Abstract
1) Animals must effectively balance the time they spend exploring the environment for new resources and exploiting them. One way that social animals accomplish this balance is by allocating these two tasks to different individuals. In honey bees, foraging is divided between scouts, which tend to explore the landscape for novel resources, and recruits, which tend to exploit these resources. Exploring the variation in cognitive and physiological mechanisms of foraging behaviour will provide a deeper understanding of how the division of labour is regulated in social insect societies.
2) Here we uncover how honey bee foraging behaviour may be shaped by predispositions in performance of latent inhibition (LI), which is a form of non-associative learning by which individuals learn to ignore familiar information.
3) We compared LI between scouts and recruits, hypothesizing that differences in learning would correlate with differences in foraging behaviour. Scouts seek out and encounter many new odours while locating novel resources, while recruits continuously forage from the same resource, even as its quality degrades.
4) We found that scouts show stronger LI than recruits, possibly reflecting their need to discriminate forage quality. We also found that scouts have significantly elevated tyramine compared to recruits. Furthermore, after associative odour training, recruits have significantly diminished octopamine in their brains compared to scouts.
5) These results suggest that individual variation in learning behaviour shapes the phenotypic behavioural differences between different types of honey bee foragers. These differences in turn have important consequences for how honeybee colonies interact with their environment. Uncovering the proximate mechanisms that influence individual variation in foraging behaviour is crucial for understanding the ecological context in which societies evolve.
Keywords: Non-Associative Learning, Latent Inhibition, Tyramine, Honey Bee, Foraging, Scout, Exploration-Exploitation Trade-Off
Graphical abstract
Graphical Abstract text: Honey bees have mastered the exploration-exploitation trade off by dividing foraging labor. Here, the authors report that underlying this division of labor is variation in learning ability: scouts ignore familiar odors while recruits readily learn novel and familiar odors. Further, scouts and recruits have different quantities of biogenic amines in their brains, modulating their behavior.

INTRODUCTION
A major question in foraging ecology is how animals balance exploring for new food sources versus exploiting known forage locations (Charnov, 1976). Animals must be able to effectively analyse when a foraging area may be running out of food and when it is time to search elsewhere. In complex societies, this trade-off between exploring for new resources and exploiting familiar ones can be resolved by allocating different individuals to each of these tasks. (Hills et al., 2015; Wilson, 1971). Foraging honey bees experience changes in resource availability as local environments change, for example, when flowering sites are depleted, and new sites are discovered. Honey bee colonies must constantly balance how many individuals gather food, with the time and energy of individuals searching for novel resources (Mosqueiro et al., 2017).
Social insect colonies can adjust quickly to changing environments because individuals vary in the tasks they perform, often referred to as ‘division of labour’. In honey bees, generally, the youngest workers are nurses, who care for larvae, while the oldest workers are foragers (Lindauer, 1952; Seeley, 1982). Foragers are further divided into scouts, who explore the landscape for novel resources, and recruits, who are signalled by scouts through waggle dances to exploit newly discovered resources (von Frisch, 1965; Seeley, 1983a). Genetic variation plays an important role in the division between scouts and recruits. Colonies with workers from multiple patrilines, which have high genetic diversity, have more scouts performing waggle dances compared to less genetically diverse, single-patriline, colonies (Mattila & Seeley, 2011). Further, more genetically diverse colonies collect food from longer distances compared to less genetically diverse colonies (Mattila & Seeley, 2011). Scouts also differ from recruits in the expression of several genes in their brains that code for behaviourally important neuromodulators, including octopamine and GABA (Liang et al., 2012). The exact behavioural and physiological mechanisms that underlie the differences between scouts and recruits are still unknown.
The ability to either explore or exploit food resources could emerge from differences between individuals in cognition. Scouts must effectively measure the food landscape by visiting many flowers, remembering them, then communicating the information they gathered back at the nest. Scouts must also remember which sites are not profitable so they do not revisit them. Further, when scouts are done performing the waggle dance to recruit other foragers, they must learn to avoid the flowers that they have already visited. Conversely, recruits must effectively follow the scout’s waggle dance, and then repeatedly visit the same foraging location (Biesmeijer & de Vries, 2001; Hills et al., 2015; von Frisch, 1952). While much work has focused on important colony-level decision making processes related to foraging (Couzin, 2009; Seeley, 1983, 1986; Visscher & Seeley, 1982), little research has explored how individual variation in cognition may explain the variation in foraging behaviour (Giurfa & Menzel, 2001; Hill, Wells, & Wells, 1997). For example, in one cognitive task, scouts exhibited increased reversal learning; they were able to learn when an odour switched from predicting a punishment to predicting a reward faster than recruits learned (Carr-Markell & Robinson, 2014). This finding indicates that scouts may also be able to learn that an environment is changing more quickly than recruits (Bitterman, 1972). Thus, the shifting ecological demands of a landscape may shape cognitive variation between scouts and recruits.
In addition to reversal learning, latent inhibition has been well studied in honey bees. Latent inhibition (LI) is a learning behaviour where individuals can learn to ignore an uninformative stimulus that they have previously experienced (Lubow, 1973). LI has been observed in numerous non-human species, including rats (Young, Joseph, & Gray, 1993), mice (Robbins, 1979), goats and sheep (Lubow & Moore, 1959), embryonic amphibians (Ferrari & Chivers, 2011), and fish (Ferrari & Chivers, 2006). Notably, some insects also exhibit LI, including fruit flies (Sudhakaran et al., 2012) and honey bees (Chandra, Hosler, & Smith, 2000). In honey bees, individuals differ in how strongly they exhibit LI. Some individuals exhibit ‘high’ LI, where they take longer to make associations between a stimulus and a reward when a familiar stimulus is reinforced in a way that normally produces robust conditioned responses (Bitterman, Menzel, Fietz, & Schafer, 1983; Chandra et al., 2000). Other individuals exhibit ‘low’ LI, where they quickly learn to make associations between familiar stimuli and reward, despite prior experience with the stimuli (Bitterman et al., 1983). Related to foraging, scouts might benefit by being able to learn to avoid flowers at a previously visited but no longer productive food location. In contrast, recruits could benefit by sustaining resource extraction even as the flowers become less rewarding.
The variation among bees in their LI response may be modulated by physiological and genetic variation. Several important regions on the honey bee genome have been found to relate to LI response (Chandra, Hunt, Cobey, & Smith, 2001). One major gene region, PLN2, shows a significant likelihood of influencing LI and also codes for a tyramine receptor (AmTYR1 for Apis mellifera tyramine receptor, Hunt et al., 2007). In invertebrates, tyramine is a biogenic amine that plays an analogous role to noradrenaline in invertebrates (Roeder, 2005). Tyramine acts as a neuromodulator and strongly influences honey bee behaviour, including the regulation of activity (Fussnecker, Smith, & Mustard, 2006), the shift from work inside the colony to external foraging activities (Lehman et al., 2006), and sucrose responsiveness (Scheiner, Plückhahn, Oney, Blenau, & Erber, 2002). This region is also associated with a division between nectar foragers and pollen foragers (Hunt, Page, Fondrk, & Dullum, 1995). Tyramine therefore likely plays a significant role in modulating the differences in learning, and potentially foraging behaviour, between scout and recruit honey bees.
Here, we hypothesize that LI plays a role in foraging behaviour. Specifically, we predict that LI is higher in scouts than recruits because scouts should be better at avoiding unrewarding stimuli. We further hypothesize that brain biogenic amines will be correlated to scouting and recruiting behaviour. We predict that tyramine will likely be the brain biogenic amine that is most different between scouts and recruits. Finally, we hypothesize that foragers that show different LI will have different biogenic amine concentrations in their brain. Again, we predict that tyramine will play a significant role in the distinction between different LI behaviours. We test these hypotheses by collecting scouts and recruits from naturally foraging colonies, then examining their LI and their brain amines. With these data, we provide a direct link between learning, neurobiology, and adaptive foraging behaviour.
MATERIALS AND METHODS
Animals
Honey bee colonies were maintained at Arizona State University’s honey bee laboratory (Mesa Arizona USA). We used Italian (n = 2 colonies from Olivarez Honey Bees Inc.) and New World Carniolan (n = 3 colonies from Olivarez Honey Bees Inc.) selected genetic backgrounds of Apis mellifera L. A colony was housed in a single or double hive body with 9 frames per hive body, thus consisting of approximately 10,000–15,000 workers each. We conducted all bee collections and behavioural assays between August and September of 2016.
Colony Moving Procedure
We used a modified version of a standard colony moving procedure to induce scouting behaviour (Carr-Markell & Robinson, 2014; Dreller, 1998; Liang et al., 2012; Mattila & Seeley, 2011). Colonies were placed in a secondary apiary ~3.8 km away from the “novel” apiary, outside the median foraging range of honey bees (1.7 km, Visscher & Seeley, 1982) for at least one week. After 7 days, 70% of foragers forget previous foraging locations (Beekman, 2005). Therefore, the novel apiary is a foraging location that is unfamiliar to foraging honey bees. We moved the colony from the secondary apiary to the novel apiary the night before forager collection and testing.
Collecting Scouts and Recruits
We collected scouts the morning after moving the colony to the novel foraging location. To collect scouts and prevent the recruitment of other foragers, we created a one-way exit that prevented returning bees from entering the colony. The exit consisted of wire mesh blocking the colony entrance, with a one-way 5cm diameter x 20cm long mesh tube which provided an exit for scouts. To distinguish scouts from undertakers and from bees performing orientation flights, we considered as scouts only the bees that were away from the colony for at least 10 min (Carr-Markell & Robinson, 2014).
Time away from the nest was determined by marking exiting bees for an hour, beginning at 8:00 am, with coloured chalk powder as they left the colony, and we switched colours every 10 minutes (See Supplemental Material for video, ESM7). We did not mark or collect individuals that were leaving the colony carrying dead bees or larvae, as they were engaged in undertaking behaviour and are typically not foragers (Breed, Williams, & Queral, 2002). Individuals that returned bearing the same colour that was being used, or returned during the next colour, were set aside in a container for the duration of the sampling and not tested. Marked individuals that were gone for at least 10 minutes (meaning at least two colours before returning to the colony) were collected for testing. We focused on nectar foragers because they have a more even distribution of LI as compared to pollen foragers, which exhibit a higher LI score (Latshaw & Smith, 2005). We therefore avoided sampling scouts that had collected pollen, distinguishable by their pollen-filled corbiculae.
After collecting scouts, we removed the one-way colony exit to allow workers to forage freely. The colony remained open for 24 h. To collect recruits, we blocked access to the colony by placing a sheet of wire mesh at the entrance. We collected returning foragers that did not carry pollen and had a distended abdomen. Giving the colony a day to become familiar with a landscape where forage was readily available reduced the chances that scouts would be captured. When colonies forage freely, most foragers (60%) receive information from the waggle dance, making them recruits (Biesmeijer & Seeley, 2005). Further, the percentage of scouts is relatively small, typically comprising 5% when forage was plentiful to 35% of foragers when less forage is available (Seeley, 1983). Forage is readily available in the fall in Arizona.
To collect foragers for LI testing, we placed an 8 mL glass vial over the bee once it landed on the mesh blocking the colony entrance and sealed the vial with a ventilated cap. Bees never spent more than 10 min in a vial before being transferred to the lab. Although collected on different days, both scouts and recruits were sampled during the same time-period, between 8:20 am and 9:00 am. For each sampling bout, we collected 20 bees to ensure use of 16 in the LI training procedure detailed below. For LI assessment, we tested scouts (n=94) and recruits (n=82) that were pre-exposed to an odour, as well as control scouts (n=14) and recruits (n=24) that were not pre-exposed. For colony of origin for these experiments, see tables S1 and S2 in ESM2.
For biogenic amine analysis, we collected scouts and recruits as they landed at the entrance using forceps and placed them directly into liquid nitrogen for snap freezing. We collected these bees at the same time as those used for LI testing.
LI and PER Assessment
LI training and proboscis extension reflex (PER) assessment were conducted in the laboratory at 25°C. We placed bees in vials on ice for 3–5 min to immobilize them. We then strapped each bee into a plastic harness and secured the bee in the harness with a strip of duct tape between the head and thorax (Chandra, Wright, & Smith, 2010; Smith & Burden, 2014). Care was taken to ensure free movement of antennae and mouthparts for later training. Once harnessed, we fed the bees with 5 µL of 1M sucrose solution then allowed them to acclimate to the harnesses for 1 h. Individuals unresponsive to the presented sucrose, 23 scouts and 20 recruits over the course of the experiment, were not used.
Bees were trained in an acrylic glass box arena connected to an automated ventilation system. An odourised airflow was dispersed by passing air through a 1 cc glass tuberculin syringe tube (with plunger removed) containing a 0.5 cm by 4 cm strip of filter paper with either 1-hexanol or 2-octanone applied to the filter paper. A subset of control scouts and recruits were exposed to a plain strip of filter paper that did not contain an odour, so that all subsequent exposures would be to novel odours to ensure that there was no effect of the LI training process on learning behaviour, and that all bees could learn both odours, irrespective of their LI phenotype. Individuals undergoing pre-exposure to a scent were pre-exposed to a scent, in order to familiarize them with an odour but to have no association of it with a reward. Comparable numbers of bees were presented with 3 µL of pure odour. These are compounds that bees readily learn but easily discriminate between them (Smith & Menzel, 1989). The tube was capped with a small silicon stopper with a hole to allow air to pass through during training while preventing the filter paper from being ejected by the airflow. If tubes were prepared in advance, they were sealed with paraffin film and placed in a 5°C refrigerator. During training, we replaced odour tubes every 25 min to avoid odour depletion with re-use (Smith & Burden, 2014). We pre-exposed each bee by dispersing forty 4-second odour bursts to its antenna at 5 min inter-stimulus intervals (Chandra et al., 2010). Tubes were placed 3 cm from the bee. The automated system was controlled by programmable logic controller (PLC: DL05 or DL105; AutomationDirect; Chandra et al., 2010) and a control screen (EZ Automation # 2 EZ3-T6C-E). The arena was connected to a ventilation system to readily remove odours (See Supplemental Material, ESM5, for photos of the training arena). After the pre-exposure, bees rested for 15 min before using the proboscis extension reflex to evaluate LI.
The PER conditioning protocol has been used extensively in many insects, including honey bees (Bitterman et al., 1983; Takeda, 1961), and was used here to evaluate LI. To perform PER, a single bee was placed into a testing arena consisting of clear acrylic glass to reduce outside disturbance and was allowed to acclimate for 20 s. After acclimation, an odourised airflow was blown directly at the bee through a glass syringe. Odour was presented for 4 s. If a bee extended its proboscis within the first 3 s, it was rewarded with a 0.4 µL droplet of 1.5M sucrose solution using a Gilmont syringe and scored as a positive response. If the bee did not extend its proboscis to the odour in this time, a 0.4 µL droplet of 1.5M sucrose solution was touched to the antenna to elicit proboscis extension, then to the proboscis for reward, during the last second of the odour delivery. After odour delivery and reward, bees remained in the testing arena for 30 s. Each individual trial lasted for 60 s. As training progressed and bees began responding to odours, they received the reward directly to their proboscis. We rewarded responses to both the pre-exposed odour and the novel odour to evaluate LI that resulted from 40 unreinforced pre-exposure phase. Timing was tracked using a stopwatch, and the precise odour and feeding interval was tracked by a programmed PLC that signalled the experimenter to feed the bee with a tone which is inaudible to the bee (For visualisation of this procedure, see Smith & Burden, 2014). The time between trials (inter-trial interval) for each individual was always 5–8 min. Testing consisted of odours presented in a pseudo-randomized order (AXXAXAAX or XAAXAXXA, with the pre-exposed odour being X and the novel odour being A). The presentation order was counterbalanced between scouts, recruits, and controls.
We calculated LI score as:
(# responses to novel odour + 1)/(# responses to pre-exposed odour + 1).
Generally, a value closer to 1 indicates there were equal numbers of responses to the novel (A) and pre-exposed (X) odours, which indicates ‘low’ LI. An LI score greater than 1 means the number of responses to A is higher than that to X, which indicates ‘high’ LI. Specifically, to qualify an LI result as high or low, we enumerated through all possible outcomes of the LI score when A and X vary from 0 to 4. The average LI score from this enumeration was 1.36. The closest observed LI score to this value was 1.33, which we used as the cut-off. This cut-off indicates more responses to the novel odour compared to the pre-exposed odour (3 responses to A and 2 responses to X, making this score “high LI”). Control individuals that did not receive pre-exposure are expected to have an LI score closer to 1, as both odours are novel during PER and they should respond similarly to both. Other expressions of LI have been either qualitative, such as percent response (Chandra et al., 2010) or did not consider the responses in the context of novel and pre-exposed (Bazhenov, Huerta, & Smith, 2013). We calculate this to consider an individual’s responses to the novel odour in context with the pre-exposed odour.
Biogenic Amine Analysis
We analysed brain and haemolymph samples from brains of individual bees with an established protocol that uses using high performance liquid chromatography (HPLC) coupled to an electrochemical detector to quantify dopamine (DA), octopamine (OA), serotonin (5-HT) and tyramine (TA) (Cook, Brent, & Breed, 2017). Individual assessment allowed the amine data to be coupled to specific behavioural responses. Additional assay details are provided in the supplemental materials (ESM1). Initially, we collected individuals that had not been exposed to training or LI assessment, to provide an unmodified comparison between scouts (n=28) and recruits (n=28). These were taken from two colonies as they landed at the entrance at the same time that individuals were collected for LI testing. Amines were also quantified in bees after LI testing, using only individuals with non-zero LI scores. This included scouts (n=33) and recruits (n=27) pre-exposed to an odour, and 11 scout and 10 recruit controls. Using forceps, the bees were placed directly into liquid nitrogen for snap freezing to avoid handling stress that may influence amine concentrations. We collected these bees at the same time as collecting bees used for LI testing. Bees were stored in a −80°C freezer until dissection. For colony of origin for these experiments, see tables S1 and S2 in ESM2. For the rest of this paper, we refer to the pre-exposed odour as the “familiar” odour for clarity.
Statistical Analysis
The LI score is not normally distributed and is a rank. Therefore, to compare the LI score of scouts, recruits, and respective controls, we performed a Kruskal-Wallis test followed by Dunn post-hoc test.
To compare the responses of scouts and recruits to the novel and familiar odour across the learning trials, we used a generalized linear mixed model (GLMM) using the “lme4” R package (Bates, Mächler, & Bolker, 2014) function glmer with binomial error distribution and logit transformation. We were specifically interested in how the response to the two odours varied between foragers characterized behaviourally as scouts and recruits in the field. The response variable was a binomial response (responded/did not respond). The fixed effects were odour (novel vs. familiar), trial (1, 2, 3, 4), and foraging group (scout vs. recruit). We included the random effects ‘individual bee’ and ‘colony of origin’, to account for individual and colony variation. We began with a full model including main effects and all 2-way and 3-way interactions. We reduced this model, beginning with the 3-way interaction, until we had the simplest model with only significant interactions and the main effects (Engqvist, 2005). We then ran post-hoc analysis on significant interaction terms using the emmeans function in the “emmeans” package in R (Lenth, 2018).
To compare the biogenic amine concentration in brains and haemolymph between scouts and recruits collected at the colony, we first performed an Anderson-Darling normality test on the estimated probability distributions of concentrations of biogenic amines. All amine concentrations were non-normally distributed (p<0.05) and each amine had a different distribution, which is why we analysed them separately. We therefore used a Mann-Whitney non-parametric test.
After LI testing, we evaluated the relationship between LI score, forager type (scout/recruit/control), and biogenic amine concentration. To do this, we performed a generalized linear mixed model with colony of origin as a random effect. Our response variable was biogenic amine concentration, while our fixed effects were LI score and forager group (scout, recruit, scout control, and recruit control). We examined q-q distribution plots and confirmed fit using goodness of fit tests from the R package “goft” (Gonzalez-Estrada & Villasenor-Alva, 2017). We determined that the octopamine and serotonin concentrations fit a normal distribution, while tyramine fit an inverse–Gaussian distribution, and dopamine fit a gamma distribution. We used the functions lmer for the normally distributed data and ‘glmer’ with a gamma family (link=inverse) for the dopamine concentration data in the lme4 R package (Bates et al. 2010) and used the inverse Gaussian family (link = “1/mu^2”) for the tyramine concentration data. To further analyse the effect of group, we performed a Tukey post-hoc test. All analyses were performed in R (version 3.4.1).
RESULTS
LI Scores for Scouts and Recruits
LI scores differed significantly across groups (Kruskal–Wallis test: N = 82, χ2 = 77.509, df = 3, p < 0.001, Figure 1). Scouts responded significantly less to the familiar odour compared to the novel odour (median LI = 2), while recruits responded equally to the familiar and novel odours (median LI = 1; Dunn Post-Hoc Test: Z = −5.869, p < 0.001). Scouts that were familiar to an odour also had significantly higher LI than control scouts (Z = 7.399, p < 0.001) and control recruits (Z = 5.564, p < 0.001), which received an odourless pre-exposure. As expected, controls had LI scores closer to 1 (mean response for combined controls = 1.04), as both odours are novel, and they learn them equally.
Figure 1: LI scores for each foraging group including controls that are not pre-exposed to the odours.
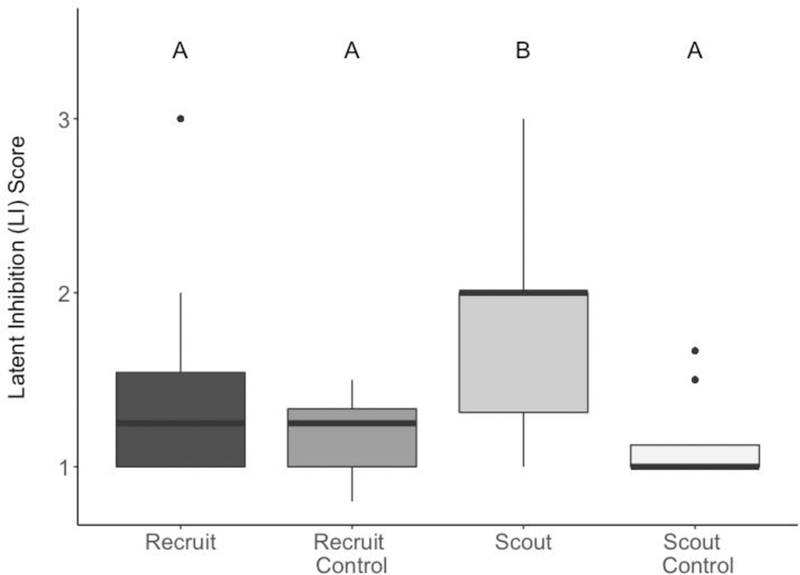
Different letters indicate significant differences based on a Dunn post-hoc test. High scores indiciate more responses to the novel odour, whereas Scores close to 1 indicate equal responses to the novel and the familiar odour, whereas higher scores indiciate more responsiveness to a novel odour. n=27 recruits, 10 recruit controls, 34 scouts, and 11 scout controls. For this boxplot and all following: thick horizontal bars are medians, boxes are 25–75th percentile, lines are 1.5 * IQR, points are outliers.
Responses of Scouts and Recruits to Odours Over Trials
Scouts and recruits respond differently to the familiar and novel odours over PER trials. As expected due to training, we found that responses significantly increased generally across trials (Type II ANOVA: χ2 = 105.4, p <0.001). Additionally, bees respond more to the novel odour compared to the familiar odour (χ2 = 44.43, p < 0.001). However, there was not a significant effect of foraging group on responses (χ2 = 0.3, p = 0.58), likely because most of the variation in foraging groups was explained by the interaction between foraging group and odour. The interaction between odour and group significantly predicted responses as the groups did differ when responding to the familiar versus the novel odour (Odour*Foraging Group, χ2 = 5.0, p = 0.025). Specifically, both scouts and recruits were more likely to respond to the novel odour compared to the familiar odour, however, comparing effect sizes, scouts were twice as likely to respond to the novel odour versus the familiar odour (Effect size: 2.2, Z = 6.44, p = <0.0001), compared to recruits (Effect size: 1.1, Z = 3.2, p = 0.001). For full model see Supplemental Material ESM6 Table S3. Qualitatively, the responses of scouts and recruits is respectively similar to the post-hoc grouping of high and low LI, where high LI individuals respond less to the familiar odour and more to the novel odour, compared to low LI bees that respond to the familiar and novel odour similarly (Figure 2).
Figure 2: Learning curves of bees tested for LI to illustrate qualitative similarities in LI.
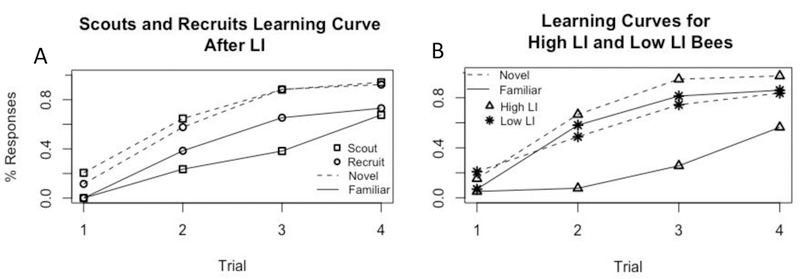
Both graphs are from the same bees, grouped by either LI score or foraging behaviour. (A) Learning curves of bees score defined as scouts and recruits when collected; n=34 scouts, 26 recruits. (B) Learning curves of bees as defined by their LI scores. High LI bees are defined as any bee with a score of 1.33 or higher (or (3 responses to a+1)/(2 responses to x+1)), n= 39 “high LI” bees, 43 “low LI” bees.
Biogenic Amines
When comparing the concentrations of biogenic amines in foragers collected directly from the colony (Figure. 3), we found brain tyramine was significantly higher in scouts than recruits (Kruskal-Wallis: N = 28 per group, W = 265.5, p = 0.039). We did not detect differences between scouts and recruits in any other amines (Octopamine: W = 329.5, p = 0.309; Dopamine: W = 437, p = 0.461; Serotonin: W = 337, p = 0.372). We also tested biogenic amines in the haemolymph of scouts and recruits, as well as the relative proportion of biogenic amines in brain and haemolymph. There were no significant differences in those analyses (for results, see Supplemental Materials ESM4).
Figure 3: Concentrations of biogenic amines in the brains of untrained scouts and recruits.
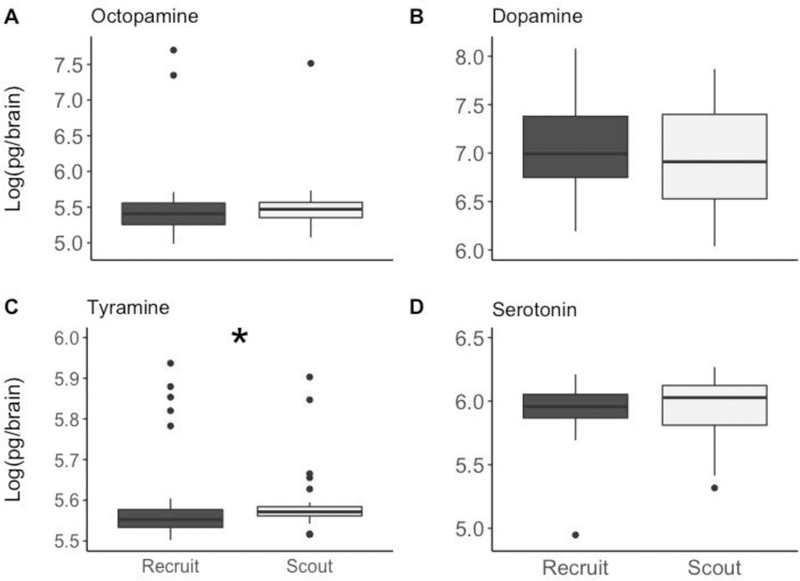
Only tyramine differs significantly between recruits (n=28) and scouts (n=28), which were collected and frozen directly from the colony and did not undergo LI testing. An asterisk indicates statistical significance at alpha = 0.05 using a Kruskal-Wallis test.
We also found that LI testing differentially impacted the amine. Octopamine was significantly different across groups (Wald Type II ANOVA: N = 82 individuals total, χ2 = 11.73, df = 3, p = 0.008; Figure 4A). Specifically, recruits had significantly lower octopamine compared to scouts (Tukey: Effect size = −55.12, Z = 2.62, p = 0.04) and recruit controls (Tukey: Effect Size = −91.2, Z = 3.04, p = 0.012, Figure 4A). There was no significant difference between scouts, recruits or controls for dopamine (GLM: χ2 = 4.352, df = 3, p = 0.226, Figure 4B), tyramine (GLM: χ2 = 1.91, df = 3, p = 0.591, Figure 4C) or serotonin (GLM: χ2 = 2.713, df = 3, p = 0.438).
Figure 4: Concentrations of biogenic amines for scouts, recruits, and controls after PER training and LI determination.
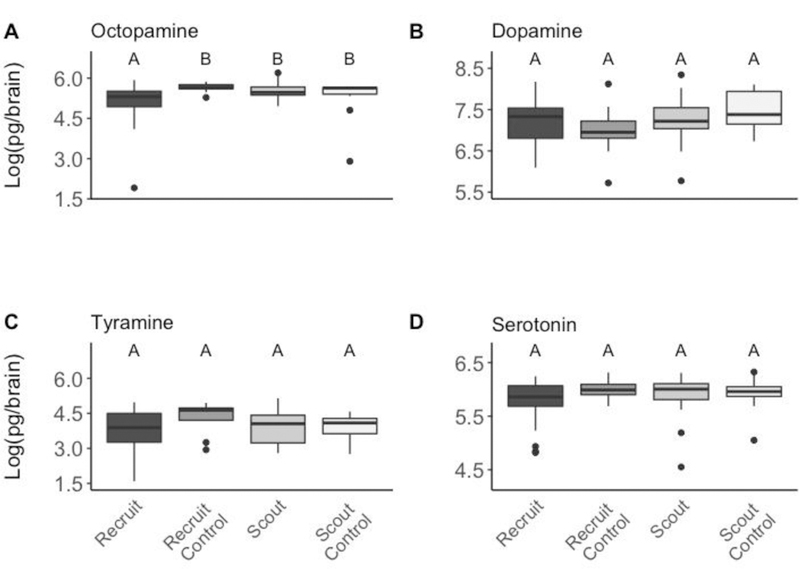
Only octopamine differs significantly between scouts (n=34), recruits (n=27), and controls (n=11 scout controls, 10 recruit controls). Controls are individuals that went through the LI procedure but received no odour during pre-exposure. Unique letters indicate statistically significant differences according to a Dunn post-hoc test.
DISCUSSION
Honey bee foragers must constantly collect food, both for their own needs and for the growth and persistence of the colony (Katz & Naug, 2016; Seeley, 1983; von Frisch, 1952). Scouting behaviour allows the foragers to sustain a consistent stream of incoming food when exploited locations become depleted (Seeley, 1983; Townsend-Mehler & Dyer, 2012). To do that, scout bees must continually seek out new floral resources as they come into bloom, while recruited foragers must continue to visit flowers with ever-diminishing resources to extract as much nectar and pollen as possible. We have found that one way that scouts avoid fixation on a single resource patch is through expressing relatively strong latent inhibition (LI). We observed that scouts exhibited stronger LI compared to recruit bees that are responsible for collecting those resources (Figure 1). In contrast, recruited foragers exhibit low LI, which we suggest disposes them to remain on diminishing resources to extract as much nectar and pollen as possible. These results suggest that the physiological and genetic mechanisms that underlie LI provide a means help solve the exploration/exploitation trade-off problem for a honeybee colony. It spurs scouts to search for novel stimuli associated with new resources and allows recruits to persist in exploitation of still productive, known resources.
As foragers are tested during PER (proboscis extension reflex), they exhibit marked differences in the way they respond to familiar and novel odours across trials. The learning curves of scouts and recruits show similar responses to a novel odour, but scouts are less likely to respond to odours to which they had been previously exposed (Figure 2). These results indicate that scouting behaviour may be a consequence of a diminished response to the familiar stimuli rather than enhanced response to a novel odour. Individuals that are likely looking for novel food sources may be better able to ignore previously visited locations in order to accommodate a temporally fluctuating landscape and allow them to find novel food resources more efficiently (Mosqueiro et al., 2017). We did find that the recruit responses to familiar odours were also significantly dampened compared to the novel odour, although not to the same extent as in scouts (Figure 2). This reduced response may reflect the inadvertent inclusion of scout-like bees among those selected as recruits (Biesmeijer & de Vries, 2001), or the recruits may retain some degree of discrimination that may help them to move on to a new source once food becomes depleted.
In addition to behavioural differences between scouts and recruits, we uncovered physiological differences as well. Scouts have elevated amounts of tyramine in their brains compared to recruits (Figure 3C) These biogenic amines have been implicated in learning and foraging behaviour (Scheiner, Entler, et al., 2017; Scheiner, Reim, Eirik, Entler, & Barron, 2017). Previous research found a QTL for learning differences using the same LI assay was linked to an specific area of the genome, Pln2 (or lrn1; 17). Variation in pln2 is associated with differences in nectar and pollen honey bee foragers, and influences pollen load size as well as sucrose responsiveness (Page et al., 2000; Page, Erber, & Fondrk, 1998; Rüppell, Pankiw, & Page, 2004). This region contains AmTAR1 (Hunt et al., 2007), a receptor for tyramine that is widely expressed in the antennal lobe and mushroom bodies of honey bees (Sinakevitch, Daskalova, & Smith, 2017). These brain regions form a critical pathway to integrate information for learning in honey bees, specifically for odour-based stimuli (Giurfa, 2003; Menzel & Müller, 1996). It remains to be determined whether variation in the AmTAR1 receptor expression is causally linked to foraging phenotype. Although we cannot rule out the possibility that bees were stressed from the move, we are confident that the 12 hours of recovery after the move was enough to diminish shifts in biogenic amines, as they change within seconds or minutes, not hours (Roeder, Seifert, Kähler, & Gewecke, 2003). Nevertheless, correlations between the genomic region that contains AmTAR1 and foraging and learning preferences, including the relationship of tyramine to these behaviours, point to a link worthy of further investigation.
Our results also show that scouts have higher concentrations of octopamine than recruits after being conditioned to an odour (Figure 4). Octopamine is a reward signal in the honey bee brain (Hammer & Menzel, 1998) and disruption of an identified octopamine receptor disrupts associative conditioning (Farooqui, Robinson, Vaessin, & Smith, 2003). Foraging behaviour has already been strongly linked to octopamine levels: foragers generally have elevated levels in their brain, specifically in their antennal lobes, compared to nurses (Schulz, Elekonich, & Robinson, 2002; Wagener-Hulme, Kuehn, Schulz, & Robinson, 1999). Scouts and recruits appear to be the same age (Seeley, 1983), so the differences in biogenic amines are likely not due to age, but to task. Treatment with octopamine promotes the onset of foraging (Barron, Schulz, & Robinson, 2002). In fact, bees treated with octopamine were significantly more likely to scout (Liang et al., 2012). Further, octopamine injected into the honey bee mushroom bodied enhances associative learning (Hammer & Menzel, 1998). Our results provide further support to the role of octopamine in associative conditioning and suggest that there might be a relationship between amine levels and foraging decisions.
There are several learning mechanisms that may be interacting with LI to influence division of foraging labour. Our learning assays show that LI plays an important role in scouting and recruiting behaviour. Specifically, our results indicate that the inability for scouts to form a conditioned response to the familiar odour shows that they have learned that the familiar odour is unrewarding. In addition to LI, scouts may prefer novel over familiar resources. Indeed, we found that high LI bees responded slightly more to the novel odour than low LI bees (Figure 2B). Other work on novelty preference found that scouts which visited more than two novel feeders had similar gene expression patterns in their brains compared to scouts collected after a hive-moving assay (Liang et al., 2012). Reward valuation may also be shaping LI and foraging. Scouts may perceive the reward during PER more strongly than recruits. This could be similar to learning behaviour observed in pollen foragers. Pollen foragers respond to lower concentrations of sucrose compared to nectar foragers (Pankiw & Page, 2000). Pollen foragers also perform better in associative learning tasks and retained that information for longer than nectar foragers (Scheiner, Erber, & Page, Jr., 1999). It remains to be determined if all of these different mechanisms attributed to scouting behaviour reflect additive, independent variation, or whether they result from a common mechanism related to the same neural and genetic pathways that control behaviour.
Learning clearly plays an important role in the division of labour of foraging honey bees. Behavioural (Liang et al., 2012; Lindauer, 1952; Oettingen-Spielberg, 1949; Seeley, 1982) and molecular (Liang et al., 2012, 2014) correlates of division of labour between scouts and recruits have been identified. Our study links controlled, laboratory-based learning to an ecologically important division of foraging labour. First, this linkage potentially allows for a more detailed evaluation and the disentangling of precise individual-level learning mechanisms – e.g. non-associative, Pavlovian, novelty seeking, reward valuation – that interact to produce the foraging decisions. Second, the linkage allows more detailed investigations of neural bases for foraging decisions. For example, a recent model integrating parts of the honey bee brain incorporated different forms of synaptic plasticity that represent variants of non-associative and associative learning (Bazhenov et al., 2013). This model also incorporated a proposed means for integrating the learning and neuroanatomical frameworks into a decision circuit. Integrating controlled laboratory experiments with field manipulations will allow refinement and testing of this theoretical model as it relates to foraging decisions. Connecting genetic, physiological, behavioural and ecological factors of individual variation brings us closer to an understanding how the individual variation in foraging behaviour has evolved. We have no competing interests.
Supplementary Material
Table 1:
Final results of generalized linear mixed model exploring how foraging group, LI group, familar or novel odour, and learning trial predicted the likelihood of response in associative learning behaviour in foraging honey bees.
| Estimate | Z-value | p-value | |
|---|---|---|---|
| Odour X:A | ‒1.14 | ‒3.24 | 0.001* |
| Trial1:Trial2 | 2.5233 | 6.256 | <0.001* |
| Trial1:Trial3 | 3.6493 | 8.605 | <0.001* |
| Trial1:Trial4 | 4.4002 | 9.777 | <0.001* |
| Foraging Group Recruit:Scout |
0.4599 | 1.277 | 0.2 |
| Odour*Foraging Group X:A, Recruit:Scout |
‒1.0672 | ‒2.239 | 0.025* |
indicates significance at alpha = 0.05.
ACKNOWLEGEMENTS
We greatly appreciate the insights and efforts of Dr. Osman Kaftanoglu, Dr. Ramon Huerta, and Sydney Ohrt. We thank Dr. Julian Resasco and two reviewers for helpful comments. Funding for this work was generously provided by the NIH grant R01GM113967 to BHS, NPW, JG and RH. Mention of trade names or commercial products in this article is solely for the purpose of providing specific information and does not imply recommendation or endorsement by the U.S. Department of Agriculture. USDA is an equal opportunity provider and employer.
Footnotes
DATA ACCESSABLITY
Data associated with this manuscript are available on FigShare: http://doi.org/10.6084/m9.figshare.5885263 (Cook et al 2018).
WORKS CITED
- Barron AB, Schulz DJ, & Robinson GE (2002). Octopamine modulates responsiveness to foraging-related stimuli in honey bees ( Apis mellifera ). J Comp Physiol A, 188, 603–610. doi: 10.1007/s00359-002-0335-5 [DOI] [PubMed] [Google Scholar]
- Bates D, Mächler M, & Bolker B (2014). Fitting linear mixed-effects models using lme4. Journal of Statistical Software …, arXiv preprint arXiv:1406.5823. Retrieved from http://arxiv.org/abs/1406.5823%5Cnhttp://listengine.tuxfamily.org/lists.tuxfamily.org/eigen/2011/06/pdfKU_S0z6LjT.pdf
- Bazhenov M, Huerta R, & Smith BH (2013). A Computational Framework for Understanding Decision Making through Integration of Basic Learning Rules. Journal of Neuroscience, 33(13), 5686–5697. doi: 10.1523/JNEUROSCI.4145-12.2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beekman M (2005). How long will honey bees (Apis mellifera L.) be stimulated by scent to revisit past-profitable forage sites? Journal of Comparative Physiology. A, Neuroethology, Sensory, Neural, and Behavioral Physiology, 191(12), 1115–1120. doi: 10.1007/s00359-005-0033-1 [DOI] [PubMed] [Google Scholar]
- Biesmeijer JC, & de Vries H (2001). Exploration and exploitation of food sources by social insect colonies : a revision of the scout-recruit concept. Behavioral Ecology and Sociobiology, 49, 89–99. [Google Scholar]
- Biesmeijer JC, & Seeley TD (2005). The use of waggle dance information by honey bees throughout their foraging careers, 133–142. doi: 10.1007/s00265-005-0019-6 [DOI]
- Bitterman ME (1972). Comparative studies of the role of inhibition in reversal learning. Inhibition and Learning (Ed. Boakes RA & Halliday MS ), 153–176. [Google Scholar]
- Bitterman ME, Menzel R, Fietz A, & Schafer S (1983). Classical Conditioning of Proboscis Extension in Honeybees (Apis mellifera). Journal of Comparative Psychology, 107–119. doi: 10.1037/0735-7036.97.2.107 [DOI] [PubMed]
- Breed MD, Williams DB, & Queral A (2002). Demand for Task Performance and Workforce Replacement : Undertakers in Honeybee , Apis mellifera , Colonies. Workforce, 15(3), 319–329. [Google Scholar]
- Carr-Markell MK, & Robinson GE (2014). Comparing Reversal-Learning Abilities, Sucrose Responsiveness, and Foraging Experience Between Scout and Non-Scout Honey bee (Apis mellifera) Foragers. Journal of Insect Behavior, 27, 736–752. doi: 10.1007/s10905-014-9465-1 [DOI] [Google Scholar]
- Chandra SBC, Hosler JS, & Smith BH (2000). Heritable Variation for Latent Inhibition and Its Correlation With Reversal Learning in Honeybees (Apis mellifera). Journal of Comparative Psychology, 114(1), 86–97. doi: 10.1037//0735-7036.114.1.86 [DOI] [PubMed] [Google Scholar]
- Chandra SBC, Hunt GJ, Cobey S, & Smith BH (2001). Quantitative Trait Loci Associated with Reversal Learning and Latent Inhibition in Honeybees (Apis mellifera). Behavior Genetics, 31(3), 275–285. [DOI] [PubMed] [Google Scholar]
- Chandra SBC, Wright GA, & Smith BH (2010). Latent inhibition in the honey bee, Apis mellifera: Is it a unitary phenomenon? Animal Cognition, 13, 805–815. doi: 10.1007/s10071-010-0329-6 [DOI] [PubMed] [Google Scholar]
- Chandra SB, Hunt GJ, Cobey S, & Smith BH (2001). Quantitative Trait Loci Associated with Reversal Learning and Latent Inhibition in Honeybees ( Apis mellifera ). Behavior Genetics, 31(3), 275–285. [DOI] [PubMed] [Google Scholar]
- Charnov EL (1976). Optimal foraging theory: the marginal value theorem. Theoretical Population Biology, 9, 129–136. doi: 10.1016/0040-5809(76)90040-X [DOI] [PubMed] [Google Scholar]
- Cook CN, Mosquiero T, Brent CS, Ozturk C, Gadau J, Pinter-Wollman N, Smith BH (2018). Data from: Individual differences in learning and biogenic amine levels influence the behavioural division between foraging honey bee scouts and recruits. FigShare: 10.6084/m9.figshare.5885263 [DOI] [PMC free article] [PubMed]
- Cook CN, Brent CS, & Breed MD (2017). Octopamine and tyramine modulate the thermoregulatory fanning response in honey bees (Apis mellifera), 1925–1930. doi: 10.1242/jeb.149203 [DOI] [PubMed]
- Couzin ID (2009). Collective cognition in animal groups. Trends in Cognitive Sciences, 13(1), 36–43. doi: 10.1016/j.tics.2008.10.002 [DOI] [PubMed] [Google Scholar]
- Dreller C (1998). Division of labor between scouts and recruits: genetic influence and mechanisms. Behavioral Ecology and Sociobiology, 43, 191–196. [Google Scholar]
- Engqvist L (2005). The mistreatment of covariate interaction terms in linear model analyses of behavioural and evolutionary ecology studies. Animal Behaviour, 70(4), 967–971. doi: 10.1016/j.anbehav.2005.01.016 [DOI] [Google Scholar]
- Farooqui T, Robinson K, Vaessin H, & Smith BH (2003). Modulation of Early Olfactory Processing by an Octopaminergic Reinforcement Pathway in the Honeybee, 23(12), 5370–5380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferrari MCO, & Chivers DP (2006). The role of latent inhibition in acquired predator recognition by fathead minnows. Canadian Journal of Zoology, 84(4), 505–509. doi: 10.1139/z06-027 [DOI] [Google Scholar]
- Ferrari MCO, & Chivers DP (2011). Learning about non-predators and safe places: The forgotten elements of risk assessment. Animal Cognition, 14(3), 309–316. doi: 10.1007/s10071-010-0363-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frisch K. von.(1965). Die Tänze der Bienen . In Tanzsprache und Orientierung der Bienen (pp. 3–330). [Google Scholar]
- Fussnecker BL, Smith BH, & Mustard JA (2006). Octopamine and tyramine influence the behavioral profile of locomotor activity in the honey bee (Apis mellifera), 52, 1083–1092. doi: 10.1016/j.jinsphys.2006.07.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giurfa M (2003). Cognitive neuroethology : dissecting non-elemental learning in a honeybee brain, 726–735. doi: 10.1016/j.conb.2003.10.015 [DOI] [PubMed]
- Giurfa M, & Menzel R (2001). Cognitive Architecture of a Mini-Brain. Adaptivity and Learning. An Interdisciplinary Debate, 5(2), 22–48. doi: 10.1016/S1364-6613(00)01601-6 [DOI] [PubMed] [Google Scholar]
- Gonzalez-Estrada E, & Villasenor-Alva JA (2017). goft: Tests of Fit for some Probability Distributions. CRAN R Project
- Hammer M, & Menzel R (1998). Multiple sites of associative odor learning as revealed by local brain microinjections of octopamine in honeybees. Learning & Memory (Cold Spring Harbor, N.Y.), 5(1–2), 146–56. doi: 10.1101/lm.5.1.146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill PSM, Wells PH, & Wells H (1997). Spontaneous flower constancy and learning in honey bees as a function of colour, 615–627. [DOI] [PubMed]
- Hills TT, Todd PM, Lazer D, Redish AD, Couzin ID, Bateson M, … Wolfe JW (2015). Exploration versus exploitation in space, mind, and society. Trends in Cognitive Sciences, 19(1), 46–54. doi: 10.1016/j.tics.2014.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Amdam GV, Schlipalius D, Emore C, Sardesai N, Guzmán-Novoa E, R. E. P. Jr (2007). Behavioral genomics of honeybee foraging and nest defense. Naturwissenschaften, 94, 247–267. doi: 10.1007/s00114-006-0183-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunt GJ, Page RE, Fondrk MK, & Dullum CJ (1995). Major quantitative trait loci affecting honey bee foraging behavior. Genetics, 141(4), 1537–45. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1206885&tool=pmcentrez&rendertype=abstract [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz K, & Naug D (2015). Energetic state regulates the exploration – exploitation trade-o ff in honeybees, 26, 1045–1050. doi: 10.1093/beheco/arv045 [DOI] [Google Scholar]
- Katz K, & Naug D (2016). Dancers and followers in a honeybee colony differently prioritize individual and colony nutritional needs. Animal Behaviour, 119, 69–74. doi: 10.1016/j.anbehav.2016.06.011 [DOI] [Google Scholar]
- Latshaw JS, & Smith BH (2005). Heritable variation in learning performance affects foraging preferences in the honey bee ( Apis mellifera ), 200–207. doi: 10.1007/s00265-004-0904-4 [DOI]
- Lehman HK, Schulz DJ, Barron AB, Wraight L, Hardison C, Whitney S, … Robinson GE (2006). Division of labor in the honey bee ( Apis mellifera ): the role of tyramine  -hydroxylase. Journal of Experimental Biology, 2774–2784. doi: 10.1242/jeb.02296 [DOI] [PubMed]
- Lenth R (2018). Emmeans: Estimated marginal means, aka least-squares means. R Package Version, 1(2). [Google Scholar]
- Liang ZS, Mattila HR, Rodriguez-zas SL, Southey BR, Seeley TD, Robinson GE, & Robinson GE (2014). Comparative brain transcriptomic analyses of scouting across distinct behavioural and ecological contexts in honeybees. Proceedings of the Royal Society B: Biological Sciences, 281, 20141868. doi: 10.1098/rspb.2014.1868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang ZS, Nguyen T, Mattila HR, Rodriguez-Zas SL, Seeley TD, & Robinson GE (2012). Molecular Determinants of Scouting Behavior in Honey Bees. Science, 335(6073), 1225–1228. doi: 10.1126/science.1213962 [DOI] [PubMed] [Google Scholar]
- Lindauer M (1952). Ein Beitrag zur Frage der Arbeitsteilung im Bienenstaat. Zeitschrift Fur Vergleichende Physiologie, 34, 299–345. [Google Scholar]
- Lubow RE (1973). Latent Inhibition. Psychological Bulletin, 79, 398–407. [DOI] [PubMed] [Google Scholar]
- Lubow RE, & Moore AU (1959). Latent inhibition: the effect of nonreinforced pre-exposure to the conditional stimulus. Journal of Comparative and Physiological Psychology, 52(October), 415–419. doi: 10.1037/h0046700 [DOI] [PubMed] [Google Scholar]
- Mattila HR, & Seeley TD (2011). Does a polyandrous honeybee queen improve through patriline diversity the activity of her colony ‘ s scouting foragers ?, 799–811. doi: 10.1007/s00265-010-1083-0 [DOI]
- Menzel R, & Müller U (1996). LEARNING AND MEMORY IN HONEYBEES: From Behavior to Neural Substrates. Annual Review of Neuroscience, 19, 379–404. [DOI] [PubMed] [Google Scholar]
- Mosqueiro T, Cook C, Huerta R, Gadau J, Smith B, & Pinter-Wollman N (2017). Task allocation and site fidelity jointly influence foraging regulation in honeybee colonies. Royal Society Open Science, 4 Retrieved from 10.6084/m9.figshare.c.384976 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oettingen-Spielberg T (1949). Über das Wesen der Suchbiene. Journal of Comparative Physiology A: Neuroethology, Sensory, Neural, and Behavioral Physiology, 31(4), 454–489. [Google Scholar]
- Page RE, Erber J, & Fondrk MK (1998). The effect of genotype on response thresholds to sucrose and foraging behavior of honey bees (Apis mellifera L.). Journal of Comparative Physiology. A, Sensory, Neural, and Behavioral Physiology, 182(4), 489–500. doi: 10.1007/s003590050196 [DOI] [PubMed] [Google Scholar]
- Page RE, Fondrk MK, Hunt GJ, Guzman-Novoa E, Humphries MA, Nguyen K, & Greene AS (2000). Genetic Dissection of Honeybee (Apis mellifera L.) Foraging Behavior. The Journal of Heredity, 91(6), 474–479. [DOI] [PubMed] [Google Scholar]
- Pankiw T, & Page RE (2000). Response thresholds to sucrose predict foraging division of labor in honeybees. Behavioral Ecology and Sociobiology, 47, 265–267. [Google Scholar]
- Robbins RJ (1979). The effect of flavor preexposure upon the acquisition and retention of poison-based taste aversions in deer mice: latent inhibition or partial reinforcement? Behavioral and Neural Biology, 25(3), 387–397. doi: 10.1016/S0163-1047(79)90459-X [DOI] [PubMed] [Google Scholar]
- Roeder T (2005). Tyramine and octopamine: ruling behavior and metabolism. Annual Review of Entomology, 50(20), 447–77. doi: 10.1146/annurev.ento.50.071803.130404 [DOI] [PubMed] [Google Scholar]
- Roeder T, Seifert M, Kähler C, & Gewecke M (2003). Tyramine and Octopamine : Antagonistic Modulators of Behavior and Metabolism, 13(April), 1–13. doi: 10.1002/arch.10102 [DOI] [PubMed] [Google Scholar]
- Rüppell O, Pankiw T, & Page RE (2004). Pleiotropy, epistasis and new QTL: The genetic architecture of honey bee foraging behavior. Journal of Heredity, 95(6), 481–491. doi: 10.1093/jhered/esh072 [DOI] [PubMed] [Google Scholar]
- Scheiner R, Entler BV, Barron AB, Scholl C, Thamm M, & Moore V (2017). The Effects of Fat Body Tyramine Level on Gustatory Responsiveness of Honeybees (Apis mellifera) Differ between Behavioral Castes. Frontiers in Systems Neuroscience, 11(August), 1–8. doi: 10.3389/fnsys.2017.00055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheiner R, Erber J, & Page RE Jr. (1999). Tactile learning and the individual evaluation of the reward in honey bees ( Apis mellifera L .). Journal of Comparative Physiology A, 185, 1–10. [DOI] [PubMed] [Google Scholar]
- Scheiner R, Plückhahn S, Oney B, Blenau W, & Erber J (2002). Behavioural pharmacology of octopamine, tyramine and dopamine in honey bees. Behavioural Brain Research, 136(2), 545–53. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12429417 [DOI] [PubMed] [Google Scholar]
- Scheiner R, Reim T, Eirik S, Entler BV, & Barron AB (2017). Learning , gustatory responsiveness and tyramine differences across nurse and forager honeybees. Journal of Experimental Biology doi: 10.1242/jeb.152496 [DOI] [PubMed]
- Schulz DJ, Elekonich MM, & Robinson GE (2002). Biogenic Amines in the Antennal Lobes and the Initiation and Maintenance of Foraging Behavior in Honey Bees ABSTRACT :, 406–416. doi: 10.1002/neu.10138 [DOI] [PubMed]
- Seeley TD (1982). Adaptive Significance of the Age Polyethism Schedule in Honeybee Colonies. Behavioral Ecology and Sociobiology, 11(4), 287–293. doi: 10.1007/BF00299306 [DOI] [Google Scholar]
- Seeley TD (1983). Division of Labor Between Scouts and Recruits in Honeybee Foraging. Behavioral Ecology and Sociobiology, 12, 253–259. [Google Scholar]
- Seeley TD (1986). Social foraging hy honeyhees : how colonies allocate foragers among patches of flowers. Behavioral Ecology and Sociobiology, 19, 343–354. [Google Scholar]
- Sinakevitch IT, Daskalova SM, & Smith BH (2017). The biogenic amine tyramine and its receptor (AmTyr1) in olfactory neuropils in the honey bee (Apis mellifera) brain. Frontiers in Systems Neuroscience, 11, 77. doi: 10.3389/fnsys.2017.00077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith BH, & Burden CM (2014). A proboscis extension response protocol for investigating behavioral plasticity in insects: application to basic, biomedical, and agricultural research. Journal of Visualized Experiments: JoVE, (91). [DOI] [PMC free article] [PubMed]
- Smith BH, & Menzel R (1989). The use of electromyogram recordings to quantify odourant discrimination in the honey bee, Apis mellifera. J Insect Physiol, 35(5), 369–375. [Google Scholar]
- Sudhakaran IP, Holohan EE, Osman S, Rodrigues V, VijayRaghavan K, & Ramaswami M (2012). Plasticity of Recurrent Inhibition in the Drosophila Antennal Lobe. Journal of Neuroscience, 32(21), 7225–7231. doi: 10.1523/JNEUROSCI.1099-12.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda K (1961). Classical Conditioned Response in the Honey Bee. Journal of Insect Physiology, 6(3), 168–179. doi: 10.1111/j.1365-2982.2005.00691.x [DOI] [Google Scholar]
- Townsend-Mehler JM, & Dyer FC (2012). An integrated look at decision-making in bees as they abandon a depleted food source. Behavioral Ecology and Sociobiology, 66(2), 275–286. doi: 10.1007/s00265-011-1275-2 [DOI] [Google Scholar]
- Visscher PK, & Seeley TD (1982). Foraging Strategy of Honeybee Colonies in a Temperate Deciduous Forest. Ecology, 63(6), 1790–1801. [Google Scholar]
- von Frisch K (1952). Die wechselseitigen Beziehungen und die Harmonie im Bienenstaat na.
- Wagener-Hulme C, Kuehn JC, Schulz DJ, & Robinson GE (1999). Biogenic amines and division of labor in honey bee colonies. Journal of Comparative Physiology. A, Sensory, Neural, and Behavioral Physiology, 184(5), 471–9. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/10377980 [DOI] [PubMed] [Google Scholar]
- Wilson E (1971). The Insect Societies Cambridge, Massachusettes: Belknap/Harvard University Press. [Google Scholar]
- Young a M., Joseph MH, & Gray J. a. (1993). Latent inhibition of conditioned dopamine release in rat nucleus accumbens. Neuroscience, 54(1), 5–9. doi:0306-4522(93)90378-S[pii] [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


