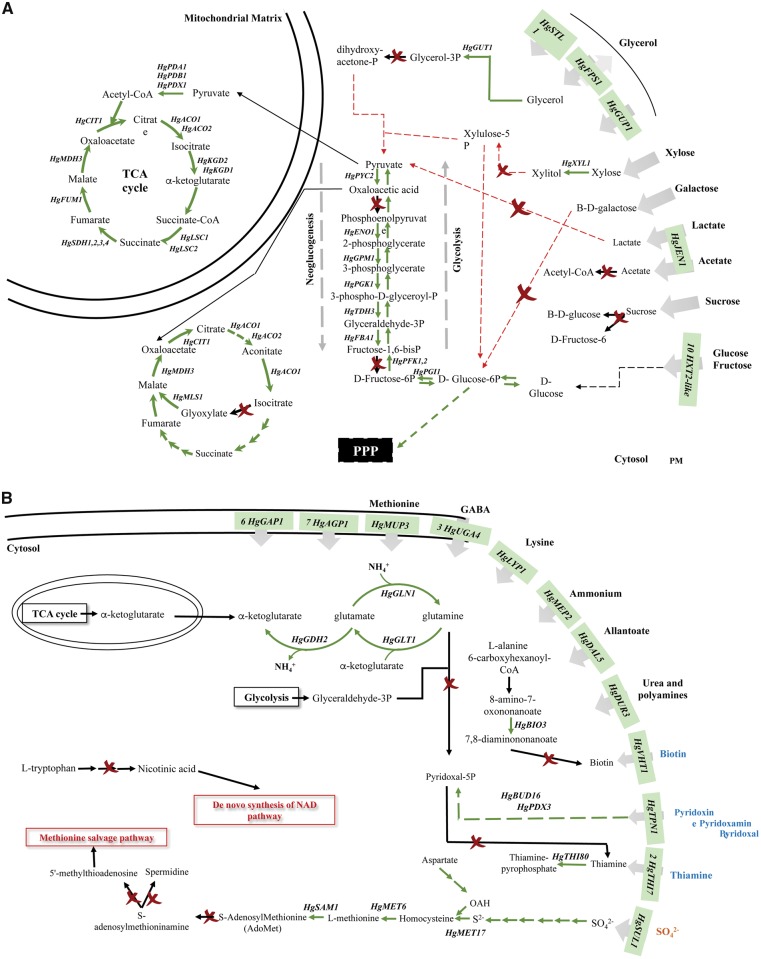
Using the KEGG Koala tool, the metabolic network of H. guilliermondii UTAD222 was reconstructed (available in Supplementary Figs S4 and S5 and briefly summarized in Fig. 5). The results show that the H. guilliermondii UTAD222 strain is equipped with genes encoding enzymes involved in all major pathways of central carbon metabolism including glycolysis, TCA cycle and pentose phosphate pathway as well as the anaplerotic enzymes, pyruvate carboxylase and malic enzyme (Supplementary Fig. S4). Notably, no genes encoding the key neoglucogenic enzymes fructose-1,6-bisphosphatase and phosphoenolpyruvate carboxykinase could be found among H. guilliermondii predicted proteins (Fig. 5 and Supplementary Fig. S4). Homologues of these proteins have also not been identified in the predicted proteomes of H. uvarum, H. opuntiae, H. vineae, H. osmophila and H. valbensys (results not shown) suggesting that the absence of a functional neoglucogenesis is a feature of the Hanseniaspora genus. This is a particular interesting trait since robust orthologues for these enzymes are present in members of all the other families that compose the Saccharomycetales order including Saccharomycetaceae, Metschnikowiaceae and Pichiaceae that, like Hanseniaspora, also harbour wine species (results not shown). Consistent with the described ability of H. guilliermondii to use fructose,18,27,56 genes involved in catabolism of this hexose were predicted in the metabolic network of the UTAD222 strain, as well as genes involved in utilization of mannose and starch (Fig. 5). Conversely, genes required for the catabolism of xylose, galactose, lactate, acetate or glycerol were absent from H. guilliermondii genetic repertoire (and also from H. uvarum and H. opuntiae) including isocitrate lyase, galactokinase and glycerol or lactate dehydrogenase (Fig. 5). The lack of these genes is consistent with the inability of UTAD222 cells to grow using these carbohydrates as the sole sources of carbon and energy (Supplementary Fig. S6). Although up to now the preferences of Hanseniaspora species concerning carbon sources have essentially focused their ability to consume glucose and/or fructose, it was recently reported the inability of H. uvarum wine isolates to use glycerol as the sole carbon source.50 Overall, the reconstruction of H. guilliermondii UTAD222 complemented with further analysis on the genome of other species having the genome sequenced shows that Hanseniaspora species appear to have a narrow range of carbon sources that can be used among those available in grape-musts, this being a factor that may contribute to reduce their competitiveness in the grape-must environment in which a strong competition for carbon sources is expected to occur.
Figure 5.
Predicted metabolic network of H. guilliermondii focused on carbon (A) or on nitrogen/sulphur metabolism (B), as suggested by in silico metabolic reconstruction of the gathered genomics data. The gene models predicted for the H. guilliermondii UTAD222 were clustered according with the biochemical pathways they are predicted to be involved in using KEGG reconstruction tool. These results (shown in Supplementary Figs S3 and S4) along with the in silico comparative proteome analyses performed with H. opuntiae, H. uvarum and S. cerevisiae (described below) were used to draw the schematic representation herein presented.
Genes encoding enzymes required for the biosynthesis of all proteinogenic amino acids could be predicted from the proteome of H. guilliermondii UTAD222, as well as those genes involved in the incorporation of ammonia to glutamine via a NAD+-dependent glutamate dehydrogenase similar to ScGdh2 (Fig. 5B and Supplementary Fig. S5). Hanseniaspora guilliermondii UTAD222 is also equipped with a functional transsulfuration pathway for sulphate assimilation, although no genes could be detected for the methionine salvage pathway (Fig. 5B and Supplementary Fig. S5). This observation is consistent with the lack of genes predicted to be involved in synthesis of polyamines (Supplementary Fig. S5) considering that one of the functions of the methionine salvage pathway is to supply precursors for the synthesis of spermine and spermidine (Fig. 5B). The fact that H. guilliermondii (and also H. opuntiae and H. uvarum) do not encode genes for synthesis of spermidine is interesting considering that this is an essential nutrient for S. cerevisiae growth.57 Spermine and spermidine are among the biogenic amines known to be present in grape-musts and they can also be produced along the alcoholic fermentation,58 which thereby turns possible for Hanseniaspora spp to eventually scavenge these nutrients from the environment rather than prompting their synthesis.
Another relevant observation that emerged from the reconstruction of the metabolic network of the UTAD222 strain was the absence of genes involved in biosynthesis of thiamine, pyridoxal, pyridoxine, biotin and for the de novo synthesis of nicotinic acid (Fig. 5B). The absence of these genes is consistent with the previously described auxotrophy of H. guilliermondii for thiamin, niacin, biotin and pyridoxine.59 The inability of Hanseniaspora species to synthesize these vitamins is likely to be a relevant factor in the described low fermentative capacity of these species, particularly in the case of thiamine whose abundance was found to determine the glycolytic flux and, consequently, the fermentation rate of S. cerevisiae cells.60,61 To respond to the depletion of thiamine, S. cerevisiae genes involved in biosynthesis of this vitamin are strongly up-regulated in stationary phase and wine yeast strains were found to evolve adaptive responses to improve the expression of these genes and, consequently, to improve the fermentation rate in grape-musts.61 Consistent with the idea that H. guilliermondii scavenges vitamins from the environment, S. cerevisiae genes involved in synthesis of thiamine (ScTHI20 and ScTHI21), biotin (ScBIO3) or pyridoxine (e.g. ScSNO1), were among those found to be more strongly up-regulated during cultivation in the presence of H. guilliermondii UTAD222.27
3.3. The ‘transportome’ of H. guilliermondii UTAD222
Around 600 genes are predicted to have a transport-related function in H. guilliermondii UTAD222, similar to the number of H. uvarum and H. opuntiae proteins predicted to have this function (Supplementary Table S2). A closer analysis into this ‘transportome’ of H. guilliermondii UTAD222, revealed that it is equipped with 22 predicted sugar transporters, 10 of these being predicted to be hexose transporters (Supplementary Table S2 and Fig. 5). All these putative H. guilliermondii hexose transporters were found to harbour a very high similarity with the high-affinity glucose transporter ScHxt262 or with ScGal2, the galactose permease that has also been shown to transport glucose with a moderate affinity.62 Although H. guilliermondii strains (including UTAD222, Supplementary Fig. S6) are fructophilic56,63,64 there was no orthologue of the fructose specific transporters Ffz1 or Fsy1.65,66 This observation is consistent with results of a previous study that pinpointed the H. guilliermondii NCYC2380 strain as exhibiting fructophilic behaviour but not harbouring Ffz-like genes.56 A large survey for Ffz-like genes in Fungi has also failed to identify robust orthologues for these transporters in the genome of H. uvarum, another reported fructophilic yeast,64,67 as well as in H. vineae and H. valbensyis.68 Interestingly, the H. guilliermondii UTAD222 protein showing a higher similarity with Z. bailii Ffz1 (e-value of the associated alignment of −56, 27% identity) is HGUI01014, predicted to be a polyamine exporter based on its high degree of similarity with the MFS multi-drug resistance transport ScTpo1 (e-value of the associated alignment of −156, 57% identity) (Supplementary Table S2). Further studies are required to better understand what could be the function of predicted hexose transporters in H. guilliermondii and, in specific, if they are somehow able to contribute for the fructophilic behaviour of the species by mediating fructose transport. Phosphorylation of fructose mediated by hexokinases is another critical parameter that was shown to determine the capacity of wine S. cerevisiae strains to use fructose in detriment of glucose.69,70 Thus, it can also be hypothesized that the two hexokinases predicted to be encoded by H. guilliermondii UTAD222 (HGUI_01771, 47% identity with glucokinase ScGlk1 and HGUI_03922, 72% identity) could have a higher affinity for fructose than for glucose. Indeed, preliminary results have shown that Z. bailii encodes a hexokinase having a preference for fructose (over glucose) phosphorylation,71 the hexokinases encoded by H. guilliermondii showing a very high similarity (99% identity at the amino acid level) with those encoded by Z. bailii strains having their genome sequenced (results not shown).
Thirty-two H. guilliermondii UTAD222 genes are predicted to encode transporters of nitrogenous compounds (Supplementary Table S3 and Fig. 5) including permeases for amino acids, ammonium, allantoin, urea and GABA (Fig. 5). Remarkably, 13 of the predicted H. guilliermondii amino acid permeases bear the highest similarity with the general amino acid permeases ScGAP1 and ScAGP1, only being possible to identify specific permeases for lysine and methionine (Supplementary Table S2 and Fig. 5). Indeed, almost all S. cerevisiae amino acid-specific permeases were found to be absent in H. guilliermondii and also in H. opuntiae and in H. uvarum (Supplementary Table S5 and Table 4). This observation suggests that Hanseniaspora species favor the utilization of permeases with a broader range of substrates in detriment of using specific permeases, which is a reasonable adaptive mechanism since the amount of nitrogen present in wine musts is largely variable and composed by different amino acids.
3.3.1. The H. guilliermondii UTAD222 ‘flavorome’
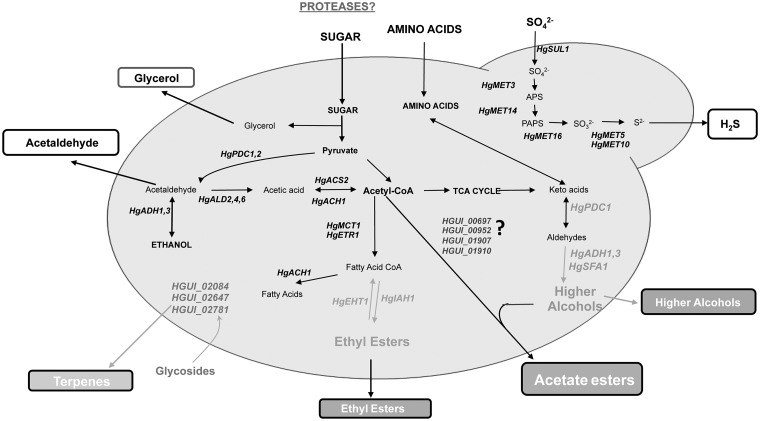
It is known that H. guilliermondii affects the production of volatile compounds of wines when in consortium with S. cerevisiae.16–18,27 Other studies have also reported the ability of H. guilliermondii itself to produce compounds having an impact in wine aroma even in single-culture including 2-phenyl ethyl acetate, ethyl acetate, isobutyric acid, ethyl esters and higher alcohols, albeit in most of these cases the production titers are below those reported for S. cerevisiae.16–19,72 Consistent with these observations, reconstruction of H. guilliermondii UTAD222 metabolic network shows that this strain is equipped with four genes encoding β-glucosidases, as well as genes required for synthesis of acetaldehyde, ethyl esters and higher alcohols (Fig. 6 and Supplementary Table S3). Notably, it was not found within the UTAD222 ORFeome proteins similar to the S. cerevisae acetyl transferases involved in synthesis of acetate esters ScATF1, ScATF2 and ScAYT1, although it is known that these are precisely the aroma compounds whose production is more strongly impacted by the presence of H. guilliermondii.17,19,72 Four proteins (HGUI_00697, HGUI_00952, HGUI_01907 and HGUI_01910) harboring motifs conserved within the alcohol acetyltransferase enzyme family were identified in the predicted proteome of H. guilliermondii UTAD222, these proteins only having orthologues in other species of the Hanseniaspora genus (Supplementary Table S3). Further studies are required to confirm if these proteins do represent a novel set of acetyl transferases involved in synthesis of aroma compounds. Another observation of remark was the absence of aryl-alcohol dehydrogenases required for synthesis of higher alcohols from corresponding aldehydes in H. guilliermondii, which could contribute for the reported lower ability of this species to produce these compounds, specially when compared with S. cerevisiae.
Figure 6.
Hanseniaspora guilliermondii UTAD222 genes predicted to be involved in formation of aroma compounds. Based on results of metabolic reconstruction and also on orthology with S. cerevisiae genes, the set of H. guilliermondii UTAD222 genes predicted to be involved in pathways leading to the production of aroma compounds was identified. Further details on these genes involved in the ‘flavorome’ of H. guilliermondii are provided in Supplementary Table S4. No genes encoding acetyl transferases involved in synthesis of acetate esters could be predicted in the H. guilliermondii UTAD222 ORFeome and comparative analysis with the proteins having such a function in S. cerevisiae also did not produce results. Nonetheless, four proteins (HGUI_00697, HGUI_00952, HGUI_01907 and HGUI_01910) with motifs found in acetyl transferases were identified in H. guilliermondii suggesting that these could represent a new class of these enzymes.
3.3.2. Comparative analysis of the predicted proteomes of H. guilliermondii, H. uvarum and H. opuntiae
To contribute for a better understanding of the differences and similarities existing among H. guilliermondii, H. uvarum and H. opuntiae, the UTAD222 predicted proteins were compared with those predicted for H. opuntiae AWRI3578 and for H. uvarum (AWRI3580 and DSM2768 strains). This comparative analysis rendered clear a very high similarity between the predicted ORFeomes of these three Hanseniaspora species, more evident for H. guilliermondii and H. opuntiae (Supplementary Fig. S7 and Supplementary Table S4). Fourteen H. guilliermondii proteins had no orthologue in H. opuntiae AWRI3578 or H. uvarum (AWRI3580 and DSM2768 strains), these being listed in Table 2. Nine of these proteins had a robust homologue in H. valbyensis indicating that their presence is not specific of the H. guilliermondii species (Table 2). The remaining five proteins could not be identified in the genome of any other species of the Hanseniaspora genus, although homologues could be identified in the genome of other fungal species such as Rhizopus microspores, Zygosaccharomyces bailii or Saitoella complicate (Table 2). These five proteins are all of viral origin including components necessary for the formation of capsid and spike and also a protein required for replication of viral dsDNA (Table 2). The fact that the genes encoding these proteins are contiguous and located in the same genomic region (contig 168) and also the observation that this genomic region exhibits a very strong homology (>95% identity at the nucleotide level) with the genomic sequences of bacteriophages, suggest that H. guilliermondii UTAD222 harbors a viral genome. The existence of viral DNA has been well documented in other wine yeast species including S. cerevisiae, H. uvarum and Z. bailii,73 this being responsible for the killer phenotype that allows cells to secrete toxin(s) to the growth medium that will cause death of sensitive species. In the cases of the three above referred yeast species, the killer phenotype is mediated by non-infectious dsRNA viruses,73 but this phenotype can also be chromosomally encoded or associated with dsDNA plasmids.73 Up to now the ability of H. guilliermondii to induce the killer phenotype has not been studied and the viral genomic sequence herein uncovered does not provide enough information to understand if a toxin can indeed be produced by UTAD222 cells. Nonetheless, this is a topic deserving further investigation considering the role that the killer phenotype has in shaping competitiveness of wine yeast species.
Table 2.
Results obtained from the comparative analysis of the predicted H. guilliermondii UTAD222 ORFeome with the set of proteins predicted for H. uvarum (AWRI3580 and DSM7210 strains) or H. opuntiae
| H. guilliermondii UTAD222 proteins not having an orthologue in both H. opuntiae and H. uvarum | ||
| ORF name | Probable function | Best ortdologue |
| HGUI_01127 | BolA-like protein | HANVADRAFT_51108 from H. valbyensis NRRL Y-1626 |
| HGUI_01165 | Ribosomal protein | HANVADRAFT_5002 from H. valbyensis NRRL Y-1626 |
| HGUI_01445 | Predicted small nuclear ribonucleo protein | HANVADRAFT_11820 from H. valbyensis NRRL Y-1626 |
| HGUI_01959 | Predicted ribonucleo protein complex subunit 3 | HANVADRAFT_53037 from H. valbyensis NRRL Y-1626 |
| HGUI_02479 | Unknown | HANVADRAFT_53473 from H. valbyensis NRRL Y-1626 |
| HGUI_02541 | Unknown | HANVADRAFT_53361 from H. valbyensis NRRL Y-1626 |
| HGUI_03386 | Unknown | HANVADRAFT_53900 from H. valbyensis NRRL Y-1626 |
| HGUI_03446 | Unknown | HANVADRAFT_23140 from H. valbyensis NRRL Y-1626 |
| HGUI_03596 | Predicted vacuolar ATPase assembly integral membrane protein | HANVADRAFT_22909 from H. valbyensis NRRL Y-1626 |
| HGUI_04019 | Minor spike protein H | CP02DC15_1109 from Chlamydia psittaci 02DC15 |
| HGUI_04020 | Replication-associated protein A | HMPREF1334_01983 from Enterococcus faecalis ERV41 |
| HGUI_04021 | External scaffolding protein D | M771_11055 from Neisseria gonorrhoeae MU_NG1 |
| HGUI_04022 | Capsid protein | M743_11545 from Neisseria gonorrhoeae NYC_2011_05_13 |
| HGUI_04023 | Major spike protein G | MPLA_2010001 from Mesorhizobium sp. ORS335 |
| H. guilliermondii UTAD222 proteins having an orthologue in H. uvarum but not in H. opuntiae | ||
| ORF name | Probable function | H. uvarum orthologue (in AWRI3580/in DSM2768) |
| HGUI_03532 | Predicted to be involved in endosome-to-Golgi intracellular trafficking | −/HuPEP8 |
| HGUI_00706 | Dolichyl pyrophosphate Man9GlcNAc2 alpha-1,3-glucosyltransferase | AWRI3580_g2240/HuALG6 |
| HGUI_03286 | Dolichyl-diphosphooligosaccharide–protein glycosyltransferase | AWRI3580_g1764/HuOST2 |
| HGUI_03882 | Dol-P-Glc: Glc(2)Man(9)GlcNAc(2)-PP-Dol alpha-1,2-glucosyltransferase | AWRI3580_g1578/- |
| HGUI_01192 | Eukaryotic translation initiation factor eIF-1 | AWRI3580_g477/- |
| HGUI_00394 | Predicted hydroxyacyl-thioester dehydratase involved in fatty acid biosynthesis | AWRI3580_g894/HuHTD2 |
| HGUI_03531 | Predicted mitochondrial import inner membrane translocase subunit | AWRI3580_g2710/HuTIM54 |
| HGUI_02284 | Predicted to be involved in intracellular protein trafficking, similar to ScSss1 | AWRI3580_g3696/- |
| HGUI_03514 | Predicted UDP-N-acetylglucosamine transferase | AWRI3580_g742/HuALG14 |
| H. guilliermondii UTAD222 proteins having an orthologue in H. opuntiae but not in H. uvarum | ||
| ORF name | Probable function | H. opuntiae orthologue |
| HGUI_02309 | Predicted chitin synthase export chaperone | AWRI3578_g694 |
| HGUI_03602 | Predicted vacuolar membrane Cu transporter | AWRI3578_g678 |
| HGUI_02076 | Cytochrome c oxidase assembly protein Cox19 | AWRI3578_g1039 |
| HGUI_01733 | Dolichyl pyrophosphate Glc1Man9GlcNAc2 alpha-1,3-glucosyltransferase | AWRI3578_g4036 |
| HGUI_01849 | Predicted to be involved in meiosis, similar to ScSpo11 | AWRI3578_g1481 |
| HGUI_02598 | Mitochondrial import inner membrane translocase subunit TIM12 | AWRI3578_g3410 |
| HGUI_01086 | Mitochondrial protein required for assembly of cytochrome c oxidase, mitochondrial | AWRI3578_g916 |
| HGUI_02427 | Predicted to be involved in protein palmitoylation, similar to ScSwf1 | AWRI3578_g3718 |
| HGUI_00194 | Predicted transcriptional regulator of the Zn-finger family, shows some similarity with ScRim101 | AWRI3578_g3297 |
| HGUI_02287 | Putative 5-hydroxyisourate hydrolase | AWRI3578_g2657 |
| HGUI_00707 | Putative glucan endo-1,3-beta-glucosidase btgC | AWRI3578_g1650 |
| HGUI_01349 | Predicted GPI-anchored protein of unknown function | AWRI3578_g2558 |
In this table, a selected set of proteins was chosen to illustrate those only found in H. guilliermondii, those found in H. guilliermondii and in H. opuntiae and those found in H. guilliermondii and H. uvarum. A more comprehensive list including all the results is provided in Supplementary Table S5.
Another aspect of relevance that emerged from the comparative analysis between the H. guilliermondii, H. opuntiae and H. uvarum proteomes was the identification of 26 proteins only shared by H. guilliermondii and H. uvarum and of 87 proteins only shared by H. guilliermondii and H. opuntiae. A full list of these proteins is provided in Supplementary Table S4 and a selected set in Table 2. From the functional point of view, the proteins apparently absent in H. opuntiae include an enzyme predicted to be involved in fatty acid biosynthesis (HGUI_00394), several proteins involved in post-translational modifications (HGUI_03286, HGUI_03882, HGUI_03882, HGUI_03514) and in intracellular protein trafficking (HGUI_03532, HGUI_03531, HGUI_02284) (Supplementary Table S3 and Table 2). Within the data set of proteins apparently absent in H. uvarum, it was possible to identify an homologue of a putative vacuolar membrane copper transporter (HGUI_03602), two proteins involved in the assembly of cytochrome c oxidase (HGUI_02076, HGUI_01086) and a putative β-glucosidase (HGUI_00707). Considering the lack of information concerning the functionality of these proteins, it is hard to link the presence/absence of these proteins with physiological traits of the different Hanseniaspora species, although this is certainly a topic deserving further attention.
3.3.3. Comparative analysis of the predicted proteomes of Hanseniaspora spp. and S. cerevisiae: emphasis on genes associated with tolerance to oenological relevant stresses
Hanseniaspora spp. are known to have multiple phenotypic traits different from those observed in S. cerevisiae wine strains (e.g. different fermentation rates and ability to use terpene-like molecules for the production of aroma compounds)4 and thus, we have hypothesized whether this could result from divergences in the proteomes of the different species. To scrutinize this, the set of H. guilliermondii UTAD222 predicted proteins was compared with those described for the S. cerevisiae wine strain EC1118, widely used for wine-making and having a well annotated genomic sequence.74 The results of this in silico proteomic analysis are detailed in Supplementary Tables S5 and S6. Approximately 1,294 H. guilliermondii proteins had no robust homologue in the S. cerevisiae strains examined (listed in Supplementary Table S5). This could occur either because the S. cerevisiae strains do not encode these proteins or because the proteins with a corresponding function are highly divergent from those one found in H. guilliermondii. These H. guilliermondii proteins that had no orthologue in S. cerevisiae were present in H. uvarum and in H. opuntiae (Supplementary Fig. S7 and Supplementary Table S5), suggesting that they comprise specific proteins of the Hanseniaspora genus. The majority of these ‘Hanseniaspora-specific’ proteins (879) have a poorly characterized function but we could identify β-glucosidases (e.g. HGUI_02084, HGUI_02647 and HGUI_02781), flocculins (e.g. HGUI_01978, HGUI_03264, HGUI_04053, HGUI_01804, HGUI_03963 and HGUI_03989), proteins involved in transcriptional regulation and in translation, as well as proteins involved in respiration and in metabolism of various carbohydrate and nitrogen compounds (Supplementary Table S5). The identification of β-glucosidase-encoding genes within the set of the putative ‘Hanseniaspora-specific’ genes was expected considering the demonstration of the influence of members of this species, including H. guilliermondii, in increasing the amount of terpenes present in wines;75 a phenotypic trait that is absent in S. cerevisiae strains.76 It was also interesting the emergence in this comparative analysis of several enzymes predicted to be involved in metabolism of tyramine, histamine, histidine or tyrosine (Supplementary Table S5), these being biogenic amines that are known to be present in grape-musts and whose content increases along alcoholic fermentation.58,77 This observation suggests that the presence of H. guilliermondii (and also H. uvarum and H. opuntiae) may play a role in controlling the levels of tyramine and histamine obtained in wines counter-acting, at least in part, the production prompted by S. cerevisiae and other microbiota present in musts.58 Consistent with this idea, previous studies have reported a decrease in the level of biogenic amines produced during the early stages of wine fermentations conducted in the presence of Kloeckera apiculata, the anamorph of H. uvarum, this effect being alleviated as the fermentation proceeds and the abundance of this non-Saccharomyces yeast is reduced.78 This is a particular relevant aspect since the level of tyramine and histamine is a critical parameter determining wine quality and consumers’ health and recommended levels for the amount of these compounds that can be present in wine has been established in a variety of countries.58
The comparative analysis performed also led to the identification of about 2,797 S. cerevisiae proteins for which we could not identify an orthologue in the predicted ORFeomes of H. guilliermondii, H. uvarum or H. opuntiae (Supplementary Table S6). This data set of S. cerevisiae proteins absent in Hanseniaspora is enriched in proteins involved in carbohydrate metabolism, in nitrogen metabolism, in cell cycle, in DNA processing, in RNA synthesis, in transcriptional regulation, in protein synthesis and in intracellular trafficking (Supplementary Table S6). Consistent with the results of the metabolic reconstruction described earlier, among the S. cerevisae genes missing in the Hanseniaspora species were ICL1 encoding isocitrate lyase, PCK1, encoding phosphoenolpyruvate carboxykinase; FBP1, encoding fructose-1,6-biphosphatase, as well as the genes involved in biosynthesis of thiamine, niacin, biotin or polyamines (Supplementary Table S6). Among the large set of S. cerevisiae proteins involved in transcriptional regulation that were not found in H. guilliermondii were Msn2 and Msn4 transcription factors, essential for control of environmental stress response in the budding yeast79 (Fig. 7 and Supplementary Table S6). No protein showing similarity to ScMsn2 or ScMsn4 could also be identified in the genome of the other Hanseniaspora species having a genome disclosed indicating that this is a feature of the genus and not specific of the H. guilliermondii species. This observation is intriguing as it leaves open the elucidation of the mechanisms by which Hanseniaspora spp respond to environmental stress, specially taking into account that robust homologues of Msn2 and of its target genes had been identified in the genome of other wine Non-Saccharomyces species such as Z. bailii.80 The existence of a general response to stress showing similarities to the one described in the budding yeast has been characterized in distant fungal species including Lachancea kluyveri, Candida albicans or Schizosaccharomyces pombe, albeit this transcriptional response was found to be mediated by regulators having little homology with ScMsn2.81–83 Thus, the absence of Msn2/Msn4 orthologues within the Hanseniaspora genus does not per se means that these yeasts will not be equipped with a general stress response.
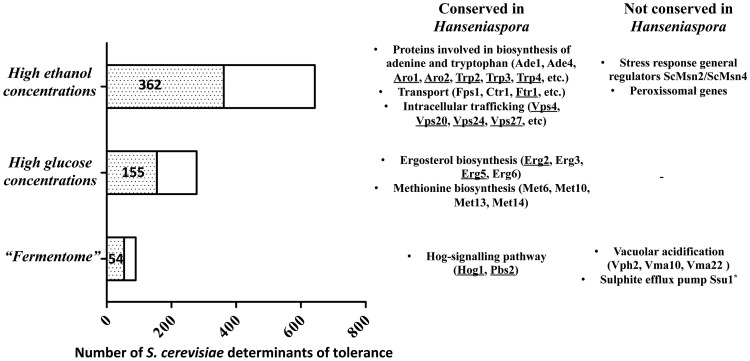
Figure 7.
Conservation in the ORFeome of H. guilliermondii UTAD222 of S. cerevisiae genes identified to be required for tolerance of this species against relevant oenological stresses. Based on results from large-scale phenotypic screenings the S. cerevisiae genes required for tolerance against high concentrations of glucose or ethanol were identified, as well, as the genes required for maximal fermentation in grape-juice medium (also called the ‘fermentome’). The predicted ORFeome of H. guilliermondii UTAD222 was searched for orthologues of these determinants of stress tolerance being the number of conserved genes identified in each horizontal bar. A selected set of the conserved and non-conserved genes is also shown while the full result is available in Supplementary Table S6. Since Hanseniaspora are highly tolerant to high-glucose concentrations it was not considered of interest to specify the set of non-conserved genes. Saccharomyces cerevisiae genes mediating multidrug resistance are underlined. *Although Ssu1 was not identified within the set of genes belonging to the fermentome, the absence of this sulphite efflux pump in the Hanseniaspora species is also highlighted in the figure since tolerance to SO2 is generally accepted as a highly relevant trait for oenological strains.
Taking advantage of the wide panoply of genetic tools available for S. cerevisiae, large-scale phenotypic screenings have been conducted to identify key players involved in tolerance to oenological relevant stresses including tolerance to ethanol84,85 or to high glucose concentrations.86,87 More recently, the S. cerevisiae genes required for fermentation in grape juice simulated medium, generally known as the ‘fermentome’, were also identified using similar chemogenomics screenings.88,89 Only half of the S. cerevisae genes required for fermentation in grape-simulated medium (48 in total of 90 genes) were found to have robust homologues in H. guilliermondii and in H. uvarum or H. opuntiae (Supplementary Table S7), while 300 ethanol-resistance and 155 ‘high-glucose’ resistance genes were identified in H. guilliermondii UTAD222. Similar numbers were also obtained for H. uvarum or H. opuntiae (Supplementary Table S6). Among the genes found to be conserved in the Hanseniaspora species is the protein kinase Hog1 (Fig. 7 and Supplementary Table S6) as well as most members of the HOG-pathway indicating that this signalling pathway, essential for S. cerevisiae response to stress induced by vinification,89 is conserved in the Hanseniaspora genus. Despite this, we could not identify orthologues within the Hanseniaspora species similar to ScMsn2/ScMsn4, ScHot1 or ScSko1, the known mediators of Hog1-dependent response (Supplementary Table S6). Proteins involved in the assembly and function of the vacuolar ATPase, also identified for S. cerevisae tolerance to vinification-stress, comprised another group of proteins for which we could not identify orthologues in H. guilliermondii (Fig. 7 and Supplementary Table S7). Among the S. cerevisiae ethanol-resistance genes that had no orthologue in H. guilliermondii are several proteins with peroxisomal function, as well as proteins of the respiratory chain and involved in intracellular protein trafficking (Fig. 7 and Supplementary Table S7). It remains to be established if the absence of these genes and eventually of others mediating tolerance to ethanol in S. cerevisae contributes for the reduced tolerance of Hanseniaspora species to stressful concentrations of ethanol. An interesting observation that emerged from this comparative analysis was the absence within the three Hanseniaspora species examined of an orthologue for the sulphite efflux pump Ssu1, an essential determinant of S. cerevisiae tolerance to SO2, which is in line with the observed low resilience of these species to SO2-stress in the phenotypic screening shown in Fig. 2 and also reported before.8
Overall, the comparative genomic analysis herein performed shows significant differences in the way by which S. cerevisiae and Hanseniaspora spp respond to environmental stress and, in particular, in the response to relevant stresses in the context of wine fermentation with emphasis on the toxicity imposed by high concentrations of ethanol. The release of the genomic sequence of H. guilliermondii as well as the annotation and the comparative analysis herein presented also focusing H. opuntiae and H. uvarum is expected to contribute for a better elucidation of the physiology and biology of these species, essentially by boosting gene and genomic functional analyses. Necessarily, the results of this research will foster the design (or re-design) of Hanseniaspora strains to be used not only in wine-making but also in other relevant biotechnological industrial processes, such as production of aroma compounds, where these species have been identified as potentially interesting cell-factories.90,91
Supplementary Material
Acknowledgements
This work was financed by FEDER through POCI-COMPETE 2020 and by Fundação para a Ciência e Tecnologia (BI/PTDC/AGR-TEC/3315/2014_2016, Project SMARTWINE-‘Smarter win fermentations: integrating omic-tools for development of novel mixed-starter cultures for tailoring wine production’), and supported by FCT to Biosystems and Integrative Sciences Institute (BioISI; FCT/UID/Multi/04046/2013) and to iBB-Institute for Bioengineering and Biosciences (through contract UID/BIO/04565/2013). Programa Operacional Regional de Lisboa 2020 is also acknowledged for its financial support to iBB (project no. 007317). I.S. is recipient of a PhD grant funded by FCT with the reference SFRH/BD/122200/2016. The authors also thank the technical support of Filipa Antunes Silva in PFGE analysis of H. guilliermondii strains.
Accession numbers
26S DNA sequences of Hanseaniaspora isoaltes used in this study were submitted to GenBank under the accession numbers MG832576 to MG832583, MG877743 and MG87744. The genome sequence of the H. guilliermondiiUTAD222 sequence has been deposited in ENA (accession numbers FQNF01000001 to FQNF01000208).
Conflict of interest
None declared.
References
- 1. Bokulich N. A., Collins T. S., Masarweh C., et al. 2016, Associations among wine grape microbiome, metabolome, and fermentation behavior suggest microbial contribution to regional wine characteristics, MBio, 7, e00631–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bokulich N. A., Thorngate J. H., Richardson P. M., Mills D. A.. 2014, Microbial biogeography of wine grapes is conditioned by cultivar, vintage, and climate, Proc. Natl. Acad. Sci. U. S. A, 111, E139–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Capozzi V., Garofalo C., Chiriatti M. A., Grieco F., Spano G.. 2015, Microbial terroir and food innovation: the case of yeast biodiversity in wine, Microbiol. Res., 181, 75–83. [DOI] [PubMed] [Google Scholar]
- 4. Jolly N. P., Varela C., Pretorius I. S.. 2014, Not your ordinary yeast: non-Saccharomyces yeasts in wine production uncovered, FEMS Yeast Res., 14, 215–37. [DOI] [PubMed] [Google Scholar]
- 5. Belda I., Ruiz J., Esteban-Fernandez A., et al. 2017, Microbial contribution to wine aroma and its intended use for wine quality improvement, Molecules, 22, 189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Wang C., Garcia-Fernandez D., Mas A., Esteve-Zarzoso B.. 2015, Fungal diversity in grape must and wine fermentation assessed by massive sequencing, quantitative PCR and DGGE, Front. Microbiol., 6, 1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Zott K., Miot-Sertier C., Claisse O., Lonvaud-Funel A., Masneuf-Pomarede I.. 2008, Dynamics and diversity of non-Saccharomyces yeasts during the early stages in winemaking, Int. J. Food Microbiol., 125, 197–203. [DOI] [PubMed] [Google Scholar]
- 8. Grangeteau C., Gerhards D., Rousseaux S., von Wallbrunn C., Alexandre H., Guilloux-Benatier M.. 2015, Diversity of yeast strains of the genus Hanseniaspora in the winery environment: what is their involvement in grape must fermentation? Food Microbiol., 50, 70–7. [DOI] [PubMed] [Google Scholar]
- 9. Sternes P. R., Lee D., Kutyna D. R., Borneman A. R.. 2017, A combined meta-barcoding and shotgun metagenomic analysis of spontaneous wine fermentation, Gigascience, 6, 1–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Grangeteau C., Gerhards D., V., Wallbrunn C., Alexandre H., Rousseaux S., Guilloux-Benatier M.. 2016, Persistence of two non-saccharomyces yeasts (Hanseniaspora and Starmerella) in the cellar, Front. Microbiol., 7, 268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Padilla B., Garcia-Fernandez D., Gonzalez B., et al. 2016, Yeast biodiversity from DOQ priorat uninoculated fermentations, Front. Microbiol., 7, 930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Clavijo A., Calderon I. L., Paneque P.. 2010, Diversity of Saccharomyces and non-Saccharomyces yeasts in three red grape varieties cultured in the Serrania de Ronda (Spain) vine-growing region, Int. J. Food Microbiol., 143, 241–5. [DOI] [PubMed] [Google Scholar]
- 13. Vigentini I., Maghradze D., Petrozziello M., et al. 2016, Indigenous Georgian wine-associated yeasts and grape cultivars to edit the wine quality in a precision oenology perspective, Front. Microbiol., 7, 352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Santiago-Urbina J. A., Arias-García J. A., Ruiz-Terán F.. 2015, Yeast species associated with spontaneous fermentation of taberna, a traditional palm wine from the southeast of Mexico, Ann. Microbiol., 65, 287–96. [Google Scholar]
- 15. Romancino D. P., Di Maio S., Muriella R., Oliva D.. 2008, Analysis of non-Saccharomyces yeast populations isolated from grape musts from Sicily (Italy), J. Appl. Microbiol., 105, 2248–54. [DOI] [PubMed] [Google Scholar]
- 16. Moreira N., Mendes F., Guedes de Pinho P., Hogg T., Vasconcelos I.. 2008, Heavy sulphur compounds, higher alcohols and esters production profile of Hanseniaspora uvarum and Hanseniaspora guilliermondii grown as pure and mixed cultures in grape must, Int. J. Food Microbiol., 124, 231–8. [DOI] [PubMed] [Google Scholar]
- 17. Lage P., Barbosa C., Mateus B., Vasconcelos I., Mendes-Faia A., Mendes-Ferreira A.. 2014, H. guilliermondii impacts growth kinetics and metabolic activity of S. cerevisiae: the role of initial nitrogen concentration, Int. J. Food Microbiol., 172, 62–9. [DOI] [PubMed] [Google Scholar]
- 18. Moreira N., Pina C., Mendes F., Couto J. A., Hogg T., Vasconcelos I.. 2011, Volatile compounds contribution of Hanseniaspora guilliermondii and Hanseniaspora uvarum during red wine vinifications, Food Control, 22, 662–7. [Google Scholar]
- 19. Rojas V., Gil J. V., Pinaga F., Manzanares P.. 2003, Acetate ester formation in wine by mixed cultures in laboratory fermentations, Int. J. Food Microbiol., 86, 181–8. [DOI] [PubMed] [Google Scholar]
- 20. de Arruda Moura Pietrowski G., dos Santos C. M., Sauer E., Wosiacki G., Nogueira A.. 2012, Influence of fermentation with Hanseniaspora sp. yeast on the volatile profile of fermented apple, J. Agric. Food Chem., 60, 9815–21. [DOI] [PubMed] [Google Scholar]
- 21. Batista N. N., Ramos C. L., Dias D. R., Pinheiro A. C., Schwan R. F.. 2016, The impact of yeast starter cultures on the microbial communities and volatile compounds in cocoa fermentation and the resulting sensory attributes of chocolate, J. Food Sci. Technol., 53, 1101–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Belda I., Navascues E., Marquina D., Santos A., Calderon F., Benito S.. 2016, Outlining the influence of non-conventional yeasts in wine ageing over lees, Yeast, 33, 329–38. [DOI] [PubMed] [Google Scholar]
- 23. Domizio P., Liu Y., Bisson L. F., Barile D.. 2014, Use of non-Saccharomyces wine yeasts as novel sources of mannoproteins in wine, Food Microbiol., 43, 5–15. [DOI] [PubMed] [Google Scholar]
- 24. Contreras A., Hidalgo C., Henschke P. A., Chambers P. J., Curtin C., Varela C.. 2014, Evaluation of non-Saccharomyces yeasts for the reduction of alcohol content in wine, Appl. Environ. Microbiol., 80, 1670–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Ciani M., Morales P., Comitini F., et al. 2016, Non-conventional yeast species for lowering ethanol content of wines, Front. Microbiol., 7, 642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tristezza M., Tufariello M., Capozzi V., Spano G., Mita G., Grieco F.. 2016, The oenological potential of Hanseniaspora uvarum in simultaneous and sequential co-fermentation with Saccharomyces cerevisiae for industrial wine production, Front. Microbiol., 7, 670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Barbosa C., Mendes-Faia A., Lage P., Mira N. P., Mendes-Ferreira A.. 2015, Genomic expression program of Saccharomyces cerevisiae along a mixed-culture wine fermentation with Hanseniaspora guilliermondii, Microbial. Cell Factories, 14, 124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Branco P., Monteiro M., Moura P., Albergaria H.. 2012, Survival rate of wine-related yeasts during alcoholic fermentation assessed by direct live/dead staining combined with fluorescence in situ hybridization, Int. J. Food Microbiol., 158, 49–57. [DOI] [PubMed] [Google Scholar]
- 29. Perez-Nevado F., Albergaria H., Hogg T., Girio F.. 2006, Cellular death of two non-Saccharomyces wine-related yeasts during mixed fermentations with Saccharomyces cerevisiae, Int. J. Food Microbiol., 108, 336–45. [DOI] [PubMed] [Google Scholar]
- 30. Pina C., Santos C., Couto J. A., Hogg T.. 2004, Ethanol tolerance of five non-Saccharomyces wine yeasts in comparison with a strain of Saccharomyces cerevisiae—influence of different culture conditions, Food Microbiol., 21, 439–47. [Google Scholar]
- 31. Albergaria H., Arneborg N.. 2016, Dominance of Saccharomyces cerevisiae in alcoholic fermentation processes: role of physiological fitness and microbial interactions, Appl. Microbiol. Biotechnol., 100, 2035–46. [DOI] [PubMed] [Google Scholar]
- 32. Branco P., Kemsawasd V., Santos L., et al. 2017, Saccharomyces cerevisiae accumulates GAPDH-derived peptides on its cell surface that induce death of non-Saccharomyces yeasts by cell-to-cell contact, FEMS Microbiol. Ecol., 93, doi: 10.1093/femsec/fix055. [DOI] [PubMed] [Google Scholar]
- 33. Seixas I., Barbosa C., Salazar S. B., et al. 2017, Genome sequence of the nonconventional wine yeast Hanseniaspora guilliermondii UTAD222, Genome Announc., 5, e01515–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Sternes P. R., Lee D., Kutyna D. R., Borneman A. R.. 2016, Genome sequences of three species of Hanseniaspora isolated from spontaneous wine fermentations, Genome Announc., 4, e01287–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Langenberg A. K., Bink F. J., Wolff L., et al. 2017, Glycolytic functions are conserved in the genome of the wine yeast Hanseniaspora uvarum, and pyruvate kinase limits its capacity for alcoholic fermentation, Appl. Environ. Microbiol., 83, e01580–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Nisiotou A. A., Nychas G. J.. 2007, Yeast populations residing on healthy or botrytis-infected grapes from a vineyard in Attica, Greece, Appl. Environ. Microbiol., 73, 2765–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Mendes Ferreira A., Climaco M. C., Mendes Faia A.. 2001, The role of non-Saccharomyces species in releasing glycosidic bound fraction of grape aroma components – a preliminary study, J. Appl. Microbiol., 91, 67–71. [DOI] [PubMed] [Google Scholar]
- 38. González J., Gallardo C., Pombar A., Rego P., Rodríguez L.. 2004, Determination of enzymatic activities in ecotypic Saccharomyces and non-Saccharomyces yeast, Electron. J. Environ. Agric. Food Chem., 3, 743–50. [Google Scholar]
- 39. Oliveira V. A., Vicente M. A., Fietto L. G., et al. 2008, Biochemical and molecular characterization of Saccharomyces cerevisiae strains obtained from sugar-cane juice fermentations and their impact in cachaca production, Appl. Environ. Microbiol., 74, 693–701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Rissman A. I., Mau B., Biehl B. S., Darling A. E., Glasner J. D., Perna N. T.. 2009, Reordering contigs of draft genomes using the Mauve aligner, Bioinformatics, 25, 2071–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Salamov A. A., Solovyev V. V.. 2000, Ab initio gene finding in Drosophila genomic DNA, Genome Res., 10, 516–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ter-Hovhannisyan V., Lomsadze A., Chernoff Y. O., Borodovsky M.. 2008, Gene prediction in novel fungal genomes using an ab initio algorithm with unsupervised training, Genome Res., 18, 1979–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Stanke M., Keller O., Gunduz I., Hayes A., Waack S., Morgenstern B.. 2006, AUGUSTUS: ab initio prediction of alternative transcripts, Nucleic Acids Res., 34, W435–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lowe T. M., Eddy S. R.. 1997, tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence, Nucleic Acids Res., 25, 955–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Walter M. C., Rattei T., Arnold R., et al. 2009, PEDANT covers all complete RefSeq genomes, Nucleic Acids Res., 37, D408–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kanehisa M., Sato Y., Morishima K.. 2016, BlastKOALA and GhostKOALA: kEGG tools for functional characterization of genome and metagenome sequences, J. Mol. Biol., 428, 726–31. [DOI] [PubMed] [Google Scholar]
- 47. Pinto C., Pinho D., Cardoso R., et al. 2015, Wine fermentation microbiome: a landscape from different Portuguese wine appellations, Front. Microbiol., 6, 905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Drumonde-Neves J., Franco-Duarte R., Lima T., Schuller D., Pais C.. 2017, Association between grape yeast communities and the vineyard ecosystems, PLoS One, 12, e0169883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Cadez N., Poot G. A., Raspor P., Smith M. T.. 2003, Hanseniaspora meyeri sp. nov., Hanseniaspora clermontiae sp. nov., Hanseniaspora lachancei sp. nov. and Hanseniaspora opuntiae sp. nov., novel apiculate yeast species, Int. J. Syst. Evol. Microbiol., 53, 1671–80. [DOI] [PubMed] [Google Scholar]
- 50. Albertin W., Setati M. E., Miot-Sertier C., et al. 2015, Hanseniaspora uvarum from winemaking environments show spatial and temporal genetic clustering, Front. Microbiol., 6, 1569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Esteve-Zarzoso B., Peris-Toran M. J., Ramon D., Quero A.. 2001, Molecular characterisation of Hanseniaspora species, Antonie Van Leeuwenhoek, 80, 85–92. [DOI] [PubMed] [Google Scholar]
- 52. Moghaieb R. E. A., Abdelhadi A. A., El-Sadawy H. A., Allam N. A. T., Baiome B. A., Soliman M. H.. 2017, Molecular identification and genetic diversity among Photorhabdus and Xenorhabdus isolates, 3 Biotech, 7, 6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Pramateftaki P. V., Kouvelis V. N., Lanaridis P., Typas M. A.. 2006, The mitochondrial genome of the wine yeast Hanseniaspora uvarum: a unique genome organization among yeast/fungal counterparts, FEMS Yeast Res., 6, 77–90. [DOI] [PubMed] [Google Scholar]
- 54. Souciet J.-L., Dujon B., Gaillardin C., et al. 2009, Comparative genomics of protoploid Saccharomycetaceae, Genome Res., 19, 1696–709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Merico A., Sulo P., Piskur J., Compagno C.. 2007, Fermentative lifestyle in yeasts belonging to the Saccharomyces complex, FEBS J., 274, 976–89. [DOI] [PubMed] [Google Scholar]
- 56. Cabral S., Prista C., Loureiro-Dias M. C., Leandro M. J.. 2015, Occurrence of FFZ genes in yeasts and correlation with fructophilic behaviour, Microbiology, 161, 2008–18. [DOI] [PubMed] [Google Scholar]
- 57. Chattopadhyay M. K., Tabor C. W., Tabor H.. 2003, Spermidine but not spermine is essential for hypusine biosynthesis and growth in Saccharomyces cerevisiae: spermine is converted to spermidine in vivo by the FMS1-amine oxidase, Proc. Natl. Acad. Sci. U. S. A., 100, 13869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Smit A., Toit W., Toit M.. 2008, Biogenic amines in wine: understanding the headache, S. Afr. J. Enol. Vitic., 29, 109–27. [Google Scholar]
- 59. Barnett J. A., Payne R. W., Yarrow D.. 1990, Yeasts: characteristics and identification.
- 60. Bataillon M., Rico A., Sablayrolles J.-M., Salmon J.-M., Barre P.. 1996, Early thiamin assimilation by yeasts under enological conditions: impact on alcoholic fermentation kinetics, J. Ferment. Bioeng., 82, 145–50. [Google Scholar]
- 61. Brion C., Ambroset C., Delobel P., Sanchez I., Blondin B.. 2014, Deciphering regulatory variation of THI genes in alcoholic fermentation indicate an impact of Thi3p on PDC1 expression, BMC Genomics, 15, 1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Maier A., Volker B., Boles E., Fuhrmann G. F.. 2002, Characterisation of glucose transport in Saccharomyces cerevisiae with plasma membrane vesicles (countertransport) and intact cells (initial uptake) with single Hxt1, Hxt2, Hxt3, Hxt4, Hxt6, Hxt7 or Gal2 transporters, FEMS Yeast Res., 2, 539–50. [DOI] [PubMed] [Google Scholar]
- 63. Ciani M., Comitini F., Mannazzu I., Domizio P.. 2010, Controlled mixed culture fermentation: a new perspective on the use of non-Saccharomyces yeasts in winemaking, FEMS Yeast Res., 10, 123–33. [DOI] [PubMed] [Google Scholar]
- 64. Ciani M., Fatichenti F.. 1999, Selective sugar consumption by apiculate yeasts, Lett. Appl. Microbiol., 28, 203–6. [DOI] [PubMed] [Google Scholar]
- 65. Leandro M. J., Cabral S., Prista C., Loureiro-Dias M. C., Sychrova H.. 2014, The high-capacity specific fructose facilitator ZrFfz1 is essential for the fructophilic behavior of Zygosaccharomyces rouxii CBS 732T, Eukaryot. Cell, 13, 1371–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Leandro M. J., Sychrova H., Prista C., Loureiro-Dias M. C.. 2013, ZrFsy1, a high-affinity fructose/H+ symporter from fructophilic yeast Zygosaccharomyces rouxii, PLoS One, 8, e68165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. De Benedictis M., Bleve G., Grieco F., Tristezza M., Tufariello M., Grieco F.. 2011, An optimized procedure for the enological selection of non-Saccharomyces starter cultures, Antonie Van Leeuwenhoek, 99, 189–200. [DOI] [PubMed] [Google Scholar]
- 68. Goncalves C., Coelho M. A., Salema-Oom M., Goncalves P.. 2016, Stepwise functional evolution in a fungal sugar transporter family, Mol. Biol. Evol., 33, 352–66. [DOI] [PubMed] [Google Scholar]
- 69. Zuchowska M., Jaenicke E., Konig H., Claus H.. 2015, Allelic variants of hexose transporter Hxt3p and hexokinases Hxk1p/Hxk2p in strains of Saccharomyces cerevisiae and interspecies hybrids, Yeast, 32, 657–69. [DOI] [PubMed] [Google Scholar]
- 70. Berthels N. J., Cordero Otero R. R., Bauer F. F., Pretorius I. S., Thevelein J. M.. 2008, Correlation between glucose/fructose discrepancy and hexokinase kinetic properties in different Saccharomyces cerevisiae wine yeast strains, Appl. Microbiol. Biotechnol., 77, 1083–91. [DOI] [PubMed] [Google Scholar]
- 71. Sutterlin K. A. 2010, Fructophilic yeasts to cure stuck fermentations in alcoholic beverages. PhD thesis, Viticulture and Oenology, University of Stellenbosch.
- 72. Viana F., Gil J. V., Genoves S., Valles S., Manzanares P.. 2008, Rational selection of non-Saccharomyces wine yeasts for mixed starters based on ester formation and enological traits, Food Microbiol., 25, 778–85. [DOI] [PubMed] [Google Scholar]
- 73. Becker B., Schmitt M. J.. 2017, Yeast killer toxin K28: biology and unique strategy of host cell intoxication and killing, Toxins, 9, 73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Novo M., Bigey F., Beyne E., et al. 2009, Eukaryote-to-eukaryote gene transfer events revealed by the genome sequence of the wine yeast Saccharomyces cerevisiae EC1118, Proc. Natl. Acad. Sci. U. S. A., 106, 16333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Lopez M. C., Mateo J. J., Maicas S.. 2015, Screening of beta-glucosidase and beta-xylosidase activities in four non-Saccharomyces yeast isolates, J. Food Sci., 80, C1696–704. [DOI] [PubMed] [Google Scholar]
- 76. Padilla B., Gil J. V., Manzanares P.. 2016, Past and future of non-saccharomyces yeasts: from spoilage microorganisms to biotechnological tools for improving wine aroma complexity, Front. Microbiol., 7, 411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Rodriguez-Naranjo M. I., Ordonez J. L., Callejon R. M., Cantos-Villar E., Garcia-Parrilla M. C.. 2013, Melatonin is formed during winemaking at safe levels of biogenic amines, Food Chem. Toxicol., 57, 140–6. [DOI] [PubMed] [Google Scholar]
- 78. Granchi L., Romano P., Mangani S.. 2005, Production of biogenic amines by wine microorganisms, Bull. O.I.V., 78, 595–609. [Google Scholar]
- 79. Gasch A. P., Spellman P. T., Kao C. M., et al. 2000, Genomic expression programs in the response of yeast cells to environmental changes, Mol. Biol. Cell, 11, 4241–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Mira N. P., Munsterkotter M., Dias-Valada F., et al. 2014, The genome sequence of the highly acetic acid-tolerant Zygosaccharomyces bailii-derived interspecies hybrid strain ISA1307, isolated from a sparkling wine plant, DNA Res., 21, 299–313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Gasch A. P. 2007, Comparative genomics of the environmental stress response in ascomycete fungi, Yeast, 24, 961–76. [DOI] [PubMed] [Google Scholar]
- 82. Chen D., Toone W. M., Mata J., et al. 2003, Global transcriptional responses of fission yeast to environmental stress,. Mol. Biol. Cell, 14, 214–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Brion C., Pflieger D., Souali-Crespo S., Friedrich A., Schacherer J.. 2016, Differences in environmental stress response among yeasts is consistent with species-specific lifestyles, Mol. Biol. Cell, 27, 1694–705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Teixeira M. C., Raposo L. R., Mira N. P., Lourenco A. B., Sa-Correia I.. 2009, Genome-wide identification of Saccharomyces cerevisiae genes required for maximal tolerance to ethanol, Appl. Environ. Microbiol., 75, 5761–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Yoshikawa K., Tanaka T., Furusawa C., Nagahisa K., Hirasawa T., Shimizu H.. 2009, Comprehensive phenotypic analysis for identification of genes affecting growth under ethanol stress in Saccharomyces cerevisiae, FEMS Yeast Res., 9, 32–44. [DOI] [PubMed] [Google Scholar]
- 86. Teixeira M. C., Raposo L. R., Palma M., Sa-Correia I.. 2010, Identification of genes required for maximal tolerance to high-glucose concentrations, as those present in industrial alcoholic fermentation media, through a chemogenomics approach, OMICS, 14, 201–10. [DOI] [PubMed] [Google Scholar]
- 87. Gonzalez R., Morales P., Tronchoni J., et al. 2016, New genes involved in osmotic stress tolerance in Saccharomyces cerevisiae, Front. Microbiol., 7, 1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Walker M. E., Nguyen T. D., Liccioli T., et al. 2014, Genome-wide identification of the Fermentome; genes required for successful and timely completion of wine-like fermentation by Saccharomyces cerevisiae, BMC Genomics, 15, 552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Novo M., Mangado A., Quiros M., Morales P., Salvado Z., Gonzalez R.. 2013, Genome-wide study of the adaptation of Saccharomyces cerevisiae to the early stages of wine fermentation, PLoS One, 8, e74086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Romano P., Suzzi G., Zironi R., Comi G.. 1993, Biometric study of acetoin production in Hanseniaspora guilliermondii and Kloeckera apiculata, Appl. Environ. Microbiol., 59, 1838–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Alamri S., Hashem M., Alrumman S., Al-Qahtani M.. 2015, Enhancement of bio-ethanol production from date molasses by non-conventional yeasts, Res. J. Microbiol., 10, 114–25. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.