Abstract
We ascertained a child with early onset cerebellar ataxia and identified a novel frameshift deletion, c.359del [p. (Pro120Leufs*2), NM_052865.2] in exon 2 of MGME1 (mitochondrial genome maintenance exonuclease 1) by exome sequencing. Variations in MGME1 have been reported to cause mitochondrial DNA (mtDNA) depletion syndrome 11 (MIM #615084) in an earlier work. The phenotype included progressive external ophthalmoplegia, emaciation, respiratory failure and late onset progressive ataxia. However, the child presented here has early onset progressive ataxia, speech delay, microcephaly, cerebellar atrophy and fundus albipunctatus. This is the second report of a mutation in MGME1 and describes a more severe phenotype.
Keywords: MGME1, Mitochondrial DNA depletion syndrome, Exome sequencing, Ataxia
1. Introduction
Genetic disorders associated with impairment of maintenance of mitochondrial genome includes two broad subgroups: multiple mtDNA deletion disorders associated with large rearrangements in the mitochondrial genome and mtDNA depletion syndromes characterized by reduction in mtDNA copy number. Fifteen sub-types of mitochondrial depletion syndromes and their genetic etiology has been described till date. Of these, loss-of-function variations in MGME1 are known to cause mtDNA depletion syndrome 11 (MTDPS11) with consistent major clinical features of progressive external ophthalmoplegia and respiratory weakness. Three families with six affected individuals are reported with multisystemic mitochondrial disease due to pathogenic variants in MGME1 till date (Kornblum et al., 2013). In this report, we describe a child with early onset progressive cerebellar ataxia, speech delay, microcephaly, cerebellar atrophy, fundus albipunctatus with a novel frameshift deletion, c.359del [p.(Pro120Leufs*2), NM_052865.2] in exon 2 of MGME1 identified through exome sequencing. Our report reaffirms loss-of-function mutations in MGME1 are responsible for mitochondrial DNA depletion syndrome 11 and expands the clinical spectrum.
2. Clinical report
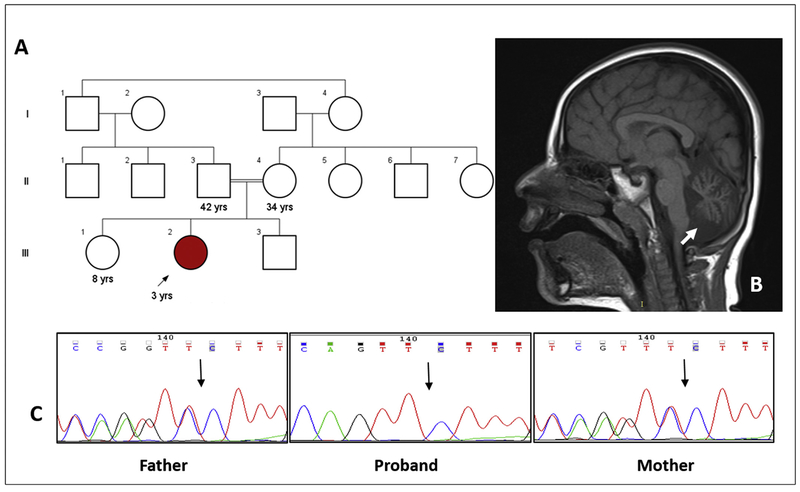
A three-years-old girl presented with early onset progressive ataxia noticed at two years of age and speech delay. She was born at full term pregnancy by normal vaginal delivery, to a third-degree consanguineous marriage (Fig. 1A), and weighed 3.45 kg at birth (normal). There were no perinatal complications. She has a healthy brother and a sister. She achieved social smile at three months, neck holding at four months, sat without support at 11 months and stood with support at and 18 months. She can walk with support at present. She spoke bisyllables by 11 months of age.
Fig. 1.
Pedigree of the family (A). Cerebellar hypoplasia can be noted on brain imaging (B; arrow). The variant (c.359del) was validated by Sanger sequencing in the proband and her parents (C).
At three years, head circumference was 42 cm (−7 SD), height was 87 cm (−2 SD), and weight was 11.5 kg (normal). Gait and truncal ataxia were present. Mild strabismus was noted. Tone was mildly decreased and deep tendon reflexes were normal. There was no significant nystagmus, emaciation, muscle atrophy, gastrointestinal symptoms, cardiac abnormalities or dysphonia observed in her. No cerebellar signs other than ataxia were noted.
Ophthalmic evaluation revealed fundus albipunctatus. Brain imaging showed cerebellar atrophy (Fig. 1B). Echocardiography, vitamin E and serum alpha fetoprotein values were observed to be normal.
2.1. Methods
Genomic DNA was extracted from the whole blood using the standard phenol-chloroform method. Genomic capture was carried out with Illumina’s Nextera Rapid Capture Exome Kit. Massively parallel sequencing was done using the NextSeq500 Sequencer (Illumina) in combination with the NextSeq™ 500 High Output Kit (2×150bp). Whole exome sequencing (WES) was carried out as described previously to achieve an average coverage depth of 100–130×, such that ~95% of the bases are covered at >20×, with a sensitivity of >90% (Girisha et al., 2016). WES raw data was processed using SeqMule and the called variants were annotated with ANNOVAR (Guo et al., 2015; Wang et al., 2010). Variants were filtered against public databases (accessed on 7th January 2017) such as 1000 Genomes Project phase 3, Exome Aggregation Consortium v.0.3.1 (ExAC), National Heart, Lung, and Blood Institute Exome Sequencing Project Exome Variant Server (ESP6500SI-V2) and an in-house exome database of 204 exomes and those with a minor allele frequency >1% were excluded. Additionally, variants flagged as low quality or putative false positives (Phred quality score <20, low quality by depth <20) were excluded from the analysis. The variants flanking 20bp from the exon were retained. Autosomal recessive conditions with homozygous variants were screened as parents are consanguineous.
Long range PCR was carried out according to the protocol and primers reported in Kornblum et al. to identify mtDNA deletions in the blood (Kornblum et al., 2013).
3. Results
A homozygous frameshift deletion, c.359del [p.(Pro120Leufs*2), NM_052865.2] was identified in MGME1 gene. The variant is a novel frameshift mutation leading to premature termination of the protein (loss of 222 amino acids) and predicted to cause nonsense mediated decay.
It is not observed in population databases like ExAC and 1000 genome project. In-silico analysis tools are consistent in predicting the variant may damage protein function. The variant was visualized on integrated genomic viewer v.2.3. The variant was validated by Sanger sequencing in proband. Segregation analysis carried out by targeted testing of parents using Sanger sequencing confirmed the carrier status in them (Fig. 1C). The variant information is submitted to ClinVar database in NCBI (Submission ID: SUB2635577). The protein is well conserved in lower organisms. Long range PCR in mtDNA did not reveal any gross deletions in the proband (Figure S1).
4. Discussion
We ascertained a child with early onset progressive ataxia, speech delay, microcephaly, cerebellar atrophy and fundus albipunctatus. A homozygous frameshift variation in MGME1 was identified in her.
The six patients reported in the literature demonstrated progressive external ophthalmoplegia, proximal weakness and generalized muscle wasting, profound emaciation and respiratory failure. The age of presentation of symptoms ranged from 10 years to 35 years. Variable intellectual disability, spinal deformities, gastrointestinal symptoms, cardiac arrhythmias, renal colic were also observed in them. Mild ataxia of adult onset was noted in a single patient with mutation in MGME1. However, early onset progressive ataxia has not been observed in the subjects with MTDPS11 till date. Intellectual disability in all three and microcephaly in two subjects from a family were reported previously. Cerebellar atrophy with or without clinical features of cerebellar disease have been observed in one family reported earlier. Retinal disease observed in our patient has not been reported in subjects with MTDPS11 previously (Kornblum et al., 2013).
The role of MGME1 as the first mitochondrial exonuclease in mtDNA replication and mtDNA maintenance is well established. One nonsense [c.456G > A, (p.Trp152*)] and missense variant [c.698A > G (p.Tyr233Cys)] each are described in MGME1 in the literature as the molecular cause for MTDPS11 (Kornblum et al., 2013). We identified a homozygous frameshift deletion in MGME1, c.359del [p.(Pro120Leufs*2), NM_052865.2], predicted to undergo premature termination of the protein leading to nonsense mediated decay. The variant is a novel frameshift mutation con-firming the loss-of-function of MGME1 implicated in MTDPS11. The other potentially pathogenic homozygous variants (Table S1) were in genes either unrelated to the proband’s phenotype or not known to cause human disease to date. Long range PCR showed multiple mitochondrial DNA deletions in the muscle biopsies of all affected individuals and in the blood and urine samples of one of the families reported by Kornblum et al. Appropriate tissue could not be obtained from the proband in our study, as the family did not consent for invasive testing. Mitochondrial deletions were not observed in her blood sample (Kornblum et al., 2013). No pathogenic variations were noted in genes implicated to cause fundus albipunctatus in our subject (RHO, PRPH2, RDH5, RLBP1).
This is the first report describing the early onset progressive ataxia, speech delay and fundus albipunctatus as a presenting feature of MTDPS11. Our report is the second study describing the pathogenic variation in MGME1. This report expands the clinical and mutational spectrum of the recently delineated condition, mitochondrial DNA depletion syndrome 11.
Supplementary Material
Acknowledgement
We thank the families who cooperated with the evaluation of the subjects and consented for participation in this study. This work was supported by National Institutes of Health funded the project titled ‘Genetic Diagnosis of Heritable Neurodevelopmental Disorders in India: Investigating the Use of Whole Exome Sequencing and Genetic Counseling to Address the High Burden of Neuro-developmental Disorders’(1R21NS094047–01).
Footnotes
Appendix A. Supplementary data
Supplementary data related to this article can be found at http://dx.doi.org/10.1016/j.ejmg.2017.07.010.
References
- Girisha KM, Shukla A, Trujillano D, Bhavani GS, Hebbar M, Kadavigere R, Rolfs A, 2016. A homozygous nonsense variant in IFT52 is associated with a human skeletal ciliopathy. Clin. Genet 90 (6), 536–539. [DOI] [PubMed] [Google Scholar]
- Guo Y, Ding X, Shen Y, Lyon GJ, Wang K, 2015. SeqMule: automated pipeline for analysis of human exome/genome sequencing data. Sci. Rep 5, 14283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kornblum C, Nicholls TJ, Haack TB, Scholer S, Peeva V, Danhauser K, Hallmann K, Zsurka G, Rorbach J, Iuso A, Wieland T, Sciacco M, Ronchi D, Comi GP, Moggio M, Quinzii CM, DiMauro S, Calvo SE, Mootha VK, Klopstock T, Strom TM, Meitinger T, Minczuk M, Kunz WS, Prokisch H, 2013. Loss-of-function mutations in MGME1 impair mtDNA replication and cause multisystemic mitochondrial disease. Nat. Genet 45 (2), 214–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang K, Li M, Hakonarson H, 2010. ANNOVAR: functional annotation of genetic variants from high-throughput sequencing data. Nucleic acids Res. 38 (16), e164. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.