Abstract
Mammary gland development is fueled by stem cell self-renewal and differentiation. External cues from the microenvironment coupled with internal cues such as post-transcriptional regulation exerted by microRNAs regulate stem cell behavior and fate. Here, we have identified a miR-205 regulatory network required for mammary gland ductal development and stem cell regeneration following transplantation into the cleared mammary fat pad. In the postnatal mammary gland, miR-205 is predominantly expressed in the basal/stem cell enriched population. Conditional deletion of miR-205 in mammary epithelial cells impairs stem cell self-renewal and mammary regenerative potential in the in vitro mammosphere formation assay and in vivo mammary reconstitution. miR-205 null transplants display significant changes in basal cells, basement membrane, and stroma. NKD1 and PTPA, which inhibit the Wnt signaling pathway, and AMOT, which causes YAP cytoplasmic retention and inactivation were identified as miR-205 downstream mediators. These studies also confirmed that miR-205 is a direct △Np63 target gene that is critical for the regulation of basal cell identity.
Keywords: miR-205, Mammary gland ductal development, Stem cell regenerative potential, YAP, Wnt
INTRODUCTON
The majority of mammary gland development occurs postnatally. The primitive gland formed during embryogenesis receives proliferative signals from systemic hormones and the extracellular matrix and initiates multiple cycles of extension, bifurcation, and lateral branching of the mammary epithelium. Terminal end buds (TEBs), which are located at the distal end of the growing ducts, are the bulb-like structures that actively proliferate and fuel the growth of the pubertal gland. Postnatally, mammary stem cells and progenitors are thought to reside initially in the outer cap cell layer [1]. By approximately 8 weeks of age, a mature mammary gland has formed, which consists of an outer basal layer enveloped by a basement membrane (BM) and an inner polarized epithelium surrounding a hollow lumen. Several transcriptional factors have been suggested to orchestrate mammary gland lineage specification. For example, Slug and △Np63 maintain the stem-like/basal state, Elf5 and GATA3 are crucial to induce luminal lineage specification during pregnancy, and Stat5 is essential for milk protein gene expression during lactation [2]. Involution is the final stage of this dynamic developmental process during which up to 80% of the alveolar epithelium undergoes massive apoptosis and the gland returns to a virgin-like state [3]. The pregnancy cycle can be repeated multiple times during the animals’ lifetime, supporting the existence of a regenerative mammary stem cell capacity in situ.
MicroRNAs (miRNAs) are a class of small noncoding RNAs that post-transcriptionally regulate multiple cellular processes through interaction with mRNAs. The 7–8 nucleotide-long seed sequence embedded in the 3’UTR of mRNAs enables target recognition by miRNAs. Minimal complementarity of the miRNAs 5’-end with the mRNA 3’UTR as well as △G value of this interaction determines the quality of the recognition [4]. Through mRNA degradation or translational repression, mRNA silencing is achieved, and tissue and stage-specific gene expression patterns are established.
Despite the potential of miRNAs to regulate large number of protein-coding genes, genetic deletion of a single miRNA usually does not cause severe developmental defects, most likely because of the redundancy of miRNA function. More recently, when analyzing the effect of germline miRNA loss on animal development, Park et al. surprisingly found that among the 11 mouse intergenic miRNAs examined, only miR-205 loss led to a perinatal lethal phenotype [5]. Although the underlying mechanism of lethality has not been fully understood, miR-205 has been shown to target negative regulators of the PI(3)K pathway in the epidermis and is essential to maintain stem cell self-renewal [6].
In previous studies from our laboratory, Greene et al. observed 80-fold higher miR-205 expression in CD24+CD29hi basal/stem cell-enriched mammary epithelial population isolated by fluorescence-activated cell sorting (FACS) analysis suggesting an important role of miR-205 in stem/progenitor cell regulation [7]. Additional studies also identified miR-205 as the top miRNA expressed in mammary epithelial progenitors isolated from the Comma-DB mammary epithelial cell (MEC) line [8]. Both of these studies suggest that similar to the epidermis, miR-205 might be critical for mammary stem cell homeostasis and mammary epithelial development. To explore this hypothesis, we utilized a miR-205 conditional knockout mouse model to examine its role in mammary gland development and stem cell regulation. Consistent with previous ex vivo observation, miR-205 is highly expressed in the mammary basal/stem cell-enriched population in vivo. Deletion of miR-205 severely impairs stem cell self-renewal capability resulting in incomplete outgrowths with altered stroma following transplantation. Loss of miR-205 results in elevated expression of negative regulators of YAP and the Wnt signaling pathway, which may be responsible for the inhibition of stem cell expansion and impairs the differentiation capacity of the basal epithelium. These studies also confirmed that miR-205 is a direct △Np63 target gene that is critical to differentially regulate basal cell identity. Together, the current data support a model where miR-205 plays an important role in specifying basal stem cell identity that is manifested during mammary reconstitution.
MATERIALS AND METHODS
Mice
The miR205 conditional knockout mouse was generated in Dr. M. McManus lab (University of California, San Francisco, CA). The FLP mouse was a generous gift from Dr. M. Dickinson (Baylor College of Medicine, Houston, TX). miR-205fl/fl; RosamTmG/mTmG [9] were kept in the C57BL6/129s mixed back-ground. SCID/beige purchased from Harlan Laboratories (Houston, TX, https://www.envigo.com/) were used to perform the cleared-fat pad transplantation assays. All mice colonies were maintained and euthanized according to the guidelines of the Institutional Animal Care and Use Committee of Baylor College of Medicine under the approved protocol AN-504.
X-Gal Staining for Whole-Mount Tissue
The fourth mammary glands were harvested from virgin to involution stages of miR-205fl/fl mice and then fixed in 2% paraformaldehyde for 3 hours at 4°C before staining in X-Gal staining solution (containing 1 M MgCl2, 5 M NaCl, 1 M pH 7.9 HEPES, 30 mM K4Fe2(CN)6•3H2O, 30 mM K3Fe2(CN)6, 10% NP-40, 1% X-Gal solution in DMF). After the 1X phosphate buffered solution wash that followed the X-Gal staining step, tissues were sequentially dehydrated, fixed (Carnoy’s fixative), and rehydrated for nuclear fast red staining. Upon detection of the red counterstain, tissues were dehydrated and cleared in Histo-Clear overnight. Whole-mount images were acquired by spreading the tissues out between two glass slides and imaging on a Leica MZ16F stereoscope.
Preparation and FACS-Purification of Primary Mammary Epithelial Cells
Primary MECs were isolated as previously described [10] with minor modification from virgin (8-wk, n = 20), early pregnant (P6, n = 8), late pregnant (P18, n = 5), lactation day 4 (n = 3), lactation day 9 (n = 3), and involution day 2( n = 8) wild type (WT) mice. Briefly, the third, fourth, and fifth mammary glands were harvested, minced, and digested in Hanks’ balanced saline solution (HBSS) containing 1 mg/ml Collagenase A (Roche, Basel, Switzerland, https://www.roche.com/) for 2 hours at 37°C followed by differential centrifugation to enrich for mammary organoids. The single cell suspension was obtained from organoids through sequential steps of trypsinization, washing, and filtration. Single cells were then resuspended in HBSS supplemented with 10 mM HEPES and 2% fetal bovine serum and incubated with anti-mouse CD24-PE (1:100, BD Biosciences, San Diego, http://www.bdbiosciences.com, 553262), anti-mouse CD24-PE-Cy7 (1:100, BioLegend, San Diego, https://www.biole-gend.com/, 101821), anti-mouse CD49f-FITC (1:100, BioLegend, 313606), and anti-mouse CD49f-Pacific Blue (1,100, BioLegend, 313620). Cells were sorted using a FACS Aria II Cell Sorter (BD) and data were processed using FlowJo version 10.0.8 (Tree Star, Ashland, OR, http://www.treestar.com/). Mammary basal cells were gated as CD24+CD49fhi, luminal cells were gated as CD24hiCD49flo, and stem cells were gated as CD24hiCD49fhi. FACS-purified cells are used later for RNA extraction, limiting dilution transplantation, and RNA-seq.
RNA Isolation, Reverse Transcription, and Real-Time Polymerase Chain Reaction (RT-PCR) miRNA Expression Analysis
Total RNAs from FACS-purified cells were isolated using miR-Neasy Minikit (QIAGEN, Hilden, Germany, https://www.qiagen.com/us/) and quantified using a NanoDrop (ThermoScientific, Waltham, MA, https://www.thermofisher.com/). Reverse transcriptions were performed using Taqman MicroRNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, www.appliedbiosystems.com) and specific stem-loop RT primers designed for miR-205 and control U6 from Taqman Small RNA Assays (Life Technologies, Rockville, MD, http://www.lifetech.com, 4427975). Probes used in RT-PCR were also from Taqman Small RNA Assays (Life Technologies) and other reagents were from Applied Biosystems. Reactions were performed according to the manufacturer’s manual and on a StepOnePlus Real Time PCR System (Applied Biosystems). Relative levels of gene expression were calculated using the 2–△△C(t) method.
Mammosphere Assay
For mixed population mammosphere assay, primary MECs were isolated from WT;RosamTmG/mTmG and miR-205fl/fl; RosamTmG/mTmG mice as described earlier [10]. Single MECs were then infected at an multiplicity of infection (MOI) of 30 with adenovirus-mediated Cre for 1 hour to induce miR-205 deletion. For recombined (green) mammosphere assay, green WT and miR-205−/− MECs were FACS purified directly from the primary cell transplants. WT and miR-205−/− MECs from both methods were then resuspended in mammosphere media and plated at a density of 10,000 cells per milliliter in a low-attachment six-well plate. The number of primary mammospheres was quantified and the mammosphere forming efficiency (MFE) was calculated after 10 days of culture. Secondary mammospheres were generated by dissociating primary mammospheres in 0.05% trypsin and re-plating at a density of 10,000 cells per milliliter in a low-attachment six-well plate. The number of secondary mammospheres was quantified and the MFE was calculated after 7 days of culture. The mammosphere media was defined as DMEM/F12 containing 1% methyl cellulose supplemented with 1X B27, 20 ng/ml basic fibroblast growth factor, 20 ng/ml epidermal growth factor, 100 ug/ml gentamycin, and 1X AA.
Immunostaining and Antibodies
Mammary glands were removed from 8-week WT and miR205−/− transplanted outgrowths and fixed in 4% paraformaldehyde overnight at 4°C before processing to be embedded in paraffin blocks. Five micrometer sections were cut from the blocks and underwent a series of steps of deparafinization and rehydration. Antigen retrieval was performed by boiling slides in Tris-EDTA buffer (10 mM Tris Base, 1 mM EDTA solution, and 0.05% Tween 20, pH 9.0) for 20 minutes in a microwave. Five percent of normal goat serum was used to block nonspecific binding and dilute primary antibodies. Mouse on Mouse (M.O.M) block and diluent were used for mouse/rat primary antibodies. The primary antibodies/dilutions/sources used were as follows: rabbit anti-△Np63 (1:200, BioLegend, 619002), rabbit anti-K5 (1:5,000, Covance, Princeton, NJ, http://www.covance.com, PRB-160P), rabbit anti-Laminin (1:200, Sigma, St. Louis, http://www.sigmaaldrich.com, L9393), rabbit anti-RFP (1:300, Rockland Immunochemicals Inc. Limerick, PA, https://rockland-inc.com/Default.aspx, 600–401-379), mouse anti-△Np63 (1:200, Living Colors, Kusatsu, Shiga Prefecture, Japan, https://www.takarabio.com/, 632381, clone number JL-8), mouse anti-SMA (1:250, Sigma, A2547), rat anti-K8 (1:250, Developmental Studies Hybridoma Bank, Iowa City, IA, http://dshb.biology.uiowa.edu/cytokeratin-Endo-A), rabbit-anti-YAP (1:500, Cell Signaling Technology, Beverly, MA, www.cellsignal.com, 4912), rabbit-anti-pYAP (1:500, Cell Signaling, 4911), rabbit anti-AMOT (1:500, Bioss, Woburn, MA, https://www.biossusa.com/, bs-4171R).
For immunohistochemistry, the Vectastain Elite ABC and diaminobensidine substrate kits (Vector Laboratories, Burlingame, CA, http://www.vectorlabs.com) were used to detect signal. Sections were then counterstained with Hematoxylin, dehydrated, and mounted using Permount (Fisher Scientific, Hampton, NH, www.fishersci.com/us/en/home.html).
Masson’s Trichrome
Sections from WT and miR-205−/− transplanted outgrowths were subjected to a series of steps of deparaffinization and rehydration then mordant in Bouin’s Solution (Millipore Sigma, St. Louis, Missouri, www.emdmillipore.com/US/en/home.html) at room temperature overnight to intensify the final coloration of the tissue. Weigert’s Iron Hematoxylin Set (Sigma) was used to stain nuclei. Sections were then put in Biebrich Scarlet-Acid Fuchsin from Accustain Trichrome Stain Kit (Sigma) for 5 minutes to stain cytoplasm and muscle red. Phosphomolyb-dic/Phosphotungstic Acid Solution (5 minutes) and Aniline Blue (5 minutes) from Accustain Trichrome Stain Kit (sigma) were used to develop collagen blue staining. Upon detection, sections were rinsed in 1% acetic acid solution for 5 minutes, dehydrated, and mounted using Permount (Fisher Scientific).
Cell and Tissue Serial Transplantation
Primary MECs were isolated from WT;RosamTmG/mTmG and miR-205fl/fl;RosamTmG/mTmG mice and infected with adenovirus mediated Cre as described earlier [10]. After washing three times with HBSS, cretreated Tomato+ WT and miR-205−/− cells were resuspended in 1:1 mixed growth factor-reduced Matrigel/HBSS. For limiting dilution transplantation, cells were diluted to 5,000, 1,000, 500, 100, 50 cells per 10 μl volume of growth factor-reduced Matrigel/HBSS for injection. Tomato+ WT and miR-205−/− cells from each cell dilution group were transplanted contralaterally to 3-week-old SCID-Beige mice (n = 5 for each dilution). Eight weeks after transplantation, fluorescent outgrowths whole-mount images were acquired as described previously. Note that since the cre-mediated recombination efficiency is not 100%, a stochastically mixed population of GFP+ and Tomato+ cells is expected.
For tissue serial transplantation, 100,000 and 1,000 cells per 10 μl volume of Matrigel/HBSS were transplanted to generate the primary outgrowths. Eight weeks after transplantation, five 1 mm fractions of epithelium were taken from the most distal tip of WT and miR205−/− outgrowths and retrains planted to 3-week-old SCID-Beige mice (n = 5 for each genotype). Eight weeks after transplantation, fluorescent outgrowths whole-mount images were taken using a Leica MZ16F fluorescent stereoscope.
In Vitro Culture of Primary WT and miR-205−/− MECs
Primary MECs were isolated and infected with Adenovirus mediated Cre as previously described. Freshly infected cells were washed two times and plated at a density of 1.5 × 106 cells in 10 cm dishes with MECs growth medium. Cells were collected 72 hours after the infection and GFP+ WT and miR-205−/− cells were FAC-sorted for quantitative RT-PCR (qRT-PCR) and Western blot analysis.
Western Blots and Antibodies
In vitro cultured primary WT and miR-205−/− MECs were tryp-sinized and collected in the lysis buffer. Protein concentrations were determined by the BCA assay. Thirty micrograms of protein from both groups were electrophoresed in 10% polyacrylamide gels and transferred to membranes overnight at 4°C. Next day, the membranes were blocked in milk at RT for 1 hour followed by primary antibody incubation overnight at 4°C. The primary antibodies and the concentrations used were: rabbit-anti-AMOT (1:200, gift from Dr. Sue-Hwa Lin at MD Anderson Cancer Center), rabbit-anti-YAP (1:1000, Cell Signaling, 4912), rabbit-anti-pYAP (1:1000, Cell Signaling, 4911), mouse-anti-PTPA (1:1000, EMD-Millipore, Burlington, MA, http://www.emdmillipore.com/US/en, 05–941), rabbit-anti-Naked1 (1:4000, Abcam, Cambridge, U.K., http://www.abcam.com, ab133650). Secondary antibody (Peroxidase affinipure goat anti-rabbit 1:10,000, Jackson ImmunoResearch Laboratories, West Grove, PA, http://www.jacksonimmuno.com, 111–035-003) was added on the third day and the image was acquired using an Amersham Imager 600.
3’UTR Luciferase Assay
A 800-bp DNA fragment flanking the pre-miR-205 hairpin was amplified from mouse genomic DNA and subsequently cloned into Sall and Hindlll sites of the PXF vector and validated by sequencing. 3’UTR fragments of RNA-seq identified potential miR205 targets were amplified from mouse cDNA. The fragments were then cloned into Sacl and Sall sites of pmirGLO vector (Promega, Madison, Wl, http://www.promega.com) and validated by sequencing. lndividual target site mutations were generated by recombinant PCR. For dual-luciferase assays, 100 ng pmirGLO reporter, 200 ng vector, or PXF-miR205 were transfected using Lipofectamine3000 reagent (lnvitrogen, Carlsbad, CA, http://www.invitrogen.com) into 293T and com-maD cells. Forty-eight hours after transfection, the relative luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega). All the primers used in cloning can be found in Supporting lnformation Table S3.
P63 BS Luciferase Assays
A 1 kb DNA fragment from miR-205 5’ upstream (–4.6 to −5.5 Kb) that encompasses the p63 BS was amplified from mouse genomic DNA, subsequently cloned into the Kpnl and Bglll sites of the pGL3 vector and validated by DNA sequencing. Point mutations and deletion of the BS were generated by recombinant PCR. The △Np63α expressing vector was obtained from Dr. Elsa Flores’ laboratory. For the luciferase assays, 100 ng pGL3-p63 BS, 100 ng △Np63α expressing vector, and 10 ng pRL Renilla control vector were transfected using Lipofectamine3000 reagent (lnvitrogen) into 293T cells. Forty-eight hours after transfection, the relative luciferase activities were determined using the Dual-Luciferase Reporter Assay System (Promega).
Lentiviral Transduction and miR-205 Inhibitor Treatment
HC11 cells were infected with the 7TCF-mCherry lentivirus at an MOl of 15 in 1 ml growth medium. mCherry+ cells were sorted and expanded 72 hours post infection. A negative control (NC) RNA (Dharmacon, lN-001005–01-05, miRlDlAN microRNA Hairpin lnhibitor Negative Control #1) and miR-205 inhibitor (Dharmacon, Lafayette, CO, https://dharmacon.horizondiscovery.com/, lH-310461–07-0005, miRlDlAN microRNA Mouse mmu-miR- 205-5p - Hairpin lnhibitor) were transfected by Lipofectamine RNAiMAX Transfection Reagent (ThermoFisher) at 200 nM to the mCherry+ cultures. Exogenous Wnt3a was added at 100 ng/ml 72 hours post inhibitor treatment. Cells were analyzed for GFP fluorescence intensity and Axin2 expression 24 hours post Wnt3a addition.
For the rescue assays, HC11 cells were infected by individual lentiviral-shRNAs (Sigma) targeting Amot, Nkd1, Ppp2r4, or in combination. Stable cell lines were generated by puromycin selection. A NC RNA and the miR-205 inhibitor were transfected at 200 nM to the stable cultures. Protein and RNA were harvested 72 hours post inhibitor treatment for analysis.
ChlP-qPCR
Mammary gland tissue was isolated from WT mice and crosslinked by 37% formaldehyde for 5 minutes at room temperature. Tissue chunks were then resuspended in cell lysis buffer (containing 10 mM TrisHCl, 0.25% Triton X-100, 10 mM EDTA, 0.1 M NaCl) and chromatin was extracted as described previously [11]. Chromatin extracts containing DNA with an average size of 500 bp were immunoprecipitated with 5 ug normal rabbit lgG (Cell Signaling, 2729) and anti-△Np63 antibody (BioLegend, 619002). The amount of DNA associated with target antibodies were determined by qPCR using ApoA4 as negative and Neuregulin –30 Kb as positive controls. See the list of primers used in Supporting lnformation Table S3.
RNA-seq
Primary MECs were isolated from WT and miR205−/− green outgrowths and sorted by flow cytometry into basal and luminal population according to different surface markers expression. RNA was extracted using the Arcturus PicoPure RNA lsolation Kit (Applied Biosystems) and submitted to the Baylor College of Medicine Genomic and RNA Profiling Core (GARP) for quality checks (QCs) and sequencing. GARP conducted sample QCs using the NanoDrop spectrophotometer and Agilent Bioanalyzer 2100, and then used NuGEN RNA-seq and lllumina DNA-Seq library preparation protocol. Paired-end reads were aligned to the mouse genome (mm9) using TopHat2. Raw counts for each gene in the Ensemble 67 gene model were measured with HTSeq. Using those raw counts, differentially expressed genes were called by edgeR with FDR 0.05. Fold changes in expression were reported by these packages after correcting for differences in library sizes.
RESULTS
miR-205 Expression Is Highly Enriched in Mammary Basal Cells and Stem Cells
Prior to studying the effects of miR-205 deletion, it was necessary to determine the expression pattern and localization of miR-205 at the different stages of mammary gland development. A miR-205 conditional knockout mouse model containing a LacZ fragment inserted upstream of the miR-205 locus (Fig. 1A) and X-gal staining were employed for these studies. During embryonic development, miR-205 is essentially uniformly expressed at E16 in the epithelium, but not the mesenchyme of the mammary anlage (Supporting lnformation Fig. S1A, arrowheads). By 5 weeks postnatal stage, miR-205 is expressed both in the TEBs and the subtending ducts, with predominant expression seen at the outer cap cell layer of TEBs and the basal cells as illustrated in both the mammary gland whole-mounts and histological sections (Fig. 1B). We also observed that miR-205 expression in the TEBs colocalizes with △Np63 (Fig. 1B, arrows) and identified miR-205/△Np63 positive cells in a small number of body cells (Fig. 1B, arrowheads) of TEBs. These △Np63 positive cells have been termed as “cap-in-body-cell,” which migrate from the cap cell layer and undergo apoptosis in the body cell compartment as detailed in a previous study [12]. A similar expression pattern is observed in the mature gland (8-week postnatal, Fig. 1C) and during early pregnancy (Fig. 1D, EP), when miR-205 is predominantly expressed in the basal layer. However in late pregnancy, while miR-205 expression is maintained in the basal layer of the ducts (Fig. 1D, LP arrows), it was not detected in the basal layer of alveoli (Fig. 1D, LP arrowheads). At day 1 of involution, miR-205 was not detectable (Fig. 1D, Inv-D1 arrowheads) but by day 3 of involution, miR-205 expression was again observed in the basal layer of the residual ducts (Fig. 1D, Inv-D3 arrowheads). At all stages of mammary gland development, miR-205 expression was undetectable in the mammary gland luminal layer and the stroma (Higher magnification images of corresponding stages are presented in Supporting Information Fig. S1B, S1C). To confirm the X-gal staining results, we performed FACS to purify basal, luminal, and stem cell-enriched populations from WT mammary glands across different developmental stages (Supporting Information Fig. S1D). qRT-PCR analysis of Keratin 14 (Krt14) and Keratin 8 (Krt8) expression confirmed the purity of each population (Supporting Information Fig. S1E). MiR-205 was found to be expressed primarily in basal/stem cells (Fig. 1E).
Figure 1.
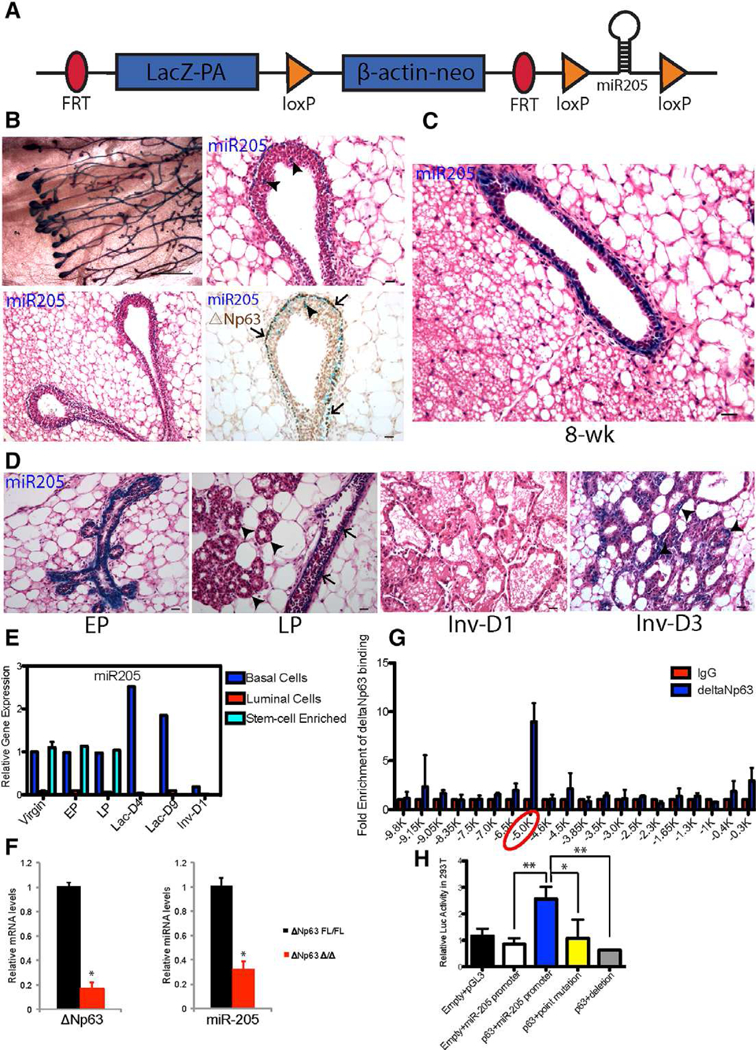
miR-205 is predominantly expressed in the basal/stem cell enriched population of the postnatal mammary gland. (A): Structure of the targeted miR-205 locus, in which the upstream lacZ fragment can be used to as a reporter of miR-205 expression. (B): MiR-205 is expressed in the outer cap cell layer of terminal end buds (TEBs) as well as the subtending ducts and co-localizes with △Np63 (arrows). There are also miR-205+/△Np63+ cells in a small number of body cells (arrowheads) of TEBs. (C): MiR-205 is expressed in the basal layer of 8-week postnatal mammary gland. (D): MiR-205 expression pattern in early pregnancy (EP: day 6 of pregnancy), late pregnancy (LP: day 18 of pregnancy), and involution day 1 and day 3. (E): MiR-205 expression pattern across different stages of development. (F): miR-205 expression displays remarkably decrease in the primary culture of Ad-cre treated △Np63fl/fl mammary epithelial cells (MECs). (G): The −5 Kb region upstream of miR-205 gene is bound by △Np63 as shown by ChIP-quantitative polymerase chain reaction (qPCR). The 20 primer sets were used to amplify the −10 Kb region upstream of miR-205 with ~500 bp intervals. (H): The miR-205 promoter region containing p63 BS was cloned into pGL3 reporter and the luciferase activity was measured with WT and mutated p63 BS. *p < .05; **p < .01 by unpaired Student’s t tests. Scale bars: (B) left panel 10 um; (B) right panel, (C), and (D) 20 um.
Both the coexpression of △Np63 and miR-205 and the observation that miR-205 was the top miRNA whose expression was altered in the conditionally deleted △Np63 epidermal stem cells [13] suggested that miR-205 might be a direct p63 gene target. Therefore, we wanted to determine if this was true in MECs. Since no outgrowths were observed following transplantation of conditionally deleted △Np63 MECs, the expression change of miR-205 was determined in primary △Np63fl/fl MECs treated with Adenovirus-Cre (Fig. 1F). These studies revealed a marked decrease in miR-205 in these shortterm primary cultures. Chromatin immunoprecipitation sequencing (ChIP) has indicated that there is a p63 consensus binding site (BS) [14] at ~5 Kb region upstream of the murine miR-205 gene. ChIP-qPCR scanning using 20 primers covering ~10 Kb upstream of miR-205 gene confirmed that there was a strong enrichment of △Np63 binding at the ~5 Kb region in mammary gland (Fig. 1G). A 1 kb DNA fragment containing the ANp63 BS (GGCATGTC) was inserted into a luciferase reporter. The expression of △Np63 resulted in a several fold induction of luciferase expression, whereas point mutation of C and G, two bases required for base recognition as well as deletion of the BS, significantly reduced luciferase activity to a basal expression level (Fig. 1H).
miR-205 Deletion Severely Impairs Mammary Gland Regenerative Potential
The pattern of basal cell expression suggested that miR-205 might play an important functional role in mammary gland development. To examine this possibility, miR-205 was conditionally deleted in MECs using Adenovirus-Cre. To facilitate these studies, we bred WT and miR-205fl/fl mice first to FLP mice resulting in deletion of the lacZ reporter and then to the mTmG reporter mice. This allowed for the transition from the Tdtomato to the GFP fluorescent reporters following recombination, which therefore facilitated visualization and isolation of both recombined and nonrecombined MECs by confocal microscopy and FACS and also acted as a surrogate for miR-205 deletion (Fig. 2A). Accordingly, miR-205 deletion in green MECs sorted from miR-205fl/fl;RosamTmG/mTmG cre+ mice was confirmed by qPCR (Fig. 2B). For the sake of simplicity, we will refer green WT;RosamTmG/mTmG cre+ and miR-205fl/fl;RosamTmG/mTmG cre+ outgrowths/MECs as WT and miR-205−/−, respectively, hereafter.
Figure 2.
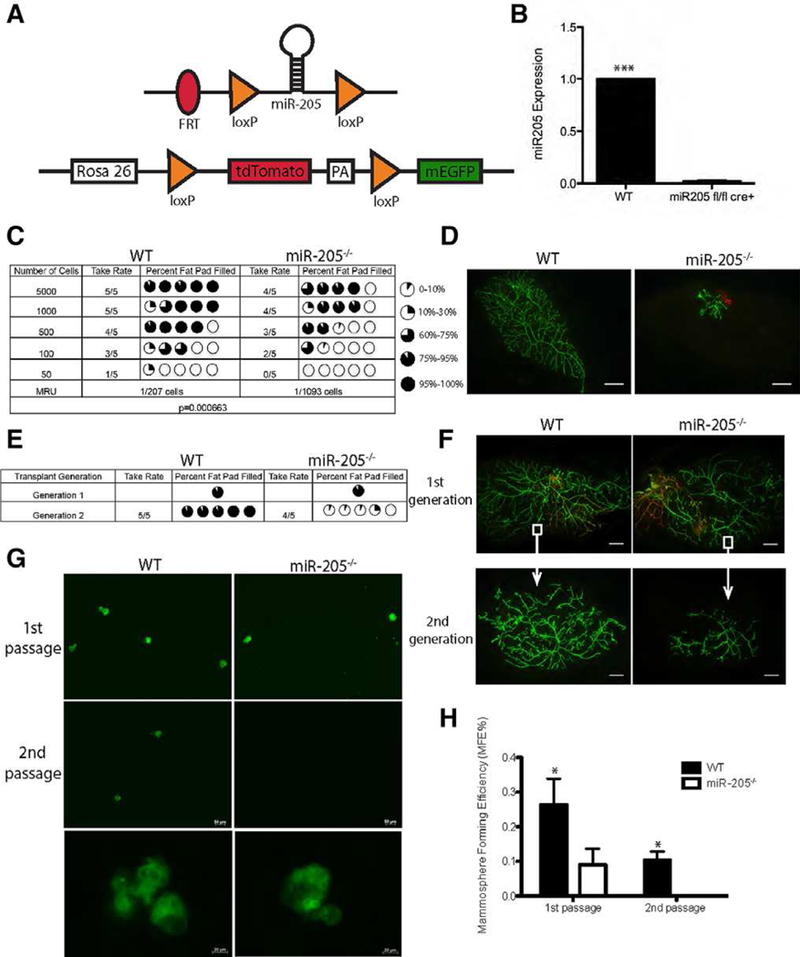
Conditional depletion of miR-205 compromises stem-cell self-renewal. (A): Structure of mTmG double fluorescent reporter and targeted miR-205 locus following the deletion of the LacZ fragment using the Flp recombinase. (B): Quantitative polymerase chain reaction (qPCR) confirms the deletion of miR-205 in green mammary epithelial cells (MECs) sorted from miR-205fl/fl;RosamTmG/mTmG cre+ mice. (C): MECs from WT and miR-205−/− mice were serially diluted and injected contralaterally at doses shown in the table. The mammary repopulating units (MRUs) were calculated based on the take rate and the percentage of fat pad filled using ELDA (extreme limiting dilution analysis) software. Note that the MRU of WT is significantly higher than that of the miR-205−/− MECs (p = .00066). (D): Representative fluorescent whole-mount images of primary outgrowths generated from transplantation of 500 WT and miR-205−/− MECs. (E): Pieces of distal tissue (white squares in panel F) from the primary WT and miR-205−/− outgrowths generated from transplantation of 100,000 cells were retransplanted contralaterally into the cleared mammary fat pads of SCID-Beige mice to generate secondary outgrowths. Summary of the outgrowths observed in WT and miR-205 null transplants are shown in the table. (F): Representative images of primary and secondary outgrowths in WT and miR-205 null transplants. (G): Representative images of primary and secondary mammo-spheres generated from WT and miR-205−/− FACS-purified recombined green MECs. Note that the size of WT and miR-205−/− mammo-spheres are similar. (H): Plot of mammosphere forming efficiency (MFE) calculated for WT and miR-205−/− MECs. MFE is defined as the number of mammosphere formed divided by the number of total cells plated. Note that the MFE% of WT mammosphere is significantly higher than that of the miR-205−/− mammosphere in both the primary and secondary mammospheres. Data shown in (B) are mean ± SD from three independent experiments. Data shown in (H) are mean ± SD from three technical replicates (source green cells are isolated from five pairs of WT and miR-205 null outgrowths). Independent biological replicates not shown here using mixed population of red and green MECs also show similar results. *p < .05; **p < .01; ***p < .001 by unpaired Student’s t tests. Scale bars: (d) and (F) 20 um; (G) 50 um.
Insertion of the lacZ-neo cassette upstream of miR-205 locus results in a hypomorphic allele. Thus, miR-205 expression was markedly decreased in mice homozygous for the LacZ knock-in (Supporting Information Fig. S2A, upper left panel). Surprisingly, the analysis of mammary gland whole-mounts from these homozygous miR-205fl/fl mice did not reveal any significant growth defects compared with the WT mammary glands, except in a few cases a decreased branching phenotype was observed in miR-205fl/fl glands (Supporting Information Fig. S2A, WT and miR-205fl/fl−1, miR-205fl/fl−2). These results suggest that miR-205 may be dispensable for in vivo mammary gland development. However, this did not preclude that miR-205 might be important for the regenerative potential of stem cells under the stress conditions present during cell dissociation and transplantation into the cleared mammary fat pad, especially under limiting dilution conditions.
To test this hypothesis, we performed cell transplantation experiments using WT and miR-205−/− MECs into the cleared mammary fat pad. No significant difference in the extent of fat pad filling was observed in the outgrowths generated from 100,000 cells between WT and miR-205−/− groups. However, when the cell number injected was reduced to 1,000, miR-205 deletion severely impaired the regenerative potential, with only tiny primitive ductal structures observed (Supporting Information Fig. S2B, S2C). Because of the cell number dependent phenotype, we performed a more systematic limiting dilution transplantation assay, the “gold standard” for measuring functional mammary stem cell regenerative activity. As illustrated in the table of Figure 2C, with decreasing number of cells injected into the fat pad, miR-205−/− cells gradually lost the capacity to generate secondary outgrowths. Even when outgrowths were observed, the miR-205−/− outgrowths filled significantly less of the fat pad as compared to WT counterparts (Fig. 2D). Mammary repopulating units (MRUs) were calculated based on the number of outgrowths generated and the percentage fat pad filled. As expected, the MRU of miR-205−/− cells was significantly lower than that of WT (1/1093 vs. 1/207, p = .00066). In light of these results, we next examined whether the same phenotype could be observed in serial tissue transplantation assays using intact tissue rather than dissociated cells. Unlike the limiting dilution transplantation assays with dissociated cells, serial tissue transplantation preserves interactions between the MECs and the microenvironment, and therefore has been suggested to better reflect the physiological condition [15]. We utilized primary WT and miR-205−/− outgrowths generated from 100,000 cells that completely filled the fat pad and performed secondary tissue transplantation assay. Consistent with the limiting dilution transplantation results, the WT tissue maintained the ability to generate secondary outgrowths from pieces of distal tissue (Fig. 2F, white squares) transplantation, while comparable miR-205−/− tissue failed to regenerate complete outgrowths (Fig. 2E, 2F). In addition to the in vivo transplantation assays, we also performed in vitro mammosphere formation assays using recombined (green) MECs specifically sorted out from WT and miR-205−/− 100,000 cells primary outgrowths. The MFE of miR-205−/− MECs was significantly reduced as compared with WT MECs in both primary and secondary mammo-spheres, but their sizes were similar (Fig. 2G, 2H). Taken together, these results support a role for miR-205 in eliciting stem cell regenerative potential during reconstitution.
miR-205 Deletion Alters Epithelium and Microenvironment Composition
We next sectioned the green WT and miR-205−/− outgrowths to determine if there were any specific alterations that could be detected at cellular level. Given that miR-205 is predominantly expressed in the basal compartment, we first stained for the basal marker keratin 5 (K5) and the luminal marker keratin 8 (K8). As expected, WT glands displayed a continuous K5 + basal layer encompassing the K8+ luminal layer in both the ductal and TEB structures. However, a partial loss of K5+ cells was observed in the miR-205−/− outgrowths (Fig. 3A, arrowheads), while most of the K8+ luminal layer remained intact. Next, we examined the cellular composition of the “yellow outgrowths” that were generated in both the WT and miR-205−/− transplants when high numbers (100,000) of cells were transplanted. When high numbers of cells were transplanted, we normally observed a mixture of red, green, and yellow outgrowths, since in the starting pool of the Ad-Cre treated miR-205fl/fl;RosamTmG/mTmG MECs, the recombination efficiency of Cre is not a 100% and as expected, there would be both WT (RFP+) and miR-205−/− (GFP+) cells present. When examined at the cellular level, we observed that GFP+ and RFP+ cells were equally distributed in both the basal and luminal compartments in the WT yellow outgrowths. However, in the miR-205−/− yellow outgrowths that completely filled the fat pad, we observed the RFP+ WT cells exclusively lining the basal layer adjacent to the luminal GFP+ miR-205−/− cells (Fig. 3B). This observation suggests that it was essential to have WT basal cells to form an intact basal layer in order for the yellow outgrowths from the miR-205−/− group to fill the fat pad. Otherwise the outgrowths were stunted as observed in the “green outgrowths” generated from low numbers of transplanted cells. Collectively, these results observed with chimeric outgrowths highlight the indispensable role of the basal layer miR-205 in mammary gland morphogenesis.
Figure 3.
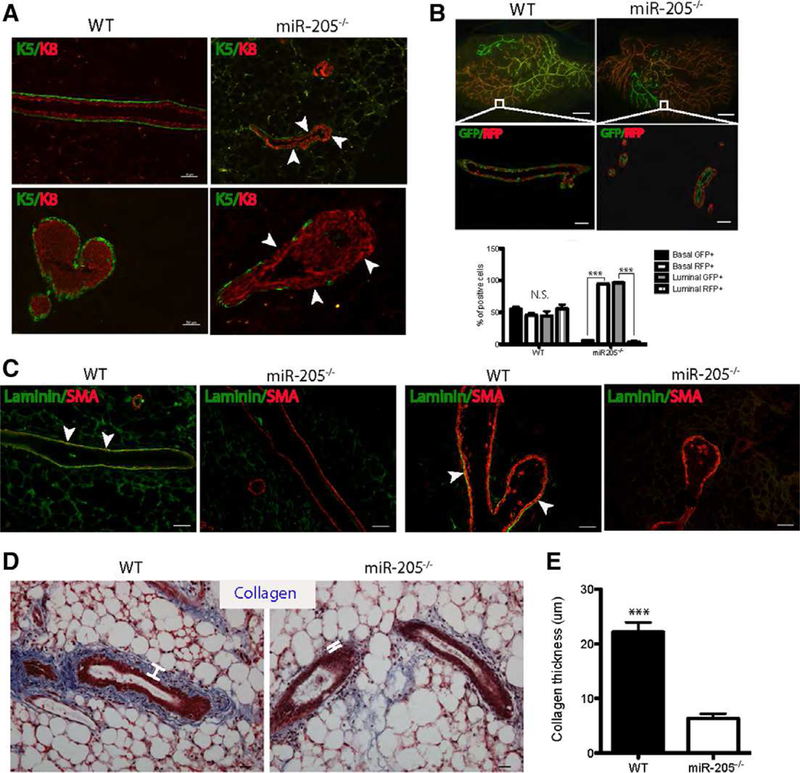
Ablation of miR-205 leads to alterations in the basal cells, extracellular matrix and stroma. (A): Immunofluorescence (IF) for K5 and K8 demonstrating the significant loss of K5+ basal cells (arrowheads) in miR-205−/− ducts (upper panel) and terminal end buds (TEBs) (lower panel) compared to that of WT. (B): Yellow WT and miR-205−/− outgrowths (white squares, generated from 100,000 MECs transplantation) were sectioned and stained with GFP and RFP. The quantification of the number of GFP/RFP+cells showed that in miR-205−/− glands, the basal compartment was mainly comprised of RFP+ cells, whereas the luminal compartment was mainly comprised of GFP+cells. (C): IF for laminin and αSMA demonstrating the loss of basement membrane in both the ducts and TEBs in miR-205−/− outgrowth compared to that of WT. (D): Trichrome staining depicting significant loss of collagen in miR-205−/− outgrowths compared to WT. White lines measured the thickness of collagen layers. (E): Collagen thickness in miR-205−/− outgrowths was significantly decreased compared to WT. Data shown in (B) and (E) are mean ± SD from three independent sections. ***p < .001 by unpaired Student’s t tests. Scale bars: (A) and (C) 50 um; lower panel in (B) 50 um; upper panel in (B) 20 um; (D) 20 um.
Knowing that BM components are mainly secreted by basal cells, we suspected that loss of basal cells might also lead to BM defects. Laminin staining confirmed the above hypothesis, in which WT outgrowths possessed an intact BM with basal cells also positive for laminin in both ducts and TEBs (Fig. 3C, arrowheads), but the miR-205−/− outgrowths lost the majority of the BM. Lastly, to examine possible changes in collagen formation around the mammary epithelium, we utilized trichrome staining and it revealed extensive loss of collagen deposition in miR-205−/− outgrowths as compared with the collagen-enriched WT outgrowths (Fig. 3D, white lines, and Fig. 3E). Thus, these studies suggest that defects in basal cells may affect the expression and deposition of BM and stromal components, which are essential to facilitate normal mammary outgrowth.
miR-205 Targets Negative Regulators of YAP and Wnt
Ribosome profiling has revealed that mRNA degradation is the dominant mechanism of miRNA-mediated gene silencing and accounts for 66%−90% of the inhibition observed in mammalian cell lines [16]. Therefore, we utilized RNA-sequencing to determine the differentially expressed genes in WT and miR-205 conditionally deleted outgrowths, which might represent potential miR-205 direct targets. To perform this analysis, we first isolated recombined green cells from pooled 40 WT and miR-205−/− 100,000 cell transplants. Green cells were further FACS-purified into CD24+CD49fhigh basal and CD24+CD49f+ luminal populations. RNA was extracted from individual population and subjected to amplification and sequencing. We identified a total of 8,427 differentially expressed genes including multiple noncoding RNAs between the WT basal and miR-205−/− basal populations. We focused on the 1,086 genes whose expression displayed more than a 25% increase in the miR-205−/− basal cells. Surprisingly, when comparing the 1,086 genes with the 696 TargetScan predicted, ingenuity pathway analysis (lPA) pathway-enriched miR-205 targets, only 57 genes overlapped between these two datasets (Fig. 4A). Amot and Nkd1 were the top two among the 57 genes, exhibiting more than 100% and 70% increase in miR-205−/− as compared to WT basal cells, respectively (Supporting lnformation Table S1).
Figure 4.
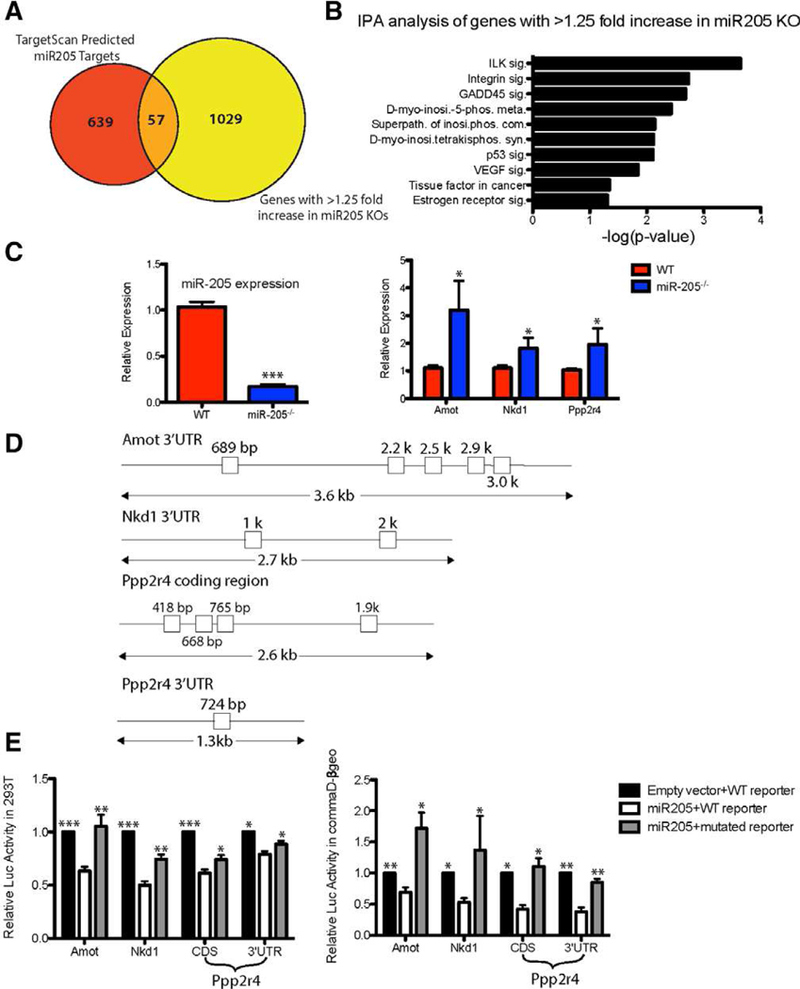
miR-205 targets negative regulators of YAP and canonical Wnt pathway. (A): RNA-seq identified 1,086 genes whose expression increased greater than 1.25-fold in the miR-205 null outgrowths. Among these, 57 were target-scan predicted miR-205 direct targets. Amot and Nkd1 were the top two targets on the list (see Supporting Information Table S1). (B): IPA analysis of the 1,086 genes whose expression increased greater than 1.25-fold in miR-205 null outgrowths. Ppp2r4 was a major component of multiple pathways, including ILK, D-myo-inositol-5-phosphate metabolism, integrin, and GADD45 signaling (also see Supporting Information Table S2). (C): Quantitative polymerase chain reaction (qPCR) confirmation of miR-205 deletion as well as targets (Amot, Nkd1, and Ppp2r4) derepression in short culture of primary WT;RosamTmG/mTmG cre+ and miR-205fl/fl;RosamTmG/mTmG cre+ MECs. (D): Seed sequences present in the 30UTR of Amot, Nkd1, Ppp2r4 and the CDS of ppp2r4. (E): 30UTR of potential targets were cloned into a luciferase reporter and the luciferase activity was measured with WT and mutated miR-205 target sites. Luciferase activity was significantly decreased when miR-205 was cotransfected with WT seed sequences in 293T cells and commaDβ-geo cells. Mutations generated in each of the seed sequences significantly restored the luciferase activity. *p < .05; **p < .01; ***p < .001 by unpaired Student’s t tests.
We also performed lPA pathway analysis on the 1,086 differentially expressed genes and identified several other pathways that are significantly enriched in miR-205−/− basal cells, including lLK, D-myo-inositol-5-phosphate metabolism, integrin, and GADD45 signaling (Fig. 4B). Ppp2r4, the gene encoding a specific phosphotyrosyl phosphatase activator (PTPA, the B56 subunit) of the dimeric form of the tumor suppressor PP2A, was present in 12 of the enriched pathways (Supporting lnformation Table S2). PP2A-B56 functions in the Wnt signaling pathway to downregulate β-catenin, possibly through dephosphorylating and activating GSK3β in the APC-axin complex [17].
qRT-PCR was performed on primary cultures of miR-205−/− MECs to validate the results obtained by RNA seq. As expected, all three identified targets displayed increased expression upon miR-205 deletion (Fig. 4C), although as expected the magnitude of these changes in the total MECs is less than that observed in purified basal cells by RNA seq. As illustrated in Supporting Information Figure S3B, qRT-PCR on purified basal cells used in the RNA-seq displayed an even greater increase in Amot in the miR-205−/− basal cells. Expression changes in AMOT at the protein level were confirmed by immunohistochemistry. Although AMOT expression was not detectable in the WT basal cells, it was observed by immunohistochemistry in most of the basal cells in the miR-205−/− sections. (Supporting Information Fig. S3C, arrowheads).
The frequently used miRNA target prediction algorithms, such as TargetScan, utilize the seed sequences present in the 3’UTR of mRNA to find potential targets. However, since Ppp2r4 did not appear to be a predicted target, we looked at both the coding sequence (CDS) and 3’UTR of Ppp2r4 for miR-205 seed sequences. Surprisingly, we found one seed sequence in the 3’UTR of Ppp2r4, along with four other seed sequences in the CDS (Fig. 4D). To test the responsiveness of the seed sequences present either in the 3’UTR or CDS of the identified targets, we cotransfected a miR-205 expression construct with luciferase reporter-fused Amot, Nkd1, and Ppp2r4 3’UTR as well as Ppp2r4 CDS and examined the changes in luciferase activity under the condition of WT and mutated miR-205 seed sequences. Luciferase activity was significantly decreased with WT seed sequences. Mutations generated in each of the seed sequences (Fig. 4D and Supporting Information Fig. S2D) significantly restored the luciferase activity (Fig. 4E).
Target expression changes at the protein level were confirmed by Western blot analysis in total MECs (Fig. 5A). As expected, AMOT, NKD1, and PTPA displayed increased expression in miR-205−/− MECs. One of the mechanisms by which AMOT may act is by regulating the oncoprotein YAP through Hippo pathway-independent and dependent mechanisms, both of which lead to YAP phosphorylation and cytoplasmic retention [18–20]. Therefore, we stained WT and miR-205−/− outgrowths to look at YAP expression and localization. As shown in Figure 5B, YAP was highly expressed in the basal layer of WT gland and displayed predominantly a nuclear localization. However, weaker and more diffused cytoplasmic YAP expression was detected in the basal cells from the miR-205−/− outgrowths, indicating the possible activation of pYAP. As shown in Figure 5C, pYAP was confined to the cytoplasm of luminal cells in the WT outgrowths. However, upon miR-205 deletion, pYAP expression was observed in the cytoplasm of basal cells, as seen in the pYAP/△Np63 and pYAP/SMA costaining. Total YAP and pYAP expression were both decreased in the primary cultures of miR-205−/− MECs, with the greatest decrease seen in total YAP expression (Fig. 5D). The specificity of anti-YAP and anti-pYAP antibodies was confirmed by Western blot analysis which revealed single bands of the expected sizes (Supporting Information Fig. S3D) as well as by immunofluorescence staining using kindly provided YAP null mammary glands as a control (Supporting Information Fig. S3E). Given the structural similarity and the established important role of TAZ in basal cells, we also examined TAZ expression in WT and miR-205−/− transplants. Total TAZ expression displayed a similar basal expression pattern as YAP, although to a lesser extent in WT. Interestingly, when miR-205 was absent, the majority of TAZ expression was only seen in luminal cells (Supporting Information Fig. S3F, arrowheads).
Figure 5.
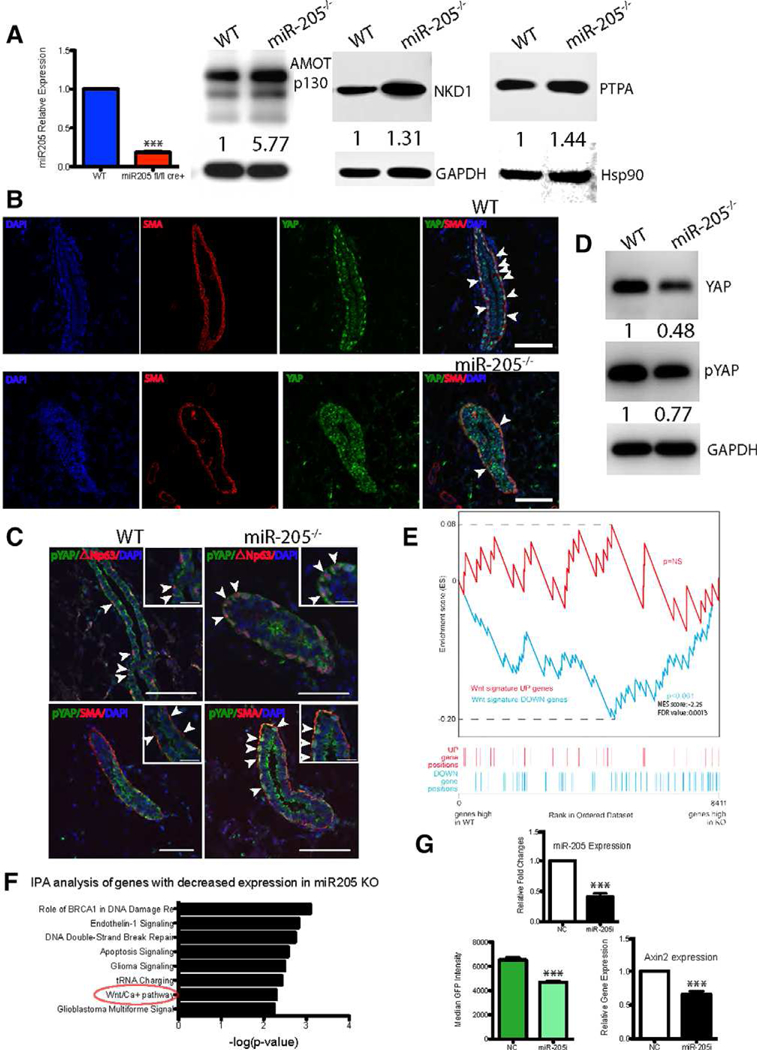
YAP and Wnt signaling alteration upon miR-205 deletion. (A): Left panel: Quantitative polymerase chain reaction (qPCR) showing there is ~80% reduction in miR-205 expression in 72 hour cultured and sorted Ad-Cre treated green miR-205−/− MECs. Right panel: Western blot analysis on the cultured MECs displaying increased AMOT, NKD1, PTPA expression in the miR-205−/− group. Note that the AMOT antibody used recognizes two AMOT isoforms. The labeled band is AM0Tp130, and the lowest band is AM0Tp80, which shows a similar extent of reduction. (B): IF for YAP and αSMA depicting loss of nuclear YAP in αSMA+ basal cells in miR205−/− outgrowths. Arrowheads show strong nuclear YAP expression in SMA+ basal cells in WT outgrowth. (C): IF for pYAP shows that the majority of pYAP expression was seen in the luminal cells in the WT outgrowth. However, pYAP expression was observed in the cytoplasm of △Np63+/αSMA+ basal cells in miR205−/− outgrowths. Scale bars are 50 um. (D): Western blot analysis on the cultured MECs displaying decreased YAP and pYAP expression in the miR205−/− group. (E): Gene set enrichment analysis showing that Wnt-down (Wnt pathway-repressed genes) signature was significantly enriched in the genes highly expressed in the miR-205 null group, but low in the WT group. (F): IPA analysis on the downregulated genes in miR-205 null outgrowths showing enrichment of Wnt/Ca2+ pathway. (G): miR-205 expression, median GFP fluorescence intensity generated from 7TGC Wnt reporter, and Axin2 expression were significantly decreased in the miR-205 inhibitor treated group compared to negative control in HC11 cells. Data shown in qPCR are mean ± SD from three biological replicates. ***p < .001 by unpaired Student’s t tests.
By directly associating with Disheveled, the branch point of canonical and noncanonical Wnt pathways, NKD1 is able to suppress the canonical Wnt pathway, but activate the noncanonical Wnt pathway through activating c-Jun-N-terminal kinase [21]. Therefore, we compared differentially expressed genes between WT and miR-205−/− outgrowths to the Wnt signature generated from the MMTV-Wntl transgenic mouse mammary gland [22] using gene set enrichment analysis. We found that Wnt pathway-repressed genes were significantly enriched in the genes highly expressed in the miR-205−/− group, but low to absent in the WT group (Fig. 5E). Moreover, the Wnt/Ca2+ pathway was enriched in genes down regulated in miR-205−/− outgrowths by IPA analysis (Fig. 5F). To confirm that miR-205 deletion represses Wnt signaling, we transduced MEC line HC11 with the lentiviral Wnt reporter 7TGC and then treated the cells with either a miR-205 inhibitor or a NC. Both the median GFP fluorescence intensity and Axin2 expression were significantly decreased in the miR-205 inhibitor treated group as compared to the NC (Fig. 5G). Thus, alterations in both the Wnt and YAP pathways are observed following deletion of miR-205.
To further confirm that Amot, Nkd1, and Ppp2r4 are indeed downstream mediators of miR-205, we performed rescue assays using Amot, Nkd1, Ppp2r4 shRNAs in the HC11 MEC line. As shown in Figure 6A, all three targets displayed increased expression in the scramble+miR-205i group as compared to that of scramble + NC, which is consistent with our observations in the miR-205−/− primary MECs. Target knock-downs were achieved in the shAmot + Nkd1 + Ppp2r4 + NC group, with a 40%, 60%, and 99% decrease in AMOT, NKD1, and PTPA expression, respectively. On the other hand, target expression was decreased in the shAmot + Nkd1 + Ppp2r4 + miR-205i group as compared to that of the scramble + miR-205i group, and therefore can be used to examine the rescue of YAP and Wnt signaling. As expected, YAP, pYAP, and Axin2 expression in the shAmot + Nkd1 + Ppp2r4 + miR-205i group increased to a comparable level to that of WT (scramble + NC) (Fig. 6B, 6C). Note that because the miR-205 inhibitor only resulted in a 60% deletion (Fig. 5G), the effects on specific targets and downstream pathways were not as significant as seen in the miR-205−/− primary MECs, which exhibit nearly 100% deletion of miR-205. Furthermore, since there are no “normal” mammary epithelial basal cell lines available for these studies and miR-205 is exclusively expressed in basal MECs, the different cell context in the luminal HC11 cells will no doubt influence the extent of rescue. Notwithstanding these caveats, together with the 3’UTR luciferase and Western blot assays showing target up-regulation in miR-205−/− MECs, we demonstrated that the identified targets are directly associated with YAP and Wnt pathways.
Figure 6.
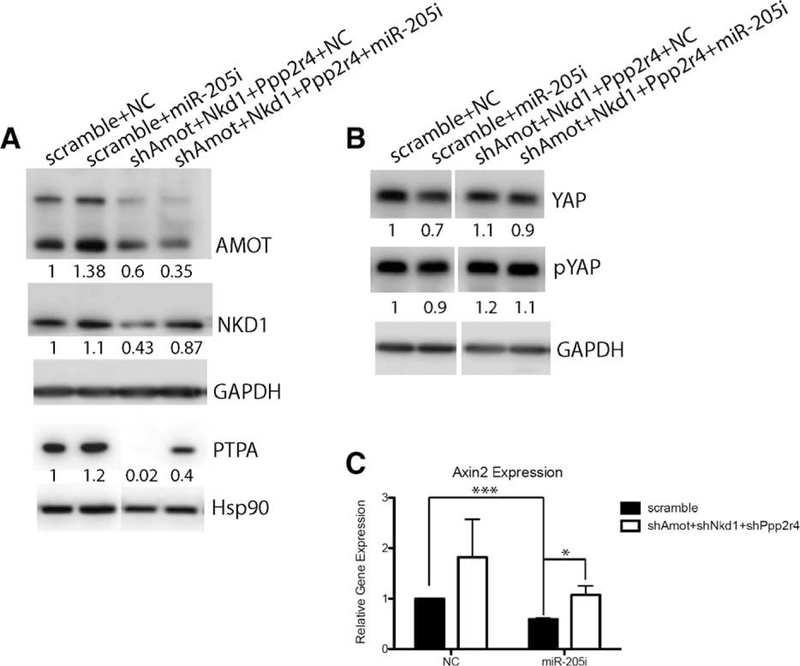
(A): Western blot depicting the efficient knockdown of Amot, Nkd1, and Ppp2r4 in the shRNAs-treated groups. (B): Western blot depicting YAP/pYAP expression in four different experimental groups. (C): Quantitative real-time polymerase chain reaction (qRT-PCR) to examine Axin2 expression in four different experimental groups. Data shown in qPCR are mean ± SD from three biological replicates. *p < .05; >***p < .001 by unpaired Student’s t tests.
miR-205 Depletion Results in Basal Cell Heterogeneity
Given the observation of basal layer alterations in the miR-205−/− transplants, we examined the expression of several basal markers from WT and miR-205−/− outgrowths by dual immunofluorescent staining. Unexpectedly, we observed several distinct subpopulations present in the basal layer of miR-205−/− outgrowths that were not detected in the WT ducts. As shown in Supporting Information Figure S4B, although WT outgrowths displayed uniform SMA+/K5+ staining across the entire basal layer, the miR-205−/− basal cells comprised doublepositive SMA+/K5+, and single-positive SMA+, K5+, and chimeric subpopulations. The chimeric subpopulations could be further separated into +/+ dominant (with <30% SMA+ cells present), +/+ dominant_2 (with <30% SMA+ and <10% K5+ cells present), SMA dominant (with <40% +/+ cells present), and K5 dominant (with <20% +/+ cells present). This surprising observation led us to speculate that these patterns might reflect different basal subpopulations, which possess distinct developmental potential.
DISCUSSIONS
This study reveals an unexpected regulatory role of a single miRNA-miR-205 on mammary gland ductal morphogenesis and stem cell regenerative potential that was revealed only under the conditions of cell transplantation. miR-205 is an intergenic epithelial-specific miRNA that when deleted may result in perinatal lethality, although this is strain dependent. Despite this striking phenotype, the underlying mechanism remains elusive. By potentially modulating the Wnt and YAP signaling pathways, miR-205, as one of the main effectors of p63 regulation, affects mammary ductal morphogenesis during mammary reconstitution, with significant defects observed in the basal cell population, BM, and collagen deposition. Coordinated regulation of both the YAP and the Wnt pathways may account for the phenotypes observed. It is conceivable that this regulatory network is not limited only to mammary gland, but may be relevant in other epithelial tissues in which p63 and miR-205 are expressed, and might help explain the partially penetrant perinatal lethality observed in the miR-205 null mice.
During puberty, miR-205 expression was observed in the cap cell layer of TEBs and basal cells (Fig. 1B). This expression pattern persists in mature glands, where miR-205 is primarily expressed in the basal cells derived from the cap cells. The body cell-derived luminal compartment contained only a few cells with detectable miR-205 expression, indicating miR-205 is essentially a basal/stem cell specific miRNA. In fact, miR-205 was identified as a direct △Np63 target gene and appears to be critical for correct basal cell specification.
Upon miR-205 deletion, MECs failed to repopulate secondary outgrowths as demonstrated both in the in vivo transplantation and the in vitro mammosphere formation assays, indicative of impaired stem cell self-renewal. Rudimentary miR-205 null transplants from the first generation transplants displayed a partial loss of the basal/stem cell population, a discontinuous BM, and decreased collagen deposition, all of which support the idea that miR-205 is essential in maintaining basal cell specification. Surprisingly, the report by Chao et al. [23] claimed that loss of miR-205 led to an expansion of mammary stem cell population. Chao et al. showed that lentivirus-generated miR-205 knockout mammary glands exhibited increased Lin–CD24+CD29hi stem cell-enriched population, ductal length, and collagen deposition. The obvious discrepancies between our study and theirs may arise from the different methodologies used to generate miR-205 null MECs. Instead of validating the miR-205 knockout phenotype by the gold standard transplantation assay into the cleared fat pad, Chao and colleagues utilized intraductal shRNA injection into 4-month-old mammary glands at day 15 of lactation. This method is problematic since intraductal shRNA injection targets primarily cells that line the lumen, and is unlikely to target cells in the basal layer, especially during lactation when tight junctions have formed to permit apical milk secretion [24]. Moreover, lactation is accompanied by a drastic expansion of luminal cells to prepare for milk production. Although the milk was depleted before injection in the above study, the expanded luminal population would render basal cells inaccessible to virus. Considering the basal-specific expression of miR205, the efficient knockdown of miR-205 in the basal cells is unlikely to have occurred using this approach.
Additionally, Chao et al. also suggested that Notch2 might be a miR-205 target through integrating the overlap among three target prediction databases (DIANSmT, miRANDA, and TargetScan). However, Notch pathway members investigated by Chao et al. were not detected in our RNA-seq analysis. Nor was the well-known epithelial to mesencymal transition (EMT) facilitator ZEB1 detected, which transcriptionally represses E-cadherin and promotes EMT. Gregory et al. showed that over-expression of miR-205 in MDCK cells reduced 50% of ZEB1 and thus proposed that miR-205 maintained an epithelial-like state through targeting ZEB1 [25]. This appears to only be a property of human but not mouse miR-205. Divergent miRNA-mRNA interaction pairs highlight the important role of cellular contexts in determining miRNA targets [26]. Under different cellular contexts (e.g., breast cancer cell lines versus normal mammary basal epithelial cells), the 3’UTR accessibility of specific mRNAs as well as the protein cofactors may be altered, which will result in context-dependent miRNA targets [27]. Therefore, how miR-205 regulates cellular processes should be evaluated in the appropriate cellular context.
Through RNA-seq comparison of WT and miR-205 null basal MECs, we identified a number of miR-205 potential targets, several of which overlap with TargetScan predicted miR-205 direct targets. Interestingly, there is one miR-205 target Ppp2r4 that shows up in the RNA-seq result but is not a TargetScan predicted miR-205 direct target. Nevertheless, decreased expression of Ppp2r4 was observed upon miR205 overexpression in a previous study using the Comma-DBgeo cell line [7].
A new miRNA-mRNA recognition paradigm may account for the above observation. There are two broad categories of miRNA targeting sites [28]. (a) The 5’ dominant sites have perfect 7mer or 8mer pairing with miRNA 5’end, and (b) the 3’ compensatory sites have weak 5’end pairing but strong 4–6 base-pairing with miRNA 3’end, just as in the case of miR-205 targeting Ppp2r4 3’UTR (Supporting Information Fig. 2D). Since current target prediction algorithm all focus on the complementarity of mRNA 3’UTR with miRNA 5’end and is only able to identify 5’ dominant sites, it is not surprising that Ppp2r4 is not a predicted target of these algorithms. Apart from the 3’ compensatory sites targeting paradigm in the 3’UTR, there is also one 5’ dominant site within the CDS of Ppp2r4. miRNA targeting CDS of mRNA has also been reported in a previous report, and less complementarity is needed for this type of regulation [29].
In our current study, a basal cell hierarchy is revealed in miR-205−/− outgrowths. It is speculated that some of the miR-205−/− basal cells were arrested at a less-differentiated state (K5+/αSMA-) and failed to differentiate. This remains to be proven using lineage tracing approaches in the WT and miR-205−/− transplants. It will also be interesting to characterize the different basal subpopulations in the context of YAP and Wnt perturbation, since it is possible that miR-205 helps coordinate basal cell differentiation through YAP and Wnt signaling.
Our studies suggest the existence of a regulatory network, starting from △Np63 and miR-205, mediated by Amot, Nkd1, and Ppp2r4, and then through YAP and the Wnt signaling pathways to affect basal cell specification and mammary ductal morphogenesis during mammary reconstitution. Chen et al. [30] reported that YAP loss did not affect virgin mammary gland development. However, YAP overexpression led to terminal differentiation defects in pregnancy. Meanwhile, by stabilizing the destruction complex through Disheveled and GSK3β, NKD1, and PTPA prevent β-catenin nuclear translocation and inhibit canonical Wnt signaling. Although we confirmed Wnt signaling inhibition in the luminal MEC line HC11, the magnitude of repression may not be representative of what might be expected in basal cells. Inhibition of β-catenin signaling has also been suggested to affect embryonic mammary gland development and alveolar formation during pregnancy [31]. In light of the present studies, it is highly possible that coordinated loss of YAP and Wnt signaling affects mammary morphogenesis to a greater extent than inhibition of either pathway alone.
The cross-regulation of YAP/TAZ and Wnt has indeed been extensively investigated in human embryonic kidney cells and in the mouse small intestine. Azzolin et al. [32] have shown that in Wnt-off cells, YAP/TAZ associated with destruction complex through binding to Axin1. The interaction between YAP/-TAZ and Axin1 not only sequesters YAP/TAZ in the cytoplasm, but also facilitates β-TCRP recruitment to the destruction complex and inhibition of downstream β-catenin signaling pathway. In Wnt-on cells, Wnt ligands induce the clustering of LRP6. Since YAP/TAZ and LRP6 compete for the same BS of Axin1, the presence of large amount of LRP6 forces YAP/TAZ to go to the nucleus and activate target genes. Meanwhile, β-catenin is also dislodged from the destruction complex and activated. Thus, this leads to a conclusion that the nuclear YAP, rather than β-catenin, mediates the effect of Wnt signaling. Consistent with this, YAP/TAZ deletion recapitulated the phenotype of LRP5 mutants, but β-catenin deletion did not. Therefore, β-catenin/TCF and YAP/TAZ may mediate Wnt in a “labor-divided” fashion, in which β-catenin/TCF mainly functions during normal homeostasis whereas YAP/TAZ effects occur during regeneration. Deletion of miR205 appears to impact both YAP and Wnt/β-catenin signaling concurrently; therefore, both normal mammary gland homeostasis and stem cell regeneration were affected. Further studies are required to determine the precise mechanisms involved in β-catenin/TCF and YAP/TAZ interactions in mammary stem cells.
Supplementary Material
SIGNIFICANCE STATEMENT.
Using a conditional knockout mouse model and mammary fat pad transplantation, we demonstrate that deletion of a single microRNA, miR-205, abrogates mammary stem cell self-renewal during mammary reconstitution. Negative regulators of YAP and Wnt signaling are identified as novel miR-205 targets. Given the conflicting results on miR-205 function in development, and its ambiguous role in cancer, our results provide additional insights on the function of miR-205 in the mouse mammary gland. Moreover, the regulation of YAP and Wnt signaling by miR-205 has broader implications since these signaling pathways are also required for the self-renewal of stem cells in several other epithelial organs.
ACKNOWLEDGMENTS
This project was supported by the Genomics and RNA Profiling Core, the Integrated Microscopy Core with funding from NIH (HD007495, DK56338, and CA125123), the Lester and Sue Smith Breast Center Pathology Core and the Cytometry and Cell Sorting Core with funding from NIH (AI036211, CA125123, and RR024574) and the expert assistance of Joel M. Sederstrom at Baylor College of Medicine. The authors would like to thank Drs. Li Ma and Sue-Hwa Lin from MD Anderson Cancer Center for providing YAP, pYAP, and AMOT antibodies, respectively. The authors would also like to thank Drs. Kevin Roarty, Deanna Acosta, Se-Jin Kim, Chi-Hsuan Chang, and Amulya Sreekumar for critical reading and editing of the manuscript. This work was supported by grants NCI R35CA197452 to E. R. Flores, 4R01CA016303–41 to J.M. Rosen, and 1U19CA179513–01 and 1R010D019128–01 to M.T. McManus.
This project was supported by grants NCI R35CA197452, 4R01CA016303–41, 1U19CA179513–01, and 1R01OD019128–01, and the Genomics and RNA Profiling Core, the Integrated Microscopy Core, the Lester and Sue Smith Breast Center Pathology Core, and the Cytometry and Cell Sorting Core.
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicated no potential conflicts of interests.
REFERENCES
- 1.Bai L, Rohrschneider LR. s-SHIP promoter expression marks activated stem cells in developing mouse mammary tissue. Genes Dev 2010;24:1882–1892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Visvader JE, Stingl J. Mammary stem cells and the differentiation hierarchy: Current status and perspectives. Genes Dev 2014;28:1143–1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gjorevski N, Nelson CM. Integrated morphodynamic signalling of the mammary gland. Nat Rev Mol Cell Biol 2011;12: 581–593. [DOI] [PubMed] [Google Scholar]
- 4.Doench JG, Sharp PA. Specificity of microRNA target selection in translational repression. Genes Dev 2004;18:504–511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Park CY, Jeker LT, Carver-Moore K et al. A resource for the conditional ablation of microRNAs in the mouse. Cell Rep 2012;1: 385–391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang D, Zhang Z, O’Loughlin E et al. MicroRNA-205 controls neonatal expansion of skin stem cells by modulating the PI(3)K pathway. Nat Cell Biol 2013;15: 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Greene SB, Gunaratne PH, Hammond SM et al. A putative role for microRNA-205 in mammary epithelial cell progenitors. J Cell Sci 2010;123:606–618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ibarra I, Erlich Y, Muthuswamy SK et al. A role for microRNAs in maintenance of mouse mammary epithelial progenitor cells. Genes Dev 2007;21:3238–3243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Muzumdar MD, Tasic B, Miyamichi K et al. A global double-fluorescent Cre reporter mouse. Genesis 2007;45:593–605. [DOI] [PubMed] [Google Scholar]
- 10.Roarty K, Shore AN, Creighton CJ et al. Ror2 regulates branching, differentiation, and actin-cytoskeletal dynamics within the mammary epithelium. J Cell Biol 2015; 208:351–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen X, Xu H, Yuan P et al. Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 2008;133:1106–1117. [DOI] [PubMed] [Google Scholar]
- 12.Paine I, Chauviere A, Landua J et al. A geometrically-constrained mathematical model of mammary gland ductal elongation reveals novel cellular dynamics within the terminal end bud. PLoS Comput Biol 2016;12: e1004839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chakravarti D, Su X, Cho MS et al. Induced multipotency in adult keratino- cytes through down-regulation of ANp63 or DGCR8. Proc Natl Acad Sci U S A 2014;111: 572–581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kouwenhoven EN, van Heeringen SJ, Tena JJ et al. Genome-wide profiling of p63 DNA-binding sites identifies an element that regulates gene expression during limb development in the 7q21 SHFM1 locus. PLoS Genet 2010;6:e1001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Rajaram RD, Buric D, Caikovski M et al. Progesterone and Wnt4 control mammary stem cells via myoepithelial crosstalk. EMBO J 2015;34:641–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Breakers Izaurralde E. and blockers—miRNAs at work. Science 2015;349:380–382.26206919 [Google Scholar]
- 17.Seeling JM, Miller JR, Gil R et al. Regulation of beta-catenin signaling by the B56 subunit of protein phosphatase 2A. Science 1999;283:2089–2091. [DOI] [PubMed] [Google Scholar]
- 18.Zhao B, Li L, Lu Q et al. Angiomotin is a novel Hippo pathway component that inhibits YAP oncoprotein. Genes Dev 2011;25:51–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paramasivam M, Sarkeshik A, Yates JR 3rd et al. Angiomotin family proteins are novel activators of the LATS2 kinase tumor suppressor. Mol Biol Cell 2011;22:3725–3733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Leung CY, Zernicka-Goetz M. Angiomotin prevents pluripotent lineage differentiation in mouse embryos via Hippo pathway-dependent and -independent mechanisms. Nat Commun 2013;4:2251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yan D, Wallingford JB, Sun TQ et al. Cell autonomous regulation of multiple Dishevelled-dependent pathways by mammalian Nkd. Proc Natl Acad Sci U S A 2001;98: 3802–3807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Huang S, Li Y, Chen Y et al. Changes in gene expression during the development of mammary tumors in MMTV-Wnt-1 transgenic mice. Genome Biol 2005;6:R84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chao CH, Chang CC, Wu MJ et al. MicroRNA-205 signaling regulates mammary stem cell fate and tumorigenesis. J Clin Invest 2014;124:3093–3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nguyen D, Parlow FA, Neville CM. Hormonal regulation of tight junction closure in the mouse mammary epithelium during the transition from pregnancy to lactation. J Endocrinol 2001;170:347–356. [DOI] [PubMed] [Google Scholar]
- 25.Gregory PA, Bert AG, Paterson EL et al. The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat Cell Biol 2008;10:593–601. [DOI] [PubMed] [Google Scholar]
- 26.Gao FB. Context-dependent functions of specific microRNAs in neuronal development. Neural Dev 2010;5:25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Didiano D, Hobert O. Perfect seed pairing is not a generally reliable predictor for miRNA-target interactions. Nat Struct Mol Biol 2006;13:849–851. [DOI] [PubMed] [Google Scholar]
- 28.Brennecke J, Stark A, Russell RB et al. Principles of microRNA-target recognition. PLoS Biol 2005;3:e85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tay Y, Zhang J, Thomson AM et al. MicroRNAs to Nanog, Oct4 and Sox2 coding regions modulate embryonic stem cell differentiation. Nature 2008;455:1124–1128. [DOI] [PubMed] [Google Scholar]
- 30.Chen Q, Zhang N, Gray RS et al. A temporal requirement for Hippo signaling in mammary gland differentiation, growth, and tumorigenesis. Genes Dev 2014;28:432–437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hatsell S, Rowlands T, Hiremath M et al. Beta-catenin and Tcfs in mammary development and cancer. J Mammary Gland Biol Neoplasia 2003;8:145–158. [DOI] [PubMed] [Google Scholar]
- 32.Azzolin L, Panciera T, Soligo S et al. YAP/TAZ incorporation in the beta-catenin destruction complex orchestrates the Wnt response. Cell 2014;158:157–170. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


