Abstract
Superoxide is a reactive oxygen species produced during aerobic metabolism in mitochondria and prokaryotes. It causes damage to lipids, proteins and DNA and is implicated in cancer, cardiovascular disease, neurodegenerative disorders and aging. As protection, cells express soluble superoxide dismutases, disproportionating superoxide to oxygen and hydrogen peroxide.
Here, we describe a membrane bound enzyme that directly oxidizes superoxide and funnels the sequestered electrons to ubiquinone in a diffusion-limited reaction. Experiments in proteoliposomes and inverted membranes show that the protein efficiently quenches superoxide generated at the membrane.
The 2.0-Å crystal structure shows an integral membrane di-heme cytochrome b poised for electron transfer from the P-side and proton uptake from the N-side. This suggests that the reaction is electrogenic and contributes to the membrane potential while also conserving energy by reducing the quinone pool.
Based on the enzymatic activity we propose that the new enzyme family is denoted superoxide oxidase (SOO).
Introduction
Molecular oxygen can undergo a series of consecutive one-electron reduction steps forming the intermediates superoxide, hydrogen peroxide and hydroxyl anion before complete reduction to water. The intermediates are collectively called reactive oxygen species (ROS). Reactive oxygen species damage various cellular components such as lipids, proteins, and DNA and are implicated in cancer, cardiovascular disease, neurodegenerative disorders and aging1–4. Superoxide is produced during aerobic metabolism in mitochondria and prokaryotes. All cells living in aerobic habitats contain soluble superoxide dismutase enzymes (SODs) disproportionating superoxide to molecular oxygen and hydrogen peroxide, which is in turn detoxified by catalases or peroxidases. SODs make up the most common biochemical defense against superoxide5,6 while in some anaerobic organisms, an enzyme reducing superoxide to hydrogen peroxide has also been identified7. Remarkably, these hydrogen peroxide-generating mechanisms remain the only identified protection against superoxide in biology.
In eukaryotic systems, superoxide is also actively produced by the immune system as a bactericide8, and an additional role as second messenger in intracellular signaling has been proposed9. A major source for ROS are the redox centers of respiratory chain enzymes that generate and maintain the electrochemical gradient during oxidative phosphorylation in mitochondria and many bacteria10,11. The quinone pool acts as the central hub for shuttling electrons between different respiratory proteins. In E. coli, for example, NDH-2 – a non-proton pumping NADH:quinone oxidoreductase – is a major contributor to both reduced quinone and superoxide11,12. In contrast to neutral oxygen, which is enriched in membranes13, superoxide is negatively charged and less soluble in the hydrophobic core of membranes14. Most superoxide is produced by flavin redox groups in the cytoplasm and quenched by SOD15, however, superoxide production at intramembranous sites has been reported for complex II and III enzymes and illuminated thylakoid membranes16–18. Moreover, release of superoxide into the periplasm via menaquinone autooxidation in the cytoplasmic membrane has also been discussed19.
In 1982, E. coli cytochrome b561 (cybB) was cloned in the Anraku laboratory20,21 and determined to encode an integral membrane protein containing two b-type hemes. In vivo experiments showed that the protein is reduced by D-lactate, suggesting that CybB is involved in the respiratory chain. It was also predicted that it may react with quinones (due to its measured midpoint potential of +20 mV)20–22. To date, the natural electron donor of CybB has not been discovered and its biological role remains unknown20. Here we show that CybB from E. coli functions as a superoxide:ubiquinone oxidoreductase, performing a previously undescribed enzymatic reaction in which superoxide is directly oxidized to molecular oxygen and the sequestered electron is funneled to ubiquinone. The protein thus functions as a superoxide oxidase (SOO). Using in vitro experiments, we unravel the electron transport process from superoxide to ubiquinone. Furthermore, we show that liposome embedded CybB efficiently reacts with superoxide generated at the membrane and reduces the amount of superoxide produced by respiring membranes. We also present the 2.0-Å crystal structure of CybB, showing an integral membrane di-heme cytochrome poised for electron transfer from the P-side to the N-side and proton uptake from the N-side. This arrangement suggests a mechanism in which superoxide quenching contributes to the generation of electrochemical potential, while avoiding generation of other reactive oxygen species like hydrogen peroxide. In contrast to SODs, CybB appears optimized for quenching superoxide produced in the membrane or at its surface.
Results
CybB is a superoxide:ubiquinone oxidoreductase
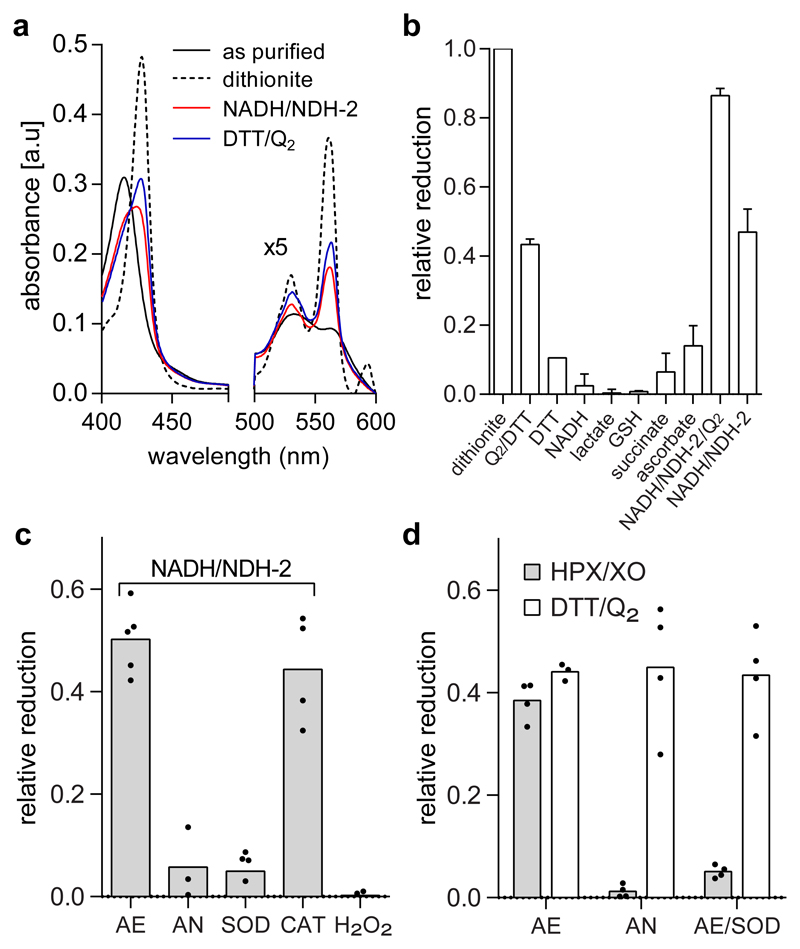
Recombinant detergent solubilized CybB displays the characteristic optical spectrum of oxidized (ferric) heme b that is fully reduced to the ferrous state by dithionite (Fig. 1a and Supplementary Fig. 1), which has been reported previously20. CybB reacted with ubiquinol Q2H2, displaying a ~50% reduction of CybB compared to dithionite (Fig. 1a+b). D-lactate alone, however, did not reduce purified CybB (as observed in vivo21). Also other cellular reductants such as NADH, succinate, glutathione, and ascorbate were only able to marginally reduce CybB after prolonged incubation (Fig. 1b). However, if NADH was combined with the the monotopic membrane protein NADH:quinone oxidoreductase NDH-223, full reduction of CybB was observed in the presence of ubiquinone Q2 (Fig. 1b and Supplementary Fig. 2). Interestingly, ~50% reduction of CybB was also observed in the absence of ubiquinone Q2, suggesting an additional, quinone independent, reduction mechanism. Kinetic analysis showed that both the quinone dependent and independent reactions are fast processes with τ1/2 < 50 ms (Supplementary Fig. 3), which is similar to the kinetics of other heme containing enzymes24. Surprisingly, the quinone independent reaction did not occur under anaerobic conditions (Figure 1c), indicating a mechanism via an intermediate oxygen-derived species. In concordance with this, the presence of superoxide dismutase (SOD) – but not catalase – abolished reduction of detergent-solubilized CybB by NADH/NDH-2. This data indicates that the superoxide anion (O2·-), but not hydrogen peroxide (H2O2) is the CybB-reducing species. This was further corroborated by the result that H2O2 was unable to reduce CybB directly (Fig. 1c). To unambiguously determine whether superoxide is indeed the CybB-reducing species, hypoxanthine (HPX) and xanthine oxidase (XO) – a prototypical superoxide producing system – was utilized. CybB was reduced under aerobic conditions (~50%), but not under anaerobic conditions or in the presence of SOD (Fig. 1d, grey bars). In contrast, CybB reduction via ubiquinol Q2H2 was not dependent on oxygen and remained unaffected by the presence of SOD, suggesting a direct electron transfer reaction (Fig.1d, open bars). In conclusion, CybB can react with both superoxide and ubiquinol, via two separate mechanisms.
Fig. 1. CybB reacts with superoxide and ubiquinol.
(a) Absorbance spectra of purified and detergent solubilized CybB (black) after reduction with dithionite (dotted), DTT/Q2 (blue), or NADH/NDH-2 (red). The spectra shown are representative of at least 3 different protein purifications with similar results. (b) Relative reduction of CybB by different electron carriers after 1 min incubation. (The average and the SD have been calculated from n≥3 biologically independent experiments). (c) Reduction of purified CybB by NADH/NDH-2 under different conditions: aerobic (AE), anaerobic (AN), AE in presence of superoxide dismutase (SOD), and AE in presence of catalase (CAT). No reduction was observed when CybB was incubated with hydrogen peroxide (H2O2). Mean values and the individual data points from n≥3 biologically independent experiments are shown. (d) Superoxide and ubiquinol reduce CybB via two different mechanisms. Comparison of CybB reduction by HPX/XO (grey bars) and DTT/Q2 (open bars) under different conditions (see 1c). Mean values and the individual data points from n≥3 biologically independent experiments are shown.
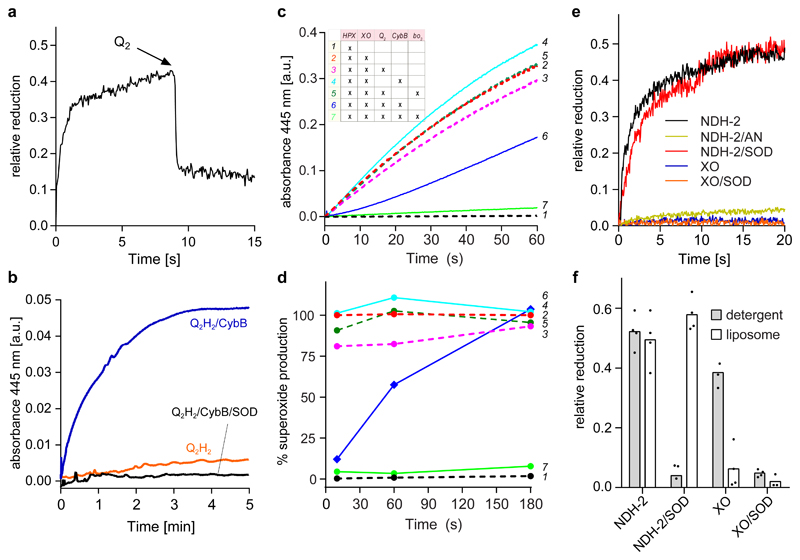
Next we investigated if CybB catalyses electron transfer between its two substrates. When superoxide-reduced CybB was mixed with an excess of ubiquinone Q2, CybB was rapidly oxidized, indicating electron transfer from CybB to ubiquinone (Fig. 2a). The reverse reaction, i.e. CybB catalysed superoxide production, was also observed when CybB was incubated with an excess of reduced ubiquinol Q2H2. This shows that electron transfer can take place in either direction, consistent with enzymatic catalysis (Fig. 2b). Considering the midpoint potentials of O2/superoxide (-160 mV)6 and ubiquinone/ubiquinol (+70 mV)25, these experiments strongly suggest that CybB is an enzyme that physiologically catalyses electron transfer from superoxide to ubiquinone, mediated by the two b hemes. This molecular mechanism was further supported by the results from multiple-turnover experiments in coupled enzyme assays, where superoxide release was monitored under various conditions (Figure 2c, inset table). The figure shows the raw data from these experiments (Fig. 2c) as well as an analysis of superoxide production rates at different time points (Fig. 2d). As expected, the HPX/XO data shows an almost linear increase in superoxide with time (Figure 2c, experiment #2), and neither separate addition of ubiquinone Q2 (experiment #3) nor purified CybB (experiment #4) affected superoxide production. However, if ubiquinone Q2 and CybB were added together (experiment #6), initial superoxide production was almost completely abolished, but increased with time reaching ~100% of the HPX/XO activity after 3 min (Fig. 2c+d). This suggests that the ubiquinone pool was fully reduced and that the electron acceptor substrate (oxidized ubiquinone) for CybB thus became rate-limiting. Indeed, when purified quinol bo3 oxidase was added in order to regenerate the quinone pool, superoxide production was completely supressed over the entire experiment (experiment #7, Fig. 2c+d), and strictly dependent on the presence of CybB (#5, Fig. 2c). The requirement for bo3 oxidase reinforces the role of ubiquinone as electron acceptor for CybB as XO does not interact directly with quinone. The coupled enzymatic assay verified the suggested biochemical reaction sequence from the pre-steady state experiments described above, and the results were consistent with a similar set of experiments using NADH/NDH-2 as the superoxide producer (Supplementary Fig. 4).
Fig. 2. CybB shows superoxide-oxidase activity in vitro.
(a) CybB was pre-reduced with HPX/XO before oxidized ubiquinone Q2 was added. The kinetic trace shown is a representative of at least 3 independent measurements. (b) Superoxide production by ubiquinol Q2H2 and CybB. The two components were mixed in the absence (blue) or in the presence (black) of SOD under aerobic conditions. Superoxide production was monitored spectrophotometrically at 445 nm following the conversion of a tetrazolium salt (WST-1) to formazan. The kinetic traces shown are representative of at least 3 independent measurements. (c) Superoxide production by HPX/XO and its suppression by CybB. See inset for the different combinations and text for further details. The kinetic traces shown are representative of at least 3 independent measurements. (d) Relative superoxide production rates at different time points during the experiments of Fig. 2c. The data are normalized to the rate of superoxide production by NADH/NDH-2 at each respective time point. (e) Membrane embedded CybB reacts preferentially with membrane derived superoxide. CybB was reconstituted into liposomes from E. coli polar lipid extract and mixed with superoxide produced either by NADH/NDH-2 or HPX/XO and either in the presence or absence of SOD under aerobic conditions. No reduction of CybB by NADH/NDH-2 was observed under anaerobic (AN) conditions. The kinetic traces shown are representative of at least 3 independent measurements. (f) Comparison of the results from Fig. 2e (open bars) with similar experiments performed with detergent solubilized CybB (grey bars). Mean values and the individual data points from n≥3 biologically independent experiments are shown.
The catalytic rate is in the diffusion-limited range
Superoxide dismutases are among the fastest enzymes known with catalytic rates in the order of 108-109 M-1s-1 26. To measure the catalytic rate of detergent solubilized CybB, we applied a modified assay described by Flint et al.27. In this assay, the amount of either SOD, cytochrome c, or CybB required to reduce the level of superoxide production by HPX/XO (assayed by WST-1) to 50% was determined by stepwise addition of the respective protein. Using the literature value for SOD (2×109 M-1s-1), rate constants for cyt c and CybB were calculated as described previously27. The results show that CybB is an extremely fast enzyme with a second order rate constant of 0.5 – 1.5×108 M-1s-1 (and thus in the diffusion-limited range) (Supplementary Table 1). In contrast, the reaction with cytochrome c was about 100 to 500 times slower. This can also be compared to other known adventitious reactions between metalloproteins and superoxide that have rate constants in the order of 104-106 6,28.
CybB reacts with membrane-proximal superoxide
CybB was also functionally characterized in an artificial membrane system. Fig. 2e shows the data from kinetic measurements in proteoliposomes and Fig. 2f shows a side-by-side comparison of experiments performed in proteoliposomes (open bars) and in detergent micelles (grey bars). Similar to the experiment in detergent micelles, reconstituted CybB mixed with NADH/NDH-2 was quickly reduced to ~50% in the presence, but not in the absence of oxygen, indicating that superoxide was the reducing agent. Strikingly, SOD did not prevent reduction of CybB by NADH/NDH-2 in membranes (proteoliposomes), whereas it completely abolished the reaction in detergent micelles (see also Fig. 1c). On the other hand, superoxide produced by HPX/XO was only able to marginally reduce membrane-reconstituted CybB, and this fractional reduction was completely abolished in the presence of SOD. The results suggest that superoxide produced in the aqueous environment (HPX/XO) cannot efficiently reach the membrane embedded CybB. We speculate that electrostatic repulsion of the anionic membrane contributes to this effect. In contrast, NDH-2 docks to the CybB containing liposomes via its C-terminal helix23 and produces superoxide at the membrane, allowing it to efficiently reach the superoxide binding site of CybB. The striking absence of an effect of SOD on the reaction between CybB and membrane proximal superoxide suggests that CybB can intercept membrane proximal superoxide before reaching the aqueous compartment.
Molecular structure of CybB
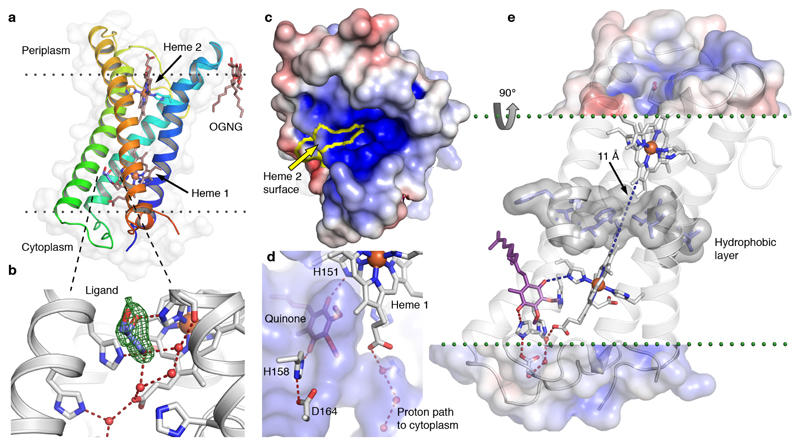
The 1.97 Å crystal structure of CybB reveals a highly distorted integral membrane four-helix bundle. The protein is well ordered and could be traced from residue 2 to 175 (out of 176) with the highest B-factors observed in the loop structures on the periplasmic side of the enzyme (the positive, P-side, of the membrane electrochemical gradient) (Supplementary Fig. 5a, Supplementary Table 2). Calculation of the positioning of CybB29 in the membrane suggests a 30 Å hydrophobic depth of insertion and a 23° insertion tilt angle. Two b-type heme groups are non-covalently bound in the core of the helical bundle, and one of the two hemes has the edge of the porphyrin ring exposed to the periplasm (Fig. 3a). The iron ions of both heme groups are octahedrally coordinated by the porphyrin nitrogen atoms and two axial histidines; originating from helix 1 and 4 for heme 1 (cytoplasmic side, N-side) and helix 2 and 4 for heme 2. Consequently, the iron ions are hexacoordinate with no free coordination positions for exogenous ligands (Supplementary Fig. 5c+d). The closest distance between the conjugated heme edges in the protein is ~11 Å; a distance that predicts electron tunneling times in the µs – ns range, which is also compatible with biochemical function30,31.
Fig. 3. High-resolution structure of CybB.
(a) 1.97 Å crystal structure of CybB colored from the N-terminus (blue) to the C-terminus (red), the protein surface is indicated (light grey). The calculated location of the hydrophobic core of the membrane is illustrated by dotted lines. (b) Binding site near the cytoplasmic heme, a glycerol molecule was modeled in the cavity based on the electron density, Fo-Fc omit map contoured at 4σ (green mesh). A chain of hydrogen bonded water molecules leads from the ligand-binding site to the cytoplasm. (c) Electrostatic potential map of the protein surface viewed from the periplasm, positive surface potential in blue, negative in red. The molecular surface contributed by the exposed heme 2 porphyrin edge, presumably acting as an electron sink, is bounded in yellow. (d) Protein surface making up the ligand binding cavity and channel to the cytoplasm in blue (viewed from inside the protein). The ubiquinone headgroup (purple) was predicted to occupy the same volume as the bound ligand with its carbonyl groups hydrogen bonded to the iron-coordinating H151 and a second completely conserved histidine (H158). (e) Proposed functional architecture of CybB. The positively charged funnel attracts superoxide to the heme edge, which serves as an electron sink and oxidizes the superoxide to molecular oxygen. The electron is subsequently tunneled to the quinone binding site (blue dashed lines). A layer of hydrophobic residues (gray surface) is located between the hemes, preventing the presence of water molecules or a proton-transfer path through the membrane. Upon quinone reduction, protons are sequestered from the cytoplasm via the indicated hydrogen-bonded path (red) to produce reduced ubiquinol.
Near the cytoplasmic heme, an exogenous ligand is hydrogen-bonded to the C-terminal metal-coordinating histidine of heme 1 (H151) and bound in a cavity delimited by helix 3, helix 4 and the heme porphyrin ring. This density is modeled as a glycerol molecule, derived from the crystallization solution, providing a good fit to the residual electron difference density (Fig. 3b).
The electrostatic potential of the protein surface on the P-side shows a pronounced positively charged funnel surrounding the exposed edge of heme 2 (Fig. 3c and Supplementary Fig. 6). This design is strikingly analogous to active site surfaces observed in all SOD classes, where the electrostatic properties attract and guide the superoxide substrate to the catalytic redox active site thereby contributing to the very high catalytic rate of SODs6,32–34.
Based on the biochemical data we hypothesize that ubiquinone binds in the membrane-exposed protein cavity near the cytosol-membrane interface. This presumption is supported by molecular docking calculations to the crystal structure (Fig. 3d). To probe the ability of the ligand-binding pocket to recognize quinones, three representative head groups were seeded into a database of 4.6 million compounds. Based on the docking energy scores, ubiquinone displayed >1000-fold enrichment over random selection (Supplementary Fig. 7 and 8).
Three histidine residues and heme 1 contribute to the quinone binding site and establish a protein channel encompassing a hydrogen-bonded solvent chain, which is potentially involved in proton uptake from the cytoplasm during quinone reduction (Fig. 3c+d). A layer of hydrophobic amino acid sidechains is located between the hemes, preventing a hydrogen-bonded proton path through the protein (Fig. 3e). The structural and electrochemical features suggest that the protein is poised to transfer superoxide-derived electrons from the P-side of the enzyme while protons would likely be sequestered from the N-side aqueous compartment. Notably, this setup would be electrogenic, contributing to the electrochemical membrane potential while reducing the quinone pool in the process (Fig. 3e).
Gene regulation of CybB
The structure and function described above suggest a potential role of CybB in superoxide detoxification under oxidative stress. We therefore probed the expression pattern of CybB under different growth conditions. Reverse transcription real-time quantitative PCR (RT-qPCR) was performed on total RNA extracted from E. coli cells sampled in logarithmic and stationary phase, under both oxic and anoxic conditions. The results show that cybB mRNA levels are ~10-20-fold higher in logarithmic phase than in stationary phase cells (Supplementary Fig. 9a) and that this pattern is not influenced by the presence or absence of oxygen. This is intriguing, as production of superoxide is expected to be significantly lower or non-existing during anoxic metabolism. However, it is known that other ROS scavenging enzymes, including E. coli FeSOD, are also produced under anaerobic conditions, possibly as a safeguard for oxygen exposure28,35.
Superoxide production of inverted membrane vesicles
Using WST-1 as indicator and NADH as electron source, we further measured the superoxide production of respiring membrane vesicles isolated from cybB-knockout and CybB-overproducing E.coli cells, as well as control cells (expressing wild-type levels of CybB). The superoxide numbers were normalized for NADH oxidation. While we could not detect a significant difference between the knockout and the control cells (Supplementary Fig. 9b), a substantial reduction of superoxide detection was observed in membranes from cells overexpressing CybB (Supplementary Fig. 9c). The absence of an effect in the former case is likely due to the experimental setup, in which the much higher concentration of the amphiphilic superoxide indicator WST-1 competes with CybB and masks the activity of the endogenously expressed CybB. Rapid scavenging of membrane produced superoxide by WST-1 is further corroborated by the observation that the presence of SOD does not reduce the detection of superoxide by WST-1 in either CybB-overexpressing or control membranes (Supplementary Fig. 9c). We note that this result and the results above also indicate the existence of a specific membrane-associated superoxide pool in slow equilibrium with soluble pools, as previously proposed for thylakoid membranes18. Such a superoxide pool could plausibly reach high effective local concentrations in this essentially two-dimensional compartment, and possibly be out of reach for globular SOD enzymes.
The CybB Protein family
Phylogenetically, E. coli CybB belongs to the NCBI cl23723 Cytochrome_b_N superfamily, and specifically to the COG3038 (Cytochrome b561) and PRK11513 protein families, comprising proteins from important human pathogens e.g. Pseudomonas aeruginosa, Salmonella enterica, Shigella dysenteriae, Vibrio cholera and Yersinia pestis (Supplementary Fig. 10). A number of other Cytochrome_b_N protein families also display the same transmembrane helix topology and similar sequence conservation patterns, including the heme coordinating histidines and residues presumably involved in proton transfer to the quinone. Many organisms encode several CybB homologs; e.g. three in E. coli, P. aeruginosa, S. enterica and Y. pestis (Supplementary Fig. 10). The functions of these homologs are presently unknown. Further biochemical and bioinformatic studies are required to define and characterize this enzyme family.
Discussion
Redox proteins are notoriously promiscuous and require significant effort to unambiguously determine the primary biochemical substrate or function. For CybB, ubiquinol and superoxide (but not hydrogen peroxide) displayed a fast reactivity, suggesting electron transfer from superoxide to ubiquinone. The measurement of the catalytic rate constant of CybB (~1 x 108 M-1s-1) shows that the reaction is in the diffusion limited range. CybB is thus what is commonly denoted a “kinetically perfect” enzyme for superoxide oxidation and it appears extremely unlikely that such an efficient enzymatic design would occur by accident. We therefore hypothesize that CybB has evolved to quench superoxide, and propose that this family of superoxide-scavenging enzymes is denoted superoxide oxidases (SOO).
The respiratory chain is a major source of superoxide28. It has been shown that approximately 25% of all superoxide is generated by membranes6 and that E. coli produces significant amounts of periplasmic superoxide during exponential growth, largely due to NDH-2 activity and menaquinone autooxidation in the membrane19. Interestingly, however, periplasmic SOD is synthesized only when cells enter stationary phase19,36. An attractive possibility is that periplasmic SOD is expressed primarily as protection against external superoxide while the primary function of CybB may be to quench membranous superoxide close to its site of production (e.g. the respiratory enzymes in the membrane). In line with this reasoning, cybB mRNA levels showed the opposite pattern to periplasmic SOD, being >20 higher during the logarithmic compared to the stationary phase.
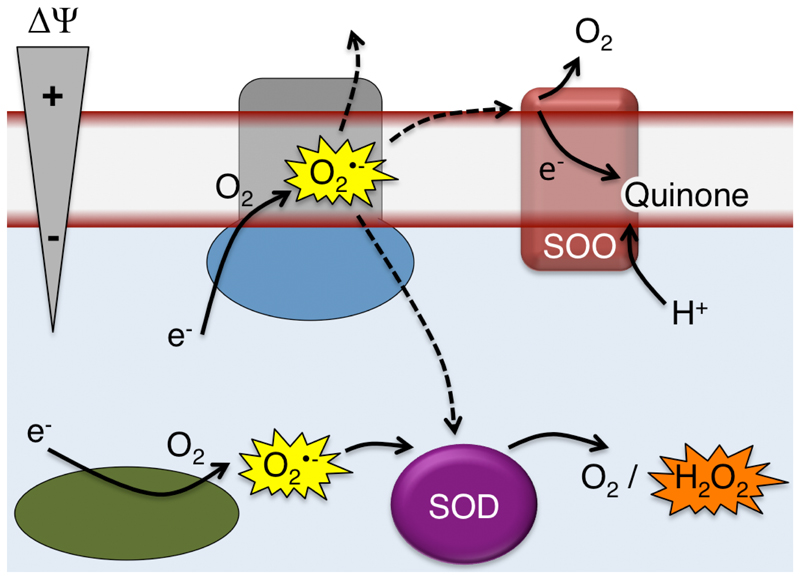
Electron leakage resulting in superoxide formation is a threat to the organism and also results in electron transfer to O2 without conservation of energy. In this study we have functionally and structurally characterized a protein that potentially alleviates both these problems. The architecture of SOO suggests electrogenic quenching of membrane-proximal superoxide by obtaining the electron from superoxide produced at, or within, the membrane and sequestering protons from the N-side; thereby creating a net charge separation while conserving energy by reducing the quinone pool in the process. A hypothetical biochemical context of SOO is depicted in Figure 4. Important future directions are to study the membrane-associated superoxide pool and the SOO mechanism in detail, as well as to establish the physiological function of the enzyme and determine if there are functional homologs of SOO in eukaryotic cells. In this context, it is interesting to note that there is a eukaryotic membrane protein family; eukaryotic Cyt b561, annotated as ascorbate- and iron-dependent oxidoreductases37. A recently solved crystal structure of this protein from Arabidopsis thaliana shows that it has a different topology and no quinone-binding site. However, it displays an analogous heme architecture and contains a positively charged patch with an exposed heme edge. It may thus be well worth to test proteins of this eukaryotic family for their reactivity with superoxide.
Fig. 4. Hypothetical functional context of SOO.
Superoxide produced by electron leakage at the membrane may escape to the cytoplasm, periplasm, or be oxidized by SOO, producing molecular oxygen, using ubiquinone as the electron acceptor. Intracellular superoxide is quenched by superoxide dismutase producing molecular oxygen and hydrogen peroxide.
Online Methods
Plasmids and expression
The expression plasmid pCybB-His was engineered from a GFP-tagged E. coli CybB construct by replacing the GFP moiety with a 8x His-tag, as described previously38. The new construct was transformed into chemically competent T7 express lysY (New England Biolabs) and grown at 37° C, either in LB supplemented with 25 mg/L kanamycin and 0.01% Antifoam 204 (for non-Se-Met labeled CybB) or in minimal media supplemented with 0.4% glucose, 2 mM MgSO4, 0.1 mM CaCl2, 1 μM MnCl2, 10 μM FeSO4, 25 mg/L kanamycin and 0.01% Antifoam 204 (for production of selenomethionine labeled protein for structural determination). At culture OD600 ~0.7, the minimal media cultures were supplemented with a mixture of amino acids (100 mg/L lysine, 100 mg/L threonine, 100 mg/L phenylalanine 50 mg/L leucine, 50 mg/L isoleucine, 50 mg/L valine and 50 mg/L selenomethionine). Expression of CybB and CybBSeMet was induced by the addition of 0.2 mM IPTG, and the culture was grown for 4 more hours. The cells were harvested by centrifugation at 7500 g in a JLA 8.1000 rotor (Beckman Coulter), transferred to 50 ml tubes and frozen at -20°C.
Membrane preparation
Membrane preparation and protein purification was performed using the same procedures for both SeMet-labeled and unlabeled protein. The frozen CybB/CybBSeMet cell pellet was thawed, resuspended in lysis buffer (50 mM phosphate buffer pH 7.0, 200 mM NaCl, 5% glycerol, 1 mg/ml lysozyme, 20 μg/ml DNase, EDTA-free cocktail protease inhibitors [Roche]) and lysed in an Emulsiflex C3 system (Avestin Inc.) or in a Maximator HPL6 (Maximator AG, Switzerland). All lysis and membrane preparation steps were performed at 4°C. The lysate was cleared by 30 min. centrifugation at 20.000 g in a Beckman JA 25.50 rotor. The supernatant was transferred to a Beckman Ti45 ultracentrifuge rotor and centrifuged at 40.000 rpm for 1 h. The membrane pellet was resuspended in membrane wash buffer (50 mM phosphate buffer pH 7.0, 200 mM NaCl, 5% glycerol). After a second round of ultracentrifugation, the washed and resuspended membranes were aliquoted and flash-frozen in liquid nitrogen.
Protein purification
Membranes were thawed and solubilized in 1% OGNG (Octyl Glucose Neopentyl Glycol) (Anatrace) for 2 hours. Insoluble material was cleared by ultracentrifugation for 30 min. at 40.000 rpm in a Beckman Ti70.1 rotor. The supernatant was incubated at 4°C with Ni2+-charged Profinity (Bio-Rad) IMAC resin for 2 h with gentle rotation (5 mM Imidazole was added after 1 h of incubation). The sample was loaded on a Econo-Pac disposable gravity flow column (Bio-Rad). The settled resin was washed with 15 CV IMAC wash buffer 1 (10 mM HEPES pH 7.0, 200 mM NaCl, 50 mM Imidazole, 5% glycerol, 0.1% OGNG) and 15 CV wash buffer 2 (10 mM HEPES pH 7.0, 200 mM NaCl, 5% glycerol, 0.1% OGNG). CybB/CybBSeMet was eluted with 3x 0.75 CV elution buffer (10 mM HEPES pH 7.0, 200 mM NaCl, 100 mM EDTA, 5% glycerol, 0.1% OGNG), concentrated to 0.5 ml using Vivaspin concentration devices with 50 kD cutoff (Sartorius) and subjected to size exclusion chromatography (10 mM HEPES pH 7.0, 200 mM NaCl, 5% glycerol, 0.1% OGNG) on a 30 ml BIOSEP-SEC-S3000 column (Phenomenex) connected to a Shimadzu Prominence HPLC system. Fractions representing the CybB/CybBSeMet peak were pooled, concentrated to 10-12 mg/ml (CybB) and 11 mg/ml (CybBSeMet) with 50 kD concentrator cut-off, and flash-frozen in liquid nitrogen.
Chemicals for functional experiments
Lipids were purchased from from Avanti Polar Lipids (Alabaster, AL, USA). WST-1 [2-(4-Iodophenyl)-3-(4-nitrophenyl)-5-(2,4-disulfophenyl)-2H-tetrazolium, monosodium salt] was purchased from Dojindo Molecular Technologies (Kumamoto, Japan). All other chemicals were purchased either from Sigma Aldrich (St. Louis, MO, USA) or Santa Cruz Biotechnology (Dallas, TX, USA).
Reduction of ubiquinone Q2
Ubiquinone Q2 was pre-reduced by addition of a few grains of sodium borohydride to a solution of 50 µM Q2 (dissolved in anhydrous EtOH). Reduction was carried out for 20 min on ice and quenched by addition of 2 µl 36% HCl (aqueous). Residual sodium borohydride was removed by centrifugation at 20’000 g for 5 min for at least three times. Reduced ubiquinol Q2H2 was stored in aliquots at -20 °C.
Preparation of liposomes
E. coli polar extract was dissolved in chloroform at 20 mg/ml and 250 µl of the solution was mixed in a 25-ml round-bottom flask. Chloroform was evaporated under a constant N2 stream, the lipid film was further dried for 4-12 h in a desiccator and resuspended at 5 mg/ml in 20 mM HEPES, pH 7.4, 20 mM KCl, and 200 mM NaCl. Unilamellar liposomes were formed by seven freeze-thaw cycles and down-sized using a Vibra-cell VCX 130 probe sonicator (Sonics & Materials, Newtown, CT, USA) at a frequency of 9 kHz for 2.5 min (30 on, 30 off) on ice. Liposomes were kept on ice until protein reconstitution.
Reconstitution of CybB into liposomes
Reconstitution of CybB was performed as described earlier for other membrane proteins39. Briefly, 20 µl 500 µM purified CybB was mixed with 250 µl sonicated liposomes (5 mg/ml) and 0.6% (w/v) sodium cholate and incubated for 30 min at RT. Detergent was removed by passing the suspension over a PD-10 gel filtration column (GE Healthcare). The final concentration of CybB in liposomes was 10 µM. Proteoliposomes were kept at 4 °C and used the same day for measurements.
Reduction of CybB
Reduction of CybB was monitored using either a Cary 60 UV-Vis Spectrophotometer (Agilent Technologies, Santa Clara, CA, USA) or a modular HR4000CG-UV-NIR Spectrophotometer (Ocean Optics, FL, USA) equipped with a HL-2000-FHSA halogen light source (Ocean Optics, Dunedin, FL, USA), respectively.
In a first set of experiments, 1 µM purified CybB in 0.5 ml assay buffer (20 mM HEPES, pH 7.4, 20 mM KCl, 200 mM NaCl, 0.05% (w/v) DDM) was mixed with different biological reductants such as 1 mM ascorbate, 1 mM succinate, 1 mM reduced glutathione (GSH), 1 mM NADH, 40 µM hydrogen peroxide (H2O2), or a mixture of 0.1 mM ubiquinone Q2 and 1 mM DTT. Alternatively, 0.2 µM purified E. coli type-II NADH:quinone oxidoreductase (NDH-2) together with 0.1 mM NADH were added. Spectra were monitored after 2 min of incubation between 400-600 nm. Reduction of 1 µM CybB by 7.6 mU/ml xanthine oxidase (XO) was measured in 100 mM sodium phosphate, pH 8.0, 0.1 mM diethylenetriaminepentaacetic acid (DETAPAC), 0.1 mM hypoxanthine (HPX), and 0.05% (w/v) DDM. ROS-mediated reduction of CybB was investigated by addition of 30 U/ml superoxide dismutase (SOD) or 6 U/ml catalase to the solutions described above. Anaerobic conditions were achieved by addition of 5 mM glucose, 60 U catalase and 2.5 U glucose oxidase after 1 min. Measurements with reconstituted CybB were done with 0.1 ml (5 mg/ml) proteoliposomes containing 10 µM CybB in 0.9 ml 20 mM HEPES, pH 7.4, 20 mM KCl, and 200 mM NaCl. Reduction of b-type hemes of CybB by different reductants was normalized to full reduction achieved by addition of 50 mM sodium dithionite. All experiments were performed at 25 °C with two separate purifications of CybB in duplicates.
Stopped-flow kinetics
Reduction of CybB by NADH/NDH-2 and pre-reduced ubiquinol Q2H2 was resolved by stopped-flow kinetics using a RX2000 Rapid Kinetics Spectrometer Accessory Unit (Applied Photophysics, Surrey, UK) with a dead-time of 8 ms. One syringe was filled with a solution of 1 µM CybB in assay buffer and the other one with either a solution of 125 µM Q2H2 or 200 µM NADH, 0.2 µM NDH-2. Shown are averaged data points from three separate measurements.
Determination of rate constants
The rate constants of detergent solubilized CybB were determined with the method described by Flint et al.27 with the following modifications. Superoxide production by HPX/XO was followed using the reduction of WST-1 at 455 nm in a buffer containing 100 mM sodium phosphate, pH 8.0 buffer containing 0.2 mM WST-1, 0.1 mM DETAPAC, 0.1 mM HPX, 0.05% (w/v) DDM, 6 U/ml catalase, 64 nM bo3 oxidase, 0.1 mM Q2. Subsequently, SOD was added stepwise until superoxide production was decreased to ~50%. This procedure was repeated in the same buffer with additions of either cytc or CybB. Using the literature value of the SOD rate constant and the respective concentrations of enzymes used to reduce superoxide production to 50%, the rate constants of cytc and CybB were calculated according to Flint27,40. Experiments were performed in triplicates with two different CybB purifications.
Preparation of E. coli inverted membrane vesicles
Overexpression of CybB in E. coli BL21/pLysS cells was achieved as described above. E. coli strains BL21/pLysS and BW25113 containing the wild-type cybB gene and the cybB KO strain (Keio designation: JW5224-141) was grown in M9 minimal medium containing 2% (w/v) glucose into the exponential phase (OD=0.5-0.7) at 37 °C, 200 rpm. Cells were collected by centrifugation (5’000 g, 4 °C 15 min), washed in 20 mM HEPES, pH 7.4, 20 mM KCl, and 200 mM NaCl and resuspended in the same buffer in the presence of 1 mM MgCl2, 1 mM phenylmethylsulfonyl fluoride and DNaseI. The solution was stirred for 20 min at 4 °C and passed twice through a Maximator HPL6 (Maximator AG, Switzerland) yielding inside-out membrane vesicles (IMVs)42. Unbroken cells were removed by low-spin centrifugation (7’500 g, 20 min, 4 °C) and membrane vesicles were washed twice in 20 mM HEPES, pH 7.4, 20 mM KCl, 200 mM NaCl by ultracentrifugation (150’00 g, 4 °C, 1h).
Purification of bo3 ubiquinol oxidase and NADH:quinone oxidoreductase NDH-2
Purification of E. coli bo3 ubiquinol oxidase and NADH:quinone oxidoreductase NDH-2 was performed as described43,44.
Superoxide assay
Production of superoxides was detected by monitoring the superoxide-mediated reduction of a tetrazolium dye (WST-1) as described by Peskin et al.45. Briefly, reduction of WST-1 was monitored at 455 nm in assay buffer, 20 mM HEPES, pH 7.4, 20 mM KCl, 200 mM NaCl containing 20 µg/ml catalase and 50 µM WST-1. NADH oxidation of IMVs from E. coli strains were adjusted to same oxidation rates before measuring superoxide production. Measurements with 200 nM purified CybB were done in assay buffer containing 0.05% DDM and one or several of the following additives: 200 µM NADH, 100 µM Q2, 2 nM NDH-2, and/or 60 nM bo3 oxidase. In another series of measurements, XO (7.6 mU/ml) instead of NDH-2 was used. The measuring buffer was 100 mM sodium phosphate, pH 8.0, 0.1 mM diethylenetriaminepentaacetic acid, 0.1 mM hypoxanthine (HPX), and 0.05% (w/v) DDM.
Bacteria strain and growth conditions for gene regulation experiments
E. coli BW25113 strain (parental strain of the Keio collection41) was used for gene regulation experiments. As control, a cybB knock-out strain (Keio designation: JW5224-1) was included. A few colonies were picked from agar plates, inoculated into 5 ml LB medium and incubated at 37 °C overnight. Cells from the overnight grown culture were harvested by centrifugation (5’000 g, 4 °C, 10 min), washed twice with PBS and resuspended in 5 ml PBS. 200 µl of the resuspended cells were used to inoculate 20 ml M9 minimal medium containing 2% (w/v) glucose as a carbon source. Cell cultures were grown at 37 °C, 200 rpm and sampling was done using half of the culture either in the mid-log phase at OD between 0.5-0.7 or in the stationary phase after overnight incubation, respectively. Cells were harvested by centrifugation (8’000 g, 25 °C, 2 min), and the pellet was directly used for RNA extraction. Anaerobic conditions were achieved using basic 1,2,3-trihydroxy-benzene (pyrogallol). Briefly, 10 ml minimal medium were poured into a test tube (14 mm inner diameter x 160 mm tall) and the medium was covered with two layers of cotton wool. To the upper layer 200 µl saturated sodium carbonate and 200 µl 20 % (w/v) pyrogallol was added dropwise and the test tube was sealed with a rubber plug and wrapped with several layers of Parafilm M. To test anaerobicity, E. coli BW25113 cells were inoculated in minimal medium containing 2% (w/v) succinate as a non-fermentable carbon source at 37°C, 200 rpm, allowing growth by oxidative phosphorylation only. No increase in OD was detected over 60 h of incubation.
RNA extraction
Total RNA was extracted as described with minor modifications46. Cell pellets were resuspended in 600 µl 100 mM Tris, pH 7.5, 10 mM EDTA, 1 M NaCl. Next, 600 µl pre-heated (65 °C) water-saturated acidic phenol (pH<7) containing 1% (w/v) SDS was immediately added and cells were lysed for 5 min at 65 °C. The lysed cells were frozen in liquid N2 and stored at – 20 °C until RNA extraction.
Frozen cell extracts were thawed at 65 °C and centrifuged (17’000 g, 4 °C, 10 min). The aqueous phase was transferred to a new tube containing equal volume of 1:1 acidic phenol : chloroform, mixed vigorously and centrifuged again. The upper phase was transferred to a new tube containing same volume of chloroform, mixed and centrifuged again. The upper phase was mixed with 700 µl ice-cold 2-propanol and incubated for 2h at -20 °C, followed by centrifugation. The RNA pellet was washed twice with 75% (v/v) ice-cold ethanol, vacuum dried for 10 min and resuspended in 50 µl DEPC-treated water. RNA extraction was performed from two independent growth studies.
cDNA synthesis and RT-qPCR
In general, 2.5 µg total RNA was used for DNA digestion and reverse transcription. DNA was removed using TURBO DNase kit (Ambion) according to the protocol of the manufacturer. Reverse transcription real-time quantitative PCR (RT-qPCR) was performed as described47. Published primer pairs of two different housekeeping genes (idnT and hcaT), which show stable expression in many different growth conditions, were used for normalization48. Amplification of a 162 bp fragment of cybB was performed using CGCAGCGTAT TGCGCAATGG AGTTTCG as forward and GGCTTAGGTA TAATCGGCGGG GTTGGG as a reverse primer. As a control cDNA from a cybB knock-out strain was included. cybB mRNA levels were measured in triplicates by RT-qPCR and normalized to the mRNA levels of idnT. The mRNA levels of cybB in the exponential phase under aerobic conditions were defined as 100%.
Crystallization
Frozen CybB-HisSeMet protein was thawed on ice and 1 mM TCEP was added. Crystallizations were set up manually in sitting drops under silicon oil (octamethyltrisiloxane, Sigma-Aldrich). A 48-well MRC Maxi crystallization plate (Molecular Dimensions) was coated with EGC-1700 (3M Science) and 0.75 μl protein and 0.25 μl well solution was added to a 4 μl silicone drop. Red, rod-shaped, rhombic cross section crystals appeared overnight in: 200 mM HEPES pH 7.5; 5% glycerol; 25% PEG 2000 MME; 50 mM MgCl2. Crystals were grown for 5 days at 20°C, and were picked straight from the drop, without additional cryoprotectant, and flash-frozen in liquid nitrogen.
Data collection and processing
Data collection was performed at beam line X06SA (PX-I) at the Swiss Light Source, Paul Scherrer Institute, Switzerland. Data was collected at the Selenium edge (0.9797 Å; 12655 keV) using a Pilatus 6M detector. The data set (Supplementary Table 1) was collected from the largest obtained crystal (~0.1x0.1x0.2 mm). A 360° dataset was collected at 100° K, with an image width of 1°. The dataset was integrated and scaled using XDS49, and the full set of 360 images was used to gain initial phase estimates by SAD, using HKL2MAP50. The protein sequence contains 11 methionine residues, 10 significant anomalous peaks were identified in the data and used for phasing.
Model building and refinement
Phenix Autosol51 was used to build an initial model. A second dataset was generated by integrating the data while merging the Friedel pairs. This dataset was used for subsequent manual model building with COOT52 and refinement with Refmac553. A few distinct volumes of experimentally determined density on the protein surface could not be accurately modeled. These presumably represent buffer molecules, ordered parts of detergent tails or (native) lipid molecules carried along through protein purification. The model refined with good stereochemistry and a Ramachandran plot distribution of 99% favored, 1% allowed and 0% outliers. The structure and crystallographic data have been deposited in the protein data bank with PDBid: 5OC0.
Crystal structure figures
Crystal structure figures were prepared using PyMol (The PyMOL Molecular Graphics System, Schrödinger, LLC.) Electrostatic surfaces were calculated including all cofactors using the Adaptive Poisson-Boltzmann Solver (APBS)54 and visualized using the APBS plugin in PyMol.
Docking
Molecular docking of ubiquinone, menaquinone and demethylmenaquinone as well as a database of commercially available compounds to the crystal structure was performed with DOCK3.6 (http://dock.compbio.ucsf.edu/) using a physics-based scoring function55–57. Ionizable residues Asp, Glu, Arg, and Lys were set to their most probable state at pH 7. Protonation states for histidines were based on analysis of hydrogen bonding network. Binding site residues H87, H151 and H158 were protonated at the Nε, Nδ, and both nitrogen positions, respectively. All compounds were docked to a rigid receptor structure using a flexible ligand sampling algorithm57. DOCK3.6 superimposes atoms of the docked molecule onto binding site matching spheres, which indicate putative ligand atom positions. Forty-five matching spheres were used to define the binding site and these were based on the binding mode of the menaquinone analog in the crystal structures of formate dehydrogenase-N (PDB code: 1KQG)58. The ligand sampling parameters bin size, bin size overlap, and distance tolerance were set to 0.8 Å, 0.1 Å, and 1.5 Å, respectively, for both the matching spheres and docked molecules. For each docked compound that passed an initial steric filer, binding energies were calculated from pre-generated grids as the sum of the receptor–ligand electrostatic and van der Waals interaction energy, corrected for compound desolvation56. AMBER force field parameters59 were used for all protein atoms except His151, for which the side chain dipole moment was increased to favor interactions with this residue. The heme group with Fe2+ was modeled based on parameters described previously60. As the docked compounds were expected to become fully desolvated by the protein and membrane environment, the full desolvation setting was used. Energy minimization of 50 steps was performed for the best scoring conformation of each compound. Only the headgroup of the quinones (Supplementary Fig. 7) were docked to the binding site as the hydrophobic tail is likely to extend into the membrane, which was not explicitly modeled. The lead-like screening library of 4.6 million commercially available compounds was retrieved from the ZINC database56. All compounds were prepared for docking using the ZINC database protocol.
Supplementary Material
Acknowledgements
This work was supported by grants from the Knut and Alice Wallenberg foundation Wallenberg Academy Fellows (2012.0233), the Swedish foundation for strategic research (FFL09-0008) and the Swedish research council (2017-04018) to M.H., and the Swiss National Science Foundation to C.v.B. (153351). The help of H. Luidalepp with RNA extraction from E_coli and A. Eberle (University of Bern) with RT-qPCR and data analysis is greatly acknowledged. The authors wish to thank the staff of beamline X06SA at the Swiss Light Source for their support with data collection.
Footnotes
Author contributions:
D.S. and M.H. conceived the study. C.A.K.L., D.S., M.B. and M.H. solved the structure. C.A.K.L., D.S., O.B., P.B., C.vB. and M.H. designed functional experiments and analyzed data. O.B., C.A.K.L., A.-L. J. and C.vB. performed functional experiments. A.R. and J.C. performed and analyzed docking experiments. C. vB. and M.H. wrote the manuscript with input from all authors.
The authors declare no competing financial interests.
Accession code:
Structure and crystallographic data have been deposited in the protein data bank with PDBid: 5OC0.
References
- 1.Dröge W. Free radicals in the physiological control of cell function. Physiol Rev. 2002;82:47–95. doi: 10.1152/physrev.00018.2001. [DOI] [PubMed] [Google Scholar]
- 2.Sabharwal SS, Schumacker PT. Mitochondrial ROS in cancer: initiators, amplifiers or an Achilles' heel? Nat Rev Cancer. 2014;14:709–721. doi: 10.1038/nrc3803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barnham KJ, Masters CL, Bush AI. Neurodegenerative diseases and oxidative stress. Nat Rev Drug Discov. 2004;3:205–214. doi: 10.1038/nrd1330. [DOI] [PubMed] [Google Scholar]
- 4.Riera CE, Merkwirth C, De Magalhaes Filho CD, Dillin A. Signaling Networks Determining Life Span. Annu Rev Biochem. 2016;85:35–64. doi: 10.1146/annurev-biochem-060815-014451. [DOI] [PubMed] [Google Scholar]
- 5.McCord JM, Fridovich I. Superoxide dismutase. An enzymic function for erythrocuprein (hemocuprein) J Biol Chem. 1969;244:6049–6055. [PubMed] [Google Scholar]
- 6.Imlay JA. Cellular defenses against superoxide and hydrogen peroxide. Annu Rev Biochem. 2008;77:755–776. doi: 10.1146/annurev.biochem.77.061606.161055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Jenney FE, Verhagen MF, Cui X, Adams MW. Anaerobic microbes: oxygen detoxification without superoxide dismutase. Science. 1999;286:306–309. doi: 10.1126/science.286.5438.306. [DOI] [PubMed] [Google Scholar]
- 8.Magnani F, et al. Crystal structures and atomic model of NADPH oxidase. Proc Natl Acad Sci U S A. 2017;114:6764–6769. doi: 10.1073/pnas.1702293114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sullivan MN, et al. Localized TRPA1 channel Ca2+ signals stimulated by reactive oxygen species promote cerebral artery dilation. Sci Signal. 2015;8:ra2. doi: 10.1126/scisignal.2005659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yankovskaya V, et al. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science. 2003;299:700–704. doi: 10.1126/science.1079605. [DOI] [PubMed] [Google Scholar]
- 11.Kerscher S, Dröse S, Zickermann V, Brandt U. The three families of respiratory NADH dehydrogenases. Results Probl Cell Differ. 2008;45:185–222. doi: 10.1007/400_2007_028. [DOI] [PubMed] [Google Scholar]
- 12.Imlay JA, Fridovich I. Superoxide production by respiring membranes of Escherichia coli. Free Radic Res Commun. 1991;12–13(Pt 1):59–66. doi: 10.3109/10715769109145768. [DOI] [PubMed] [Google Scholar]
- 13.Windrem DA, Plachy WZ. The diffusion-solubility of oxygen in lipid bilayers. Biochim Biophys Acta. 1980;600:655–665. doi: 10.1016/0005-2736(80)90469-1. [DOI] [PubMed] [Google Scholar]
- 14.Mao GD, Poznansky MJ. Electron spin resonance study on the permeability of superoxide radicals in lipid bilayers and biological membranes. FEBS Lett. 1992;305:233–236. doi: 10.1016/0014-5793(92)80675-7. [DOI] [PubMed] [Google Scholar]
- 15.Messner KR, Imlay JA. Mechanism of superoxide and hydrogen peroxide formation by fumarate reductase, succinate dehydrogenase, and aspartate oxidase. J Biol Chem. 2002;277:42563–42571. doi: 10.1074/jbc.M204958200. [DOI] [PubMed] [Google Scholar]
- 16.Guo J, Lemire BD. The ubiquinone-binding site of the Saccharomyces cerevisiae succinate-ubiquinone oxidoreductase is a source of superoxide. J Biol Chem. 2003;278:47629–47635. doi: 10.1074/jbc.M306312200. [DOI] [PubMed] [Google Scholar]
- 17.Husen P, Solov'yov IA. Spontaneous Binding of Molecular Oxygen at the Qo-Site of the bc1 Complex Could Stimulate Superoxide Formation. J Am Chem Soc. 2016;138:12150–12158. doi: 10.1021/jacs.6b04849. [DOI] [PubMed] [Google Scholar]
- 18.Takahashi M, Asada K. Superoxide production in aprotic interior of chloroplast thylakoids. Arch Biochem Biophys. 1988;267:714–722. doi: 10.1016/0003-9861(88)90080-x. [DOI] [PubMed] [Google Scholar]
- 19.Korshunov S, Imlay JA. Detection and quantification of superoxide formed within the periplasm of Escherichia coli. J Bacteriol. 2006;188:6326–6334. doi: 10.1128/JB.00554-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Murakami H, Kita K, Anraku Y. Purification and properties of a diheme cytochrome b561 of the Escherichia coli respiratory chain. J Biol Chem. 1986;261:548–551. [PubMed] [Google Scholar]
- 21.Murakami H, Kita K, Anraku Y. Cloning of cybB, the gene for cytochrome b561 of Escherichia coli K12. Mol Gen Genet. 1984;198:1–6. doi: 10.1007/BF00328692. [DOI] [PubMed] [Google Scholar]
- 22.Murakami H, Kita K, Oya H, Anraku Y. Chromosomal location of the Escherichia coli cytochrome b556 gene, cybA. Mol Gen Genet. 1984;196:1–5. doi: 10.1007/BF00334084. [DOI] [PubMed] [Google Scholar]
- 23.Villegas JM, Volentini SI, Rintoul MR, Rapisarda VA. Amphipathic C-terminal region of Escherichia coli NADH dehydrogenase-2 mediates membrane localization. Arch Biochem Biophys. 2011;505:155–159. doi: 10.1016/j.abb.2010.10.001. [DOI] [PubMed] [Google Scholar]
- 24.Antonini E, Brunori M, Colosimo A, Greenwood C, Wilson MT. Oxygen ‘pulsed’ cytochrome c oxidase: functional properties and catalytic relevance. Proc Natl Acad Sci U S A. 1977;74:3128–3132. doi: 10.1073/pnas.74.8.3128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ingledew WJ, Poole RK. The respiratory chains of Escherichia coli. Microbiol Rev. 1984;48:222–271. doi: 10.1128/mr.48.3.222-271.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Abreu IA, Cabelli DE. Superoxide dismutases-a review of the metal-associated mechanistic variations. Biochim Biophys Acta. 2010;1804:263–274. doi: 10.1016/j.bbapap.2009.11.005. [DOI] [PubMed] [Google Scholar]
- 27.Flint DH, Tuminello JF, Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem. 1993;268:22369–22376. [PubMed] [Google Scholar]
- 28.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lomize MA, Pogozheva ID, Joo H, Mosberg HI, Lomize AL. OPM database and PPM web server: resources for positioning of proteins in membranes. Nucleic Acids Res. 2012;40:D370–6. doi: 10.1093/nar/gkr703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Moser CC, Chobot SE, Page CC, Dutton PL. Distance metrics for heme protein electron tunneling. Biochim Biophys Acta. 2008;1777:1032–1037. doi: 10.1016/j.bbabio.2008.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Winkler JR, Gray HB. Electron flow through metalloproteins. Chem Rev. 2014;114:3369–3380. doi: 10.1021/cr4004715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Getzoff ED, et al. Electrostatic recognition between superoxide and copper, zinc superoxide dismutase. Nature. 1983;306:287–290. doi: 10.1038/306287a0. [DOI] [PubMed] [Google Scholar]
- 33.Getzoff ED, et al. Faster superoxide dismutase mutants designed by enhancing electrostatic guidance. Nature. 1992;358:347–351. doi: 10.1038/358347a0. [DOI] [PubMed] [Google Scholar]
- 34.Tainer JA, Getzoff ED, Richardson JS, Richardson DC. Structure and mechanism of copper, zinc superoxide dismutase. Nature. 1983;306:284–287. doi: 10.1038/306284a0. [DOI] [PubMed] [Google Scholar]
- 35.Fee JA. Regulation of sod genes in Escherichia coli: relevance to superoxide dismutase function. Mol Microbiol. 1991;5:2599–2610. doi: 10.1111/j.1365-2958.1991.tb01968.x. [DOI] [PubMed] [Google Scholar]
- 36.Benov LT, Fridovich I. Escherichia coli expresses a copper- and zinc-containing superoxide dismutase. J Biol Chem. 1994;269:25310–25314. [PubMed] [Google Scholar]
- 37.Lu P, et al. Structure and mechanism of a eukaryotic transmembrane ascorbate-dependent oxidoreductase. Proc Natl Acad Sci U S A. 2014;111:1813–1818. doi: 10.1073/pnas.1323931111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sjöstrand D, Diamanti R, Lundgren CAK, Wiseman B, Högbom M. A rapid expression and purification condition screening protocol for membrane protein structural biology. Protein Sci. 2017;8:1653–1666. doi: 10.1002/pro.3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.von Ballmoos C, Biner O, Nilsson T, Brzezinski P. Mimicking respiratory phosphorylation using purified enzymes. Biochim Biophys Acta. 2016;1857:321–331. doi: 10.1016/j.bbabio.2015.12.007. [DOI] [PubMed] [Google Scholar]
- 40.Klug D, Rabani J, Fridovich I. A direct demonstration of the catalytic action of superoxide dismutase through the use of pulse radiolysis. J Biol Chem. 1972;247:4839–4842. [PubMed] [Google Scholar]
- 41.Baba T, et al. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: the Keio collection. Mol Syst Biol. 2006;2 doi: 10.1038/msb4100050. 2006.0008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Futai M. Orientation of membrane vesicles from Escherichia coli prepared by different procedures. J Membr Biol. 1974;15:15–28. doi: 10.1007/BF01870079. [DOI] [PubMed] [Google Scholar]
- 43.Frericks HL, Zhou DH, Yap LL, Gennis RB, Rienstra CM. Magic-angle spinning solid-state NMR of a 144 kDa membrane protein complex: E. coli cytochrome bo3 oxidase. J Biomol NMR. 2006;36:55–71. doi: 10.1007/s10858-006-9070-5. [DOI] [PubMed] [Google Scholar]
- 44.Björklöf K, Zickermann V, Finel M. Purification of the 45 kDa, membrane bound NADH dehydrogenase of Escherichia coli (NDH-2) and analysis of its interaction with ubiquinone analogues. FEBS Lett. 2000;467:105–110. doi: 10.1016/s0014-5793(00)01130-3. [DOI] [PubMed] [Google Scholar]
- 45.Peskin AV, Winterbourn CC. Assay of superoxide dismutase activity in a plate assay using WST-1. Free Radical Biol Med. 2017;103:188–191. doi: 10.1016/j.freeradbiomed.2016.12.033. [DOI] [PubMed] [Google Scholar]
- 46.Khodursky AB, et al. Escherichia coli spotted double-strand DNA microarrays: RNA extraction, labeling, hybridization, quality control, and data management. Methods Mol Biol. 2003;224:61–78. doi: 10.1385/1-59259-364-X:61. [DOI] [PubMed] [Google Scholar]
- 47.Nicholson P, Joncourt R, Mühlemann O. Analysis of nonsense-mediated mRNA decay in mammalian cells. Curr Protoc Cell Biol. 2012;Chapter 27:Unit27.4–27.4.61. doi: 10.1002/0471143030.cb2704s55. [DOI] [PubMed] [Google Scholar]
- 48.Zhou K, et al. Novel reference genes for quantifying transcriptional responses of Escherichia coli to protein overexpression by quantitative PCR. BMC Mol Biol. 2011;12:18. doi: 10.1186/1471-2199-12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kabsch W. XDS. Acta Crystallogr Sect D-Biol Crystallogr. 2010;66:125–132. doi: 10.1107/S0907444909047337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pape T, Schneider TR, IUCr HKL2MAP: a graphical user interface for macromolecular phasing with SHELX programs. J Appl Crystallogr. 2004;37:843–844. [Google Scholar]
- 51.Adams PD, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr D Biol Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot. Acta Crystallogr D Biol Crystallogr. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vagin AA, et al. REFMAC5 dictionary: organization of prior chemical knowledge and guidelines for its use. Acta Crystallogr D Biol Crystallogr. 2004;60:2184–2195. doi: 10.1107/S0907444904023510. [DOI] [PubMed] [Google Scholar]
- 54.Baker NA, Sept D, Joseph S, Holst MJ, McCammon JA. Electrostatics of nanosystems: application to microtubules and the ribosome. Proc Natl Acad Sci U S A. 2001;98:10037–10041. doi: 10.1073/pnas.181342398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Irwin JJ, et al. Automated docking screens: a feasibility study. J Med Chem. 2009;52:5712–5720. doi: 10.1021/jm9006966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Mysinger MM, Shoichet BK. Rapid context-dependent ligand desolvation in molecular docking. J Chem Inf Model. 2010;50:1561–1573. doi: 10.1021/ci100214a. [DOI] [PubMed] [Google Scholar]
- 57.Lorber DM, Shoichet BK. Hierarchical docking of databases of multiple ligand conformations. Curr Top Med Chem. 2005;5:739–749. doi: 10.2174/1568026054637683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Jormakka M, Törnroth S, Byrne B, Iwata S. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science. 2002;295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 59.Weiner SJ, Kollman PA, Case DA. A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc. 1984;106:765–784. [Google Scholar]
- 60.Brenk R, Vetter SW, Boyce SE, Goodin DB, Shoichet BK. Probing molecular docking in a charged model binding site. J Mol Biol. 2006;357:1449–1470. doi: 10.1016/j.jmb.2006.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.