Abstract
The 2014–2015 Ebola epidemic affected several African countries, claiming more than 11,000 lives and leaving thousands with ongoing sequelae. Safe and effective vaccines could prevent or limit future outbreaks. The recombinant vesicular stomatitis virus–vectored Zaire Ebola (rVSV-ZEBOV) vaccine has shown marked immunogenicity and efficacy in humans but is reactogenic at higher doses. To understand its effects, we examined plasma samples from 115 healthy volunteers from Geneva who received low-dose (LD) or high-dose (HD) vaccine or placebo. Fifteen plasma chemokines/cytokines were assessed at baseline and on days 1, 2 to 3, and 7 after injection. Significant increases in monocyte-mediated MCP-1/CCL2, MIP-1β/CCL4, IL-6, TNF-α, IL-1Ra, and IL-10 occurred on day 1. A signature explaining 68% of cytokine/chemokine vaccine-response variability was identified. Its score was higher in HD versus LD vaccinees and was associated positively with vaccine viremia and negatively with cytopenia. It was higher in vaccinees with injection-site pain, fever, myalgia, chills, and headache; higher scores reflected increasing severity. In contrast, HD vaccinees who subsequently developed arthritis had lower day 1 scores than other HD vaccinees. Vaccine dose did not influence the signature despite its influence on specific outcomes. The Geneva-derived signature associated strongly (ρ = 0.97) with that of a cohort of 75 vaccinees from a parallel trial in Lambaréné, Gabon. Its score in Geneva HD vaccinees with subsequent arthritis was significantly lower than that in Lambaréné HD vaccinees, none of whom experienced arthritis. This signature, which reveals monocytes’ critical role in rVSV-ZEBOV immunogenicity and safety across doses and continents, should prove useful in assessments of other vaccines.
Introduction
A vaccine’s safety is a core element in its development and acceptance, yet there is little information on how vaccine-induced responses determine adverse outcomes. Despite recent progress in discovery of molecular signatures of vaccine-induced immune responses in humans offered by novel, cutting-edge technologies and systems biology approaches, biomarkers of vaccine safety and immunogenicity have yet to be identified for most vaccines.
There are currently no approved vaccines against Ebola virus disease (EVD). In 2014, an EVD outbreak affecting several African countries triggered international collaboration in the testing of EVD vaccine candidates (1). The most advanced in its development is the replication-competent recombinant vesicular stomatitis virus (rVSV)–based vector vaccine expressing the glycoprotein (GP) of the Zaire Ebola virus (rVSV-ZEBOV) (2), which conferred a high protection rate in the ring vaccination trial conducted in Guinea (3). The phase 1/2 studies were performed in 2014–2015 in the United States (4) and in Africa and Europe, with trials in the latter two continents led by a World Health Organization (WHO)–coordinated consortium [VSV-Ebola Consortium (VEBCON)] (5). In healthy adults, rVSV-ZEBOV was immunogenic but reactogenic. In phase 1 trials, vaccine doses ranged from 3 × 105 to 1 × 108 plaque-forming units (pfu), and both reactogenicity and immunogenicity proved to be dose-dependent (4–6), although the frequency and intensity of adverse events (AEs) were variable. In the Geneva randomized controlled trial (RCT) comparing low-dose (LD) (3 × 105 pfu) or high-dose (HD) (1 × 107 or 5 × 107 pfu) vaccine to placebo, 97% of vaccinees experienced reactogenicity (6). Characterized by early-onset local and systemic inflammation, it was transient and generally well tolerated (6). In the second week after immunization, rVSV-ZEBOV–associated arthritis was identified in 13 of 51 LD and 11 of 51 HD vaccinees (24%) (6). Although early reactogenicity was similar at other sites, arthritis was rarely reported (4, 5). The underlying mechanisms of rVSV-ZEBOV–induced AE remain unknown; further investigation is required to determine vaccine safety in vulnerable populations such as children, pregnant women, and the immunocompromised and to inform the clinical development of other rVSV-based vaccines (7–9). The Innovative Medicine Initiative 2 (IMI2) Joint Undertaking–supported VSV-EBOVAC project is examining the mechanisms underlying the immunogenicity and safety of rVSV-ZEBOV by using cutting-edge omics and state-of-the-art technologies (10).
Inflammation results from coordinated vaccine-specific and non-specific biochemical and cellular events reflecting cell migration and activation triggered early after infection or vaccination. Chemokines attract immune cells such as monocytes, granulocytes, or lymphocytes to infected or inflamed tissues (11, 12). Upon activation, these cells locally release mediators such as cytokines and chemokines (11), which play a key role in EVD (13). Because Ebola virus GP mediates cell tropism in EVD, we postulated that vaccination with rVSV-ZEBOV might involve similar target cells.
To study the immunological basis of rVSV-ZEBOV–induced AE and the influence of the vaccine dose on these immune responses, we quantified selected chemokines and cytokines in the plasma of Geneva vaccinees before and after LD or HD immunization (5, 6). We investigated whether a composite pattern of interconnected mediators might be identified. A distinct plasma signature emerged, composed of six markers whose up-regulation was vaccine dose–dependent and significantly correlated with vaccine-related viremia, cytopenia, and AE—including rVSV-ZEBOV–associated arthritis. Extending our analyses to vaccinees from Lambaréné, Gabon confirmed the signature’s validity across different genetic backgrounds and environmental settings.
Results
The Geneva derivation cohort identifies vaccine-induced, dose-dependent changes in specific plasma markers
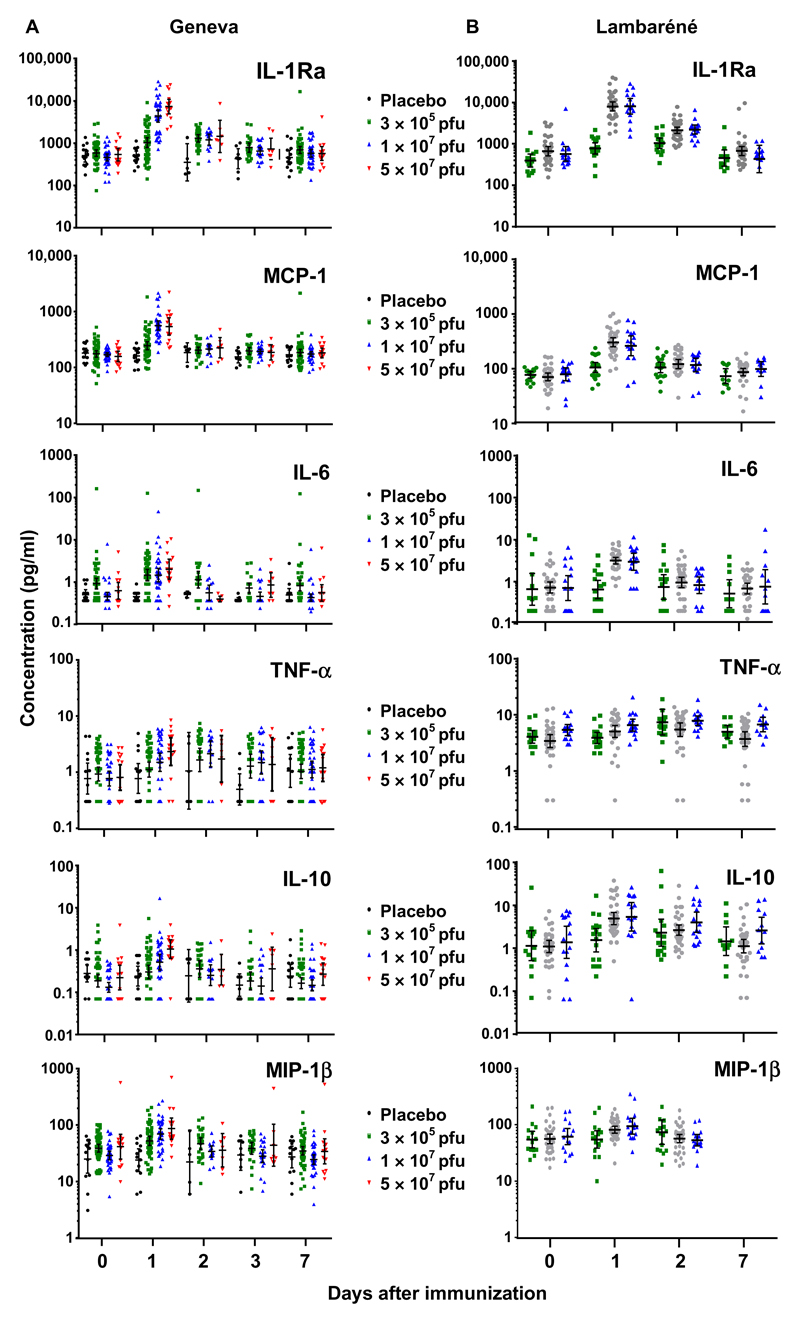
Plasma samples were collected on days 0, 1, 2 or 3, and 7 in the 115 participants of the Geneva RCT (fig. S1) and were subjected to multiplex analysis of 15 chemokines/cytokines with documented involvement in responses to Ebola (13–19), VSV (20, 21), or other viral vaccines (22, 23). These included monocyte [monocyte chemoattractant protein-1 (MCP-1/CCL2), macrophage inflammatory protein-1α (MIP-1α/CCL3), and MIP-1β/CCL4]– or granulocyte [interleukin-8 (IL-8/CXCL8) and epithelial-derived neutrophil-activating peptide 78 (ENA-78/CXCL5)]–attracting chemokines, growth factors [granulocyte-macrophage colony-stimulating factor (GM-CSF) and granulocyte colony-stimulating factor (G-CSF)], proinflammatory [tumor necrosis factor–α (TNF-α) and interferon-γ (IFN-γ)] and anti-inflammatory [IL-1 receptor antagonist (IL-1Ra) and IL-10] cytokines, and some T cell cytokines (IL-2, IL-4, IL-6, and IL-17). The geometric mean concentrations (GMCs) of granulocyte-attracting chemokines, growth factors, T cell cytokines, IL-1β, and MIP-1α/CCL3 remained unchanged after immunization (table S1). In contrast, a synchronized day 1 GMC increase was observed for two monocyte-attracting chemokines (MCP-1/CCL2 and MIP-1β/CCL4) and two proinflammatory (IL-6 and TNF-α) and two anti-inflammatory (IL-1Ra and IL-10) cytokines (Table 1 and Figs. 1A and 2, A and B). This peak was followed by a rapid return to baseline, identifying the ratio of day 1/day 0 GMCs as the optimal marker of vaccine responses (table S2). In HD vaccinees, the largest fold increases were observed for IL-6 [13.5 (95% CI, 8.3 to 21.9)], IL-1Ra [10.6 (95% CI, 8.4 to 13.4)], and IL-10 [7.1 (95% CI, 4.7 to 10.7)], followed by TNF-α [4.0 (95% CI, 2.4 to 6.5)], MCP-1/CCL2 [3.4 (95% CI, 3.0 to 3.8)], and MIP-1β/CCL4 [2.3 (95% CI, 2.1 to 2.6)]. Although weaker in magnitude, significant changes were also observed in LD vaccinees (table S2).
Table 1. GMCs of up-regulated markers in Geneva participants.
Analyses included all subjects and the indicated numbers of plasma samples. ANOVA, analysis of variance; CI, confidence interval.
| Markers | Placebo | LD (3 × 105 pfu) | HD (107 or 5 × 107 pfu) | P values for comparison between dose groups ANOVA with post hoc Scheffe | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Days | n | GMC (pg/ml) (95% CI) |
n | GMC (pg/ml) (95% CI) |
n | GMC (pg/ml) (95% CI) |
Global | Placebo versus LD |
Placebo versus HD |
LD versus HD |
|
| IL-1Ra | 0 | 13 | 505.5 (371.1–688.7) | 51 | 581.3 (488.5–691.9) | 51 | 480.3 (410.7–561.6) | 0.27 | 0.76 | 0.96 | 0.28 |
| 1 | 13 | 492.3 (385.4–628.8) | 49 | 1029.3 (797.3–1328.9) | 51 | 5093.9 (3941.5–6583.1) | <0.001 | 0.031 | <0.001 | <0.001 | |
| 2 | 5 | 352.8 (171.9-724) | 28 | 1307.6 (1079.7–1583.5) | 21 | 1279.2 (960.7–1703.2) | <0.001 | <0.001 | <0.001 | 0.99 | |
| 3 | 8 | 428.3 (272.3–673.7) | 23 | 784.4 (620.6–991.6) | 30 | 670.3 (558.6–804.3) | 0.035 | 0.035 | 0.14 | 0.59 | |
| 7 | 13 | 457.5 (321.8–650.3) | 51 | 689.1 (572.6–829.3) | 51 | 574 (482.3–683.1) | 0.099 | 0.14 | 0.54 | 0.37 | |
| P = 0.14 | P < 0.001 | P < 0.001 | |||||||||
| IL-6 | 0 | 13 | 0.1 (0–0.4) | 51 | 0.4 (0.2–0.8) | 51 | 0.1 (0–0.2) | 0.002 | 0.19 | 0.89 | 0.002 |
| 1 | 13 | 0.1 (0–0.3) | 49 | 0.8 (0.4–1.4) | 51 | 1.2 (0.8–1.9) | <0.001 | 0.003 | <0.001 | 0.50 | |
| 2 | 5 | 0.2 (0–1.1) | 28 | 0.6 (0.3–1.3) | 21 | 0.1 (0–0.3) | 0.027 | 0.67 | 0.77 | 0.028 | |
| 3 | 8 | 0 (0–0.1) | 23 | 0.3 (0.1–0.7) | 29 | 0.1 (0–0.2) | 0.056 | 0.077 | 0.48 | 0.26 | |
| 7 | 13 | 0.1 (0–0.2) | 51 | 0.3 (0.2–0.6) | 51 | 0.1 (0–0.1) | 0.001 | 0.035 | 0.90 | 0.004 | |
| P = 0.21 | P < 0.001 | P < 0.001 | |||||||||
| TNF-α | 0 | 13 | 0.2 (0–0.8) | 51 | 0.3 (0.1–0.6) | 51 | 0.3 (0.1–0.5) | 0.87 | 0.88 | 0.95 | 0.96 |
| 1 | 13 | 0.3 (0.1–1) | 49 | 0.4 (0.2–0.7) | 51 | 1 (0.6–1.8) | 0.047 | 0.90 | 0.18 | 0.099 | |
| 2 | 5 | 0.3 (0–4) | 28 | 0.6 (0.2–1.6) | 21 | 1.4 (0.6-3) | 0.30 | 0.84 | 0.43 | 0.46 | |
| 3 | 8 | 0.1 (0–0.3) | 23 | 0.4 (0.1–1.2) | 30 | 0.6 (0.3–1.5) | 0.081 | 0.21 | 0.081 | 0.81 | |
| 7 | 13 | 0.3 (0.1–1.3) | 51 | 0.3 (0.1–0.6) | 51 | 0.5 (0.3–1) | 0.46 | 1 | 0.74 | 0.50 | |
| P = 0.097 | P = 0.11 | P < 0.001 | |||||||||
| IL-10 | 0 | 13 | 0.2 (0.1–0.5) | 51 | 0.1 (0.1–0.1) | 51 | 0.1 (0–0.1) | 0.17 | 0.31 | 0.17 | 0.86 |
| 1 | 13 | 0.2 (0.1–0.4) | 49 | 0.2 (0.1–0.3) | 51 | 0.5 (0.3–0.8) | 0.010 | 0.96 | 0.11 | 0.021 | |
| 2 | 5 | 0.2 (0–0.8) | 28 | 0.2 (0.1–0.4) | 21 | 0.2 (0.1–0.4) | 0.92 | 0.95 | 0.92 | 0.99 | |
| 3 | 8 | 0.1 (0–0.2) | 23 | 0.1 (0–0.2) | 30 | 0.1 (0–0.1) | 0.83 | 0.94 | >0.99 | 0.84 | |
| 7 | 13 | 0.1 (0–0.4) | 51 | 0.1 (0–0.1) | 51 | 0.1 (0–0.1) | 0.52 | 0.52 | 0.66 | 0.93 | |
| P = 0.10 | P = 0.003 | P < 0.001 | |||||||||
| MCP-1 | 0 | 13 | 180.4 (151–215.6) | 51 | 175.1 (154.1–199) | 51 | 165.9 (153.6–179.3) | 0.68 | 0.97 | 0.78 | 0.77 |
| 1 | 13 | 160.6 (132.9–194) | 49 | 246.1 (209.4–289.1) | 51 | 556 (474.1–652) | <0.001 | 0.053 | <0.001 | <0.001 | |
| 2 | 5 | 184.2 (138.7–244.6) | 27 | 204 (180.7–230.3) | 21 | 218.1 (185.4–256.5) | 0.58 | 0.83 | 0.62 | 0.80 | |
| 3 | 8 | 154.6 (126.2–189.3) | 24 | 197 (172.4–225.1) | 30 | 191.4 (174.5–210) | 0.13 | 0.14 | 0.20 | 0.94 | |
| 7 | 13 | 169.4 (142.3–201.8) | 51 | 184.1 (160.5–211.3) | 51 | 176 (161.8–191.5) | 0.75 | 0.81 | 0.96 | 0.86 | |
| P = 0.42 | P < 0.001 | P < 0.001 | |||||||||
| MIP-1β | 0 | 13 | 23.6 (13.4–41.6) | 51 | 38.8 (33–45.5) | 51 | 32 (26.7–38.4) | 0.055 | 0.070 | 0.36 | 0.37 |
| 1 | 13 | 22.7 (14.1–36.6) | 49 | 52.5 (43.8–62.9) | 51 | 74.1 (61.5–89.1) | <0.001 | 0.001 | <0.001 | 0.048 | |
| 2 | 5 | 19.3 (6.3–59.1) | 27 | 46.7 (37.2–58.7) | 21 | 34.5 (27.6-43) | 0.020 | 0.028 | 0.21 | 0.29 | |
| 3 | 8 | 29.3 (17.5-49) | 24 | 38.7 (30.7–48.8) | 30 | 31.6 (24.2–41.1) | 0.45 | 0.61 | 0.96 | 0.55 | |
| 7 | 13 | 27.1 (18.2–40.4) | 51 | 34.8 (29.5-41) | 51 | 26.8 (22.1–32.6) | 0.13 | 0.49 | >0.99 | 0.15 | |
| P = 0.16 | P < 0.001 | P < 0.001 | |||||||||
Fig. 1. Up-regulated plasma markers after rVSV-ZEBOV immunization.
Individual values are expressed in picograms per milliliter for each subject in each dose group and at each time point assessed, before and after rVSV-ZEBOV immunization or placebo injection in Geneva (A) or Lambaréné (B). The number of samples assessed at each time point is given in Table 1 and table S12. Samples with undetectable concentrations were arbitrarily given 50% of the specific minimal detection dose, as described in Materials and Methods.
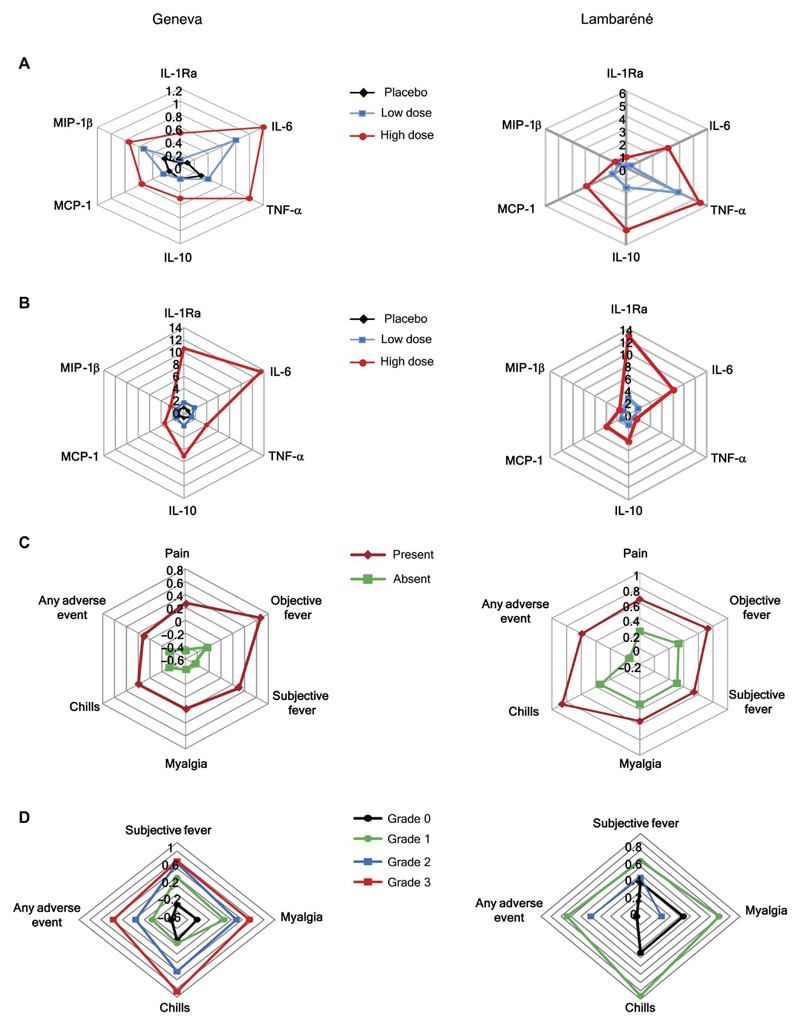
Fig. 2. Plasma chemokines/cytokines and signatures in rVSV-ZEBOV vaccinees from Geneva and Lambaréné.
(A) Day 1 GMCs are expressed in picograms per milliliter (IL-6, TNF-α, and IL-10),1 × 10−2 (MIP-1β/CCL4), 1 × 10−3 (MCP-1/CCL2), or 1 × 10−4 (IL-1Ra) and illustrated for Geneva (left) and Lambaréné (right) vaccinees. (B) Vaccine responses, expressed by the day 1/day 0 ratios, are illustrated for each chemokine and cytokine for which up-regulation was observed after immunization in Geneva (left) or Lambaréné (right). (C) The plasma signatures are illustrated for Geneva (left) and Lambaréné (right) vaccinees with and without AEs. (D) The plasma signatures are expressed for Geneva (left) and Lambaréné (right) vaccinees reporting grade 0 to 3 AEs.
Correlated plasma chemokine/cytokine responses define a specific rVSV-ZEBOV signature
We found that the chemokine and cytokine responses in the plasma of vaccinees correlated significantly in a dose-independent manner (table S3, A to C). The strongest associations among day 1 GMCs were between MCP-1/CCL2 and MIP-1β/CCL4 (Spearman’s correlation coefficient ρ = 0.71, P < 0.001) and between IL-6 and TNF-α (ρ = 0.6, P < 0.001). Strong correlations were similarly identified for the day 1/day 0 GMC ratios (table S4). Cronbach’s α values were 0.89, 0.86, and 0.91 for LD, HD, and all vaccinees, respectively, indicating that the variability in vaccine-induced responses is largely based on a common trait. Principal components analysis (PCA) identified the contribution of each marker to the variability of responses and defined this common trait. A significant association was observed in placebo recipients between MCP-1/CCL2 and MIP-1β/CCL4 (ρ = 0.66, P < 0.01) (table S4), indicating that this variance was not associated with immunization. MIP-1β/CCL4, contributing less to the variability, was thus excluded from the equation, without affecting Cronbach’s α (0.88 in all vaccinees). The single principal component with an Eigen value greater than 1 (3.38), explaining 68% of the variability of the day 1/day 0 chemokine/cytokine ratios, was retained. After normalization and standardization, the equation of the signature was defined by “0.266 × MCP-1STD + 0.265 × IL-1RaSTD + 0.211 × TNF-αSTD + 0.228 × IL-10STD + 0.242 × IL-6STD,” where each marker is expressed by its log10-transformed day 1/day 0 ratio and is standardized.
The score of this equation, henceforth, the Geneva rVSV-ZEBOV signature, was significantly influenced by vaccine dose (overall, P < 0.001): It was higher in HD versus LD vaccinees [mean, 0.59 (SD, ±0.80) versus −0.61 (SD, ±0.81); P < 0.001] and lowest in placebo recipients [mean, −1.12 (SD, ±0.16); P < 0.001 versus HD; fig. S2]. To define the influence of the dose on the signature’s equation, we reassessed it separately within the LD and HD vaccinees. Similar signatures were obtained (see Supplementary Materials), indicating that the equation defining the vaccine signature is dose-independent.
The signature is strongly associated with hematological, virological, and immunological outcomes
rVSV-ZEBOV immunization triggers transient, dose-dependent viremia and hematological changes (5, 6). We observed strong positive associations between the rVSV-ZEBOV signature and peak (ρ = 0.68, P < 0.001), day 1 (ρ = 0.64, P < 0.001), and day 3 (ρ = 0.61, P < 0.001) viremia. In contrast, strong negative associations were identified with day 1 lymphopenia (ρ = −0.79, P < 0.001) and thrombocytopenia (ρ = −0.55, P < 0.001) and day 3 thrombocytopenia (ρ = −0.49, P < 0.001) and neutropenia (ρ = −0.42, P < 0.001). Thus, higher viral loads were associated with higher signature scores, which, in turn, correlated with more pronounced cytopenia. Multivariate analyses indicated that the signature’s influences on biological outcomes largely persisted independent of vaccine dose (Table 2).
Table 2. Multivariate analyses of the determinants of biological outcomes in Geneva.
Multivariate analyses were conducted to assess the association between the signature and biological outcomes adjusting for the vaccine dose. Linear regression models were used. For the factor dose, placebo was the reference category (denoted “Ref”): The regression coefficients for LD and HD represent the mean difference in the biological outcome compared with placebo, adjusted for the signature. The P value parallel to the reference category is the P value for testing the global association between the vaccine dose (all doses) and the outcomes. The regression coefficients for the signature represent the mean increase in biological outcomes for an increase in signature of one unit, adjusted for the vaccine dose. For some biological outcomes (peak VSV and VSV viremia at day 1 and at day 3), a log10 transformation was applied to fulfill the assumptions of a linear regression model. n denotes the number of volunteers included in the analyses (that is, those without any missing data for the outcomes and the signature).
| Outcomes | Predictors | Estimate (SE) | P | |
|---|---|---|---|---|
|
Peak VSV (log10) (n = 112) |
Dose | Placebo | Ref | <0.001 |
| Low | 0.04 (0.16) | 0.78 | ||
| High | 1.03 (0.19) | <0.001 | ||
| Signature | Per unit | 0.20 (0.07) | 0.003 | |
| VSV viremia (log10) | ||||
| Day 1 (n = 113) | Dose | Placebo | Ref | <0.001 |
| Low | 0.01 (0.17) | 0.95 | ||
| High | 0.97 (0.21) | <0.001 | ||
| Signature | Per unit | 0.15 (0.07) | 0.035 | |
| Day 3 (n = 112) | Dose | Placebo | Ref | <0.001 |
| Low | −0.01 (0.17) | 0.97 | ||
| High | 0.68 (0.21) | 0.001 | ||
| Signature | Per unit | 0.24 (0.07) | 0.001 | |
| Lymphopenia | ||||
| Day 1 (n = 113) | Dose | Placebo | Ref | <0.001 |
| Low | −0.12 (0.06) | 0.060 | ||
| High | −0.26 (0.08) | <0.001 | ||
| Signature | Per unit | −0.19 (0.03) | <0.001 | |
| Day 3 (n = 113) | Dose | Placebo | Ref | 0.014 |
| Low | −0.14 (0.07) | 0.043 | ||
| High | −0.01 (0.08) | 0.88 | ||
| Signature | Per unit | −0.04 (0.03) | 0.13 | |
| Thrombocytopenia | ||||
| Day 1 (n = 113) | Dose | Placebo | Ref | 0.55 |
| Low | 0.00 (0.02) | 0.92 | ||
| High | −0.02 (0.03) | 0.53 | ||
| Signature | Per unit | −0.04 (0.01) | <0.001 | |
| Day 3 (n = 113) | Dose | Placebo | Ref | 0.022 |
| Low | −0.07 (0.02) | 0.008 | ||
| High | −0.08 (0.03) | 0.013 | ||
| Signature | Per unit | −0.03 (0.01) | 0.003 | |
| Monocytosis | ||||
| Day 1 (n = 113) | Dose | Placebo | Ref | 0.14 |
| Low | 0.23 (0.13) | 0.088 | ||
| High | 0.32 (0.16) | 0.049 | ||
| Signature | Per unit | −0.04 (0.06) | 0.48 | |
| Day 3 (n = 113) | Dose | Placebo | Ref | 0.048 |
| Low | 0.32 (0.14) | 0.025 | ||
| High | 0.18 (0.17) | 0.30 | ||
| Signature | Per unit | −0.04 (0.06) | 0.45 | |
| Neutropenia | ||||
| Day 1 (n = 112) | Dose | Placebo | Ref | 0.59 |
| Low | 0.10 (0.13) | 0.44 | ||
| High | 0.02 (0.15) | 0.88 | ||
| Signature | Per unit | 0.15 (0.05) | 0.006 | |
| Day 3 (n = 111) | Dose | Placebo | Ref | <0.001 |
| Low | −0.26 (0.07) | <0.001 | ||
| High | −0.34 (0.09) | <0.001 | ||
| Signature | Per unit | −0.05 (0.03) | 0.10 | |
Given that innate responses are considered to affect subsequent adaptive responses to vaccination, we asked whether this day 1 signature correlated with anti–EBOV-GP immunoglobulin G (IgG) antibody titers. Significant positive associations were observed at 1 month (ρ = 0.44, P < 0.001) and 6 months (ρ = 0.45, P < 0.001) after immunization (fig. S3) (5, 6). In contrast to biological outcomes, associations with antibody responses essentially reflected the influence of the vaccine dose.
Signature scores correlate with the type and severity of vaccine-induced AEs
We next assessed associations between the rVSV-ZEBOV signature and clinical characteristics. We found lower scores in females (−0.33 versus 0.04, P = 0.047) and no association with age (ρ = 0.12, P = 0.21). As reported, AEs occurred in most (97%) HD vaccinees, with dose-dependent incidence and severity (5, 6). Strongly positive associations (Table 2) were observed between the score of the chemokine/cytokine signature and injection site pain and swelling, fever, myalgia, chills, and even headaches but not early-onset arthralgia, fatigue, or other AEs (Table 3, table S5, and Fig. 2C). Scores were significantly higher in subjects with “any” (at least one) AEs. These AEs correlatedmost strongly with changes in the GMCs of the MCP-1/CCL2 and MIP-1β/CCL4 chemokines and in IL-1Ra and IL-6 (Table 3). TNF-α elevations were seen only in subjects with myalgia or “any AEs.” Thus, local and systemic inflammatory rVSV-ZEBOV–triggered AEs presumably reflect the recruitment and activation of specific cells, predominantly monocytes, given the role of their chemokines and products.
Table 3. Associations between the signature, chemokines/cytokines, and early AEs in Geneva participants.
AEs were included if reported within 14 days of VSV-ZEBOV immunization.
| Markers* | Local pain (n = 53) | No local pain (n = 62) | Difference/ratio† | P‡§ |
|---|---|---|---|---|
| Signature | 0.27 (0.00 to 0.54) | −0.46 (−0.68 to −0.23) | 0.73 (0.37 to 1.08) | 0.001 |
| IL-1Ra | 6.89 (4.98 to 9.54) | 2.21 (1.64 to 2.99) | 3.11 (1.99 to 4.87) | <0.001 |
| IL-6 | 7.83 (4.17 to 14.69) | 2.31 (1.44 to 3.69) | 3.39 (1.53 to 7.51) | 0.003 |
| TNF-α | 2.61 (1.58 to 4.32) | 1.84 (1.17 to 2.91) | 1.42 (0.71 to 2.82) | 0.32 |
| IL-10 | 4.73 (2.71 to 8.27) | 2.17 (1.38 to 3.41) | 2.18 (1.05 to 4.52) | 0.036 |
| MCP-1 | 2.75 (2.27 to 3.34) | 1.50 (1.29 to 1.75) | 1.83 (1.43 to 2.36) | <0.001 |
| MIP-1β | 2.03 (1.76 to 2.35) | 1.36 (1.18 to 1.56) | 1.50 (1.22 to 1.83) | 0.001 |
| Objective fever (n = 14) | No objective fever (n = 101) | |||
| Signature | 0.67 (0.25 to 1.09) | −0.24 (−0.43 to −0.05) | 0.91 (0.41 to 1.40) | 0.001 |
| IL-1Ra | 10.60 (5.77 to 19.46) | 3.19 (2.48 to 4.09) | 3.33 (1.64 to 6.74) | 0.002 |
| IL-6 | 14.06 (4.14 to 47.71) | 3.36 (2.23 to 5.06) | 4.19 (1.04 to 13.88) | 0.045 |
| TNF-α | 3.21 (1.40 to 7.35) | 2.04 (1.41 to 2.95) | 1.57 (0.60 to 4.15) | 0.34 |
| IL-10 | 13.29 (4.66 to 37.96) | 2.51 (1.74 to 3.62) | 5.30 (1.60 to 17.60) | 0.009 |
| MCP-1 | 3.59 (2.56 to 5.04) | 1.81 (1.58 to 2.08) | 1.98 (1.34 to 2.93) | 0.002 |
| MIP-1β | 2.10 (1.61 to 2.73) | 1.57 (1.40 to 1.76) | 1.33 (0.98 to 1.81) | 0.066 |
| Subjective fever (n = 49) | No subjective fever (n = 66) | |||
| Signature | 0.31 (−0.02 to 0.63) | −0.45 (−0.63 to −0.27) | 0.76 (0.38 to 1.13) | <0.001 |
| IL-1Ra | 7.15 (4.92 to 10.39) | 2.27 (1.74 to 2.96) | 3.15 (1.98 to 5.01) | <0.001 |
| IL-6 | 8.34 (3.90 to 17.83) | 2.33 (1.63 to 3.33) | 3.57 (1.52 to 8.40) | 0.004 |
| TNF-α | 3.28 (1.71 to 6.28) | 1.58 (1.14 to 2.19) | 2.07 (0.99 to 4.34) | 0.053 |
| IL-10 | 4.73 (2.39 to 9.36) | 2.25 (1.57 to 3.21) | 2.11 (0.96 to 4.61) | 0.062 |
| MCP-1 | 2.72 (2.23 to 3.32) | 1.56 (1.33 to 1.82) | 1.75 (1.35 to 2.26) | <0.001 |
| MIP-1β | 1.95 (1.64 to 2.32) | 1.43 (1.26 to 1.62) | 1.37 (1.10 to 1.70) | 0.048 |
| Myalgia (n = 57) | No myalgia (n = 58) | |||
| Signature | 0.18 (−0.11 to 0.47) | −0.43 (−0.63 to −0.24) | 0.61 (0.25 to 0.97) | 0.001 |
| IL-1Ra | 5.29 (3.68 to 7.61) | 2.60 (1.93 to 3.51) | 2.03 (1.26 to 3.27) | 0.004 |
| IL-6 | 7.19 (3.73 to 13.85) | 2.26 (1.50 to 3.39) | 3.19 (1.46 to 6.97) | 0.004 |
| TNF-α | 3.54 (1.95 to 6.45) | 1.32 (1.01 to 1.74) | 2.68 (1.37 to 5.22) | 0.004 |
| IL-10 | 4.25 (2.33 to 7.75) | 2.25 (1.52 to 3.32) | 1.89 (0.92 to 3.90) | 0.085 |
| MCP-1 | 2.43 (2.01 to 2.94) | 1.61 (1.35 to 1.91) | 1.51 (1.17 to 1.96) | 0.002 |
| MIP-1β | 1.86 (1.59 to 2.17) | 1.43 (1.25 to 1.64) | 1.30 (1.05 to 1.60) | 0.015 |
| Chills (n = 44) | No chills (n = 71) | |||
| Signature | 0.20 (−0.12 to 0.51) | −0.32 (−0.54 to −0.11) | 0.52 (0.12 to 0.91) | 0.011 |
| IL-1Ra | 5.92 (3.93 to 8.92) | 2.80 (2.11 to 3.72) | 2.12 (1.28 to 3.51) | 0.004 |
| IL-6 | 6.22 (3.15 to 12.27) | 3.09 (1.91 to 5.01) | 2.01 (0.86 to 4.69) | 0.104 |
| TNF-α | 2.65 (1.46 to 4.79) | 1.91 (1.27 to 2.88) | 1.39 (0.67 to 2.88) | 0.38 |
| IL-10 | 5.18 (2.62 to 10.24) | 2.27 (1.53 to 3.36) | 2.28 (1.03 to 5.09) | 0.043 |
| MCP-1 | 2.55 (2.03 to 3.19) | 1.70 (1.45 to 1.98) | 1.50 (1.14 to 1.98) | 0.005 |
| MIP-1β | 1.87 (1.59 to 2.19) | 1.50 (1.31 to 1.72) | 1.24 (1.00 to 1.54) | 0.047 |
| Arthralgia (n = 17) | No arthralgia (n = 98) | |||
| Signature | −0.10 (−0.44 to 0.24) | −0.13 (−0.34 to 0.08) | 0.03 (−0.39 to 0.45) | 0.89 |
| IL-1Ra | 3.96 (2.15 to 7.30) | 3.65 (2.80 to 4.76) | 1.09 (0.54 to 2.19) | 0.81 |
| IL-6 | 4.30 (1.80 to 10.28) | 3.96 (2.54 to 6.17) | 1.09 (0.39 to 3.04) | 0.87 |
| TNF-α | 1.91 (1.15 to 3.16) | 2.21 (1.50 to 3.25) | 0.86 (0.45 to 1.67) | 0.66 |
| IL-10 | 2.68 (1.66 to 4.33) | 3.16 (2.09 to 4.79) | 0.85 (0.44 to 1.63) | 0.61 |
| MCP-1 | 2.15 (1.67 to 2.76) | 1.94 (1.67 to 2.26) | 1.11 (0.82 to 1.50) | 0.50 |
| MIP-1β | 1.62 (1.35 to 1.94) | 1.63 (1.45 to 1.84) | 0.99 (0.79 to 1.24) | 0.93 |
| Headache (n = 52) | No headache (n = 63) | |||
| Signature | 0.12 (−0.20 to 0.44) | −0.33 (−0.53 to −0.14) | 0.45 (0.07 to 0.84) | 0.021 |
| IL-1Ra | 5.56 (3.82 to 8.09) | 2.64 (1.97 to 3.55) | 2.10 (1.30 to 2.41) | 0.003 |
| IL-6 | 8.36 (4.18 to 16.73) | 2.19 (1.48 to 3.24) | 3.82 (1.70 to 8.58) | 0.002 |
| TNF-α | 3.05 (1.54 to 6.02) | 1.62 (1.27 to 2.07) | 1.88 (0.90 to 3.93) | 0.092 |
| IL-10 | 3.06 (1.67 to 5.64) | 3.10 (2.02 to 4.76) | 0.99 (0.46 to 2.10) | 0.98 |
| MCP-1 | 2.23 (1.80 to 2.77) | 1.78 (1.51 to 2.10) | 1.25 (0.95 to 1.65) | 0.11 |
| MIP-1β | 1.71 (1.43 to 2.04) | 1.57 (1.38 to 1.78) | 1.09 (0.87 to 1.36) | 0.46 |
| Fatigue (n = 69) | No fatigue (n = 46) | |||
| Signature | −0.07 (−0.34 to 0.19) | −0.21 (−0.45 to 0.03) | 0.14 (−0.22 to 0.50) | 0.45 |
| IL-1Ra | 3.88 (2.81 to 5.37) | 3.43 (2.38 to 4.96) | 1.13 (0.69 to 1.86) | 0.62 |
| IL-6 | 5.27 (3.00 to 9.26) | 2.65 (1.60 to 4.38) | 1.99 (0.93 to 4.27) | 0.078 |
| TNF-α | 2.41 (1.43 to 4.08) | 1.82 (1.35 to 2.46) | 1.32 (0.72 to 2.45) | 0.37 |
| IL-10 | 2.81 (1.71 to 4.61) | 3.55 (2.14 to 5.90) | 0.79 (0.39 to 1.62) | 0.52 |
| MCP-1 | 2.04 (1.70 to 2.45) | 1.87 (1.54 to 2.28) | 1.09 (0.83 to 1.43) | 0.53 |
| MIP-1β | 1.70 (1.47 to 1.96) | 1.53 (1.31 to 1.78) | 1.11 (0.90 to 1.38) | 0.33 |
| Any AEs (n = 106) | No AEs (n = 9) | |||
| Signature | −0.08 (−0.27 to 0.12) | −0.70 (−1.07 to −0.34) | 0.62 (0.17 to 1.08) | 0.011 |
| IL-1Ra | 3.93 (3.04 to 5.08) | 1.83 (1.11 to 2.99) | 2.15 (1.17 to 3.97) | 0.018 |
| IL-6 | 4.46 (2.92 to 6.80) | 1.17 (0.60 to 2.26) | 3.83 (1.64 to 8.94) | 0.004 |
| TNF-α | 2.32 (1.61 to 3.33) | 0.94 (0.69 to 1.28) | 2.47 (1.51 to 4.04) | 0.001 |
| IL-10 | 3.27 (2.22 to 4.80) | 1.59 (0.86 to 2.95) | 2.05 (0.93 to 4.50) | 0.071 |
| MCP-1 | 2.03 (1.76 to 2.34) | 1.42 (0.99 to 2.04) | 1.43 (0.92 to 2.22) | 0.098 |
| MIP-1β | 1.67 (1.49 to 1.87) | 1.25 (1.00 to 1.57) | 1.33 (1.00 to 1.76) | 0.048 |
Mean values (95% CI).
Mean difference (signature) and mean ratios (chemokines/cytokines) between participants with and without specific AEs.
Signature: t tests were conducted to compare the mean signature in participants with or without specific AEs.
Chemokines/cytokines: P value for the comparison of the day 1/day 0 ratio of plasma markers between participants with and without each AE. t tests were conducted on log10-transformed data.
The standardized grading of the severity of rVSV-ZEBOV–induced AEs (24) identified 63 vaccinees (62%) with at least one grade ≥2 AE. The scores correlated with the severity of subjective fever (P < 0.001), myalgia (P = 0.008), chills (P = 0.003), and any grade ≥2 AEs (P = 0.031) (Fig. 2C and table S6).
Although the signature correlated independently with vaccine dose and biological/clinical outcomes (Table 2), multivariate analyses indicated that reactogenicity was jointly influenced by the dose and the signature (Table 4). As an example, restricting analyses to HD vaccinees identified significantly lower IL-10 responses in subjects with early-onset arthralgia (table S7), a finding not identified when including all vaccinees (Table 2).
Table 4. Multivariate analyses of the determinants of clinical outcomes in Geneva vaccinees (n = 102).
Multivariate analyses were conducted to assess the association between the signature and AEs adjusting for the vaccine dose. Logistic regression models were used. The reported adjusted odds ratios (ORs) capture the increase in risk of an AE compared with the reference category (denoted “Ref”).
| AEs (outcome) | Predictors | Adjusted OR (95% CI) | P | |
|---|---|---|---|---|
| Pain | Dose | Low | Ref | |
| High | 13.98 (4.01–48.72) | <0.001 | ||
| Signature | <0 | Ref | ||
| ≥0 | 1.05 (0.30–3.64) | 0.94 | ||
| Objective fever | Dose | Low | Ref | |
| High | 8.42 (0.84–84.42) | 0.070 | ||
| Signature | <0 | Ref | ||
| ≥0 | 3.00 (0.51–17.54) | 0.22 | ||
| Subjective fever | Dose | Low | Ref | |
| High | 3.36 (1.16–9.68) | 0.025 | ||
| Signature | <0 | Ref | ||
| ≥0 | 1.97 (0.69–5.66) | 0.21 | ||
| Myalgia | Dose | Low | Ref | |
| High | 2.36 (0.83–6.66) | 0.11 | ||
| Signature | <0 | Ref | ||
| ≥0 | 1.60 (0.56–4.54) | 0.38 | ||
| Arthralgia | Dose | Low | Ref | |
| High | 0.94 (0.23–3.88) | 0.94 | ||
| Signature | <0 | Ref | ||
| ≥0 | 1.62 (0.39–6.64) | 0.51 | ||
| Chills | Dose | Low | Ref | |
| High | 3.84 (1.25–11.74) | 0.018 | ||
| Signature | <0 | Ref | ||
| ≥0 | 0.73 (0.24–2.20) | 0.57 | ||
| Headache | Dose | Low | Ref | |
| High | 1.87 (0.67–5.28) | 0.23 | ||
| Signature | <0 | Ref | ||
| ≥0 | 1.78 (0.63–5.00) | 0.28 | ||
| Fatigue | Dose | Low | Ref | |
| High | 1.11 (0.37–3.29) | 0.85 | ||
| Signature | <0 | Ref | ||
| ≥0 | 0.55 (0.18–1.61) | 0.27 |
HD vaccinees with subsequent arthritis had lower day 1 signature scores
We previously reported viral arthritis at similar frequencies (20 to 25%) in Geneva HD and LD vaccinees, identifying age as a determinant only in LD vaccinees and suggesting that distinct mechanisms were at play (6). We thus sought to investigate associations between the signature and viral arthritis in each dose group. Similar baseline GMCs were observed in subjects with or without subsequent arthritis. In HD vaccinees who later developed arthritis, the day 1 score was significantly lower than in their counterparts (Table 5). This reflected significantly lower IL-1Ra, IL-6, TNF-α, MCP-1/CCL2, and MIP-1β/CCL4 responses. In contrast, LD vaccinees who later developed arthritis had a higher composite score but similar GMCs of cytokines/chemokines as vaccinees without arthritis. Thus, the association between rVSV-ZEBOV–induced innate responses and subsequent arthritis is dose-dependent.
Table 5. Innate responses (day 1/day 0 ratios of geometric means) and association with rVSV-ZEBOV arthritis.
Analyses included all vaccinees (n = 102).
| HD group | Arthritis (n = 11) | No arthritis (n = 40) | P* | Difference of ratios (95% CI)† |
|---|---|---|---|---|
| Signature | −0.16 (−0.12 to 0.44) | 0.70 (0.45 to 0.96) | 0.009 | −0.54 (−0.94 to −0.14) |
| IL-1Ra | 6.23 (4.27 to 9.10) | 12.28 (9.15 to 16.47) | 0.011 | 0.51 (0.31 to 0.84) |
| IL-6 | 4.37 (1.44 to 13.25) | 18.37 (9.14 to 36.89) | 0.045 | 0.24 (0.06 to 0.97) |
| TNF-α | 2.01 (1.16 to 3.47) | 4.80 (2.73 to 8.45) | 0.037 | 0.42 (0.19 to 0.95) |
| MCP-1 | 2.59 (2.20 to 3.06) | 3.59 (2.99 to 4.33) | 0.014 | 0.72 (0.56 to 0.93) |
| MIP-1β | 1.91 (1.70 to 2.18) | 2.44 (2.12 to 2.80) | 0.017 | 0.78 (0.64 to 0.96) |
| IL-10 | 6.03 (2.09 to 17.44) | 7.40 (4.27 to 12.82) | 0.74 | 0.82 (0.21 to 2.98) |
| LD group | Arthritis (n = 13) | No arthritis (n = 38) | P value* | Difference of ratios (95% CI)† |
| Signature | −0.19 (−0.62 to 0.25) | −0.75 (−1.00 to −0.50) | 0.042 | 0.56 (0.02 to 1.10) |
| IL-1Ra | 2.43 (1.47 to 4.03) | 1.58 (1.17 to 2.14) | 0.18 | 1.54 (0.81 to 2.946) |
| IL-6 | 4.25 (1.35 to 13.39) | 1.37 (0.93 to 2.02) | 0.099 | 3.11 (0.78 to 12.38) |
| TNF-α | 2.68 (0.86 to 8.34) | 1.03 (0.56 to 1.89) | 0.18 | 2.60 (0.62 to 10.87) |
| MCP-1 | 1.67 (1.37 to 2.04) | 1.33 (1.11 to 1.59) | 0.12 | 1.26 (0.94 to 1.68) |
| MIP-1β | 1.56 (1.23 to 1.97) | 1.23 (1.02 to 1.48) | 0.14 | 1.27 (0.92 to 1.75) |
| IL-10 | 5.28 (1.78 to 15.63) | 1.36 (0.75 to 2.49) | 0.053 | 3.87 (0.98 to 15.34) |
t test on the signature and on log10-transformed ratios for cytokines/chemokines.
Mean difference in signature or ratios of the geometric mean ratio (cytokines/chemokines) in vaccinees with or without arthritis.
The plasma signature of rVSV-ZEBOV likely originates outside the blood compartment
We next used the dual-color reverse transcriptase multiplex ligation-dependent probe amplification (dcRT-MLPA) assay (25) to assess changes in mRNA expression of the signature markers in whole-blood RNA samples. Baseline gene expression occasionally differed between groups (Table 6), perhaps due to successive rather than simultaneous recruitment of HD and LD vaccinees, but was compensated for by using the day 1/day 0 ratio. Although significant up-regulation of MCP-1/CCL2 and IL-1Ra/IL-1Rn transcripts was observed in the blood, the expression of MIP-1β/CCL4, TNF-α, IL-6, and IL-10 remained unexpectedly unchanged (Table 6, table S8, and fig. S4). Thus, the plasma vaccine signature likely reflects vaccine-induced innate responses occurring largely outside of the blood compartment.
Table 6. Relative gene expression levels (log2) of markers comprising the plasma signature in rVSV-ZEBOV vaccinees or placebo recipients of Geneva.
Analyses included all vaccinees (n = 102) and placebo controls (n = 13).
| Placebo | LD (3 × 105 pfu) | HD (107 or 5 × 107 pfu) | P values for comparison between dose groups ANOVA with post hoc Scheffe | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Days | n | GMC* (95% CI) | n | GMC* (95% CI) | n | GMC* (95% CI) | P versus LD | P versus HD | LD versus HD | |
| IL-1RN (IL-1Rα) | 0 | 13 | 13.59 (13.08–14.09) | 51 | 14.17 (13.99–14.36) | 51 | 12.39 (12.02–12.75) | 0.18 | 0.001 | <0.001 |
| 1 | 13 | 13.93 (13.39–14.48) | 51 | 15.04 (14.71–15.37) | 51 | 15.81 (15.64–15.98) | 0.001 | <0.001 | <0.001 | |
| IL-6 | 0 | 13 | 8.61 (7.74–9.48) | 51 | 8.33 (8.14–8.51) | 51 | 7.95 (7.83–8.07) | 0.43 | 0.012 | 0.028 |
| 1 | 13 | 8.06 (7.68–8.45) | 51 | 7.97 (7.76–8.17) | 51 | 7.73 (7.64–7.81) | 0.87 | 0.17 | 0.11 | |
| TNF-α | 0 | 13 | 11.20 (11.08–11.31) | 51 | 11.16 (11.08–11.25) | 51 | 11.06 (10.97–11.15) | 0.93 | 0.31 | 0.19 |
| 1 | 13 | 11.22 (11.02–11.42) | 51 | 11.26 (11.18–11.34) | 51 | 11.49 (11.37–11.60) | 0.95 | 0.056 | 0.006 | |
| IL-10 | 0 | 13 | 7.83 (7.61–8.04) | 51 | 7.76 (7.67–7.85) | 51 | 7.89 (7.66–8.15) | 0.95 | 0.96 | 0.62 |
| 1 | 13 | 7.66 (7.61–7.71) | 51 | 7.74 (7.64–7.84) | 51 | 7.94 (7.72–8.16) | 0.91 | 0.30 | 0.22 | |
| MCP-1 | 0 | 13 | 7.81 (7.59–8.03) | 51 | 7.91 (7.78–8.04) | 51 | 7.65 (7.63–7.66) | 0.64 | 0.31 | 0.001 |
| 1 | 13 | 7.78 (7.64–7.92) | 51 | 10.52 (10.13–10.91) | 51 | 11.66 (11.44–11.89) | <0.001 | <0.001 | <0.001 | |
| MIP-1β | 0 | 13 | 10.97 (10.67–11.27) | 51 | 10.65 (10.44–10.87) | 51 | 10.99 (10.89–11.09) | 0.21 | >0.99 | 0.017 |
| 1 | 13 | 10.95 (10.62–11.28) | 51 | 10.88 (10.76–11.01) | 51 | 11.04 (10.92–11.16) | 0.90 | 0.79 | 0.20 | |
We report the GMC of log2-transformed relative expression levels in whole-blood samples normalized against glyceraldehyde-3-phosphate dehydrogenase (GAPDH).
The rVSV-ZEBOV Geneva and Lambaréné signatures correlate strongly
Vaccine responses may be modulated by genetic and environmental influences. We thus asked whether innate responses to rVSV-ZEBOV differ between the Geneva cohort and a distinct cohort from an African setting with potential Ebola virus exposure (26, 27). Cytokines/chemokines were quantified in cryopreserved plasma samples of 75 subjects immunized in Lambaréné (fig. S1). Up-regulated markers included the same chemokines/cytokines that defined the Geneva signature, peaking on day 1 (Figs. 1B and 2, A and B, and table S9). At baseline, higher TNF-α (15-fold), IL-10 (12-fold), and MIP-1β/CCL4 (1.7-fold) concentrations were observed in Lambaréné than in Geneva, whereas MCP-1/CCL2 levels were twofold higher in Geneva (table S10). Nevertheless, similar vaccine responses were observed in both sites, except for TNF-α and MIP-1β/CCL4, whose higher baseline concentrations blunted responses in Lambaréné (Fig. 1B and tables S11 to S13).
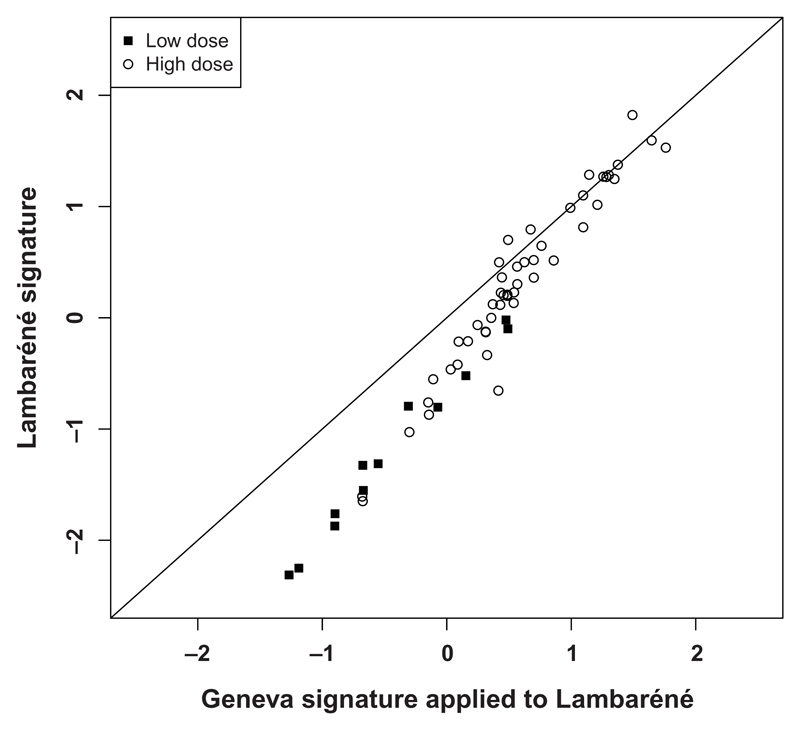
To define whether the Geneva signature could predict rVSV-ZEBOV responses elicited elsewhere, we applied an independent PCA to the Lambaréné data (see Materials and Methods). The main component explained 48.4% of the variability of responses, included the same markers, and was translated by a similar signature (0.242 × MCP-1STD + 0.344 × IL-1RaSTD + 0.247 × TNF-αSTD + 0.340 × IL-10STD + 0.244 × IL-6STD). Its mean scores differed significantly between LD (−1.22 ± 0.78) and HD (0.30 ± 0.80, P < 0.001) vaccinees (fig. S5). We next asked whether applying the Geneva signature to the Lambaréné data (fig. S6) would generate similar results. Both signatures correlated strongly (ρ = 0.97, P < 0.001; Fig. 3) but are not equal. In Lambaréné vaccinees, PCA identified an additional component including TNF-α and IL-10, the two cytokines with high baseline concentrations, explaining a further 23.5% of the variability of responses. The Lambaréné signature can be derived from the Geneva signature and applied to the Lambaréné data (Geneva/Lambaréné signature) by the following equation: Lambaréné signature = −0.53 + 1.40 × Geneva/Lambaréné signature.
Fig. 3. Correlation between the Lambaréné signature and the Geneva signature applied to Lambaréné vaccinees.
Representation of the signature (in arbitrary units) calculated directly in vaccine recipients from Lambaréné (vertical axis) and when applying the Geneva signature to the Lambaréné data (horizontal axis).
When the Geneva signature was applied to the Lambaréné data, significant positive and negative associations were again observed with day 1 viremia (ρ = 0.36, P = 0.002) and lymphopenia (ρ = −0.50, P < 0.001) but not thrombocytopenia (table S14). We found significant associations with the occurrence (P = 0.001; Table 7 and Fig. 2C) and severity (P = 0.001; Fig. 2D and table S14) of any AEs and of injection site pain (P = 0.003; Table 7). For other AEs, scores and chemokine/cytokine GMCs were higher in affected subjects but did not reach statistical significance. Similarly, responses did not significantly differentiate Lambaréné vaccinees with grade 1 or 2 AEs (Fig. 2D and table S14). Although significantly fewer Lambaréné HD vaccinees reported fever, chills, and myalgia (tables S15 and S16), similar day 1/day 0 cytokine/chemokine responses were observed in both sites (Tables 3 and 7). Baseline IL-10 concentrations, higher in Lambaréné, were not associated with reduced reactogenicity. The signature score of GenevaHDvaccinees with subsequent arthritis [0.16 (95% CI, 0.12 to 0.44)] was significantly lower than that of Lambaréné HD vaccinees [0.58 (95% CI, 0.42 to 0.74); P = 0.021; fig. S7], none of whom experienced arthritis.
Table 7. Associations between the signature, chemokines/cytokines and main vaccine-related AEs in Lambaréné vaccinees.
AEs were included if reported within 14 days of VSV-ZEBOV immunization. “Missing” indicates the number of subjects without available day 0 or day 1 plasma samples, that is, not included.
| Markers | Local pain (n = 42) | No local pain (n = 33) | Difference/ratio* | P†‡ |
|---|---|---|---|---|
| Missing | 10 | 5 | ||
| Signature | 0.65 (0.44 to 0.86) | 0.13 (−0.12 to 0.38) | 0.52 (0.18 to 0.85) | 0.003 |
| IL-1Ra | 11.53 (8.36 to 15.90) | 7.83 (5.14 to 11.94) | 1.47 (0.86 to 1.06) | 0.16 |
| IL-6 | 10.73 (4.66 to 24.71) | 3.71 (1.74 to 7.91) | 2.90 (0.92 to 9.16) | 0.070 |
| TNF-α | 1.43 (1.29 to 1.59) | 1.29 (1.08 to 1.53) | 1.12 (0.90 to 1.38) | 0.30 |
| IL-10 | 6.35 (3.63 to 11.12) | 2.34 (1.25 to 4.35) | 2.72 (1.16 to 6.39) | 0.022 |
| MCP-1 | 4.14 (3.39 to 5.06) | 2.47 (1.92 to 3.17) | 1.68 (1.21 to 2.32) | 0.003 |
| MIP-1β | 1.66 (1.42 to 1.94) | 1.22 (1.01 to 1.48) | 1.36 (1.05 to 1.75) | 0.020 |
| Markers | Objective fever (n = 7) | No objective fever (n = 68) | Difference/ratio* | P†‡ |
| Missing | 1 | 14 | ||
| Signature | 0.73 (0.20 to 1.27) | 0.33 (0.15 to 0.52) | 0.40 (−0.30 to 1.10) | 0.22 |
| IL-1Ra | 13.99 (6.87 to 28.46) | 8.97 (6.70 to 12.01) | 1.56 (0.61 to 3.96) | 0.30 |
| IL-6 | 16.03 (1.56 to 165.07) | 5.47 (3.05 to 9.79) | 2.93 (0.14 to 61.77) | 0.42 |
| TNF-α | 1.30 (1.02 to 1.64) | 1.36 (0.21 to 1.53) | 0.95 (0.70 to 1.30) | 0.73 |
| IL-10 | 7.12 (3.15 to 16.11) | 3.47 (2.15 to 5.58) | 2.05 (0.69 to 6.12) | 0.17 |
| MCP-1 | 4.09 (3.15 to 5.33) | 3.05 (2.53 to 3.69) | 1.34 (0.93 to 1.93) | 0.10 |
| MIP-1β | 1.39 (1.02 to 1.89) | 1.41 (1.22 to 1.63) | 0.99 (0.66 to 1.48) | 0.93 |
| Markers | Subjective fever (n = 18) | No subjective fever (n = 67) | Difference/ratio* | P†‡ |
| Missing | 1 | 14 | ||
| Signature | 0.54 (0.24 to 0.85) | 0.31 (0.09 to 0.52) | 0.23 (−0.15 to 0.62) | 0.22 |
| IL-1Ra | 11.73 (7.26 to 18.95) | 8.59 (6.18 to 11.93) | 1.37 (0.75 to 2.50) | 0.30 |
| IL-6 | 7.43 (2.74 to 20.14) | 5.62 (2.79 to 11.33) | 1.32 (0.37 to 4.68) | 0.66 |
| TNF-α | 1.44 (1.21 to 1.72) | 1.32 (1.16 to 1.50) | 1.09 (0.87 to 1.37) | 0.44 |
| IL-10 | 6.66 (2.93 to 15.12) | 2.96 81.78 to 4.92) | 2.25 (0.82 to 6.15) | 0.11 |
| MCP-1 | 3.39 (2.55 to 4.51) | 3.05 (2.46 to 3.79) | 1.11 (0.77 to 1.61) | 0.57 |
| MIP-1β | 1.54 (1.27 to 1.87) | 1.36 (1.15 to 1.61) | 1.13 (0.87 to 1.47) | 0.36 |
| Markers | Myalgia (n = 12) | No myalgia (n = 63) | Difference/ratio* | P†‡ |
| Missing | 0 | 15 | ||
| Signature | 0.55 (0.26 to 0.84) | 0.33 (0.12 to 0.54) | 0.22 (−0.16 to 0.59) | 0.24 |
| IL-1Ra | 11.17 (6.65 to 18.77) | 9.98 (6.55 to 12.32) | 1.24 (0.65 to 2.37) | 0.49 |
| IL-6 | 4.58 (2.41 to 8.73) | 6.53 (3.25 to 13.14) | 0.70 (0.26 to 1.87) | 0.47 |
| TNF-α | 1.38 (1.18 to 1.61) | 1.35 (1.19 to 1.53) | 1.03 (0.83 to 1.27) | 0.81 |
| IL-10 | 6.95 (2.65 to 18.25) | 3.19 (1.96 to 5.18) | 2.18 (0.68 to 6.98) | 0.18 |
| MCP-1 | 4.08 (2.99 to 5.56) | 2.95 (2.41 to 3.60) | 1.38 80.93 to 2.05) | 0.10 |
| MIP-1β | 1.46 (1.15 to 1.84) | 1.40 (1.20 to 1.63) | 1.04 (0.78 to 1.40) | 0.77 |
| Markers | Chills (n = 4) | No chills (n = 71) | Difference/ratio* | P†‡ |
| Missing | 0 | 15 | ||
| Signature | 0.86 (0.27 to 1.46) | 0.34 (0.16 to 0.52) | 0.52 (−0.40 to 1.45) | 0.18 |
| IL-1Ra | 23.42 (10.15 to 54.01) | 8.79 (6.65 to 11.62) | 2.67 (0.73 to 9.66) | 0.10 |
| IL-6 | 8.72 (0.91 to 83.93) | 5.93 (3.27 to 10.77) | 1.47 (0.04 to 50.97) | 0.77 |
| TNF-α | 1.68 (1.12 to 2.50) | 1.33 (1.19 to 1.49) | 1.26 (0.67 to 2.35) | 0.35 |
| IL-10 | 7.69 (2.88 to 20.58) | 3.54 (2.23 to 5.61) | 2.17 (0.50 to 9.55) | 0.23 |
| MCP-1 | 4.81 (2.93 to 7.90) | 3.05 (2.54 to 3.66) | 1.58 (0.74 to 3.37) | 0.17 |
| MIP-1β | 1.27 (0.79 to 2.03) | 1.42 (1.24 to 1.63) | 0.89 (0.43 to 1.86) | 0.68 |
| Markers | Any AEs (n = 45) | No AEs (n = 30) | Difference/ratio* | P†‡ |
| Missing | 5 | 10 | ||
| Signature | 0.59 (0.40 to 0.78) | −0.06 (−0.35 to 0.24) | 0.65 (0.28 to 1.01) | 0.001 |
| IL-1Ra | 11.82 (8.94 to 15.62) | 5.91 (3.40 to 10.29) | 2.00 (1.05 to 3.82) | 0.037 |
| IL-6 | 8.18 (4.24 to 15.79) | 3.37 (1.14 to 9.95) | 2.43 (0.65 to 9.03) | 0.18 |
| TNF-α | 1.40 (1.28 to 1.54) | 1.26 (0.98 to 1.64) | 1.11 (0.83 to 1.48) | 0.47 |
| IL-10 | 6.15 (3.87 to 9.78) | 1.37 (0.63 to 2.96) | 4.49 (1.76 to 11.45) | 0.003 |
| MCP-1 | 3.83 (3.19 to 4.60) | 2.12 (1.55 to 2.89) | 1.81 (1.24 to 2.63) | 0.003 |
| MIP-1β | 1.56 (1.36 to 1.79) | 1.16 (0.88 to 1.51) | 1.35 (0.98 to 1.85) | 0.064 |
We report the mean difference (signature) and mean ratios (chemokines/cytokines) between participants with and without specific AEs.
Signature: t tests were conducted to compare the signature in participants with or without specific AEs.
Chemokines/cytokines: P value for the comparison of the day 1/day 0 ratio of plasma markers between participants with and without each AE. t tests were conducted on log10-transformed data.
Discussion
Herein, we report a safety biosignature for rVSV-ZEBOV, identified in the plasma as a specific composition of interconnected chemokines/cytokines and validated in two distinct cohorts across various vaccine doses and continents. This signature reveals previously uncharacterized innate responses to rVSV-ZEBOV and provides insights into their contribution to the onset and severity of biological and clinical outcomes.
Among the 15 plasma markers selected for their potential involvement in responses to rVSV-ZEBOV (13–23), a subset of only six chemokines/cytokines differentiated placebo recipients from vaccinees and LD from HD vaccinees. In contrast to the predominantly lymphoid markers composing a recently identified H1N1 influenza vaccine signature (28), vaccine responses here are monocytic: The two chemokines are monocyte chemoattractants (29), and the induced pro- and anti-inflammatory cytokines are monocyte products (30–32). The finding that rVSV-ZEBOV preferentially elicits monocyte responses is not unexpected: Ebola virus targets monocytes/macrophages (19, 33), and we previously reported their dose-dependent activation by rVSV-ZEBOV (6). Despite the abundance of blood monocytes, the transcript expression of most of these chemokines/cytokines was not increased in blood cells. Although blood cells may contribute to the response by releasing proteins such as TNF-α and IL-6 from storage vesicles (34, 35), we postulate that this plasma signature is generated mostly in extravascular, vaccine-targeted cells and tissues. Responses to rVSV-ZEBOV, initiated at the site of injection and in the draining lymph nodes, might be orchestrated by type I IFNs: In mice, VSV RNA induces high-level production of IFN-α and IFN-β (36), known to trigger the six markers (37, 38). In humans, studies initially demonstrated that live vaccines such as yellow fever vaccine induce genes regulating virus innate sensing and type I IFN production (39, 40). Subsequently, type I IFN was confirmed as a pivotal marker of vaccine responses, even in response to inactivated influenza vaccines, in both adults (41) and young children (42). Further studies are required to characterize type I IFN responses to rVSV-ZEBOV, as well as the relative contribution of rVSV [which triggers reactogenicity (21) and transient lymphopenia in mice (43) and induces MCP-1/CCL2 production by human monocytes (20)] and EBOV-GP [which also triggers immune reactivity (14, 15, 44–50)]. These early innate responses likely play a critical role in the large, cross-reactive post-exposure efficacy observed in experimental animal models given rVSV-ZEBOV (51, 52). Their potential contribution to the early effectiveness observed in the Guinea ring immunization trial (53) is intriguing.
The signature score increased with VSV viremia yet independent of vaccine dose: Its positive association, persisting through day 3, suggests a minor and/or insufficient role in the control of primary VSV viremia. In contrast, negative dose-independent associations with lymphopenia, neutropenia, and thrombocytopenia point to protective effects against virus-induced cytopenia; whether innate responses reduce cell egress from the blood compartment and/or limit virus-mediated cell destruction remains unknown. Although innate immune responses set the stage for adaptive vaccine responses (42, 54, 55) and were associated with EBOV-GP antibody responses, these were associated with the dose rather than with plasma cytokines. It will be of significant interest to define whether (or not) gene expression analyses will reveal a direct influence of specific responses [such as type I IFN (36)] on antibody titers.
Vaccine reactogenicity has long been thought to reflect innate responses and inflammation. Local pain was associated with MCP-1/CCL2, MIP-1β/CCL4, IL-1Ra, and IL-6, likely reflecting monocyte recruitment and activation at the injection site. The same markers correlated with vaccine-induced fever and “flu-like symptoms,” a finding that again points to the preferential activation of monocytes/macrophages by rVSV-ZEBOV (19). IL-1Ra has anti-inflammatory properties but is produced as an acute-phase protein also reflecting monocyte activation (56). TNF-α was significantly elevated only in myalgia, and early-onset arthralgia did not coincide with myalgia but was associated with lower IL-10 responses, possibly reflecting the protective role of IL-10 in joint inflammation (57). These influences were jointly influenced by the vaccine dose, indicating associations between dose, innate responses, and reactogenicity. Comparing the signature of replicating and nonreplicating Ebola vaccine candidates (45–50) of rVSV with distinct envelope genes and of rVSV vectors with distinct viral properties could shed light on mechanisms underlying their relative safety profile and be used in various populations. The acute reactogenicity of rVSV-ZEBOV was transient and did not prevent its use in the field (53), and vaccine safety was subsequently confirmed in thousands of vaccinees (3). Thus, the levels of reactogenicity and attendant plasma chemokine and cytokine responses described here may also inform the development of standards and standardized templates assessing the risks and benefits of live virus vaccines (58).
The pathophysiology leading to the dissemination of rVSV-ZEBOV into the joints and to arthritis (5, 6) remained undefined; arthritis after wild-type VSV infection or rVSV vaccination with non–EBOV-GP inserts has not been reported (59, 60). Here, we demonstrate significantly lower scores and weaker day 1 innate responses in HD vaccinees who later developed arthritis. Although higher VSV viremia is associated with higher scores (Table 2), days 1 to 3 viral loads were similar in subjects with or without subsequent arthritis (6). This suggests that at a high viral inoculum, such as the current dose of 2 × 107 pfu, strong early innate responses do not reduce early VSV viremia but may contribute to limit the duration of viral replication or viral dissemination in peripheral tissues and subsequent enhanced risks of rVSV-ZEBOV–induced viral arthritis and dermatitis (5, 6). In contrast, in LD vaccinees who later developed arthritis, early cytokine/chemokine concentrations were similar, and the score was higher compared to that of their LD counterparts. This confirms that distinct mechanisms are at play in HD and LD vaccinees, where age is an independent factor (6). Further studies are thus needed to define the relative contribution of the magnitude and duration of immune responses, the relative role of monocytes in infection control versus viral dissemination, and the age-associated joint vulnerability (5, 6); these may then help to explain the markedly higher reporting rates of arthritis in Geneva than in Lambaréné.
Applying the Geneva signature to an African population at risk of Ebola exposure was key to its validation. In Lambaréné, rVSV-ZEBOV up-regulated the same markers, with similar responses despite distinct baseline TNF-α, IL-10, and MCP-1/CCL2 levels. The Geneva signature correlated highly with that of the independently derived Lambaréné signature.
The frequency of self-reported, vaccine-induced AEs is notably lower in African settings (61). We demonstrate here that this lower incidence (5) does not reflect weaker innate responses nor higher baseline concentrations of anti-inflammatory cytokines such as IL-10; innate responses were similar in European and African volunteers. The fact that the signature score of Geneva HD vaccinees with subsequent arthritis is significantly lower than that of Lambaréné vaccinees may provide a first explanation for the discrepant reporting of arthritis among centers (5). It also emphasizes the importance of assessing vaccine safety in the settings where they will be used.
Our study has limitations. Samples were collected in rapidly implemented clinical trials, resulting in some missing samples. Further, we preselected markers of potential interest to increase the likelihood of detection of vaccine-associated changes. Although this proved successful, additional biomarkers are likely to be identified and eventually refine this first plasma signature. The measure of 63 markers in an exposed health care worker identified the same and additional up-regulated markers (62), some of which were not confirmed in our controlled study. Finally, only Geneva participants were randomized; a few of them are open-label. Thus, some changes might have been overlooked by distinct baseline values.
Nonetheless, a vaccine signature’s independence from genetic and environmental influences could be demonstrated. This signature of the cytokine pattern underlying vaccine reactogenicity offers the first lead to understanding the pathophysiology of rVSV-ZEBOV, a complex chimeric vaccine with great potential against a deadly disease (53) but with reactogenicity at high doses. It strongly suggests the direct influence of innate responses on biological and clinical outcomes, including viral dissemination and arthritis. This prompts further work toward the use of early signatures to predict subsequent AEs and the reassessment of the strategy to use lower doses of rVSV-ZEBOV in subjects with weaker immunity, such as young children. It will also be useful in the development of distinct rVSV-EBOV constructs or of rVSV-based vaccines. The signature’s relevance extends beyond the rVSV-ZEBOV vaccine: It highlights the critical contribution of monocyte recruitment and activation to both immunogenicity and safety and the possibility that blood transcriptomics may fail to identify responses elicited outside the blood compartment.
Materials and Methods
The VEBCON Consortium
In August 2014, the Public Health Agency of Canada donated 800 vials of the rVSV-ZEBOV vaccine to WHO, which created VEBCON to initiate dose-ranging phase 1 clinical trials. These phase 1 trials were initiated in Germany (NCT02283099), Kenya (NCT02296983), Gabon (PACTR2014000089322), and Switzerland [a phase 1/2 RCT; NCT02287480] and were supported financially through WHO by grants from the Wellcome Trust Foundation, the Bill and Melinda Gates Foundation, and the German Center for Infection Research.
The VEBCON Consortium includes the following members: S. T. Agnandji (Centre de Recherches Médicales de Lambaréné, Gabon; Institut für Tropenmedizin, Universitätsklinikum Tübingen, Germany) and S. Krishna (St George’s University of London, U.K.; Institut für Tropenmedizin, Universität Tübingen, Germany; Centre de Recherches Médicales de Lambaréné Lambarene, Gabon); P. G. Kremsner and J. S. Brosnahan (Institut für Tropenmedizin, Universität Tübingen, Germany; Centre de Recherches Médicales de Lambaréné, Gabon); P. Bejon and P. Njuguna (Kenya Medical Research Institute, Kilifi, Kenya); M. M. Addo (University Medical Center Hamburg, Germany); S. Becker and V. Krähling (Institute of Virology, Marburg, Germany); C.-A. Siegrist and A. Huttner (Geneva University Hospitals); and P. Fast (WHO, Geneva, Switzerland).
The EBOVAC Consortium
The VSV-EBOVAC Consortium was constituted at the launch of the VSV-EBOVAC IMI2 project to include its core members. The VSV-EBOVAC Consortium includes the following members (in alphabetical order): S. T. Agnandji, R. Ahmed, J. Anderson, F. Auderset, L. Borgianni, J. Brosnahan, A. Ciabattini, O. Engler, M. C. Haks, G. Heppner, A. Gerlini, P. G. Kremsner, S. Leib, T. Monath, F. Ndungu, P. Njuguna, G. Pozzi, F. Santoro, and C.-A. Siegrist.
Study design, population, and key previous outcomes
We performed a prospective derivation and validation cohort study nested within the phase 1 Geneva (randomized) and Lambaréné (dose-escalation) trials (registration nos. NCT02287480 and PACTR201411000919191, respectively), whose biological and clinical outcomes have been reported elsewhere (5, 6). All participants with available plasma samples (n = 190) were included; these had received an LD [3 × 105 pfu, n = 71 (Geneva, 51; Lambaréné, 20)] or HD [Geneva, 1 × 107 or 5 × 107 pfu (n = 51); Lambaréné, 3 × 106 or 2 × 107 pfu (n = 55)] of the same batch of rVSV-ZEBOV or placebo (Geneva, n = 13) (fig. S1). Although vaccine reactogenicity, viremia, and ZEBOV-GP–specific IgG enzyme-linked immunosorbent assay antibody titers were dose-dependent in Geneva and Lambaréné, vaccine-induced arthritis was reported only in Geneva. In that RCT, age emerged as a risk factor for rVSV-ZEBOV arthritis in LD but not in HD vaccinees (6).
Multiplex analyses of chemokines and cytokines
The kinetics of 15 markers were quantified in cryopreserved plasma using a specifically designed 15-nucleotide oligomer multiplex (Fluorokine MAP Multiplex Human Cytokine Panel, R&D Systems) according to the supplier’s instructions (see Supplementary Materials). Briefly, beads conjugated to the analyte-specific capture antibodies, samples, standards, and controls were incubated at room temperature for 3 hours. Biotinylated detector antibodies and R-phycoerythrin–conjugated streptavidin (SAPE) were subsequently added. The mean fluorescence intensity of each analyte was read on the Bio-Plex 200 array reader (Bio-Rad Laboratories) using the Luminex xMAP Technology (Luminex Corporation). Sample concentrations were calculated using a five-parameter logistic regression curve (Bio-Plex Manager 6.0). Interassay variation coefficients were monitored using internal controls. These were below 15% for all. Values below each assay’s cutoff were arbitrarily valued at 0.01 pg/ml to use a single negative threshold for each marker. Alternatively, we used 50% of the minimal detection dose of each specific marker, as provided by the manufacturer, to display individual results in Fig. 1. The use of either method did not affect the definition of the signature.
Reverse transcriptase multiplex ligation-dependent probe amplification assay
Total RNA from PAXgene blood collection tubes was extracted using the PAXgene Blood RNA kit (BD Biosciences) including on-column deoxyribonuclease digestion according to the manufacturer’s protocol. RNA was quantified by a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Gene expression profiles of IL-1RN (IL-1Ra), IL-6, TNF-α, IL-10, MCP-1/CCL2, and MIP-1β/CCL4 were determined using dcRT-MLPA, as described previously (25, 63). Briefly, for each target-specific sequence, a specific reverse transcriptase primer was designed, located immediately downstream of the left- and right-hand half-probe target sequence. After reverse transcription of 125 ng of RNA using M-MLV reverse transcriptase (Promega), left- and right-hand half-probes were hybridized to the complementary DNA at 60°C overnight. Annealed half-probes were ligated and subsequently amplified by polymerase chain reaction (PCR) (33 cycles for 30 s at 95°C, 30 s at 58°C, and 60 s at 72°C, followed by 1 cycle for 20 min at 72°C). Primers and probes were from Sigma-Aldrich Chemie, and MLPA reagents were from MRC-Holland. PCR amplification products were diluted in a 1:10 ratio in Hi-Di formamide containing 400HD ROX size standard and analyzed on an Applied Biosystems 3730 capillary sequencer in GeneScan mode (BaseClear). Trace data were analyzed using the GeneMapper software package 5.0 (Applied Biosystems). Signals below the threshold value for noise cutoff in GeneMapper (log2-transformed peak area, ≤7.64) were assigned the threshold value for noise cutoff. Subsequently, results from target genes were normalized to the average signal of housekeeping gene GAPDH and assigned the threshold value if below 7.64.
Results from target genes were calculated relative to the average signal of reference gene GAPDH. After data normalization, signals below the assay cutoff (log2-transformed peak area, ≤7.64) were assigned the threshold value for noise cutoff.
Identification of the Geneva signature
Cronbach’s α was used to assess whether the variation of cytokines/chemokines up-regulated between days 0 and 1 measured a single trait. Postulating that day 1 changes were triggered by immunization, a PCA was conducted in Geneva participants to identify components explaining the variability in cytokines/chemokines. The day 1/day 0 ratios of the selected cytokines/chemokines were introduced into the model. Ratios were transformed (log10 function) to normalize data and were standardized so that the means and the SD equaled 0.
The Eigen values for the components were 3.38, 0.70, 0.53, 0.29, and 0.11. The single component with the Eigen value greater than 1 was therefore retained. This component explained 68% of the variability of the ratios. The equation of this principal component was as follows:
The ratio of cytokines/chemokines introduced in the equation was also standardized. This standardization involves the mean and SD of each ratio in the Geneva samples:
The SD of the signature in participants was 1.84 (and the mean was 0). The coefficient in the equation was then divided by 1.84 so that the SD of the signature equals 1. Thus, the final equation for the signature was as follows:
The Kaiser-Meyer-Olkin measure of adequacy (64) was assessed to check the adequacy of our model. The obtained values were 0.73 for IL-1Ra, 0.83 for IL-6, 0.80 for TNF-α, 0.84 for IL-10, and 0.71 for MCP-1; the overall measure of adequacy was 0.77. A measure between 0.70 and 0.79 is interpreted as “middling” and between 0.80 and 0.89 as “meritorious” (64). We concluded that our model was adequate. The signature was assessed in n = 113 participants because two participants had at least one cytokine missing.
Identification of the Lambaréné plasma signature
After the same approach used for the Geneva data, we conducted a PCA with the Lambaréné data. The ratio of the five selected cytokines/chemokines was transformed (log10 function) and standardized.
The Eigen values for the components were 2.42, 1.18, 0.77, 0.41, and 0.22. The two components with an Eigen value greater than 1 were retained. The first component explained 48% of the variability of ratios of cytokines/chemokines, whereas the second components explained 24% of the variability. The equations of these components were as follows:
The ratio of cytokines/chemokines introduced in the equations was also standardized. This standardization involved the mean and SD of each ratio in the Lambaréné samples:
The standardization is not the same as in the Geneva sample because the distribution of the ratios of cytokines/chemokines is different in Lambaréné and Geneva.
Interpreting the first components as the signature, the SD of the signature was 1.56 (and the mean was 0). The coefficient in the equation was then divided by 1.56 so that the SD of the signature equaled 1. Finally, the equation for the signature was as follows:
The Kaiser-Meyer-Olkin values were 0.60 for IL-1Ra, 0.71 for IL-6, 0.53 for TNF-α, 0.56 for IL-10, and 0.58 for MCP-1. The overall measure of adequacy was 0.59. A measure between 0.50 and 0.59 is interpreted as “miserable,” between 0.60 and 0.69 as “mediocre,” and between 0.70 and 0.79 as “middling.” The PCA model in the Lambaréné samples was thus less adequate than with the Geneva samples. However, this model identified a similar principal component to the signature determined with the Geneva samples and explaining by itself 48% of the variability in ratios of cytokines/chemokines. The signature was assessed in n = 60 vaccinees; in the remaining 15 vaccinees, at least one cytokine was missing.
Statistical analyses
Chemokines and cytokines were reported by vaccine dose and study day using GMCs (log10). GMCs were compared between independent groups using t tests or ANOVA (with Scheffe’s correction for multiplicity of tests and post hoc analyses) and over time using linear regression models with mixed effects to account for repeated measures. The association between the signature and biological outcomes/AEs was assessed using linear and logistic regression models with adjustment for the dose. The type I error level was 0.05, and all statistical tests were two-sided. Analyses were conducted in R 3.2.2 (R Foundation for Statistical Computing, version 2.15.2) and STATA 14.0 IC (StataCorp LP).
Supplementary Material
www.sciencetranslationalmedicine.org/cgi/content/full/9/385/eaaj1701/DC1
Acknowledgments
We thank D. Pejovski (WHO Collaborating Center for Vaccine Immunology, Geneva, Switzerland) for contribution to cytokine/chemokine data analysis; M. Dhawan (Leiden University Medical Center, Netherlands) for help in isolating RNA and dcRT-MLPA; M.-P. Kieny (WHO) for critical contribution to the organization and funding of VEBCON trials; O. Lapujade, V. Moorthy, B. Savarese (WHO), S. A. Dubey (Merck), S. Leib, and M. Strasser (Federal Office for Civil Protection, Spiez Laboratory, Switzerland) for support of the VSV-EBOVAC project; A. Ciabattini (Sclavo Vaccines Association) for graphical support; and the Clinical Research Center of the University Hospital and Faculty of Medicine, Geneva.
Funding: The VSV-EBOVAC project is supported by the IMI2 Joint Undertaking (www.imi.europa.eu) within the IMI Ebola+program launched in response to the EVD outbreak in Western Africa in 2014 (grant agreement no. 115842). This Joint Undertaking receives support from the European Union’s Horizon 2020 research and innovation program and from the European Federation of Pharmaceutical Industries and Associations. The phase 1 clinical trials in Geneva and Lambaréné were funded by the Wellcome Trust Foundation through a grant awarded to WHO. The Bill and Melinda Gates Foundation contributed to the funding of the Lambaréné trial and the coordination of the VEBCON Data Safety Monitoring Board. A grant from the Trafigura Foundation supported the work of the Center for Vaccinology of Geneva. The VSV-EBOVAC project is coordinated by the Sclavo Vaccines Association (www.sclavo.org), a nonprofit organization devoted to support research and development of human vaccines.
Footnotes
Author contributions: This work was carried out in collaboration between all authors. C.-A.S. had full access to all study data and takes responsibility for its integrity and for the accuracy of its analysis. All authors read and gave final approval of the version to be submitted and any revised version. Study concept and design: A.H., C.C., S.T.A., J.B., A.M.H., L.K., D.M., P.R.-L., VSV-EBOVAC, P.G.K., T.H.M.O., and C.-A.S. Acquisition, analysis, or interpretation of data: A.H., C.C., S.G., M.C.H., E.Q., C.M., S.T.A., J.B., J.-A.D., P.R.-L., P.G.K., T.H.M.O., and C.-A.S. Drafting of the manuscript: A.H., C.C., and C.-A.S. Critical revision of the manuscript for important intellectual content: A.H., C.C., M.C.H., S.T.A., J.B., A.M.H., L.K., D.M., T.M., VEBCON, VSV-EBOVAC, P.R.-L., P.G.K., T.H.M.O., and C.-A.S. Administrative, technical, or material support: A.H., C.C., S.G., M.C.H., E.Q., C.M., S.T.A., J.B., J.-A.D., P.R.-L., VEBCON, VSV-EBOVAC, P.G.K., T.H.M.O., and C.-A.S. Study supervision: P.G.K. (Lambaréné), T.H.M.O. (Leyden), and C.-A.S. (Geneva).
Competing interests: T.M. is an employee of NewLink Genetics Corporation. All other authors declare that they have no competing interests.
Data and materials availability: Data are available upon request.
References
- 1.Kanapathipillai R, Henao Restrepo AM, Fast P, Wood D, Dye C, Kieny M-P, Moorthy V. Ebola vaccine—An urgent international priority. N Engl J Med. 2014;371:2249–2251. doi: 10.1056/NEJMp1412166. [DOI] [PubMed] [Google Scholar]
- 2.Garbutt M, Liebscher R, Wahl-Jensen V, Jones S, Moller P, Wagner R, Volchkov V, Klenk H-D, Feldmann H, Stroher U. Properties of replication-competent vesicular stomatitis virus vectors expressing glycoproteins of filoviruses and arenaviruses. J Virol. 2004;78:5458–5465. doi: 10.1128/JVI.78.10.5458-5465.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Henao-Restrepo AM, Camacho A, Longini IM, Watson CH, Edmunds WJ, Egger M, Carroll MW, Dean NE, Diatta I, Doumbia M, Draguez B, et al. Efficacy and effectiveness of an rVSV-vectored vaccine in preventing Ebola virus disease: Final results from the Guinea ring vaccination, open-label, cluster-randomised trial (Ebola Ça Suffit!) Lancet. 2017;389:505–518. doi: 10.1016/S0140-6736(16)32621-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Regules JA, Beigel JH, Paolino KM, Voell J, Castellano AR, Muñoz P, Moon JE, Ruck RC, Bennett JW, Twomey PS, Gutiérrez RL, et al. A recombinant vesicular stomatitis virus ebola vaccine. N Engl J Med. 2015;376:330–341. doi: 10.1056/NEJMoa1414216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Agnandji ST, Huttner A, Zinser ME, Njuguna P, Dahlke C, Fernandes JF, Yerly S, Dayer J-A, Kraehling V, Kasonta R, Adegnika AA, et al. Phase 1 trials of rVSV ebola vaccine in Africa and Europe. N Engl J Med. 2016;374:1647–1660. doi: 10.1056/NEJMoa1502924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huttner A, Dayer J-A, Yerly S, Combescure C, Auderset F, Desmeules J, Eickmann M, Finckh A, Goncalves AR, Hooper JW, Kaya G, et al. The effect of dose on the safety and immunogenicity of the VSV Ebola candidate vaccine: A randomised double-blind, placebo-controlled phase 1/2 trial. Lancet Infect Dis. 2015;15:1156–1166. doi: 10.1016/S1473-3099(15)00154-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lichty BD, Power AT, Stojdl DF, Bell JC. Vesicular stomatitis virus: Re-inventing the bullet. Trends Mol Med. 2004;10:210–216. doi: 10.1016/j.molmed.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 8.Hastie E, Grdzelishvili VZ. Vesicular stomatitis virus as a flexible platform for oncolytic virotherapy against cancer. J Gen Virol. 2012;93:2529–2545. doi: 10.1099/vir.0.046672-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buijs PRA, Verhagen JHE, van Eijck CHJ, van den Hoogen BG. Oncolytic viruses: From bench to bedside with a focus on safety. Hum Vaccin Immunother. 2015;11:1573–1584. doi: 10.1080/21645515.2015.1037058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Medaglini D, Harandi AM, Ottenhoff THM, Siegrist CA, VSV-EBOVAC Consortium Ebola vaccine R&D: Filling the knowledge gaps. Sci Transl Med. 2015;7:317ps24. doi: 10.1126/scitranslmed.aad3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Melchjorsen J. Learning from the messengers: Innate sensing of viruses and cytokine regulation of immunity—Clues for treatments and vaccines. Viruses. 2013;5:470–527. doi: 10.3390/v5020470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Melchjorsen J, Sorensen LN, Paludan SR. Expression and function of chemokines during viral infections: From molecular mechanisms to in vivo function. J Leukoc Biol. 2003;74:331–343. doi: 10.1189/jlb.1102577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bixler SL, Goff AJ. The role of cytokines and chemokines in filovirus infection. Viruses. 2015;7:5489–5507. doi: 10.3390/v7102892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Escudero-Pérez B, Volchkova VA, Dolnik O, Lawrence P, Volchkov VE. Shed GP of Ebola virus triggers immune activation and increased vascular permeability. PLOS Pathog. 2014;10:e1004509. doi: 10.1371/journal.ppat.1004509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Okumura A, Pitha PM, Yoshimura A, Harty RN. Interaction between Ebola virus glycoprotein and host Toll-like receptor 4 leads to induction of proinflammatory cytokines and SOCS1. J Virol. 2010;84:27–33. doi: 10.1128/JVI.01462-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leroy EM, Baize S, Volchkov VE, Fisher-Hoch SP, Georges-Courbot MC, Lansoud-Soukate J, Capron M, Debré P, McCormick JB, Georges AJ. Human asymptomatic Ebola infection and strong inflammatory response. Lancet. 2000;355:2210–2215. doi: 10.1016/s0140-6736(00)02405-3. [DOI] [PubMed] [Google Scholar]
- 17.Hill-Batorski L, Halfmann P, Marzi A, Lopes TJS, Neumann G, Feldmann H, Kawaoka Y. Loss of interleukin 1 receptor antagonist enhances susceptibility to Ebola virus infection. J Infect Dis. 2015;212(Suppl. 2):S329–S335. doi: 10.1093/infdis/jiv335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McElroy AK, Erickson BR, Flietstra TD, Rollin PE, Nichol ST, Towner JS, Spiropoulou CF. Ebola hemorrhagic fever: Novel biomarker correlates of clinical outcome. J Infect Dis. 2014;210:558–566. doi: 10.1093/infdis/jiu088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Martinez O, Johnson JC, Honko A, Yen B, Shabman RS, Hensley LE, Olinger GG, Basler CF. Ebola virus exploits a monocyte differentiation program to promote its entry. J Virol. 2013;87:3801–3814. doi: 10.1128/JVI.02695-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bußfeld D, Nain M, Hofmann P, Gemsa D, Sprenger H. Selective induction of the monocyte-attracting chemokines MCP-1 and IP-10 in vesicular stomatitis virus-infected human monocytes. J Interferon Cytokine Res. 2000;20:615–621. doi: 10.1089/107999000414781. [DOI] [PubMed] [Google Scholar]
- 21.Athearn K, Sample CJ, Barefoot BE, Williams KL, Ramsburg EA. Acute reactogenicity after intramuscular immunization with recombinant vesicular stomatitis virus is linked to production of IL-1β. PLOS ONE. 2012;7:e46516. doi: 10.1371/journal.pone.0046516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hacker UT, Jelinek T, Erhardt S, Eigler A, Hartmann G, Nothdurft HD, Endres S. In vivo synthesis of tumor necrosis factor-α in healthy humans after live yellow fever vaccination. J Infect Dis. 1998;177:774–778. doi: 10.1086/517806. [DOI] [PubMed] [Google Scholar]
- 23.Zhang J-S, Zhao Q-M, Zuo S-Q, Jia N, Guo X-F. Cytokine and chemokine responses to Japanese encephalitis live attenuated vaccine in a human population. Int J Infect Dis. 2012;16:e285–e288. doi: 10.1016/j.ijid.2011.12.010. [DOI] [PubMed] [Google Scholar]
- 24.U.S. Food and Drug Administration (FDA) Guidance for Industry: Toxicity Grading Scale for Healthy Adult and Adolescent Volunteers Enrolled in Preventive Vaccine Clinical Trials (FDA) 2007 www.fda.gov/downloads/BiologicsBloodVaccines/GuidanceComplianceRegulatoryInformation/Guidances/Vaccines/ucm091977.pdf.
- 25.Haks MC, Goeman JJ, Magis-Escurra C, Ottenhoff THM. Focused human gene expression profiling using dual-color reverse transcriptase multiplex ligation-dependent probe amplification. Vaccine. 2015;33:5282–5288. doi: 10.1016/j.vaccine.2015.04.054. [DOI] [PubMed] [Google Scholar]
- 26.Becquart P, Wauquier N, Mahlakõiv T, Nkoghe D, Padilla C, Souris M, Ollomo B, Gonzalez J-P, De Lamballerie X, Kazanji M, Leroy EM. High prevalence of both humoral and cellular immunity to Zaire ebolavirus among rural populations in Gabon. PLOS ONE. 2010;5:e9126. doi: 10.1371/journal.pone.0009126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Heffernan RT, Pambo B, Hatchett RJ, Leman PA, Swanepoel R, Ryder RW. Low seroprevalence of IgG antibodies to Ebola virus in an epidemic zone: Ogooué-Ivindo region, Northeastern Gabon, 1997. J Infect Dis. 2005;191:964–968. doi: 10.1086/427994. [DOI] [PubMed] [Google Scholar]
- 28.Sobolev O, Binda E, O’Farrell S, Lorenc A, Pradines J, Huang Y, Duffner J, Schulz R, Cason J, Zambon M, Malim MH, et al. Adjuvanted influenza-H1N1 vaccination reveals lymphoid signatures of age-dependent early responses and of clinical adverse events. Nat Immunol. 2016;17:204–213. doi: 10.1038/ni.3328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Deshmane SL, Kremlev S, Amini S, Sawaya BE. Monocyte chemoattractant protein-1 (MCP-1): An overview. J Interferon Cytokine Res. 2009;29:313–326. doi: 10.1089/jir.2008.0027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Clahsen T, Schaper F. Interleukin-6 acts in the fashion of a classical chemokine on monocytic cells by inducing integrin activation, cell adhesion, actin polymerization, chemotaxis, and transmigration. J Leukoc Biol. 2008;84:1521–1529. doi: 10.1189/jlb.0308178. [DOI] [PubMed] [Google Scholar]
- 31.Iwamoto S, Iwai S-i, Tsujiyama K, Kurahashi C, Takeshita K, Naoe M, Masunaga A, Ogawa Y, Oguchi K, Miyazaki A. TNF-α drives human CD14+ monocytes to differentiate into CD70+ dendritic cells evoking Th1 and Th17 responses. J Immunol. 2007;179:1449–1457. doi: 10.4049/jimmunol.179.3.1449. [DOI] [PubMed] [Google Scholar]
- 32.Perrier S, Darakhshan F, Hajduch E. IL-1 receptor antagonist in metabolic diseases: Dr Jekyll or Mr Hyde? FEBS Lett. 2006;580:6289–6294. doi: 10.1016/j.febslet.2006.10.061. [DOI] [PubMed] [Google Scholar]
- 33.Feldmann H, Bugany H, Mahner F, Klenk H-D, Drenckhahn D, Schnittler H-J. Filovirus-induced endothelial leakage triggered by infected monocytes/macrophages. J Virol. 1996;70:2208–2214. doi: 10.1128/jvi.70.4.2208-2214.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yáñez-Mó M, Siljander PR-M, Andreu Z, Zavec AB, Borrás FE, Buzas EI, Buzas K, Casal E, Cappello F, Carvalho J, Colás E, et al. Biological properties of extracellular vesicles and their physiological functions. J Extracell Vesicles. 2015;4 doi: 10.3402/jev.v4.27066. 27066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Stanley AC, Lacy P. Pathways for cytokine secretion. Physiology. 2010;25:218–229. doi: 10.1152/physiol.00017.2010. [DOI] [PubMed] [Google Scholar]
- 36.Kawai T, Akira S. Innate immune recognition of viral infection. Nat Immunol. 2006;7:131–137. doi: 10.1038/ni1303. [DOI] [PubMed] [Google Scholar]
- 37.Dowell NG, Cooper EA, Tibble J, Voon V, Critchley HD, Cercignani M, Harrison NA. Acute changes in striatal microstructure predict the development of interferon-alpha induced fatigue. Biol Psychiatry. 2016;79:320–328. doi: 10.1016/j.biopsych.2015.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hokeness KL, Kuziel WA, Biron CA, Salazar-Mather TP. Monocyte chemoattractant protein-1 and CCR2 interactions are required for IFN-α/β-induced inflammatory responses and antiviral defense in liver. J Immunol. 2005;174:1549–1556. doi: 10.4049/jimmunol.174.3.1549. [DOI] [PubMed] [Google Scholar]
- 39.Luiza-Silva M, Campi-Azevedo AC, Batista MA, Angela Martins M, Sathler Avelar R, da Silveira Lemos D, Camacho LAB, de Menezes Martins R, de Lourdes de Sousa Maia M, Guedes Farias RH, da Silva Freire M, et al. Cytokine signatures of innate and adaptive immunity in 17DD yellow fever vaccinated children and its association with the level of neutralizing antibody. J Infect Dis. 2011;204:873–883. doi: 10.1093/infdis/jir439. [DOI] [PubMed] [Google Scholar]
- 40.Querec TD, Akondy RS, Lee EK, Cao W, Nakaya HI, Teuwen D, Pirani A, Gernert K, Deng J, Marzolf B, Kennedy K, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, Li G-M, et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43:1186–1198. doi: 10.1016/j.immuni.2015.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nakaya HI, Clutterbuck E, Kazmin D, Wang L, Cortese M, Bosinger SE, Patel NB, Zak DE, Aderem A, Dong T, Del Giudice G, et al. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc Natl Acad Sci USA. 2016;113:1853–1858. doi: 10.1073/pnas.1519690113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schattner A, Meshorer A, Wallach D. Involvement of interferon in virus-induced lymphopenia. Cell Immunol. 1983;79:11–25. doi: 10.1016/0008-8749(83)90046-1. [DOI] [PubMed] [Google Scholar]
- 44.Ledgerwood JE, DeZure AD, Stanley DA, Novik L, Enama ME, Berkowitz NM, Hu Z, Joshi G, Ploquin A, Sitar S, Gordon IJ, et al. Chimpanzee adenovirus vector Ebola vaccine. N Engl J Med. 2017;376:928–938. doi: 10.1056/NEJMoa1410863. [DOI] [PubMed] [Google Scholar]
- 45.Zhu F-C, Hou L-H, Li J-X, Wu S-P, Liu P, Zhang G-R, Hu Y-M, Meng F-Y, Xu J-J, Tang R, Zhang J-L, et al. Safety and immunogenicity of a novel recombinant adenovirus type-5 vector-based Ebola vaccine in healthy adults in China: Preliminary report of a randomised, double-blind, placebo-controlled, phase 1 trial. Lancet. 2015;385:2272–2279. doi: 10.1016/S0140-6736(15)60553-0. [DOI] [PubMed] [Google Scholar]
- 46.Kibuuka H, Berkowitz NM, Millard M, Enama ME, Tindikahwa A, Sekiziyivu AB, Costner P, Sitar S, Glover D, Hu Z, Joshi G, et al. Safety and immunogenicity of Ebola virus and Marburg virus glycoprotein DNA vaccines assessed separately and concomitantly in healthy Ugandan adults: A phase 1b, randomised, double-blind, placebo-controlled clinical trial. Lancet. 2015;385:1545–1554. doi: 10.1016/S0140-6736(14)62385-0. [DOI] [PubMed] [Google Scholar]
- 47.Ewer K, Rampling T, Venkatraman N, Bowyer G, Wright D, Lambe T, Imoukhuede EB, Payne R, Fehling SK, Strecker T, Biedenkopf N, et al. A monovalent chimpanzee adenovirus Ebola vaccine boosted with MVA. N Engl J Med. 2016;374:1635–1646. doi: 10.1056/NEJMoa1411627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Milligan ID, Gibani MM, Sewell R, Clutterbuck EA, Campbell D, Plested E, Nuthall E, Voysey M, Silva-Reyes L, McElrath MJ, De Rosa SC, et al. Safety and immunogenicity of novel adenovirus type 26- and modified vaccinia ankara-vectored Ebola vaccines: A randomized clinical trial. JAMA. 2016;315:1610–1623. doi: 10.1001/jama.2016.4218. [DOI] [PubMed] [Google Scholar]
- 49.De Santis O, Audran R, Pothin E, Warpelin-Decrausaz L, Vallotton L, Wuerzner G, Cochet C, Estoppey D, Steiner-Monard V, Lonchampt S, Thierry A-C, et al. Safety and immunogenicity of a chimpanzee adenovirus-vectored Ebola vaccine in healthy adults: A randomised, double-blind, placebo-controlled, dose-finding, phase 1/2a study. Lancet Infect Dis. 2016;16:311–320. doi: 10.1016/S1473-3099(15)00486-7. [DOI] [PubMed] [Google Scholar]
- 50.Tapia MD, Sow SO, Lyke KE, Haidara FC, Diallo F, Doumbia M, Traore A, Coulibaly F, Kodio M, Onwuchekwa U, Sztein MB, et al. Use of ChAd3-EBO-Z Ebola virus vaccine in Malian and US adults, and boosting of Malian adults with MVA-BN-Filo: A phase 1, single-blind, randomised trial, a phase 1b, open-label and double-blind, dose-escalation trial, and a nested, randomised, double-blind, placebo-controlled trial. Lancet Infect Dis. 2016;16:31–42. doi: 10.1016/S1473-3099(15)00362-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marzi A, Robertson SJ, Haddock E, Feldmann F, Hanley PW, Scott DP, Strong JE, Kobinger G, Best SM, Feldmann H. VSV-EBOV rapidly protects macaques against infection with the 2014/15 Ebola virus outbreak strain. Science. 2015;349:739–742. doi: 10.1126/science.aab3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Marzi A, Ebihara H, Callison J, Groseth A, Williams KJ, Geisbert TW, Feldmann H. Vesicular stomatitis virus–based Ebola vaccines with improved cross-protective efficacy. J Infect Dis. 2011;204(Suppl. 3):S1066–S1074. doi: 10.1093/infdis/jir348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Henao-Restrepo AM, Longini IM, Egger M, Dean NE, Edmunds WJ, Camacho A, Carroll MW, Doumbia M, Draguez B, Duraffour S, Enwere G, et al. Efficacy and effectiveness of an rVSV-vectored vaccine expressing Ebola surface glycoprotein: Interim results from the Guinea ring vaccination cluster-randomised trial. Lancet. 2015;386:857–866. doi: 10.1016/S0140-6736(15)61117-5. [DOI] [PubMed] [Google Scholar]
- 54.Querec T, Bennouna S, Alkan S, Laouar Y, Gorden K, Flavell R, Akira S, Ahmed R, Pulendran B. Yellow fever vaccine YF-17D activates multiple dendritic cell subsets via TLR2, 7, 8, and 9 to stimulate polyvalent immunity. J Exp Med. 2006;203:413–424. doi: 10.1084/jem.20051720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li S, Rouphael N, Duraisingham S, Romero-Steiner S, Presnell S, Davis C, Schmidt DS, Johnson SE, Milton A, Rajam G, Kasturi S, et al. Molecular signatures of antibody responses derived from a systems biology study of five human vaccines. Nat Immunol. 2014;15:195–204. doi: 10.1038/ni.2789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gabay C, Smith MF, Eidlen D, Arend WP. Interleukin 1 receptor antagonist (IL-1Ra) is an acute-phase protein. J Clin Invest. 1997;99:2930–2940. doi: 10.1172/JCI119488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Finnegan A, Kaplan CD, Cao Y, Eibel H, Glant TT, Zhang J. Collagen-induced arthritis is exacerbated in IL-10-deficient mice. Arthritis Res Ther. 2003;5:R18–R24. doi: 10.1186/ar601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Clarke DK, Hendry RM, Singh V, Rose JK, Seligman SJ, Klug B, Kochhar S, Mac LM, Carbery B, Chen RT, Brighton Collaboration Viral Vector Vaccines Safety Working Group Live virus vaccines based on a vesicular stomatitis virus (VSV) backbone: Standardized template with key considerations for a risk/benefit assessment. Vaccine. 2016;34:6597–6609. doi: 10.1016/j.vaccine.2016.06.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Fields BN, Hawkins K. Human infection with the virus of vesicular stomatitis during an epizootic. N Engl J Med. 1967;277:989–994. doi: 10.1056/NEJM196711092771901. [DOI] [PubMed] [Google Scholar]
- 60.Fuchs JD, Frank I, Kochar N, Elizaga M, Allen M, Carter DK, Frahm N, Kalams SA, Mulligan M, Sheets R, Pensiero M, et al. paper presented at the 6th IAS Conference on HIV Pathogenesis, Treatment and Prevention; Rome. 17 July 2011. [Google Scholar]
- 61.Muyanja E, Ssemaganda A, Ngauv P, Cubas R, Perrin H, Srinivasan D, Canderan G, Lawson B, Kopycinski J, Graham AS, Rowe DK, et al. Immune activation alters cellular and humoral responses to yellow fever 17D vaccine. J Clin Invest. 2014;124:3147–3158. doi: 10.1172/JCI75429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lai L, Davey R, Beck A, Xu Y, Suffredini AF, Palmore T, Kabbani S, Rogers S, Kobinger G, Alimonti J, Link CJ, Jr, et al. Emergency postexposure vaccination with vesicular stomatitis virus-vectored Ebola vaccine after needlestick. JAMA. 2015;313:1249–1255. doi: 10.1001/jama.2015.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Joosten SA, Goeman JJ, Sutherland JS, Opmeer L, de Boer KG, Jacobsen M, Kaufmann SH, Finos L, Magis-Escurra C, Ota MO, Ottenhoff TH, et al. Identification of biomarkers for tuberculosis disease using a novel dual-color RT-MLPA assay. Genes Immun. 2012;13:71–82. doi: 10.1038/gene.2011.64. [DOI] [PubMed] [Google Scholar]
- 64.Kaiser HF. An index of factor simplicity. Psychometrika. 1974;39:31–36. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.