Abstract
Late embryogenesis abundant (LEA) proteins include eight multigene families that are expressed in response to water loss during seed maturation and in vegetative tissues of desiccation tolerant species. To elucidate LEA proteins evolution and diversification, we performed a comprehensive synteny and phylogenetic analyses of the eight gene families across 60 complete plant genomes. Our integrated comparative genomic approach revealed that synteny conservation and diversification contributed to LEA family expansion and functional diversification in plants. We provide examples that: 1) the genomic diversification of the Dehydrin family contributed to differential evolution of amino acid sequences, protein biochemical properties, and gene expression patterns, and led to the appearance of a novel functional motif in angiosperms; 2) ancient genomic diversification contributed to the evolution of distinct intrinsically disordered regions of LEA_1 proteins; 3) recurrent tandem-duplications contributed to the large expansion of LEA_2; and 4) dynamic synteny diversification played a role on the evolution of LEA_4 and its function on plant desiccation tolerance. Taken together, these results show that multiple evolutionary mechanisms have not only led to genomic diversification but also to structural and functional plasticity among LEA proteins which have jointly contributed to the adaptation of plants to water-limiting environments.
Keywords: LEA proteins, gene family evolution, abiotic stress adaptation, desiccation tolerance, intrinsic disorder
Introduction
When plants colonized land 450 Ma, they developed a wide range of adaptations including physiological, structural, and regulatory mechanisms to cope with variable environments. Land plants (embryophytes) evolved from streptophyte algae, a paraphyletic group of green algae believed to be physiologically preadapted to terrestrial environments due to their fresh water origin (Kenrick and Crane 1997; Becker and Marin 2009; Wodniok et al. 2011).
As they colonized the land, plants also developed desiccation tolerance (DT) which is the ability to survive the removal of almost all cellular water without irreparable damage. DT is recurrent in reproductive structures of most vascular plants (e.g., during embryogenesis), in the vegetative body of nonvascular plants and in a few angiosperms species commonly known as “resurrection plants” (Oliver et al. 2000; Illing et al. 2005; Leprince and Buitink 2010; Farrant and Moore 2011; Gaff and Oliver 2013). Several genes that are thought to be important for DT are common among nonvascular and vascular plants, and are also present in their ancestral streptophyte algae (Rensing et al. 2008; Wodniok et al. 2011).
Within the conserved actors of cellular protection involved in DT, a common group named late embryogenesis abundant (LEA) proteins, has received considerable attention. LEA proteins were originally associated with the acquisition of DT in plant embryos due to the high gene expression and protein accumulation in the later stages of seed maturation (Galau et al. 1986; Dure et al. 1989; Espelund et al. 1992; Manfre et al. 2005; Delahaie et al. 2013). In vegetative tissues, LEA genes were found to accumulate under abiotic stresses such as drought, salinity, heat and freezing, and under desiccation in resurrection plants (Hoekstra et al. 2001; Cuming et al. 2007; Amara 2014; Stevenson et al. 2016). Interestingly, LEA genes are also found outside the plant kingdom, suggesting a common mechanism of DT across distinct life forms (Browne et al. 2002; Tunnacliffe et al. 2005; Kikawada et al. 2006; Gusev et al. 2014).
LEA proteins exhibit peculiar biochemical properties such as a high proportion of polar amino acids, high hydrophilicity, and the presence of intrinsically disordered regions (IDRs) (Dure et al. 1989; Garay-Arroyo et al. 2000; Goyal et al. 2005; Battaglia et al. 2008). Intrinsically disordered proteins (IDPs) have been proposed as critical for plant adaptation in new environments because of their ability to perform more than one function, the so called “moonlighting” activity (Covarrubias et al. 2017). This property allows LEA proteins to perform antiaggregation, protein stabilization, as well as molecular chaperone-like activities (Chakrabortee et al. 2007, 2012; Battaglia et al. 2008; Kovacs et al. 2008; Hincha and Thalhammer 2012; Cuevas-Velazquez et al. 2017).
Several studies have attempted to identify, classify, and assess LEA proteins function in plants (Battaglia et al. 2008; Hundertmark and Hincha 2008; Shih et al. 2008; Amara 2014), however, a comprehensive understanding of the evolutionary history and its relationship with the high diversification of sequence and function of LEA proteins in plants is still elusive.
With the increasing number of plant genomes available, a comprehensive analysis of the evolution and functional diversification of LEA gene families is now possible. The reconstruction of the evolutionary history of a protein family in an entire lineage involves homology identification by comparative genome analysis among different taxa, and provide a deeper understanding of the evolution of genomic complexity and lineage-specific adaptations (Koonin 2005). Phylogenomic analysis (i.e., phylogenetic analysis at the genome scale) has often been employed in order to identify cross-species homologs and predict gene function by reconstructing the evolutionary history (Eisen 1998).
In this study, we performed a large-scale phylogenomic analysis across 60 complete genomes, combining synteny network and phylogenetic analysis, in order to identify LEA genes and investigate their origin and evolution in plants. Our synteny analysis reveals independent evolutionary patterns that shaped synteny diversification of LEA genes in plants, and illustrates resultant functional novelties related to water-stress adaptation. Our work provides compelling opportunities for further functional classification and discovery of new LEA functions in plants.
Materials and Methods
Identification of LEA Proteins in 60 Genomes
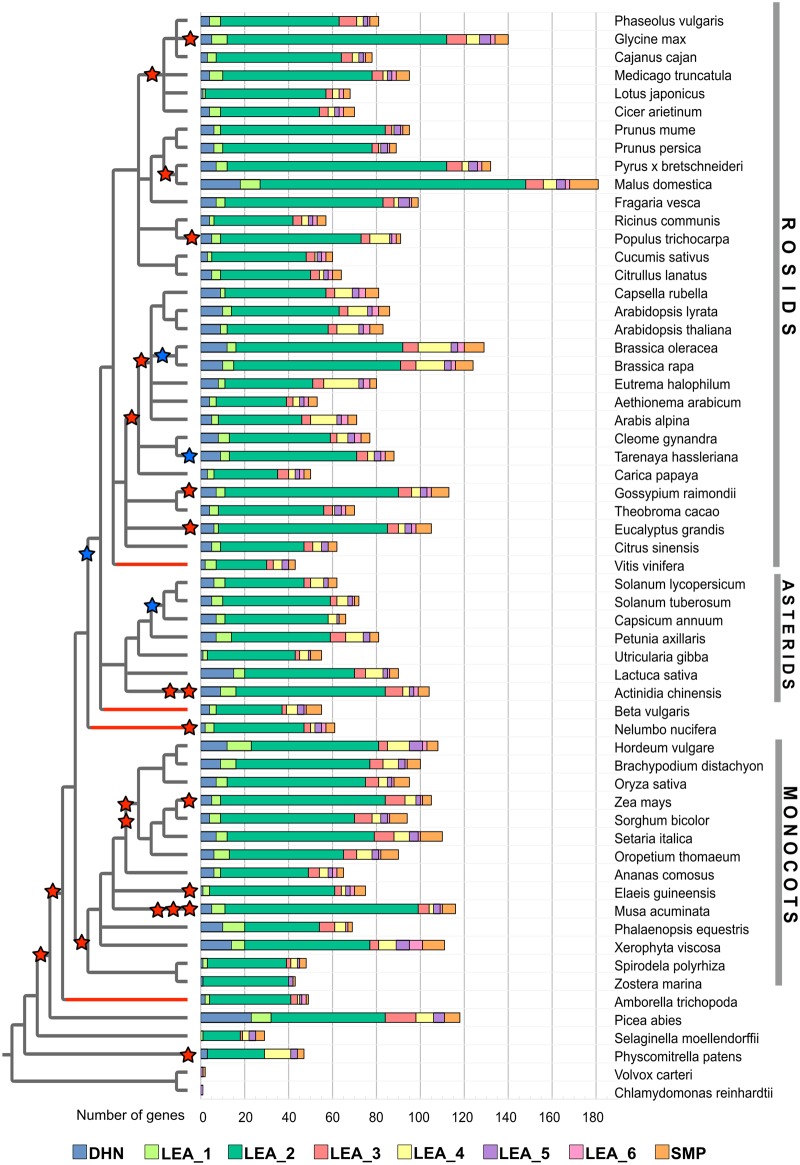
We used 60 fully sequenced genomes available in Phytozome (Goodstein et al. 2012) (https://phytozome.jgi.doe.gov/; last accessed November 13, 2017), and the recently published genome of Xerophyta viscosa (Costa et al. 2017). Our species list includes representative species belonging to green algae, mosses, lycophytes, gymnosperms, early angiosperms, monocots, early eudicots, asterids, and rosids (fig. 1 and supplementary table S1, Supplementary Material online).
Fig. 1.
—Species phylogeny and number of LEA genes identified in plant genomes. The species tree was inferred using NCBI Taxonomy Browser (https://www.ncbi.nlm.nih.gov/taxonomy; last accessed September 27, 2017). Each LEA family is represented by a specific color (see also supplementary table S1, Supplementary Material online). The red branches in the phylogenetic tree indicate the basal rosid Vitis vinifera, the basal eudicots Beta vulgaris and Nelumbo nucifera, and the basal angiosperm Amborella trichopoda. The red and blue stars on the phylogenetic tree indicate whole-genome duplication (WGD), and whole-genome triplication (WGT), respectively.
Several classifications have been proposed for LEA proteins (for a review, see Battaglia et al. 2008). Here, we used the Pfam annotation for protein families (Bateman et al. 2002) (http://pfam.xfam.org/; last accessed November 13, 2017) based on conserved protein domains (Hundertmark and Hincha 2008). This annotation classifies LEA proteins into eight Pfams: Dehydrin (DHN) (PF00257), LEA_1 (PF03760), LEA_2 (PF03168), LEA_3 (PF03242), LEA_4 (PF02987), LEA_5 (PF00477), LEA_6 (PF10714), and Seed Maturation Protein (SMP) (PF04927). Hidden Markov Models (HMM) retrieved from the Pfam 3.0 database (http://Pfam.xfam.org) were queried against the 60 plant genomes to identify LEA proteins for each family using the program “hmmscan” of the HMMER3.0 package (Finn et al. 2011). All proteins with significant hits (e-value < 0.001) were used in this analysis.
Synteny Network Construction and Community Detection
We used the Synets method (Zhao and Schranz 2017; Zhao et al. 2017) for syntenic block calculations, network construction, and community detection (https://github.com/zhaotao1987/SynNet-Pipeline). In summary, pairwise all-against-all comparisons were performed using RAPSearch (Zhao et al. 2012). Synteny block detection was performed with MCScanX software (Wang et al. 2012) with default parameters (minimum collinear block size = 5 genes, maximum gaps = 25 genes). The syntenic blocks containing the identified LEA sequences were used to build synteny networks (Synets) that were visualized and edited with Cytoscape 3.3.0 (Shannon et al. 2003) and Gephi 0.9.1 (Bastian et al. 2009) (https://gephi.org/). Infomap (Rosvall and Bergstrom 2008) was used to find communities within the synteny networks, which is implemented under “igraph” package in R (http://igraph.org/r/doc/cluster_infomap.html). All synteny communities were numbered according to the largest to the smallest number of genes, and later renamed per LEA family accordingly (supplementary table S2, Supplementary Material online). The synteny communities were further analyzed with a phylogenetic profiling. Phylogenetic profiling allows the visualization of the synteny communities that are lineage-specific or shared among different species. All synteny communities were decomposed into numbers of involved syntenic gene copies in each genome. Dissimilarity index of all clusters was calculated using the “Jaccard” method of the vegan package (Dixon 2003), hierarchically clustered by “ward.D,” and visualized by “pheatmap.”
Phylogenetic Analysis
Multiple sequence alignments (MSAs) were built for each of the eight LEA families using MAFFT v.7 (Katoh et al. 2002). We used the automated method for the Pfam LEA_2 due to the large number of sequences, and the method G-INS-l for all other LEA Pfams. Phyutility 2.2.6 (Smith and Dunn 2008) was used to trim gaps and maintain 75% the consensus alignment. The final MSAs were edited and displayed with Jalview 2.10.3 (Waterhouse et al. 2009). IQ-TREE v.1.5.1 (Nguyen et al. 2015) was used to infer Maximum Likelihood (ML) trees with 1,000 bootstraps for each alignment. All phylogenetic trees were edited and displayed with the online tool iTOL (Letunic and Bork 2016).
Physicochemical Properties and Expression Data of Dehydrin
The hydrophilicity index of Dehydrin proteins was calculated with the online GRAVY calculator (http://www.gravy-calculator.de/; last accessed January 6, 2018). More hydrophilic proteins have a more negative GRAVY score, and more hydrophobic proteins have a more positive GRAVY score. In order to reveal hydrophylin-type proteins (GRAVY < −1 and Gly >6%), individual GRAVY scores were plotted against the percentage of Glycine (Gly) per protein sequence (Garay-Arroyo et al. 2000; Battaglia et al. 2008). Absolute gene expression values were retrieved from the e-Northern tool provided by the Bio-Array Resource for Arabidopsis Functional Genomics (http://bar.utoronto.ca/; last accessed May 11, 2018) as well as from the data sets of seed and silique development, dry seed, drought, and heat shock of Hundertmark and Hincha (2008).
Results
Distinct Origins of LEA Families in Plants
We performed a genome-wide sequence homology search to identify the complete repertoires of LEA genes across 60 genomes of diverse plant species (fig. 1). For that we used the most widely employed classification of LEA proteins that defines eight multigene protein families (Pfam): Dehydrin (DHN), LEA_1, LEA_2, LEA_3, LEA_4, LEA_5, LEA_6, and Seed Maturation Protein (SMP) (Hundertmark and Hincha 2008). Based on the conservation of Hidden Markov Model (HMM) profiles of the eight LEA protein families we identified a total of 4,836 genes, with variable copy number distribution among the LEA families and the genomes investigated (fig. 1 and supplementary table S1, Supplementary Material online). Only single genes belonging to SMP and LEA_5 were found in algal genomes, suggesting an ancestral origin of these families. The Dehydrin, LEA_2 and LEA_4 families were identified in the bryophyte clade (Physcomitrella patens) and LEA_1 and LEA_3 families appeared in the lycophyte lineage (Selaginella moelendorffii). The LEA_6 family only emerged in early angiosperms (Amborella trichopoda), likely representing the most recent LEA family in plants. Overall, the LEA_2 family was the most abundant with 3,126 genes, which are multicopy in genomes of both angiosperms and lower plants. LEA_6, on the other hand, represents the smallest family with a total of 89 identified genes, with copy-number varying from 0 to 3, with the exception of the resurrection plant Xerophyta viscosa in which six LEA_6 genes were identified. The variable copy-number between different taxa suggests independent loss or duplication of genes in individual genomes. The underrepresentation of LEA genes in Zostera marina and Spirodela polyrhiza (Olsen et al. 2016), and the overrepresentation in X. viscosa (Costa et al. 2017) have already been reported and correlated with the respective desiccation-sensitive and -tolerant lifestyle of these species, suggesting that the evolution of LEA genes contributed to water stress adaptation in plants.
Differential Conservation of LEA Genes in Angiosperms
We used a synteny-based method to identify homology between the proteins and to explore the evolutionary history of LEA genes in plants. Homologous genes comprise orthologs and paralogs, which are corresponding genes in different species that evolved from the same ancestral gene, and to genes duplicated within the same genome, respectively (Koonin 2005; Gabaldon and Koonin 2013). Generally, orthologs have equivalent functions in different taxa, while paralogs may display functional diversification and specialization, although paralogs within the same organism may perform more similar functions than orthologs in distinct organisms at the same diversification level (Koonin 2005; Gabaldon and Koonin 2013). Synteny homologs (syntelogs) are localized in similar genomic regions and have similar genomic context in different species, and likely evolved from a common ancestor gene (Zhao and Schranz 2017; Zhao et al. 2017). Syntelogs were inferred with the Synteny Network (Synets) method (Zhao and Schranz 2017; Zhao et al. 2017) which enables detection of homologs in corresponding chromosomes in different species, as well as paralogs within a species. The output is a network in which the nodes represent anchor genes in a syntenic block and the edges indicate synteny similarity (supplementary fig. S1, Supplementary Material online). Synteny communities can be detected in synteny networks using community detection methods, and indicate genes that are located in the same genomic regions in distantly related species (Zhao et al. 2017). Table 1 summarizes the percentage of syntelogs identified per LEA family as well as the number of synteny communities detected in each network (detailed information in supplementary table S2, Supplementary Material online).
Table 1.
Summary of Syntenic Genes and Synteny Communities Identified Per LEA Protein Family
| Pfam | Total Genes | Syntelogs (%) | Synteny Communities |
|---|---|---|---|
| DHN | 365 | 62.2 | 12 |
| LEA_1 | 251 | 63.3 | 10 |
| LEA_2 | 3,126 | 76.0 | 130 |
| LEA_3 | 274 | 79.9 | 16 |
| LEA_4 | 298 | 77.2 | 18 |
| LEA_5 | 153 | 67.3 | 4 |
| LEA_6 | 89 | 76.4 | 8 |
| SMP | 280 | 59.3 | 11 |
| Total | 4,836 | 209 |
The variable percentage of syntenic genes and number of synteny communities suggest independent evolution between and within the LEA protein families. Genes not incorporated in synteny communities by our clustering method are likely to be species-specific singletons. Only a few “in-paralogs” (paralogs from the same species) were detected in the basal species Sellaginela moellendorffii and Physcomitrella patens (supplementary table S2, Supplementary Material online). Considering the large evolutionary distance between the species analyzed, we hypothesize that ancient and independent synteny diversification between LEA families may have played important roles in their functional diversification.
Phylogenetic Profiling Reveals Angiosperm-Wide and Lineage-Specific LEA Genes
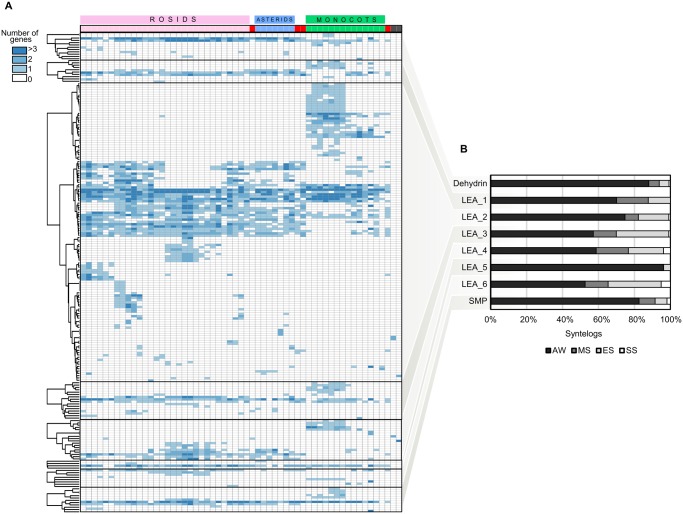
We further analyzed the origin of the synteny communities detected with Synets in order to obtain information on the evolutionary conservation and diversification of LEA genes in angiosperms. Presence or absence of a species syntelog in a community of the synteny network can be visualized as a phylogenetic profile, enabling inference of the origin, expansions, and contractions of the gene family in each clade of their phylogenetic tree (fig. 2A).
Fig. 2.
—Phylogenetic profile and evolutionary categorization of syntenic LEA genes in the genomes analyzed. (A) Phylogenetic profile showing the number and distribution of syntenic LEA genes in plants. Rows represent synteny communities and columns indicate species. The colors on top of the profile indicate rosids (pink), asterids (blue), monocots (green), basal angiosperm species (red) and Physcomitrella patens and Selaginella moelendorffii (dark gray). The species were ordered from the most recent to the most ancient, from the left to the right. (B) Distribution of syntenic genes in each evolutionary category. AW, angiosperm-wide; MS, monocot-specific; ES, eudicot-specific; SS, species-specific (see also supplementary table S3, Supplementary Material online).
We subdivided the synteny communities into four main evolutionary categories: angiosperm-wide (AW), monocot-specific (MS), eudicot-specific (ES), and species-specific (SS) (fig. 2B and supplementary table S3, Supplementary Material online). Angiosperm-wide are synteny communities that contain genes of at least one monocot and one eudicot species. Monocot-specific includes synteny communities containing only monocot genes, and eudicot-specific includes communities comprising eudicot genes only. Species-specific correspond to paralogs duplicated in an individual genome, also named ohnologs.
AW communities were found in all LEA families and encompasses the largest number of the syntelogs identified (fig. 2B), indicating that the majority of LEA genes have a common origin in angiosperms and are likely located in a more ancestral genomic context. The angiosperm-wide conservation of LEA genes is particularly observed in the families DHN, LEA_5, and SMP, where >80% of the syntelogs identified are shared among angiosperm species. Lineage-specific duplications (MS and ES) have also significantly contributed to the repertoire of LEA genes in plants, especially in LEA_3 and LEA_6 families, where >40% of the syntelogs are distributed over these two categories. SS paralogs were overall underrepresented or absent in the genomes investigated, likely due to low frequency of local gene duplications, or the duplicated copies were more likely to be lost in individual genomes. The finding of lineage-specific and species-specific synteny suggests that duplication events other than whole genome duplications (WGD) have significantly contributed to the expansion of LEA families in plant genomes.
The fact that LEA_5 has the smallest number of synteny communities and that the majority of the genes belong to AW conserved genomic context indicates that this is the most conserved LEA family in plants. On the other hand, the large number of LEA_2 syntelogs in AW communities indicates that this is the most diverse LEA family in the plant lineage.
Structural and Functional Diversification Contributed to LEA Proteins Evolution
Duplication events may introduce a gene copy into a new regulatory context, leading to differential evolutionary and regulatory constraints, which is one of the main sources driving functional innovation within a gene family (Conant and Wolfe 2008; Flagel and Wendel 2009). Therefore, in the next sections we provide a few examples from our synteny analysis of remarkable structural and functional innovations within LEA families resulting from differential evolution of the genomic context.
Dehydrin: Biochemical, Structural, and Expression Pattern Innovations during Angiosperm Evolution
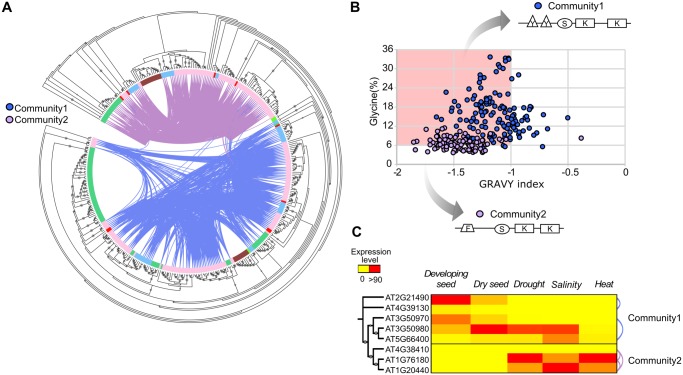
Dehydrin (DHN) is classified as a LEA family due to the gene expression during late seed embryogenesis and ability to perform “classical” chaperone-like activity, preventing heat-induced protein aggregation and inactivation in vitro (Kovacs et al. 2008; Liu et al. 2017). In our data set, we found that DHN genes are distributed across two main angiosperm-wide synteny communities and a maximum likelihood tree supports the phylogenetic separation of these communities in angiosperms (fig. 3A).
Fig. 3.
—Characteristics of Dehydrin synteny communities. (A) Maximum likelihood tree of all DHN genes found in the genome of 60 species. The inner circle indicates species belonging to monocots (green), rosids (pink), asterids (blue), basal species (red), the gymnosperm Picea abies (brown), and the bryophyte Physcomitrella patens (light green). The connections between the branches indicate synteny between the gene pairs. Synteny communities 1 and 2 are indicated (blue and pink connections, respectively), dots on the branches represent bootstrap support values (>85). The larger the dots the higher the bootstrap values. (B) Glycine (Gly) content and GRAVY index plot (Gly/GRAVY plot) showing the distribution of hydrophylins (highlighted in red) between community 1 and 2. The arrows indicate a schematic representation of the consensus sequence of proteins of community 1 and 2, respectively. The F-, Y-, S-, and K-protein segments are indicated according to their position in the protein sequences. (C) Expression levels of DHN genes in Arabidopsis thaliana. The expression data were retrieved from the Bio-Array Resource for Arabidopsis Functional Genomics (http://bar.utoronto.ca/) and from Hundertmark and Hincha (2008). The dots on the branches of the phylogenetic tree indicate bootstrap support values (>75). Connections between the rows represent synteny relationships.
Some of the DHNs are called “hydrophylins” because of their specific response to osmotic stress (Garay-Arroyo et al. 2000; Jaspard and Hunault 2014). Hydrophylins play important roles in protecting cell components from the adverse effects caused by low water availability due to their biochemical properties such as high Glycine (Gly) content (> 6%) and low grand average hydropathy (GRAVY) (< −1) (Garay-Arroyo et al. 2000; Battaglia et al. 2008; Reyes et al. 2008). In order to investigate the distribution of hydrophylins in angiosperms, we analyzed the Gly content and GRAVY index of each protein within the two largest angiosperm-wide DHN communities (fig. 3B). Although both communities contain proteins with hydrophylin properties, community 1 contains proteins with more variable Gly/GRAVY composition than community 2 proteins which have a more homogeneous Gly/GRAVY distribution. These findings indicate that, even though hydrophylin-type proteins do not form an isolated synteny community, there is a clear biochemical divergence between proteins that evolved in distinct genomic contexts.
DHN proteins have been functionally subdivided into four to five main architectures based on the presence and organization of specific motifs called Y-, S- or K-segments (Close 1996; Hunault and Jaspard 2010; Banerjee and Roychoudhury 2016; Malik et al. 2017). We performed multiple sequence alignments of proteins from the DHN synteny communities 1 and 2 in order to investigate the diversification of the different functional motifs (supplementary fig. S2A and B, Supplementary Material online). Our data indicate that the majority of proteins of community 1 comprises Y(n)SK(n) types (fig. 3B andsupplementary fig. S2A, Supplementary Material online), while community 2 contains mainly SK(n)-type proteins, lacking the Y-segment at the N-terminus (supplementary fig. S2B, Supplementary Material online). While lacking the Y-segment, proteins from community 2 possess a new conserved segment at the N-terminus (DRGLFDFLGKK). This motif is named F-segment, and it was recently characterized as an overlooked motif in angiosperms and gymnosperms, with potential functional roles in membrane and protein binding (Strimbeck 2017). Interestingly, genes encoding proteins belonging to community 1 are expressed mainly during seed development in Arabidopsis thaliana, and some of the genes can be induced by abiotic stress (fig. 3C). On the other hand, genes encoding the F-type DHN proteins of community 2 seem to be specifically induced by abiotic stresses such as drought, heat, and salinity. The combined results indicate that the ancient synteny diversification of DHN in angiosperms has resulted in protein biochemical and sequence innovations, and likely changes in expression patterns that may be related to functional specificity within this protein family, although further experimental information is necessary in order to draw stronger conclusions. To date, this is the first documentation of the evolution and diversification of the F-segment in angiosperms, and its association with abiotic stress.
LEA_1: Ancient Diversification of IDPs in Angiosperms
LEA_1 proteins, also known as Group 4, accumulate in the plant cell in response to water stress and have been proposed as model to study IDPs in plants (Olvera-Carrillo et al. 2010; Cuevas-Velazquez et al. 2017). This family has been subdivided into two main subclasses based on protein sequence features (Battaglia et al. 2008). One of the subgroups, named group 4A, comprises smaller proteins (80–124 residues) and the second group, 4B, has longer representatives (108–180 residues). Both subclasses possess a variable C-terminal region, and a conserved portion at the N-terminal region predicted to form alpha-helices under water limiting conditions, which is a characteristic found in many IDPs (Cuevas-Velazquez et al. 2017).
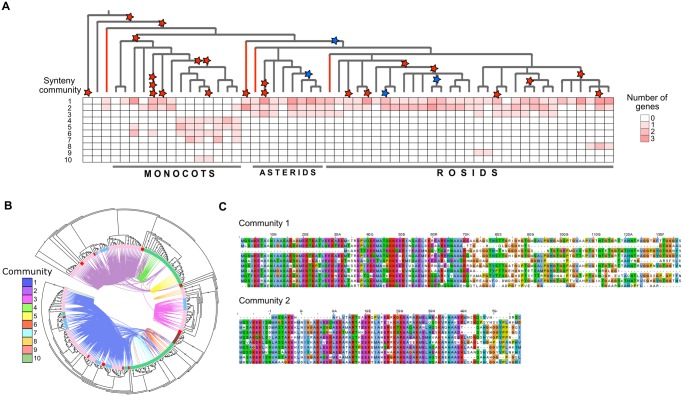
Our data indicate that LEA_1 members are distributed in ten synteny communities, and 70% of the homologs identified with Synets belong to two angiosperm-wide (AW) communities (figs. 2B and 4A). The absence of clear synteny and phylogenetic separation in the phylogenetic tree suggests that some of the ES and MS communities have originated through duplication or transposition of genes from AW communities (fig. 4B). We found differences between the consensus sizes of the multiple sequence alignments of proteins from the two AW communities (fig. 4C and supplementary fig. S3A and B, Supplementary Material online), that indicates that AW community 1 represents the subclass 4B of longer protein sequences, whereas community 2 contains members of subclass 4A of smaller proteins. The multiple sequence alignments of subgroups 4A and 4B revealed that the N-terminal region able to form alpha-helices is also variable (fig. 4C and supplementary fig. S3A and B, Supplementary Material online). This suggests that the diversification of intrinsically disordered regions (IDRs) able to fold into alpha-helices under water deficit conditions in LEA_1 occurred before the origin of monocots and eudicots, and that these protein types have been conserved in angiosperm genomes during evolution.
Fig. 4.
—Phylogenetic and synteny characteristics of LEA_1. (A) Phylogenetic profile of LEA_1 indicating the distribution of the synteny communities detected in the species phylogenetic tree. The red and blue stars indicate whole-genome duplication (WGD) and whole-genome triplication (WGT), respectively. (B) Maximum likelihood tree of the LEA_1 family. The circle inside the tree indicates species belonging to monocots (green), rosids (pink), asterids (blue), basal species (red), the gymnosperm Picea abies (brown), the bryophyte Physcomitrella patens (light green), and the lycophyte Selaginella moellendorffii (olive green). The connections between the branches indicate synteny between the gene pairs, and dots on the branches represent bootstrap support values (>85).The larger the dots the higher the bootstrap values. (C) Partial representation of the multiple sequence alignments of amino acid sequences of the communities 1 and 2 (top ten sequences).
LEA_2: Expansion and Diversification through Recurrent Tandem Duplications
LEA_2 is the largest LEA family, and has been considered atypical because it contains proteins with more hydrophobic amino acids and more defined secondary structure in solution compared with the other LEA families (Singh et al. 2005; Hundertmark and Hincha 2008). Members of this family have been associated with the hypersensitive response (HR) after microbial and parasitic nematode infection, which also differs from the other families (VanderEycken et al. 1996; Escobar et al. 1999; Ciccarelli and Bork 2005). However, functions associated with salinity, freezing, heat, UV radiation, osmotic, and oxidative stress in vitro have also been documented for LEA_2 proteins (He et al. 2012; Jia et al. 2014; Jiang et al. 2017).
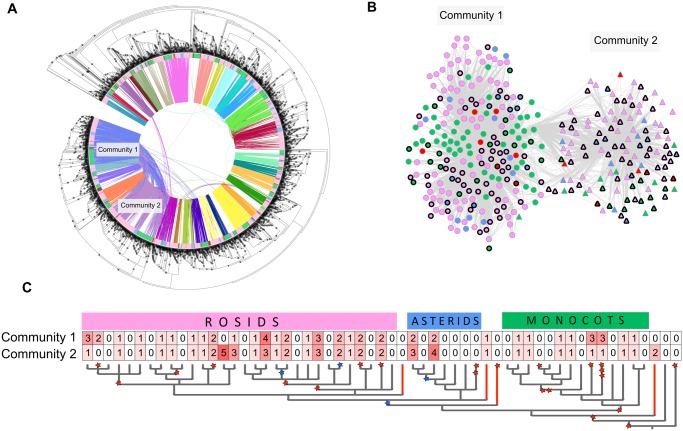
Despite the large number of members, in general, synteny and phylogeny of the LEA_2 are in agreement, with highly supported branches in the phylogenetic tree connecting genes that belong to the same synteny community (fig. 5A). Interestingly, there is an evident interconnection between two of the largest LEA_2 synteny communities (fig. 5A and B). We found that these communities contain several tandem duplicates widespread in monocots and eudicots (fig. 5B and C). In fact, we also found several other tandem duplicates across other LEA_2 communities containing monocots and eudicots genes (supplementary table S4, Supplementary Material online). These results indicate that tandem duplications have significantly contributed to the expansion and diversification of the large LEA_2 family in angiosperms, and may be one of the causes of the diversified functionality of this atypical LEA family.
Fig. 5.
—Tandem duplications of the LEA_2 family. (A) Maximum likelihood tree containing all LEA_2 genes identified. The colors displayed in the inner circle indicate genes belonging to monocots (green), rosids (pink), asterids (blue), basal species (red), the gymnosperm Picea abies (brown), the bryophyte Physcomitrella patens (light green), and the lycophyte Selaginella moellendorffii (olive green). The connections between the branches indicate synteny between the gene pairs, and all the communities with at least 100 syntenic genes are displayed in different colors. Synteny communities 1 and 2 are indicated. The dots on the branches indicate bootstrap support values (>85). The larger the dots the higher the bootstrap values. (B) Synteny network of genes belonging to community 1 (circles) and community 2 (triangles). The colors displayed in the nodes represent the clades as indicated in (A). Tandem genes are indicated by a thicker black border. (C) Summary of the number of tandem duplicates in the synteny communities 1 and 2 (see also supplementary table S4, Supplementary Material online). The tree is a simplified version of the species tree presented in figure 1. Red stairs indicate WGD and blue stars indicate WGT.
LEA_4: Dynamic Synteny in Plant Desiccation Tolerance
LEA_4 genes, also known as group 3, are also found in nonplant organisms that display DT such as rotifers, arthropods, nematodes, and tardigrades (Browne et al. 2002; Tunnacliffe et al. 2005; Kikawada et al. 2006; Gusev et al. 2014) suggesting an association with the evolution of DT. In plants, LEA_4 is strongly associated with DT in basal and angiosperm resurrection species via an ancient conserved ABA signaling pathway (Cuming et al. 2007; Hundertmark and Hincha 2008; Shinde et al. 2012; Delahaie et al. 2013; Stevenson et al. 2016). Our species set contained two desiccation tolerant species, the bryophyte Physcomitrella patens and the monocot Xerophyta viscosa; however, synteny cannot be detected between these species due to the large evolutionary distance. Nevertheless, we found that LEA_4 genes are distributed across several AW, MS, and ES synteny communities that are phylogenetically separated, suggesting a dynamic evolutionary history of this gene family in angiosperms (fig. 2B and supplementary fig. S4, Supplementary Material online). In X. viscosa, the LEA_4 family has expanded as compared with other monocot species (Costa et al. 2017), and its upregulation during dehydration was correlated with a stronger desiccation response. Interestingly, only one of the eight LEA_4 genes identified in X. viscosa shares synteny with other angiosperm species, all the other duplicates are singletons or in-paralogs (supplementary table S5, Supplementary Material online). These results suggest that species-specific duplications were important for LEA_4 family expansion in X. viscosa and likely contributed to their role in DT (Costa et al. 2017).
Discussion
How does the plant genome adapt to environmental stress? This question has been addressed frequently in recent years. It has been proposed that adaptation to novel or stressful environments is correlated with the retention of duplicated genes (Flagel and Wendel 2009; Jiao et al. 2011; Kondrashov 2012; Panchy et al. 2016). Genes which products should be rapidly or constantly produced at high level in response to environmental stress are thought to be more prone to selection after duplication (Conant and Wolfe 2008; Innan and Kondrashov 2010; Kondrashov 2012).
In plants, the group of LEA proteins, composed of eight multigene families (Dehydrin, LEA_1, LEA_2, LEA_3, LEA_4, LEA_5, LEA_6, and SMP), have been shown to play roles in water stress tolerance, and may represent a conserved and indispensable component of regulatory networks involved in environmental stress adaptation that allowed plants to endure the constraints associated with land adaptation (Shih et al. 2008; Hincha and Thalhammer 2012; Amara 2014). Evidence suggests that there is functional variability between and within each of the eight families (Hundertmark and Hincha 2008), which raises questions pertaining to the sources of functional variations, the precise biological functions of each family, if and how LEA families work as one entity, and which LEA genes are involved in plant development and stress tolerance.
To address these questions we interrogated 60 whole genomes, ranging from green algae to angiosperms and analyzed the ancestry, conservation, and diversification of LEA proteins in plants. We found that LEA proteins belonging to LEA_5 and SMP families have arisen early in the plant lineage, while the other families appeared at later instants during plant evolution (fig. 1). Previous studies have already shown the presence and expression of ancient LEA proteins in algal genomes (Joh et al. 1995; Wodniok et al. 2011), corroborating the hypothesis that the ancestral fresh water lineages were preadapted to terrestrial environments, and the evolution of pre-existing and new gene families, including LEA gene families, may have facilitated the colonization of land (Rensing et al. 2008; Becker and Marin 2009). It seems possible that later LEA families expanded and diversified in embryophytes as a result of the evolution of more specialized cells, tissues and organs such as spores and seeds, that required a better control of water retention and protection against desiccation and other stresses.
Synteny homology analysis indicated that the majority of LEA genes are located in angiosperm-wide conserved genomic regions, while the finding of clade-specific as well as species-specific gene copies indicates that the continuing expansion and diversification of angiosperm genomes contributed to the evolution of LEA gene families (fig. 2). Stress-regulated genes retained after duplication events are more likely to neofunctionalize instead of inheriting the ancestral function, which might be in part related to changes in biochemical function and in cis-regulatory regions (Conant and Wolfe 2008; Zou et al. 2009; Arsovski et al. 2015). As a result of these changes, complete or partial diversification of the interaction and regulatory networks in which the duplicated genes are involved might also occur. It is likely that the genes belonging to the same synteny community (positional homologs) display similar functions, and genes in different communities are likely to display functional innovations (Dewey 2011).
We identified highly conserved synteny between LEA_5 genes in most genomes investigated, suggesting evolutionary constraints on maintaining the stability of their genomic context. These constraints may include the correct functioning of the maturation-induced desiccation program, where LEA_5 genes of A. thaliana were shown to play important roles (Manfre et al. 2005), and appeared conserved across all angiosperm species that produce desiccation tolerant seeds (also called orthodox seeds).
We also found several examples of correlation between synteny diversification and functional innovations. Genes from the largely studied Dehydrin (DHN) family are localized in two distinct synteny communities across the angiosperm lineage (fig. 3A). Presumably, new regulatory elements were acquired in the duplicated copies, and differential evolutionary forces may have driven protein diversification, resulting in distinct biochemical properties (fig. 3B). The consequent differential gene expression (developmental or stress induced) may have allowed the preservation of duplicated copies in the different genomes, and amplified the stress tolerance response. The finding of functionally diverse Dehydrin types in Physcomitrella patens suggests that the colonization of land was one of the forces driving Dehydrin evolution (Ruibal et al. 2012; Agarwal 2017). Similarly, LEA_1 have evolved into two angiosperm-wide synteny communities composed by two protein types containing different protein consensus size (fig. 4C). Our findings point toward an ancient functional divergence among LEA_1 members, which would explain their structural plasticity and “moonlighting” properties associated with multiple abiotic stresses (Covarrubias et al. 2017; Cuevas-Velazquez et al. 2017).
Another source of evolutionary adaptations to environmental stress is gene family expansion via recurrent tandem duplications (Cannon et al. 2004; Hanada et al. 2008). Tandem duplications offers a pool of targets for evolutionary selection contributing to the maintenance of large gene families. These large gene families are enriched with genes important for rapid environmental adaptation such as biotic stress-responsive genes (Cannon et al. 2004; Hanada et al. 2008). We found several tandem duplicates in the synteny network of LEA_2 distributed across all angiosperm lineage (fig. 5). This supports the atypical structured and hydrophobic nature of LEA_2 proteins and its broader spectra of gene expression in response to biotic and abiotic stresses (Ciccarelli and Bork 2005; Singh et al. 2005; Hundertmark and Hincha 2008).
Most of the LEA gene expression during seed development and environmental stresses is regulated via abscisic acid (ABA)-signaling pathways (Galau et al. 1986; Espelund et al. 1992; Shinde et al. 2012; Delahaie et al. 2013; Stevenson et al. 2016). The desiccation-induced LEA gene expression via ABA-responsive pathways is conserved across basal and angiosperm resurrection species (Cuming et al. 2007; Shinde et al. 2012; Stevenson et al. 2016). It seems that the acquisition of new genomic contexts by desiccation-related LEA genes of the resurrection monocot Xerophyta viscosa is an important footprint of DT, and suggests a conserved regulation of these duplicates in order to assure cellular protection under desiccation conditions.
Resurrection plants are species adapted to live in environments with low water availability, displaying specific molecular and genomic adaptations of DT (Oliver et al. 2000; Mundree 2002; Illing et al. 2005; Farrant and Moore 2011; Gaff and Oliver 2013). The concept of DT is different from drought tolerance because drought tolerance refers to the tolerance to moderate water removal without removal of the bulk of cytoplasmic water (Shih et al. 2008), while DT refers to the tolerance to a further dehydration with an increased removal of the water shell and the capacity to survive long periods in the dry state (Hoekstra et al. 2001). Understanding the mechanisms underlying DT can help to improve drought tolerance in crops (Mundree 2002; Leprince and Buitink 2010; Costa et al. 2017). Several crops from the grass family (Poaceae) constitute major contributors to global food security that have become targets of genomic programs aiming at improved drought tolerance. In grasses, overexpression of LEA genes has already been shown to enhance tolerance to drought and other stresses (Babu 2004; Fu et al. 2007; Xiao et al. 2007; Chen et al. 2015). We believe that comprehending the impact of synteny diversification in functional innovations in the LEA families may offer an extra powerful tool to select candidates for engineering drought and desiccation tolerant crops.
These data also provide hypothesis-driven fundamental and experimental questions about the various functions of LEA proteins, and the role of the diversification of the genomic context in plant evolution and adaptation to environmental stresses. Deciphering the evolution of eight gene families, with variable protein structure and diversified expression patterns over billions of years, is a challenging task. Despite the general association of LEA proteins with water stress response, our work provides strong examples of a clear evolutionary divergence resulting in differential protein evolution. The diversity of LEA families in angiosperms is a result of extensive and dynamic synteny evolution, which indicates that the complexity of these gene families goes beyond their protein sequences.
Supplementary Material
Supplementary data are available at Genome Biology and Evolution online.
Supplementary Material
Acknowledgments
M.A.S.A. received financial support from CAPES—Brazilian Federal Agency for Support and Evaluation of Graduate Education (BEX 0857/14-9). T.Z. was supported by China Scholarship Council.
Author Contributions
M.A.S.A, T.Z., W.L., M.E.S., and H.W.M.H. planned and designed the research. M.A.S.A and T.Z. performed the research and analyzed the data. M.A.S.A interpreted the data and wrote the article with contributions of T.Z., W.L., M.E.S., and H.W.M.H. All authors edited and commented on the article.
Literature Cited
- Agarwal T. 2017. Different dehydrins perform separate functions in Physcomitrella patens. Planta 245(1):101–118. [DOI] [PubMed] [Google Scholar]
- Amara I, et al. 2014. Insights into late embryogenesis abundant (LEA) proteins in plants: from structure to the functions. Am J Plant Sci. 05(22):3440–3445. [Google Scholar]
- Arsovski AA, Pradinuk J, Guo XQ, Wang S, Adams KL.. 2015. Evolution of cis-regulatory elements and regulatory networks in duplicated genes of Arabidopsis. Plant Physiol. 169(4):2982–2991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Babu RC, et al. 2004. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci. 166:855–862. [Google Scholar]
- Banerjee A, Roychoudhury A.. 2016. Group II late embryogenesis abundant (LEA) proteins: structural and functional aspects in plant abiotic stress. Plant Growth Regul. 79(1):1–17. [Google Scholar]
- Bastian M, Heymann S, Jacomy M.. 2009. Gephi: an open source software for exploring and manipulating networks. Proc. Int. AAAI Conf. Weblogs Soc. Media 8:361–362. [Google Scholar]
- Bateman A, et al. 2002. The Pfam protein families database. Nucleic Acids Res. 30(1):276–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battaglia M, Olvera-Carrillo Y, Garciarrubio A, Campos F, Covarrubias AA.. 2008. The enigmatic LEA proteins and other hydrophilins. Plant Physiol. 148(1):6–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker B, Marin B.. 2009. Streptophyte algae and the origin of embryophytes. Ann Bot. 103(7):999–1004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne J, Tunnacliffe A, Burnell A.. 2002. Anhydrobiosis – plant desiccation gene found in a nematode. Nature 416(6876):38–38. [DOI] [PubMed] [Google Scholar]
- Cannon SB, Mitra A, Baumgarten A, Young ND, May G.. 2004. The roles of segmental and tandem gene duplication in the evolution of large gene families in Arabidopsis thaliana. BMC Plant Biol. 4(1):10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, et al. 2007. Hydrophilic protein associated with desiccation tolerance exhibits broad protein stabilization function. Proc Natl Acad Sci U S A. 104(46):18073–18078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chakrabortee S, et al. 2012. Intrinsically disordered proteins as molecular shields. Mol Biosyst. 8(1):210–219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen YS, et al. 2015. A late embryogenesis abundant protein HVA1 regulated by an inducible promoter enhances root growth and abiotic stress tolerance in rice without yield penalty. Plant Biotechnol J. 13(1):105–116. [DOI] [PubMed] [Google Scholar]
- Ciccarelli FD, Bork P.. 2005. The WHy domain mediates the response to desiccation in plants and bacteria. Bioinformatics 21(8):1304–1307. [DOI] [PubMed] [Google Scholar]
- Close TJ. 1996. Dehydrins: emergence of a biochemical role of a family of plant dehydration proteins. Physiol Plantarum. 97(4):795–803. [Google Scholar]
- Conant GC, Wolfe KH.. 2008. Turning a hobby into a job: how duplicated genes find new functions. Nat Rev Genet. 9(12):938–950. [DOI] [PubMed] [Google Scholar]
- Costa M-CD, et al. 2017. A footprint of desiccation tolerance in the genome of Xerophyta viscosa. Nat Plants. 3:17038. [DOI] [PubMed] [Google Scholar]
- Covarrubias AA, Cuevas-Velazquez CL, Romero-Pérez PS, Rendón-Luna DF, Chater CCC.. 2017. Structural disorder in plant proteins: where plasticity meets sessility. Cell Mol Life Sci. 74(17):3119–3147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuevas-Velazquez CL, Reyes JL, Covarrubias AA.. 2017. Group 4 late embryogenesis abundant proteins as a model to study intrinsically disordered proteins in plants. Plant Signal Behav. 12(7):e1343777.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cuming AC, Cho SH, Kamisugi Y, Graham H, Quatrano RS.. 2007. Microarray analysis of transcriptional responses to abscisic acid and osmotic, salt, and drought stress in the moss, Physcomitrella patens. New Phytol. 176(2):275–287. [DOI] [PubMed] [Google Scholar]
- Delahaie J, et al. 2013. LEA polypeptide profiling of recalcitrant and orthodox legume seeds reveals ABI3-regulated LEA protein abundance linked to desiccation tolerance. J Exp Bot. 64(14):4559–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dewey CN. 2011. Positional orthology: putting genomic evolutionary relationships into context. Brief Bioinform. 12(5):401–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dixon P. 2003. VEGAN, a package of R functions for community ecology. J Veg Sci. 14(6):927–930. [Google Scholar]
- Dure L, et al. 1989. Common amino-acid sequence domains among the Lea proteins of higher-plants. Plant Mol Biol. 12(5):475–486. [DOI] [PubMed] [Google Scholar]
- Eisen JA. 1998. Phylogenomics: improving functional predictions for uncharacterized genes by evolutionary analysis. Genome Res. 8(3):163–167. [DOI] [PubMed] [Google Scholar]
- Escobar C, et al. 1999. Isolation of the LEMMI9 gene and promoter analysis during a compatible plant-nematode interaction. Mol Plant Microbe Interact. 12(5):440–449. [DOI] [PubMed] [Google Scholar]
- Espelund M, et al. 1992. Late embryogenesis-abundant genes encoding proteins with different numbers of hydrophilic repeats are regulated differentially by abscisic-acid and osmotic-stress. Plant J. 2(2):241–252. [PubMed] [Google Scholar]
- Farrant JM, Moore JP.. 2011. Programming desiccation-tolerance: from plants to seeds to resurrection plants. Curr Opin Plant Biol. 14(3):340–345. [DOI] [PubMed] [Google Scholar]
- Finn RD, Clements J, Eddy SR.. 2011. HMMER web server: interactive sequence similarity searching. Nucleic Acids Res. 39(Suppl):W29–W37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flagel LE, Wendel JF.. 2009. Gene duplication and evolutionary novelty in plants. New Phytol. 183(3):557–564. [DOI] [PubMed] [Google Scholar]
- Fu D, Huang B, Xiao Y, Muthukrishnan S, Liang GH.. 2007. Overexpression of barley hva1 gene in creeping bentgrass for improving drought tolerance. Plant Cell Rep. 26(4):467–477. [DOI] [PubMed] [Google Scholar]
- Gabaldon T, Koonin EV.. 2013. Functional and evolutionary implications of gene orthology. Nat Rev Genet. 14:360–366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaff DF, Oliver M.. 2013. The evolution of desiccation tolerance in angiosperm plants: a rare yet common phenomenon. Funct Plant Biol. 40(4):315–328. [DOI] [PubMed] [Google Scholar]
- Galau GA, Hughes DW, Dure L.. 1986. Abscisic-acid induction of cloned cotton late embryogenesis-abundant (LEA) messenger-RNAs. Plant Mol Biol. 7(3):155–170. [DOI] [PubMed] [Google Scholar]
- Garay-Arroyo A, Colmenero-Flores JM, Garciarrubio A, Covarrubias AA.. 2000. Highly hydrophilic proteins in prokaryotes and eukaryotes are common during conditions of water deficit. J Biol Chem. 275(8):5668–5674. [DOI] [PubMed] [Google Scholar]
- Goodstein DM, et al. 2012. Phytozome: a comparative platform for green plant genomics. Nucleic Acids Res. 40(D1):D1178–D1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goyal K, Walton LJ, Tunnacliffe A.. 2005. LEA proteins prevent protein aggregation due to water stress. Biochem J. 388(1):151–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gusev O, et al. 2014. Comparative genome sequencing reveals genomic signature of extreme desiccation tolerance in the anhydrobiotic midge. Nat Commun. 5:4784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanada K, Zou C, Lehti-Shiu MD, Shinozaki K, Shiu SH.. 2008. Importance of lineage-specific expansion of plant tandem duplicates in the adaptive response to environmental stimuli. Plant Physiol. 148(2):993–1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He S, et al. 2012. Molecular characterization and functional analysis by heterologous expression in E. coli under diverse abiotic stresses for OsLEA5, the atypical hydrophobic LEA protein from Oryza sativa L. Mol Genet Genomics. 287(1):39–54. [DOI] [PubMed] [Google Scholar]
- Hincha DK, Thalhammer A.. 2012. LEA proteins: iDPs with versatile functions in cellular dehydration tolerance. Biochem Soc Trans. 40(5):1000–1003. [DOI] [PubMed] [Google Scholar]
- Hoekstra FA, Golovina EA, Buitink J.. 2001. Mechanisms of plant desiccation tolerance. Trends Plant Sci. 6(9):431–438. [DOI] [PubMed] [Google Scholar]
- Hunault G, Jaspard E.. 2010. LEAPdb: a database for the late embryogenesis abundant proteins. BMC Genomics 11(1):221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hundertmark M, Hincha DK.. 2008. LEA (Late Embryogenesis Abundant) proteins and their encoding genes in Arabidopsis thaliana. BMC Genomics 9(1):118.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Illing N, Denby KJ, Collett H, Shen A, Farrant JM.. 2005. The signature of seeds in resurrection plants: a molecular and physiological comparison of desiccation tolerance in seeds and vegetative tissues. Integr Comp Biol. 45(5):771–787. [DOI] [PubMed] [Google Scholar]
- Innan H, Kondrashov F.. 2010. The evolution of gene duplications: classifying and distinguishing between models. Nat Rev Genet. 11(2):97–108. [DOI] [PubMed] [Google Scholar]
- Jaspard E, Hunault G.. 2014. Comparison of amino acids physico-chemical properties and usage of late embryogenesis abundant proteins, hydrophilins and WHy domain. PLoS One 9(10):e109570.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia FJ, et al. 2014. Overexpression of late embryogenesis abundant 14 enhances Arabidopsis salt stress tolerance. Biochem Biophys Res Commun. 454(4):505–511. [DOI] [PubMed] [Google Scholar]
- Jiang SJ, et al. 2017. DrwH, a novel WHy domain-containing hydrophobic LEA5C protein from Deinococcus radiodurans, protects enzymatic activity under oxidative stress. Sci Rep. 7:9281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiao Y, et al. 2011. Ancestral polyploidy in seed plants and angiosperms. Nature 473(7345):97–100. [DOI] [PubMed] [Google Scholar]
- Joh T, et al. 1995. Molecular-cloning and expression of hardening-induced genes in Chlorella vulgaris c-27 – the most abundant clone encodes a late embryogenesis abundant protein. Plant Cell Physiol. 36(1):85–93. [PubMed] [Google Scholar]
- Katoh K, Misawa K, Kuma K, Miyata T.. 2002. MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30(14):3059–3066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kenrick P, Crane PR.. 1997. The origin and early evolution of plants on land. Nature 389(6646):33–39. [Google Scholar]
- Kikawada T, et al. 2006. Dehydration-induced expression of LEA proteins in an anhydrobiotic chironomid. Biochem Biophys Res Commun. 348(1):56–61. [DOI] [PubMed] [Google Scholar]
- Kondrashov FA. 2012. Gene duplication as a mechanism of genomic adaptation to a changing environment. Proc R Soc B Biol Sci. 279(1749):5048–5057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koonin EV. 2005. Orthologs, paralogs, and evolutionary genomics. Annu Rev Genet. 39(1):309–338. [DOI] [PubMed] [Google Scholar]
- Kovacs D, Kalmar E, Torok Z, Tompa P.. 2008. Chaperone activity of ERD10 and ERD14, two disordered stress-related plant proteins. Plant Physiol. 147(1):381–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leprince O, Buitink J.. 2010. Desiccation tolerance: from genomics to the field. Plant Sci. 179(6):554–564. [Google Scholar]
- Letunic I, Bork P.. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res. 44(W1):W242–W245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Song QP, Li DX, Yang XH, Li DQ.. 2017. Multifunctional roles of plant dehydrins in response to environmental stresses. Front Plant Sci. 8:1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malik AA, Veltri M, Boddington KF, Singh KK, Graether SP.. 2017. Genome analysis of conserved dehydrin motifs in vascular plants. Front Plant Sci. 8:709.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manfre AJ, Lanni LM, Marcotte WR.. 2005. The Arabidopsis group 1 late embryogenesis abundant protein ATEM6 is required for normal seed development. Plant Physiol. 140(1):140–149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mundree SG, et al. 2002. Physiological and molecular insights into drought tolerance. Afr J Biotechnol. 1:28–38. [Google Scholar]
- Nguyen LT, Schmidt HA, von Haeseler A, Minh BQ.. 2015. IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol Biol Evol. 32(1):268–274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oliver MJ, Tuba Z, Mishler BD.. 2000. The evolution of vegetative desiccation tolerance in land plants. Plant Ecol. 151(1):85–100. [Google Scholar]
- Olsen JL, et al. 2016. The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea. Nature 530(7950):331. [DOI] [PubMed] [Google Scholar]
- Olvera-Carrillo Y, Campos F, Reyes JL, Garciarrubio A, Covarrubias AA.. 2010. Functional analysis of the group 4 late embryogenesis abundant proteins reveals their relevance in the adaptive response during water deficit in Arabidopsis. Plant Physiol. 154(1):373–390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchy N, Lehti-Shiu M, Shiu SH.. 2016. Evolution of gene duplication in plants. Plant Physiol. 171(4):2294–2316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rensing SA, et al. 2008. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science 319(5859):64–69. [DOI] [PubMed] [Google Scholar]
- Reyes JL, et al. 2008. Functional dissection of hydrophilins during in vitro freeze protection. Plant Cell Environ. 31(12):1781–1790. [DOI] [PubMed] [Google Scholar]
- Rosvall M, Bergstrom CT.. 2008. Maps of random walks on complex networks reveal community structure. Proc Natl Acad Sci U S A. 105(4):1118–1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruibal C, et al. 2012. Differential contribution of individual dehydrin genes from Physcomitrella patens to salt and osmotic stress tolerance. Plant Sci. 190:89–102. [DOI] [PubMed] [Google Scholar]
- Shannon P, et al. 2003. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 13(11):2498–2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shih MD, Hoekstra FA, Hsing YIC.. 2008. Late embryogenesis abundant proteins. Adv Bot Res. 48:211–255. [Google Scholar]
- Shinde S, Nurul Islam M, Ng CK.. 2012. Dehydration stress-induced oscillations in LEA protein transcripts involves abscisic acid in the moss, Physcomitrella patens. New Phytol. 195(2):321–328. [DOI] [PubMed] [Google Scholar]
- Singh S, et al. 2005. Solution structure of a late embryogenesis abundant protein (LEA14) from Arabidopsis thaliana, a cellular stress-related protein. Protein Sci. 14(10):2601–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith SA, Dunn CW.. 2008. Phyutility: a phyloinformatics tool for trees, alignments and molecular data. Bioinformatics 24(5):715–716. [DOI] [PubMed] [Google Scholar]
- Stevenson SR, et al. 2016. Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE, a regulator of ABA responses unique to basal land plants and required for desiccation tolerance. Plant Cell 28(6):1310–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strimbeck GR. 2017. Hiding in plain sight: the F segment and other conserved features of seed plant SKn dehydrins. Planta 245:1061–1066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tunnacliffe A, Lapinski J, McGee B.. 2005. A putative LEA protein, but no trehalose, is present in anhydrobiotic bdelloid rotifers. Hydrobiologia 546(1):315–321. [Google Scholar]
- VanderEycken W, Engler JD, Inze D, VanMontagu M, Gheysen G.. 1996. A molecular study of root-knot nematode-induced feeding sites. Plant J. 9:45–54. [DOI] [PubMed] [Google Scholar]
- Wang YP, et al. 2012. MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40(7):e49–e49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ.. 2009. Jalview version 2 – a multiple sequence alignment editor and analysis workbench. Bioinformatics 25(9):1189–1191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wodniok S, et al. 2011. Origin of land plants: do conjugating green algae hold the key? BMC Evol Biol. 11:104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xiao B, Huang Y, Tang N, Xiong L.. 2007. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor Appl Genet. 115(1):35–46. [DOI] [PubMed] [Google Scholar]
- Zhao T, et al. 2017. Phylogenomic synteny network analysis of MADS-Box Transcription factor genes reveals lineage-specific transpositions, ancient tandem duplications, and deep positional conservation. Plant Cell 29:1278–1292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao T, Schranz ME.. 2017. Network approaches for plant phylogenomic synteny analysis. Curr Opin Plant Biol. 36:129–134. [DOI] [PubMed] [Google Scholar]
- Zhao YA, Tang HX, Ye YZ.. 2012. RAPSearch2: a fast and memory-efficient protein similarity search tool for next-generation sequencing data. Bioinformatics 28(1):125–126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou C, Lehti-Shiu MD, Thomashow M, Shiu SH.. 2009. Evolution of stress-regulated gene expression in duplicate genes of Arabidopsis thaliana. PLoS Genet. 5(7):e1000581.. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.