Abstract
Parent specific-DNA methylation is the genomic imprint that induces mono-allelic gene expression dependent on parental origin. Resetting of DNA methylation in the germ line is mediated by a genome-wide re-methylation following demethylation known as epigenetic reprogramming. Most of our understanding of epigenetic reprogramming in germ cells is based on studies in mice, but little is known about this in marsupials. We examined genome-wide changes in DNA methylation levels by measuring 5-methylcytosine expression, and mRNA expression and protein localization of the key enzyme DNA methyltransferase 3 L (DNMT3L) during germ cell development of the marsupial tammar wallaby, Macropus eugenii. Our data clearly showed that the relative timing of genome-wide changes in DNA methylation was conserved between the tammar and mouse, but in the tammar it all occurred post-natally. In the female tammar, genome-wide demethylation occurred in two phases, I and II, suggesting that there is an unidentified demethylation mechanism in this species. Although the localization pattern of DNMT3L in male germ cells differed, the expression patterns of DNMT3L were broadly conserved between tammar, mouse and human. Thus, the basic mechanisms of DNA methylation-reprogramming must have been established before the marsupial-eutherian mammal divergence over 160 Mya.
Keywords: DNA methylation, epigenetic reprogramming, DNMT3L, germ cell, 5-methylcytosine
1. Introduction
Genomic imprinting results in parent-of-origin expression of genes and controls normal development and maternal behaviour.1–3 Disruption of imprinting leads to developmental defects in humans.4 Imprinting occurs in both groups of therian mammals, the eutherians and marsupials, but there is, as yet, no evidence for imprinting in monotremes.5,6 DNA methylation (5-methylcytosine; 5 mC) is the best known epigenetic mark that plays a critical role in genomic imprinting and regulates retrotransposon activity.7,8 This DNA modification is catalysed by the well characterized DNA methyltransferases (DNMT) family of enzymes including DNMT1, DNMT3A, DNMT3B and DNMT3L.9–11 The DNMT3 family is present in eutherians, marsupials and all non-mammalian vertebrates examined so far.5,12 DNMT3L has no catalytic domain but it is needed to establish DNA methylation.13–15 It is thought to act as a cofactor with other DNMT3 family members to establish DNA methylation and genomic imprinting.16,17
Genome-wide reprogramming of DNA methylation can be observed during both gametogenesis and early embryogenesis in mammals.11 These changes are mediated by a global re-methylation following demethylation known as epigenetic reprogramming. In early embryogenesis, demethylation is necessary for acquisition of totipotency.18–22 Gametogenesis requires global demethylation to ensure the erasure of parent-specific imprinting.7,23 This allows resetting of sex-specific imprinting marks in the male and female germ cells.2,7,23 Most of our understanding of epigenetic reprogramming in germ cells is based on studies in mice, but much less is known about this in marsupials. In mice there is only a short pre-natal period when germ cells proliferate, but in marsupials there is an extended period of post-natal development. In the tammar wallaby, Macropus eugenii, germ cells continue proliferation for at least 25 days after birth before beginning to enter meiotic and mitotic arrest,24,25 giving an extended window of time in which to examine the relative roles of the somatic environment versus intrinsic germ cell programming in controlling the imprinting. Using a marsupial that diverged from eutherian mammals over 160 Mya to highlight the similarities and dissimilarities in the epigenetic control of germ cell maturation and epigenetic reprogramming can provide new understanding of the evolution and control of mammalian epigenetic processes. As yet, just one gene, H19, has been examined for epigenetic reprogramming of germ cells. This study established that the sequence of events and the relative timing of epigenetic reprogramming of H19 in males is conserved between these mammals.26 However, genome-wide changes in methylation levels in either sex of marsupials are still unknown.
In this study, we hypothesized that germ-line methylation is an intrinsic and conserved property of mammalian germ cells and is independent of germ cell migration, proliferation or absolute time of birth. We also hypothesized that the timing of 5 mC emergence and DNMT3L expression coincides with critical stages of germ cell development and maturation and not with the time of birth. To test these hypotheses, we examined the timing of 5 mC emergence in marsupial male and female germ cells and measured the gene expression and localization patterns of DNMT3L in developing tammar gonads of both sexes throughout the duration of their post-natal development in the pouch.
2. Materials and methods
2.1. Animals and tissue collection
Tammar wallabies (Macropus eugenii) originally from Kangaroo Island were collected from our colony at the University of Melbourne. Post-natal samples were collected at appropriate times of postpartum (pp) and were aged based on head length growth curves.27 Pouch young (PY) smaller than 5 g are heterothermic,28 and were cooled on ice then killed by decapitation. PYs over 5 g were anaesthetized with Zoletil (Virbac, Australia) i.m. before killing by decapitation. Gonads were dissected immediately after death and one gonad was snap-frozen in liquid nitrogen for expression analysis and the other was fixed in 4% paraformaldehyde (PFA) in phosphate buffered saline (PBS) (140 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4, pH7.4) for immunohistochemistry. All animal experimentation followed the Australian National Health and Medical Research Council (2013) guidelines and were approved by the University of Melbourne Animal Experimental Ethics Committees.
2.2. Immunofluorescence (IF)
Extensive testing of different antibody concentrations and pre-treatments was performed on tammar tissue to optimize each antibody. The DNMT3L antibody chosen (Ab3493, Abcam, Cambridge, UK) was based on the BLAST search of amino acid sequence that was specific to DNMT3L. PFA fixed samples were washed in 1×PBS and re-hydrated through an ethanol series before being embedded in paraffin. Embedded samples were serially sectioned at 6 μm on a Rotary Microtome CUT 4060 microtome (microTec Laborgeräte GmbH, Waldorf, Germany) and mounted on poly-lysine coated slides (Thermo Fisher scientific, Massachusetts, USA). Sections were de-waxed, rehydrated through an ethanol-PBS series and then incubated in 0.1% (v/v) Triton X-100 in PBS (PBST) for 15 min at room temperature to permeabilize the tissue. Sections of testes and ovaries of young aged between day 10 and 70 pp were then treated for 3 min with 4 M HCl, for 10 min at day 100–120 pp and for 20 min at day 200 pp to adult, then washed in PBS and treated for 1.5 min with 0.1% (w/v) trypsin in young aged day 10–25 pp, for 2 min at day 45–70 pp and for 4 min at day 100 pp–adult. Next, the sections were incubated for 1 h with 10% (w/v) horse serum diluted in PBST for blocking. After blocking, sections were incubated with primary antibody solution at 4 °C for 16 h (Table 1). For SOX9 and 5 mC double-staining, we treated sections with 4 M HCl for 10 min after boiling Tris-EDTA (pH 8.0) treatment for 20 min. The following day, sections were washed three times with PBST and then incubated with fluorescent secondary antibodies for 1 h (Table 1). Sections were washed three times with PBST again and then incubated for 10 min with 4’, 6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, Missouri, USA). DAPI treated sections were washed with PBST and mounted with DAKO fluorescence mounting solution (Sigma-Aldrich, Missouri, USA). The control for all treatments was no primary antibody. These slides were treated in the same manner as described above except primary antibody treatment was omitted. Images were collected on an Olympus DP70 Microscope Digital Camera System (Olympus, Tokyo, Japan).
Table 1.
Antibodies used for this study
| Target | Host | Product code | Concentration (mg/ml) | Dilution |
|---|---|---|---|---|
| Primary antibody | ||||
| 5-methylcytosine (5mC) | Mouse | ab10805 (Abcam) | 1 | 1/100 (Testes) |
| 1/50 (ovaries) | ||||
| Human DNMT3L | Rabbit | ab3493 (Abcam) | 1.11 | 1/50 |
| SOX9 | Rabbit | AB5535 (Merck) | 1 | 1/100 |
| Secondary antibody | ||||
| Mouse immunogen | Goat | Alexa Fluor 568 | 2 | 1/200 |
| Rabbit immunogen | Donkey | Alexa Fluor 488 | 2 | 1/200 |
2.3. Quantitation of 5 mC staining
Genome-wide changes in DNA methylation levels were investigated by immunofluorescence (IF) using a DNA methylation specific antibody to 5-methylcytosine (5 mC). We also performed H&E staining using the same samples to distinguish germ cells from somatic cells on the basis of their distinctive morphology as described previously.25,29–31 Intensity of fluorescence was quantified using ImageJ (https://imagej.nih.gov/ij/). The intensity of 5 mC staining in germ cells was normalized to that of somatic cells in adjacent areas of the section: Sertoli cells in males and pre-granulosa/granulosa cells in females. Differences in levels of 5 mC between stages were analysed by Tukey–Kramer’s honestly significant difference (HSD) test after analysis of variance (ANOVA) with R (https://cran.r-project.org/bin/windows/base/).
2.4. Gonadal RNA extraction and cDNA synthesis
Snap-frozen gonads were used for RNA extraction using the GenElute Mammalian total RNA Miniprep Kit (Sigma-Aldrich, Missouri, USA) following the manufacturer’s instructions. The extracted RNA was treated with the DNA free DNase treatment and removal kit (Thermo Fisher scientific, Massachusetts, USA) to remove residual genomic DNA and then used as a template for cDNA synthesis using SuperScript III First strand Synthesis System (Invitrogen, Carlsbad, USA).
2.5. Sequencing of cDNA encoding the partial tammar DNMT3L
A BLAST search of the new tammar genome database (Wallabase 4.0 O’Neill, Fujiyama, Heider, Renfree, Pask et al. personal communication) was performed using the mouse Dnmt3l sequence and then a putative DNMT3L gene was identified. Tammar DNMT3L specific primers were made on the basis of the putative sequence (Table 2). PCR reactions were performed using GoTaq Green master mix (Promega Corporation, USA). PCR products were purified using QIAquick PCR purification kit (QIAGEN, Hilden, Germany) and directly sequenced (Micromon Sequencing Facility, Monash University, Clayton) using the PCR primers.
Table 2.
Primers used for this study
| Primer | Sequence | Tm (°C) |
|---|---|---|
| For sequencing | ||
| Forward | 5ʹ-GATGGGTATCGTCCTCTTTG-3ʹ | 52.5 |
| Reverse | 5ʹ-CCTTAGGCTTTGCGTATTGT-3ʹ | 53.0 |
| For quantitative RT-PCR | ||
| DNMT3L | ||
| Forward | 5ʹ-ATGGACTACTGAAGAAGAGGAG-3ʹ | 53.2 |
| Reverse | 5ʹ-AGGGACAGAACTTGGATTGG-3ʹ | 54.6 |
| TBP | ||
| Forward | 5ʹ-GGACAAACTGAAGCAAAGGGACC-3ʹ | 58.5 |
| Reverse | 5ʹ-AGGGCATCATTGGGCTAAAGATAG-3ʹ | 56.8 |
| HMBS | ||
| Forward | 5ʹ-ACCTGACTGGAGGAGTATGGAGT-3ʹ | 58.8 |
| Reverse | 5ʹ-TGGGCTAAGATGTTGACGGTTGT-3ʹ | 58.7 |
2.6. Bioinformatics analysis
DNA sequences of various mammalian DNMT3s (koala, grey short-tailed opossum, Tasmanian devil, mouse and human) and human DNMT2 were obtained by BLAST (https://blast.ncbi.nlm.nih.gov/Blast.cgi). Amino acid sequences retrieved from DDBJ/EMBL/GenBank/RefSeq database were used to generate a phylogenetic tree. The alignment of the translated sequences was performed by Muscle with MEGA7 (http://www.megasoftware.net/). Genetic distances were calculated using the Poisson model. A phylogenetic tree of the DNMT3 family members was produced with MEGA7 using the Neighbour joining method. Accession numbers were: Phascolarctos cinereus predicted DNMT3L, RefSeq Accession XM_020995934.1; Monodelphis domestica predicted DNMT3L, RefSeq Accession XM_007493298.2; Sarcophilus harrisii predicted DNMT3L, RefSeq Accession XM_012544408.2; Mus musculus Dnmt3a, AF068625; M. musculus Dnmt3b, BC083147; M. musculus Dnmt3l, BC083147; Homo sapiens DNMT3A, AF331856; H. sapiens DNMT3B, AF176228; H. sapiens DNMT3L, BC002560 and H. sapiens DNMT2, AF045888.
2.7. Reverse-transcription quantitative RT-PCR
Reverse-transcription quantitative RT-PCR (RT-qPCR) primers (Table 2) were designed from the known tammar partial sequence and amplified a 133 base pairs product. RT-qPCR was performed on a MX3000P (STRATAGENE, California, USA) on gonadal cDNA of both sexes from day 10 pp to adult using the SYBR Green PCR kit (QIAGEN, Hilden, Germany). TATA-box binding protein (TBP) and hydroxymethylbilane synthase (HMBS) were selected as the housekeeping genes. The DNMT3L gene expression levels were normalized to the geometric mean of the expression levels of housekeeping genes. Differences in the gene expression levels between stages were analysed by Tukey–Kramer’s honestly significant difference (HSD) test after ANOVA in R as described above.
3. Results
3.1. Genome-wide reprogramming of DNA methylation during spermatogenesis
Male germ cells in developing testes migrated from the centre to the periphery of tubules, while Sertoli cells were always located around the periphery of tubules. In H&E and DAPI staining, nuclei of Sertoli cells had a more intense and darker staining than that of germ cells. The nuclei of germ cells were large and ovoid. The nuclei of Sertoli cells were positive for 5 mC at all developmental stages. At day 10 postpartum (pp), the intensity of 5 mC staining in germ cells was almost identical with Sertoli cells (Fig. 1A). At day 25 pp, the intensity of 5 mC in germ cells decreased so that some of germ cells were negative for 5 mC (Fig. 1B). At day 50 pp, all germ cells were stained and the intensity of 5 mC in the germ cells had increased (Fig. 1C). By day 70 pp, germ cell staining was similar to that seen at day 50 pp (Fig. 1D). From day 100 pp onwards, approx. 54% of germ cells were intensely stained (Fig. 1E). The average number of intensely stained germ cells increased by 34% at around day 200 pp (Fig. 1F). In adults, all germ cells and Sertoli cells were clearly stained by anti-5 mC antibody, especially the nuclei of spermatozoa (Fig. 1G and H).
Figure 1.
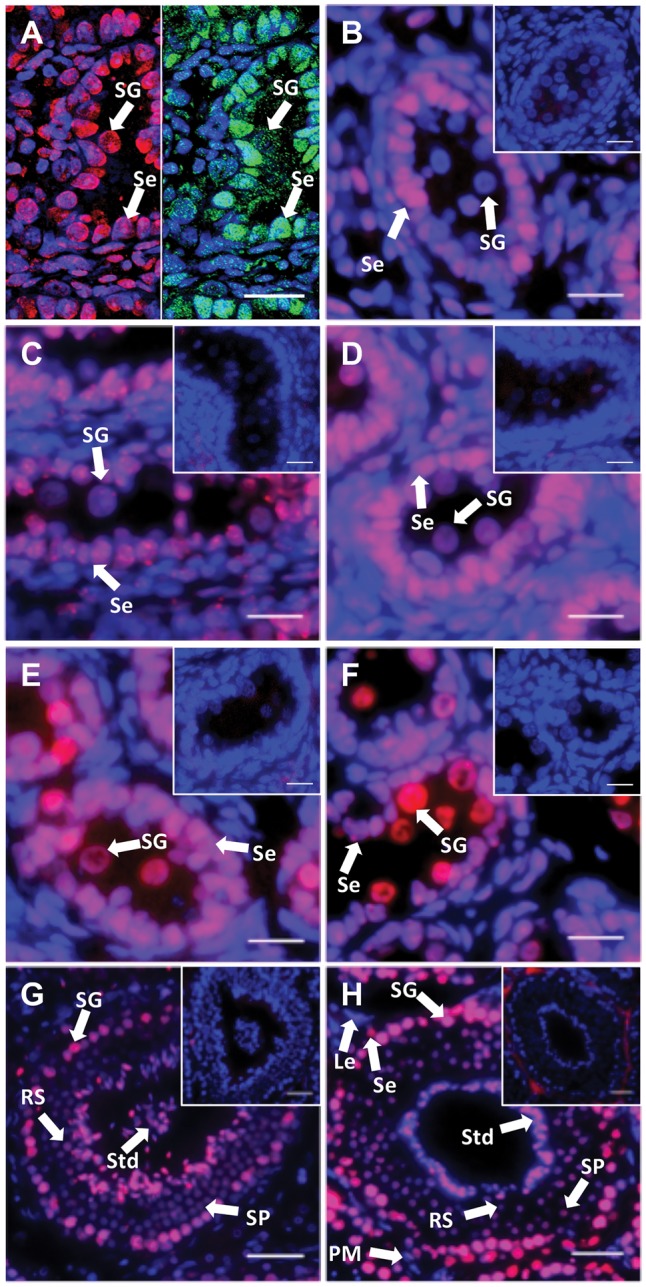
5 mC localization during spermatogenesis. DNA methylation was investigated by 5 mC immunohistochemical staining. DAPI and 5 mC stain nuclei blue and red, respectively (grey and white in black and white (BW) respectively). (A) Day 10 pp. Left-hand side: 5 mC staining (red); right-hand side SOX9 staining (green). (B) Day 25 pp. (C) Day 50 pp. (D) Day 70 pp. (E) Day 100 pp. (F) Day 200 pp. (G) and (H) Adult. All insets are negative controls. (A) Scale bar 8 μm. (B–F) Scale bars 20 μm. (G and H) Scale bars 50 μm. Abbreviations: Le, Leydig cells; PM, peritubular myoid cells; RS, Round spermatids; Se, Sertoli cells; SG, spermatogonia; SP, spermatocyte; Std, spermatid.
3.2. Genome-wide reprogramming of DNA methylation during oogenesis
Female germ cells had large and ovoid nuclei. From day 10 pp to day 50 pp, germ cells formed germ cell nests. Somatic cell nuclei stained darker with both H&E and DAPI. DNA methylation of developing ovaries were also tested by IF with the 5 mC specific antibody. Around day 10 pp, nuclei of oogonia were more intensely stained by 5 mC antibody than those of pre-granulosa cells (Fig. 2A). From day 25 pp to day 50 pp, intensities of 5 mC staining in germ cells were similar to each other (Fig. 2B and C). At day 70 pp, several primordial follicles were observed and nuclei of oogonia were still intensely stained at this stage (Fig. 2D). Around day 120 pp, intensities of 5 mC staining in germ cells were weaker than in pre-granulosa and granulosa cells (Fig. 2E). At this stage, there was a larger number of primary follicles. Around day 200 pp, nuclei of oocytes were intensely stained by 5 mC antibody (Fig. 2F). The intensity of 5 mC in oocyte nuclei increased between day 120 and day 200 pp, and granulosa cells were also stained. In adults, there were some secondary follicles and tertiary follicles and 5 mC staining in germ cells was intense (Fig. 2G and H).
Figure 2.
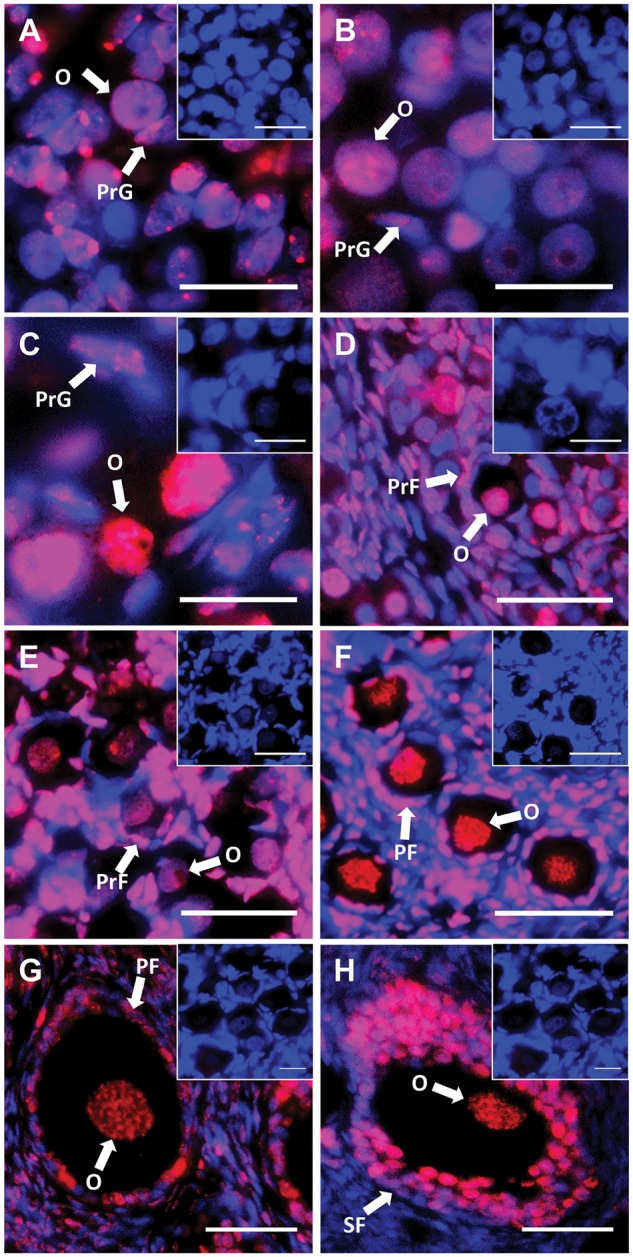
5mC localization during oogenesis. DNA methylation was investigated by 5 mC immunohistochemical staining. DAPI and 5 mC stain nuclei blue (grey in BW) and red (white in BW), respectively. (A) Day 10 pp. (B) Day 25 pp. (C) Day 50 pp. (D) Day 70 pp. (E) Day 100 pp. (F) Day 200 pp. (G) and (H) Adult. All insets are negative controls. (A–C) Scale bars 20 μm. (D–H) Scale bars 50 μm. Abbreviations: O, oogonia; PrF, primordial follicles; PrG, pre-granulosa cells; PF, primary follicles; SF, secondary follicles.
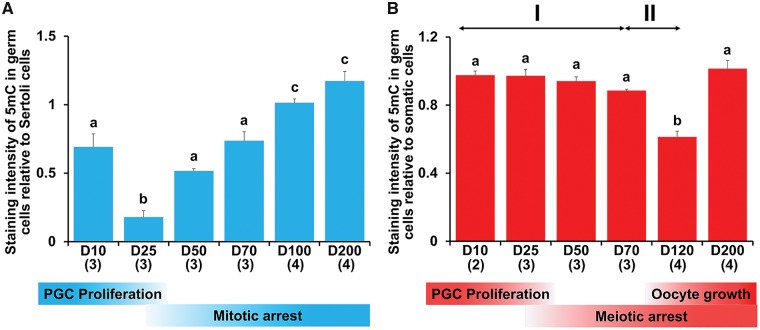
3.3. Quantitation of DNA methylation levels during gametogenesis
Regardless of sex, levels of 5 mC in late-stage germ cells were relatively high. In males during testicular differentiation, 5 mC in germ cells significantly decreased between day 10 pp and day 25 pp (P < 0.05) and then gradually increased to day 200 pp (Fig. 3A). In females during ovarian differentiation, levels of 5 mC in germ cells were constant until day 25 pp and then slowly demethylated. At that time, levels of 5 mC in somatic cells decreased slightly and there was no significant difference between stages (Supplementary Fig. S1A). Although there was no significant difference in levels of 5 mC in germ cells between day 10 pp and day 70 pp, there was significant demethylation (P < 0.05) by linear regression analysis based on the ages of the individual PYs (Supplementary Fig. S2). The speed of demethylation was effectively increased in female germ cells after day 70 pp. Levels of 5mC significantly decreased between day 70 pp and day 120 (P < 0.05). Compared with day 25 pp, methylation levels in germ cells had decreased by ∼36% between day 70 pp and day 120 pp. Based on the two different demethylation rates, we divided DNA demethylation into two phases, I and II. After day 120 pp when oogenesis and follicular formation was beginning, levels of 5 mC in germ cells then increased by day 200 pp (Fig. 3B). At these stages, levels of 5 mC in somatic cells were almost identical (Supplementary Fig. S1B).
Figure 3.
Quantitation of 5mC during gametogenesis. DNA methylation levels were estimated from 5 mC staining of germ cells versus adjacent somatic cells using ImageJ. (A) Male germ cells. (B) Female germ cells. Demethylation in female germ cells appears to be biphasic (see text). Mean + SE are shown, and sample numbers of each stage are represented as the number in parentheses. Bars labelled with same letters represent values that do not differ significantly by Tukey–Kramer’s multiple comparison test at P < 0.05. The corresponding developmental stages of male and female tammar germ cells are shown at the bottom.
3.4. Isolation and sequencing of the partial tammar DNMT3L
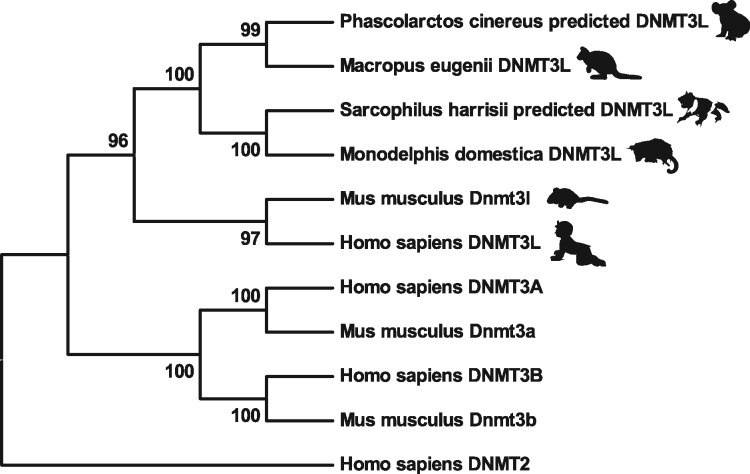
Overall 1245 bp of full tammar DNMT3L candidate sequence was identified from BLAST. The tammar DNMT3L appears to be highly conserved with that of other mammals. Although we could not amplify the full coding sequence of the predicted tammar DNMT3L, a partial sequence was obtained by PCR with DNMT3L primers described in Table 2. The partial nucleotide sequence was identical with our predicted tammar DNMT3L, and the amino acid sequence included partial ATRX-DNMT3-DNMT3L (ADD) domain. The amino acid sequence of predicted full tammar DNMT3L was generated (Supplementary Fig. S3), and it was 60% identical with Homo sapiens DNMT3L. Tammar DNMT3L lacks the consensus residues which are necessary for the formation of the catalytic centre of cytosine methyltransferases, similar to mouse Dnmt3l. Amino acid sequences were aligned against several DNMT3s including both human and mouse DNMT3 family members to produce a phylogenetic tree of the family members (Fig. 4). All the marsupial DNMT3Ls clustered into one branch, clearly distinct from the eutherian species such as mouse and human. Within the marsupials, the South American didelphid Monodelphis grouped with the Australian dasyurid Sarcophilus. The dasyurids are thought to be close to the ancestral Australian marsupials, from which the diprotodont species (tammar and koala, Phascolarctos) derived. The tammar DNMT3L clustered together with that of another marsupial, the koala, Phascolarctos cinereus predicted DNMT3L. DNMT3A, DNMT3B and DNMT2 all occupy separate branches of this tree.
Figure 4.
Sequencing and phylogenetic analysis of DNMT3L. Phylogenetic tree analysis of mammalian DNMTs. The values represented at the branch nodes are percentage bootstrap values (10,000 replicates), and the length of branches are proportional to the number of amino acid substitutions. All the marsupial DNMT3Ls clustered into one branch and DNMT3A, DNMT3B and DNMT2 all occupy separate branches of this tree.
3.5. Gene expression patterns of tammar DNMT3L during gametogenesis
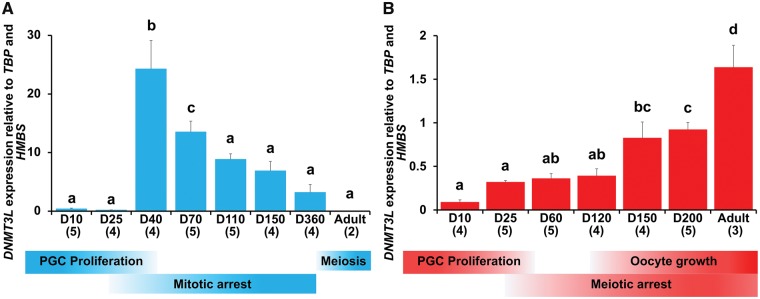
During tammar testicular differentiation, DNMT3L gene expression was significantly up-regulated at day 40 pp (P < 0.05) and gradually down-regulated from 40 pp to adult. In adults, DNMT3L expression was low (Fig. 5A). In contrast, during ovarian differentiation DNMT3L gene expression was gradually up-regulated from day 25 pp to day 120 pp and then rapidly up-regulated from day 120 pp to adult (Fig. 5B).
Figure 5.
Profiling of tammar DNMT3L expression during gametogenesis. Tammar DNMT3L expression in (A) Developing testes and (B) developing ovaries based on RT-qPCR of DNMT3L gene expression. Mean + SE are shown and the numbers in parentheses indicate the sample numbers at each developmental stage. Bars labelled with the same letters represent values that do not differ significantly by Tukey–Kramer’s multiple comparison test at P < 0.05. The corresponding developmental stages of male and female tammar germ cells are shown at the bottom.
3.6. Immunolocalization of tammar DNMT3L
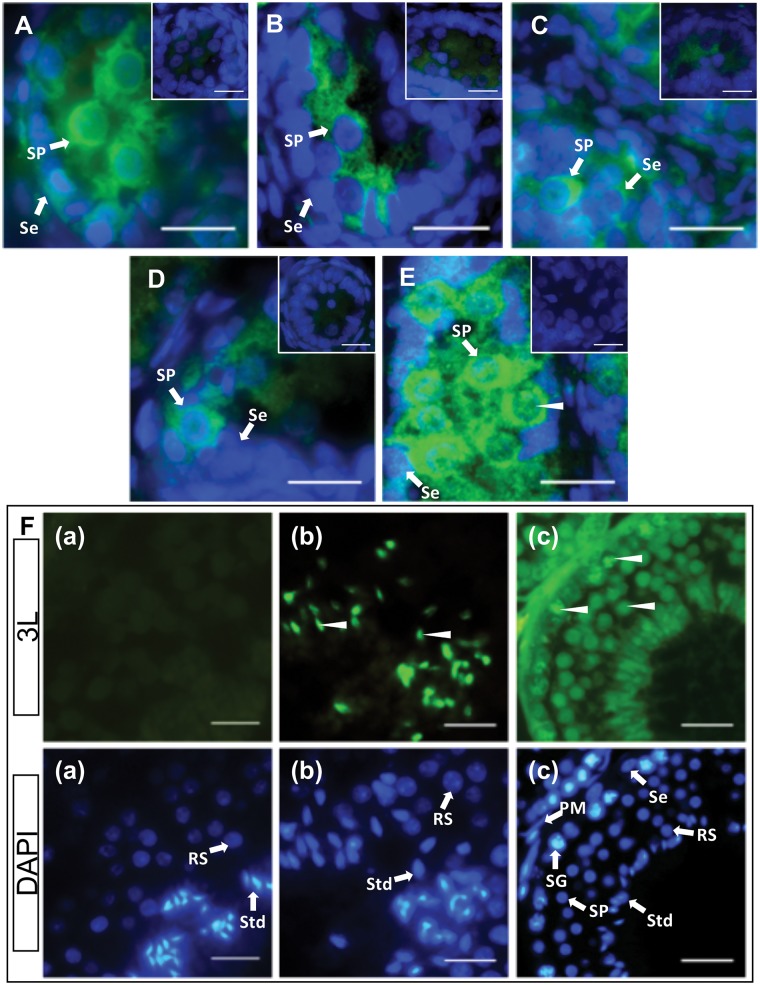
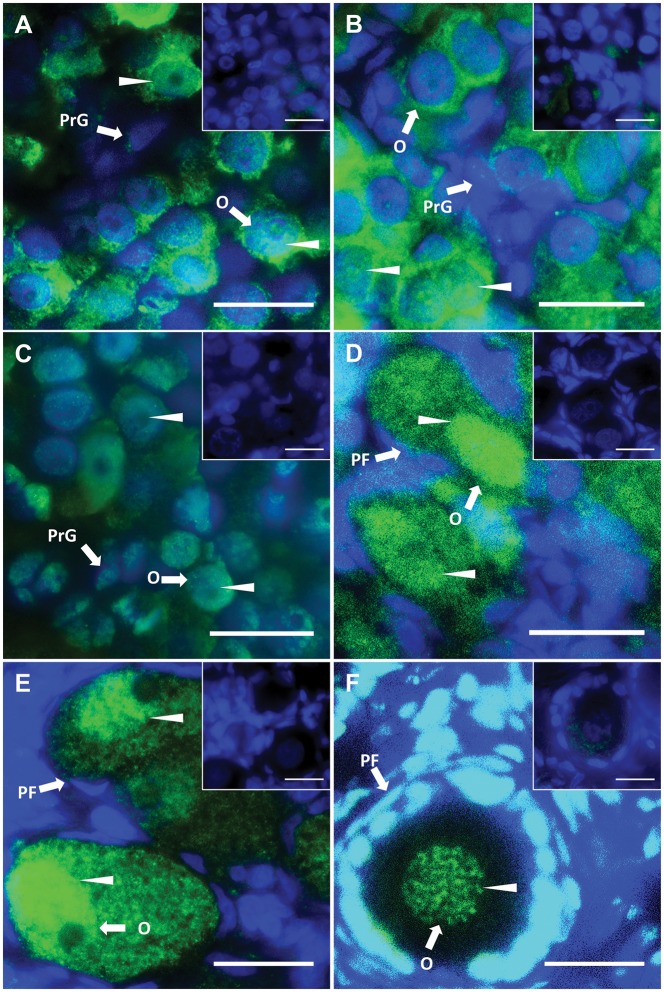
A single band was detected between 37 kDa and 50 kDa of molecular weight marker in western blotting. Thus, the predicted size of full length tammar DNMT3L is approx. 45 kDa, similar to eutherian DNMT3L. During male testicular differentiation, DNMT3L was predominantly cytoplasmic in male germ cells (Fig. 6A–D). By day 200 pp, weak DNMT3L staining was detected in the nuclei of germ cells (Fig. 6E). Interestingly, DNMT3L localized in the nuclei of germ cells and heads of spermatozoa in the adult testis (Fig. 6F). Nuclei of female germ cells stained more intensively than those of males. In the early stages, up to day 50 pp, DNMT3L was predominantly cytoplasmic in female germ cells and nuclear staining was detected in a few germ cells (Fig. 7A and B). By day 70 pp, DNMT3L staining was now predominant in the nuclei of germ cells (Fig. 7C). From day around 100 pp, DNMT3L staining in the nuclei was quite intense (Fig. 7D–F).
Figure 6.
DNMT3L localization during spermatogenesis. Immunofluorescent localization of tammar DNMT3L protein in developing and adult testes. DAPI stains nuclei blue (grey in BW) and DNMT3L is stained green (white in BW). Arrow heads represent nuclear localization. (A) Day 25 pp. (B) Day 50 pp. (C) Day 70 pp. (D) Day 100 pp. (E) Day 200 pp. (F) Adult. All insets and F(a) are negative controls. Scale bars 20 μm. Abbreviations: PM, peritubular myoid cells; RS, Round spermatids; Se, Sertoli cells; SG, spermatogonia; SP, spermatocyte; Std, spermatid.
Figure 7.
DNMT3L localization during oogenesis. Immunofluorescent localization of tammar DNMT3L protein in developing and adult ovaries. DAPI stains nuclei blue (grey in BW) and DNMT3L is stained green (white in BW). Arrow heads represent nuclear localization. (A) Day 25 pp. (B) Day 50 pp. (C) Day 70 pp. (D) Day 110 pp. (E) Day 200 pp. (F) Adult. All insets are negative controls. Scale bars 20 μm. Abbreviations: O, oogonia; PrG, pre-granulosa cells; PF, primary follicles.
4. Discussion
Global DNA methylation levels in the developing germ cells in the developing tammar changed dynamically during the extended period of gametogenesis in this marsupial. In addition, the timing of methylation events differed between males and females. This correlated with sex-specific patterns for both mRNA expression and protein localization of the key DNA-methyltransferase DNMT3L. This is the first study to report the expression patterns of the DNMT3L gene in the developing gonads of any marsupial and suggests that DNA methylation-reprogramming in tammar is mediated by processes that have been highly conserved for at least 160 million years. The phylogenetic tree also suggests that the marsupial and eutherian genes had a common ancestor at the time of divergence of the therian mammals.
In the tammar, most primordial germ cells (PGCs) migrate to the gonadal ridge just before birth.25 Female germ cells do not begin to enter meiosis until 25 days postpartum (pp) and all have not entered meiotic arrest until around day 70 pp. Similarly, male germ cells do not enter mitotic arrest until 25 days pp and this process is also not complete until after day 70 pp.25,32 At day 10 and day 25 pp, 5 mC expression levels were similar in female germ cells, but it rapidly decreased in male germ cells between day 10 and day 25 pp. Rapid PGC proliferation occurs in both sexes at this stage24 so it is possible that DNA replication-dependent passive demethylation was operative at this time in males but not in females. During mouse spermatogenesis, genome-wide demethylation occurs during PGC migration and is complete before mitotic arrest.33 Tammar male germ cells also showed genome-wide demethylation before mitotic arrest, indicating that the relative timing of demethylation during spermatogenesis is conserved between the mouse and tammar. DNA methylation levels in male germ cells rapidly increased from day 25 pp to day 200 pp. Male germ cells enter mitotic arrest from day 25 pp.32 During mouse spermatogenesis, DNA re-methylation begins during mitotic arrest,33 similar to the tammar. Before oocyte growth, mouse female germ cells have very low levels of DNA methylation.34 In the female tammar, rapid loss of DNA methylation was detected between day 70 pp and day 120 pp, before oocyte growth and ovarian maturation, when the methylation level was significantly lower than other stages. Thereafter, DNA methylation levels rapidly increased between day 120 pp and day 200 pp. During tammar oogenesis, rapid oocyte growth and follicle expansion begins around day 120 and continues to day 250 pp.35 In mice, re-establishing DNA methylation also occurs during oocyte growth.34,36 Thus, the relative timing of completion of demethylation and initiation of re-methylation during oogenesis is also conserved between the tammar and mouse, but the duration is greatly extended in the tammar. The genome-wide changes in DNA methylation occurs during post-natal stages of tammar, while it occurs during the very short prenatal stages of mouse, but the patterns and relative timing of demethylation and re-methylation are identical as described above.
During mouse gametogenesis, demethylation of the H19 imprinted gene locus is mediated by both active and passive mechanisms and active demethylation is necessary for the completion of imprint erasure.37 Due to the rapid development of mice, the genome-wide demethylation process is complete within a few days, making it difficult to observe the active and passive mechanisms separately. In contrast, the timing of demethylation in female germ cells is extended in the tammar. A significant but small linear decrease in 5 mC was detected between day 25 pp and day 70 pp in the female tammar. At that time, somatic 5 mC levels also decreased slightly, but there was no significant difference between the developmental stages. This data coincided with previous reports about DNA methylation of gonadal somatic cells in developing ovaries in mouse and human.38,39 Between day 70 pp and day 120 pp, when almost all germ cells have entered meiotic arrest, there was a significant demethylation. The two different demethylations observed before and after day 70 pp suggest that demethylation in female tammar germ cells occurs in two phases, I and II. The first demethylation phase occurred during germ cell proliferation and onset of meiosis when this process may be mediated by DNA replication-dependent passive demethylation. The second demethylation phase occurred after the germ cells were in meiotic arrest and before oogenesis, suggesting this process may be mediated by DNA replication-independent active demethylation mechanisms. In mice, there is an enzyme-linked active demethylation mechanism during gametogenesis,40–42 and the putative poly (ADP-ribose) polymerase (PARP) pathway may act as a candidate mechanism through transcriptional regulation and/or histone modification.37,43 We hypothesize that two demethylation phases in tammar reflects a passive phase followed by an active phase, possibly controlled by a PARP pathway.
Mouse Dnmt3l is clearly involved in the re-establishment of DNA methylation patterns in the germ cells.13,15,44 However, expression patterns of other DNMT3 family members such as Dnmt3a and Dnmt3b are not correlated with methylation dynamics or germline development in mice45 so it is difficult to compare the relative timing between mouse and tammar based on developmental events. Thus, from expression studies observed here, it is likely that tammar DNMT3L also has a critical role for re-establishing DNA methylation. During spermatogenesis, tammar DNMT3L was highly expressed around day 40 pp, at which time a global re-methylation occurred in male germ cells and then expression levels were gradually decreased. Similarly, mouse and rat Dnmt3l is up-regulated at the time of re-methylation and then expression levels are gradually decreased during spermatogenesis.44,46 In contrast, during tammar oogenesis, both genome-wide DNA re-methylation and up-regulation of tammar DNMT3L expression in female germ cells coincided with the time of rapid oocyte growth that occurs from day 12035 in a similar way to that in the mouse.47 In the mouse, these sex-specific expression patterns are regulated by sex-specific promoters.48 Since the expression pattern of tammar DNMT3L was similar to that of the mouse, it is likely that sex-specific promoters may be conserved between the mouse and tammar. Analysis of promoter regions of tammar DNMT3L would be interesting to discover whether the regulatory mechanisms have also been conserved though mouse Dnmt3l has some sex-specific isoforms.48
Several studies report that mouse Dnmt3l localizes in the nuclei of germ cells in the perinatal testes,49,50 but tammar DNMT3L was detected predominantly as a cytoplasmic protein until the later stages of pouch life in the tammar developing testis, consistent with the greatly extended duration of post-natal development compared to that of the mouse. DNMT3L was also strongly localized in the heads of spermatozoa in the adult. During early ovarian differentiation, DNMT3L protein was predominantly localized in the cytoplasm of germ cells and its nuclear localization was detected only in a few germ cells up to day 50 pp. After day 50 pp, DNMT3L became predominantly localized in the nuclei of female germ cells. During mouse oogenesis, Dnmt3l is essential for the establishment of maternal imprints and acts as an accessary protein of Dnmt3a to recruit target gene loci15,16 presumably from a nuclear location. The gene expression patterns of tammar DNMT3L corresponded to the time of re-methylation during oogenesis, and significantly the protein in germ cells was strongly nuclear at the time of re-methylation. These results suggest that tammar DNMT3L may act in the establishment of maternal imprints during oogenesis as in other mammals.
Although the expression patterns of tammar DNMT3L in male germ cells corresponded with the timing of re-methylation, tammar DNMT3L was predominantly cytoplasmic in male germ cells. This raises the possibility that not only tammar DNMT3L but also other molecular machineries are necessary for re-establishing methylation in developing testes. Although mouse Dnmt3l is required for the establishment of normal paternal methylation imprints during spermatogenesis, there is some methylation present in Dnmt3l deficient male germ cells but not in female germ cells.51 Therefore, a DNMT3L-independent methylation pathway may exist in male germ cells during spermatogenesis. The present results support the idea that the DNMT3L-independent mechanism may act in re-methylation processes during spermatogenesis. Interestingly, DNMT3L changed its localization and became nuclear in adult germ cells, suggesting it might regulate methylation in later stages of spermatogenesis. Dnmt3l deficient male mice have meiotic ‘catastrophes’ during which there is reactivation of retrotransposons in the male germ cells.13 In addition, mouse Dnmt3l establishes DNA methylation not only at paternally imprinted gene loci but also at non-pericentric heterochromatic sequences and interspersed repeats, and it affects chromatic compaction in later stage testes.51 In Dnmt3l deficient adult mice, there is a loss of chromatin compaction which leads to apoptosis.51,52 Since Dnmt3l transcripts are almost absent in adults, it was assumed that there is no Dnmt3l protein in adults.13 Although the expression levels of tammar DNMT3L were very low, similar to that of mouse Dnmt3l, tammar DNMT3L was detected in the nuclei of germ cells in adults. Therefore, it is possible that tammar DNMT3L has an un-documented function which may be required for the chromatin condensation in differentiating male germ cells.
Our findings clearly show that the basic mechanisms of epigenetic reprogramming must have been established before the marsupial-eutherian split over 160 Mya and open new perspectives for identifying a novel active demethylation mechanism in female germ-line. In addition to these, our data point towards a major role of a key enzyme, DNMT3L, in the process of epigenetic reprogramming and gametogenesis with respect to its protein localization. Since epigenetic reprogramming in the tammar occurs over such an extended period of time and whilst the young are accessible in the mother’s pouch, further study of the marsupial will allow for more precise manipulation of the processes to elucidate the mechanisms of control.
Supplementary Material
Acknowledgements
The authors thank Chris Lucas for assistance with the animals, all members of the wallaby research group for help with the animals and the dissections, and Dr. Stephen Frankenberg for assistance with the analysis of the tammar DNMT3L sequence.
Funding
This study was supported by funds from the Norma Hilda Schuster (nee Swift) Scholarship from the University of Melbourne to T.I. and the School of BioSciences, The University of Melbourne.
Conflict of interest
None declared.
References
- 1. Ferguson-Smith A.C. 2011, Genomic imprinting: the emergence of an epigenetic paradigm, Nat. Rev. Genet., 12, 565–75. [DOI] [PubMed] [Google Scholar]
- 2. Reik W., Walter J.. 2001, Genomic imprinting: parental influence on the genome, Nat. Rev. Genet., 2, 21–32. [DOI] [PubMed] [Google Scholar]
- 3. Reik W., Walter J.. 2001, Evolution of imprinting mechanisms: the battle of the sexes begins in the zygote, Nat. Genet., 27, 255–6. [DOI] [PubMed] [Google Scholar]
- 4. Khavari D.A., Sen G.L., Rinn J.L.. 2010, DNA methylation and epigenetic control of cellular differentiation, Cell Cycle, 9, 3880–3. [DOI] [PubMed] [Google Scholar]
- 5. Renfree M.B., Hore T.A., Shaw G., Graves J.A.M., Pask A.J.. 2009, Evolution of genomic imprinting: insights from marsupials and monotremes, Annu. Rev. Genomics Hum. Genet., 10, 241–62. [DOI] [PubMed] [Google Scholar]
- 6. Pask A.J., Papenfuss A.T., Ager E.I., McColl K.A., Speed T.P., Renfree M.B.. 2009, Analysis of the platypus genome suggests a transposon origin for mammalian imprinting, Genome Biol., 10, R1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bartolomei M.S., Ferguson-Smith A.C.. 2011, Mammalian genomic imprinting, Cold Spring Harb. Perspect. Biol, 3, a002592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Vasilyev S.A., Tolmacheva E.N., Lebedev I.N.. 2016, Epigenetic regulation and role of LINE-1 retrotransposon in embryogenesis. Russ. J. Genet., 52, 1219–26. [Google Scholar]
- 9. Sadakierska-Chudy A., Kostrzewa R.M., Filip M.. 2015, A comprehensive view of the epigenetic landscape part I: dNA methylation, passive and active DNA demethylation pathways and histone variants, Neurotox. Res., 27, 84–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Auclair G., Weber M.. 2012, Mechanisms of DNA methylation and demethylation in mammals, Biochimie, 94, 2202–11. [DOI] [PubMed] [Google Scholar]
- 11. Saitou M., Kagiwada S., Kurimoto K.. 2012, Epigenetic reprogramming in mouse pre-implantation development and primordial germ cells, Development, 139, 15–31. [DOI] [PubMed] [Google Scholar]
- 12. Yokomine T., Hata K., Tsudzuki M., Sasaki H.. 2006, Evolution of the vertebrate DNMT3 gene family: a possible link between existence of DNMT3L and genomic imprinting, Cytogenet. Genome Res., 113, 75–80. [DOI] [PubMed] [Google Scholar]
- 13. Bourc’his D., Bestor T.H.. 2004, Meiotic catastrophe and retrotransposon reactivation in male germ cells lacking Dnmt3L, Nature, 431, 96–9. [DOI] [PubMed] [Google Scholar]
- 14. Kato Y., Kaneda M., Hata K., et al. 2007, Role of the Dnmt3 family in de novo methylation of imprinted and repetitive sequences during male germ cell development in the mouse, Hum. Mol. Genet., 16, 2272–80. [DOI] [PubMed] [Google Scholar]
- 15. Bourc’his D., Xu G.L., Lin C.S., Bollman B., Bestor T.H.. 2001, Dnmt3L and the establishment of maternal genomic imprints, Science, 294, 2536–9. [DOI] [PubMed] [Google Scholar]
- 16. Hata K., Okano M., Lei H., Li E.. 2002, Dnmt3L cooperates with the Dnmt3 family of de novo DNA methyltransferases to establish maternal imprints in mice, Development, 129, 1983–93. [DOI] [PubMed] [Google Scholar]
- 17. Margot J.B., Ehrenhofer-Murray A.E., Leonhardt H.. 2003, Interactions within the mammalian DNA methyltransferase family, BMC Mol. Biol., 4, 7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Xu G.L., Wong J.. 2015, Oxidative DNA demethylation mediated by Tet enzymes, Nat. Sci. Rev., 2, 318–28. [Google Scholar]
- 19. Wossidlo M., Nakamura T., Lepikhov K., et al. 2011, 5-Hydroxymethylcytosine in the mammalian zygote is linked with epigenetic reprogramming, Nat. Commun., 2, 241. [DOI] [PubMed] [Google Scholar]
- 20. Surani M.A. 1998, Imprinting and the initiation of gene silencing in the germ line, Cell, 93, 309–12. [DOI] [PubMed] [Google Scholar]
- 21. Cheng Y., Xie N., Jin P., Wang T.. 2015, DNA methylation and hydroxymethylation in stem cells, Cell Biochem. Funct., 33, 161–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhou L., Dean J.. 2015, Reprogramming the genome to totipotency in mouse embryos, Trends Cell Biol., 25, 82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hsu F.-M., Clark A., Chen P.-Y.. 2015, Epigenetic reprogramming in the mammalian germline, Oncotarget, 6, 35151–2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Alcorn G. T., Robinson E. S.. 1983, Germ cell development in female pouch young of the tammar wallaby (Macropus eugenii), J. Reprod. Fertil., 67, 319–25. [DOI] [PubMed] [Google Scholar]
- 25. Ullmann S.L., Shaw G., Alcorn G.T., Renfree M.B.. 1997, Migration of primordial germ cells to the developing gonadal ridges in the tammar wallaby Macropus eugenii, J. Reprod. Fertil., 110, 135–43. [DOI] [PubMed] [Google Scholar]
- 26. Suzuki S., Shaw G., Renfree M.B.. 2013, Postnatal epigenetic reprogramming in the germline of a marsupial, the tammar wallaby, Epigenet. Chromatin., 6, 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Poole W.E., Simms N.G., Wood J.T., Lubulwa M.. 1991, Tables for Age Determination of the Kangaroo Island Wallaby (Tammar) Macropus eugenii. CSIRO (Wildlife and Ecology) Technical memorandum 32 Lyneham ACT 2600 Australia [Google Scholar]
- 28. Renfree M. 2002, Hypothermic anaesthesia of early postnatal marsupial pouch young, ANZCCART, 15, 125–7. [Google Scholar]
- 29. Renfree M.B., Fenelon J., Wijiyanti G., Wilson J.D., Shaw G.. 2009, Wolffian duct differentiation by physiological concentrations of androgen delivered systemically, Dev. Biol., 334, 429–36. [DOI] [PubMed] [Google Scholar]
- 30. Hickford D.E., Frankenberg S., Pask A.J., Shaw G., Renfree M.B.. 2011, DDX4 (VASA) is conserved in germ cell development in marsupials and monotremes, Biol. Reprod., 85, 733–43. [DOI] [PubMed] [Google Scholar]
- 31. Jiang F.X., Clark J., Renfree M.B.. 1997, Ultrastructural characteristics of primordial germ cells and their amoeboid movement to the gonadal ridges in the tammar wallaby, Anat. Embryol. (Berl)., 195, 473–81. [DOI] [PubMed] [Google Scholar]
- 32. Renfree M.B., Shaw G.. 2001, Germ cells, gonads and sex reversal in marsupials, Int. J. Dev. Biol., 45, 557–67. [PubMed] [Google Scholar]
- 33. Sasaki H., Matsui Y.. 2008, Epigenetic events in mammalian germ-cell development: reprogramming and beyond, Nat. Rev. Genet., 9, 129–40. [DOI] [PubMed] [Google Scholar]
- 34. Smallwood S.A., Tomizawa S.-I., Krueger F., et al. 2011, Dynamic CpG island methylation landscape in oocytes and preimplantation embryos, Nat. Genet., 43, 811–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Mattiske D., Shaw G., Shaw J.M.. 2002, Influence of donor age on development of gonadal tissue from pouch young of the tammar wallaby, Macropus eugenii, after cryopreservation and xenografting into mice, Reproduction, 123, 143–53. [DOI] [PubMed] [Google Scholar]
- 36. Tomizawa S.I., Nowacka-Woszuk J., Kelsey G.. 2012, DNA methylation establishment during oocyte growth: mechanisms and significance, Int. J. Dev. Biol., 56, 867–75. [DOI] [PubMed] [Google Scholar]
- 37. Kawasaki Y., Lee J., Matsuzawa A., Kohda T., Kaneko-Ishino T., Ishino F.. 2014, Active DNA demethylation is required for complete imprint erasure in primordial germ cells, Sci. Rep., 4, 3658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Guo H., Hu B., Yan L.. 2017, DNA methylation and chromatin accessibility profiling of mouse and human fetal germ cells, Cell Res. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Guo F., Yan L., Guo H., et al. 2015, The transcriptome and DNA methylome landscapes of human primordial germ cells, Cell, 161, 1437–52. [DOI] [PubMed] [Google Scholar]
- 40. Hill P.W.S., Amouroux R., Hajkova P.. 2014, DNA demethylation, Tet proteins and 5-hydroxymethylcytosine in epigenetic reprogramming: an emerging complex story, Genomics, 104, 324–33. [DOI] [PubMed] [Google Scholar]
- 41. Popp C., Dean W., Feng S., et al. 2010, Genome-wide erasure of DNA methylation in mouse primordial germ cells is affected by AID deficiency, Nature, 463, 1101–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Messerschmidt D.M. 2016, A twist in zygotic reprogramming, Nat. Cell Biol., 18, 139–40. [DOI] [PubMed] [Google Scholar]
- 43. Ciccarone F., Klinger F.G., Catizone A., et al. 2012, Poly(ADP-ribosyl)ation acts in the DNA demethylation of mouse primordial germ cells also with DNA damage-independent roles. PLoS One,7(10): e46927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. La Salle S., Oakes C.C., Neaga O.R., Bourc’his D., Bestor T.H., Trasler J.M.. 2007, Loss of spermatogonia and wide-spread DNA methylation defects in newborn male mice deficient in DNMT3L, BMC Dev. Biol., 7, 104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Maksakova I.A., Mager D.L., Reiss D.. 2008, Endogenous retroviruses–Keeping active endogenous retroviral-like elements in check: the epigenetic perspective, Cell. Mol. Life Sci., 65, 3329–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Xu H.X., Qin J.Z., Zhang K.Y., Zeng W.X.. 2015, Dynamic expression profile of DNA methyltransferases in rat testis development, Pol. J. Vet. Sci., 18, 549–56. [DOI] [PubMed] [Google Scholar]
- 47. Lucifero D., La Salle S., Bourc’his D., Martel J., Bestor T.H., Trasler J.M.. 2007, Coordinate regulation of DNA methyltransferase expression during oogenesis, BMC Dev. Biol., 7, 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Shovlin T.C., Bourc’his D., La Salle S., et al. 2007, Sex-specific promoters regulate Dnmt3L expression in mouse germ cells, Hum. Reprod., 22, 457–67. [DOI] [PubMed] [Google Scholar]
- 49. Sakai Y., Suetake I., Shinozaki F., Yamashina S., Tajima S.. 2004, Co-expression of de novo DNA methyltransferases Dnmt3a2 and Dnmt3L in gonocytes of mouse embryos, Gene Expr. Patterns, 5, 231–7. [DOI] [PubMed] [Google Scholar]
- 50. Nimura K., Ishida C., Koriyama H., et al. 2006, Dnmt3a2 targets endogenous Dnmt3L to ES cell chromatin and induces regional DNA methylation, Genes Cells, 11, 1225–37. [DOI] [PubMed] [Google Scholar]
- 51. Webster K.E.O’Bryan M.K.Fletcher S., et al. 2005, Meiotic and epigenetic defects in Dnmt3L-knockout mouse spermatogenesis, Proc. Natl. Acad. Sci. USA, 102, 4068–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Nicklas R.B., Ward S.C., Gorbsky G.J.. 1995, Kinetochore chemistry is sensitive to tension and may link mitotic forces to a cell cycle checkpoint, J. Cell Biol., 130, 929–39. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.