SUMMARY
Viral-genetic tracing techniques have enabled mesoscale mapping of neuronal connectivity by teasing apart inputs to defined neuronal populations in regions with heterogeneous cell types. We previously observed input biases to output-defined ventral tegmental area dopamine (VTA-DA) neurons. Here, we further dissect connectivity in the VTA by defining input-output relations of neurochemically and output-defined neuronal populations. By expanding our analysis to include input patterns to subtypes of excitatory (vGluT2-expressing) or inhibitory (GAD2-expressing) populations, we find that the output site, rather than neurochemical phenotype, correlates with whole-brain inputs of each subpopulation. Lastly, we find that biases in input maps to different VTA neurons can be generated using publicly available whole-brain output mapping datasets. Our comprehensive dataset and detailed spatial analysis suggest that connection specificity in the VTA is largely a function of the spatial location of the cells within the VTA.
In Brief
Beier et al. comprehensively identify inputs to different cell types in the ventral tegmental area, defined by neurochemical phenotype and/or output site. They find that neurochemical phenotype has little relation to input specificity, whereas the output site determines input patterns through the spatial definition of cell bodies within the midbrain.
Graphical Abstract
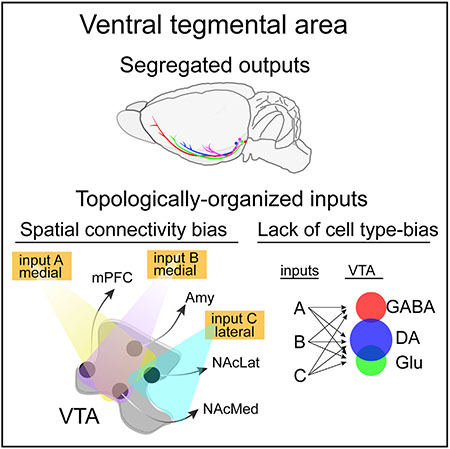
INTRODUCTION
A comprehensive anatomical map of neuronal connections is the foundation for understanding brain function (Alivisatos et al., 2012). Classical tracers and dyes have been used to map out the major pathways in the brain. These include the retrograde tracers horseradish peroxidase (Kristensson and Olsson, 1971), wheat germ agglutinin (Schwab et al., 1978), and fluorogold (Schmued and Fallon, 1986), which label neurons projecting to the site of injection, as well as anterograde tracers, such as Phaseolus vulgaris-leucoagglutinin (PHA-L) (Gerfen and Sawchenko, 1984), biocytin (King et al., 1989), and biotinylated dextran amine (BDA) (Veenman et al., 1992), that fill axons from neurons near the site of injection. However, these methods rely on bulk injection of micro- or macromolecules into anatomically defined brain sites that are then taken up by any neuron at or projecting to the site of injection. Therefore, these approaches lack the ability to specifically label inputs to or outputs from cell types that differ by gene expression. They also do not resolve how connectivity at the level of major tracts relates to connectivity at the level of individual cell types. Recently, the development of the rabies-based monosynaptic input tracing technique has enabled specific labeling of the direct monosynaptic inputs onto genetically defined cell populations (Wickersham et al., 2007). Although this method has been used to successfully map inputs to cell types in many brain regions (Callaway and Luo, 2015), it has yet to be used to comprehensively investigate patterns of connectivity between multiple cell types defined by a combination of gene expression and output site.
The VTA is an ideal brain site in which to test the wiring patterns onto distinct cell types intermingled within a common nucleus. The VTA comprises heterogeneous cell types with diverse projections. The global inputs to multiple different cell types have been reported (Beier et al., 2015;Faget et al., 2016; Lammel et al., 2012; Menegas et al., 2015; Watabe-Uchida et al., 2012). Most studies have found differences in inputs between spatially separated DA populations in the midbrain (Beier et al., 2015;Lammel et al., 2012; Lerner et al., 2015;Menegas et al., 2015), though the extent of these differences varies between studies. Here, we have expanded our previous analysis of the input-output connectivity patterns of specific subtypes of VTA-DA neurons based on their projection targets (Beier et al., 2015). Specifically, we have constructed a comprehensive, high-resolution input-output map of the VTA by standardizing viral reagents and tracing inputs to unique VTA cell types, defined both by their neurochemical identity and specific projection targets. DAergic, GABAergic, and glutamatergic neurons in the VTA were genetically isolated using recombinase-expressing mouse lines enabling analysis of inputs either to all Cre-expressing neurons in the VTA or a subset of each defined by output site using the cTRIO technique (cell-type-specific tracing the relationship between input and output; Beier et al., 2015;Schwarz et al., 2015).
RESULTS
Largely Segregated Outputs of Four VTA-DA Subpopulations
We previously showed that by expressing a membrane-tagged GFP (mGFP) specifically in DA neurons defined by a projection to either the medial (NAcMed) or lateral (NAcLat) shells of the nucleus accumbens that these neurons had largely segregated global projection targets (Beier et al., 2015). As retrograde mapping of DA neurons has suggested the existence of at least five unique VTA-DA subpopulations (Lammel et al., 2008), with additional DA subpopulations located in the adjacent substantia nigra (Lerner et al., 2015; Menegas et al., 2015, 2018), we extended our analysis to include global output patterns of additional VTA-DA neurons projecting to the medial prefrontal cortex (mPFC) and amygdala (Amy). We used the same strategy to express mGFP in projection-defined VTA-DA neurons (Beier et al., 2015; Schwarz et al., 2015). Briefly, a canine adenovirus expressing Cre-dependent Flp recombinase (CAV-FLExloxP-Flp) that labels neurons projecting to the targeted site was injected into either the mPFC or Amy. During the same surgery, an Flp-dependent adeno-associated virus (AAV) expressing mGFP (AAVDJ-hSyn1-FLExFRT-mGFP) was injected into the VTA (Figure 1A).
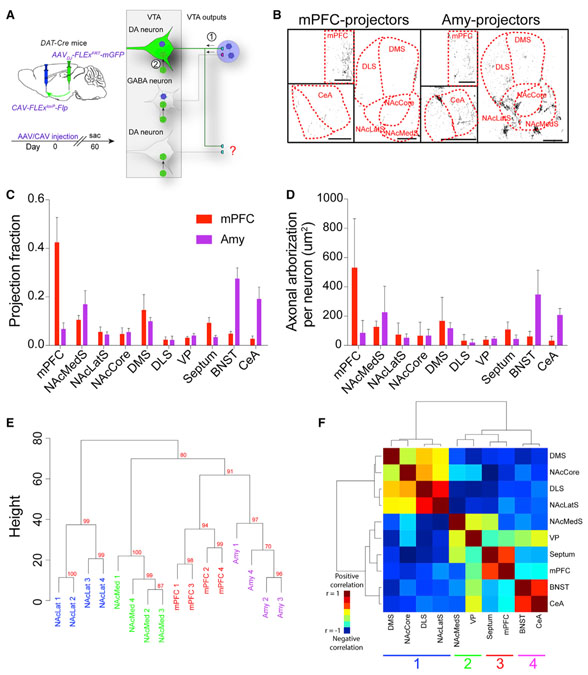
Figure 1. Axon Collateralization Patterns from VTA-DA Subpopulations.
(A) Experimental schematic. CAV-FLExloxP-Flp was injected into either the mPFC or Amy, and an Flp-dependent AAV-expressing mGFP was injected into the VTA of DAT-Cre mice. Axons were imaged throughout the brain. (B) Sample images of projections from VTA-DA subpopulations targeted by mPFC and Amy injections. Scale bar, 500 mm. (C) Projection fraction of each subtype to ten different brain regions. (D) Average axonal arborization per labeled VTA-DA neuron in each brain region. (E) Hierarchical clustering and bootstrapping based on outputs. Each sample segregates by targeted output site. The approximately unbiased (AU) p value was calculated and is shown in red for each branch. An AU p value higher than 95% indicates that the cluster is highly supported by the data. (F) Covariance analysis of the ten quantified output sites using data from each of the four targeted VTA-DA subpopulations here and in Beier et al. (2015). There are four distinct clusters which correspond to the different VTA-DA subpopulations. mPFC n = 5; Amy n = 4. Error bars, SEM. See Figure S1 for related data.
The majority of mGFP expression was observed in the region targeted with CAV-FLExloxP-Flp, as expected (Figures 1B and 1C). However, for each population, we also observed fluorescently labeled collaterals in other brain regions (Figures 1B and 1C). These collateralization patterns appeared stereotyped. For example, the bed nucleus of the stria terminalis (BNST) received collaterals mostly from Amy-projecting VTA-DA neurons (Figures 1C and 1D). For comparison purposes, previously published data for DA neurons projecting to the NAcMed and NAcLat (Beier et al., 2015) is included in Figure S1. Each VTA-DA neuron subtype had a preferential projection site but also sent collaterals to many other brain regions. The NAcLat-projecting VTA-DA neurons demonstrated the largest overall arborization per neuron, with the other three populations having a similar overall arborization area (Figure S1D).
As each of the four output-defined VTA-DA subpopulations expressed varying degrees of overlap in their collateralization to other brain regions, we performed a statistical analysis to test the null hypothesis that output distribution is independent of CAV injection site. A two-way ANOVA indicated that, as expected, output patterns from VTA-DA neurons were indeed distinct (interaction factor p < 0.0001). To test the relationships between the axonal projections of each VTA-DA subpopulation, we performed unsupervised hierarchical clustering and bootstrapping of these data for each of the sixteen animals. Each replicate segregated appropriately based on the site of CAV-FLExloxP-Flp injection (NAcLat approximately unbiased, or AU p value = 0.99 [where clusters with values >0.95 are strongly supported by the data], NAcMed AU p = 1, mPFC AU p = 0.94, and Amy AU p = 0.97; Figure 1E). To identify relationships between output sites targeted by VTA-DA neurons, we next performed a covariance analysis of the entire axonal arborization of labeled VTADA neurons in each of the sixteen animals, followed by hierarchical clustering. Highly correlated clusters of brain sites indicate regions that tend to be targeted by the same populations of VTA-DA neurons. We observed four clusters of collateralized outputs: (1) nucleus accumbens core (NAcCore), NAcLatS, and the dorsomedial (DMStr) and dorsolateral (DLStr) striatum, (2) NAcMedS and ventral pallidum (VP), (3) mPFC and septum, and (4) central amygdala (CeA) and BNST (Figure 1F), which generally correspond to the major outputs of (1) NAcLat-projecting, (2) NAcMed-projecting, (3) mPFC-projecting, and (4) Amy-projecting VTA-DA neurons (Beier et al., 2015; Figures 1C and S1). These results further confirm that the four output-defined VTA-DA subpopulations have unique global output patterns.
Projection, Not Neurochemical Phenotype, Defines Input Patterns
Our previous work suggested that the input patterns onto NAcLat-projecting VTA-DA neurons are quantitatively distinct from the inputs onto the three other DA cell populations studied, which have similar global input patterns (Beier et al., 2015). To further understand input organization to the VTA, we examined differences in inputs to VTA cell types defined neurochemically or by output site. We previously showed that when defined neurochemically, DA and GABA cells in the VTA received largely similar inputs (Beier et al., 2015). Here, we expanded our analysis to glutamatergic neurons in the VTA, defined by expression of the vesicular glutamate transporter vGluT2. VTA-vGluT2 neurons are heterogeneous in their projection targets (Hnasko et al., 2012), and some co-transmit DA or GABA (Kawano et al., 2006; Root et al., 2014). We found that all three populations receive inputs from the same regions in quantitatively similar proportions (Figure S2A), consistent with previous reports (Faget et al., 2016).
While VTA cell populations solely defined by neurochemical phenotype do not display differences in inputs, it is possible that each neurochemically defined class contains subpopulations of neurons with different input patterns. We previously observed that VTA-DA neurons projecting to different forebrain sites had biased inputs (Beier et al., 2015). To test whether this observation generalized to other VTA cell types, we subdivided VTA-GABA and VTA-vGluT2 populations based on output site using cTRIO, as previously done for VTA-DA neurons (Beier et al., 2015; Figures 2A and 2B). We were able to isolate DA and vGluT2 neurons projecting to the NAcLat, NAcMed, mPFC, and Amy, and VTA-GABA neurons projecting to the NAcLat, mPFC, and Amy, demonstrating that both vGluT2-Cre and GAD2-Cre neurons in the VTA project to multiple forebrain sites. As only a very small number of VTA-GABA neurons projecting to the NAcMed could be labeled, these cells were excluded from our analysis.
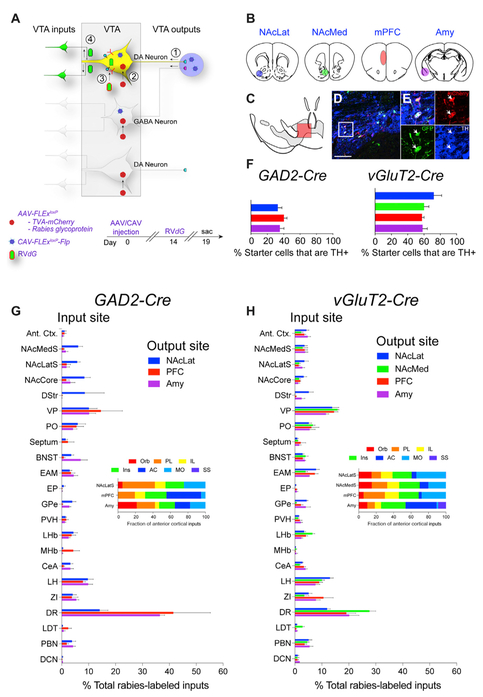
Figure 2. Tracing Inputs to VTA-GABA and VTA-vGluT2 Neurons Defined by Output Site.
(A) cTRIO schematic. CAV-FLExloxP-Flp was injected into an output site of VTA neurons, and Flp-dependent AAVs expressing TC and G were injected into the VTA, followed 2 weeks later by RVdG. (B) The four output sites targeted with CAVFLExloxP-Flp injection were NAcLat, NAcMed, mPFC, and Amy. (C) Schematic midbrain image showing location of cells shown in (D) and (E). (D and E) Sample image of TH co-localization at low (D) and high (E) magnification from a cTRIO experiment in a vGluT2-Cre animal; output site = NAcMedS. Scale bar, 150 μm. (F) Percentage of starter cells co-staining with TH for GAD2-Cre and vGluT2-Cre cTRIO experiments. (G and H) Whole-brain input patterns from GAD2-Cre (G) or vGluT2-Cre (H) cTRIO experiments are shown. n = 4 for each condition. Insets show the fraction of anterior cortical inputs from the seven quantified cortical subregions. n = 4 for all conditions. Error bars, SEM. See Figures S2, S3, and S4 for related data.
To estimate the proportion of VTA-GABA and VTA-vGluT2 cells with long-range projections that are also DAergic, we calculated the percentage of these neurons expressing the DA marker tyrosine hydroxylase, TH (Figures 2C – 2F). Thirty-three to forty percent of VTA-GABA neurons isolated by output site co-stained with TH, and 58%–72% of all VTA-vGluT2 neurons labeled by output site expressed TH (Figure 2F). Thus, there is significant overlap in the neurotransmitters released from VTA cells isolated by projection site, with the extent of overlap depending on the Cre driver line used.
cTRIO experiments revealed similar whole-brain input patterns for GAD2-Cre and vGluT2-Cre populations (Figures 2G and 2H). Subpopulations of VTA neurons projecting to the NAcLat received more inputs from striatal sites (NAcLat, NAcCore, DStr) and fewer inputs from the dorsal raphe (DR) (Figures 2G and 2H). Similar results were reported previously for VTA-DA neurons (Beier et al., 2015: for comparison, previously published data from DAT-Cre populations are shown in Figure S2B). These data suggest that while the neurochemical phenotype of a given VTA population has little correlation with global input distributions, the output site may be related to input patterns.
Statistical Analysis of the Determinants of Input Specificity
We noted previously from VTA-DA neurons isolated by output site that input patterns appeared to be related to the spatial distribution of each VTA-DA subpopulation within the VTA (Beier et al., 2015). The inclusion of cTRIO data generated using VTA-GABA and VTA-vGluT2 subpopulations enabled us to more thoroughly test the hypothesis that spatial location of starter cells within the VTA was an important determinant of whole-brain input patterns. To test for a relationship between spatial location of VTA starter cells and labeled inputs, we performed a linear regression between the inputs and either the corrected starter cell center of mass (the geometric mean of the spatial location of all starter cells, center of mass [COM]) (Figures S3 and S4), the Cre driver line, or both the COM and the Cre driver line, for each animal included in this work and previous studies (Beier et al., 2015). The COM explained input site variation to different extents, reaching statistical significance for about half the input sites with roughly equal contribution from the medial/lateral and dorsal/ventral coordinates (Table S1). The anterior/posterior coordinate explained little variation, presumably because cells were counted from nine midbrain sections, and thus there were only nine possible z-coordinates. In contrast to the COM, the mouse driver line essentially explained no input site variation, either on its own or in addition to the COM (Figure 3A). When we ran a similar regression of the percent of inputs from each site against either the COM, defined output site, or both, we observed that the output site and the COM explained input site variation similarly well and that the performance was only marginally improved when both conditions were combined, suggesting that these variables carry redundant information (Figure 3B).
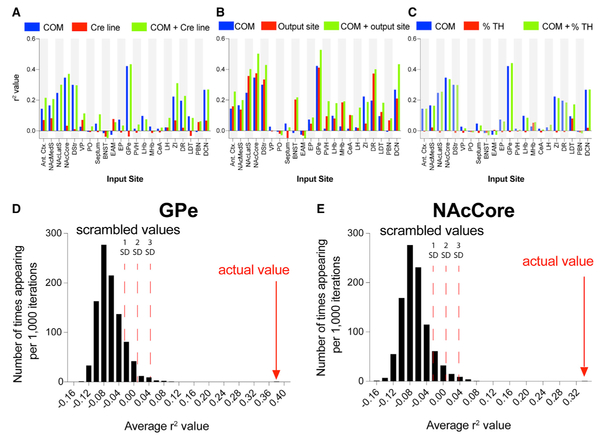
Figure 3. Statistical Analysis of Input Biases to VTA Neurons.
(A) Linear regression between the percentage of total rabies-labeled inputs from each of the 22 input sites for each animal (n = 76) and the starter cell COM, the mouse Cre driver line, or both the COM and the Cre driver line. (B) Similar regressions were performed like in (A), but inputs were run against the COM, output site, or both. (C) Regressions were run against the COM, the percentage of starter cells immunostaining for TH, or both. (D and E) A linear regression model was trained on a subset of the 76 brains and tested on the remainder of the dataset. An r2 value was calculated over 500 randomized training/testing set divisions and compared to control sets where the link between COM and inputs was scrambled. The obtained r2 value for the GPe (D) was 0.38 and was 0.33 for the NAcCore (E). Each graph shows 1, 2, and 3 SD from the mean in the positive direction. See Table S1 for related data.
To assess whether cell-type overlap contributed to the observed results, we extended our statistical test to include the percentage of TH+ cells in the starter populations as an additional predictor. We performed a regression of each input against the percentage of starter cells co-labeling with TH, the COM of starter cells, or both, for all 76 brains in the dataset (Figure 3C). We observed that, similar to the observation that the Cre line provided little information about the whole-brain input patterns, the percentage of starter neurons co-staining with TH also was largely unrelated to the whole-brain input patterns observed in our data.
To test the COM’s predictive power to generate the percentage of input from a particular brain region, we trained a linear regression model on a subset of the data and evaluated the prediction on the rest of the dataset using r2 as the metric. We tested this on GPe and NAcCore input sites, as these sites had the strongest relationship between labeled inputs and starter cell COM (Figure 3A). We calculated an average r2 over 500 randomized training/testing set divisions. As a comparison, we generated 999 control sets by scrambling the link between the COM and inputs and performed the same procedure to generate a distribution of the corresponding average r2 values. For the GPe and NAcCore inputs, the average r2 values for the actual data were 0.38 and 0.33, respectively, which were >10 standard deviations above chance (Figures 3D and 3E). Taken together, these data provide strong evidence for a topological organization of inputs into the VTA.
Anterograde Tract Tracing Data Can Be Used to Predict Differences in Whole-Brain Input Patterns
The lack of input specificity to VTA populations defined by different Cre driver lines argues against a dominant role of cell type-specific connectivity in the VTA. If there is no clear difference in the number of rabies-labeled cells from an input site when starter cells are defined by neurochemical phenotype, but there is a difference when starter populations are defined by spatial location, one simple hypothesis is that an input site may differentially innervate spatially distinct regions of the brain, and the differences in connectivity are related to this variance in intensity of innervation. Testing this hypothesis in the VTA requires labeling of fibers from a site projecting to the VTA and quantifying relative intensity of projection across the VTA. Fortunately, these data exist in the publicly available Allen Mouse Brain Connectivity Atlas (Oh et al., 2014). This atlas was created by injecting an AAV expressing GFP into hundreds of anatomically distinct regions in the brain and imaging labeled neurons and fibers throughout the brain.
We selected injections into each of twenty input sites quantified here for rabies input maps, excluding the DStr and EP as these injections had few to no detectable-labeled fibers within the VTA. We then obtained three images in the VTA from the Allen Mouse Brain Connectivity Atlas from each experiment. Criteria for selection of experiments were (1) non-Cre-dependent AAVs were used (with the exception of striatal injections, where viruses targeting D1-Cre neurons were also used as these neurons constitute nearly the entire striatal output to the ventral midbrain, as well as the DR, where experiments using Cre-dependent AAVs injected into Sert-Cre mice were used); (2) infected neurons were predominately restricted to the targeted site; and (3) fibers from labeled neurons could be visualized in the VTA.
To account for differences in viral injection volume and the number of neurons labeled at the injection site in the Allen Mouse Brain Connectivity Atlas images, the relative intensity of axons across the mediodorsal to ventrolateral dimensions of the VTA from the midline to the medial lemniscus was quantified for each image (Figure 4A). The relative intensity value was obtained at the COM for each VTA-DA subpopulation, as calculated from our cTRIO experiments. This relatively intensity value was then compared to the relative amount of labeled inputs generated using cTRIO, compared across each of the four VTA-DA subpopulations. Representative images from the Allen Mouse Brain Connectivity Atlas showing labeled fibers in the VTA originating from five different input sites are shown in Figures 4B – 4F. With a few exceptions (e.g., a higher proportion of GPe inputs to Amy-projecting VTA-DA neurons, Figure 4B), biases (or lack thereof) in inputs identified using cTRIO were very similar to those identified from labeled axons in the VTA (Figures 4B – 4F; Tables S2 and S3).
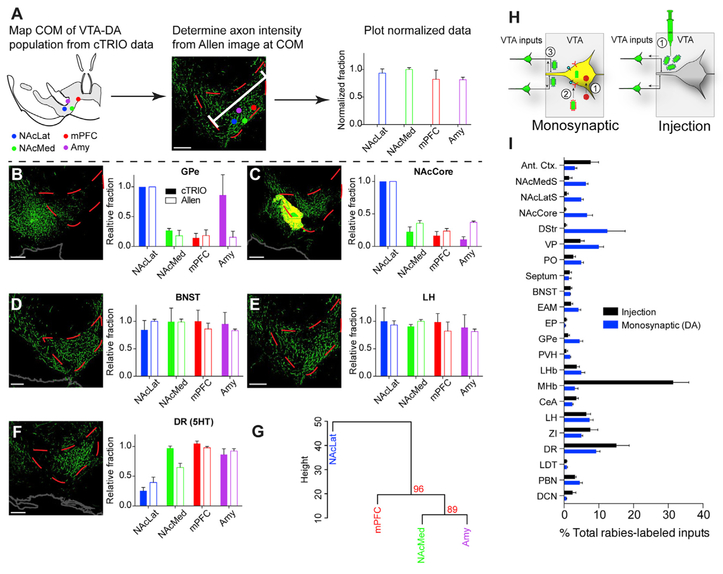
Figure 4. Relating VTA-DA Inputs to Whole-Brain Output Tracing Datasets.
(A) Schematic of experimental flow. Three representative images from the Allen Mouse Brain Connectivity Atlas were taken in the VTA for each of the 20 input sites analyzed. Each image contained GFP-labeled fibers from an AAV injection into a targeted brain site that projects to the VTA. The axon intensity of GFP+ fibers in the VTA was measured as a function of the dorsomedial-ventrolateral distance from the midline to the medial lemniscus through the VTA. The axon intensity value was obtained at the COM of each VTA-DA subpopulation that was calculated from published DAT-Cre cTRIO data (Beier et al., 2015). These data were normalized as a fraction of the maximum value among the VTA-DA subpopulations. (B–F) Inputs from each VTA-DA subpopulation (cTRIO) were compared to data generated using the publicly available output dataset from the Allen Mouse Brain Connectivity Atlas (Allen). Data from two inputs showing a lateral bias onto NAcLat-projecting VTA-DA neurons (B and C), two inputs showing no spatial bias (D and E), and one input showing a medial bias (F) are shown. (G) When results from axonal projections from each of the 20 quantified input sites were averaged and hierarchically clustered/bootstrapped, we observed that axon projection patterns to the COM of NAcMed-, mPFC-, and Amy-projecting VTA-DA neurons were more similar to one another than to NAcLat-projecting VTADA neurons, recapitulating our observation using cTRIO from these populations (Beier et al., 2015). The AU p value was calculated and is shown in red for each branch. (H) Experimental schematics comparing monosynaptic input tracing from VTA-DA neurons to bulk injection of RVdG+G to label neurons projecting to the VTA. (I) The monosynaptic spread of rabies from VTA-DA neurons labeled a different pattern of input as compared to direct injection of RVdG+G. n = 3 (A–G) and n = 4 (I) for both conditions. Scale bar, 200 mm (all panels). Error bars, SEM. All images in this figure were taken from the Allen Mouse Brain Connectivity Atlas (Oh et al., 2014). Image credit: Allen Institute for Brain Science. See Tables S2 and S3 for related data.
We noted earlier that VTA neurons tend to segregate into two groups based on input: those projecting to the NAcLat, and those projecting to the NAcMed, mPFC, or Amy (Beier et al., 2015; Figure 2). To test whether the same two groups would emerge based on the relative projection intensity of inputs in the VTA, we performed hierarchical clustering and bootstrapping of averaged data from three individual AAV1-GFP injections from each of the 20 different input sites. We used the COM of VTA-DA subpopulations as a test case. We again observed that NAcMed-, mPFC-, and Amy-projecting VTA-DA neurons share a closer relationship with one another than with NAcLat-projecting VTA neurons (AU p = 0.96; Figure 4G), further supporting the observation that input biases generated using the rabies virus are related to the relative intensity of projection from a given input site.
Verifying Specificity of Transsynaptic Labeling
If rabies input maps refiect the presence of axons passing through the VTA, one possible interpretation is that rabies transmission is not restricted to connected neurons but rather spreads non-specifically to local axons quantitatively in proportion to axonal coverage. If true, then an injection of a G-deleted RVdG coated with its own glycoprotein (RVdG+G; Figure 4H) directly into the VTA should produce similar results. However, this was not observed: rather, we found roughly 10-fold fewer inputs from each subregion of the striatum, and 10-fold more inputs from the medial habenula (MHb), when RVdG+G was directly injected into the VTA (Figure 4I). As the MHb sends a strong projection to the interpeduncular nucleus (IPN) with axons passing through the VTA, the enhanced labeling of MHb inputs using RVdG+G is likely a consequence of infection of en passant axons. Conversely, most striatal inputs predominately target the adjacent substantia nigra pars reticulata (SNr) (e.g., Figure 4C). The soma of midbrain DA neurons are not located in the SNr; however, these neurons can extend processes into the SNr, where they receive up to ~40% of their afferent synapses (Henny et al., 2012). Therefore, viral transmission from infected VTA-DA neurons to striatal inputs likely occurred through dendritic viral release. That such large differences in labeled inputs from the MHb and striatum exist argues that RVdG monosynaptic spread does not significantly label passing axons. Thus, these data support the interpretation that rabies virus spread is restricted to synaptically connected populations. Together, the data also strongly support the conclusion that the soma location of starter populations (COM) is the major determinant of input connectivity in the VTA.
DISCUSSION
Our viral-genetic approach has enabled a detailed dissection of input-output relationships of heterogeneous VTA populations. We demonstrated that VTA-DA subpopulations have largely segregated outputs but share some collaterals to a variety of forebrain sites. Biased input patterns were observed to output-defined VTA-DA, VTA-GABA, and VTA-vGluT2 neurons depending on the site to which they projected. When we investigated the factors influencing input patterns, we found that the output site of a given subpopulation correlated with global inputs by spatially restricting the location of starter cells. Lastly, we showed that the relative density of axonal projections from input sites in the VTA recapitulated differences in inputs to VTA-DA subpopulations, which in turn was related to the spatial location of each VTA-DA subpopulation.
While our data suggest a topological model of connectivity, rabies-generated input maps must be interpreted with caution. For example, the level of activity in input populations may influence rabies transsynaptic transmission (Beier et al., 2017). Furthermore, while these maps reflect physical synaptic connectivity, they reveal little information about the strength of synaptic connections. We recently found that the GPe→VTA/SNr-GABA functional connection was much stronger than the GPe→VTADA connection despite a similar quantitative input as measured using the rabies virus (Beier et al., 2017). Thus, a conservative interpretation is that the number of connections identified by the rabies virus does not equate to strength of functional connections, but rather appears to reflect the presence of connections between neurons. Furthermore, it remains to be determined if the rabies virus retrograde monosynaptic tracing technique exhibits preference/tropism for specific types of synaptic inputs, for example synapses releasing glutamate, GABA, monoamines, or neuropeptides.
While the mapping conducted in this study was focused on the VTA, the data from our work and that of others supports the general conclusions that heterogeneous, intermingled cell types share qualitatively similar inputs. Similar observations have been reported with other rabies-generated maps in diverse brain sites, such as dopamine receptor 1- and 2-expressing medium spiny neurons in the dorsal striatum (Guo et al., 2015; Wall et al., 2013), and excitatory and inhibitory neurons in the cortex (Wall et al., 2016) and in the hippocampus (Sun et al., 2014). We do not believe these results reflect non-synapse-specific transmission of virus, as transsynaptic specificity has been demonstrated in slice cultures (Wickersham et al., 2007), synaptic connections have been validated in vivo (Beier et al., 2015; DeNardo et al., 2015;Pollak Dorocic et al., 2014; Lerner et al., 2015; Weissbourd et al., 2014), and retrograde non-synapse-specific tracing from the VTA using RVdG yields very different input patterns than monosynaptically restricted viral spread (Figure 4I). However, it is important to note that significant overlap exists between our VTA populations expressing DAT-Cre, GAD2-Cre, or vGluT2-Cre. It is not clear how inputs from these co-expressing populations may differ from populations expressing only one of these genes.
Interestingly, we observed significant overlap of GABAergic and glutamatergic neurons in cTRIO experiments with the dopaminergic marker TH. While these numbers are higher than co-labeling reported in past studies examining all VTA neurons (Qi et al., 2016; Tritsch et al., 2014), our measurements were made when defining VTA populations by output site. Therefore, our observations are not inconsistent with previously published results. In fact, while we observed that 33%–40% output-defined GAD2-Cre neurons in the VTA co-stained with TH, we found that only 4% of total GABAergic neurons were TH+ (Beier et al., 2015). One likely possibility is that most of the GAD2-Cre+ VTA cells are local interneurons and thus not captured by our projection target-specific approach.
In summary, our comprehensive input-output analysis of VTA populations using the rabies virus suggests that connectivity is topologically organized within the VTA. However, the fact that rabies does not appear to reflect important factors such as synaptic strength suggests that other complementary methods assaying synaptic strength, such as ex vivo slice electrophysiology and neuronal activity, including in vivo methods, are required to take the next step in elucidating the functional connectivity and outputs of these circuits.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Kevin Beier (kbeier@uci.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Mice
Generation and characterization of DAT-Cre (Bäckman et al., 2006; JAX strain 006660), GAD2-Cre (Taniguchi et al., 2011; JAX strain 010802), and vGluT2-Cre (Vong et al., 2011; JAX strain 016963) mice has been previously described. Wild-type littermates not containing the Cre transgene were used for TRIO experiments. 6–12 week old males and females were both used in approximately equal proportions throughout, and mice were kept on a mixed C57BL/6 and CD1 background. All mice were group housed with littermates. Mice were housed in plastic cages with disposable bedding on a 12 hours light/dark cycle with food and water available ad libitum, and all experiments were done during the light phase. All surgeries were done under isoflurane or ketamine/xylazine anesthesia, and procedures followed animal care and biosafety guidelines approved by Stanford University’s Administrative Panel on Laboratory Animal Care and Administrative Panel of Biosafety.
METHOD DETAILS
Stereotaxic surgeries
Mice were anesthetized and placed in a stereotaxic apparatus (Kopf Instruments). For virus injection, the following coordinates (in mm) were used, in mm:
NAcMed: AP +1.78, LM 0.4, DV –4.1
NAcLat: AP +1.45, LM 1.75, DV –4.0
Amy: AP –1.43, LM 2.5, DV –4.5
mPFC: two injections of 500 nL, one at AP +2.15, LM 0.27, DV –2.1 and another at AP +2.15, L0.27, DV –1.6 VTA: AP –3.2, LM 0.4, DV –4.2
AP and LM is relative to bregma; DV is relative to the brain surface.
The titers of viruses, based on quantitative PCR analysis, were as follows:
AAV5 -CAG-FLExloxP-TC, 2.4 × 1013 genome copies (gc)/mL;
AAV8 -CAG-FLExloxP -G, 1.0 × 1012 gc/mL;
AAV5 -CAG-FLExFRT-TC, 2.6 × 1012 gc/mL;
AAV8-CAG-FLExFRT-G, 1.3 × 1012 gc/mL;
AAVDJ-hSyn1-FLExFRT-mGFP (actually AAVDJ-hSyn1-FLExFRT-mGFP-2A-synaptophysin-mRuby), 2.9 × 1013 gc/mL;
CAV-Cre, 2.5 × 1012 gc/mL;
CAV-FLExloxP-Flp, 5.0 × 1012 gc/mL.
The titer of RVdG was estimated to be 5.0 × 108 colony forming units (cfu)/mL based on serial dilutions of the viral stock followed by infection of the 293-TVA800 cell line.
Transsynaptic tracing
Transsynaptic tracing studies were carried out as previously described (Beier et al., 2015). Mice were anaesthetized and injected with either 100 nL of a 1:1 mixture of AAV5 -CAG-FLExloxP -TC and AAV8 -CAG-FLExloxP-G into the VTA for non-TRIO and non-cTRIO experiments, or 500 nL for TRIO experiments. For cTRIO experiments. 500 nL of a 1:1 mixture of AAV5-CAG-FLExFRT-TC and AAV8 -CAG-FLExFRT -G was injected into the VTA. For TRIO experiments, 250 nL (NAcMed and NAcLat), 500 nL (Amy) or 1 mL (mPFC) of CAV2-Cre was injected into an output site of VTA-DA neurons. For cTRIO experiments, the same volume of CAV2-FLExloxP-Flp was injected, using coordinates described above.
After recovery, mice were housed in a BSL2 facility. Two weeks later, 500 nL RVdG was injected into the VTA using the procedure described above. After recovery, mice were housed in a BSL2 facility for 5 days before euthanasia. Cell counting was performed manually. We did not adjust for the possibility of double counting cells from consecutive sections, which would result in overestimation, the extent of which would depend on the size of the cells in the regions quantified.
Histology and imaging
Animals were perfused transcardially with phosphate buffered saline (PBS) followed by 4% formaldehyde. Brains were dissected, post-fixed in 4% formaldehyde for 12–24 hours at 4°C, then placed in 30% sucrose in PBS for 24–48 hours. They were then embedded in Optimum Cutting Temperature (OCT, ThermoFisher) and stored at −80°C until sectioning.
For rabies tracing analysis, consecutive 60-mm coronal sections were collected onto Superfrost Plus slides and stained for NeuroTrace Blue (NTB, Invitrogen). For NTB staining, slides were washed 1×5 min in PBS, 2×10 min in PBS with 0.3% Triton X-100 (PBST), incubated for 2–3 hours at RT in (1:500) NTB in PBST, washed 1×20 min with PBST and 1×5min with PBS. Sections were additionally stained with DAPI (1:10,000 of 5 mg/mL, Sigma-Aldrich), which was included in the last PBST wash of NTB staining. Whole slides were then imaged with a 5x objective using a Leica Ariol slide scanner with the SL200 slide loader.
For starter cell identification, sections were unmounted after slide scanning, blocked in PBST and 10% normal goat serum (NGS) for 2–3 hours at room temperature, and incubated in rat anti-mCherry antibody (1:2000, Life Sciences) and rabbit anti-TH antibody (1:1000, Millipore) at 4°C for four nights. After primary antibody staining, sections were washed 3×10 min in PBST, and secondary antibodies (donkey anti-rat Cy3 and donkey anti-rabbit 647, Jackson ImmunoResearch) were applied for two nights at 4°C, followed by 3×10min washes in PBST and remounting. Confocal z stacks were acquired using a 20x objective on a Zeiss LSM 780 confocal microscope.
For analysis of DA neuron output, every 60-mm coronal section was collected sequentially into PBS. Sections were washed 2×10 min in PBS and blocked for 2–3 hours at room temperature (RT) in 10% NGS in PBST. Primary antibody (chicken anti-GFP, Aves Labs, 1:1000) was diluted in 5% NGS in PBST and incubated for four nights at 4°C. After 3×10 min washes in PBST, secondary antibodies were applied for two nights at 4°C (donkey anti-chicken AlexaFluor488, 1:250, Jackson ImmunoResearch), followed by 3×10min washes in PBST. Sections were additionally stained with DAPI. All images were acquired using a 5x objective on the Leica Ariol slide scanner, and processed using NIH ImageJ software.
Axon quantification
Quantification of axons from VTA-DA neurons in forebrain sites was performed as previously described (Beier et al., 2015). After sectioning and antibody staining, sections were mounted and imaged, as for input tracing analysis. For each region, five 60-mm sections were chosen for analysis from within a defined anterior-posterior segment (all coordinates are relative to bregma in mm): anterior 1.41 to 0.85 for nucleus accumbens and striatum, 0.37 to 0.01 for BNST and VP, 1.87 to 1.53 for mPFC, and posterior 0.95 to 1.79 for the CeA. For the septum, 10 sections were analyzed, 5 each from 1.41 to 0.85 and 0.37 to 0.01. The entire regions within each section were taken, and boundaries were defined based on DAPI staining and on the Franklin and Paxinos mouse brain atlas, 3rd ed. (Franklin & Paxinos, 2007). For analysis, the background was first subtracted, and the mean of local background after subtraction was multiplied by a constant value (4, for all but one sample targeting NAcLat-projecting neurons that had sparser labeling, where a value of 3 was used). This value was then set as the threshold, with pixels above this gray-scale value being interpreted as positive signal from VTA-DA neuron axons. The threshold value was kept constant for all sections analyzed within a brain. The total area and area fraction covered by the above-threshold axon signal was measured for each region and averaged across the 5 (or 10) sections.
Starter cell center of mass (COM)
Determination of starter cell COM and corrections was performed as previously described (Beier et al., 2015). To assess the spatial distribution of starter cells in our experiments, nine 60-mm coronal sections through the VTA were quantified (corresponding to one tissue section per atlas section in the Franklin and Paxinos Atlas). These sections were collected in order through the VTA of experimental animals and processed as described above in Histology and Imaging. Starter cells were considered as all cells that were both GFP+ and TC+. The location of these cells was manually mapped onto digital reference images from the Franklin and Paxinos atlas using the Cell Counter plugin in ImageJ. x, y, and z coordinates were obtained for each starter cell in each experiment. Diagrams displaying the spatial distribution of starter cells (Figure S4) are collapsed along the anterior–posterior (z) axis, and therefore underestimate the true distinction in spatial distribution of starter neurons. The spatial distribution for the starter cells was calculated for the x and y dimensions separately.
As we injected two AAVs to express TC and G separately, it was possible that some cells expressed one gene, but not the other. Cells that expressed TC, but not G, would therefore appear red (TC) and green (RVdG) and would be considered a starter cell, but as they would lack expression of G, no inputs would be labeled from these “false” starter cells. Corrections for false starter cells (Figure S3) were made using the observed spread of AAV5-CAG-FLExloxP-TC and AAV8-CAG-FLExloxP-G. Native mCherry fluorescence was used to visualize TC; for G, 60-mm tissue sections were stained for the rabies glycoprotein (mouse anti-rabies glycoprotein, Millipore, 1:500, followed by donkey anti-mouse AlexaFluor488, 1:500, Jackson ImmunoResearch). The distance from the targeted center of the injection site in the VTA to the edge of observed fluorescence for three sections located near the injection site, and three sections located approximately 600 μm posterior to the injection site was quantified. False starter cells were considered as those that expressed TC, but not G. The Euclidian distance from the center of injection to the edge of spread was then calculated for each AAV, and as the radius of spread from the injection site as visualized in both anterior and posterior sections approximated a sphere, a virtual sphere was created with a radius equal to the spread distance of AAV8-CAG-FLExloxP-G (which was smaller than that of AAV5-CAG-FLExloxP-TC). These values (for either 0.1 μL, in the case of DAT-Cre, GAD2-Cre, and vGluT2-Cre input tracing, or 0.5 μL, for TRIO and cTRIO) were then used to exclude starter cells that were located outside of the sphere of TC and G co-expression. The false starter cell correction, as well as the center of mass (COM; the mean location of starter cells) and standard deviation of the starter cell spatial distribution calculations were made using custom MATLAB code based on the x, y, and z coordinates for each starter cell.
For input quantification, not every section was counted for starter cell quantification: while the VTA spanned about twenty sections of 60-μm thickness, only nine sections containing the VTA were quantified, as this corresponded to the number of unique sections containing the VTA that are represented in the Franklin and Paxinos atlas. In this way, starter cells from one 60-μm section would correspond to one digital image. Therefore, when total numbers of starter cells were reported (Figure S4), the total number of starter cells for each brain was multiplied by 20/9.
Regression analysis
Linear regression model fitting was performed in MATLAB, where input site percentages were dependent variables, COM and percentage of TH+ cells were predictors, and driver strain as well as output site were categorical predictors. Models with several different combinations of predictors were assessed (COM, driver, output, percentage of TH+ cells, COM + driver, COM + output, COM + percentage of TH+ cells). The fitted models were then analyzed using ANOVA to report the adjusted r2 to indicate how well the percentage for each input site could be explained by each combination of predictor variables.
To demonstrate the predictive power of COM for input site pattern, for the actual data, 80% was chosen randomly to train a linear regression model, using input site percentages as dependent variables, and COM as predictors. The model was then used to predict the other 20%, with r2 calculated as 1 – SSE/SST. Such randomization-training-testing was repeated 500 times, to derive an average r2. For each one of the 999 controls, the link between COM and input site in the dataset was scrambled, and an average r2 was calculated from 500 randomized training/testing regressions, following the exact procedure as for the actual data. The histogram of 999 scrambled average r2 values was then plotted, with the actual r2 indicated on the x axis. The 76 brains used in this dataset are as follows:
Cre-dependent tracing (16):
DAT-Cre (4), replicates of DAT-Cre (4), GAD2-Cre (4), vGluT2-Cre (4)
TRIO (16):
NAcLat (4), NAcMed (4), mPFC (4), Amy (4)
cTRIO DAT-Cre (16):
NAcLat (4), NAcMed (4), mPFC (4), Amy (4)
cTRIO GAD2-Cre (12):
NAcLat (4), mPFC (4), Amy (4)
cTRIO vGluT2-Cre (16):
NAcLat (4), NAcMed (4), mPFC (4), Amy (4)
In situ hybridization
In situ experiments were conducted using GAD2 and vGluT2 probes as described previously (Weissbourd et al., 2014). To make ISH probes, DNA fragments of 400–1000 bp containing the coding or untranslated region sequences were amplified by PCR from mouse whole brain cDNA (Zyagen) and subcloned into pCR-BluntII-topo vector (Life Technologies, cat# K2800–20). A T3 RNA polymerase recognition site (AATTAACCCTCACTAAAGGG) was added to the 3′ end of the PCR product. Primer sets used in the present study are listed below.
Plasmids were then amplified, the insert removed via EcoRI (New England Biolabs, cat#R0101L) digest, and purified using a PCR purification kit (QIAGEN, cat#28104). 500–1000 ng of the DNA fragment was then used for in vitro transcription by using DIG RNA labeling mix (cat#11277073910) and T3 RNA polymerase (cat#11031163001) according to the manufacture’s instruction (Roche Applied Science). After DNase I (Roche Applied Science, cat#04716728001) treatment for 30 min at 37°C, the RNA probe was purified by ProbeQuant G-50 Columns (GE Healthcare, cat# 28-9034-08) according to the manufacturer’s instructions.
60-μm consecutive sections were collected onto Superfrost slides (no-coating, Fisher Scientific, cat#22-034-980), dried, and stored at −80°C until use. Specific slides containing the VTA were then thawed and viewed on a Zeiss compound fluorescence microscope, and the sections containing regions of interest were recorded. Those sections were then floated off using PBS into wells of a 24-well plate for use with multiple probes. The sections were fixed for 15 min in 4% formaldehyde in PBS at room temperature, rinsed with PBS, and incubated with 7 mg/ml Proteinase K (Life Technologies, cat#25530–049) in 10 mM Tris-Cl, pH 7.4, 1 Mm EDTA for 10 min at 37°C. After fixing again with 4% formaldehyde in PBS for 10 min and rinsing with PBS, the sections were incubated with 0.25% acetic anhydride in 0.1 M triethanolamine, pH 8.0, for 15 min and washed with PBS. Probes were diluted (~1:1000) with the hybridization buffer (50% formamide, 10mM Tris-Cl pH 8.0, 200 mg/ml tRNA, 10% Dextran Sulfate, 1x Denhalt’s solution, 600mM NaCl, 0.25% SDS), mixed well, preheated at 85°C for 5 min, and applied to each well (300–500 μl/well). After 16–20h of incubation at 50°C, the sections were washed, first with 2x SSC-50% formamide, then with 2x SSC, and finally with 0.2x SSC twice for 20 min at 65°C. After blocking for 1–2h with a 1% blocking reagent (Roche Applied Science, cat#10057177103), sections were incubated with an alkaline phosphatase-conjugated anti-DIG antibody (1:1000, Roche Applied Science, cat# 1093274) and a rat anti-mCherry antibody (1:500) overnight at 4°C.
After washing with Roche Wash Buffer (cat#11585762001) three times for 15 min followed by rinsing with the detection buffer (100mM Tris-Cl pH8.0, 100mM NaCl, 10mM MgCl2), probe-positive cells were detected by Fast Red TR/Naphthol AS-MX Tablets (Sigma-Aldrich, cat#F4523). After being washed with Roche Wash Buffer three times for 10 min, sections were incubated with mCherry-conjugated donkey anti-rat Alexa 488 antibody (1:200; Jackson ImmunoResearch) for an additional 1–2h, and washed with PBS three times for 10 min. Finally, the sections were treated with PBS containing 4’,6-diamidino-2-phenylindole dihydrochloride (DAPI; Sigma-Aldrich, Cat#D8417) for 20 min and mounted with cover glass using Fluorogel (Electron Microscopy Sciences, Cat#17985–10). Sections were imaged by confocal microscopy (Zeiss 780). Images were processed in ImageJ. We used FIJI and the cell counter plugin to quantify overlap.
vGlut2
5′-CTCCCCCATTCACTACCTGA;
5′- AATTAACCCTCACTAAAGGGGGTCAGGAGTGGTTTGCATT
Gad1
5′-CACAAACTCAGCGGCATAGA;
5′- AATTAACCCTCACTAAAGGGGGACGAGCAACATGCTATGG
Gad2
5′-GGGATGTCAACTACGCGTTT;
5′- AATTAACCCTCACTAAAGGGTGCATCAGTCCCTCCTCTCT
5′-CTCCAATCCCCTTCTTCTCC;
5′- AATTAACCCTCACTAAAGGGTGTGCATCCTTTGTCCATGT
Analysis of fluorescent axonal labeling from Allen Mouse Brain Connectivity Atlas
For analysis of projections from AAV1-GFP tracing experiments in the Allen Mouse Brain Connectivity Atlas, data from three separate injections for each VTA-DA input site, excluding the DStr and EP, were analyzed. Injections into wild-type C57JBl/6 mice were chosen with the following exceptions: DStr, NAcLat, NAcMed, or NAcCore, where C57BL/6 or D1-Cre mice where used, or DR, where Sert-Cre mice were used. Experiments also had to show GFP labeling in the VTA.
For each experiment, three images of the VTA, each spaced approximately two sections apart, were captured at screen resolution. In ImageJ, using the line tool at a thickness of 100, a line was drawn from the midline to the end of the medial lemniscus running ventro-lateral through the VTA, and the gray value (a metric of axon coverage) was obtained as a function of distance from the midline. To normalize these values, data were run through custom MATLAB code to segment data into 100 bins and normalized to the maximum intensity value for that image. The COM of each VTA-DA population obtained using cTRIO was plotted on a coronal image, and the same tools in ImageJ were used to identify the coordinate for COM. The obtained COM values were rounded to the nearest whole number (NAcLat 73; NAcMed 58; mPFC 25; Amy 47), and the corresponding value of axonal coverage at that distance from the midline was obtained for each image. These were then normalized to the maximum value obtained from the four points.
The images shown in Figures 4B – 4F can be found at the following URLs:
The full list of URLs can be found in Table S3.
QUANTIFICATION AND STATISTICAL ANALYSIS
Quantification of anatomical data is described in the corresponding sections of the text and Method Details. All statistical tests and data analyses were performed using MATLAB, GraphPad Prism, and R. Data were expressed as means ± SEMs in the figures and text. All distributions of data meet the normality assumption, as assed by Bartlett’s test for equal variance. Significance was defined as p < 0.05. Sample sizes were chosen based results from Beier et al. (2015) and similar manuscripts. Heatmaps and dendrograms were generated in R using K-means clustering. Bootstrapping was conducted using pvclust, an R package for hierarchical clustering with p values (Suzuki and Shimodaira, 2006). After bootstrap re-sampling (100,000 iterations), the approximately unbiased (AU) p value was calculated and is shown for each branch. An AU p value higher than 95% indicates that the cluster is highly supported by the data.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Antibodies | ||
| Anti-GFP chicken polyclonal antibody | Aves | Cat# GFP-1020; RRID: AB_10000240 |
| Anti-rabies glycoprotein mouse monoclonal antibody | Millipore | Cat# MAB8727; RRID: AB_571110 |
| Anti-tyrosine hydroxylase rabbit polyclonal antibody | Millipore | Cat# AB152; RRID: AB_390204 |
| Anti-mCherry rat polyclonal antibody | Life Technologies (ThermoFisher) | Cat# M11217; RRID: AB_2536611 |
| Anti-DIG sheep polyclonal antibody | Roche Applied Science (Sigma-Aldrich) | Cat# 1093274; RRID: AB_514496 |
| Donkey anti-rabbit alexa 647 | Jackson ImmunoResearch | Cat# 711-605-152; RRID: AB_2492288 |
| Donkey anti-rat Cy3 | Jackson ImmunoResearch | Cat# 712-165-153; RRID: AB_2340667 |
| Donkey anti-chicken alexa 488 | Jackson Immunoresearch | Cat# 703-545-155; RRID: AB_2340375 |
| Donkey anti-mouse alexa 488 | Jackson Immunoresearch | Cat# 703-545-155; RRID: AB_2340375 |
| Bacterial and Virus Strains | ||
| AAV5-CAG-FLExloxP-TC | UNC Vector Core; Beier et al., 2015 | N/A |
| AAV8-CAG-FLExloxP-G | UNC Vector Core; Beier et al., 2015 | N/A |
| AA V5-CAG-FLExFRT-TC | UNC Vector Core; Beier et al., 2015 | N/A |
| AAV8-CAG-FLExFRT- G | UNC Vector Core; Beier et al., 2015 | N/A |
| AA VDJ-hSyn1-FLExFRT-mGFP-2A-synaptophysin-mRuby | Stanford Viral Core; Beier et al., 2015 | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Isoflurane | Henry Schein Animal Health | CAS# 26675-46-7; CHEBI:6015 |
| DAPI | ThermoFisher Scientific | D1306 |
| Pentobarbital | Vortech Pharmaceuticals | NDC 0298-9373-68 |
| Tissue-plus O.C.T. Compound | ThermoFisher Scientific | Cat# 23-730-571 |
| NeuroTrace blue | Invitrogen (ThermoFisher) | Cat#N21479 |
| Experimental Models: Organisms/Strains | ||
| Mouse: B6.SJL-SLc6a3tm1.1(cre)Bkmn/J (DAT-Cre) | The Jackson Laboratory | JAX:006660 |
| Mouse: GAD2tm2(cre)zjh/J (GAD2-Cre) | The Jackson Laboratory | JAX: 010802 |
| Mouse: Slc17a6tm2(cre)Lowl/J (vGluT2-Cre) | The Jackson Laboratory | JAX: 016963 |
| Software and Algorithms | ||
| ImageJ (Fiji) software | NIH | N/A |
| MATLAB | MathWorks | https://www.mathworks.com/ |
| GraphPad Prism | GraphPad | https://www.graphpad.com/ |
| R; pvclust | The R Foundation | https://www.r-project.org/ |
Highlights.
Ventral tegmental area dopamine neurons have largely segregated outputs
Input patterns to ventral tegmental area neurons do not depend on cell type
Input biases are related to spatial location of cell bodies
Input biases can be predicted from the location of axons from input populations
ACKNOWLEDGMENTS
A special thanks to Takeru Matsuda for assistance with clustering and bootstrapping analysis and Nic Berns for the MATLAB code used to bin axonal projection data. Research was funded by an NIH K99 grant (DA041445) and an NIH F32 grant (DA038913) from NIDA (to K.T.B.) and the Howard Hughes Medical Institute (to L.L.).
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information includes four figures and three tables and can be found with this article online at https://doi.org/10.1016/j.celrep.2018.12.040.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Alivisatos AP, Chun M, Church GM, Greenspan RJ, Roukes ML, and Yuste R (2012). The brain activity map project and the challenge of functional connectomics. Neuron 74, 970–974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bäckman CM, Malik N, Zhang Y, Shan L, Grinberg A, Hoffer BJ, West-phal H, and Tomac AC (2006). Characterization of a mouse strain expressing Cre recombinase from the 3′ untranslated region of the dopamine transporter locus. Genesis 44, 383–390. [DOI] [PubMed] [Google Scholar]
- Beier KT, Steinberg EE, DeLoach KE, Xie S, Miyamichi K, Schwarz L, Gao XJ, Kremer EJ, Malenka RC, and Luo L (2015). Circuit architecture of VTA dopamine neurons revealed by systematic input-output mapping. Cell 162, 622–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beier KT, Kim CK, Hoerbelt P, Hung LW, Heifets BD, DeLoach KE, Mosca TJ, Neuner S, Deisseroth K, Luo L, and Malenka RC (2017). Rabies screen reveals GPe control of cocaine-triggered plasticity. Nature 549, 345–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Callaway EM, and Luo L (2015). Monosynaptic circuit tracing with glycoprotein-deleted rabies viruses. J. Neurosci 35, 8979–8985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeNardo LA, Berns DS, DeLoach K, and Luo L (2015). Connectivity of mouse somatosensory and prefrontal cortex examined with trans-synaptic tracing. Nat. Neurosci 18, 1687–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faget L, Osakada F, Duan J, Ressler R, Johnson AB, Proudfoot JA, Yoo JH, Callaway EM, and Hnasko TS (2016). Afferent inputs to neuro-transmitter-defined cell types in the ventral tegmental area. Cell Rep. 15, 2796–2808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franklin KBJ, and Paxinos G (2007). The Mouse Brain in Stereotaxic Coordinates, Third Edition (Elsevier; ). [Google Scholar]
- Gerfen CR, and Sawchenko PE (1984). An anterograde neuroanatomical tracing method that shows the detailed morphology of neurons, their axons and terminals: immunohistochemical localization of an axonally transported plant lectin, Phaseolus vulgaris leucoagglutinin (PHA-L). Brain Res. 290, 219–238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Q, Wang D, He X, Feng Q, Lin R, Xu F, Fu L, and Luo M (2015). Whole-brain mapping of inputs to projection neurons and cholinergic interneurons in the dorsal striatum. PLoS ONE 10, e0123381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henny P, Brown MTC, Northrop A, Faunes M, Ungless MA, Magill PJ, and Bolam JP (2012). Structural correlates of heterogeneous in vivo activity of midbrain dopaminergic neurons. Nat. Neurosci 15, 613–619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hnasko TS, Hjelmstad GO, Fields HL, and Edwards RH (2012). Ventral tegmental area glutamate neurons: electrophysiological properties and projections. J. Neurosci 32, 15076–15085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawano M, Kawasaki A, Sakata-Haga H, Fukui Y, Kawano H, Nogami H, and Hisano S (2006). Particular subpopulations of midbrain and hypothalamic dopamine neurons express vesicular glutamate transporter 2 in the rat brain. J. Comp. Neurol 498, 581–592. [DOI] [PubMed] [Google Scholar]
- King MA, Louis PM, Hunter BE, and Walker DW (1989). Biocytin: a versatile anterograde neuroanatomical tract-tracing alternative. Brain Res. 497, 361–367. [DOI] [PubMed] [Google Scholar]
- Kristensson K, and Olsson Y (1971). Retrograde axonal transport of protein. Brain Res. 29, 363–365. [DOI] [PubMed] [Google Scholar]
- Lammel S, Hetzel A, Ha¨ckel O., Jones I, Liss B, and Roeper J (2008). Unique properties of mesoprefrontal neurons within a dual mesocorticolimbic dopamine system. Neuron 57, 760–773. [DOI] [PubMed] [Google Scholar]
- Lammel S, Lim BK, Ran C, Huang KW, Betley MJ, Tye KM, Deisseroth K, and Malenka RC (2012). Input-specific control of reward and aversion in the ventral tegmental area. Nature 491, 212–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner TN, Shilyansky C, Davidson TJ, Evans KE, Beier KT, Zalocusky KA, Crow AK, Malenka RC, Luo L, Tomer R, and Deisseroth K. (2015). Intact-brain analyses reveal distinct information carried by SNc dopamine subcircuits. Cell 162, 635–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Bergan JF, Ogawa SK, Isogai Y, Umadevi Venkataraju K, Osten P, Uchida N, and Watabe-Uchida M (2015). Dopamine neurons projecting to the posterior striatum form an anatomically distinct subclass. eLife 4, e10032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menegas W, Akiti K, Amo R, Uchida N, and Watabe-Uchida M (2018). Dopamine neurons projecting to the posterior striatum reinforce avoidance of threatening stimuli. Nat. Neurosci 21, 1421–1430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh SW, Harris JA, Ng L, Winslow B, Cain N, Mihalas S, Wang Q, Lau C, Kuan L, Henry AM, et al. (2014). A mesoscale connectome of the mouse brain. Nature 508, 207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pollak Dorocic I, Fürth D, Xuan Y, Johansson Y, Pozzi L, Silberberg G, Carlén M, and Meletis K (2014). A whole-brain atlas of inputs to serotonergic neurons of the dorsal and median raphe nuclei. Neuron 83, 663–678. [DOI] [PubMed] [Google Scholar]
- Qi J, Zhang S, Wang HL, Barker DJ, Miranda-Barrientos J, and Morales M (2016). VTA glutamatergic inputs to nucleus accumbens drive aversion by acting on GABAergic interneurons. Nat. Neurosci 19, 725–733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Root DH, Mejias-Aponte CA, Zhang S, Wang HL, Hoffman AF, Lupica CR, and Morales M (2014). Single rodent mesohabenular axons release glutamate and GABA. Nat. Neurosci 17, 1543–1551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmued LC, and Fallon JH (1986). Fluoro-Gold: a new fluorescent retrograde axonal tracer with numerous unique properties. Brain Res. 377, 147–154. [DOI] [PubMed] [Google Scholar]
- Schwab ME, Javoy-Agid F, and Agid Y (1978). Labeled wheat germ agglutinin (WGA) as a new, highly sensitive retrograde tracer in the rat brain hippo-campal system. Brain Res. 152, 145–150. [DOI] [PubMed] [Google Scholar]
- Schwarz LA, Miyamichi K, Gao XJ, Beier KT, Weissbourd B, DeLoach KE, Ren J, Ibanes S, Malenka RC, Kremer EJ, and Luo L (2015). Viral-genetic tracing of the input-output organization of a central noradrenaline circuit. Nature 524, 88–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun Y, Nguyen AQ, Nguyen JP, Le L, Saur D, Choi J, Callaway EM, and Xu X (2014). Cell-type-specific circuit connectivity of hippocampal CA1 revealed through Cre-dependent rabies tracing. Cell Rep. 7, 269–280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki R, and Shimodaira H (2006). Pvclust: an R package for assessing the uncertainty in hierarchical clustering. Bioinformatics 22, 1540–1542. [DOI] [PubMed] [Google Scholar]
- Taniguchi H, He M, Wu P, Kim S, Paik R, Sugino K, Kvitsiani D, Fu Y, Lu J, Lin Y, et al. (2011). A resource of Cre driver lines for genetic targeting of GABAergic neurons in cerebral cortex. Neuron 71, 995–1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tritsch NX, Oh WJ, Gu C, and Sabatini BL (2014). Midbrain dopamine neurons sustain inhibitory transmission using plasma membrane uptake of GABA, not synthesis. eLife 3, e01936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veenman CL, Reiner A, and Honig MG (1992). Biotinylated dextran amine as an anterograde tracer for single- and double-labeling studies. J. Neurosci. Methods 41, 239–254. [DOI] [PubMed] [Google Scholar]
- Vong L, Ye C, Yang Z, Choi B, Chua S Jr., and Lowell BB (2011). Leptin action on GABAergic neurons prevents obesity and reduces inhibitory tone to POMC neurons. Neuron 71, 142–154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Callaway EM, and Kreitzer AC (2013). Differential innervation of direct- and indirect-pathway striatal projection neurons. Neuron 79, 347–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wall NR, De La Parra M, Sorokin JM, Taniguchi H, Huang ZJ, and Call-away EM (2016). Brain-wide maps of synaptic input to cortical interneurons. Neurosci J. 36, 4000–4009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watabe-Uchida M, Zhu L, Ogawa SK, Vamanrao A, and Uchida N (2012). Whole-brain mapping of direct inputs to midbrain dopamine neurons. Neuron 74, 858–873. [DOI] [PubMed] [Google Scholar]
- Weissbourd B, Ren J, DeLoach KE, Guenthner CJ, Miyamichi K, and Luo L (2014). Presynaptic partners of dorsal raphe serotonergic and GABAergic neurons. Neuron 83, 645–662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wickersham IR, Lyon DC, Barnard RJO, Mori T, Finke S, Conzel-mann K-K, Young JAT, and Callaway EM (2007). Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 53, 639–647. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.