Abstract
As personalized medicine becomes more integrated into healthcare, the rate at which human genomes are being sequenced is rising quickly together with a concomitant acceleration in compute and storage requirements. To achieve the most effective solution for genomic workloads without re-architecting the industry-standard software, we performed a rigorous analysis of usage statistics, benchmarks and available technologies to design a system for maximum throughput. We share our experiences designing a system optimized for the “Genome Analysis ToolKit (GATK) Best Practices” whole genome DNA and RNA pipeline based on an evaluation of compute, workload and I/O characteristics. The characteristics of genomic-based workloads are vastly different from those of traditional HPC workloads, requiring different configurations of the scheduler and the I/O subsystem to achieve reliability, performance and scalability. By understanding how our researchers and clinicians work, we were able to employ techniques not only to speed up their workflow yielding improved and repeatable performance, but also to make more efficient use of storage and compute resources.
General Terms: Measurement, performance, design, reliability, management
Keywords: High performance, high throughput and data-intensive computing, parallel file systems, genomic sequencing, scheduling and resource management, performance analysis, benchmarking, GPFS, LSF and flash memory
Categories and Subject Descriptors
B.4.3 Interconnections (Subsystems) Parallel I/O, B.4.4 Performance Analysis and Design Aids (Verification), B.8 Performance and Reliability, C.4 Performance of Systems, C.5.1 Large and Medium Computers, D.2.10 Design Methodologies, D.4.2 Storage Management, D.3.4 Processors, D.4.3 File Systems Management, E. Data J.3 Life and Medical Sciences, K.6.4 System Management
1. INTRODUCTION
In May 2012, we deployed Minerva, Mount Sinai’s first supercomputer. It was swiftly accepted as an essential and integral part of the scientific discovery process and used by a variety of disciplines. Researchers immediately exploited Minerva to devise more complex experiments and higher fidelity simulations to expand and accelerate progress on their scientific investigations. The usage has directly translated into new understanding of, and therapies for, a wide spectrum of disease categories including autism, insulin resistance in diabetics, schizophrenia and related behavioral disorders, cardiac care, the origins of drug addiction and depression, and cancer progression [1–8].
In its first year of operation, Minerva served 339 total users (277 from Sinai and 62 from external institutions) across 161 projects, and 11 departments. Eight million jobs consumed over 40 million core hours. Minerva had a 99% uptime rate and was utilized 84% of the time with frequent extended periods over 90%. The usage was split with two-thirds of the total core hours used for genomic sequencing, and one third for molecular modeling simulations.
Although we collected user requirements before designing the system, the actual usage of the genomics pipeline was very different from what we anticipated: massive numbers of extremely small files and extremely short running jobs on single cores. This caused both our parallel file system and scheduler to exhibit unexpected behavior and cause the system to be less reliable from a usability perspective, and unreliable from a consistent I/O performance perspective. Through the creation of a new monitoring tool, benchmarking, and the advent of new technology, we designed and deployed a new system scalable and reliable for our genomics workload, optimized for high throughput and consistent I/O performance. Today, Minerva is comprised of 12,864 cores, 50 terabytes of RAM, 160 terabytes of flash memory, and 11 petabytes of storage capacity. There are over 1,000 user accounts, including over 200 external users. The system reliably handles the necessary workload of 500,000 jobs in the queue, providing consistent high I/O performance for hundreds of millions of tiny files, and computes nearly five times faster on the genomic pipeline workflow.
2. GENOMICS PIPELINE WORKFLOW
The Broad Institute’s “GATK Best Practices” pipelines provide step-by-step recommendations of the most widely used DNAseq and RNAseq software. While the name of workflow itself includes the words “Best Practices”, we are not asserting that it is a best practice, however, it is the “industry standard” with over 20,000 users [9–12, 14]. This paper is focused on sharing our experiences architecting a system to optimally run this workflow and not on the quality of this workflow from a bioinformatics perspective. There are three major steps in the variant discovery pipeline: data pre-processing, variant discovery and evaluation [15]. Within each of these steps, the specific software tools used may vary depending on what is necessary for a particular analysis. For instance, a specific variant caller might be preferred depending on the degree of specificity needed for your specific dataset [16,17]. The “GATK Best Practices” pipeline recommendations are developed by the GATK team in collaboration with research groups and are constantly updated as new and improved methodology is developed. The paper is based on the “GATK Best Practices” pipeline that was deployed at Mount Sinai. This so-called “Best Practices” pipeline is widely regarded in the community to produce the best analysis results possible from NGS high-throughput sequencing data for the most common use cases and data types. Other options for whole pipeline analysis include accelerated appliances for specific mutations such as Genalice and alternative approaches such as Johns Hopkins University’s and Beijing Genomics Institute’s SOAPdenovo [18–21]. Here, we extensively test the performance of Mount Sinai’s implementation of the Broad Institute’s “GATK Best Practices” pipeline for variant discovery in high throughput sequenced DNA data [13], and we describe the performance and tuning of our supercomputer resource, Minerva.
The genomics workflow for both RNA and DNA analysis typically proceeds as a three-step process. First, the primary analysis involves concatenating and de-multiplexing the data that comes from the genomic sequencers into Next Generation Sequencer (NGS) FASTQ files. This step is computationally trivial, however, the storage requirements are significant: a typical DNA exome sequence generates 20 to 40 million 100 nucleotide reads per subject, which, including quality scores, uses 4 to 8 gigabytes. Typical genomic studies involve the exomes of tens to thousands of subjects, bringing the total storage requirements of a single study to terabytes [23–26]. Secondary analysis is the process of sequence alignment to a reference genome and variant calling. This step is the most computationally intense portion of the genomics workflow for both RNA and DNA. Execution of a typical whole genome pipeline to process one sample of RNA or DNA takes 1,000 jobs of varying duration and a total time to solution of approximately one week after scheduling and dependencies are taken into account.
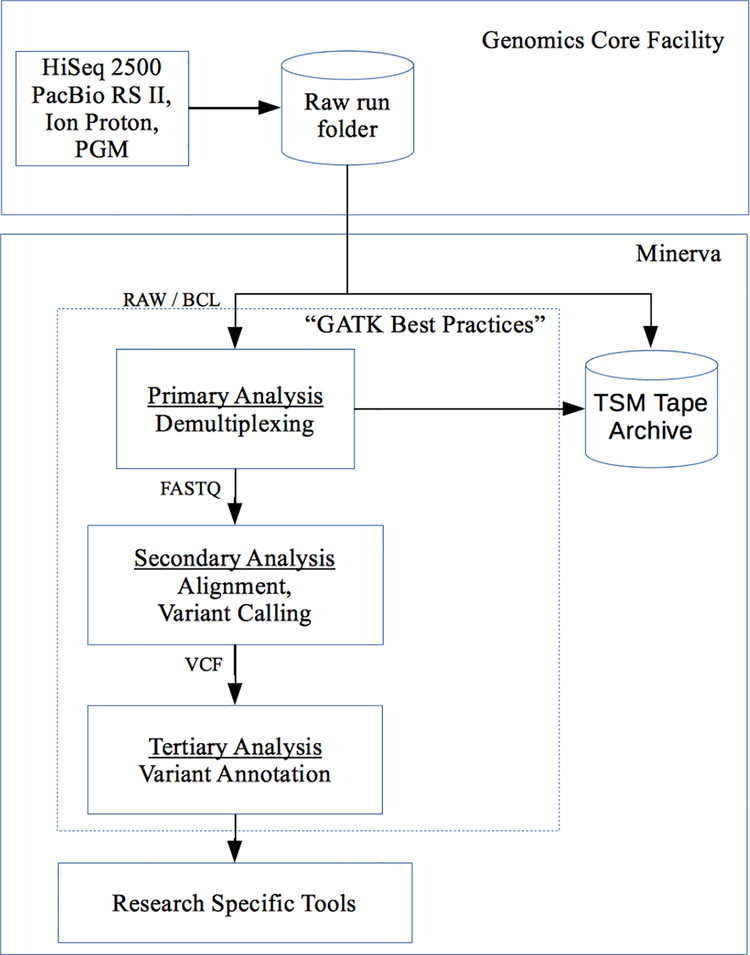
The Mount Sinai Genomics Core Facility (GCF) is a Clinical Laboratory Improvement Amendments (CLIA) certified laboratory which enables Mount Sinai investigators to carry out basic and translational genomics research. It helps investigators and clinicians analyze samples, identify genetic variants contributing to disease risk, and reveal complex mechanisms involved in human disease. Refer to Figure 1 for an overview of the workflow. Its capabilities include: (1) sequencing with the Illumina HiSeq 2500 and MiSeq, (2) single molecule sequencing with the Pacific Biosciences RS II, (3) Ion Proton for development of whole-exome and whole-genome sequencing using Life Technologies sequencing chemistry for rapid-turnaround NGS sequencing, (4) Ion Personal Genome Machine for targeted research and clinical assays with high throughput and intermediate length reads with rapid turnaround time, (5) next-generation sequencing library preparation from RNA and DNA, (6) Sanger sequencing with capillary electrophoresis, (7) genotyping, gene expression profiling and epigenetics using the high-throughput Illumina BeadArray platform, and (8) real-time polymerase chain reaction (PCR) using the Applied Biosystems 7900HT. Mount Sinai currently has nine Illumina sequencers each running several times per week. Combining with other sequencers, the GCF generates 6 terabytes of raw data per week. After analysis and post processing, and including the raw data, the data footprint is 20 terabytes per week.
Figure 1.
Genomics sequencing pipeline at Mount Sinai
The application workload is be grouped into four categories: (1) primary analysis of samples processed in the genomics core sequencing facility, (2) secondary analysis (primarily genome alignment and variant calling) for these samples performed by the genomics sequencing core or other groups at Mount Sinai, (3) tertiary analysis (annotation and variant classification) from these samples performed by the genomics sequencing core, and (4) biomedical interpretation with many different styles of statistical and network analysis. All of the biomedical interpretation analyses are performed with the aim of identifying genes and networks of genes/proteins that associate with human disease and/or disrupted animal models. While steps 1–3 are usually very predictable in terms of their computational and storage requirements, the costs of step 4, biomedical interpretation, can vary and grow substantially. These steps can involve simulations and network analyses of high computational complexity.
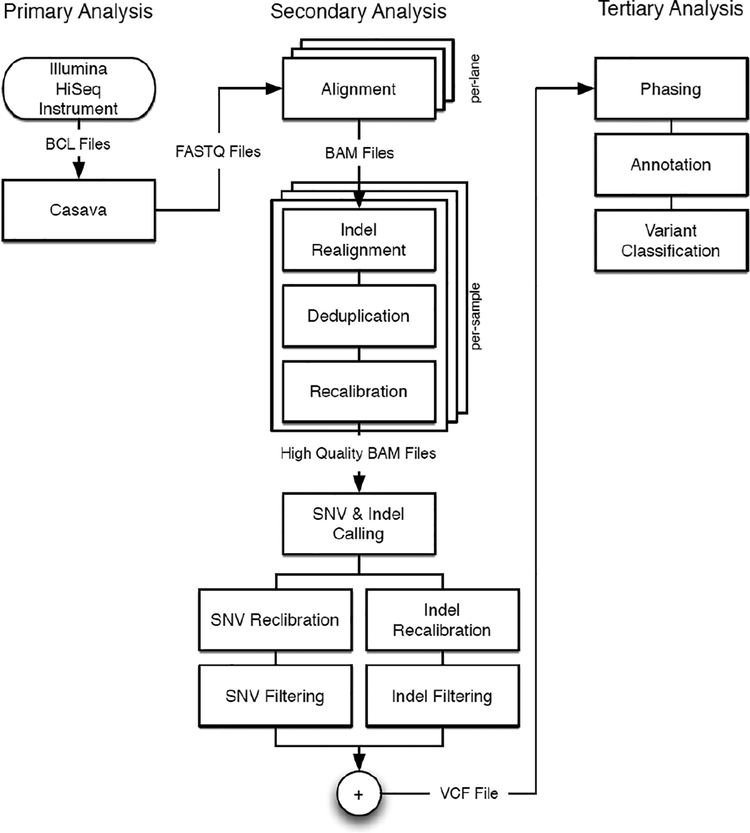
A schematic of the DNA pipeline can be found in Figure 2. These diagrams show the sets of tools used at each stage in the analysis pipeline (sequencing, secondary, and tertiary). In each case, primary sequencing involves the transfer and pre-processing of raw data collected from our Illumina and PacBio sequencers. These raw data are converted to the FASTQ format by either proprietary, vendor supplied programs or custom scripts. These converted data become the primary analysis data and inputs to the secondary stage of the pipelines. The secondary stage is the process of creating the alignment or BAM files, iteratively improving the quality of those BAM files, and calling variants. This is the most computationally intense step of both the DNA (and RNA) pipelines. This analysis step involves the execution of many programs, the collection of which comprises a highly heterogeneous compute pipeline. At various stages during this pipeline, each of I/O, compute, or memory bandwidth is the dominant limiting factor. For secondary analysis, however, the procedure is generally fixed for all samples that come off the sequencers. In contrast, further downstream analysis involves computational algorithms that directly answer the scientific questions at hand and are varied in their computational requirements. For example, one such question might be: “What are the rare variations in a genome sequence which correlate to patients with schizophrenia or autism?” Our researchers have developed their own algorithms for these questions [1–2, 23].
Figure 2.
DNA workflow [22]
3. INITIAL MINERVA DESIGN
Minerva’s components are connected using a single tier of Infiniband in a fat-tree topology. Although genomic workflows do not typically employ any distributed parallelism, this network is used for high performance I/O and for MPI in molecular dynamics simulations.
Most of the genomics workflow does not require double precision floating point calculation, so we opted to purchase the largest number of cores possible, knowing that the calculations that need double precision, e.g. molecular dynamics (MD) simulations, could reserve extra cores on the whole nodes. Lacking historical workflow information from our constituency, but knowing that many of the jobs would need single cores, we operated the cluster in a shared node configuration, utilizing cpusets to enforce core assignment to jobs. The initial scheduler, TORQUE 2.5.11, had built-in support for assigning cpusets to tasks within a job. The TORQUE implementation at the time of deployment however, was unable to enforce memory limits per job. Instead, we enforced this through Moab’s resource limit policies. The scheduling initially utilized TORQUE 2.5.11 and Moab 6.1.5. The resource manager and workload manager were subsequently upgraded to TORQUE 4.1.5 followed by TORQUE 4.2.4 and Moab 7.2.2. The original queue configuration was based on job size for long running multi-core SMP and MPI jobs. A concession was made for short jobs and single core jobs by instituting express and serial queues.
The storage subsystem was composed of two sets of disks: (1) a Data Direct Networks (DDN) SFA10K controller couplet with 3 terabyte 7,200 RPM NL-SAS drives grouped in 60 LUNs of 8+2p RAID6 totaling 1.8 petabytes of raw storage and used for file system data, and (2) a DotHill/DDN EF3015 controller pair serving 450 gigabyte 15,000 RPM 6G SAS drives grouped in 6 LUNs of RAID1 used for the file system metadata. We separated the underlying metadata storage from the data storage to provide better performance for the expected large number of small files [27]. The system had four storage servers with two of them also attached to the DDN EF3015 controller with two 8 gigabit fibre channel paths per server, providing several data paths for both load balancing and redundancy. We selected IBM’s General Parallel File System (GPFS) 3.4 over Lustre because it has a number of advantages that are specifically useful for this workload such as parallel metadata servers, tiered storage, and subblock allocation [27]. GPFS has been renamed to “Spectrum Scale” but we will refer to it as GPFS throughout this paper.
4. USAGE AND ANALYSIS
To understand how to optimize our configuration for our actual computational and data needs, we performed a rigorous data collection and an analysis of our workload. We also had anecdotal experience that informed our analysis. After the first year, we collected the usage statistics shown in Table 1. We were surprised by the sheer number of files, the tiny size of the vast majority of them, and the short job run time. We also experienced unexpected issues with scheduling and the file system due to unpredicted use cases. This workload is vastly different than a typical HPC workload where there may be only a couple of jobs using all of the system’s cores and writing to only a few files [49–53].
Table 1.
Minerva usage year 1
| # of files | 54,026,071 |
| Total storage used (TB) | 770.7 |
| Average file size (MB) | 15 |
| Median file size (bytes) | 29 |
| Maximum file size (TB) | 1.3 |
| Minimum file size (bytes) | 0 |
| 80% of files smaller than (KB) | 10 |
| # of zero-length files | 1,731,148 |
| Amount of archival storage used (PB) | 1 |
| Maximum # of core hours used/job | 22,161 |
| Minimum # of core hours used/job | 0 |
| Average # of core hours used/job | 4.2 |
| Median # of core hours used/job | 10.8 |
| Maximum job run time (hours) | 205 |
| Minimum job run time (hours) | 0 |
| Average job run time (hours) | 1.6 |
| Median job run time (hours) | 0.08 |
The GATK pipelines typically involve the use of many different types of parallelism. In the most compute intensive steps, such as the alignment phase implemented using BWA-MEM (and benchmarked in detail below), shared-memory thread-based parallelism is implemented and achieves nearly linear scaling well beyond 16 cores for all of our benchmarked systems. In most other steps of the pipeline, parallelism is implemented with a scatter-gather methodology, where the full problem is broken into many embarrassingly parallel serial jobs which are submitted all at once to the cluster. The next step of the pipeline is then executed only when all scatter-gather jobs for the current step are complete. A control process running on a head node is responsible for all job submission and monitoring. Communication between jobs is handled exclusively on disk, in many cases using zero length or very short files to indicate the status of a given scatter-gather job. The “GATK Best Practices” Queue (GATK-Queue) control process is responsible for managing failure of individual jobs. If one scatter-gather job fails, for example, during the variant calling step, the control process is signaled and resubmits the job immediately before the next stage in the pipeline can proceed. This mode of execution is very similar to MapReduce applications such as Hadoop, in that it reruns failed tasks, and avoids the need for a system-level checkpoint restart mechanism such as Berkeley Lab Checkpoint/Restart (BLCR). If the pipeline fails some number of times, the control process will terminate the pipeline, while leaving it in a re-startable state. An example of this type of parallelism, the most common in the “GATK Best Practices” pipeline, is found in the HaplotypeCaller. One user, who executed analysis of 100 exomes simultaneously had over 1.2 million files open at one time with 1,400 jobs on 1,400 cores during the HaplotypeCaller step. Portions of the “GATK Best Practices” pipeline such as the HaplotypeCaller can be run in embarrassingly parallel fashion via the gather functionality of GATK-Queue. Since the initial Minerva and most standard HPC clusters are not designed for this use case, we had reliability and performance issues with both the scheduler and the file system.
It quickly became evident that the workflow on Minerva was predominantly single core, short jobs. Our statistics for 2013 show that 99% of jobs used only one node and took 77% of the utilized core hours. Furthermore, 62% of the jobs used only 1 core and took 17% of the utilized core hours. 67% of jobs completed in less than 24 hours and represented only 2% of all core hours used.
When analyzing the usage we observed on Minerva, it is tempting to conclude that an HPC-style resource is simply not a good fit for the disk-intensive and data-parallel jobs inherent in the “GATK Best Practices” pipeline. In fact, many groups have and are currently working on implementing certain components of the DNAseq pipeline on alternative architectures, such as Hadoop-style clusters [9,28,29]. However, these implementations have not found success in production use for many reasons, and the GATK toolkit available from the Broad Institute remains an HPC-oriented codebase. Perhaps most importantly, while the DNAseq pipeline is in some portions data-parallel, it remains very compute-intensive during both shared-memory-parallel portions (e.g., alignment) and during scatter-gather parallel portions (e.g., the HaplotypeCaller, where HPC-style SIMD optimization, implemented in C intrinsics by Intel and introduced in GATK version 3.1, have significant improved the overall performance of the pipeline). Hadoop-style implementations of these tools have been attempted, though they are in their infancy, have not been adopted by the community at large, and have relatively unknown result quality. Exciting development in this area is forthcoming, though it is an area of current research and tools have not been broadly integrated into an overall pipeline [28, 43, 44].
4.1. Impact on the Scheduler
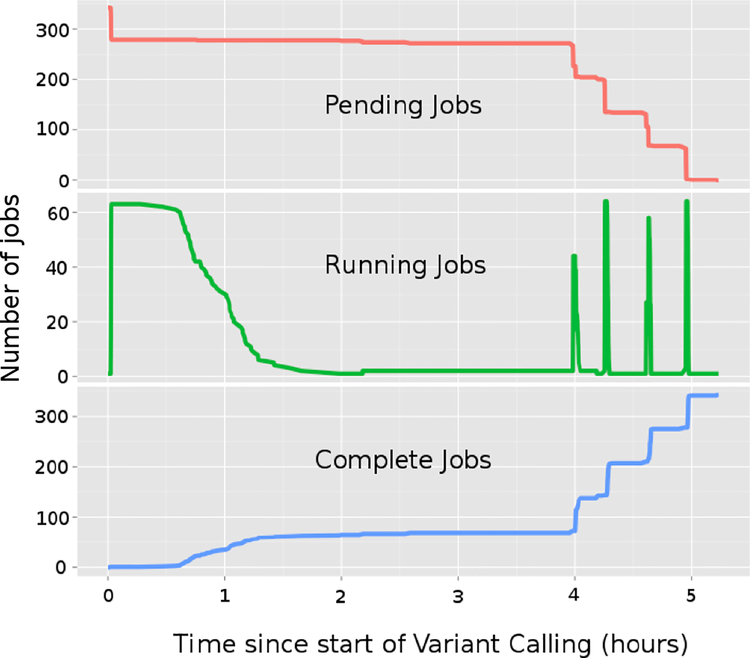
Each instance of the genomic pipeline workflow submits 10–20,000 short jobs (run time is less than 10 minutes per job) at a time. The GATK-Queue control process manages each workflow by querying the queue for job status constantly to determine if the next batch of jobs should be submitted. Refer to Figure 3 for the job profile for the variant calling portion of the “GATK Best Practices” pipeline. During normal production on Minerva, many users run their own pipelines. TORQUE and Moab had difficulty managing the large bolus of jobs each user submitted along with the constant querying by the GATK-Queue control process, and quickly degraded into an unresponsive state. Often, the scheduling of the jobs took longer than the amount of the time it took for the jobs to actually run. Users with longer-running molecular dynamics simulations became less productive due to inaccurate job start times caused by genomics jobs submitted with the maximum wall clock time of six days. Genomics users submitted jobs with maximum wall clock times to compensate for the highly variable run-times of their jobs due to the fluctuations in the I/O performance. 67% of jobs submitted with six day long wall clock time completed in less than 24 hours. Employing a bit of social engineering, we configured high priority queues with 24 hours maximum wall clock time to encourage more realistic job runtime estimations through faster turnaround time. We implemented severe limits on the number of jobs per user in the queues, use batched Moab scheduling, and restrict the number of jobs to be scheduled in scheduling cycle to achieve scheduler stability. To ameliorate the load imposed by the GATK-Queue control processes, we used a command wrapper to return pre-cached qstat information.
Figure 3.
“GATK Best Practices” variant calling job profile
TORQUE 2.5.11 core dumped at least once a month, which led us to set a cron job to restart TORQUE and Moab automatically. For ten hours per month on average, TORQUE would not start new jobs, though jobs already running were not affected. New jobs would not start unless the job control and log files were manually removed. An upgrade to TORQUE 4.1.5 introduced more instability due to bugs with job arrays, so we disabled this capability in TORQUE. This version also core dumped on a daily basis and we were not able to schedule jobs for 30 hours per month on average. The situation improved in TORQUE 4.2, but it still was not reliable. With custom patches from Adaptive, we were able to run job arrays and increase the number of jobs in the queues to 45,000 with TORQUE 4.2.4. Although this was helpful, we still continued to limit the number of jobs each user could submit to control the overall number of jobs in the queue to prevent further scheduler instability.
4.2. Impact on the File System
As we discovered, genomic analyses created an inordinate number of tiny files. Refer to Table 2 for the distribution of files from the genomics-based workload on Minerva in August 2013. A typical I/O workflow for one genome created a directory with 0.5–1.2 million files and 10,000 folders, although many files are removed after the job completes. In normal production, thousands of jobs randomly accessed many tiny files less than 1 kilobyte in size. Each job created directory trees with inefficient scripting methods that added to the load on the metadata server (for instance, running “ls –l” in a directory with 10,000 files in a “for” loop). This is vastly different than a typical HPC workload where a few large files would be accessed [49–53]. This created file system and compute performance challenges for several reasons: (1) the file system cannot take advantage of speed from parallel striped data LUNs as the tiny files do not span more than one file system subblock, (2) storage space is wasted because files are significantly smaller than our minimum allocable block of 32 kilobytes, (3) job runtime and interactive command line directory listings are slow due to an overloaded metadata server that is looking up information on tens of thousands of files for each job (multiplied by 10s of these kinds of jobs on the system at any one time), and (4) high latency responses from a large number of recursive directories force the metadata server to lookup chained inodes.
Table 2.
Distribution of file sizes
| Decile | Max Size | # Files |
|---|---|---|
| 10% | 3 B | 5.5 M |
| 20% | 6 B | 32 M |
| 30% | 10 B | 17 M |
| 40% | 39 B | 20 M |
| 50% | 340 B | 19 M |
| 60% | 2 KB | 19 M |
| 70% | 3.7 KB | 19 M |
| 80% | 17 KB | 19 M |
| 90% | 84 KB | 19 M |
| 100% | 5 TB | 19 M |
In a typical month during 2013, there were approximately 160 million files less than 20 kilobytes in size consuming 300 gigabytes, and 30 million files greater than 20 kilobytes in size consuming 1,000 terabytes. For the files smaller than 20 kilobytes, about 100 million files were of zero length. 50% of the files for the genomics workload were 29 bytes or smaller and 80% of the files were smaller than 10 kilobytes. Files in the larger category were unremarkably distributed. The largest file was 1.3 terabytes. 93 million out of 200 million files were less than 1 kilobyte and consumed 0.007% of the total disk space in aggregate. All files less than 1 megabyte consumed only 1 terabyte in aggregate. Nine million out of 18 million directories only contained one file. Upwards of 5,500 directories had greater than 10,000 files in them, with several exceeding 500,000 files. Although the GATK-Queue control process has an option to clean up intermediate files automatically when the pipeline has completed successfully, it appeared that not all files were removed.
Our researchers exhausted the initial 1.5 petabyte (usable) storage capacity by mid-2013. GPFS began to perform poorly when more than 80% full because the distribution of files became highly fragmented. We limited the maximum footprint of files stored on the machine to be less than 1.2 petabytes to guarantee reasonable performance. In September 2013, genomics users accounted for 707 terabytes of actively used research data on the file system, while the remaining 560 terabytes were generated by users in other departments such as Structural and Chemical Biology. Although we had regular purging in place, we still had to ask users to delete or move their files to archival storage to make room for new runs. Obviously this was not the most efficient use of our researchers’ time, and more storage was needed urgently.
Although the file system performed adequately during benchmarks and under normal use cases, we began to see issues as the load increased. We, along with our users, occasionally noticed a slow response when traversing or listing directories in real-time on the command line and we experienced variable job run time. Directory operations were especially challenging, as they required multiple and/or chained metadata operations to retrieve all required information to complete the command. Genomic workloads that performed directory listings with on-disk hash tables for looking up references or updating the date/time on files used as semaphores added to the metadata server load.
4.2.1. New GPFS-storage correlation tool
To find the root cause of our periodic slow downs, we needed to have an overall picture of what was happening on each component of the I/O subsystem at the same time. In 2013, we found several tools that showed detailed information for specific components including a GPFS Ganglia Plugin and a GPFS Monitoring Suite by NCAR but nothing that could track events from end-to-end in real-time to replay an event step-by-step [30,31]. To understand our system holistically, we wrote our own tool to gather metrics from each of the following major components: (1) GPFS “waiters,” the list of outstanding I/O requests between nodes, (2) GPFS mmpmon, internal file system counters, (3) the DDN SFA 10K storage controller virtual disk statistics, and (4) the DDN EF3015 metadata controller load statistics [32–34]. Each of these components can be monitored through vendor-provided scripts, but they only output a single point-in-time reading at the user’s request. While they can be looped, it is not a good long-term solution for bottleneck assessment, nor does it allow troubleshooting after the fact. Our tool enabled us to capture data in real-time continuously across all of these components and allowed us to “play-back” what was occurring on each of these components during a file system slow down.
The back-end architecture of the tool stores and retrieves metrics, and does not display end user graphs in any specific way. With this approach, it is easy to import metrics into tools such as R or Matlab for further analysis. For the front-end, we deployed a webpage using HTML, HighCharts, and JQuery [35]. The webpage is completely client-side and requires no rendering from the server side, only a REST API written in PHP. The REST interface passes through selected metrics from the database for the user requested time period. We implemented start and end parameters to limit the data range so that the browser can easily render the dataset and make it easy to read. We chose PHP script and MySQL as the backend because it is easy to use, ubiquitous, has many import/export choices with unlimited full-resolution storage capability.
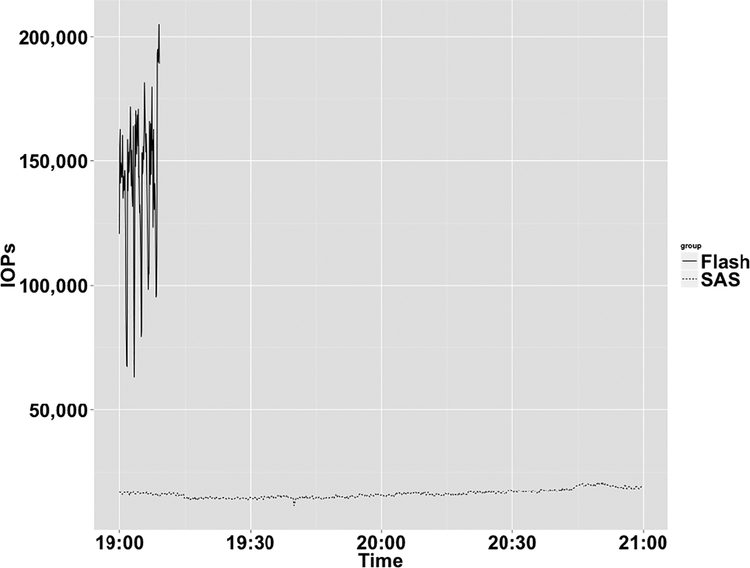
Using this tool, we collected over 137 million data points in three months. We examined the dashboard and aggregate metrics and found the following themes: (1) regular achievement of the maximum possible Input/output Operations Per second (IOPs) on both the metadata and data controllers, (2) throttling of the data controller due to high load on the metadata controller, and (3) thrashing between high and low load conditions. In addition to user workloads, we utilized GPFS’s Integrated Lifecycle Management (ILM) to purge older files, which triggered whole file system scans, and compounded the metadata load. Since the ILM scans were a good example of a “high load” condition, we selected it as a benchmark. Given our findings, we realized we had to design for a much higher metadata IOPs capability across the entire metadata disk subsystem and a much lower latency for the metadata controller. Latency is especially important because GPFS utilizes indirect on-disk inode pointers for directories with large numbers of files [31]. One industry standard method for achieving high IOPs is to implement a cache, however, as our metadata workload style was a high load of chained random I/O across the entire metadata array, it was likely that a cache would only be drained and refilled continuously, and thus not aid performance. With flash having the lowest latency of all storage options, we decided to incorporate it into the expanded Minerva design. In subsequent tests using ILM scans in the same system conditions, flash outperformed SAS by 15 times as shown in Figure 4. We found that our genomic workload requires a high volume of random, chained I/O operations, and that flash storage significantly increased and provided reliable I/O performance for these metadata-intensive operations.
Figure 4.
IOPs for flash vs. SAS under high load
5. PROJECTED USAGE
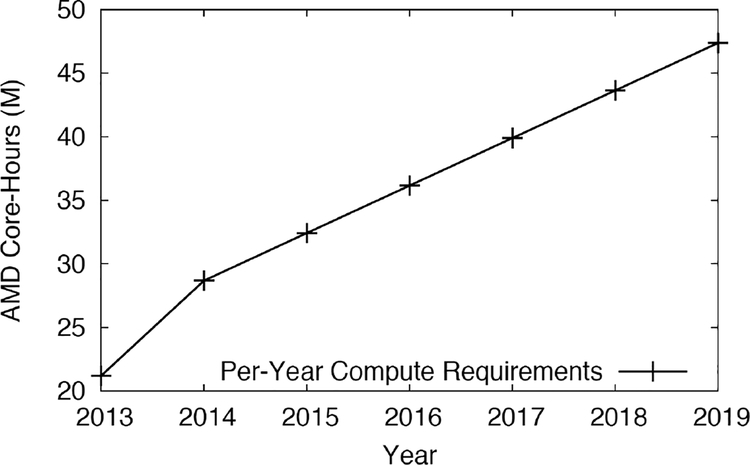
We would be remiss if we didn’t explore future compute and storage usage and incorporate it into our new design. We wanted the new system to accommodate expected storage and compute growth for the next 3–4 years. The need for dramatic growth in compute and storage directly corresponded to the growth of genomics projects, and we combined our actual usage from the previous section with projections from our users to determine our future needs. Refer to Figures 5 and 6 for our projections. In 2013, the Mount Sinai GCF had five Illumina sequencers generating data. By 2015, nine Illumina sequencers were in production. By 2019, Mount Sinai expects that number to increase to at least 12, which will require the equivalent of an additional 48 million core-hours on our older, AMD-based Minerva compute nodes. This will more than double the number of simultaneous samples to be processed and analyzed over a five-year period. This is equivalent to approximately 32.9 million core hours at 3.5 GHz using a string linear clock frequency comparison, far beyond our current capacity in 2013.
Figure 5.
Projected genomics compute usage
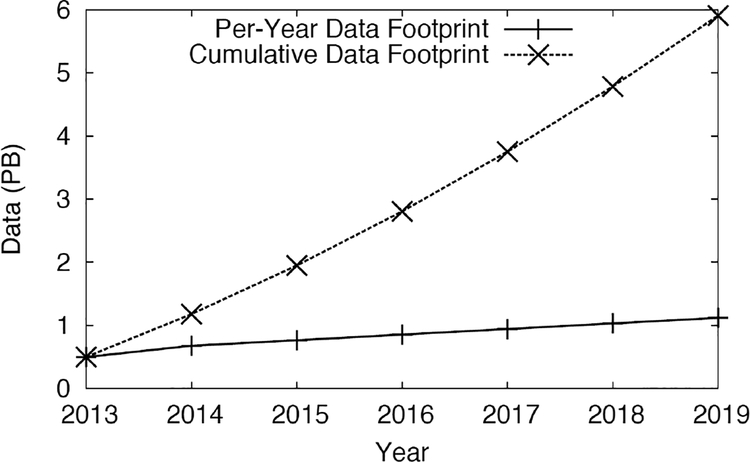
Figure 6.
Projected genomics data usage
Additionally, data from previous analysis cannot simply be discarded. As newer and more accurate/efficient techniques are invented, entire cohorts of samples are re-analyzed. The cumulative storage requirements for new sequencing projects are expected to approach 6 petabytes by 2019. With these continuously growing datasets, and increased rate of sequencing due to additional projected sequencers and re-analysis of existing data due to updated algorithms, we needed to add a long-term archival storage solution to our system. For this, we selected IBM’s Tivoli Storage Manager (TSM).
6. WHOLE-PIPELINE BENCHMARKING
In addition to providing a reliable system using the lessons learned from section 4, and providing enough capacity for the future as explained in section 5, we also wanted to decrease the time to solution by using the fastest components for our workload. By increasing the computational speed of the pipeline, we would also be increasing the overall throughput, thus enabling more science to be done in the same period of time.
Therefore, we embarked on a series of benchmarks on the “GATK Best Practices” 3.1.1 whole-genome pipeline in production at Mount Sinai. We initially aimed for a 24-hour turnaround for most pipeline runs, and aimed for even faster results as we tuned the system and software.
The underlying GATK toolset included alignment by BWA-MEM in a multithreaded configuration, and variant calling using the now-standard Haplotype caller including Intel’s AVX-extension vectorization where possible. We used a GATK-Queue scatter-gather width of 64 in all pipeline benchmarks, and ran BWA alignment at 16 threads on our original AMD Opteron processors and 12 threads on our trial 12-core Intel Ivy Bridge Xeon processors. Two underlying storage subsystems were also benchmarked: our original DDN 10K system, and a trial system based on IBM flash storage and IBM GPFS Storage Server (GSS). These new trial processor and storage subsystem configurations are described in detail below, in section 7.
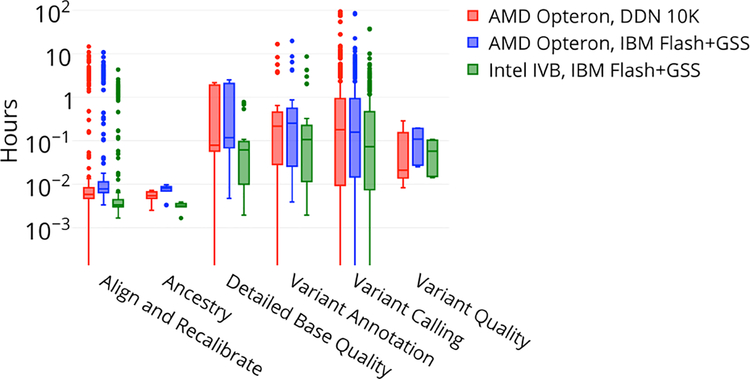
Figure 7 shows a log plot of CPU-hour consumption for the six major categories of GATK pipeline jobs, presented as box plots including median, interquartile range, and outlier points. Unsurprisingly, the largest number of CPU-intensive jobs was found in the alignment, recalibration and variant calling steps of the pipeline. Both the BWA-MEM and Haplotype caller are computationally intensive components of the pipeline in which CPU architecture and speed completely determine efficiency and time-to-solution. Our analysis of CPU architecture therefore naturally focused on these components of the pipeline.
Figure 7.
Distribution of GATK CPU time
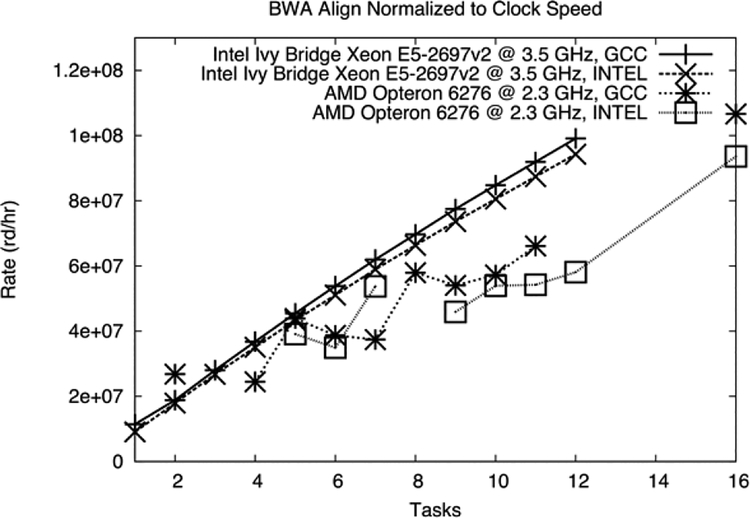
Figure 8 shows the strong scaling performance of two CPU configurations during BWA-ALN alignment for our two trial CPU architectures. The performance variability of the AMD Opteron chips was immediately apparent. This originated from our desire to maximally utilize our very wide, 64-way AMD Opteron nodes. Here, each alignment job was given 16 threads in a cgroup (as described in the initial Minerva design above), and other jobs may be scheduled on the remaining cores. In contrast, the 12-core Intel Ivy Bridge nodes were scheduled as a whole, limiting this performance variability. We found very early on that both scheduling and performance vastly improved as we reduced node core width, so that CPU-intensive portions of the genomics pipeline utilized all of a node’s resources without wasting cycles. Although the performance improvement for 3.5 GHz Ivy Bridge chips is clear in Figure 8, these processors also operated at a 52% higher clock rate. As described in the section above on scheduler performance, the vast majority of genomics pipeline jobs are single core, short jobs implemented in relatively poor-performing, high-level languages. In these cases, maximizing clock speed makes intuitive sense, as the codes are unable to take advantage of the feature sets present in modern CPU architectures. Alignment, however, is perhaps the only traditional-style HPC job in the genomics pipeline, consisting of highly optimized C code with excellent scaling performance. For this reason we normalized Figure 8 for clock speed, to indicate architectural improvement for BWA alignment. This result was fundamental to our choice of next-generation CPU for Minerva. Returning to the overall results of Figure 8, we found a maximum alignment time on 12 Ivy Bridge cores of 4.33 hours, compared to 10.86 using 16 AMD Opteron cores, with the same underlying IBM Flash+GSS file system. Similarly we found maximum job lengths of 37.87 and 86.07 hours for Intel Ivy Bridge over AMD Opteron Interlagos, respectively, during the variant calling procedure. Similar reductions in maximum and median wall time were apparent throughout all components of the pipeline. In total, the total performance improvement for the whole pipeline was 2.51 times when using small nodes of Intel Ivy Bridge over our wide AMD Opteron cores (471 vs. 1,182 hours, respectively).
Figure 8.
Strong scaling on BWA
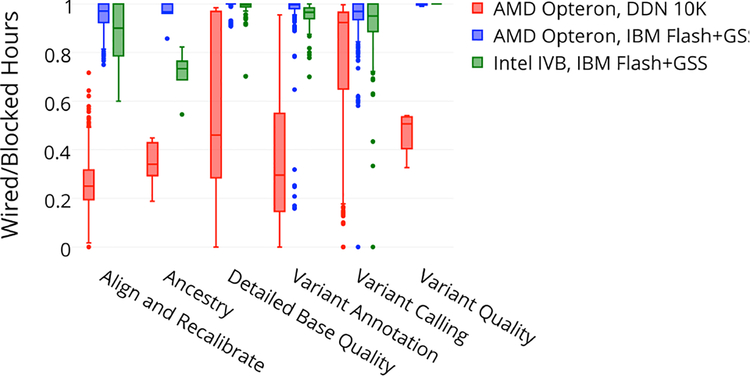
Further overall performance improvement was observed when considering the underlying storage subsystem. Figure 9 shows box plots of CPU efficiency for the six major categories of “GATK Best Practice” jobs. Here, efficiency is computed as the total CPU cycles consumed by the job (“wired” hours) divided by the total CPU-hours the job was resident on the cores (“blocked” hours, or CPU cycles produced during that time). It was clear that significant efficiency increases were gained by moving to the flash storage backed by IBM GSS. This is discussed further in section 7.
Figure 9.
GATK CPU efficiency
These results strongly suggest that the genomics pipelines were starved of IOPs when backed exclusively by spinning disks. While this improvement resulted in only a 5% decrease in CPU-time on the new storage subsystem (1,182 vs. 1,247 hours, respectively), it yielded an incredible 1.91 times improvement in the overall wall time (1,412 vs. 2,702 hours, respectively). Combining both our CPU and file system benchmark results, we observed a total wall time performance improvement of 4.8 times (562 vs. 2,702 hours, respectively). These results directly informed our design of the expanded Minerva, described next.
7. EXPANDED MINERVA DESIGN
Our overarching goal was to accelerate scientific discovery by (1) providing a stable and reliable platform with repeatable performance, (2) increasing to the maximal affordable size of storage to accommodate future growth, (3) improving the computational throughput through a tuned architecture, and (4) employing the best new technologies to boost our overall throughput. We selected our new design by reviewing our previous system performance in the context of our users’ application performance and projected usage. In section 4, we discussed the workflow and requirements for the scheduler and in section 5, we explored the projected compute and storage usage along with the results of our benchmarking as discussed in section 6. We then architected a system to meet a combination of the expected and actual user requirements using a selection of new technologies. We expanded Minerva in two phases: first in February 2014 funded by Mount Sinai, and the second in February 2015 funded by the NIH to build a Big Omics Data Engine (BODE). In the first expansion, we added 2,508 Intel Ivy Bridge compute cores, 160 terabytes of flash and 3 petabytes of IBM GSS storage. In the second expansion, we added an additional 2,484 Intel Haswell compute cores and 5 petabytes of DDN storage. The aggregate configuration is displayed in Table 3. The main interconnect for the cluster is Infiniband with both Quad Data Rate (QDR) and Fourteen Data Rate (FDR) connections between the nodes depending on their age.
Table 3.
Expanded Minerva configuration
| Minerva system | |
| Peak speed (TF) | 325 |
| # of core hours avail/year (millions) | 110 |
| # of cores | 12,864 |
| # of nodes | 536 |
| Amount of memory (TB) | 57 |
| Interconnection | QDR & FDR |
| Total/usable storage (PB) | 11.1/8.5 |
| Storage bandwidth (GB/s) | 130 |
| Parallel file system | GPFS |
| Resource manager/scheduler | LSF |
| Initial Minerva nodes (Dell) | |
| # of sockets | 4 |
| # of cores | 64 |
| Type of cores | AMD Interlagos |
| Speed of cores (GHz) | 2.3 |
| Amount of memory (GB) | 256 |
| Expanded Minerva nodes (IBM) | |
| # of sockets | 2 |
| # of cores | 12 |
| Type of cores | Intel Ivy Bridge |
| Speed of cores (GHz) | 3.5 |
| Amount of memory (GB) | 64 |
| Big Omics Data Engine (Cray) | |
| # of sockets | 2 |
| # of cores | 12 |
| Type of cores | Intel Haswell |
| Speed of cores (GHz) | 2.4 |
| Amount of memory (GB) | 64 |
7.1. Improved Scheduling
As we designed the expansion in 2013, it was evident to us that the high rates of job ingest and job querying made TORQUE a non-contender at that time. Therefore, we evaluated Slurm 2.6 and IBM’s Load Sharing Facility (LSF) 9.1. Although Slurm was designed to handle the scalability that we needed, it had several key missing features, such as high-speed accounting and DRMAA support. LSF had demonstrated the ability to schedule large numbers of jobs efficiently and reliably, allowed extensive tuning, and included support for all of our required functionality with a comprehensive set of programming APIs, including Python LSF wrappers and official DRMAA support. Based on our requirements, we selected LSF 9.1.2. We customized the LSF/Gold integration to support accounting for our high job load on Minerva. We also changed our queue structure from one that was based on job size to one based on self-determined priority. This enabled our users to have control over how quickly their jobs run and naturally balanced “urgent” clinical tasks with research projects to keep overall utilization high. Jobs submitted to higher priority queues are charged at a rate 1.5 times higher than allocated projects while low priority scavenger jobs are not charged at all. User-based fairshare policies are configured on all of the queues.
The HIGH_THROUGHPUT job template was used for the initial configuration. After the initial installation and configuration, we performed the vendor’s recommended tuning for large clusters. We then added customizations for cgroup support, increased the timeout for connections to the LIM API, Gold accounting integration and BLCR. The NEWJOB_REFRESH parameter was turned on to allow users see their job immediately after submission, and CONDENSE_PENDING_REASONS was disabled so that users see detailed information on why their jobs are pending. There has been no impact on scheduler performance from these two parameter changes. We also customized the “esub” and “eexec” scripts for LSF/Gold integration. A custom patch for DRMAA/LSF was needed to fix a missing job status parameter to charge a user’s Gold account for a job’s run time. Our utilization averaged about 84% for 2014, with long periods over 90%. Any LSF stability issues have been inconsequential compared with TORQUE.
We also enforced memory and core usage by creating a cgroup “container” for each job, binding cores and memory for that job’s exclusive use. Cgroups provided an interface to assign a cgroup to a linux PID and its child processes. LSF utilized this interface to create a new cgroup “container” for each job and place the main PID within that container.
LSF scaled for our workload due to (1) the multithreading of the “mbatchd” daemon, (2) the ability to utilize the increased number of allocated file descriptors on the master host, (3) the use of a local LIM for host status and load information, and (4) the limit on the number of job status queries. These features enabled LSF to support a high job submission rate and still remain responsive [36]. With LSF in production for over a year, we have not had any LSF-related crashes, and it has been responsive even with 500,000 jobs in the queue. We removed the limits on the number of jobs that each user can submit. LSF is not open source software and therefore any troubleshooting involves IBM support. Overall, we found LSF to be a highly configurable and stable scheduler capable of handling our demands of workload ingestion in large payloads.
7.2. Optimized File System
To improve the performance and reliability of the metadata server and the overall file system, we employed flash to hold the metadata and small files due to its low latency and high IOPs. Unfortunately flash is expensive so we only implemented it for the most necessary components in the storage subsystem.
GPFS offers the capability to place a file, based on its attributes, into a specific location that best addresses that file’s access requirements. This capability is called “tiering”. For example, small files that are accessed frequently may be placed on faster disks for faster retrieval, thus decreasing the job run time. This feature of GPFS is implemented using ILM policies. These policies are SQL-like statements that place a file initially at a location based on an attribute, (or on time, size, and an attribute), and then later move the file to a new location. We used ILM policies to specify a default initial file placement on the flash to quickly capture incoming files, both large and small. We wrote rules that run hourly to move files from flash to spinning disk based on owner, file size, and age. Our typical daily data ingest rate is between 1 and 10 terabytes. With this rate, our flash array accommodates a week’s worth of data. To ensure there is enough space to ingest new data, we migrate between 100 gigabytes to 1 terabytes data hourly to disk in approximately 30 minutes.
In GPFS 3.5, and later, IBM added support for large (4 kilobyte) inodes and the storage of “extra small” files in a file’s extended attributes. Typically inodes only store a file’s location in the file system and assigned data blocks, however, in recent years extended attributes have allowed administrators to add more specific metadata to files, such as ACLs and archiving status. By increasing the size of the inodes, the file system is providing better support for additional and more advanced uses of extended attributes. IBM chose to leverage this feature by placing the file data content into the extended attributes. With 4 kilobyte inodes, approximately 130 million files or over half of the files on our file system fit into the inode itself, thus saving us storage and improving I/O performance. This feature is extremely important for systems such as ours where we have millions of files that fit into these large inodes. The latency to lookup the file content was removed, as the content was delivered from the inode itself. Additionally, as the minimum allocation for a data block is one subblock (32 kilobytes on the 2013 system), even for zero size files, this new metadata feature saves approximately 4 terabytes of space on the 2013 system, or 16 terabytes on the 2014 system (128 kilobyte subblocks).
8. RELATED WORK
There has been significant recognition of the computational and data challenges posed by genomic sequencing [24,26,29,37,44,61,62]. Much of the work centers on the evaluation of existing tools and the optimization of existing tools and platforms. Researchers are especially interested in the evaluation of tools such as variant callers and their associated algorithms, as each tool interprets the genomic data with differing levels of specificity and sensitivity [16,17,21,25]. Many efforts have been made to optimize the underlying algorithms and I/O for many of these tools to reduce the time to solution. In our experience, the individual software optimizations are not accepted by the broader computational genomics community unless they are incorporated into the “GATK Best Practices” pipeline. Some have customized pipelines for their specific environments [24,37].
Projects such as Galaxy [59], Globus Genomics [40], SparkSeq [43], Google Genomics [38], ClusterK [39], Atlas2 Cloud [60], DNA Nexus, SeqInCloud [29] all provide the “GATK Best Practices” pipeline as a service backed by the cloud. This approach removes the need for local computation and storage resources. Many of the user interfaces for these services are graphical which some find easier to use.
Although various methods exist to collect small jobs together for more efficient scheduling and processing [65–68] including HTCondor [69] and DAGMan [70], integration with the GATK-Queue control process would require new plugins to manage individual job failures and other aspects of the pipeline.
ADAM is a new approach to improving data portability and scalability for genomic data built on Apache Spark [44]. Cloudburst uses Hadoop to map next generation sequence data [28]. SeqInCloud uses Microsoft Hadoop on Azure. The “GATK Best Practices” pipeline uses a proprietary MapReduce framework [9].
There has been significant research in the area of characterizing [41] and optimizing scientific workflows [42,54–58]. In addition, there has been work on characterizing scientific I/O patterns [51] and sharing best practices [52]. There has been some work evaluating flash storage for scientific workflows that did not find significant improvements in I/O performance [53]. This was not the case in our work, and helps to emphasize the importance of characterizing and designing systems around specific workflows. Other historical work includes optimizations for small I/O patterns [63] and estimates of batch queue delays [64].
Work describing actual HPC user and system metrics include papers on scheduling HPC workloads [46–48], parallel file systems [50] and reliability [45]. To the best of our knowledge, no prior work shares the actual usage and impact of the “GATK Best Practices” pipeline on the underlying computing system.
9. CONCLUSIONS
To accelerate scientific throughput on our computing resources, we performed a comprehensive analysis of user requirements, system metrics and benchmarks to architect an expanded system best matched to our actual usage patterns. In total, our expanded Minerva design provided nearly a five times speedup for our “GATK Best Practices” workflows, with slow downs due to storage subsystem and scheduler saturation eliminated.
We improved Minerva’s reliability, performance and utility for our users by studying the characteristics of our job mix and combining it with a new scheduler and optimized queue structure. We identified I/O bottlenecks and usage patterns with a new tool. We architected a new storage subsystem with flash, tiering and data stored in inodes that performed consistently, reliably and better for the large number of tiny files that the GATK-Queue workflow generated. We saved storage space by storing data within inodes. We benchmarked the most computationally intensive portions of the pipeline and selected a processor and node type tuned for our genomics workload. Our choices facilitated more jobs through the queue and a shorter job turn around time for our researchers.
We believe our strategy for collecting requirements through direct discussions with scientists coupled with data analysis from actual and projected usage is an effective approach to architect a system. A further step that might be helpful would be to “replay” our actual workload on a scheduler simulator to experiment with how changes to our scheduling policies might improve throughput. Another technique for increasing the throughput of the workflow might be to further explore the work done with threading and resource provisioning and workflow scheduler optimization [37,41,42,54–58].
Ultimately, our experience made clear the vastly different style of workflow required by genomics researchers, which required a critical re-thinking of the standard, large and wide job paradigm most typical of HPC workloads. Our thorough understanding of the actual usage patterns empowered us to build a system optimized for increased scientific productivity.
10. ACKNOWLEDGMENTS
Warm thanks to Ray Paden at IBM for helping us with GPFS and its underlying hardware, Frank Leers at DDN for the automated scripting of DDN commands, Larry Adams at IBM for LSF and Gold integration and Paolo Narvaez at Intel. Thanks to Elizabeth Hand for expert editing, Michael Linderman, Arthur Goldberg and Hardik Shah for their expertise with our “GATK Best Practices” pipeline, and Devesh Tiwari for his support. Thanks to Jonathon Anderson for his contributions to the Minerva system and data gathering scripts. We are grateful to Geraldine Van der Auwera, Eric Banks and from the Broad Institute for their advice. Special thanks to Mark Roberts and Troy Baer for their insight.
Research reported in this paper was supported by the Office of Research Infrastructure of the National Institutes of Health under award number S10OD018522. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
11. REFERENCES
- [1].Fromer M et al. 2014. De novo mutations in schizophrenia implicate synaptic networks. Nature 506 (2014), 179–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Purcell S et al. 2014. A polygenic burden of rare disruptive mutations in schizophrenia. Nature 506 (2014), 185–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Gaugler T et al. 2014. Most genetic risk for autism resides with common variation. Nat. Genet 46 (2014), 881–885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Neale B et al. 2012Patterns and rates of exonic de novo mutations in autism spectrum disorders. Nature 485 (2012), 242–245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Tu Z et al. 2012. Integrative Analysis of a Cross-Loci Regulation Network Identifies App as a Gene Regulating Insulin Secretion from Pancreatic Islets. PLoS Genet. 8 (2012), e1003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Deloukas P et al. 2012. Large-scale association analysis identifies new risk loci for coronary artery disease. Nat Genet 45 (2012), 25–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Gomes I et al. 2013. Identification of a μ-δ opioid receptor heteromer-biased agonist with antinociceptive activity. Proc. Natl. Acad. Sci. U. S. A 110, (2013), 12072–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gandy S, Haroutunian V et al. CR1 and the vanishing amyloid hypothesis of Alzheimer’s disease. Biol. Psychiatry 73 (2013), 393–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].McKenna A, Hanna M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. 2010. Genome Research 20 (2010), 1297–303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].DePristo M, et al. 2011. A framework for variation discovery and genotyping using next-generation DNA sequencing data. Nature Genetics 43 (2011), 491–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Van der Auwera G, et al. 2013. From FastQ Data to High-Confidence Variant Calls: The Genome Analysis Toolkit Best Practices Pipeline. Current Protocols in Bioinformatics 43 (2013), 11.10.1–11.10.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].GATK Best Practices, 2012. Retrieved July 1, 2012, from Broad Institute: https://www.broadinstitute.org/gatk/guide/best-practices.
- [13].Goldsmith P Broad Institute, Google Genomics combine bioinformatics and computing expertise to expand access to research tools. Retrieved July 15, 2015, from Broad Institute: https://www.broadinstitute.org/news/6994.
- [14].Introduction to GATK, 2012. Retrieved July 1, 2012, from Broad Institute: https://www.broadinstitute.org/gatk/about/.
- [15].RNASeq Overview GATK Best Practices. Retrieved July 1, 2012 from Broad Institute: https://www.broadinstitute.org/gatk/guide/best-practices?bpm=RNAseq.
- [16].Pabinger S, et al. 2014. A survey of tools for variant analysis of next-generation genome sequencing data. Brief Bioinform (2014), 15(2), 256–278. DOI: 10.1093/bib/bbs086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Liu X, et al. 2013. Variant callers for next-generation sequencing data: a comparison study. PLoS One (Sep. 2013), 8(9): e75619 DOI: 10.1371/journal.pone.0075619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].GENALICE, Technology for People and Science. Retrieved June 15, 2015 from GENALICE: http://www.genalice.com/.
- [19].Langmead B, Schatz M, Lin J, Pop M, Salzberg S 2009. Searching for SNPs with cloud computing. Genome Biology (2009), 0:R134 DOI: 10.1186/gb-2009-10-11-r134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Xie Y, Wu G, et al. 2014. SOAPdenovo-Trans: de novo transcriptome assembly with short RNA-Seq reads. Bioinformatics (Jun. 2014),15;30(12):1660–6 DOI: 10.1093/bioinformatics/btu077. [DOI] [PubMed] [Google Scholar]
- [21].O’Rawe J, Jiang T, et al. 2013. Low concordance of multiple variant-calling pipelines: practical implications for exome and genome sequencing. Genome Medicine (2013), 5:28DOI: 10.1186/gm432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Linderman MD; Brandt T; Edelmann L et al. 2014. Analytical Validation of Whole Exome and Whole Genome Sequencing for Clinical Applications. BMC Med Genomics (Apr. 2014), 7:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].De Rubeis S, He X, Goldberg A, et al. 2014. Synaptic, transcriptional and chromatin genes disrupted in autism. Nature (2014), 515(7526): 209–215. DOI: 10.1038/nature13772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Puckelwartz MJ et al. 2014. Supercomputing for the parallelization of whole genome analysis. Bioinformatics (2014), 30, 1508–1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Chapman B 2013. Scaling variant detection pipelines for whole genome sequencing analysis. Blue Collar Bioinformatics, May 22, 2013. Retrieved June 15, 2015 from http://bcb.io/2013/05/22/scaling-variant-detection-pipelines-for-whole-genome-sequencing-analysis/.
- [26].Baker M 2010. Next-generation sequencing: adjusting to data overload. Nature Methods (2010) 7:495–499. [Google Scholar]
- [27].Schmuck F, Haskin R 2002. GPFS: A Shared-Disk File System for Large Computing Clusters, USENIX File and Storage Technologies Conference (San Jose, CA, 2002). [Google Scholar]
- [28].Schatz M 2009. CloudBurst: Highly sensitive read mapping with MapReduce. Bioinformatics (2009), 25 (11), 1,363–1,369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Mohamed N, Lin H, Feng W 2013. Accelerating Data-Intensive Genome Analysis in the Cloud. Proceedings from the 5th International Conference on Bioinformatics and Computational Biology (2013). [Google Scholar]
- [30].qinyuan/ganglia-gpfs-plugin. Retrieved December 15, 2013 2013. from GitHub: https://hithub.com/qinyuan/ganglia-gpfsplugin.
- [31].The Glade Environment. Retrieved December 15, 2013 from GPFS User Group: http://files.gpfsug.org/presentations/2013/SC13GPFSUserForum.pdf.
- [32].Interpreting GPFS Waiter Information. Retrieved December 15, 2013 from IBM developerWorks: https://www.ibm.com/developerworks/community/wikis/home?lang=en#!/wiki/General%20Parallel%20File%20System%20%28GPFS%29/page/Interpreting%20GPFS%20Waiter%20Information
- [33].IBM Corp. 2013. General Parallel File System Administration and Programming Reference, Version 3 Release 5.0.11, 386–390. [Google Scholar]
- [34].Monitoring GPFS I/O performance with the mmpmon command. Retrieved December 15, 2013 from IBM Knowledge Center. http://www-01.ibm.com/support/knowledgecenter/SSFKCN_3.5.0/com.ibm.cluster.gpfs.v3r5.gpfs200.doc/bl1adv_mmpmonch.htm
- [35].Highcharts Products. Retrieved December 30, 2015 from Highcharts: http://www.highcharts.com/
- [36].CERN IT Facility Planning and Procurement. Retrieved January 15, 2014 from Indico: https://indico.cern.ch/event/92498/session/7/contribution/7/material/slides/0.pdf.
- [37].Kawalia A, Motameny S, et al. 2015. Leveraging the Power of High Performance Computing for Next Generation Sequencing Data Analysis: Tricks and Twists from a High Throughput Exome Workflow. PLoS ONE (2015). 10(5): e0126321. doi: 10.1371/journal.pone.0126321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Broad Institute GATK on Google Cloud Platform. Retrieved June 15, 2015 from: Google Cloud Platform. https://cloud.google.com/genomics/gatk?hl=en
- [39].Proffitt, A. Playing the Markets: ClusterK Launches Cloud Scheduler, Open Source GATK Pipeline. Retrieved June 15, 2015, from Bio-IT World: http://www.bioitworld.com/2015/2/18/playing-markets-clusterk-launches-cloud-scheduler-gatk-pipeline.html.
- [40].Bhuvaneshwar K, et al. A case study for cloud based high throughput analysis of NGS data using the globus genomics system. Computational and Structural Biotechnology Journal, 13 (2015). 64–74. DOI: 10.1016/j.csbj.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Juve G, et al. 2013. Characterizing and profiling scientific workflows. Future Generation Computer Systems, 29 (2013), 682–692. [Google Scholar]
- [42].Singh G, Deelman E The interplay of resource provisioning and workflow optimization in scientific applications. Concurrency and Computation: Practice and Experience, 23, (January 2011), 1969–1989. [Google Scholar]
- [43].Wiewiorka M, et al. 2014. SparkSeq: fast, scalable, cloud-ready tool for the interactive genomic data analysis with nucleotide precision. Bioinformatics (May 19, 2014). DOI: 10.1093/bioinformatics/btu343. [DOI] [PubMed] [Google Scholar]
- [44].Massie M, et al. ADAM: Genomics Formats and Processing Patterns for Cloud Scale Computing. University of California, Berkeley Technical Report, No. UCB/EECS-2013–207 (Dec. 15, 2013).
- [45].Kovatch P, et al. 2011. The Malthusian Catastrophe is upon us! Are the largest HPC machines ever up? Euro-Par 2011: Parallel Processing Workshops (Bordeaux, France, August 29 – September 2, 2011), Lecture Notes in Computer Science, 7156 211–220. Berlin/Heidelberg: Springer; DOI: 10.1007/978-3-642-29740-3_25. [DOI] [Google Scholar]
- [46].Andrews P, Kovatch P, et al. 2010. Scheduling a 100,000-core supercomputer for maximum utilization and capability. 39th International Conference on Parallel Processing Workshops (ICPPW), (San Diego, California, September 13–16, 2010). 421–427. DOI: 10.1109/ICPPW.2010.63. [DOI] [Google Scholar]
- [47].Samuel T, Baer T, Brook R, Ezell M, Kovatch P 2011. Scheduling diverse high performance computing systems with the goal of maximizing utilization. 18th International Conference on High Performance Computing (HiPC), (Bangalore, India, December 18–21, 2011), 1–6. DOI: 10.1109/HiPC.2011.6152723. [DOI] [Google Scholar]
- [48].Margo M, Yoshimoto K, Kovatch P, Andrews P 2008. Impact of reservations on production job scheduling Job Scheduling Strategies for Parallel Processing, Lecture notes in Computer Science, 4942, 116–131. Berlin/Heidelberg: Springer; DOI: 10.1007/978-3-540-78699-3_7. [DOI] [Google Scholar]
- [49].Andrews P, Banister B, Kovatch P, et al. 2005. Scaling a global file system to the greatest possible extent, performance, capacity and number of users. Twenty-Second IEEE / Thirteenth NASA Goddard Conference on Mass Storage Systems and Technologies (Monterey, California, April 11–14, 2005). 109–117. DOI: 10.1109/MSST.2005.30. [DOI] [Google Scholar]
- [50].Andrews P, Kovatch P, Jordan C 2005. Massive high performance global file systems for grid computing. Supercomputing, 2005. Proceedings of the ACM/IEEE SC 2005 Conference (May 12–18, 2005). 53–66. DOI: 10.1109/SC.2005.44. [DOI] [Google Scholar]
- [51].Carns P, et al. , 2011. Understanding and improving computational science storage access through continuous characterization. MSST’11 Proceedings of the 2011 IEEE 27th Symposium on Mass Storage Systems and Technologies (Denver, Colorado, May 23–27, 2011). 1–14. [Google Scholar]
- [52].Oral S, et al. 2014. Best practices and lessons learned from deploying and operating large-scale data-centric parallel file systems. SC’14 Proceedings of the International Conference for High Performance Computing, Networking, Storage and Analysis (New Orleans, Louisiana, November 16–21, 2014). 217–228. [Google Scholar]
- [53].Park S, et al. 2012. A performance evaluation of specific I/O workloads on flash-based SSDs. Proceedings from the Workshop on Interfaces and Abstractions for Scientific Data Storage (Beijing, China, September 24–28, 2012). [Google Scholar]
- [54].Ilyushkin A, et al. 2015. Towards a realistic scheduler for mixed workloads with workflows. Proceedings from the 15th IEEE/ACM International Symposium on Cluster, Cloud and Grid Computing, (Shenzhen, Guangdong, China, May 4–7, 2015). 653–756. [Google Scholar]
- [55].Mattoso M, Dias J, Ocana K, Ogasawara E, Costa F, Horta F, Silva V Oliveira D 2015. Dynamic steering of HPC scientific workflows: a survey. Future Generation Computer Systems (May 2015), 46, 100–113. [Google Scholar]
- [56].Chen W, da Silva R, Deelman E, Sakellariou R 2015. Using imbalance metrics to optimize task clustering in scientific workflow executions. Future Generation Computer Systems (May 2015), 46, 69–84. [Google Scholar]
- [57].Ferreira da Silva R et al. 2014. A unified approach for modeling and optimization of energy, makespan and reliability for scientific workflows on large-scale computing Infrastructures. Workshop on Modeling & Simulation of Systems and Applications (Seattle, Washington, August 13–14, 2014). [Google Scholar]
- [58].Maheshwari K, et al. 2014. Improving multisite workflow performance using model-based scheduling. 43rd International Conference on Parallel Processing, (Minneapolis, MN, September 9–12, 2014), 131–140. [Google Scholar]
- [59].Blankenberg D, et al. 2010. Galaxy: a web-based genome analysis tool for experimentalists. Current Protocols in Molecular Biology (Jan. 2010). 89:19.10.1–19.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Evani U, et al. 2012. Atlas2 Cloud: a framework for personal genome analysis in the cloud. BMC Genomics (2012), 13(Suppl. 6). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Wandelt S, et al. 2012. Data management challenges in next generation sequencing. Datenbank-Spektrum (Nov. 2012), 12(3), 161–171. [Google Scholar]
- [62].El-Metwally S, et al. 2013. Next-generation sequence assembly: four stages of data processing and computational challenges. PLoS Computational Biology (Dec. 2013). DOI: 10.1371/journal.pcbi.1003345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Carns P, et al. 2009. Small-file access in parallel file systems. IPDPS 2009. Proceedings of the 2009 IEEE International Parallel & Distributed Processing Symposium (Rome, May 23–29, 2009), 1–11. [Google Scholar]
- [64].Nurmi D, et al. 2007. QBETS: queue bounds estimation from time series. 13th Workshop on Job Scheduling Strategies for Parallel Processing (June, 2007). [Google Scholar]
- [65].High Throughput Computing—Looking Under the Hood of Nitro, 2015. Retrieved July 1, 2015 from Adaptive Computing: http://www.adaptivecomputing.com/products/high-throughput-nitro/
- [66].Anisimov V The aggregate job launcher of single-core or single-node applications on HPC sites, 2015. Retrieved July 30, 2015 from GitHub, Inc.: https://github.com/ncsa/Scheduler
- [67].PBS Tools: parallel-command-processor, 2013. Retrieved July 30, 2015 from The National Institute for Computational Sciences: http://www.nics.tennessee.edu/~troy/pbstools/man/parallel-command-processor.1.html
- [68].Parallel-command-processor, 2009. Retrieved July 1, 2015 from the Ohio Supercomputer Center: http://archive.osc.edu/supercomputing/software/apps/parallel.shtml
- [69].Thain D, et al. 2005. Distributed computing in practice: the condor experience. Concurrency-Practice and Experience, volume 17, number 2–4, 323–356. [Google Scholar]
- [70].Couvares P, et al. 2007. Workflow in Condor In Workflows for e-Science, Editors: Taylor I, Deelman E, Gannon D, Shields M, Springer Press, January 2007 (ISBN: 1-84628-519-4). [Google Scholar]